Abstract
An understanding of how communities are organized is a fundamental goal of ecology but one which has historically been elusive for microbial systems. We used a bar-coded pyrosequencing approach targeting the V3 region of the bacterial small-subunit rRNA gene to address the factors that structure communities along the thermal gradients of two alkaline hot springs in the Lower Geyser Basin of Yellowstone National Park. The filtered data set included a total of nearly 34,000 sequences from 39 environmental samples. Each was assigned to one of 391 operational taxonomic units (OTUs) identified by their unique V3 sequence signatures. Although the two hot springs differed in their OTU compositions, community resemblance and diversity changed with strikingly similar dynamics along the two outflow channels. Two lines of evidence suggest that these community properties are controlled primarily by environmental temperature. First, community resemblance decayed exponentially with increasing differences in temperature between samples but was only weakly correlated with physical distance. Second, diversity decreased with increasing temperature at the same rate along both gradients but was uncorrelated with other measured environmental variables. This study also provides novel insights into the nature of the ecological interactions among important taxa in these communities. A strong negative association was observed between cyanobacteria and the Chloroflexi, which together accounted for ∼70% of the sequences sampled. This pattern contradicts the longstanding hypothesis that coadapted lineages of these bacteria maintain tightly cooccurring distributions along these gradients as a result of a producer-consumer relationship. We propose that they instead compete for some limiting resource(s).
Elucidating how biodiversity is distributed and the mechanisms underlying those patterns is a central goal of ecology. Although microorganisms make critical contributions to ecosystem function through their participation in biogeochemical cycles, we still have only a limited understanding of the factors that control the spatial structure and diversity of microbial communities (37). For example, although it is clear that microbial community composition is influenced by environmental variation (11, 17, 18, 28), the question of how diversity changes along environmental gradients remains generally unresolved. Of particular interest is the relationship between microbial diversity and temperature, as this environmental variable is strongly correlated with diversity over a broad range of spatial scales for many plant and animal taxa in terrestrial, freshwater, and marine ecosystems (1, 12, 36, 39, 43). In addition, because spatially resolved abundance data for individual microbial taxa are scarce, we have only limited information regarding the patterns of association among microorganisms. Consequently, microbial ecology has developed with little clarity regarding the potential roles of either negative or positive biotic interactions for structuring communities or whether microbial community organization along environmental gradients conforms to either individualistic (e.g., see references 13 and 20) or organismal (e.g., see reference 9) community ecology paradigms developed for macroscopic organisms.
Hot spring microbial ecosystems present an excellent opportunity to investigate fundamental questions regarding community organization. Steep temperature gradients enable the explicit investigation of the importance of environmental variation for structuring diversity while controlling for other factors that typically vary across sampling locations on larger geographic scales, such as solar energy availability and geological history. Decades of biological research on the alkaline-silica hot springs of Yellowstone National Park (reviewed in reference 49) further inform predictions regarding the nature of the ecological interactions among abundant community members.
These systems are particularly notable for the presence of ecologically diverse groups of cyanobacteria and photosynthetic green nonsulfur bacteria (i.e., the Chloroflexi). The former group includes lineages of Synechococcus, the most thermotolerant of which delimits the thermal maximum for photosynthesis, whereas the latter includes divergent “green” (the genus Chloroflexus and relatives) and “red” (the genus Roseiflexus) clades. The conventional view is that interactions among cyanobacteria and the Chloroflexi are generally positive. Specifically, it has been proposed that coadapted lineages of Synechococcus and Chloroflexi maintain tightly cooccurring distributions due to a producer-consumer relationship in which the Chloroflexi grow as photoheterotrophs on low-molecular-weight organic compounds excreted by the cyanobacteria (48, 49). In turn, the filamentous Chloroflexi were previously suggested to provide a matrix within which Synechococcus cells become stably embedded (5). According to this model, we would expect to observe coincident peaks in abundance between coadapted lineages of cyanobacteria and Chloroflexi along alkaline hot spring gradients as well as a general positive correlation between the abundances of both groups. However, other evidence raises the possibility that these groups may not share a strict cross-feeding relationship. The genus Chloroflexus is metabolically flexible in laboratory culture, and certain strains have been grown as photoautotrophs with hydrogen sulfide or hydrogen as an electron donor (16, 26, 30). Although Roseiflexus is yet to be grown autotrophically in the laboratory, comparative genomics of laboratory strains and metagenomic data from microbial mat communities have also revealed the presence of Roseiflexus genes involved in the autotrophic hydroxypropionate pathway (24). In addition, stable carbon isotope data (46, 47) suggest that certain members of the Chloroflexi may have the capacity to grow autotrophically in situ. The issue of the nature of the ecological interactions between cyanobacteria and the Chloroflexi therefore requires clarification.
In the present study, we investigated the patterns of distribution of bacteria along the temperature gradients of two alkaline-silica hot springs in Yellowstone National Park to determine how community properties changed in response to temperature and whether the realized distributions of cyanobacteria and the Chloroflexi meet the prediction of the coadaptation hypothesis. To do so, we used bar-coded mass parallel pyrosequencing of the V3 variable region of the bacterial small-subunit (SSU) rRNA gene to simultaneously interrogate the sequence diversity of environmental samples from multiple locations along both hot springs. This strategy is distinct from recent applications of next-generation DNA sequencing technology for investigating microbial diversity, as previous efforts focused principally on the deep sampling of one or a few discrete habitats for the purpose of quantifying the magnitude of diversity (e.g., see references 21, 38, and 41). We report that community properties changed with similar dynamics in response to temperature despite differences between hot springs in water chemistries and taxon composition, and we reject the coadaptation hypothesis for these communities based on a strong negative association in abundances of cyanobacteria and Chloroflexi.
MATERIALS AND METHODS
Field methods.
Microbial mat samples were collected in June 2006 from along the outflow channels of two alkaline-silica hot springs located approximately 2 km from each other in the Lower Geyser Basin of Yellowstone National Park: White Creek (Universal Transverse Mercator 516823E 4930490N [location of White Creek site 10]) and Rabbit Creek (Universal Transverse Mercator 515009E 4929570N [location of Rabbit Creek site 10]). Triplicate samples of approximately 0.5 g (wet weight) were sterilely collected from 10 locations along each channel covering a similar thermal gradient, ranging from approximately 39°C to the respective upstream boundaries for visible microbial growth (Table 1). The sampling sites spanned distances of approximately 1.5 km at White Creek and 0.4 km at Rabbit Creek, respectively. Samples were cryopreserved on-site and returned to the University of Montana. Water samples sterilely collected in June 2008 at five of the sampling sites along each creek channel were analyzed by anion chromatography, inductively coupled plasma analysis, and flow injection analysis (SEAL Analytical) of low-level inorganic nitrogen and phosphorus by the Montana State University Microbial Biogeochemistry Laboratory.
TABLE 1.
Temperature and water chemistry data summary for sampling sitesa
| Site | Temp (°C) | Concn
|
||||||
|---|---|---|---|---|---|---|---|---|
| NH4 (μM) | NO3 (μM) | NO2 (μM) | PO4 (μM) | Mg (μM) | As (μM) | Na (mM) | ||
| WC1 | 39 | 6.71 | 9.14 | 0.49 | 2.60 | 3.7 | 6.4 | 5.90 |
| WC2 | 43 | — | — | — | — | — | — | — |
| WC3 | 46.5 | 0.35 | 0.35 | 0.08 | 0.97 | 5.8 | 1.5 | 3.85 |
| WC4 | 51 | — | — | — | — | — | — | — |
| WC5 | 54 | 0.32 | 0.38 | 0.08 | 0.52 | 8.6 | BD | 2.97 |
| WC6 | 57 | — | — | — | — | — | — | — |
| WC7 | 61 | 7.77 | 9.22 | 0.14 | 0.67 | 4.5 | BD | 3.10 |
| WC8 | 64 | — | — | — | — | — | — | — |
| WC9 | 67.5 | — | — | — | — | — | — | — |
| WC10 | 72.5 | 1.03 | 0.89 | 0.21 | 0.51 | 3.8 | BD | 2.90 |
| RC1 | 38.5 | 0.44 | 1.50 | 0.85 | 8.14 | BD | 17.8 | 14.55 |
| RC2 | 47 | — | — | — | — | — | — | — |
| RC3 | 51 | 2.31 | 4.81 | 1.08 | 8.66 | BD | 17.4 | 14.42 |
| RC4 | 53 | — | — | — | — | — | — | — |
| RC5 | 55 | 4.49 | 8.70 | 1.18 | 7.53 | BD | 17.0 | 14.69 |
| RC6 | 61 | — | — | — | — | — | — | — |
| RC7 | 63.5 | 2.24 | 1.96 | 1.46 | 9.40 | BD | 15.8 | 13.50 |
| RC8 | 63.5 | — | — | — | — | — | — | — |
| RC9 | 67.5 | — | — | — | — | — | — | — |
| RC10 | 69 | 0.49 | 0.67 | 1.34 | 5.08 | BD | 16.6 | 14.20 |
Temperature estimates for White Creek site 1 (WC1) to White Creek site 5 represent the mean annual temperatures for data collected at 10-min intervals between September 2004 and September 2005. All other estimates are point estimates. —, not sampled; BD, below the detection limit; RC1, Rabbit Creek site 1.
PCR amplification and environmental sequencing.
Community DNA was isolated from each environmental sample by a modification of a method described previously by Nübel et al. (35). Primers for bar-coded mass parallel sequencing of the V3 variable region of the bacterial SSU rRNA gene (see Table S1 in the supplemental material) were produced by adding a unique bar code of 5 nucleotides between the sequences of 454 Life Sciences adapter A (27) and reverse primer U529R (10). We focused on the diversity of the Bacteria, because the Archaea and Eukarya are known to be comparatively rare in the alkaline-silica hydrothermal systems of Yellowstone National Park (49; our unpublished data). To construct environmental libraries, each sample was amplified using forward primer U341F (15) and a reverse primer with a different bar code. The engineering of reverse primers with unique bar codes enabled amplification products from the different samples to be pooled by equal mass, sequenced in the reverse direction via massively parallel sequencing by synthesis (454 Life Sciences), and then reassigned to their original sample based on primer bar code identification. All PCRs were conducted with 25-μl volumes containing 1 μl of environmental DNA (∼10 ng/μl), 1 unit of Taq polymerase, and final concentrations of 30 mM Tricine, 50 mM KCl, 2 mM MgCl2, 0.1 mM each deoxynucleoside triphosphate, and 0.32 mM each primer. Reactions were performed using an MJ Research (Watertown, MA) PTC-100 thermal cycler with an initial denaturation step at 95°C for 1 min, followed by 30 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s and a final extension step at 72°C for 3 min. For each sample, six replicate PCRs (and two negative control reactions) were conducted, and the five PCR products exhibiting the strongest amplification were pooled by equal volumes for sequencing. Pooled PCR products were purified prior to sequencing using an Agencourt AMPure PCR purification kit. Two samples from each of the 10 White Creek and Rabbit Creek collection locations were processed by these methods, with the exception of White Creek site 10, for which amplification was successful for only one sample. These 39 samples were pooled for pyrosequencing (27) by 454 Life Sciences (Branford, CT). For the White Creek site 10 sample, a 2× (by mass) contribution to the sample pool was made. Because samples were pooled by equal mass, variation in the number of sequences recovered from each sample likely reflects slight biases in PCR efficiency among primer bar codes.
Identification of OTUs.
Using customized Perl scripts, raw sequence reads with a valid bar code were identified and subsequently filtered to minimize the effects of poor sequence quality and sequencing errors by removing (i) sequences with greater than one ambiguous base call, (ii) sequences that were shorter than 85 nucleotides, and (iii) sequences that were longer than 135 nucleotides. We next grouped sequences sharing 99% sequence identity into contigs by CAP3 sequence assembly (19). For each contig, the SSU rRNA gene sequence(s) producing the alignment with greatest E value was determined with a local BLAST similarity search (2) of the Michigan State University Center for Microbial Ecology Ribosomal Database Project II (RDP II) database (downloaded on 13 October 13 2006 from http://rdp.cme.msu.edu). Contigs sharing the same top alignment accession number were provisionally binned together for direct sequence alignment comparison using BioEdit, version 2.0 (14). For the purposes of this study, contigs of unequal sequence length which were identical over the region of sequence overlap were grouped together as an operational taxonomic unit (OTU), as were singleton contigs differing from the consensus sequence solely by an insertion or deletion (typically, this involved the inferred length of a homopolymer tract). With respect to the first criterion, any contig sequence that was too short to distinguish specific OTU membership (i.e., information at a diagnostic nucleotide position was missing) was excluded from the abundance matrix (see below). However, we were still able to assign about 97% of the sequences to OTUs. We applied the second criterion because indels appear to comprise the majority of pyrosequencing errors, particularly with respect to the resolution of homopolymer length (23). Frequency distributions of contig relative abundances for individual OTUs were typically dominated by a single contig, suggesting that these minor indel variants (frequently, these were singletons recovered from a single sample) were likely sequence read errors (data not shown). Contig sequences which shared top alignment scores but which did not meet either of the above-described criteria were separated into different OTUs.
To determine the abundance (i.e., the number of sequences recovered) of individual OTUs in each sample, we wrote Perl scripts to identify the contig membership of each sequence by performing a local BLAST similarity search against the CAP3 contig database. Each sequence was subsequently assigned to its corresponding OTU on the basis of its contig identity. The resultant 391 (OTUs) × 39 (sample) abundance data matrix was used for subsequent analyses.
Assignment of OTUs to taxonomic groups.
We attempted to assign OTUs to taxonomic groups (i.e., bacterial divisions) on the basis of our BLAST sequence similarity searches against the RDP II database. To ascertain OTU resolving power, we generated frequency distributions of pairwise V3 sequence similarities among 100 randomly selected strains represented in the RDP II database for four bacterial divisions inferred to be abundant in our data set (Cyanobacteria, Chloroflexi, Cytophaga-Flavobacter-Bacteroidetes [CFB]-Chlorobi, and Proteobacteria). For the Chloroflexi, the complete set of 47 strains was used. The mean similarity within divisions was variable, exceeding 84% for the Chloroflexi and Cyanobacteria, but was estimated to be 76% for the CFB-Chlorobi. As expected, the interdivision pairwise sequence similarity was much lower. For example, the mean V3 sequence similarity among interdivision pairwise comparisons between the Cyanobacteria and Chloroflexi was approximately 74%, with >99.8% of pairwise comparisons having less than 85% similarity. We therefore assigned an OTU to a division if it exhibited sequence similarity to a cultured representative of greater than 85%. However, the percent similarity was typically much greater. For example, all OTUs that were assigned to the Cyanobacteria or Chloroflexi (approximately 70% of the data set) were at least 91% similar to a laboratory strain. This resolving power is comparable to that achieved by use of the V3 region in simulations (45).
Community richness and diversity.
Raw taxonomic richness (S) was calculated by summing the number of OTUs (including singletons) observed in each sample. To account for differences in sampling effort (i.e., the number of sequences obtained), we also estimated richness with the Chao1 estimator (7), with the coverage-based estimator ACE (8), and by rarefaction. We calculated Chao1 with EstimateS, version 8.0 (purl.oclc.org/estimates). Because the coefficient of variation of the abundance data was greater than 0.5 for some samples, which results in an imprecision of the bias-corrected formula, we used the classic version of the estimator according to the author's recommendation. Estimates of the mean richness for 500 individuals drawn randomly from each sample were obtained with EcoSim, version 7.0 (N. J. Gotelliand and G. L. Entsminger, Acquired Intelligence Inc. and Kesey-Bear), by 1,000 iterations of rarefaction using the independent sampling algorithm. Shannon's diversity {eΣpi[ln(pi)]} was also calculated for each sample, where pi is the frequency of occurrence of each OTU. Fisher's alpha log series was calculated by iterating the equation S/N = [(1 − x)/x][−ln(1 − x)], where N is the number of sequences in the sample. Once x was solved, the diversity index alpha (α) was calculated as N(1 − x)/x.
Ordination methods.
Nonmetric multidimensional scaling plots were generated with Primer, version 5.0 (Plymouth Marine Laboratory, United Kingdom), from a resemblance matrix of Bray-Curtis similarities calculated for each pair of community samples from the square-root-transformed OTU abundance matrix. The best two-dimensional solution presented in this study resulted in a minimum stress value of 0.09 and occurred in 23 of 25 runs (each of 100 iterations maximum). The proportion of variance (r2) among community samples represented by each axis was estimated by the regression of distance in ordination space of Bray-Curtis similarity for each pair of samples (29). Rates of community composition change (i.e., OTU turnover or beta diversity) were estimated by the regression of Bray-Curtis similarity on both geographic distance and the difference in the estimated environmental temperature of community samples. For all models, the most linear relationship and the most homoscedastic distribution of data points were achieved by logarithmic transformation of Bray-Curtis similarity.
RESULTS
Physical and chemical properties of study sites.
Although both are examples of alkaline-silica hydrothermal systems common within the Yellowstone National Park caldera, White Creek and Rabbit Creek differed both in the rates of temperature change along their channels and in water chemistries (Table 1). The thermal gradient of Rabbit Creek was much steeper (corresponding to an average change of 7.5°C per 100 m [r2 = 0.98; F1,8 = 387.62; P < 0.0001]) than that of White Creek (2°C per 100 m [r2 = 0.93; F1,8 = 100.06; P < 0.0001]). In addition, the creeks systematically differed in the concentrations of several nutrients and trace elements, including phosphate, magnesium, sodium, and arsenic (Table 1). Temperature did not covary with any measured nutrient or trace element, including nitrate (F1,8 = 0.51; P = 0.50), ammonium (F1,8 = 0.14; P = 0.72), and phosphate (F1,8 = 0.16; P = 0.70).
Pyrosequencing data set.
A total of 47,267 sequences with a valid bar code were obtained from a total of 39 samples collected from 10 sites along the thermal gradients of each hot spring. The data set was reduced to 33,863 sequences (including 32,018 sequences with no ambiguous base calls) after filtering of poor-quality sequences (see Materials and Methods). This approximately 28% reduction is comparable to that described previously Sogin et al. (41). Groups of sequences sharing 99% sequence identity were next determined by CAP3 sequence assembly. A total of 1,424 sequence groups were identified. Groups with low E values (representing 169 sequence reads) when queried against the RDP II database were removed from the data set at this stage, as subsequent NCBI GenBank BLAST analyses revealed that they were not bacterial V3 sequences (these included sequences with no match against the database, mitochondrial V3 sequences, and other nonhomologous loci). From the remaining contigs, we identified a total of 391 distinct OTUs (see Materials and Methods) representing 33,140 sequences (see Tables S2 and S3 in the supplemental material), the eight most abundant of which comprised 50% of the sequences recovered. Rarefaction analyses (see Fig. S1 in the supplemental material) indicated that all samples had reached the curvilinear phase of sampling effort and that the curve had started to plateau for White Creek site 10, the sampling site with the highest temperature.
Decay dynamics of community similarity.
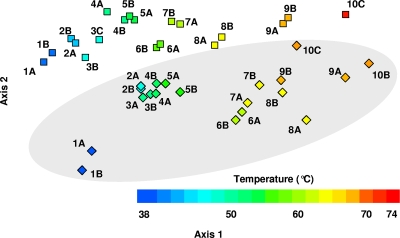
To estimate community resemblance among samples, Bray-Curtis similarities (i.e., the shared abundance between communities) were first calculated for each pair of community samples from the square-root-transformed 391 × 39 OTU abundance matrix. A nonmetric multidimensional scaling plot in two-dimensional taxon space (Fig. 1) represented most (∼86%) of the variation in community structure among samples (74.2% for axis 1 and 10.7% for axis 2, respectively, based on regression analyses; P < 0.0001 and n = 741 for each). The stress value was low (0.09), and the addition of a third dimension did not substantially improve the model.
FIG. 1.
Nonmetric multidimensional scaling plot of hot spring community samples. Color coding of samples from White Creek (squares) and Rabbit Creek (diamonds, shaded in gray) indicate the temperatures of sampling locations.
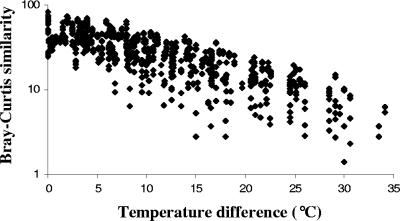
Community samples from each hot spring were generally oriented sequentially along axis 1 (Fig. 1). Samples collected from the same location were typically near each other in ordination space. Still, contingency analyses indicated that the null hypothesis of no difference in community composition could be rejected for most within-site pair comparisons (for 6 of 9 White Creek locations and for 8 of 10 Rabbit Creek locations) (see Fig. S2 in the supplemental material). Axis 1 values were strongly correlated (r = −0.93; F1,37 = 254.64; P < 0.0001) with the estimated environmental temperature from which samples were collected. Community similarity (estimated between all pairwise combinations of samples) decreased exponentially with an increasing difference in the temperature of the sample collection locations regardless of the creek of origin for the respective samples (Fig. 2). Rates of community change along the gradients were indistinguishable (means ± standard errors of −0.075 ± 0.0030 at White Creek and −0.076 ± 0.0032 at Rabbit Creek). In contrast, community similarity was only weakly associated with the physical distance between samples (r = −0.21; F1,739 = 35.33; P < 0.0001), suggesting that contemporary environmental variation primarily shapes the observed spatial decay dynamics.
FIG. 2.
Exponential decay dynamics of community similarity among White Creek and Rabbit Creek samples. The negative correlation (r = −0.80) between log-transformed Bray-Curtis similarity and the difference in environmental temperature for all pairwise community sample comparisons was highly significant by a Mantel test (P < 0.0001; n = 741).
Although they share similar community resemblance decay dynamics, White Creek and Rabbit Creek did exhibit broad differences in taxon composition that distinguish their respective samples as roughly parallel clusters in ordination space (Fig. 1). In addition to differences in the relative abundances among shared OTUs (see below), the majority of OTUs were observed only in a single spring. A total of 162 OTUs were unique to White Creek, and 67 were unique to Rabbit Creek. Although nearly 75% (n = 170) of these OTUs were rare (<10 sequences recovered) and together accounted for only about 10% of the data set (3,440 sequences), several represent abundant community members (e.g., OTU 54) (see Table S3 in the supplemental material).
Richness and diversity decrease with increasing temperature.
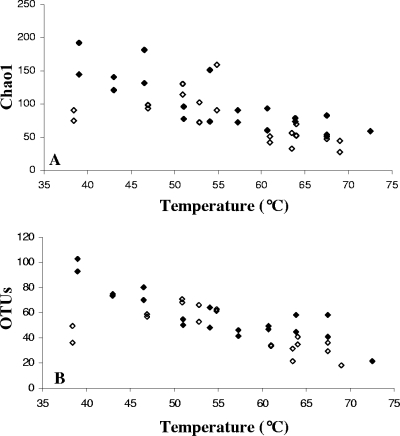
The two creeks exhibited strikingly similar decreases in richness and diversity with increasing environmental temperature (Fig. 3; see Fig. S3 in the supplemental material). All data fit a linear model (with adjusted r2 values ranging between 0.56 and 0.62 for analyses of covariance with creek as a factor and temperature as a covariate; P < 0.0001 for all models). The rate of decrease in diversity was not significantly different between springs for any diversity measure (i.e., the creek × temperature interaction was not significant for any model, with P values ranging between 0.11 and 0.26). The estimated slopes (b ± standard errors) for different richness estimators ranged between −1.7% ± 0.29% (rarefaction) and −3.3% ± 0.36% (Chao1); that is, richness decreased by roughly 8 to 16 OTUs per 5°C increase in temperature. Comparison of within-creek marginal means additionally indicated that the expected richness at a particular temperature was generally highly similar between creeks (data not shown). Estimates of richness and diversity for the most diverse community samples in our data set (Fig. 3; see Fig. S3 in the supplemental material) were approximately 100-fold lower than recent estimates of bacterial richness for communities from a hypersaline microbial mat (25), deep seawater and hydrothermal vents (21, 41), and soils (38).
FIG. 3.
Community OTU richness decreased with increasing environmental temperature. Richnesses at White Creek (closed diamonds) and Rabbit Creek (open diamonds) exhibited similar temperature responses by Chao1 (A) and rarefaction (B) (per 500 sequences sampled). (See Fig. S3 in the supplemental material for analyses with other diversity measures.)
Although the above-described conclusions are robust, we note that results were clearly sensitive to data collected for Rabbit Creek site 1 samples (Fig. 3; see Fig. S3 in the supplemental material), which were statistical outliers for all diversity measures, and that the above-described patterns were even stronger if the data obtained for these samples were excluded from the analyses (model r2 values of 0.62 to 0.76 and interaction P values ranging between 0.38 and 0.82).
Community composition.
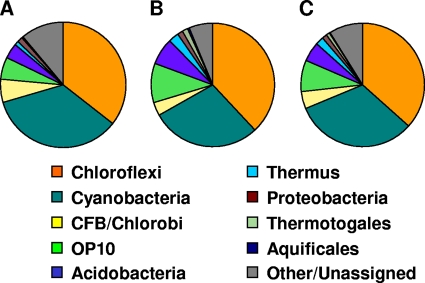
We could assign greater than 90% of OTUs to a taxonomic group (i.e., a bacterial division) (see Materials and Methods). Cyanobacteria and Chloroflexi accounted for nearly 70% of the sequences obtained (Fig. 4). Other abundant taxa included CFB-Chlorobi, Acidobacteria, and candidate division OP10 (22). The latter two groups are of particular interest. At temperatures below 65°C in both creeks, we observed several OTUs related to a recently described, novel phototrophic member of the Acidobacteria (“Candidatus Chloracidobacterium thermophilum”) (6), and the sorting of these lineages along the gradients suggest that they have diverged in thermotolerance (Fig. 5). OP10, a group with at present only two cultured representatives but which is frequently observed in community diversity surveys of hot springs and other environments (44), was recovered in all samples but was more abundant at temperatures greater than 60°C (data not shown).
FIG. 4.
Relative abundances of bacterial divisions in White Creek (A), Rabbit Creek (B), and the combined data set (C).
FIG. 5.
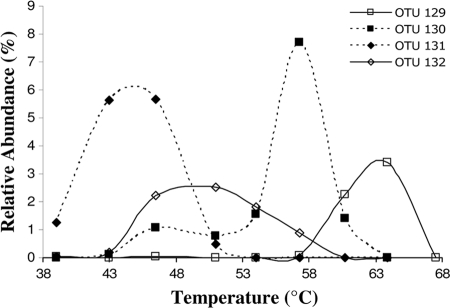
Distribution of “Candidatus Chloracidobacterium thermophilum”-like OTUs along the White Creek temperature gradient.
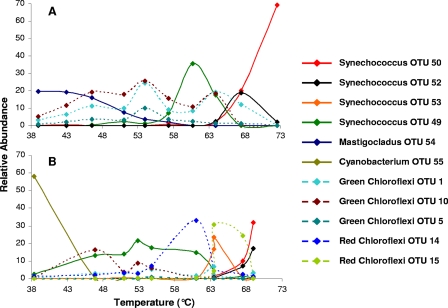
The V3 sequences were sufficient to resolve distinct lineages of cyanobacteria (see Fig. S4 in the supplemental material), including four Synechococcus OTUs which composed at least 10% of the sequences recovered from at least one sample. On the basis of sequence similarity with cultured strains and environmental clones, three OTUs (OTUs 50, 52, and 53) belonged to the A clade of Synechococcus (49), whereas a fourth (OTU 49) was identical in sequence to members of the B clade. Analysis of the relative abundances of these OTUs along the two gradients (Fig. 6) corroborates previously reported observations that members of the A clade are more thermotolerant and are generally more abundant at higher temperatures than are B clade lineages (e.g., see references 32 and 49) but at a much higher level of resolution than was previously reported.
FIG. 6.
Distribution of major cyanobacteria and Chloroflexi OTUs along the thermal gradients of White Creek (A) and Rabbit Creek (B). Relative abundance represents the percentage of sequences of an OTU recovered from a particular community sample.
The V3 sequence signatures likewise distinguished divergent “green” clades (Chloroflexus and relatives) and “red” clades (Roseiflexus) of Chloroflexi (see Fig. S4 in the supplemental material). Green clade OTUs (OTUs 1, 5, and 10) were the predominant Chloroflexi sequences recovered throughout White Creek and from Rabbit Creek at lower temperatures (Fig. 6). Red clade taxa (OTUs 14 and 15) were the principal Chloroflexi found at higher temperatures in Rabbit Creek.
Negative association between cyanobacteria and Chloroflexi.
If different lineages of cyanobacteria and Chloroflexi have coadapted along these thermal gradients, we would expect to observe tight niche overlap between members of these groups. However, peaks in the abundances of individual OTUs of cyanobacteria and Chloroflexi did not exhibit the tightly cooccurring distributions predicted by the coadaptation hypothesis. Rather, they were generally offset with respect to each other in both creeks (Fig. 6). In fact, in contrast with the positively correlated abundances characteristic of cooccurring organisms, we instead observed a strong negative correlation between the relative abundances of cyanobacteria and Chloroflexi at both White Creek (r = −0.90; F1,17 = 69.51; P < 0.0001) and Rabbit Creek (r = −0.76; F1,18 = 24.24; P < 0.0001). We therefore reject the coadaptation hypothesis for these communities. This general conclusion also applies to the relationships between the distribution patterns of cyanobacteria and the individual green and red clades of Chloroflexi, respectively. Negative correlations with cyanobacterial abundance were observed for green Chloroflexi and red Chloroflexi at both White Creek (r = −0.85, F1,18 = 45.17, and P < 0.0001 for green Chloroflexi; r = −0.63, F1,18 = 10.99, and P < 0.005 for red Chloroflexi) and Rabbit Creek (r = −0.62, F1,18 = 11.08, and P < 0.005 for green Chloroflexi; r = −0.63, F1,18 = 9.66, and P < 0.01 for red Chloroflexi).
DISCUSSION
An understanding of the mechanisms that determine the spatial organization of microbial diversity is essential for resolving the relative contributions of historical events and current conditions to contemporary distribution patterns and can inform predictions regarding ecosystem response to future environmental change. Despite recent interest in the biogeography of microorganisms (reviewed in reference 28), we still have only a limited understanding of the factors that shape the structure and distribution of microbial communities. Advances in high-throughput DNA sequencing technologies (e.g., see reference 27) have enabled a deeper sampling of microbial communities than was previously possible and therefore offer the potential to more powerfully address fundamental questions regarding the spatial distribution of microbial diversity. In the present study, a relatively modest but strategic allocation of sequencing effort along known environmental gradients has enabled novel insights into how microbial communities respond to environmental variation and suggests the additional importance of biotic interactions for community organization.
Community response to changing temperature.
Because community similarity was strongly associated with the similarity of environmental temperature (Fig. 2) but only weakly associated with geographic distance, we conclude that contemporary environmental variation is more important than distance per se for shaping the observed decay of community similarities, in agreement with data from other recent studies of microbial communities at different landscape scales (e.g., see references 11 and 18). We further believe that taxon sorting along these hot springs is principally a response to temperature rather than water chemistry, because environmental temperature did not covary with the concentration of any measured nutrient or trace element (Table 1). The decay of similarity that we observed is rapid compared with rates of spatial turnover that were previously reported for communities of macroorganisms (34, 42) and likely reflects the steepness of the thermal gradients in these structured environments.
Our observation that both richness and diversity decrease with increasing temperature at indistinguishable rates in White Creek and Rabbit Creek (Fig. 3; see Fig. S3 in the supplemental material) suggests the existence of similar temperature-imposed constraints operating on community properties. We propose that this may include an energetic constraint. The thin Synechococcus-dominated biofilms that develop near the thermal limit for photosynthesis persist under conditions of chronic stress at temperatures greater than approximately 65°C (31), and there is a clear productivity gradient as these channels cool, supporting more biomass, more available energy, and a potentially more-complex food web. Such a taxon-energy relationship is one of many hypotheses that have been put forward to explain biodiversity patterns observed for macroscopic organisms on larger geographic scales, including the well-known latitudinal gradient of diversity observed for many taxa (1, 12, 36, 39, 43). Here, however, the relationship would not be mediated directly by the availability of solar energy, which is controlled across sampling locations, but rather by the physiology of primary producers.
The similarity of community responses to temperature in these hot springs further motivates several questions regarding the generality of organizing principles for microbial communities. How common are the observed exponential decay dynamics of community similarity? Are they restricted to photosynthesis-based hydrothermal systems, or do they apply more broadly? How variable are the rates of change of community resemblance and diversity along different environmental gradients, and what determines these differences? Such questions remain to be explored in detail, although we note that Fierer and Jackson (11) did not observe a relationship with temperature (but rather a curvilinear increase in diversity with increasing pH) for North American soil microbial communities. Future investigations for different kinds of environmental gradients over a range of spatial scales promise a fresh perspective on the nature of the rules by which microbial communities are structured.
Although the biological explanation for the observed outlier behavior at Rabbit Creek site 1 (Fig. 3; see Fig. S3 in the supplemental material) is not clear, we suggest that we may have sampled a disturbed, nonequilibrium community disproportionately represented by a cyanobacterium that was abundant only at this location (OTU 55) (Fig. 6). Because the main channel of Rabbit Creek did not present a suitable collection site comparable in temperature to the 39°C microbial mat boundary in White Creek, we instead sampled from a smaller, cooler side channel. If the low level of diversity that we observed in these samples can be attributed ultimately to their derivation from a disturbed community, it raises the prospect that such outlier behavior might be generally useful for identifying disturbed systems.
Differences in taxon compositions between hot springs.
Several factors could in principle contribute to the observed differences in community structure between White Creek and Rabbit Creek (Fig. 1 and 6; see Table S3 in the supplemental material). We suggest that water chemistry may play a role. For example, at temperatures below ∼55°C, the hot springs differed markedly in the availability of combined nitrogen (Table 1). At these temperatures, the heterocyst-forming, nitrogen-fixing cyanobacterium Mastigocladus laminosus (OTU 54) was the most abundant cyanobacterium in White Creek, but no M. laminosus sequences were recovered from Rabbit Creek (Fig. 6; see Table S3 in the supplemental material). Other possible explanations for differences in taxon composition between the two hot springs include dispersal limitations, differences in the outcome of interactions among taxa, and/or failure to recover certain rare OTUs from a particular hot spring at our level of sampling.
Ecological interactions between cyanobacteria and Chloroflexi.
Because simply describing the distribution patterns of microorganisms has historically been a technological challenge, there remain several unresolved questions regarding the organization of microbial communities, which recapitulate longstanding debates in ecological theory developed for communities of macroorganisms. For example, are microbial communities simply chance assemblages of individuals with similar environmental requirements (13), or, alternatively, do the outcomes of ecological interactions among taxa contribute to community structure? Do these include negative interactions such as competition, or, rather, do microbial systems consist primarily of closed communities of positively associated populations that are discretely distributed along an environmental gradient (9)?
The distributions of cyanobacteria and both green and red Chloroflexi (Fig. 6) suggest an important role for biotic interactions in community organization although not in the manner predicted by the coadaptation hypothesis. We do not believe that the strongly negative association between these taxa can be explained solely by differences among community members in environmental tolerance (i.e., by niche-based processes). For example, the realized distributions of Synechococcus and Mastigocladus OTUs are much more narrow than the thermal performance curves of strains grown in the laboratory (32, 33). We therefore conclude that cyanobacteria and Chloroflexi likely compete with each other in these communities for habitat (i.e., physical space) and/or a limiting nutrient.
The nature of the potential competitive interactions between cyanobacteria and Chloroflexi remains to be determined. Although a producer-consumer relationship would not exclude competition for a limiting resource, another possibility is that these groups may instead compete within the same trophic level as autotrophs for carbon dioxide (16, 24, 26, 30). Although it was previously shown that light stimulates the uptake of organic carbon within these mat communities in situ (3, 4, 40), stable carbon isotope signatures in Chloroflexi lipids have deviated from the expectation for cross-feeding alone (46, 47). Future investigations will seek to clarify the basis of the competition between these groups and whether the observed negative association between these groups may be generalized to other microbial mat communities.
Supplementary Material
Acknowledgments
We thank Arpiar Saunders and Julie Rose for their generous assistance and advice with the writing of Perl scripts for OTU identification and with the ordination analyses, respectively. We also thank Bill Inskeep and Rich Macur of Montana State University for performing the water chemistry analyses, Christie Hendrix and the NPS for supporting our research in Yellowstone National Park, and Winsor Lowe and two anonymous reviewers for comments on an earlier version of the manuscript.
This work was supported by the National Science Foundation EPSCoR Ecological Genomics Program at Kansas State University and by National Science Foundation grant MCB-0347627 (to S.R.M.).
Footnotes
Published ahead of print on 8 May 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Allen, A. P., J. H. Brown, and J. F. Gillooly. 2002. Global biodiversity, biochemical kinetics, and the energetic-equivalence rule. Science 297:1545-1548. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. Myers, and D. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, K. L., T. A. Tayne, and D. M. Ward. 1987. Formation and fate of fermentation products in hot spring cyanobacterial mats. Appl. Environ. Microbiol. 53:2343-2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bateson, M. M., and D. M. Ward. 1988. Photoexcretion and fate of glycolate in a hot spring cyanobacterial mat. Appl. Environ. Microbiol. 54:1738-1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brock, T. D. 1978. Thermophilic microorganisms and life at high temperatures. Springer-Verlag, New York, NY.
- 6.Bryant, D. A., A. M. G. Costas, J. A. Maresca, A. G. M. Chew, C. G. Klatt, M. M. Bateson, L. J. Tallon, J. Hostetler, W. C. Nelson, J. F. Heidelberg, and D. M. Ward. 2007. Candidatus Chloracidobacterium thermophilum: an aerobic phototrophic acidobacterium. Science 317:523-526. [DOI] [PubMed] [Google Scholar]
- 7.Chao, A. 1984. Non-parametric estimation of the number of classes in a population. Scand. J. Statist. 11:265-270. [Google Scholar]
- 8.Chao, A., and S.-M. Lee. 1992. Estimating the number of classes via sample coverage. J. Am. Stat. Assoc. 87:210-217. [Google Scholar]
- 9.Clements, F. E. 1916. Plant succession. Carnegie Inst. Wash. Publ. 242:1-512. [Google Scholar]
- 10.DasSarma, S., and E. F. Fleischmann. 1995. Archaea: a laboratory manual, p. 269-272. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 11.Fierer, N., and R. B. Jackson. 2006. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. USA 103:626-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaston, K. J. 2000. Global patterns in biodiversity. Nature 405:220-227. [DOI] [PubMed] [Google Scholar]
- 13.Gleason, H. A. 1917. The structure and development of the plant association. Bull. Torrey Bot. Club 43:463-481. [Google Scholar]
- 14.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 15.Hansen, M. C., T. Tolker-Neilson, M. Givskov, and S. Molin. 1998. Biased 16S rDNA PCR amplification caused by interference from DNA flanking template region. FEMS Microbiol. Ecol. 26:141-149. [Google Scholar]
- 16.Holo, H., and R. Sirevåg. 1986. Autotrophic growth and CO2 fixation in Chloroflexus aurantiacus. Arch. Microbiol. 145:173-180. [Google Scholar]
- 17.Horner-Devine, M. C., K. M. Carney, and B. J. M. Bohannan. 2004. An ecological perspective on bacterial biodiversity. Proc. Biol. Sci. 271:113-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horner-Devine, M. C., M. Lage, J. B. Hughes, and B. J. M. Bohannan. 2004. A taxa-area relationship for bacteria. Nature 432:750-753. [DOI] [PubMed] [Google Scholar]
- 19.Huang, X., and A. Madan. 2006. CAP3: a DNA sequence assembly program. Genome Res. 9:868-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hubbell, S. P. 2001. The unified neutral theory of biodiversity and biogeography. Princeton University Press, Princeton, NJ.
- 21.Huber, J. A., D. B. M. Welch, H. G. Morrison, S. M. Huse, P. R. Neal, D. A. Butterfield, and M. L. Sogin. 2007. Microbial population structures in the deep marine biosphere. Science 318:97-100. [DOI] [PubMed] [Google Scholar]
- 22.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huse, S. M., J. A. Huber, H. G. Morrison, M. L. Sogin, and D. M. Welch. 2007. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 8:R143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klatt, C. G., D. A. Bryant, and D. M. Ward. 2007. Comparative genomics provides evidence for the 3-hydroxypropionate autotrophic pathway in filamentous anoxygenic phototrophic bacteria and in hot spring microbial mats. Environ. Microbiol. 9:2067-2078. [DOI] [PubMed] [Google Scholar]
- 25.Ley, R. E., J. K. Harris, J. Wilcox, J. R. Spear, S. R. Miller, B. M. Bebout, J. A. Maresca, D. A. Bryant, M. L. Sogin, and N. R. Pace. 2006. Unexpected diversity and complexity of the Guerrero Negro hypersaline microbial mat. Appl. Environ. Microbiol. 72:3685-3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Madigan, M. T., and T. D. Brock. 1975. Photosynthetic sulfide oxidation by Chloroflexus aurantiacus, a filamentous, photosynthetic, gliding bacterium. J. Bacteriol. 122:782-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Margulies, M., M. Egholm, W. E. Altman, S. Attiya, J. S. Bader, L. A. Bemben, J. Berka, M. S. Braverman, Y. J. Chen, Z. Chen, S. B. Dewell, L. Du, J. M. Fierro, X. V. Gomes, B. Z. C. Godwin, W. He, S. Helgesen, C. H. Ho, G. P. Irzyk, S. C. Jando, M. L. I. Alenquer, T. P. Jarvie, K. B. Jirage, J. B. Kim, J. R. Knight, J. R. Lanza, J. H. Leamon, S. M. Lefkowitz, M. Lei, J. Li, K. L. Lohman, H. Lu, V. B. Makhijani, K. E. McDade, M. P. McKenna, E. W. Myers, E. Nickerson, J. R. Nobile, R. Plant, B. P. Puc, M. T. Ronan, G. T. Roth, G. J. Sarkis, J. F. Simons, J. W. Simpson, M. Srinivasan, K. R. Tartaro, A. Tomasz, K. A. Vogt, G. A. Volkmer, S. H. Wang, Y. Wang, M. P. Weiner, P. Yu, R. F. Begley, and J. M. Rothberg. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martiny, J. B. H., B. J. M. Bohannan, J. H. Brown, R. K. Colwell, J. A. Fuhrman, J. L. Green, M. C. Horner-Devine, M. Kane, J. A. Krumins, C. R. Kuske, P. J. Morin, S. Naeem, L. Ovreas, A.-L. Reysenbach, V. H. Smith, and J. T. Staley. 2006. Microbial biogeography: putting microorganisms on the map. Nat. Rev. Microbiol. 4:102-112. [DOI] [PubMed] [Google Scholar]
- 29.McCune, B., and J. B. Grace. 2002. Analysis of ecological communities. MjM Software Design, Gleneden Beach, OR.
- 30.Menendez, C., Z. Bauer, H. Huber, N. Gad'on, K.-O. Stetter, and G. Fuchs. 1999. Presence of acetyl coenzyme A (CoA) carboxylase and propionyl-CoA carboxylase in autotrophic Crenarchaeota and indication for operation of a 3-hydroxypropionate cycle in autotrophic carbon fixation. J. Bacteriol. 181:1088-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller, S. R., C. E. Wingard, and R. W. Castenholz. 1998. Effects of visible light and UV radiation on photosynthesis in a population of a hot spring cyanobacterium, a Synechococcus sp., subjected to high-temperature stress. Appl. Environ. Microbiol. 64:3893-3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller, S. R., and R. W. Castenholz. 2000. Evolution of thermotolerance in hot spring cyanobacteria of the genus Synechococcus. Appl. Environ. Microbiol. 66:4222-4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller, S. R., R. W. Castenholz, and D. Pedersen. 2007. Phylogeography of the thermophilic cyanobacterium Mastigocladus laminosus. Appl. Environ. Microbiol. 73:4751-4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nekola, J. C., and P. S. White. 1999. The distance decay of similarity in biogeography and ecology. J. Biogeograph. 26:867-878. [Google Scholar]
- 35.Nübel, U., F. Garcia-Pichel, M. Kuhl, and G. Muyzer. 1999. Quantifying microbial diversity: morphotypes, 16S rRNA genes, and carotenoids of oxygenic phototrophs in microbial mats. Appl. Environ. Microbiol. 65:422-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oberdorff, T., J.-F. Guégan, and B. Hugueny. 1995. Global scale patterns of fish species richness in rivers. Ecography 18:345-352. [Google Scholar]
- 37.Prosser, J. I., B. J. M. Bohannan, T. P. Curtis, R. J. Ellis, M. K. Firestone, R. P. Freckleton, J. L. Green, L. E. Green, K. Killham, J. J. Lennon, A. M. Osborn, M. Solan, C. J. van der Gast, and J. P. W. Young. 2007. The role of ecological theory in microbial ecology. Nat. Rev. Microbiol. 5:384-392. [DOI] [PubMed] [Google Scholar]
- 38.Roesch, L. F. W., R. R. Fulthorpe, A. Riva, G. Casella, A. K. M. Hadwin, A. D. Kent, S. H. Daroub, F. A. O. Camargo, W. G. Farmerie, and E. W. Triplett. 2007. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 1:283-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roy, K., D. Jablonski, J. W. Valentine, and G. Rosenberg. 1998. Marine latitudinal diversity gradients: tests of causal hypotheses. Proc. Natl. Acad. Sci. USA 95:3699-3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sandbeck, K. A., and D. M. Ward. 1981. Fate of immediate methane precursors in low-sulfate, hot-spring algal-bacterial mats. Appl. Environ. Microbiol. 41:775-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sogin, M. L., H. G. Morrison, J. A. Huber, D. M. Welch, S. M. Huse, P. R. Neal, J. M. Arrieta, and G. J. Herndl. 2006. Microbial diversity in the deep sea and the underexplored “rare biosphere.” Proc. Natl. Acad. Sci. USA 103:12115-12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soininen, J., R. McDonald, and H. Hillebrand. 2007. The distance decay of similarity in ecological communities. Ecography 30:3-12. [Google Scholar]
- 43.Stevens, G. C. 1989. The latitudinal gradient in geographical range: how so many species coexist in the tropics. Am. Nat. 133:240-256. [Google Scholar]
- 44.Stott, M. B., M. A. Crowe, B. W. Mountain, A. V. Smirnova, S. Hou, M. Alam, and P. F. Dunfield. 2008. Isolation of novel bacteria, including a candidate division, from geothermal soils in New Zealand. Environ. Microbiol. 10:2030-2041. [DOI] [PubMed] [Google Scholar]
- 45.Sundquist, A., S. Bigdeli, R. Jalili, M. Druzin, S. Waller, K. Pullen, Y. El-Sayed, M. M. Taslimi, S. Batzoglou, and M. Ronaghi. 2007. Bacterial flora-typing with targeted, chip-based pyrosequencing. BMC Microbiol. 7:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van der Meer, M. T. J., S. Schouten, J. W. de Leeuw, and D. M. Ward. 2000. Autotrophy of green non-sulphur bacteria in hot spring microbial mats: biological explanations for isotopically heavy organic carbon in the geological record. Environ. Microbiol. 2:428-435. [DOI] [PubMed] [Google Scholar]
- 47.van der Meer, M. T. J., S. Schouten, J. S. Sinninghe Damsté, J. W. de Leeuw, and D. M. Ward. 2003. Compound-specific isotopic fractionation patterns suggest different carbon metabolisms among Chloroflexus-like bacteria in hot-spring microbial mats. Appl. Environ. Microbiol. 69:6000-6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ward, D. M., T. A. Tayne, K. L. Anderson, and M. M. Bateson. 1987. Community structure and interactions among community members in hot spring cyanobacterial mats. Symp. Soc. Gen. Microbiol. 41:179-210. [Google Scholar]
- 49.Ward, D. M., M. J. Ferris, S. C. Nold, and M. M. Bateson. 1998. A natural view of microbial diversity within hot spring cyanobacterial mat communities. Microbiol. Mol. Biol. Rev. 62:1353-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.