Abstract
Staphylococcus aureus is responsible for numerous food poisonings due to the production of enterotoxins by strains contaminating foodstuffs, especially dairy products. Several parameters, including interaction with antagonistic flora such as Lactococcus lactis, a lactic acid bacterium widely used in the dairy industry, can modulate S. aureus proliferation and virulence expression. We developed a dedicated S. aureus microarray to investigate the effect of L. lactis on staphylococcal gene expression in mixed cultures. This microarray was used to establish the transcriptomic profile of S. aureus in mixed cultures with L. lactis in a chemically defined medium held at a constant pH (6.6). Under these conditions, L. lactis hardly affected S. aureus growth. The expression of most genes involved in the cellular machinery, carbohydrate and nitrogen metabolism, and stress responses was only slightly modulated: a short time lag in mixed compared to pure cultures was observed. Interestingly, the induction of several virulence factors and regulators, including the agr locus, sarA, and some enterotoxins, was strongly affected. This work clearly underlines the complexity of L. lactis antagonistic potential for S. aureus and yields promising leads for investigations into nonantibiotic biocontrol of this major pathogen.
Staphylococcus aureus is a major human pathogen that causes a wide range of diseases, such as septicemia, meningitis, endocarditis, osteomyelitis, and toxic shock syndrome (77). S. aureus is also a major causative agent of food poisonings (41). Staphylococcal food poisonings (SFP) are due to the production of staphylococcal enterotoxins (SEs) by S. aureus strains contaminating foodstuffs. Milk and dairy products are often incriminated in SFP, especially in France (19, 20, 35). During the cheese-making process, contamination by S. aureus may come from several sources. Raw milk (notably milk from mastitic cows) may be a source of contamination; however, strains endemic in the processing plant environment (present on fomites and in biofilms), as well as strains present in healthy human carriers, may also contaminate dairy products. A complex and balanced ecosystem is required throughout the cheese-making process for the development of the organoleptic properties of the final product. The inhibition of S. aureus growth and of the production of SE in cheeses must thus be achieved by nonantibiotic means so that the biodiversity and correct microbial ecology of the environment may be maintained, yielding a satisfactory final product. To this end, the appropriate and rational use of lactic acid bacteria (LAB) appears to be a promising strategy in the fight against S. aureus.
Interactions between S. aureus and LAB in several ecosystems, including fermented foodstuffs but also nasal and vaginal environments, have been explored for years (11). However, apart from offering general observations, studies have not yet comprehensively described the mechanism of inhibition of S. aureus by LAB. LAB have frequently been considered to be a factor that influences S. aureus growth by physically and chemically changing the environment. Several possibilities have been proposed to explain the inhibition of S. aureus by LAB, including the production of bacteriocins (2) and hydrogen peroxide (2, 58), competition for nutrients (28), and needless to say, acidification (4, 20, 52), even if the impact of the latter mechanism has been questioned (12). Studies of the inhibition of S. aureus virulence expression by LAB, including the inhibition of SE production, are quite scarce. Few studies have described the inhibition of enterotoxin production in the presence of LAB, and none have unraveled the mechanisms involved in such antagonism (26, 29, 50, 51, 57, 68).
Virulence expression in S. aureus is controlled by complex regulatory networks that include two-component systems (e.g., AgrAC, SrrAB, SaeRS, and ArlRS) and transcription factors (e.g., SarA and its homologs and Rot) but also the Clp proteolytic complex and the alternative sigma factor σB (5, 13, 15, 21, 47, 48, 54, 60, 64, 72). Among these regulatory systems, the accessory gene regulator (agr) is a key modulater of virulence expression. It combines a two-component system and a quorum-sensing system (55, 64, 78). The agr system comprises two transcripts, RNAII and RNAIII, which are divergently transcribed. RNAII encodes AgrBDCA, the structural components of the quorum-sensing system. RNAIII is the effector molecule of the agr system but also encodes the delta-hemolysin (32, 55). The accessory gene regulator is responsible for the post-exponential-phase induction of several virulence factors, including exoproteins (21, 54, 62, 64, 72). The activity of agr itself can be modulated by other regulatory systems and in response to environmental variations.
Exploring the interactions between positive microbiota and S. aureus and understanding the mechanisms involved in the inhibition of S. aureus growth or virulence will enable the setting of rational screening criteria for positive microbiota to be used in food preservation and will also yield new strategies for fighting against this major human pathogen. Such an endeavor is, however, complex and requires the employment of new approaches. DNA microarrays are powerful tools for the investigation of these multifactorial interactions. The microarray technique has been widely used to investigate the global responses of S. aureus to several stimuli (5, 10, 17, 21, 38, 59, 65, 76). Nonetheless, a transcriptomic analysis of S. aureus in interaction with other bacterial species has, as of yet, not been reported. In this study, we have developed a dedicated S. aureus-specific microarray, usable for the study of mixed cultures of S. aureus and LAB. This microarray was used to investigate the transcriptomic response of S. aureus to the presence of Lactococcus lactis in a chemically defined medium (CDM) at a constant pH. Surprisingly, while L. lactis hardly affected S. aureus growth and the expression of genes involved in central metabolism processes under these conditions, it was able to dramatically impair the expression of several virulence genes.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
L. lactis subsp. lactis biovar diacetylactis LD61 (kindly provided by R. Perrin, Soredab, La Boissière Ecole, France) was used throughout this work. The LD61 strain contains plasmids that enable optimal growth in milk (by facilitating lactose, protease, and citrate utilization) and is thus a highly acidifying strain which is industrially used in dairy fermentations. L. lactis LD61 does not produce any detectable bacteriocin active against S. aureus (data not shown). The previously sequenced strain S. aureus MW2 was chosen for the interaction experiments for the following reasons. The complete genome sequence of MW2 is publicly available, which ensures access to information on the gene content of the strain. Of note, MW2 possesses genes encoding several toxins, including six enterotoxins, of which the sea and sec toxins (3) are frequently involved in SFP (41). The expression of some enterotoxin genes, including sec, is known to be under agr control (62), which is functional in this strain (74) in contrast to other strains (e.g., N315 and RN4220). Furthermore, S. aureus MW2 was preferred over other sequenced strains because it is a typical community-acquired strain of S. aureus. It is actually more probable for community rather than clinical strains to contaminate foodstuffs. Moreover, contamination with S. aureus strains of human origin is reportedly prevalent in SFP outbreaks (37), and the MW2 strain belongs to one of the major clonal lineages found among human isolates of S. aureus (43).
Mixed cultures of S. aureus and L. lactis and pure cultures (used as controls) were grown on CDM at 30°C and a constant pH (6.6) in a 2-liter fermentor as described elsewhere (53). Both strains were inoculated at 106 CFU ml−1. The kinetics of growth, consumption of glucose, and formation of fermentation products are discussed elsewhere (53).
Two additional S. aureus strains, clonally unrelated to the MW2 strain (which belongs to sequence type 1 [ST1] and clonal complex 1) (43), were used in complementary interaction experiments: SH1000 (30), a derivative of strain NCTC 8325 (ST8, clonal complex 8; agrI), and RN4850 (ST not available; agrIV [kindly provided by G. Lina, CNR des Staphylococcies, Lyon, France]) (33).
RNA extraction and purification.
Volumes of culture each corresponding to 6 mg of cells (dry weight) were harvested and frozen immediately in liquid nitrogen. Samples were stored at −80°C until RNA extraction. Samples were thawed on ice and centrifuged (at 4°C for 5 min at 6,500 × g), and cell pellets were resuspended in 500 μl of Tris-EDTA buffer. Cells in tubes containing a mixture of 0.6 g of zirconium beads, 25 μl of sodium dodecyl sulfate (20%), 3.5 μl of β-mercaptoethanol, and 500 μl of phenol (pH 4.7) were disrupted for 4 min at 30 Hz by using a Retsch MM301 high-speed mixer mill (Grosseron, France). After the addition of 200 μl of chloroform and centrifugation (at 4°C for 25 min at 15,500 × g), the aqueous phase was extracted using a Nucleospin RNA L kit according to the instructions of the manufacturer (Macherey Nagel, Hoerdt, France). After precipitation with isopropanol, the RNA sample was incubated with RNase-free DNase I from a DNA-free kit (Applied Biosystems, Warrington, United Kingdom). RNA was quantified and its contamination by proteins was assessed using an ND-1000 spectrophotometer (NanoDrop Technologies, Inc., Rockland, DE), and the quality of the preparation was evaluated using a model 2100 bioanalyzer (Agilent Technologies, Santa Clara, CA). Noncontamination of the RNA sample by genomic DNA (gDNA) was confirmed by quantitative PCR.
gDNA extraction and purification.
gDNA was prepared from overnight cultures grown in tryptic soy broth with agitation at 37°C (S. aureus) or grown under static conditions on M17 supplemented with 0.5% glucose at 30°C (L. lactis). Briefly, cells were centrifuged (at 4°C for 5 min at 6,500 × g), washed in 1 ml of Tris-HCl (50 mM; pH 8), and resuspended in 450 μl of Tris-EDTA buffer. Cells were disrupted with 0.6 g of zirconium beads for 4 min at 30 Hz by using an MM301 high-speed mixer mill. After centrifugation (at 4°C for 25 min at 15,500 × g), the lysate was incubated at 37°C with RNase (50 μg μl−1) for 30 min and proteinase K (0.4 mg ml−1) for one additional hour. Following extraction with phenol-chloroform-isoamyl alcohol (25/24/1) and chloroform, gDNA was precipitated with isopropanol and resuspended in water. DNA was quantified and contamination by proteins was assessed using a NanoDrop ND-1000 spectrophotometer.
Construction of the S. aureus microarray.
A dedicated microarray containing 420 probes specific for S. aureus and 27 probes specific for L. lactis IL1403 was designed. The complete list of target genes is available in the supplemental material. The design of oligonucleotides (60-mers) was carried out using OligoArray 2.1 software (66, 67) and was based on the genome sequence of strain Mu50 (BA000017), which is one of the strains for which the complete genome sequence was publicly available at the beginning of this work and for which the annotation is regularly updated (http://www.bio.nite.go.jp/dogan/Top). The design took into account the use of the microarray in the context of mixed cultures with L. lactis (and possibly other LAB). Oligonucleotides generated by using OligoArray 2.1 were selected based on thermodynamic parameters and their specificities with regard to S. aureus, as determined by a BLAST analysis comparing the oligonucleotide sequences against genome sequences of S. aureus Mu50, LAB (L. lactis IL1403 [AE005176], Streptococcus thermophilus CNRZ1066 [CP000024], Lactobacillus acidophilus NCFM [CP000033], and Lactobacillus plantarum WCFS1 [AL935263]), and Bifidobacterium longum NCC2705 (AE014295). The specificity of the microarray with regard to S. aureus MW2 was further validated by a BLAST analysis comparing the oligonucleotide sequences against the MW2 genome sequence: 92.6% of oligonucleotides hybridized in silico with strain MW2. All remaining oligonucleotides which did not hybridize in silico corresponded to genes carried by a genomic island or mobile genetic elements in strain Mu50 and absent from strain MW2. The 5′-end-aminated oligonucleotides (Sigma Genosys, Suffolk, United Kingdom) were spotted onto Slide E epoxysilane slides (Schott Nexterion, Jena, Germany) in sextuplicate with a Biorobotics MicroGrid II as specified by the manufacturer (Genomic Solutions Ltd., Cambridgeshire, United Kingdom). This spotting step was carried out through the transcriptomic platform at Ouest Genopole (Rennes, France).
Fluorescent labeling.
Five micrograms of total RNA was labeled with Cy3- or Cy5-dCTP (Perkin Elmer, Waltham, MA) by using a ChipShot kit according to the recommendations of the manufacturer (Promega, Madison, WI). Labeled cDNA was dried out and stored at −20°C until use. gDNA was labeled basically as described previously (23) with modifications as follows: gDNA was digested overnight with restriction endonuclease HinPII, precipitated with isopropanol, and resuspended in water. Samples of 400 ng of digested gDNA were denatured at 95°C for 5 min and employed as a template in a 50-μl reaction mixture containing reaction buffer (1×); dATP, dGTP, and dTTP (each at 20 μM); Cy3-dCTP (0.5 mM); bovine serum albumin (0.4 mg ml−1); and a Klenow fragment (5 U) for the direct incorporation of Cy3 or Cy5-dCTP by using the Prime-a-Gene labeling system (Promega, Madison, WI). The reaction mixture was incubated for 6 h at 22°C, and the reaction was stopped by adding EDTA (20 mM; pH 8.0). Unincorporated nucleotides were removed using a Nucleospin Extract II kit (Macherey Nagel, Hoerdt, France). The reaction mixture was supplemented with 4 volumes of NTC buffer (Macherey Nagel) and loaded onto a column. Labeled gDNA was dried out and stored at −20°C until use, according to the specifications of the labeling system manufacturer.
Hybridization and scanning of microarrays.
Hybridization and scanning of microarrays were performed using the transcriptomic platform of the IFR 140 Génétique Fonctionnelle, Agronomie et Santé (Université Rennes 1, France). The specificity of the S. aureus microarray was tested by hybridizing either labeled cDNA or gDNA from S. aureus MW2 and N315 and L. lactis IL1403, separately or in competition. The expression profile of S. aureus in pure cultures or mixed cultures with L. lactis was determined by hybridizing Cy5-labeled cDNA corresponding to each sample of RNA with Cy3-labeled gDNA of S. aureus MW2 and N315 (50:50). gDNA was used as a reference for normalization, as described previously (25, 70). Microarray analyses at all experimental time points were performed in triplicate (using three independent biological replicates).
Labeled cDNA and gDNA were resuspended in 200 μl of ChipHybe 80 (reference no. 760-127; Ventana). Hybridizations were carried out in a Discovery station at 42°C for 8 h according to the recommendations of the manufacturer (Ventana, Illkirch, France). Slides were then washed twice manually in RiboWash solution (Ventana, Illkirch, F) and once in 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), dried by centrifugation, and scanned with a Genepix 4000B scanner (Axon Instruments, Foster City, CA). Image analysis was performed with GenePix Pro4 software (Axon Instruments, Foster City, CA).
Data analysis.
The median intensity of each spot was used as the signal value. To test species specificity, spots were considered to be detected when the signal value was higher than a background value, calculated as the mean intensity of spots containing buffer plus 2 standard deviations.
To determine the expression profiles for S. aureus in pure and mixed cultures, data were analyzed with R software (63). Oligonucleotides with very low signal values for gDNA (corresponding to a signal/noise ratio of less than 2 in more than 75% of microarrays) were flagged and removed from further analysis. Data were first normalized per spot (cDNA signal values were divided by corresponding gDNA signal values) and log2 transformed and then normalized per chip (to the 50th percentile). Statistical analyses were performed using the analysis of variance (ANOVA) test with the criteria that P values and false-discovery rates (FDRs) be lower than 0.05. Genes showing significant changes in expression in either pure or mixed cultures and a ≥3-fold change were considered to be differentially expressed.
qRT-PCR.
To confirm microarray data, expression profiles of agrA, hld, sigB, asp23, clpL, capA, katA, sel, sea, MW2096, and MW802 were determined by quantitative reverse transcription-PCR (qRT-PCR) analyses. Expression profiles of sarA, spa, hla, and genes encoding enterotoxins not included in the S. aureus microarray, namely, seh, sec4, sek, and seg, were also determined by qRT-PCR. cDNA was synthesized using the high-capacity cDNA archive kit as recommended by the manufacturer (Applied Biosystems, Warrington, United Kingdom). Quantitative real-time PCR was performed using an Opticon 2 real-time PCR detector (Bio-Rad, Hercules, CA). The reaction mixture contained power Sybr green PCR master mix (1×; Applied Biosystems, Warrington, United Kingdom), each primer (0.5 μM; sequences are given in Table 1), and a cDNA template. Thermal cycling consisted of 10 min at 95°C, followed by 40 cycles of 15 s at 95°C and 60 s at 60°C. Each PCR product was further analyzed by generating melting curves to ensure the specificity of the assay. qRT-PCR analyses for all experimental time points were performed in triplicate (using three independent biological replicates). Standard curves were generated to calculate the copy number for each gene in each sample. The hu gene was used as an internal standard for normalization as described previously (25). Gene expression levels are thus reported relative to that for hu and were calculated by using following the relation: (number of copies of each gene in each cDNA preparation/number of copies of hu in each cDNA preparation) ×10. Statistical analysis was performed using the ANOVA test with the criterion of a P value lower than 0.05 to identify genes showing significant changes in expression in either pure or mixed cultures over time. Similarly, statistical analysis was performed using the t test with the criterion of a P value lower than 0.05 to identify, for each time point, genes showing significant changes in expression in pure versus mixed cultures.
TABLE 1.
Oligonucleotides used in this study for quantitative real-time PCR
| Gene identification no. | Gene name(s) | Forward primer | Reverse primer |
|---|---|---|---|
| MW0051 | seh | TGGTCAATATAATCACCCATTCA | TCAAATCATTGCCACTATCACC |
| MW0084 | spa | TTAAAGACGATCCTTCAGTGAGC | TGTTGTTGTCTTCCTCTTTTGGT |
| MW0124 | cap8A | GCGCTATTGTTACATTTTTCGTC | TCTTGTGCCATAAACTGAGGATT |
| MW0759 | sec4 | AAACATGAAGGAAACCACTTTGA | TTTGCACTTCAAAAGAAATTGTG |
| MW0760 | sel2 | GGTTACCGCACAAGAAATAGATG | TGCCGTATTCTTTACCTTTACCA |
| MW0802 | TCTAAGGGTCAACCTCAAGAACA | TCCAACCTACTCGTCTCAATTTC | |
| MW1044 | hla | ATGGATAGAAAAGCATCCAAACA | TTTCCAATTTGTTGAAGTCCAAT |
| MW1362 | hu | AGAAGCTGGTTCAGCAGTAGATG | TACCTCAAAGTTACCGAAACCAA |
| MW1889 | sea | TAATCGATTGACCGAAGAGAAAA | ATAACGTCTTGCTTGAAGATCCA |
| MW1937 | seg2 | TACGATTTGTTTTACACCGGAAC | TCCAAATGAAAATTCTCTGCATC |
| MW1938 | sek2 | CTACACAGGAGATGATGGGCTAC | CATCCAAATGGAATTTCTCAGAC |
| MW1959 | RNAIII/hld | TAAGGAAGGAGTGATTTCAATGG | GTGAATTTGTTCACTGTGTCGAT |
| MW1963 | agrA | CCTCGCAACTGATAATCCTTATG | ACGAATTTCACTGCCTAATTTGA |
| MW2108 | asp23 | AGACATGAAAGGTGGCTTAACTG | GCTTGTTTTTCACCAACTTCAAC |
| MW2469 | clpL | AAGATGCACTGATTCGACTTGAT | TGTCATCATAACCGACATAACCA |
Microarray data accession number.
The microarray data were deposited in the public repository database Array Express under the accession number E-MEXP-1837.
RESULTS
Development and validation of an S. aureus microarray usable with mixed cultures.
The goal of this study was to investigate S. aureus gene expression profiles during S. aureus interaction with L. lactis in mixed cultures. Bacterial populations of each species could not be separated or differentially lysed prior to retrotranscription and cDNA labeling. Probe specificity was thus a crucial parameter. The strategy for the design of S. aureus-specific probes focused on having only labeled cDNA generated from S. aureus RNA out of a bulk of total RNAs of both species that would hybridize on the microarray. Each probe had to be specific to only one gene within the S. aureus genome, and cross-hybridization with genes of the L. lactis genome had to be minimized as well. Oligonucleotides (60-mers) were preferred to PCR fragments due to the potential of the latter for cross-hybridization and the consideration of oligonucleotides, due to their short length, to be more specific. The 420 S. aureus genes selected for the design corresponded to genes involved in virulence and associated regulation pathways, stress adaptation, peptide transport, central metabolism (for carbohydrates and amino acids), cellular machinery (for replication, transcription, and translation), and cell wall biosynthesis and turnover (see the supplemental material). Twenty-seven oligonucleotide probes specific to L. lactis IL1403 (as determined by genome subtraction) were included as controls. Oligonucleotides were designed and validated in silico as described in Materials and Methods to minimize the risk of cross-hybridization with LAB cDNA. The specificity of the microarray for S. aureus was experimentally validated by performing hybridizations with cDNAs or gDNA of L. lactis LD61 and S. aureus MW2 and/or N315, a strain closely related to strain Mu50 (39). A mixture of gDNA of strains MW2 and N315 was used in order to maximize the detection of oligonucleotides: 92.6 and 98.6% of oligonucleotides were indeed detected in silico (as determined by performing a BLAST analysis) with strains MW2 and N315, respectively. Major differences between strains corresponded to oligonucleotides targeting genes located on genomic islands or mobile genetic elements present in strain Mu50 but absent in other strains, as observed previously (3). Of the oligonucleotides designed based on S. aureus Mu50 genes, 76% were detected using a mixture of labeled gDNA extracts from S. aureus N315 and MW2. This yield slightly decreased to 70.4% with gDNA from S. aureus MW2 alone, as expected from in silico hybridization results. In vitro yields of hybridization were in good agreement with those generally observed with gDNA, which range from 65 to 80%, depending on the preparation of gDNA and labeling and hybridization conditions (70). In parallel, total RNA was extracted from MW2 or N315 cells harvested in post-exponential phase. Hybridization with cDNA obtained from these samples showed that 72 to 75% of S. aureus oligonucleotides were detected. In hybridization experiments using L. lactis LD61 cDNA and/or gDNA, only 16 of the 420 oligonucleotides (3.8%) targeting S. aureus genes gave a signal (see the supplemental material). These 16 oligonucleotides were removed from the analysis when the microarray was used further for mixed cultures. Among the oligonucleotides designed as positive controls for L. lactis, 78% were detected using L. lactis LD61 cDNA or gDNA. Altogether, these in vitro results validated the S. aureus specificity of the dedicated microarray developed in this study.
Time lag in the expression of S. aureus genes involved in the cellular machinery, carbohydrate metabolism, and stress responses in mixed cultures.
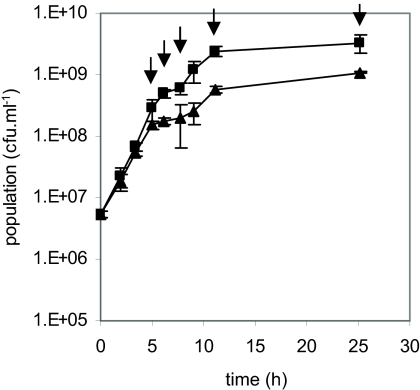
As a first approach to study interactions between S. aureus and L. lactis, we investigated the transcriptomic responses of S. aureus MW2 in pure and mixed cultures with L. lactis LD61 at 30°C in CDM at a constant pH (6.6) with agitation to homogenize and allow moderate aeration of the medium. These conditions allowed us to monitor the kinetics of substrate and product formation and to control environmental parameters such as a drop in pH, which may have masked other phenomena. Furthermore, the effect of acidification on S. aureus expression has already been documented (7, 76). S. aureus growth rates at the beginning of the exponential phase in pure and mixed cultures were similar. In the pure culture, S. aureus MW2 growth slowed down and a transient plateau of growth appeared at the end of the exponential phase (Fig. 1). In the mixed culture, the growth rate decrease occurred earlier and the plateau lasted longer. Altogether, the microbiological analyses showed that, under the conditions used, L. lactis hardly affected S. aureus growth and the final staphylococcal population in the mixed culture was only threefold lower than that in the pure culture.
FIG. 1.
Kinetics of growth of S. aureus MW2 in pure cultures (▪) and mixed cultures with L. lactis LD61 (▴) on CDM medium at 30°C and a constant pH. Data are the averages of results from three independent experiments. Arrows indicate the time points at which samples were harvested for transcriptome analyses.
S. aureus MW2 gene expression profiles in the pure culture and in the mixed culture with L. lactis LD61 were investigated. Samples were harvested at different time points (5, 6.2, 7.8, 11.1, and 25 h after inoculation) in order to monitor the kinetics of gene expression under both growth conditions. Genes whose expression varied significantly (≥3-fold) in the pure and/or the mixed culture were identified by an ANOVA test as described in Materials and Methods (Tables 2, 3, and 4).
TABLE 2.
Expression profiles of genes involved in cellular machinery, cell envelope biosynthesis or turnover, or metabolism of carbohydrates, nitrogen, nucleotides, nucleic acids, or iron that exhibited significant variations in pure and/or mixed culturesa
| ORF (Mu50)b | Corresponding ORF (MW2)c | Gene named | Description of gene product or ORFe | cDNA/gDNA ratiof for pure culture at h:
|
cDNA/gDNA ratiof for mixed culture at h:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5 | 6.2 | 7.8 | 11.1 | 25 | 5 | 6.2 | 7.8 | 11.1 | 25 | ||||
| Transcription, translation, and cellular division genes | |||||||||||||
| SAV0535 | MW0491 | nusG | Transcription antitermination protein | 4.0 | 1.8 | 1.1 | 1.3 | 1.1 | |||||
| SAV0547 | MW0502 | fus | Translational elongation factor G | 25.8 | 48.3 | 30.0 | 20.3 | 1.5 | 7.7 | 14.0 | 5.2 | 4.3 | 2.3 |
| SAV0548 | MW0503 | tufA | Translational elongation factor TU | 20.2 | 27.5 | 17.0 | 15.6 | 1.3 | 8.6 | 10.0 | 4.3 | 4.2 | 2.1 |
| SAV1678 | MW1622 | rplT | 50S ribosomal protein L20 | 4.4 | 3.8 | 4.6 | 3.1 | 0.4 | 1.9 | 1.8 | 1.2 | 0.5 | 0.6 |
| SAV1679 | MW1623 | rpmI | 50S ribosomal protein L35 | 5.0 | 8.3 | 6.5 | 3.3 | 0.4 | 3.5 | 2.6 | 1.3 | 0.6 | 0.6 |
| SAV1128 | MW1010 | rpmF | Ribosomal protein L32 | 5.2 | 4.4 | 2.0 | 1.7 | 1.3 | |||||
| IGS2 | Intergenic sequence (16S to 23S) | 14.4 | 25.9 | 13.6 | 10.2 | 1.0 | 20.7 | 9.7 | 3.1 | 1.1 | 0.9 | ||
| SAV1180 | MW1063 | ftsL | Cell division protein | 1.4 | 2.4 | 2.0 | 2.2 | 0.7 | |||||
| SAV0511 | MW0466 | ftsH | Cell division protein | 8.3 | 9.4 | 5.8 | 6.3 | 1.1 | |||||
| Cell envelope biosynthesis and turnover genes | |||||||||||||
| SAV0307 | MW0284 | Similar to outer membrane protein precursor | 2.7 | 2.8 | 2.0 | 3.0 | 0.4 | 1.9 | 1.4 | 0.8 | 0.9 | 0.6 | |
| SAV1433 | MW1323 | Similar to cell wall enzyme EbsB | 0.3 | 0.4 | 0.3 | 0.3 | 1.0 | ||||||
| SAV1474 | MW1363 | gpsA | Glycerol-3-phosphate dehydrogenase | 4.1 | 6.2 | 5.5 | 4.5 | 0.8 | |||||
| SAV1261 | MW1144 | cdsA | Phosphatidate cytidylyltransferase | 2.9 | 3.0 | 2.0 | 1.7 | 0.4 | |||||
| SAV1228 | MW1111 | Transcription factor FapR (fatty acid and phospholipid biosynthesis regulator) | 2.8 | 3.7 | 2.1 | 2.2 | 0.6 | ||||||
| SAV0465 | MW0419 | Probable autolysin (N-acetylmuramoyl-l-alanine amidase) | 2.8 | 2.8 | 10.2 | 4.7 | 0.6 | 2.3 | 1.5 | 1.8 | 0.7 | 0.6 | |
| SAV2090 | MW2013 | Lipoprotein precursor | 2.1 | 3.4 | 2.1 | 1.4 | 0.5 | ||||||
| SAV0464 | MW0418 | Lactococcal lipoprotein | 2.9 | 2.6 | 1.8 | 1.0 | 0.8 | ||||||
| SAV0642 | MW0604 | pbp4 | Penicillin binding protein 4 | 2.9 | 3.0 | 2.1 | 1.7 | 0.5 | |||||
| SAV1754 | MW1697 | Spore cortex protein homolog | 1.5 | 1.5 | 1.7 | 1.8 | 0.5 | ||||||
| SAV0935 | MW0817 | dltD | Poly(glycerophosphate chain) d-alanine transfer protein | 5.6 | 1.0 | 1.1 | 0.6 | 0.5 | |||||
| SAV2644 | MW2565 | Similar to autolysin precursor | 0.7 | 0.4 | 0.8 | 1.0 | 2.1 | ||||||
| Carbohydrate metabolism genes | |||||||||||||
| SAV1698 | MW1642 | pfk | 6-Phosphofructokinase | 7.5 | 7.3 | 9.2 | 10.4 | 2.3 | 7.5 | 3.9 | 3.2 | 2.3 | 1.6 |
| SAV1697 | MW1641 | pykA | Pyruvate kinase | 4.9 | 4.0 | 6.5 | 6.1 | 1.3 | 3.2 | 2.3 | 1.8 | 1.2 | 1.0 |
| SAV2609 | MW2528 | Acetate-CoA ligase | 1.6 | 1.2 | 1.0 | 1.8 | 18.6 | 0.8 | 0.5 | 0.7 | 1.8 | 4.2 | |
| SAV0605 | MW0568 | adh1 | Alcohol dehydrogenase I | 8.3 | 4.8 | 10.0 | 17.0 | 45.9 | |||||
| SAV1149 | MW1032 | sdhB | Iron sulfur subunit of succinate dehydrogenase | 1.9 | 1.8 | 2.2 | 2.0 | 7.4 | |||||
| SAV0269 | MW0245 | rbsD | Ribose permease | 0.2 | 0.3 | 0.4 | 0.3 | 1.0 | |||||
| SAV0214 | MW0190 | Similar to maltose/maltodextrin transport system | 0.5 | 0.5 | 0.3 | 0.3 | 1.1 | ||||||
| SAV2158 | MW2084 | mtlA | PTS | 0.9 | 0.9 | 0.5 | 0.6 | 7.3 | |||||
| SAV0189 | MW0163 | glcA | PTS enzyme II | 10.2 | 9.1 | 1.9 | 3.1 | 0.4 | 14.7 | 2.6 | 0.6 | 0.5 | 0.5 |
| SAV2323 | MW2244 | glvC | PTS | 0.3 | 0.4 | 0.3 | 0.3 | 4.2 | |||||
| SAV0474 | MW0428 | treP | Phosphoenolpyruvate-dependent and trehalose-specific PTS enzyme II | 0.3 | 0.4 | 0.2 | 0.3 | 1.1 | |||||
| SAV0771 | MW0733 | gapR | Glycolytic operon regulator | 17.8 | 24.7 | 5.9 | 13.9 | 9.1 | 10.5 | 10.4 | 4.1 | 1.3 | 1.6 |
| Energetic metabolism genes | |||||||||||||
| SAV2607 | MW2526 | mqo2 | Malate/quinone oxidoreductase | 6.1 | 4.8 | 5.9 | 5.0 | 1.2 | 2.7 | 2.0 | 1.8 | 1.3 | 1.0 |
| SAV1116 | MW0999 | ctaB | Cytochrome caa3 oxidase homolog | 6.5 | 6.3 | 3.9 | 4.7 | 2.3 | 2.2 | 3.0 | 1.5 | 1.3 | 0.9 |
| SAV0623 | MW0586 | Na+/H+ antiporter | 4.8 | 6.2 | 5.1 | 4.1 | 0.8 | 2.9 | 2.1 | 0.9 | 1.2 | 0.9 | |
| SAV0625 | MW0588 | MnhD homolog, similar to Na+/H+ antiporter subunit | 5.7 | 5.9 | 3.6 | 3.2 | 1.3 | ||||||
| SAV1832 | MW1772 | hemY | Protoporphyrinogen oxidase | 2.5 | 2.4 | 3.1 | 2.4 | 0.7 | |||||
| SAV0627 | MW0590 | Similar to Na+/H+ antiporter | 1.0 | 0.9 | 0.8 | 0.9 | 6.4 | ||||||
| Nitrogen metabolism genes | |||||||||||||
| SAV2056 | MW1980 | ilvC | Alpha-keto-beta-hydroxylacil reductoisomerase | 0.1 | 0.2 | 0.2 | 0.1 | 0.7 | |||||
| SAV1751 | MW1694 | Xaa-His dipeptidase homolog | 2.0 | 3.0 | 3.3 | 3.0 | 0.9 | 1.5 | 1.1 | 1.1 | 0.9 | 0.5 | |
| SAV0986 | MW0868 | oppB | Oligopeptide transport system permease protein | 0.8 | 1.2 | 8.6 | 6.9 | 2.1 | |||||
| SAV1380 | MW1268 | Oligopeptide transport ATPase | 0.5 | 0.8 | 0.9 | 0.7 | 4.3 | ||||||
| SAV1382 | MW1270 | Oligopeptide transporter membrane permease domain | 2.1 | 2.3 | 1.6 | 1.4 | 0.4 | ||||||
| SAV0727 | MW0689 | Ditripeptide ABC transporter | 3.9 | 3.0 | 4.7 | 4.3 | 0.5 | 7.2 | 1.6 | 1.8 | 0.6 | 0.4 | |
| SAV0989 | MW0871 | oppF | OppF homolog | 0.6 | 0.8 | 6.9 | 4.1 | 0.5 | |||||
| SAV0205 | MW0181 | oppF | Oligopeptide transport ATP binding protein | 0.2 | 0.2 | 0.2 | 0.2 | 0.6 | |||||
| SAV0010 | MW0010 | Amino acid permease | 0.3 | 0.3 | 0.6 | 0.4 | 1.5 | ||||||
| SAV2384 | MW2304 | gltT | Proton/sodium glutamate symport protein | 9.3 | 8.6 | 11.0 | 7.0 | 1.0 | |||||
| SAV1255 | MW1138 | codY | Pleiotropic transcription repressor | 3.2 | 4.4 | 2.9 | 3.8 | 0.6 | |||||
| Nucleotide and nucleic acid metabolism genes | |||||||||||||
| SAV1065 | MW0948 | purK | Phosphoribosylaminoimidazole carboxylase | 0.3 | 0.4 | 0.8 | 2.3 | 0.6 | |||||
| SAV0521 | MW0476 | nupC | Pyrimidine nucleoside transport protein | 4.7 | 4.6 | 5.3 | 5.9 | 0.7 | 2.6 | 2.2 | 1.4 | 0.9 | 0.4 |
| DNA replication, recombination, and repair genes | |||||||||||||
| SAV1146 | MW1029 | uvrC | Excinuclease ABC subunit C | 0.7 | 0.8 | 0.7 | 0.7 | 3.2 | |||||
| SAV1252 | MW1135 | xerC | Site-specific recombinase XerC homolog | 1.9 | 2.8 | 1.7 | 1.8 | 0.5 | 3.1 | 0.7 | 0.5 | 0.8 | 0.9 |
| SAV0432 | MW0393 | hsdS | Probable restriction modification system specificity subunit | 0.1 | 0.1 | 0.1 | 0.1 | 0.3 | |||||
| Secretion genes | |||||||||||||
| SAV0964 | MW0846 | spsA | Type I signal peptidase | 1.9 | 1.5 | 1.4 | 0.9 | 0.6 | 3.7 | 4.6 | 5.6 | 0.9 | 0.7 |
| SAV2230 | MW2149 | secY | Preprotein translocase SecY subunit | 32.2 | 69.4 | 35.6 | 33.1 | 6.3 | 23.5 | 16.2 | 8.3 | 5.0 | 3.0 |
| SAV0778 | MW0740 | secG | Probable protein export membrane protein | 5.7 | 7.8 | 5.7 | 4.2 | 2.0 | |||||
| Iron metabolism genes | |||||||||||||
| SAV1893 | MW1834 | Ferritin | 6.5 | 7.2 | 7.6 | 9.6 | 48.3 | ||||||
| SAV0225 | MW0200 | Similar to periplasmic iron binding protein BitC | 0.5 | 0.5 | 0.6 | 0.6 | 3.1 | ||||||
| SAV2550 | MW2471 | feoB | Ferrous iron transport protein B homolog | 0.4 | 0.4 | 2.3 | 1.0 | 1.5 | |||||
| SAV0113 | MW0086 | sirC | Lipoprotein | 0.2 | 0.2 | 0.2 | 0.2 | 0.5 | |||||
| Other genes | |||||||||||||
| SAV1837 | MW1777 | ecsA ABC transporter homolog | 6.2 | 7.4 | 7.3 | 7.6 | 1.0 | ||||||
| SAV2698 | MW2617 | Similar to high-affinity nickel transport protein | 0.2 | 0.2 | 0.2 | 0.3 | 1.0 | ||||||
| SAV0643 | MW0605 | ABC transporter A | 0.7 | 0.7 | 0.8 | 1.2 | 2.1 | ||||||
| SAV1321 | MW1208 | Similar to two-component sensor histidine kinase | 1.3 | 2.1 | 1.8 | 1.5 | 7.4 | ||||||
| SAV2146 | MW2070 | czrB | Cation efflux system membrane protein homolog | 1.0 | 0.9 | 3.4 | 0.8 | 0.6 | |||||
| SAV2631 | MW2552 | Similar to transcription regulator Crp/Fnr family protein | 0.3 | 0.3 | 0.4 | 0.2 | 0.6 | 1.0 | 0.3 | 0.3 | 0.8 | 0.4 | |
| SAV1026 | MW0906 | Competence transcription factor | 0.1 | 0.2 | 0.1 | 0.2 | 0.5 | ||||||
| SAV0070 | MW2003 | kdpE | Transcription regulator protein KdpE | 0.1 | 0.1 | 0.1 | 0.1 | 0.4 | |||||
| SAV2078 | MW2002 | kdpD | Sensor protein | 0.2 | 0.2 | 0.2 | 0.2 | 0.6 | |||||
| SAV0909 | Cell wall hydrolase (encoded within bacteriophage φMu1) | 0.6 | 0.7 | 0.8 | 0.9 | 33.6 | 1.1 | 1.3 | 1.6 | 2.4 | 8.1 | ||
| SAV2462 | MW2386 | Antibiotic resistance protein | 2.2 | 2.0 | 1.5 | 1.6 | 0.7 | ||||||
See Materials and Methods.
Identification number of S. aureus Mu50 open reading frame (ORF) targeted on the microarray.
Identification number of corresponding S. aureus MW2 ORF.
Previously published gene name.
Previously reported description of gene product or ORF. CoA, coenzyme A; PTS, phosphotransferase system.
Expression levels in pure and mixed cultures at 5, 6.2, 7.8, 11.1, and 25 h, detected with the S. aureus microarray and presented as normalized cDNA/gDNA ratios. Only genes that showed significant variations in expression levels in pure and/or mixed cultures over time (as determined by the ANOVA test with the criterion of an FDR of <0.05) and a degree of change of ≥3-fold were defined as differentially expressed.
TABLE 3.
Expression profiles of stress response genes showing significant variations in pure and/or mixed culturesa
| ORF (Mu50)b | Corresponding ORF (MW2)c | Gene named | Description of gene producte | cDNA/gDNA ratiof for pure culture at h:
|
cDNA/gDNA ratiof for mixed culture at h:
|
σB controlg | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5 | 6.2 | 7.8 | 11.1 | 25 | 5 | 6.2 | 7.8 | 11.1 | 25 | |||||
| SAV1253 | MW1136 | clpQ | Heat shock protein HslV | 3.0 | 4.5 | 3.1 | 3.3 | 0.8 | ||||||
| SAV1254 | MW1137 | clpY | Heat shock protein HslU | 2.6 | 4.3 | 2.6 | 2.9 | 0.8 | ||||||
| SAV0380 | MW0356 | ahpF | Alkyl hydroperoxide reductase subunit F | 2.1 | 1.7 | 4.5 | 6.3 | 4.8 | ||||||
| SAV1553 | MW1505 | sodA | Superoxide dismutase | 2.5 | 2.6 | 2.1 | 2.9 | 8.7 | ||||||
| SAV1334 | MW1221 | katA | Catalase | 3.4 | 5.5 | 5.6 | 3.4 | 62.8 | + | |||||
| SAV0551 | MW0506 | Chaperone protein HchA (Hsp31) | 3.3 | 3.2 | 2.1 | 2.4 | 21.2 | |||||||
| SAV2029 | MW1953 | groEL | GroEL protein | 1.5 | 1.4 | 1.0 | 1.2 | 5.1 | ||||||
| SAV0975 | MW0857 | clpB | ClpB chaperone homolog | 0.6 | 0.7 | 0.6 | 0.5 | 5.4 | ||||||
| SAV0525 | MW0480 | clpC | Endopeptidase | 2.6 | 2.7 | 2.1 | 2.0 | 7.0 | ||||||
| SAV2182 | MW2108 | asp23 | Alkaline shock protein 23 | 5.3 | 2.8 | 2.3 | 8.0 | 141.0 | 3.3 | 2.3 | 2.8 | 25.8 | 51.2 | + |
| SAV1739 | MW1682 | General stress protein-like protein | 5.3 | 4.9 | 6.3 | 7.6 | 59.6 | + | ||||||
| SAV2548 | MW2469 | clpL | ATP-dependent Clp proteinase chain | 4.1 | 2.4 | 1.5 | 4.1 | 58.8 | 3.2 | 0.9 | 1.0 | 5.5 | 19.2 | + |
| SAV0828 | MW0781 | Similar to general stress protein | 1.0 | 1.0 | 0.7 | 1.5 | 5.9 | + | ||||||
| SAV0498 | MW0453 | spoVG | Stage V sporulation protein G homolog | 1.3 | 1.1 | 1.4 | 8.6 | 21.3 | + | |||||
| SAV0704 | MW0666 | Similar to CsbB stress response protein | 1.1 | 1.0 | 0.9 | 1.6 | 10.4 | 1.0 | 0.8 | 0.7 | 2.1 | 4.4 | + | |
| SAV1348 | MW1236 | opuD | Glycine betaine transporter | 4.7 | 5.1 | 3.6 | 4.1 | 0.7 | 3.5 | 1.9 | 1.1 | 0.6 | 0.6 | |
| SAV1339 | MW1226 | lexA | SOS regulatory LexA protein | 1.6 | 2.5 | 2.9 | 6.3 | 15.7 | ||||||
| SAV0522 | MW0477 | ctsR | Transcription repressor of class III stress gene homologs | 1.1 | 1.3 | 0.7 | 1.1 | 6.2 | ||||||
| SAV1491 | MW1445 | srrB | Staphylococcal respiratory response protein | 1.5 | 1.8 | 2.6 | 1.9 | 0.8 | ||||||
| SAV1492 | MW1446 | srrA | Staphylococcal respiratory response protein | 3.1 | 3.4 | 3.4 | 2.9 | 1.0 | ||||||
See Materials and Methods.
Identification number of S. aureus Mu50 open reading frame (ORF) targeted on the microarray.
Identification number of corresponding S. aureus MW2 ORF.
Previously published gene name.
Previously reported description of gene product.
Expression levels in pure and mixed cultures at 5, 6.2, 7.8, 11.1, and 25 h, detected with the S. aureus microarray and presented as normalized cDNA/gDNA ratios. Only genes that showed significant variations in expression levels in pure and/or mixed cultures over time (as determined by the ANOVA test with the criterion of an FDR of <0.05) and a degree of change of ≥3-fold were defined as differentially expressed.
+ indicates that σB exerted positive control (5).
TABLE 4.
Expression profiles of virulence genes showing significant variations in pure and/or mixed culturesa
| ORF (Mu50)b | Corresponding ORF (MW2)c | Gene name(s)d | Description of gene producte | cDNA/gDNA ratiof for pure culture at h:
|
cDNA/gDNA ratiof for mixed culture at h:
|
agr controlg | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5 | 6.2 | 7.8 | 11.1 | 25 | 5 | 6.2 | 7.8 | 11.1 | 25 | |||||
| Adhesin genes | ||||||||||||||
| SAV0811 | MW0764 | fnb | Fibrinogen binding protein | 0.8 | 0.9 | 1.0 | 2.9 | 37.7 | 1.1 | 0.6 | 1.4 | 2.3 | 10.1 | |
| SAV1159 | MW1041 | Fibrinogen binding protein precursor | 3.4 | 1.8 | 2.4 | 1.2 | 1.0 | |||||||
| SAV1208 | MW1091 | Fibrinogen binding protein | 1.0 | 0.7 | 0.8 | 0.5 | 0.3 | |||||||
| SAV1481 | MW1369 | ebpS | Elastin binding protein | 1.5 | 1.2 | 1.0 | 1.4 | 7.9 | ||||||
| SAV2371 | MW2293 | Similar to products for attachment to host cells and virulence | 0.2 | 0.2 | 0.1 | 0.2 | 0.5 | |||||||
| Antigen genes | ||||||||||||||
| SAV2569 | MW2490 | isaAh | Immunodominant antigen A | 7.2 | 4.9 | 6.8 | 5.3 | 0.6 | 5.0 | 3.1 | 1.3 | 1.2 | 1.0 | |
| SAV2638 | MW2559 | isaB | Immunodominant antigen B | 3.5 | 3.1 | 10.0 | 6.5 | 10.3 | ||||||
| SAV0665 | MW2217 | Secretory antigen SsaA homolog | 2.3 | 2.0 | 4.6 | 6.2 | 0.5 | |||||||
| Capsule genes | ||||||||||||||
| SAV0149 | MW0124 | capA | Capsular polysaccharide synthesis enzyme Cap5A | 0.2 | 0.2 | 0.2 | 0.4 | 15.5 | ||||||
| SAV0164 | MW0139 | capP | Capsular polysaccharide synthesis enzyme Cap5P | 0.2 | 0.3 | 0.2 | 0.3 | 2.3 | ||||||
| Enzyme genes | ||||||||||||||
| SAV0815 | MW0769 | nuc | Staphylococcal nuclease | 0.9 | 2.2 | 5.0 | 4.8 | 0.5 | ||||||
| SAV1324 | MW1211 | nuc | Thermonuclease | 6.0 | 6.0 | 7.3 | 4.8 | 0.6 | ||||||
| SAV1811 | MW1753 | splC | Serine protease | 0.4 | 0.6 | 0.3 | 0.6 | 1.8 | + | |||||
| SAV1813 | MW1755 | splA | Serine protease | 17.1 | 16.2 | 17.0 | 9.3 | 0.9 | 8.7 | 7.2 | 8.5 | 1.9 | 0.9 | + |
| SAV1046 | MW0930 | sspC | Cysteine protease | 0.2 | 0.3 | 0.3 | 0.3 | 2.2 | 1.7 | 2.3 | 1.7 | 0.6 | 0.9 | + |
| SAV0320 | MW0297 | geh | Glycerol ester hydrolase | 0.5 | 0.4 | 1.2 | 0.8 | 1.6 | + | |||||
| Hemolysin genes | ||||||||||||||
| SAV2035 | MW1959 | hld/RNAIII | Delta-hemolysin | 1.6 | 1.8 | 20.0 | 39.0 | 96.7 | + | |||||
| SAV0919 | MW0802 | Hemolysin | 0.4 | 0.5 | 0.3 | 0.5 | 11.4 | |||||||
| SAV2170 | MW2096 | Hemolysin III | 1.4 | 2.0 | 1.5 | 2.3 | 5.5 | |||||||
| Toxin genes | ||||||||||||||
| SAV1948 | MW1889 (sea) | sep | Enterotoxin P (encoded within bacteriophage φ Sa 3Mu) | 2.2 | 1.7 | 1.6 | 1.6 | 0.6 | ||||||
| SAV2008 | MW0760 | sel | Extracellular enterotoxin L | 1.5 | 1.5 | 1.8 | 2.1 | 5.4 | ||||||
| SAV1819 | MW1767 | lukD | Leukotoxin F subunit | 0.1 | 0.1 | 0.1 | 0.2 | 0.4 | + | |||||
| SAV0422 | MW0382 (set16) | set6 | Exotoxin 6 | 0.3 | 0.3 | 0.2 | 0.4 | 1.4 | ||||||
| SAV0423 | MW0383 (set17) | set7 | Exotoxin 7 | 0.1 | 0.1 | 0.1 | 0.1 | 0.3 | ||||||
| SAV0424 | MW0384 (set18) | set8 | Exotoxin 8 | 0.1 | 0.1 | 0.1 | 0.1 | 0.3 | ||||||
| SAV0428 | MW0390 (set24) | set13 | Exotoxin 13 | 0.4 | 0.4 | 0.4 | 0.6 | 2.1 | ||||||
| Virulence regulation genes | ||||||||||||||
| SAV2039 | MW1963 | agrA | Accessory gene regulator A | 0.6 | 0.8 | 4.1 | 14.1 | 15.0 | ||||||
| SAV1884 | MW1824 | vraR | Two-component response regulator | 3.4 | 3.1 | 1.1 | 1.9 | 0.6 | ||||||
| SAV0661 | MW0623 | vraF | ABC transporter ATP binding protein | 1.1 | 0.8 | 1.3 | 1.5 | 2.7 | ||||||
See Materials and Methods.
Identification number of S. aureus Mu50 open reading frame (ORF) targeted on the microarray.
Identification number of corresponding S. aureus MW2 ORF.
Previously published gene name.
Previously reported description of gene product.
Expression levels in pure and mixed cultures at 5, 6.2, 7.8, 11.1, and 25 h, detected with the S. aureus microarray and presented as normalized cDNA/gDNA ratios. Only genes that showed significant variations in expression levels in pure and/or mixed cultures over time (as determined by the ANOVA test with the criterion of an FDR of <0.05) and a degree of change of ≥3-fold were defined as differentially expressed.
+ indicates that agr in strain MW2 exerted positive control (62).
isaA was found to be positively regulated by σB, as its expression decreased in a σB mutant (5). However, its expression decreased in late log and stationary phases in both wild-type and mutant strains, as observed in this experiment.
A general survey of the data indicated that the number of genes whose expression varied significantly in the pure culture was ∼3-fold higher than that in the mixed culture. This pattern was emphasized when the analysis was restricted to virulence-related genes: the expression of 29 genes varied in the pure culture, versus that of only 4 genes in the mixed culture. Such a difference was related, at least partially, to the loss of a signal in the mixed culture. Hybridizations were indeed carried out with cDNAs resulting from a constant amount of total RNAs (i.e., a mixture of lactococcal and staphylococcal cDNAs in a mixed culture). The amount of S. aureus cDNA in mixed culture hybridizations was lower than that in pure-culture hybridizations, resulting in lower signals and a subsequent loss of sensitivity. Total RNA extracted from pure cultures gave a signal (signal/noise ratio of ≥2) in 90% of the spots retained for statistical analysis (see Materials and Methods), whereas total RNAs extracted from mixed cultures gave a signal in 64% of the spots. However, general trends in pure cultures, as well as in mixed cultures, were observed.
Several genes involved in the cellular machinery, cell division, cell envelope biosynthesis and turnover, carbohydrate and nitrogen metabolism, stress responses, and virulence displayed differential expression levels over the time course of the cultures (Tables 2, 3, and 4). The expression profiles of some genes belonging to various functional categories were clearly related to growth: genes involved in the translational machinery, cell division, the biosynthesis and turnover of the cell envelope, secretion, and the metabolism of nucleotides and nucleic acids maintained high levels of expression in the pure culture until h 11, when cells entered stationary phase. Likewise, the expression of genes associated with energetic metabolism (e.g., mnhD and ctaB) decreased slightly during the first slowing down of growth and more drastically in the stationary phase. Similar variations in the expression of genes in these functional categories were observed in the mixed culture. Of note, the decrease of expression occurred earlier in the mixed culture than in the pure culture. This finding correlates well with the premature slowing down of S. aureus growth observed in the mixed culture. Hence, the expression of genes encoding transcriptional and translational machinery, as well as genes associated with energetic metabolism, the metabolism of nucleotides and nucleic acids, and secretion, started decreasing from the 7.8-h time point onward and even earlier for some genes (e.g., nusG and xerC). The premature slowing down of growth in the mixed culture may also account for the earlier induction of genes belonging to the σB regulon under this condition, as observed for asp23 and clpL (confirmed by qRT-PCR) (data not shown).
The expression of genes involved in carbohydrate metabolism and transport was linked to growth kinetics but also to glucose availability. The levels of expression of pfk and pykA seemed to be maintained until glucose exhaustion, which occurred in the pure culture after 11 h and in the mixed culture after ∼8 h (53). In the pure culture, the exhaustion of glucose resulted in the upregulation of the expression of genes encoding several transporters for other carbon sources, like ribose (rbsD), maltose and maltodextrin (MW0190 [mtlA]), and trehalose (treP). Such regulation resulting from catabolic repression in several gram-positive bacteria, including Staphylococcus species, was reported previously (31, 71, 75).
Selective repression of enterotoxin expression in mixed cultures.
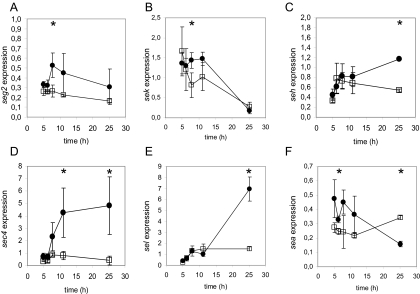
In a food context, S. aureus is particularly undesirable because of the ability of some strains to produce SEs, which are the causative agents of SFP. Six enterotoxins are encoded within the S. aureus MW2 genome, including the two classical enterotoxins SE type A (SEA) and SEC and four SE-like (SEl) proteins: SElG, SElH, SElK, and SElL. SEls have not been well characterized, and their involvement in food poisoning has not yet been established (42). Indeed, their emetic properties, except for those of SEH and SEG, still need to be evaluated (34, 49, 56, 69). Transcriptomic analyses revealed changes in the expression of enterotoxins A and L (Table 4). The expression of all enterotoxin genes harbored in strain MW2 was further investigated by qRT-PCR to confirm and complete the microarray data. L. lactis barely affected the expression of enterotoxins G, K, and H (Fig. 2A, B, and C; Table 5): the expression of seg2 remained constant in both pure and mixed cultures. The expression of sek was maximal during the exponential phase and decreased in the stationary phase under both conditions. A moderate induction of seh in the stationary phase of the pure culture was observed, whereas seh expression was slightly impaired in the mixed culture. In contrast, the induction of sec4 and sel expression in the exponential and/or post-exponential phase was dramatically affected or possibly delayed by the presence of L. lactis, as observed for genes of other functional categories (Fig. 2D and E; Table 5). sea was expressed mainly during the exponential phase in the pure culture, and its expression decreased in the stationary phase (Fig. 2F; Table 5), as observed previously (8, 18). The level of expression of sea during the exponential phase in the mixed culture was slightly lower than that in the pure culture. However, in the mixed culture, sea expression remained constant during the stationary phase (to h 25) and was thus slightly (∼2-fold) higher than that in the pure culture. Altogether, these results showed that the presence of L. lactis strongly reduced the expression of two of the six enterotoxin and enterotoxin-like proteins identified so far in S. aureus MW2.
FIG. 2.
Transcription profiles of genes encoding S. aureus MW2 enterotoxins as determined by qRT-PCR. Transcript levels in pure (•) and mixed (□) cultures of S. aureus between 5 and 25 h were measured. Data points were plotted as relative intensity values (see Materials and Methods). Profiles of transcript titers for enterotoxin type G2 (seg2), enterotoxin K (sek), enterotoxin H (seh), enterotoxin type C4 (sec4), enterotoxin L (sel), and enterotoxin A (sea) genes are presented. Statistical analyses (by an ANOVA test with the criterion of a P value of <0.05) indicated that these genes showed significant changes in expression in pure and mixed cultures over time, except for sea and seh in mixed cultures (P > 0.05). Time points at which genes showed significant changes in expression in pure versus mixed cultures, as determined by a t test (P < 0.05), are indicated by asterisks, and ratios of expression levels are presented in Table 5.
TABLE 5.
Ratios of expression levels in pure versus mixed culturesa
| Gene | Ratio of expression in pure culture to expression in mixed culture at:
|
||||
|---|---|---|---|---|---|
| 5 h | 6.2 h | 7.8 h | 11.1 h | 25.2 h | |
| seg | − | − | 2.0 | − | − |
| sek | − | − | 1.8 | − | − |
| seh | − | − | − | − | 2.2 |
| sec | − | − | − | 5.2 | 11.6 |
| sel | − | − | − | − | 4.7 |
| sea | − | 1.3 | − | − | 0.5 |
| agrA | − | − | − | − | 4.7 |
| RNAIII | − | − | − | 9.2 | 73.9 |
| MW802 | 1.8 | 2.3 | − | − | 5.6 |
| capA | − | − | − | 0.2 | 7.4 |
| hla | 2.8 | 1.7 | − | − | − |
| spa | − | − | − | − | 0.4 |
| sarA | 1.5 | 1.9 | 4.7 | 2.5 | 6.4 |
Ratios were calculated by using qRT-PCR for genes showing significant changes in expression levels (as determined by a t test with the criterion of a P value of <0.05). − indicates the absence of an effect of L. lactis on gene expression.
Modified expression of other S. aureus virulence genes in mixed cultures.
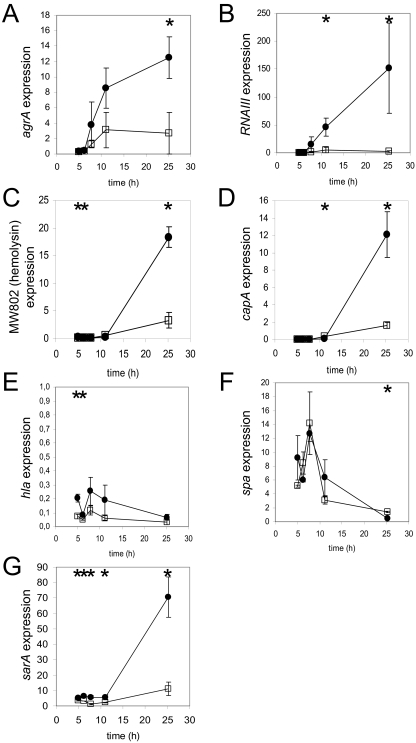
Virulence gene expression is tightly controlled in S. aureus by complex and intricate systems, of which agr and sar are central elements (9, 54). As expected, the expression of several virulence genes in the pure culture of S. aureus MW2 was modified over time (Table 4). Genes encoding exoproteins (such as geh, splC, and sspC), hemolysins (MW0802 and MW2096), toxins (such as sel and lukD), and exotoxins (MW0382 to MW0384 and MW0390) and genes involved in capsule synthesis (capA and capP) were induced in the post-exponential and stationary phases of growth. This upregulation of exoprotein production was concomitant with the induction of the agr system. The expression of agrA and RNAIII was indeed 25- and 60-fold higher, respectively, in the stationary phase than in the exponential phase. Accordingly, some genes encoding cell surface proteins, such as MW1041 and MW1091 (encoding fibrinogen binding proteins), isaA (encoding immunodominant antigen A), and MW2217 (encoding a secretory antigen SsaA homolog) were repressed when cells entered the stationary phase (Table 4).
Strikingly, in the mixed culture, the induction of exoprotein genes and the agr system was not observed. To confirm that these observations were not due to the lower sensitivity of the microarray hybridization analysis for mixed-culture samples, qRT-PCR experiments with several virulence-related genes were carried out (Fig. 3). In the pure culture, agrA and RNAIII expression was dramatically induced (37- and 1,555-fold, respectively) in post-exponential and stationary phases of growth, whereas it was strongly altered in the mixed culture, with maximum inductions of 12- and 25-fold, respectively, at 11 h (Fig. 3A and B; Table 5). Similarly, the expression of genes encoding MW0802 and Cap5A hemolysins was strongly induced in post-exponential and stationary phases in the pure culture, whereas only a moderate induction was observed in the mixed culture (Fig. 3C and D; Table 5). The expression of hla and spa, two well-characterized genes encoding alpha-hemolysin and protein A, respectively, was also assayed by qRT-PCR (Fig. 3E and F; Table 5). As observed previously (21, 24), the expression of spa was maximal during the exponential phase and decreased when cells entered post-exponential growth in the pure culture. A similar spa expression profile in the mixed culture was observed. Surprisingly, hla was not induced in the stationary phase, either in the pure culture, as observed previously in other strains (21), or in mixed cultures.
FIG. 3.
Transcription profiles of virulence genes as determined by qRT-PCR. Transcript levels in pure (•) and mixed (□) cultures of S. aureus between 5 and 25 h were measured. Data points were plotted as relative intensity values (see Materials and Methods). Profiles of transcript titers for accessory gene regulator A (agrA), delta-hemolysin (RNAIII/hld), hemolysin (MW802), capsular polysaccharide synthesis enzyme Cap5A (capA), alpha-hemolysin (hla), protein A (spa), and staphylococcal accessory regulator (sarA) genes are presented. Statistical analyses (by an ANOVA test with the criterion of a P value of <0.05) indicated that these genes showed significant changes in expression in pure and mixed cultures over time, except for agrA and RNAIII in mixed cultures (P > 0.05). Time points at which genes showed significant changes in expression in pure versus mixed cultures, as determined by a t test (P < 0.05), are indicated by asterisks, and ratios of expression levels are presented in Table 5.
In order to complete the microarray data with regard to virulence regulation, the expression of sarA, one of the major and pleiotropic regulators of virulence in S. aureus (together with the agr system) (9, 21, 54), was assayed by qRT-PCR. The expression of sarA was constant throughout the exponential phase of the pure culture, as well as in the mixed culture, although at a slightly lower level (Fig. 3G; Table 5). However, strong (13-fold) induction of sarA expression in the stationary phase of the pure culture was observed, as reported previously (45), whereas the level of induction in the mixed culture (4-fold) was significantly lower. In conclusion, L. lactis was able to alter the induction of agr and sarA, the two main regulators of virulence in S. aureus.
Supernatant from an L. lactis culture does not affect RNAIII expression.
RNAIII downregulation may be due to the release of an inhibitory molecule(s) into the culture supernatant by L. lactis. If so, supernatants from a pure culture of L. lactis LD61 or those from a mixed culture would have an impact on the expression of hld/RNAIII. To test this hypothesis, S. aureus MW2 was grown in pure cultures on CDM supplemented with supernatants harvested at 24 h from pure or mixed cultures of L. lactis (1 volume of supernatant was added to 1 volume of 2× CDM). The expression of RNAIII was investigated by qRT-PCR as described in Materials and Methods. The addition of a supernatant did not alter the kinetics of RNAIII expression, as similar profiles with and without supernatants were observed (data not shown). Moreover, the addition of supernatants obtained from pure or mixed cultures of L. lactis to a pure S. aureus culture in the post-exponential phase of growth, which corresponds to the apex of RNAIII induction, did not affect RNAIII expression (data not shown). Altogether, these results suggest that the downregulation of agr locus expression by L. lactis does not rely on the release of an inhibitory molecule or, alternatively, that the inhibitory molecule is labile.
L. lactis downregulates RNAIII expression in other S. aureus strains belonging to various agr groups.
The downregulation of a key virulence regulator like the agr system by L. lactis is of great interest. We therefore wondered whether this effect was restricted to strain MW2 or if it could be observed in other strains, notably strains belonging to different agr groups. We thus investigated the effect of L. lactis LD61 on two additional strains, namely, strain SH1000 (derived from the well-characterized strain NCTC 8325, of agr group I) and RN4850 (the prototype strain of agr group IV). The strains were grown in mixed cultures under conditions similar to those used for strain MW2, and the expression of RNAIII was monitored by qRT-PCR (data not shown). While RNAIII expression in both strains was strongly induced in post-exponential and stationary phases of growth in pure cultures, it was strongly altered in mixed cultures (>100-fold downregulation of expression in mixed cultures compared to that in pure cultures had occurred at 25 h). This result clearly indicates that the observed downregulation of RNAIII in strain MW2 by L. lactis is not unique, even though we cannot exclude the possibility that some S. aureus strains may be resistant to this inhibitory effect. Furthermore, it does not seem to be restricted to one agr group.
DISCUSSION
Successful exploration of the S. aureus transcriptome in mixed cultures with L. lactis.
Interactions between S. aureus and other bacterial species, notably LAB, arouse more and more interest (1, 11, 40). A better understanding of bacterial interference phenomena will indeed lead to promising nonantibiotic means to control the growth of and/or pathogenesis by S. aureus (or other undesirable microorganisms) in ecosystems where keeping a complex and balanced microbial community is crucial (e.g., the vaginal ecosystem and food fermentation environments). Bacterial interactions involved in such contexts are quite complex and require global molecular approaches, such as transcriptome analyses. Only a few studies have investigated the behavior of S. aureus during interaction with a host or other microorganisms. One recent study reports the transcriptome analysis of S. aureus in interaction with (internalized by) human epithelial host cells (25). Another study reports a transcriptional analysis of the interaction between S. aureus and Pseudomonas aeruginosa, yet only the P. aeruginosa transcriptome was analyzed (46). We report here the first transcriptome analysis of S. aureus in interaction with another bacterial species, namely, L. lactis. A dedicated S. aureus-specific microarray was successfully developed and validated in order to minimize the cross-hybridizations that may result from hybridization with a pool of labeled cDNAs from two species. Genomes of other LAB species were taken into account in designing the microarray so that S. aureus gene expression profiles during interaction with other LAB ecosystems (e.g., that of a complex starter culture in the cheese-making process or vaginal lactobacilli) can be analyzed.
Culture with L. lactis induces a time lag in the kinetics of S. aureus gene expression.
The inhibitory effect of L. lactis on S. aureus growth is widely documented (1, 12, 28, 36; for a review, see reference 11); however, its effect on the global physiology of S. aureus and on virulence expression had never been explored. Under the conditions tested in this work, i.e., a chemically defined culture medium at a constantly regulated pH, L. lactis hardly affected S. aureus growth and the final population of S. aureus in the presence of L. lactis was only threefold smaller than that in the pure culture. Despite the moderate effect on growth, the kinetics of S. aureus gene expression in the mixed culture with L. lactis was modified compared to that in the pure culture. For instance, most functions associated with growth (translation and cell envelope biosynthesis) showed reduced expression earlier in the mixed culture than in the pure culture, which may be related to the premature slowing down of growth in the mixed culture. Likewise, the kinetics of expression of genes involved in carbohydrate metabolism was modified in the presence of L. lactis. Altogether, our results showed that coculturing with L. lactis LD61 inflicted a time lag on S. aureus transcriptomic profiles rather than wild variations in the whole transcriptomic profile. The altered profiles correlated with environmental changes provoked by the presence of L. lactis, such as the premature exhaustion of glucose (53).
Kinetic follow-up of S. aureus gene expression reveals an alteration in the expression of several virulence genes in mixed cultures with L. lactis.
In the present study, we showed that L. lactis was able to impair the expression of several S. aureus virulence genes. Staphylococcal virulence is tightly controlled and temporally regulated. During the post-exponential and stationary phases of growth, many exoproteins, including some enterotoxins, exofoliative toxins, alpha-, beta-, gamma-, and delta-hemolytic toxins, toxic shock syndrome toxin 1, and several secreted enzymes, are upregulated whereas many cell surface-exposed factors, such as protein A, immunodominant antigen A, coagulase, and fibronectin binding protein, are downregulated (5, 21, 48, 54, 55, 64, 72). Accordingly, we found that several exoproteins, hemolysins, toxins, and proteins involved in capsule biosynthesis were induced in the post-exponential and stationary phases of growth in the pure culture of S. aureus MW2. Surprisingly, the expression of hla was rather constant throughout the time course of the culture, suggesting that the complex regulation of hla expression established in other strains may not apply to strain MW2 (21, 72). Furthermore, we observed that some cell surface protein genes, including isaA, spa, and genes encoding fibrinogen binding proteins, were repressed in the post-exponential phase, although no general downregulation of genes encoding cell surface proteins was observed. Thus, the expression profiles of virulence genes in strain MW2 fitted the temporal regulation patterns generally observed in other S. aureus strains. However, the gene expression profile was very much altered in the presence of L. lactis with, in particular, no general upregulation of many exoproteins.
L. lactis hinders S. aureus virulence expression through the alteration of agr expression.
In a pure culture of S. aureus MW2, we found that the induction of several exoprotein genes, including geh, sspC, capA, and sec4, was concomitant with the activation of the agr system. The agr regulon in strain MW2 was very recently characterized (62), revealing that virulence regulation by agr in strain MW2 partially fits the current model established for well-characterized laboratory strains (derived from strain NCTC 8325) (54): while several toxins and other exoproteins are upregulated by agr, the global downregulation of surface binding proteins by agr observed in other strains does not occur in strain MW2. Such discrepancies in the agr regulon profiles indeed explain why in our study, no general downregulation of surface binding protein expression occurred in the post-exponential phase of the MW2 pure culture. Strikingly, only transient and feeble induction of the agr locus was observed in the mixed culture, suggesting that the lack of induction of exoprotein expression in the mixed culture was, at least in part, due to the nonactivation of the agr system.
It is noteworthy that agr downregulation was not restricted to S. aureus strain MW2 and occurred in other S. aureus strains belonging to different agr groups. Hence, although the ability of L. lactis to downregulate the agr system cannot be generalized to all S. aureus strains without further investigations, the downregulation in MW2 is not an isolated case. This result strongly reinforces the interest in the observed inhibitory capacity of L. lactis, the mechanism of which has now to be identified. We did not observe any significant downregulation of RNAIII expression when S. aureus MW2 was grown in CDM supplemented with L. lactis pure- or mixed-culture supernatants. This finding suggests that agr downregulation was not due to the production of an inhibitory molecule or the release of an autoinducing-peptide-modifying or -degrading enzyme by L. lactis, although we cannot totally exclude the possibility that the inhibitory molecule(s)/enzyme is highly unstable. The production of an inhibitory molecule (metabolite or peptide) that interacts with the two-component system within the agr system and blocks signal transduction in different species of the Staphylococcus genus (27, 73) and in other gram-positive or negative bacteria (40, 61) has already been described. In particular, Laughton and coworkers showed that Lactobacillus reuteri has the potential to repress the expression of the superantigen-like protein 11 (SSL11) and RNAIII via the secretion of a soluble factor which still remains to be identified (40). Our study suggests that the signaling pathways involve the transcriptional regulator SarA. SarA acts as a global regulator of virulence expression in S. aureus (21, 44, 45) and is, in particular, required for the full expression of agr (6, 14, 16, 21). The lower level of sarA expression in the mixed culture with L. lactis than in the pure culture may thus account for the weaker induction of agr locus expression in the mixed culture. Further investigations will be necessary to unravel both the lactococcal factor and the signaling pathway responsible for agr downregulation.
Impact of L. lactis on S. aureus enterotoxigenesis.
Among virulence factors, enterotoxins play a particular role and are the main threat in foodstuffs. Despite the importance of enterotoxins, the regulation of enterotoxins and enterotoxin-like protein expression is still poorly documented. We here report the expression profiles of the six SE and SEl proteins of S. aureus MW2. All enterotoxin genes were found to be expressed in strain MW2. sea was expressed mainly during the exponential phase, as established previously (8), as was sek, whereas the expression of seg2 was constant throughout the culturing. In contrast, sec, sel, and seh were induced in the post-exponential and stationary phases of growth. The induction of sec expression in a pure culture of S. aureus, concomitant with the activation of the agr locus, corroborated the results of previous studies, as sec expression is controlled by RNAIII (62). Accordingly, sec induction was impaired in the presence of L. lactis, and the impairment correlated with agr downregulation. The impact of L. lactis on enterotoxin expression was enterotoxin type dependent. While L. lactis strongly affected the expression of sec and sel, it barely affected the expression of sek, seg2, and seh and even slightly favored the maintenance of sea expression in the stationary phase. This finding is of particular interest, as SEA is the main enterotoxin identified in SFP outbreaks (22, 37). The high prevalence of SEA involvement in SFP outbreaks associated with dairy products may be related to the lack of inhibition of sea expression by L. lactis while the expression of other enterotoxin genes, including sec, is affected.
Conclusions.
This study reports a transcriptomic analysis of S. aureus in interaction with another bacterial species through the development and use of a dedicated species-specific microarray. Together with Nouaille et al. (53), we have provided the first reports of studies of bacterial interaction at the transcriptome level in which both partners are comprehensively analyzed. We showed that some staphylococcal virulence genes were selectively downregulated in a mixed culture. The inhibition of virulence expression correlated with the repression of the virulence-associated regulators agr and sarA in S. aureus MW2. To our knowledge, this is the first study that demonstrates the capability of L. lactis, the model LAB, of inhibiting agr, sarA, and some of the virulence genes in S. aureus under conditions in which S. aureus growth is not dramatically impaired. This study is a first step toward the identification of molecular mechanisms involved in the inhibition of S. aureus by LAB and opens avenues for the biocontrol of S. aureus contamination and/or virulence expression.
Supplementary Material
Acknowledgments
Spotting and hybridization services were provided, respectively, by the transcriptomic platform of the IFR 140 GFAS (Rennes, France) and the transcriptomic platform of Ouest Genopole (Rennes, France). We are grateful to Aurélie Le Cam, Jérome Monfort, Amandine Etchevery, and Régis Bouvet for their help and technical assistance during this work. We thank John McCulloch for critical reading of the manuscript.
Cathy Charlier was the recipient of a Ph.D. fellowship from INRA and Région Bretagne. Marina Cretenet is the recipient of a CIFRE Ph.D. fellowship from the Centre National Interprofessionnel de l'Economie Laitière. This research was supported by grants from Région Bretagne (PRIR Adaptome) and the Agence Nationale de la Recherche (GenoFerment Project).
Footnotes
Published ahead of print on 8 May 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Alomar, J., P. Loubiere, C. Delbes, S. Nouaille, and M. C. Montel. 2008. Effect of Lactococcus garvieae, Lactococcus lactis and Enterococcus faecalis on the behaviour of Staphylococcus aureus in microfiltered milk. Food Microbiol. 25:502-508. [DOI] [PubMed] [Google Scholar]
- 2.Ammor, S., G. Tauveron, E. Dufour, and I. Chevallier. 2006. Antibacterial activity of lactic acid bacteria against spoilage and pathogenic bacteria isolated from the same meat small-scale facility. 2. Behaviour of pathogenic and spoilage bacteria in dual species biofilms including a bacteriocin-like-producing lactic acid bacteria. Food Control 17:462-468. [Google Scholar]
- 3.Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819-1827. [DOI] [PubMed] [Google Scholar]
- 4.Barber, L. E., and R. H. Deibel. 1972. Effect of pH and oxygen tension on staphylococcal growth and enterotoxin formation in fermented sausage. Appl. Microbiol. 24:891-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bischoff, M., P. Dunman, J. Kormanec, D. Macapagal, E. Murphy, W. Mounts, B. Berger-Bachi, and S. Projan. 2004. Microarray-based analysis of the Staphylococcus aureus σB regulon. J. Bacteriol. 186:4085-4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blevins, J. S., K. E. Beenken, M. O. Elasri, B. K. Hurlburt, and M. S. Smeltzer. 2002. Strain-dependent differences in the regulatory roles of sarA and agr in Staphylococcus aureus. Infect. Immun. 70:470-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bore, E., S. Langsrud, O. Langsrud, T. M. Rode, and A. Holck. 2007. Acid-shock responses in Staphylococcus aureus investigated by global gene expression analysis. Microbiology 153:2289-2303. [DOI] [PubMed] [Google Scholar]
- 8.Borst, D., and M. Betley. 1993. Mutations in the promoter spacer region and early transcribed region increase expression of staphylococcal enterotoxin A. Infect. Immun. 61:5421-5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bronner, S., H. Monteil, and G. Prevost. 2004. Regulation of virulence determinants in Staphylococcus aureus: complexity and applications. FEMS Microbiol. Rev. 28:183-200. [DOI] [PubMed] [Google Scholar]
- 10.Chang, W., D. A. Small, F. Toghrol, and W. E. Bentley. 2006. Global transcriptome analysis of Staphylococcus aureus response to hydrogen peroxide. J. Bacteriol. 188:1648-1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Charlier, C., M. Cretenet, S. Even, and Y. Le Loir. 2009. Interactions between Staphylococcus aureus and lactic acid bacteria: an old story with new perspectives. Int. J. Food Microbiol. 131:30-39. [DOI] [PubMed] [Google Scholar]
- 12.Charlier, C., S. Even, M. Gautier, and Y. Le Loir. 2008. Acidification is not involved in the early inhibition of Staphylococcus aureus growth by Lactococcus lactis in milk. Int. Dairy J. 18:197-203. [Google Scholar]
- 13.Cheung, A. L., A. S. Bayer, G. Zhang, H. Gresham, and Y. Q. Xiong. 2004. Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Immunol. Med. Microbiol. 40:1-9. [DOI] [PubMed] [Google Scholar]
- 14.Cheung, A. L., M. G. Bayer, and J. H. Heinrichs. 1997. sar genetic determinants necessary for transcription of RNAII and RNAIII in the agr locus of Staphylococcus aureus. J. Bacteriol. 179:3963-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheung, A. L., J. M. Koomey, C. A. Butler, S. J. Projan, and V. A. Fischetti. 1992. Regulation of exoprotein expression in Staphylococcus aureus by a locus (sar) distinct from agr. Proc. Natl. Acad. Sci. USA 89:6462-6466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chien, Y., A. C. Manna, and A. L. Cheung. 1998. SarA level is a determinant of agr activation in Staphylococcus aureus. Mol. Microbiol. 30:991-1001. [DOI] [PubMed] [Google Scholar]
- 17.Cui, L., J.-Q. Lian, H. Neoh, E. Reyes, and K. Hiramatsu. 2005. DNA microarray-based identification of genes associated with glycopeptide resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 49:3404-3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Czop, J. K., and M. S. Bergdoll. 1974. Staphylococcal enterotoxin synthesis during the exponential, transitional, and stationary growth phases. Infect. Immun. 9:229-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Buyser, M. L., B. Dufour, M. Maire, and V. Lafarge. 2001. Implication of milk and milk products in food-borne diseases in France and in different industrialised countries. Int. J. Food Microbiol. 67:1-17. [DOI] [PubMed] [Google Scholar]
- 20.Delbes, C., J. Alomar, N. Chougui, J. F. Martin, and M. C. Montel. 2006. Staphylococcus aureus growth and enterotoxin production during the manufacture of uncooked, semihard cheese from cows' raw milk. J. Food Prot. 69:2161-2167. [DOI] [PubMed] [Google Scholar]
- 21.Dunman, P. M., E. Murphy, S. Haney, D. Palacios, G. Tucker-Kellogg, S. Wu, E. L. Brown, R. J. Zagursky, D. Shlaes, and S. J. Projan. 2001. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J. Bacteriol. 183:7341-7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.European Commission. 2003. Opinion of the Scientific Committee on Veterinary Measures Relating to Public Health on staphylococcal enterotoxins in milk products, particularly cheeses. Health & Consumer Protection Directorate, Brussels, Belgium.
- 23.Fitzgerald, J. R., D. E. Sturdevant, S. M. Mackie, S. R. Gill, and J. M. Musser. 2001. Evolutionary genomics of Staphylococcus aureus: insights into the origin of methicillin-resistant strains and the toxic shock syndrome epidemic. Proc. Natl. Acad. Sci. USA 98:8821-8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao, J., and G. C. Stewart. 2004. Regulatory elements of the Staphylococcus aureus protein A (Spa) promoter. J. Bacteriol. 186:3738-3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garzoni, C., P. Francois, A. Huyghe, S. Couzinet, C. Tapparel, Y. Charbonnier, A. Renzoni, S. Lucchini, D. P. Lew, P. Vaudaux, W. L. Kelley, and J. Schrenzel. 2007. A global view of Staphylococcus aureus whole genome expression upon internalization in human epithelial cells. BMC Genomics 8:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gonzalez-Fandos, E., A. Otero, M. Sierra, M. L. Garcia-Lopez, and M. Prieto. 1994. Effect of three commercial starters on growth of Staphylococcus aureus and enterotoxins (A-D) and thermonuclease production in broth. Int. J. Food Microbiol. 24:321-327. [DOI] [PubMed] [Google Scholar]
- 27.Gov, Y., A. Bitler, G. Dell'Acqua, J. V. Torres, and N. Balaban. 2001. RNAIII inhibiting peptide (RIP), a global inhibitor of Staphylococcus aureus pathogenesis: structure and function analysis. Peptides 22:1609-1620. [DOI] [PubMed] [Google Scholar]
- 28.Haines, W. C., and L. G. Harmon. 1973. Effect of selected lactic acid bacteria on growth of Staphylococcus aureus and production of enterotoxin. Appl. Microbiol. 25:436-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haines, W. C., and L. G. Harmon. 1973. Effect of variations in conditions of incubation upon inhibition of Staphylococcus aureus by Pediococcus cerevisiae and Streptococcus lactis. Appl. Microbiol. 25:169-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horsburgh, M. J., J. L. Aish, I. J. White, L. Shaw, J. K. Lithgow, and S. J. Foster. 2002. σB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 184:5457-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jankovic, I., and R. Bruckner. 2007. Carbon catabolite repression of sucrose utilization in Staphylococcus xylosus: catabolite control protein CcpA ensures glucose preference and autoregulatory limitation of sucrose utilization. J. Mol. Microbiol. Biotechnol. 12:114-120. [DOI] [PubMed] [Google Scholar]
- 32.Janzon, L., and S. Arvidson. 1990. The role of the delta-lysin gene (hld) in the regulation of virulence genes by the accessory gene regulator (agr) in Staphylococcus aureus. EMBO J. 9:1391-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jarraud, S., G. J. Lyon, A. M. Figueiredo, L. Gerard, F. Vandenesch, J. Etienne, T. W. Muir, and R. P. Novick. 2000. Exfoliatin-producing strains define a fourth agr specificity group in Staphylococcus aureus. J. Bacteriol. 182:6517-6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jarraud, S., M. A. Peyrat, A. Lim, A. Tristan, M. Bes, C. Mougel, J. Etienne, F. Vandenesch, M. Bonneville, and G. Lina. 2001. egc, a highly prevalent operon of enterotoxin gene, forms a putative nursery of superantigens in Staphylococcus aureus. J. Immunol. 166:669-677. [DOI] [PubMed] [Google Scholar]
- 35.Jorgensen, H. J., T. Mork, and L. M. Rorvik. 2005. The occurrence of Staphylococcus aureus on a farm with small-scale production of raw milk cheese. J. Dairy Sci. 88:3810-3817. [DOI] [PubMed] [Google Scholar]
- 36.Kao, C. T., and W. C. Frazier. 1966. Effect of lactic acid bacteria on growth of Staphylococcus aureus. Appl. Microbiol. 14:251-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kerouanton, A., J. A. Hennekinne, C. Letertre, L. Petit, O. Chesneau, A. Brisabois, and M. L. De Buyser. 2007. Characterization of Staphylococcus aureus strains associated with food poisoning outbreaks in France. Int. J. Food Microbiol. 115:369-375. [DOI] [PubMed] [Google Scholar]
- 38.Koessler, T., P. Francois, Y. Charbonnier, A. Huyghe, M. Bento, S. Dharan, G. Renzi, D. Lew, S. Harbarth, D. Pittet, and J. Schrenzel. 2006. Use of oligoarrays for characterization of community-onset methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 44:1040-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 40.Laughton, J. M., E. Devillard, D. E. Heinrichs, G. Reid, and J. K. McCormick. 2006. Inhibition of expression of a staphylococcal superantigen-like protein by a soluble factor from Lactobacillus reuteri. Microbiology 152:1155-1167. [DOI] [PubMed] [Google Scholar]
- 41.Le Loir, Y., F. Baron, and M. Gautier. 2003. Staphylococcus aureus and food poisoning. Genet. Mol. Res. 2:63-76. [PubMed] [Google Scholar]
- 42.Lina, G., G. A. Bohach, S. P. Nair, K. Hiramatsu, E. Jouvin-Marche, and R. Mariuzza. 2004. Standard nomenclature for the superantigens expressed by Staphylococcus. J. Infect. Dis. 189:2334-2336. [DOI] [PubMed] [Google Scholar]
- 43.Lindsay, J. A., and M. T. Holden. 2006. Understanding the rise of the superbug: investigation of the evolution and genomic variation of Staphylococcus aureus. Funct. Integr. Genomics 6:186-201. [DOI] [PubMed] [Google Scholar]
- 44.Manna, A., and A. L. Cheung. 2001. Characterization of sarR, a modulator of sar expression in Staphylococcus aureus. Infect. Immun. 69:885-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Manna, A. C., M. G. Bayer, and A. L. Cheung. 1998. Transcriptional analysis of different promoters in the sar locus in Staphylococcus aureus. J. Bacteriol. 180:3828-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mashburn, L. M., A. M. Jett, D. R. Akins, and M. Whiteley. 2005. Staphylococcus aureus serves as an iron source for Pseudomonas aeruginosa during in vivo coculture. J. Bacteriol. 187:554-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Michel, A., F. Agerer, C. R. Hauck, M. Herrmann, J. Ullrich, J. Hacker, and K. Ohlsen. 2006. Global regulatory impact of ClpP protease of Staphylococcus aureus on regulons involved in virulence, oxidative stress response, autolysis, and DNA repair. J. Bacteriol. 188:5783-5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morfeldt, E., D. Taylor, A. von Gabain, and S. Arvidson. 1995. Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J. 14:4569-4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Munson, S. H., M. T. Tremaine, M. J. Betley, and R. A. Welch. 1998. Identification and characterization of staphylococcal enterotoxin types G and I from Staphylococcus aureus. Infect. Immun. 66:3337-3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Niskanen, A., and E. Nurmi. 1976. Effect of starter culture on staphylococcal enterotoxin and thermonuclease production in dry sausage. Appl. Environ. Microbiol. 31:11-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Noleto, A. L., L. M. Malburg Junior, and M. S. Bergdoll. 1987. Production of staphylococcal enterotoxin in mixed cultures. Appl. Environ. Microbiol. 53:2271-2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Notermans, S., and C. J. Heuvelman. 1983. Combined effect of water activity, pH and sub-optimal temperature on growth and enterotoxin production of Staphylococcus aureus. J. Food Sci. 48:1832-1835. [Google Scholar]
- 53.Nouaille, S., S. Even, C. Charlier, Y. Le Loir, M. Cocaign-Bousquet, and P. Loubière. 2009. Transcriptomic response of Lactococcus lactis in mixed culture with Staphylococcus aureus. Appl. Environ. Microbiol. 75:4473-4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Novick, R. P. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48:1429-1449. [DOI] [PubMed] [Google Scholar]
- 55.Novick, R. P., H. F. Ross, S. J. Projan, J. Kornblum, B. Kreiswirth, and S. Moghazeh. 1993. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 12:3967-3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Orwin, P. M., D. Y. Leung, H. L. Donahue, R. P. Novick, and P. M. Schlievert. 2001. Biochemical and biological properties of staphylococcal enterotoxin K. Infect. Immun. 69:360-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Otero, A., M. C. Garcia, M. L. Garcia, and B. Moreno. 1988. Effect of growth of a commercial starter culture on growth of Staphylococcus aureus and thermonuclease and enterotoxins (C1 and C2) production in broth cultures. Int. J. Food Microbiol. 6:107-114. [DOI] [PubMed] [Google Scholar]
- 58.Otero, M. C., and M. E. Nader-Macias. 2006. Inhibition of Staphylococcus aureus by H2O2-producing Lactobacillus gasseri isolated from the vaginal tract of cattle. Anim. Reprod. Sci. 96:35-46. [DOI] [PubMed] [Google Scholar]
- 59.Pane-Farre, J., B. Jonas, K. Forstner, S. Engelmann, and M. Hecker. 2006. The sigmaB regulon in Staphylococcus aureus and its regulation. Int. J. Med. Microbiol. 296:237-258. [DOI] [PubMed] [Google Scholar]
- 60.Peng, H. L., R. P. Novick, B. Kreiswirth, J. Kornblum, and P. Schlievert. 1988. Cloning, characterization, and sequencing of an accessory gene regulator (agr) in Staphylococcus aureus. J. Bacteriol. 170:4365-4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Qazi, S., B. Middleton, S. H. Muharram, A. Cockayne, P. Hill, P. O'Shea, S. R. Chhabra, M. Camara, and P. Williams. 2006. N-Acylhomoserine lactones antagonize virulence gene expression and quorum sensing in Staphylococcus aureus. Infect. Immun. 74:910-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Queck, S. Y., M. Jameson-Lee, A. E. Villaruz, T. H. Bach, B. A. Khan, D. E. Sturdevant, S. M. Ricklefs, M. Li, and M. Otto. 2008. RNAIII-independent target gene control by the agr quorum-sensing system: insight into the evolution of virulence regulation in Staphylococcus aureus. Mol. Cell 32:150-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.R Development Core Team. 2007. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria.
- 64.Recsei, P., B. Kreiswirth, M. O'Reilly, P. Schlievert, A. Gruss, and R. P. Novick. 1986. Regulation of exoprotein gene expression in Staphylococcus aureus by agar. Mol. Gen. Genet. 202:58-61. [DOI] [PubMed] [Google Scholar]
- 65.Rogasch, K., V. Ruhmling, J. Pane-Farre, D. Hoper, C. Weinberg, S. Fuchs, M. Schmudde, B. M. Broker, C. Wolz, M. Hecker, and S. Engelmann. 2006. Influence of the two-component system SaeRS on global gene expression in two different Staphylococcus aureus strains. J. Bacteriol. 188:7742-7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rouillard, J. M., C. J. Herbert, and M. Zuker. 2002. OligoArray: genome-scale oligonucleotide design for microarrays. Bioinformatics 18:486-487. [DOI] [PubMed] [Google Scholar]
- 67.Rouillard, J. M., M. Zuker, and E. Gulari. 2003. OligoArray 2.0: design of oligonucleotide probes for DNA microarrays using a thermodynamic approach. Nucleic Acids Res. 31:3057-3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sameshima, T., C. Magome, K. Takeshita, K. Arihara, M. Itoh, and Y. Kondo. 1998. Effect of intestinal Lactobacillus starter cultures on the behaviour of Staphylococcus aureus in fermented sausage. Int. J. Food Microbiol. 41:1-7. [DOI] [PubMed] [Google Scholar]
- 69.Su, Y. C., and A. C. Wong. 1995. Identification and purification of a new staphylococcal enterotoxin, H. Appl. Environ. Microbiol. 61:1438-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Talaat, A. M., S. T. Howard, W. Hale, R. Lyons, H. Garner, and S. A. Johnston. 2002. Genomic DNA standards for gene expression profiling in Mycobacterium tuberculosis. Nucleic Acids Res. 30:e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Titgemeyer, F., and W. Hillen. 2002. Global control of sugar metabolism: a gram-positive solution. Antonie van Leeuwenhoek 82:59-71. [PubMed] [Google Scholar]
- 72.Vandenesch, F., J. Kornblum, and R. P. Novick. 1991. A temporal signal, independent of agr, is required for hla but not spa transcription in Staphylococcus aureus. J. Bacteriol. 173:6313-6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vieira-da-Motta, O., P. D. Ribeiro, W. Dias da Silva, and E. Medina-Acosta. 2001. RNAIII inhibiting peptide (RIP) inhibits agr-regulated toxin production. Peptides 22:1621-1627. [DOI] [PubMed] [Google Scholar]
- 74.Wang, R., K. R. Braughton, D. Kretschmer, T. H. Bach, S. Y. Queck, M. Li, A. D. Kennedy, D. W. Dorward, S. J. Klebanoff, A. Peschel, F. R. DeLeo, and M. Otto. 2007. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat. Med. 13:1510-1514. [DOI] [PubMed] [Google Scholar]
- 75.Warner, J. B., and J. S. Lolkema. 2003. CcpA-dependent carbon catabolite repression in bacteria. Microbiol. Mol. Biol. Rev. 67:475-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Weinrick, B., P. M. Dunman, F. McAleese, E. Murphy, S. J. Projan, Y. Fang, and R. P. Novick. 2004. Effect of mild acid on gene expression in Staphylococcus aureus. J. Bacteriol. 186:8407-8423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wertheim, H. F., D. C. Melles, M. C. Vos, W. van Leeuwen, A. van Belkum, H. A. Verbrugh, and J. L. Nouwen. 2005. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect. Dis. 5:751-762. [DOI] [PubMed] [Google Scholar]
- 78.Yarwood, J. M., and P. M. Schlievert. 2003. Quorum sensing in Staphylococcus infections. J. Clin. Investig. 112:1620-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.