Abstract
Trypanosoma cruzi, the causative agent of Chagas disease, is surrounded by a mucin coat that plays important functions in parasite survival/invasion and is extensively O-glycosylated by Golgi and cell surface glycosyltransferases. The addition of the first sugar, α-N-acetylglucosamine (GlcNAc) linked to Threonine (Thr), is catalyzed by a polypeptide α-GlcNAc-transferase (pp-αGlcNAcT) which is unstable to purification. Here, a comparison of the genomes of T. cruzi and Dictyostelium discoideum, an amoebazoan which also forms this linkage, identified two T. cruzi genes (TcOGNT1 and TcOGNT2) that might encode this activity. Though neither was able to complement the Dictyostelium gene, expression in the trypanosomatid Leishmania tarentolae resulted in elevated levels of UDP-[3H]GlcNAc:Thr-peptide GlcNAc-transferase activity and UDP-[3H]GlcNAc breakdown activity. The ectodomain of TcOGNT2 was expressed and the secreted protein was found to retain both activities after extensive purification away from other proteins and the endogenous activity. Product analysis showed that 3H was transferred as GlcNAc to a hydroxyamino acid, and breakdown was due to hydrolysis. Both activities were specific for UDP-GlcNAc relative to UDP-GalNAc and were abolished by active site point mutations that inactivate a related Dictyostelium enzyme and distantly related animal pp-αGalNAcTs. The peptide preference and the alkaline pH optimum were indistinguishable from those of the native activity in T. cruzi microsomes. The results suggest that mucin-type O-glycosylation in T. cruzi is initiated by conserved members of CAZy family GT60, which is homologous to the GT27 family of animal pp-αGalNAcTs that initiate mucin-type O-glycosylation in animals.
Keywords: Chagas disease, Dictyostelium, GlcNAcT, polypeptide αGlcNAc-transferase, trypanosomatid
Introduction
Trypanosoma cruzi is the causative agent of Chagas disease (American trypanosomiasis) that affects millions of humans in endemic areas of Latin America (Tarleton et al. 2007). The acute phase often shows parasitemia, prior to a chronic phase that may be asymptomatic or exhibit varying clinical features including myocarditis or pathology of the digestive and peripheral nervous systems (Coura 2007). Throughout its life cycle in both the triatomine insect vector and the mammalian host (Brener 1973), most of the parasite is covered by glycosylphosphatidylinositol (GPI)-anchored glycoproteins and free glycoinositolphospholipids (McConville et al. 2002; Previato et al. 2004). These glycoconjugates likely support survival in the wide range of extracellular and intracellular environments confronted by T. cruzi during its life cycle.
The abundant GPI-anchored mucin-like glycoproteins are extensively modified by O-glycans attached via GlcNAcα1-Thr linkages. The UDP-GlcNAc:polypeptide N-acetyl-α-glucosaminyltransferase (pp-αGlcNAcT, EC 2.4.1.) activity that initiates this glycosylation has been characterized biochemically (Previato et al. 1998), but has thus far been too unstable to purify. Depending on the parasite strain, β-galactopyranose (Galp) and/or β-galactofuranose (Galf) residues are linked respectively to the O-6 and/or O-4 positions of the GlcNAc residue (Previato et al. 1994; Todeschini et al. 2001; Jones et al. 2004) and can be extended by other βGal residues before the addition of terminal αGalp or sialic acid residues (Almeida et al. 1994; Mendonça-Previato et al. 2005). The dense arrays of mucins may protect epimastigotes from proteases in the intestinal tract of the insect vector (Mortara et al. 1992) and support survival of metacyclic trypomastigotes and their penetration of the gastric mucosal epithelium during acute infection after oral ingestion by the mammalian host (Hoft et al. 1996). Mucin sialoglycans may also protect bloodstream forms from complement attack (Tomlinson and Raper 1998). Mucin O-glycans have been implicated in T-cell suppression possibly through binding l-selectin (Argibay et al. 2002; Alcaide and Fresno 2004). Antibodies against mucin-like O-glycan Gal epitopes inhibit host-cell invasion (Yoshida et al. 1989; Ruiz et al. 1993), and anti-sialic acid Abs inhibit trypomastigote attachment to host cells (Schenkman et al. 1991), suggesting that the sialoglycans play a direct role in parasite-host cell attachment. Sialic acid levels are associated with both reduced and increased infectivity; the issue is complicated because the two major phylogenetic groups of T. cruzi appear to utilize distinct mechanisms of infection, and infection processes change in different life cycle stages (Yoshida 2006).
The addition of GlcNAc and that of Gal likely occur according to conventional mechanisms in the Golgi (Morgado-Diaz et al. 2001), and sialic acids are transferred from host glycans by a cell-surface trans-sialidase (Previato et al. 1985; Schenkman et al. 1991), with mucin glycoproteins being the sole acceptors on the parasite's cell surface (Schenkman et al. 1993). Transfer of Gal is mediated by potentially dozens of genes (El-Sayed et al. 2005), probably reflecting the extensive structural diversity observed in T. cruzi O-glycans (Mendonça-Previato et al. 2005), and the trans-sialidases and mucins are encoded by a large gene family with potentially hundreds of members (Frasch 2000; Acosta-Serrano et al. 2001; Buscaglia et al. 2006), making a genetic analysis of these processes intractable using current methods. The addition of αGlcNAc to Thr represents the first step and a possible “bottleneck” for subsequent O-glycosylation. In higher eukaryotes, the biosynthesis of mucin-type O-linked glycans is initiated by a family of pp-αGalNAcTs (EC 2.4.1.41) which transfer GalNAc from UDP-GalNAc to protein substrates in the Golgi (Ten Hagen et al. 2003). Genomic and functional analyses of pp-αGalNAcTs derived from humans, Caenorhabditis elegans, Drosophila melanogaster, and the unicellular parasite Toxoplasma gondii have revealed the presence of 20, 9, 14, and 5 genetic isoforms of the transferase, respectively (Ten Hagen et al. 2003; Wojczyk et al. 2003; Hang and Bertozzi 2005). However, the genetic basis of the O-GlcNAc addition in T. cruzi is unknown.
A seemingly unrelated glycosylation pathway in the cytoplasm of the social amoeba Dictyostelium discoideum has provided a clue for the genetic basis of the initial transfer of GlcNAc in T. cruzi. The first sugar added to the recipient protein, Skp1, is a GlcNAc α-linked to hydroxyproline (Teng-umnuay et al. 1998), and the HyPro-pp-αGlcNAcT (DdGnt1) has been identified (Teng-umnuay et al. 1999; van der Wel et al. 2002). Bioinformatics studies yield three classes of related sequences in other genomes: (1) other predicted soluble cytoplasmic pp-αGlcNAcTs in eukaryotes and prokaryotes, (2) putative membrane-bound Golgi pp-αGlcNAcTs, and (3) the animal Golgi pp-αGalNAcTs (West et al. 2004). Examination of the only class 2 sequence in Dictyostelium (DdGnt2) showed (Wang et al. 2003) it to be the anticipated pp-αGlcNAcT for cell surface proteins (Zachara et al. 1996), and the enzyme is inhibited by two neutral uridine analogs that inhibit class 3 pp-αGalNAcTs (Ercan and West 2005). All known trypanosomatid genomes harbor three class 2-like genes related to DdGnt2, though the GlcNAcα1-Thr linkage has only been observed in T. cruzi. If the predicted T. cruzi homologs mediate this activity, the low complexity of the gene family, compared to subsequent processing steps, may enable future genetic analysis of the role of mucin-type O-glycosylation in Chagas disease. Expression studies reported here show that at least one of the T. cruzi genes possesses this expected biochemical function, but may exhibit a more complex biochemical mechanism than its Dictyostelium counterpart.
Results
pp-αGlcNAcT-like sequences in the T. cruzi genome
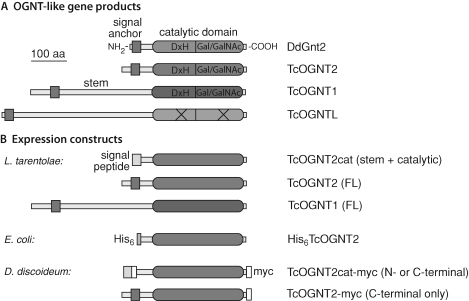
The T. cruzi CL-Brener genome contains three open reading frames related to the catalytic domain of DdGnt2, a CAZy family GT60 (Cantarel et al. 2009) sequence, based on BLAST analysis (Table I). All three genes are predicted to encode type 2 membrane proteins with topology similar to that of DdGnt2 and other eukaryotic Golgi-associated glycosyltransferases, with their catalytic domains oriented toward the Golgi lumen (Figure 1). The predicted sequences of TcOGNT1 and TcOGNT2 are 38–46% identical and 61–81% similar to the 250-amino-acid catalytic domain of DdGnt2 and are more distantly related to the family GT27 pp-αGalNAcTs that initiate mucin-type O-glycosylation in animals (supplementary Figure 1). Similarity was highest at positions thought to be important for catalytic activity in both the NRD2 and so-called Gal/GalNAc-like regions. In contrast, TcOGNTL lacked the DxD-like DxH motif of the NRD2 domain and key motifs of the Gal/GalNAc domain characteristic of family GT60 and GT27 sequences (West et al. 2004) and was only 27–31% identical and 51–57% similar to 174 amino acids of the catalytic domains of TcOGNT2 and TcOGNT1, respectively (supplementary Figure 1). TcOGNT2, like DdGnt2, has a short predicted stem region separating its catalytic and transmembrane domains, in contrast to the long sequence of TcOGNT1 (Figure 1). The predicted catalytic domain of TcOGNT2 possesses three pairs of Cys residues in similar (but not identical) positions to Cys residues in DdGnt2 and may form conserved disulfide bonds. Its two N-glycosylation sequons are not conserved with the single one in DdGnt2. The TcOGNT2 open reading frame is represented in the GeneDB genomic sequence database by two genomic fragments, whose nt and amino acid sequences are 98% identical (supplementary Figure 2). These probably represent the two haplotypes present in the hybrid CL-Brener strain (El-Sayed et al. 2005). Eight of the 11-amino-acid differences are in the transmembrane and short cytoplasmic domains; of the other three, one is a conservative N to D substitution and the other two (M–S and L–V) are in the less-conserved C-terminal region.
Table I.
Characteristics of predicted OGNT-related coding regions in trypanosomatids
| Organism | Locus | Nuc. (bp) | Amino acids | Mr (calc.) | Pred. pI | GeneDB identifiera |
|---|---|---|---|---|---|---|
| T. cruzi | TcOGNT2-1 | 1443 | 480 | 55.1 | 6.8 | Tc00.1047053511309.70 |
| TcOGNT2-2 | 1443 | 480 | 55.3 | 7.0 | Tc00.1047053511759.30 | |
| TcOGNT1-1 | 2175 | 724 | 84.2 | 8.2 | Tc00.1047053503511.10 | |
| TcOGNT1-2 | 2175 | 724 | 84.0 | 7.8 | Tc00.1047053508741.340 | |
| TcOGNTL-1 | 2445 | 814 | 90.6 | 6.7 | Tc00.1047053503509.10 | |
| TcOGNTL-2 | 2442 | 813 | 90.6 | 6.7 | Tc00.1047053508741.360 | |
| L. major | LmOGNT2 | 1917 | 638 | 71.6 | 7.9 | LmjF17.1020 |
| LmOGNT1 | 3036 | 1011 | 110.7 | 9.1 | LmjF02.0240 | |
| LmOGNTL | 3438 | 1145 | 123.3 | 6.4 | LmjF02.0250 | |
| T. brucei | TbOGNT2 | 1503 | 500 | 56.5 | 7.2 | Tb927.5.2350 |
| TbOGNT1 | 2250 | 749 | 85.2 | 7.6 | Tb927.2.2400 | |
| TbOGNTL | 2625 | 874 | 96.6 | 8.7 | Tb927.2.2380 |
Fig. 1.
Domain analysis and expression of pp-αGlcNAcT-like sequences from D. discoideum and T. cruzi. (A) The lead sequence DdGnt2 is a type 2 membrane protein whose catalytic domain, connected via a “stem” domain to its transmembrane (signal anchor) domain, is oriented toward the lumen of the Golgi. The genome of T. cruzi (CL-Brener) has three predicted genes with similar sequences, as described further in Table I, Figure 2 and supplementary Figure 1. In contrast to TcOGNT1 and TcOGNT2, TcOGNTL is expected to be catalytically inactive. (B) Expression constructs of TcOGNT2 and TcOGNT1 employed in this study. Position of the DxH motif, subjected to mutagenesis, is indicated in A. Organism for expression is listed at the left.
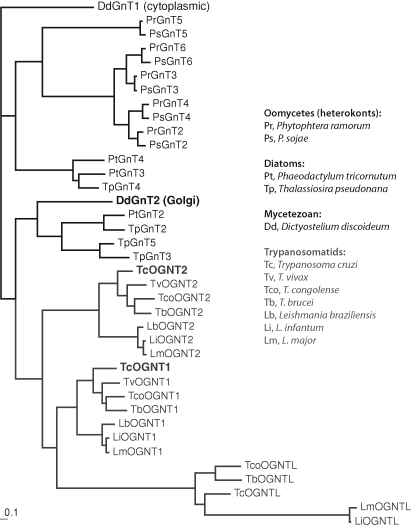
Sequences related to those of TcOGNT1, TcOGNT2, and TcOGNTL are found in the genomes of T. brucei and L. major (Table I), and in all other trypanosomatids for which genomic sequence information is available. Related putative genes are present in other protists including plant pathogens in the oomycete group and diatoms. These sequences, including a cytoplasmic pp-αGlcNAcT (DdGnt1) which appears to have descended along a separate lineage tracing back to an early common eukaryotic ancestor (West et al. 2004), were aligned and a maximum likelihood method was used to infer evolutionary relationships. The best tree, rooted with DdGnt1, indicates that TcOGNT1, TcOGNT2, and TcOGNTL derive from gene duplications early in the trypanosomatid lineage (Figure 2). These genes exhibit conserved synteny (supplementary Figure 3) in all examined trypanosomatid genomes. The sequence of TcOGNTL is closer to that of TcOGNT1 than TcOGNT2, which correlates with their chromosomal proximity (supplementary Figure 3). The five DdGnt2-like sequences in the Phytophthora species diversified in a separate clade, indicative of independent, lineage-specific diversification of the oomycete genes (Figure 2). In contrast, the multiple diatom sequences (from T. pseudonana and P. tricornutum) are represented in both the oomycete and trypanosomatid-containing clades, suggesting an early duplication in the common ancestor of all these organisms. The results suggest that trypanosomatids retained and diversified one of these sequences, oomycetes retained and diversified the other sequence, Dictyostelium retained (but did not diversify) the trypanosomatid type, and diatoms retained and diversified both types.
Fig. 2.
Phylogenetic analysis of putative Golgi pp-αGlcNAcT sequences from protists. Conceptually translated protist nucleotide sequences related to DdGnt2 (dark bold) and predicted to encode Golgi-associated pp-αHexNAcTs, and the cytoplasmic pp-αGlcNAcT DdGnt1, were aligned at the amino acid level. The phylogenetic relationship of their putative catalytic domains was examined using a maximum likelihood method, and the tree was rooted with DdGnt1. Species of origin are listed in the key. The predicted pp-αGlcNAcTs of T. cruzi are in gray bold. The bar represents the fractional rate of amino acid substitutions.
TcOGNT2 and TcOGNT1 expression in T. cruzi
T. cruzi coding RNA is typically spliced at its 5′-end to a shared miniexon. Total RNA from epimastigotes was amplified by reverse transcription and PCR using primers corresponding to the miniexon and a TcOGNT2-specific reverse primer. Analysis of 10 clones (supplumentary Figure 4) revealed at least three splice donor sites 70–109 nt upstream of the predicted ATG start codon, to the 3′-side of typical consensus ag dinucleotide motifs (shown in supplementary Figure 2). These findings indicate that TcOGNT2 mRNA accumulates in parasites, confirm the position of the predicted start codon, and suggest that TcOGNT2 is a functional gene. Similar studies indicated that TcOGNT1-2 (Table I) is also a functional gene with a 5′-UTR splice-acceptor site located 88 nt upstream of its predicted start codon (data not shown).
Expression of TcOGNTs in Dictyostelium
Dictyostelium was initially used to test the function of TcOGNT2 because a previous study showed that expression of the DdGnt2 ectodomain complemented deletion of the DdGnt2 locus (Wang et al. 2003). TcOGNT2 was examined first because it is more similar to DdGnt2 than TcOGNT1. Strain DL118, which harbors a null DdGnt2 allele and expresses no detectable endogenous Golgi pp-αGlcNAcT activity, was used as the host. The predicted ectodomain (stem and catalytic regions) of TcOGNT2 (TcOGNT2cat) (Figure 1B) was expressed with either an N- or C-terminal c-myc epitope tag under control of the semi-constitutive discoidin 1γ promoter (Figure 1B). Both strains accumulated substantial levels of a novel myc-tagged protein at the expected apparent Mr position in cells (data not shown). However, no protein was detected in the conditioned media fraction in contrast with the ability of D. discoideum to secrete truncated DdGnt2-myc (Wang et al. 2003). Cell fractionation revealed that the myc-tagged TcOGNT2cat proteins could be released from microsomes by sonication (data not shown), consistent with normal folding compatible with solubility though not secretion. However, no changes in transfer of 3H from UDP-[3H]GlcNAc to an acceptor peptide or in UDP-[3H]GlcNAc hydrolysis activity relative to the parental extracts were detected in these soluble fractions, using optimized conditions described below (supplementary Figure 5 and data not shown). In addition, these constructs did not consistently restore DdGnt2-dependent glycoepitopes normally recognized by mAb 54.2 or WGA (data not shown), indicating the absence of pp-αGlcNAcT activity in vivo. Furthermore, no activity or glycoepitope complementation was observed when a full-length TcOGNT2 construct (Figure 1B) was expressed (data not shown). Similar negative results were obtained using TcOGNT1, or when TcOGNTL or TcOGNT1 was coexpressed with TcOGNT2, despite successful protein accumulation (unpublished data). Though these negative results have multiple possible explanations, we speculated that either the T. cruzi proteins required specific posttranslational modifications that do not occur in Dictyostelium, that Dictyostelium exerted a negative modification on the T. cruzi proteins, that the expressed proteins do not tolerate either N- or C-terminal epitope tags, or that these polypeptides require an additional factor(s) during biosynthesis or for activity.
Expression of TcOGNT2 and TcOGNT1 in Leishmania tarentolae microsomes
L. tarentolae is a trypanosomatid distantly related to T. cruzi and is an effective host for expressing heterologous proteins (Breitling et al. 2002). DNA corresponding exactly to the full-length coding regions of TcOGNT2 and TcOGNT1 were cloned into commercially available expression vectors (see Materials and Methods). TcOGNT2cat was also subcloned downstream of DNA encoding an N-terminal cleavable signal peptide from secreted acid phosphatase. No epitope tag was incorporated to avoid potential interference given the negative results from the Dictyostelium expression studies. Their RNA polymerase II-dependent transcripts were designed to undergo appropriate 5′-trans-splicing with spliced leader RNA for stable expression in the axenically grown promastigote stage. Incorporation of linearized DNAs from these plasmids, expected to integrate as tandem arrays into the ssu locus, was selected for using G418 or hygromycin B according to the vector employed. Drug-resistant control strains were generated by transfection with empty expression vectors.
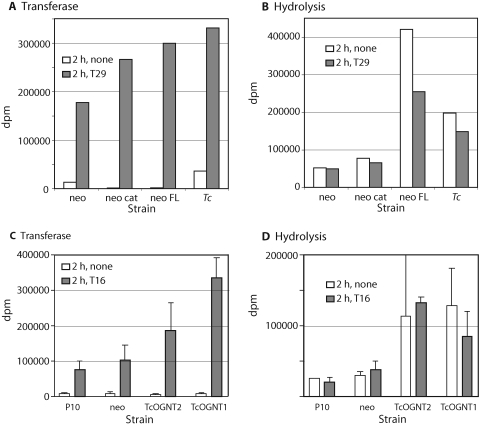
Microsomal fractions were analyzed initially for pp-αGlcNAcT-like activity using peptide T29 (Table II), modeled from a Dictyostelium cell surface protein with a mucin domain. Activity was based on transfer of 3H from UDP-[3H]GlcNAc to peptide T29. Microsomes from parental P10 or empty vector control strains (referred to as neo or hyg) exhibited substantial activity that was 10-fold higher than zero-time (not shown) and zero-peptide controls (Figure 3A). The existence of peptide and time-dependent activity was consistent with bioinformatics evidence that all trypanosomatids possess a TcOGNT2-like gene. A 67% higher level was measured in microsomes from cells expressing full-length TcOGNT2, suggesting that TcOGNT2 may catalyze transfer of GlcNAc from UDP-GlcNAc to Thr residues in the T16 peptide. An intermediate increase was observed in the truncated TcOGNT2cat expressing strain. The highest level did not match the activity detected in microsomes from T. cruzi, where the gene originated. Therefore, though a consistent increase in secreted activity was detected, the difference was small and could be explained by modulation of endogenous activity.
Table II.
Peptide acceptor substrates (Ercan and West 2005)
| Name | Origin | Sequence |
|---|---|---|
| T29 peptide-1 | Dd-SP29 tandem repeats | TVTP TVTP TVTP TPTN |
| TPNP TPSQ TS | ||
| T16 peptide-2 | Dd-SP85 tandem repeats | TYPP TQPP TQPP TYPP |
| T2 | TcMUC I tandem repeats | KPPTTTT TTTTKPP |
Fig. 3.
Expression of TcOGNT2 and TcOGNT1 in microsomes of stably transfected L. tarentolae cells. (A) Microsomes (150 μg protein) were assayed for UDP-[3H]GlcNAc transferase activity in the absence or presence of 200 μM T29 peptide for 2 h at 28°C. Strains: neo, L. tarentolae P-10 stably transfected with empty neo vector; neo cat, transfected with TcOGNT2cat; neo FL, transfected with full-length TcOGNT2; Tc, T. cruzi CL-Brener. (B) The same reactions were assayed for UDP-[3H]GlcNAc breakdown activity. Similar trends were observed in each of four independent trials. (C and D) Transferase (C) and hydrolysis (D) activities were detected using 200 μM T16 peptide in microsomes from L. tarentolae cells (P10), or P-10 cells stably transfected with empty neo vector (neo), full-length TcOGNT2, or full-length TcOGNT1. dpm values represent the mean ± SD of two independent microsomal preparations.
Some glycosyltransferases also hydrolyze sugar nucleotides (e.g., Hagen and Nehrke 1998; Gerken et al. 2002). Hydrolysis-like activity was assayed by passing the C18-SepPak eluate of the transferase assay through a Dowex-1 anion exchanger column to remove negatively charged UDP-[3H]GlcNAc. Cells expressing full-length TcOGNT2 exhibited up to 8-fold greater level of hydrolysis activity than control strains and this activity was not dependent on T29 peptide (Figure 3B). Less hydrolysis activity in the presence of peptide could represent competitive transferase activity. Interestingly, T. cruzi microsomes exhibited 3- to 4-fold higher specific hydrolysis activity than control L. tarentolae membranes. Analysis of a strain expressing full-length TcOGNT1 but using T16 as an acceptor peptide (Table II), modeled from a Dictyostelium spore coat protein, yielded results similar to TcOGNT2 (Figure 3C and D). The stimulation of hydrolysis activity was more dramatic than that of transferase activity, and were together consistent with the idea that the expressed proteins have intrinsic enzymatic activities.
Secretion of truncated TcOGNT2cat from L. tarentolae
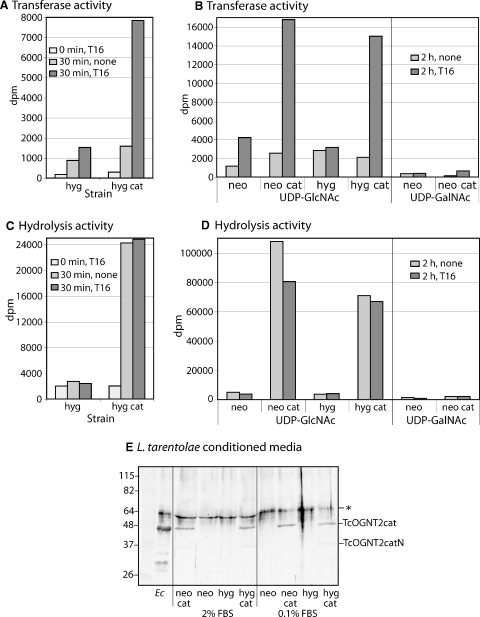
Conditioned media from the cell lines were examined for evidence of secreted activity. Individual clones were grown in parallel to the same density for the same period of time in 0.1–2% FBS, and the culture media were concentrated 50-fold. Low but reproducible incorporation of 3H into peptide T16 was observed for samples from parental cells (P-10; not shown) or an empty vector control strain (neo or hyg), relative to zero-time controls (Figure 4A). Incorporation was also detected in the absence of peptide, suggesting a low rate of transfer to endogenous protein that could be eluted from the C18-SepPak. The amount of activity was approximately 100-fold less than that found in the corresponding microsomal fraction, indicating that endogenous activity is efficiently retained in the cell. Conditioned media from cells transfected with TcOGNT2cat exhibited 4- to 8-fold higher levels of peptide-dependent incorporation. Strains transfected with expression plasmids carrying either G418 or hygromycin resistance markers yielded similar results (Figure 4B). No increase was observed in cells expressing full-length TcOGNT2 (data not shown), indicating that the full-length protein is stably compartmentalized intracellularly. As expected, both endogenous and TcOGNT2cat-dependent activities were specific for UDP-GlcNAc relative to UDP-GalNAc (Figure 4B).
Fig. 4.
Secretion of TcOGNT2cat in conditioned media. (A) Conditioned media from cultures of L. tarentolae grown in 2% FBS were concentrated 50-fold by centrifugal ultrafiltration and assayed for 0 or 30 min at 22°C for the transfer of 3H from UDP-[3H]GlcNAc to 0 or 200 μM T16 peptide. hyg, L. tarentolae transfected with empty hyg-resistance plasmid; hyg cat, cells transfected with TcOGNT2cat. (B) Similar assay of independent 50-fold concentrated samples of L. tarentolae grown in 0.1% FBS, assayed for 0 or 2 h. neo, L. tarentolae transfected with empty neo-resistance plasmid; neo cat, cells transfected with TcOGNT2cat. (C and, D) The same reactions described in panels A–B were assayed in tandem for conversion of 3H of UDP-[3H]GlcNAc into a neutral species (hydrolysis). Note different ordinate scales. (E) Western blot analysis of samples analyzed in panels A and B. Concentrated conditioned media from cells grown in 2% or 0.1% FBS were probed using a 1:500 dilution of anti-TcOGNT2cat (bleed 5). Ec, soluble fractions from E. coli expressing His6TcOGNT2cat. The position of TcOGNT2cat and that of a nonspecific band (*) not recognized by a later antiserum bleed (Figure 7A) are indicated at the right. A faint lower Mr band in the TcOGNT2cat expressing cells represents a C-terminal breakdown product (see Figure 7). Mr standards with values in kD are shown at the left. The results are representative of three independent experiments.
The TcOGNT2cat conditioned media exhibited higher levels of UDP-GlcNAc hydrolysis-like activity than transferase activity (Figure 4C and D). Under these conditions (0.2 mM T16), hydrolysis activity was 3- to 6-fold greater than transferase activity (compare to Figure 4A and B, which were performed in tandem on the same sample). A similar amount of neutral radioactivity was released in the absence of peptide. Negligible neutral radioactivity was formed in the control strains lacking TcOGNT2cat, suggesting that UDP-GlcNAc hydrolysis was a novel activity of TcOGNT2cat. The reduced level of apparent hydrolysis in the presence of peptide acceptor was probably due to consumption of UDP-GlcNAc since these advanced reactions consumed the majority of sugar nucleotide. Since elevated hydrolysis-like activity was also observed in microsomes from L. tarentolae expressing the full-length protein, breakdown was probably not a consequence of the N-terminal truncation.
To confirm expression of TcOGNT2, an antiserum was prepared against His6TcOGNT2 expressed in E. coli (Figure 1B). His6-TcOGNT2, found in inclusion bodies with an apparent Mr of 49,000 as expected for this construct (data not shown), was solubilized, purified by Ni++-column chromatography and preparative SDS–PAGE, and used to immunize rabbits as described in Materials and Methods. The antiserum (bleed 5) recognized a band corresponding to TcOGNT2 in E. coli expressing this protein (Figure 4, lane Ec) but not nontransfected E. coli (data not shown), and a second nonspecific band was seen in all samples that might correspond to keratin. A specific band at a similar position was observed in conditioned media from L. tarentolae expressing TcOGNT2cat, but not from cells that lacked TcOGNT2 (Figure 4E). Similar levels were observed in conditioned media containing 2% or 0.1% FBS. The band was not recognized by preimmune sera (data not shown). A faint slightly more rapidly migrating species also recognized by the antibody represents a C-terminally truncated breakdown product (see below). Therefore, pp-αGlcNAcT-like activity corresponded with the presence of the TcOGNT2cat protein. The antiserum did not reproducibly react with other bands, suggesting that it does not cross-react with the predicted endogenous LtOGNT2, though the possibility that a cross-reactive soluble fragment of LtOGNT2 is present below the detection limit is not excluded.
Characterization of the transferase and hydrolysis reaction products
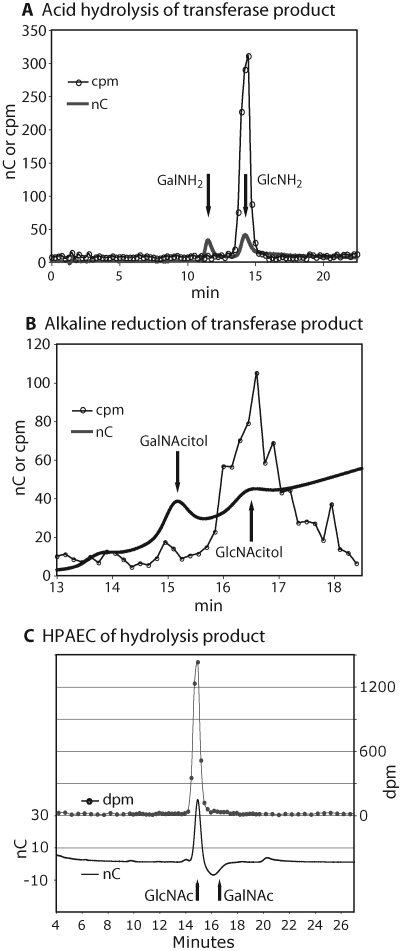
The radioactive peptide fraction from the transferase reaction was acid hydrolyzed using suitable conditions for cleaving sugar-peptide linkages. Chromatography on a Dionex CarboPac PA-1 column showed that the radioactivity co-eluted with GlcNH2 (Figure 5A). GlcNH2 likely derived from GlcNAc as a result of acid-catalyzed N-deacetylation. This was confirmed by analysis of the product of alkaline degradation, which yielded radioactivity that co-eluted with N-acetylglucosaminitol on a Dionex MA-1 column (Figure 5B), consistent with release by β-elimination. Therefore, GlcNAc appeared to be attached to the peptide in O-linkage to the hydroxyl side chains of Thr residues.
Fig. 5.
Reaction product characterizations. (A and B) The peptide product of the transferase reaction was subjected to acid hydrolysis in 6 N HCl (A), or alkaline degradation in 100 mM NaOH and 1 M NaBH4 (B), as described in Materials and Methods. The acid hydrolysate was dried, supplemented with 1 nmol each of GlcNH2 and GalNH2, and analyzed on a Dionex-PA10 HPAEC column. Internal standards were detected by pulsed amperometry (nC), and 3H was detected in fractions using scintillation counting (cpm). Alkali-treated material was supplemented with 1 nmol each of GlcNAcitol and GalNAcitol and analyzed using a Dionex MA-1 HPAEC column. (C) The breakdown product from a reaction with UDP-[14C]GlcNAc, collected as the flow-through fraction from C18-SepPak and Dowex-1 and -50 columns, was supplemented with 1 nmol of GlcNAc, and analyzed on a Dionex PA-1 column by HPAEC. The position of GalNAc from a parallel run is indicated. Standards and 14C were detected as above. The results are representative of three (A, B) or two (C) independent experiments.
For analysis of the breakdown reaction, the Dowex-1 flow through fraction from a UDP-[14C]GlcNAc reaction was observed to flow through a Dowex-50 (H+-form) column (96% recovery), confirming its neutrality. Further analysis by chromatography on a Dionex PA-1 column showed it to co-elute with αGlcNAc and βGlcNAc standards, which are not resolved (Figure 5C, and data not shown). This confirms that UDP-GlcNAc breakdown results from hydrolysis, as expected.
Temperature dependence of the activities
The functional significance of sugar nucleotide hydrolysis is not understood. A possible explanation is that it reflects an abnormal state of the enzyme protein, possibly due to aberrant posttranslational modification, incomplete folding or absence of a modulatory factor, as might occur during heterologous expression. To assess the potential physiological relevance of the hydrolase activity, its temperature dependence was examined. As expected, both peptide-dependent and -independent endogenous transferase activities approximately doubled as the temperature was raised from 22°C to 32.5°C, slightly above the physiological temperature of T. cruzi epimastigotes, but declined to the 22°C level at 37°C (supplementary Figure 6A). The increase is consistent with Q10 values (activity coefficient for a 10°C increase) of 2–3 for many enzymes (e.g., Trasar-Cepeda et al. 2007) until they reach their Tm, when activity declines. Surprisingly, activity in the TcOGNT2cat samples was not stimulated over this temperature range, even though the data also presumably include 20–30% contribution from endogenous activity. In contrast, the hydrolase activity of TcOGNT2cat was stimulated at 32.5°C (supplementary Figure 6B). The endogenous activity was below the detection threshold. The lack of temperature stimulation for the transferase activity in the TcOGNT2 sample is unusual and, in comparison with the endogenous activity, suggests a deficiency during heterologous expression.
Mutagenesis of TcOGNT2 activities
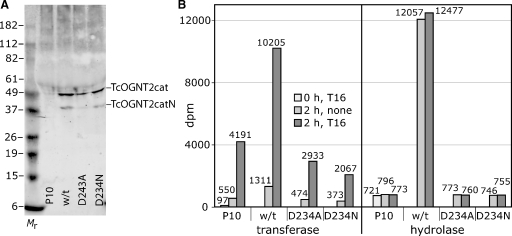
To address whether the TcOGNT2cat-dependent activities were mediated directly by TcOGNT2cat, point mutations in the DxD-like DxH motif required for catalytic activity of DdGnt1 (van der Wel et al. 2002) were introduced. Western blotting of concentrated conditioned media prepared in parallel showed that TcOGNT2cat(D234N) was expressed at nearly the same level as normal TcOGNT2cat, whereas the D234A mutant was expressed at a somewhat lower level (Figure 6A). A lower Mr breakdown product (see below) was detected in all preparations. The mutant samples exhibited no more transferase or hydrolysis activity than the nonexpressing control strains (Figure 6B). This pointed to a direct catalytic function for normal TcOGNT2cat in both transferase and hydrolysis activities.
Fig. 6.
Analysis of mutant TcOGNT2cat. Concentrated (50-fold) conditioned media (0.1% FBS) from L. tarentolae expressing native TcOGNT2cat, TcOGNT2cat(D234N), or TcOGNT2cat(D234A) were analyzed as described in Figure 4. (A) Western blot analysis using 1:500 anti-TcOGNT2 (bleed 7). Positions of TcOGNT2cat and an N-terminal fragment (TcOGNT2catN; see the text) are indicated. (B) Transferase and UDP-GlcNAc hydrolysis activities. P10, normal parental strain; wt, P10 expressing wild-type TcOGNT2cat; D234A and D234N, point mutations at Asp234. Similar results were obtained in an independent trial.
Chromatographic resolution of the endogenous and TcOGNT2cat-dependent activities
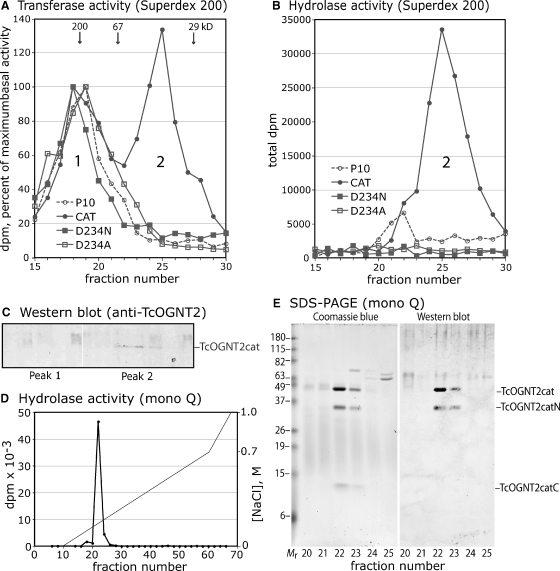
Based on contrasting temperature dependence and the importance of catalytic amino acids, the TcOGNT2-dependent activities are hypothesized to be independent of, rather than derived from the endogenous activities. This was examined further by gel filtration of concentrated conditioned media (Figure 7A). Endogenous transferase activity eluted monodispersely within the included volume (peak 1). Conditioned media from TcOGNT2cat-expressing cells exhibited, in addition, a later-eluting novel activity peak (peak 2), which co-eluted with the TcOGNT2cat protein as determined by Western blotting (Figure 7C). Based on the comparison with globular protein standards, the activity eluted at a position roughly consistent with the monomeric Mr of 48,000 as estimated by SDS–PAGE. Peak 2 also exhibited abundant hydrolase activity (Figure 7B), confirming that both activities were associated with TcOGNT2cat. Both activities were recovered at high yield relative to the sample load (data not shown). No hydrolase activity was found in peak 1 though a low level of unknown origin was observed at an intermediate position. Similar analysis of the mutant TcOGNT2 proteins confirmed the absence of peak 2 activities (Figure 7A and B).
Fig. 7.
Chromatographic purification of TcOGNT2 activities from endogenous activities. Conditioned media (50-fold concentrated) from the strains described in Figure 6, collected at cell densities within ±20% of each other, were analyzed on a Superdex 200 gel filtration column. Fractions were analyzed for (A) transferase activity, (B) hydrolase activity, (C) TcOGNT2cat protein, and total protein (not shown), as in Figure 6. Negligible activity was detected outside the fractions shown. (A) Results are normalized to the maximum dpm at the peak of the endogenous activity (fraction 18 or 19). Actual values ranged from 3360–13,700 dpm. Peak positions of Mr markers are shown. (B) Actual dpm, after subtraction of a background value of 6580 dpm, are shown. (C) Western blot analysis of corresponding fractions of a separate gel filtration experiment (not shown) which also exhibited peaks 1 and 2 activities. Expected position of TcOGNT2cat is shown at right. (D) Peak 2 fractions from a Superdex 200 separation (as in A) were applied to an anion exchange column and eluted with a salt gradient as indicated. Alternate fractions were assayed for hydrolysis activity. (E) Fractions in the vicinity of the activity peak were analyzed by SDS–PAGE and Coomassie blue staining for protein, or Western blotting for TcOGNT2 as in panel C. The three major Coomassie blue bands from fraction 22 were excised and trypsinized and are labeled according to their identification by MS and MS-MS sequencing (supplementary Table II). All results are representative of two or more independent experiments.
To examine whether TcOGNT2cat is alone capable of catalyzing transferase and hydrolysis reactions, the Superdex 200 peak 2 activity pool was further purified on a mono-Q anion exchange column. Only a single major OD280 peak (not shown) and activity peak (Figure 7D) was observed, at good yield (data not shown), and analysis by SDS–PAGE showed it to contain three predominant bands that stained with Coomassie blue. The upper two bands reacted with the anti-TcOGNT2 antibody. The gel slices were trypsinized and analyzed by MALDI-TOF-TOF-MS. Based on MS and MS-MS analysis, the upper band was confirmed to be full-length TcOGNT2cat, and the middle and lower bands were confirmed to be derived from the N-terminal and C-terminal regions of TcOGNT2cat (supplementary Table II and Supplementary Figure 2). The cleavage event was localized between aa 371 and 432. Since the three bands were not resolved by gel filtration, it is likely that cleavage does not dissociate the N- and C-terminal regions. The ratio of the upper band at the Mr 48,000 position, corresponding to intact TcOGNT2cat, to the lower bands at Mr 35,000 and 13,000, varied between experiments, but the ratio of transferase to hydrolysis activities did not (data now shown), suggesting that cleaved TcOGNT2cat is not the source of hydrolysis activity. Since no other discrete bands were detected by Coomassie blue or silver staining (data not shown), though a broad smear of OD280-absorbing material was noted in the central region of the gel of most fractions, TcOGNT2cat likely acts alone to catalyze the transferase and hydrolysis activities.
Further characterization of TcOGNT2cat activities and comparison with microsomal activity of T. cruzi
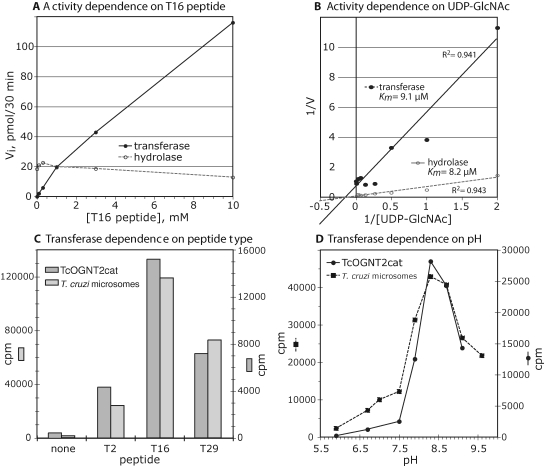
The concentration dependence of the highly purified activity toward the T16 peptide was determined in the presence of 50 μM UDP-GlcNAc under initial velocity conditions (Figure 8A). Transferase activity increased nearly linearly up to the highest concentration tested, 10 mM (or 40 mM with respect to potential acceptor Thr residues), indicating a high but undeterminable Km with respect to this substrate. In contrast, hydrolysis activity was only slightly reduced at the higher concentrations of T16 tested. Thus, hydrolysis activity predominated below 1 mM T16, at which the previous assays were performed, whereas transferase activity prevailed above 1 mM. The purified enzyme showed the expected hyperbolic dependence on UDP-GlcNAc at 0.2 mM T16, with indistinguishable Km values of 8–9 μM for the transferase and hydrolysis activities (Figure 8B). Based on enzyme protein amounts estimated from OD280 values and Coomassie blue staining intensity relative to known standards, the optimized transferase activity at 50 μM UDP-GlcNAc and 10 mM T16 was approximately 0.5 mol product/mol enzyme/min, at least 100-fold slower than typical glycosyltransferases.
Fig. 8.
Characterization of the highly purified activities and comparison with T. cruzi microsomal transferase activity. (A) Highly purified preparation of TcOGNT2cat (fraction 22 in Figure 7D and E) was analyzed for dependence of its transferase and hydrolase activities on the concentration of T16 peptide. The standard assay was conducted for 30 min in the presence of 50 μM UDP-GlcNAc. (B) Double-reciprocal plot of similar reactions conducted over a series of UDP-GlcNAc concentrations in the presence of 0.2 mM T16 peptide. (C) Microsomes from T. cruzi epimastigotes or TcOGNT2cat concentrated from L. tarentolae culture supernatants (as in Figure 4) were assayed for transferase activity in the absence (none) or presence of 0.2 mM T2, T16, or T29 peptides, at pH 8.0. (D) Independent samples were assayed for pH dependence in the 0.1 M Tris-maleate buffer using 0.2 mM T16.
To begin to evaluate acceptor substrate preference, peptides (Table II) corresponding to a T. cruzi mucin sequence (T2) and from the two D. discoideum mucin sequences (T16 and T29) were compared at a 0.2 mM concentration. The peptides exhibited the following order of activity using TcOGNT2 from the cell culture supernatant: T16>T29>T2 (Figure 8C). Analysis of a crude microsomal preparation from T. cruzi exhibited an almost identical ratio of activities; in contrast, D. discoideum microsomes were inactive toward T2 (unpublished data). Analysis of the pH dependence revealed an unusually high optimum of 8.3–8.5 (Figure 8D), which contrasts with the broad pH optimum of DdGnt2 centered at 7.5 (Wang et al. 2003), at which TcOGNT2 exhibits <10% maximal activity. A similar preference for alkaline pH was observed for the endogenous microsomal activity, in accord with a previous report (Previato et al. 1998). The similar and novel acceptor substrate and pH preferences suggest that recombinant TcOGNT2 represents at least part of the native enzyme detected in microsomes.
Discussion
The first step in mucin-type O-glycosylation in T. cruzi involves formation of the GlcNAcα1-Thr linkage in the Golgi apparatus. This report establishes TcOGNT2 as a prime candidate for catalyzing this reaction. Sequence analysis shows that TcOGNT2 is the T. cruzi gene most similar to the Golgi-associated pp-αGlcNAcT from the mycetozoan D. discoideum, DdGnt2. A soluble expression construct of TcOGNT2, deleted of its transmembrane anchor to allow secretion as for DdGnt2 (Wang et al. 2003), conferred increased pp-GlcNAcT activity to the conditioned medium of cells of the distantly related trypanosomatid L. tarentolae, based on acid hydrolysis and β-elimination analyses of the glycopeptide product of the reaction. The expressed activity was chromatographically purified from endogenous pp-GlcNAcT-like activity to near homogeneity and consisted of the primary polypeptide and lesser amounts of a proteolytic cleavage product. The dependence of activity on the Asp residue of the DxH motif, which lies in the active site based on the comparison with the distant homolog Mm pp-αGalNAcT2 (Fritz et al. 2006), indicates that TcOGNT2 directly contributes the catalytic function rather than activates a potential minor component in the purified preparation. The considerable sequence similarity with DdGnt2 indicates that, as for DdGnt2 and Mm pp-αGalNAcT2, TcOGNT2 attaches the HexNAc in an α-linkage. The enzyme preparations were, however, insufficiently active to produce enough product for chemical confirmation.
As known for some other glycosyltransferases (e.g., Hagen and Nehrke 1998; Gerken et al. 2002), secreted TcOGNT2cat also expresses a robust sugar nucleotide hydrolysis activity. Hydrolysis was confirmed based on chromatographic coelution of the product with GlcNAc. This distinctive activity was a useful correlate during initial screening for the transferase activity. The activity copurified at a similar ratio to transferase activity throughout the purification. Hydrolase activity was inactivated together with transferase activity by the Asp mutations, confirming the involvement of TcOGNT2. Each activity exhibited a similar Km for UDP-GlcNAc (Figure 8B), consistent with a related catalytic mechanism. Whereas the transferase activity was dependent on the T16 peptide concentration up to the highest concentration tested (10 mM), hydrolase activity was relatively insensitive to T16 (Figure 8A). This resulted in hydrolase activity prevailing below 1 mM T16 peptide. TcOGNT2 was partially cleaved during purification, but the similar ratio of hydrolase to transferase activities in different preparations indicated that the cleaved polypeptide is not responsible for hydrolase activity. Although it is not known if the hydrolase activity is biologically relevant, it is noted that microsomes prepared from T. cruzi, where TcOGNT2 is probably normally expressed, exhibit higher UDP-GlcNAc hydrolysis activity than native L. tarentolae microsomes, though not as high as microsomes from transfected L. tarentolae (Figure 3).
The pp-αGlcNAcT activity assigned to the secreted TcOGNT2 construct offers an attractive mechanism for initiating mucin-type O-glycosylation in T. cruzi. In support of this model, a fusion protein containing the N-terminal region of TbOGNT2, the presumptive ortholog of TcOGNT2 in T. brucei (Table I), accumulates in the Golgi apparatus (He et al. 2004) where the pp-αGlcNAcT activity resides in T. cruzi (Morgado-Diaz et al. 2001). TcOGNT2 mRNA accumulates in epimastigotes (supplementary Figure 4) consistent with protein expression. Heterologous expression of full-length TcOGNT2 confers additional transferase activity to microsomes of L. tarentolae such that their activity approaches the native activity of T. cruzi microsomes (Figure 3). A comparison of the pp-αGlcNAcT activity of T. cruzi microsomes with that of TcOGNT2cat secreted from L. tarentolae cells reveals a striking similarity with respect to acceptor peptide preferences and narrow alkaline pH optima (Figure 8C and D). TcOGNT2 is distinctive in these characteristics relative to DdGnt2, which has a broad pH optimum, is inactive toward T2, and exhibits substrate inhibition with T16 (Wang et al. 2003; Ercan and West 2005). Each activity also requires divalent cations (Previato et al. 1998; data not shown). As mentioned above, both preparations exhibit substantial UDP-GlcNAc hydrolysis activity though the source of activity in microsomes is not known. Limited analysis of the related predicted protein, TcOGNT1, indicates that its mRNA is also expressed in epimastigotes and may contribute transferase and hydrolase activities (Figure 3). Genetic studies enabled by the present work can now be planned to confirm the predicted and potentially essential in vivo roles of TcOGNT2 and TcOGNT1 in cells, which might target separate classes of mucins in different life cycle stages (Buscaglia et al. 2004, 2006).
Although TcOGNT2 possesses the pp-αGlcNAcT activity predicted by its sequence similarity to DdGnt2, characteristics of the in vitro activity raise critical questions about its function in cells. The sharp alkaline pH optimum of TcOGNT2cat, observed also in T. cruzi microsomes (Figure 8D), suggests that the enzyme will have low activity in the more neutral pH environment expected for the Golgi apparatus. Even under optimized conditions, the transferase activity of the purified enzyme is low (∼0.5 mol/mol/min). Physiological acceptor substrates, which remain to be identified, may have greater activity. Though peptide acceptors are generally active for this enzyme class (Ercan and West 2005), some pp αGalNAcTs prefer partially modified substrates (Ten Hagen et al. 2003). Alternatively, the enzyme protein may not be correctly posttranslationally modified in the heterologous expression system, or is abnormal without its transmembrane anchor. Nevertheless, the level of protein secreted from L. tarentolae was surprisingly low compared to other recombinant proteins (Breitling et al. 2002), consistent with the deficiency of an unknown limiting factor in this heterologous system. The good recovery of enzyme activity during purification (data not shown) suggests that low activity is not due to instability or separation from a critical factor during purification itself.
A comparison of transgenic TcOGNT2cat with the endogenous activity is potentially informative. Peak 1 shown in Figure 7A likely derived from an LtOGNT, considering the widespread distribution of the genes in trypanosomatids (Figure 2). Proteolytically cleaved but active Golgi glycosyltransferases are commonly secreted at low levels from cells (e.g., Cho et al. 1997). Unlike the endogenous transferase activity, TcOGNT2cat was not activated by temperature in the physiological range (supplementary Figure 6), exhibited substantial UDP-GlcNAc hydrolysis activity, and eluted later on the gel filtration column (Figure 7A and B). The low Q10 value of unity could be explained by imperfect folding or the absence of a critical cofactor. Hydrolysis might be more stable than transferase activity, as described for bovine pp-αGalNAcT1 (Gerken et al. 2002). Since additional proteins were not observed in the purified TcOGNT2cat protein preparation (Figure 7E), the retarded elution on a gel filtration column relative to the endogenous activity (Figure 7A) is consistent with the absence of such a protein cofactor needed for optimal activity. Dependence on a cofactor might help explain the difficulty of purifying native pp-αGlcNAcT activity from microsomes of T. cruzi. In future studies, the possible importance of TcONGT1 and TcOGNTL will be examined by co-expression. Alternatively, a missing factor might serve as a chaperone during biosynthesis as exemplified by the role of Cosmc for T-synthase (Ju and Cummings 2002), which might be restricted to trypanosomatids and/or saturated by heterologous overexpression. These differences raise the possibility that regulation of TcOGNT2 is more complex than that of other members of the GT60 and GT27 pp-αHexNAcT families, whose known examples appear to function in the absence of heterologous subunits (Teng-umnuay et al. 1999; Ten Hagen et al. 2003; Wang et al. 2003).
CAZy family GT60 pp-αGlcNAcT-like sequences are broadly distributed in protists and prokaryotes (Cantarel et al. 2009). Examples include predicted soluble cytoplasmic and Golgi-associated type 2 membrane proteins (West et al. 2004). A phylogenetic analysis of the sequences suggested that an ancient eukaryotic gene duplication enabled population of the Golgi lumen with this enzyme activity while retaining a cytoplasmic copy, inherited from prokaryotes possibly for the modification of cytosolic and nuclear proteins. Both genes were retained by Dictyostelium, the diatoms, green algae, and the oomycetes. The gene encoding the Golgi enzyme duplicated, with one form retained by the trypanosomatids and Dictyostelium, the other form retained by the oomycetes, and both forms retained by the diatoms (Figure 2). The trypanosomatids multiplied their Golgi enzyme gene early on and now harbor three related genes. A subsequent gene duplication is postulated to have allowed evolution of the family GT27 class of sequences which encode pp-αGalNAcTs in Toxoplasma gondii and other apicomplexans (Wojczyk et al. 2003), and in animals. However, family membership may not predict donor–acceptor substrate specificity because the animal Caenorhabditis elegans, which has multiple GT27 but no GT60 sequences, apparently forms both GalNAcα1-Thr/Ser and GlcNAcα1-Thr/Ser linkages (Guérardel et al. 2001), whereas an unconfirmed study suggests that T. cruzi also forms a GalNAcα1-Thr linkage (Freire et al. 2003) in addition to GlcNAcα1-Thr (Previato et al. 1998). Though most family GT27 sequences possess a discrete C-terminal lectin domain that supports acceptor substrate targeting (Fritz et al. 2006), it is not required for in vitro activity. Further studies are therefore required to understand the relationship between sequence and selection of donor and acceptor substrates for these enzymes. Nevertheless, it is evident that mucin-type O-glycosylation represents an ancient class of glycosylation that potentially changed from an αGlcNAc in early protists to an αGalNAc linkage in animals as epimerases and transporters co-evolved to increase sugar diversity. Mucin-type O-glycosylation is involved in cell–cell adhesion, cell sorting, and spore coat integrity in Dictyostelium (West et al. 2004), and recent studies have identified critical functions for specific pp-αGalNAcTs in animals (e.g., Kato et al. 2006; Tian and Ten Hagen 2007). If the prediction that TcOGNT2 and TcOGNT1 solely mediate the first step in mucin-type O-glycosylation in T. cruzi is correct, it should be practical to test the function of mucin-type O-glycosylation genetically, where indirect evidence indicates a role in virulence, and in other trypanosomatids, where the genes are present (Table I, Figure 2) though mucin-type O-glycosylation has not yet been described.
Materials and methods
Cells
Epimastigote forms of T. cruzi strain CL-Brener were cultured at 28°C in 3.7% (w/v) brain–heart infusion (Difco) medium supplemented with 5% (v/v) fetal bovine serum (FBS) (Atlanta Biologicals), 10 μg/mL hemin (Sigma) and 20 μg/mL folic acid (Sigma) in T25 flasks, and split 1:10 every 6 days. For scale-up, parasites were inoculated at 5–10 × 105 cells/mL in the same medium in Erlenmeyer flasks and maintained on a gyratory shaker at ∼200 rpm and 22°C until stationary phase (5–10 × 107 cells/mL). Leishmania tarentolae strain P-10 (Jena Bioscience, Germany) was maintained at 26°C in M199 medium (US Biologicals) containing 5% FBS and Pen-Strep (Gibco) as above, and split 1:10 every 4 days. The serum level was reduced by serial splitting at 1:10 in brain–heart infusion (Difco) medium containing 5 μg/mL hemin. Large-scale cultures were prepared as above. Cultures were renewed from frozen stocks every 2 months. D. discoideum strains Ax3 (normal) and DL118 (modB−) were grown axenically and developed as described (Wang et al. 2003).
Phylogenetic analysis of sequences
Amino acid sequences related to that of the catalytic domain of DdGnt2 were identified at NCBI (www.ncbi.nlm.nih.gov/), the Wellcome Trust Sanger Institute Pathogen Sequencing Unit (www.genedb.org/), and the Joint Genome Institute at the US Department of Energy (www.jgi.doe.gov/), using tBLASTn, PSI-BLAST, and PHI-BLAST. For the phylogenetic analysis of the aligned sequences, Modelgenerator (Whelan and Goldman 2001) was used to find the best fitting amino acid substitution model. A WAG+G+I substitution model (Keane et al. 2006) was selected consisting of the Whelan and Goldman model with a gamma rate distribution (eight categories, alpha = 2.099) and a proportion of invariable sites. This model was implemented in a maximum likelihood framework, executed using several runs of the program PhyML (Guindon and Gascuel 2003) to search for the best tree.
Mapping the 5′-region of the TcOGNT transcripts
Total RNA from CL-Brener epimastigotes was reverse transcribed using internal reverse primers TcE5AS415 or TcE2NAS (supplementary Table I) and SuperScript® III Reverse Transcriptase (Invitrogen), according to the manufacturer's instructions. The reactions were amplified by PCR with Taq DNA polymerase after the addition of the mini-exon primer TcME33 (Nepomuceno-Silva et al. 2001) and additional internal primer, over 30 cycles of 95°C, 30 s; 63°C, 45 s; 72°C, 1 min, and a final extension for 7 min at 72°C. The amplified products were cloned into InstaClone pTZ57R (Fermentas) and transfected into chemically competent DH5α cells. Plasmids from ampicillin-resistant clones were sequenced using M13F and M13R primers.
Recombinant expression of TcOGNT2 in D. discoideum
TcOGNT2 DNA was amplified from CL-Brener gDNA prepared with a Rapidprep genomic DNA isolation kit (Pharmacia). PCR reactions were performed in 50 μL of 20 mM Tris-HCl (pH 8.8), 2 mM MgSO4, 10 mM KCl, 10 mM (NH4)2SO4, 0.1% (v/v) Tx-100, 1 mg/mL nuclease-free BSA, 1–10 ng template DNA, 0.4 mM dNTPs, 0.8 μM of primers, and a mixture of 2.5 U Taq (Eppendorf) and 0.25 U Pfu (Stratagene) DNA polymerases. The full-length TcOGNT2 coding region was amplified using primers E5SATG2 and E5AS2 (supplementary Table I), over 30 cycles of 94°C, 45 s; 65°C, 45 s; 68°C, 3 min, followed by 68°C at 7 min. The DNA was cloned into pCR4TOPO, released by digestion with KpnI and BglII, and ligated into similarly digested pVS4 expression plasmid (Zhang et al. 1999) yielding pVS4OGNT2. Primers E5-U2 and E5L1 (supplementary Table I) were used to amplify DNA for expression of N-terminally truncated TcOGNT2cat as mentioned above, except that the annealing temperature was 60°C. Coding DNA was cloned into pCR4TOPO, excised using BglII, and ligated in the forward orientation into pVS4 predigested with the same enzyme, yielding pVS4OGNT2cat.C-myc. An N-terminal myc-tagged construct was prepared using BamH1-digested pVS4.
Plasmids were electroporated and expression clones were isolated essentially as described (Zhang et al. 1999). Clonal isolates grown in the HL-5 axenic medium were screened by Western blotting with mAb 9E10 for expression of the c-myc epitope-tagged protein.
Recombinant expression of TcOGNT2 and TcOGNT1 in L. tarentolae
The full-length TcOGNT2 coding region was amplified using primers E5S1 and E5AS1 (supplementary Table 1) as mentioned above, using an annealing temperature of 57°C. The amplified fragment was cloned into pCR4TOPO (Invitrogen). After digestion with BglII and XhoI, the released DNA was ligated into similarly digested pF4SPmX1.4neo (Jena), yielding pF4SPlmpOGNT2.neo. Full-length TcOGNT1 was created similarly using primers E2S1 and E2AS1 (supplementary Table I) using an annealing temperature of 62°C. After digestion with NcoI and XhoI, the released DNA was ligated into similarly digested pF4SPmX1.4neo, yielding pF4SPlmpOGNT1.neo. DNA encoding truncated TcOGNT2 lacking the transmembrane signal anchor domain and short cytoplasmic extension was amplified using primers E5S2 and E5AS1. Coding DNA was excised using KasI and XhoI and ligated separately into similarly digested pF4SPlmsapX1.4neo and pF4SPlmsapX1.4hyg, yielding the expression plasmids pF4SPlmsapOGNT2cat.neo and pF4SPlmsapOGNT2cat.hyg. Amplified sequences were confirmed by sequencing and found to match sequences in the Sanger genome database.
Expression plasmids (5–10 μg) were digested with 10 U of SwaI, EtOH precipitated, and resuspended in 50 μL 2 mM Tris-HCl (pH 8.5). L. tarentolae P-10 cells were transfected essentially as described (Breitling et al. 2002). After 24 h in the M199 culture medium, G418 or hygromycin B were added to 50 or 100 μg/mL, respectively. Non-DNA control cells were killed in 7–9 days. Resistant cultures were split (1:5) 7 days after the drug addition and again after 4 days. Cells were cloned by limiting dilution in 50 μL in 96-well plate wells.
Site-directed mutagenesis of TcOGNT2cat
The amino acid sequence of TcOGNT2cat was altered by site-directed mutagenesis of pF4SPlmsapOGNT2cat.neo (van der Wel et al. 2002). The D234A substitution was generated using primers TcE5D234Afor and TcE5D234Arev (supplementary Table I), resulting in the point mutations A701C and C702T and a novel NheI restriction site; D234N was generated using primers TcE5D234Nfor and TcE5D234Nrev, which yielded G700A and C702T, and a novel AseI restriction site (supplementary Figure 2). DNA was amplified as mentioned above using an annealing temperature of 65–70°C, digested with DpnI for 1 h at 37°C, heat inactivated, and used to transform chemically competent TOP10 E. coli cells. After sequence confirmation, the plasmids were introduced into L. tarentolae as mentioned above.
Conditioned media and cell extracts
Stationary phase cultures of T. cruzi (108 cells/mL) or L. tarentolae (2–4 × 108 cells/mL) were centrifuged as mentioned above. The supernatants were supplemented with protease inhibitors (10 μg/mL leupeptin, 10 μg/mL aprotinin, 4 μg/mL n-α-p-tosyl-l-lysine-chloromethyl ketone, and 5 mM benzamidine), clarified further by centrifugation at 16,000 g × 15 min at 4°C, and concentrated 50-fold in a YM-30 Centricon centrifugal concentrator (Amicon, Inc.) at 4°C. Microsomes were prepared by resuspending the cell pellets in the sucrose buffer (100 mM HEPES-NaOH (pH 7.4), 10 mM MgCl2, 50 mM KCl, 0.5 M sucrose, protease inhibitors) by probe sonication as described (Heise et al. 1996). The P100 pellet was resuspended in 5 mL of sucrose buffer and stored at −80°C. Protein was quantified using a micro BCA protein assay kit (Pierce).
S100 and P100 fractions were prepared from exponentially growing D. discoideum cells as described (Teng-umnuay et al. 1999). The P100 fraction was resuspended in 5 mL of the above sucrose buffer by probe sonication and centrifuged again at 100,000 g × 1 h at 4°C. The supernatant (SP100) was removed, and the pellet (PP100) was resuspended in the same volume of sucrose buffer; each was stored at −80°C.
TcOGNT2cat purification
Concentrated cell culture supernatants were chromatographed on a Superdex 200 PC3.2/30 or HiLoad 16/60 gel filtration column (Pharmacia Biotech) pre-equilibrated in 50 mM Tris-HCl (pH 7.5), 15% (v/v) glycerol, 1 mM MnCl2, 0.1 mM EDTA. Active fractions were chromatographed on a 5/5 mono-Q column (Pharmacia) at 1 mL/min using a 0–0.7 M NaCl gradient in 25 mM Tris-HCl (pH 7.5), and 0.5 mL fractions were collected.
SDS–PAGE and Western blotting
SDS–PAGE and Western blotting were performed as described (Wang et al. 2003; van der Wel et al. 2005). Apparent Mr values were estimated using the Benchmark prestained protein ladder from Invitrogen. Wheat germ agglutinin-reactive proteins were detected using 0.125 μg/mL Alexa Fluor 680-conjugated WGA (Molecular Probes) in 0.5% (w/v) hemoglobin in 50 mM Tris-HCl (pH 7.5), 100 mM NaCl.
Protein identification
Slices from a Coomassie blue stained 7–20% SDS–polyacrylamide gel were vacuum dried and rehydrated in a solution of sequence grade trypsin (12 ng/μL; Promega), as described (Hellman et al. 1995). The resulting peptides were profiled in positive ion mode on an Ultraflex MALDI-TOF-TOF MS (Bruker Daltonics, Bremen, Germany) by co-crystallizing with an HCCA matrix. Peptide masses were determined in MS-reflector mode, and MS-MS was carried out in LIFT mode.
Enzyme activity assays
The standard transferase assay was carried out for 1 h at 28°C in a final volume of 100 μL and contained 0.5 μCi UDP-[6-3H]GlcNAc (40–60 Ci/mmol; American Radiochemical), 50 mM Tris-HCl (pH 8.5), 5 mM MgCl2, 5 mM MnCl2, 0.2% (w/v) CHAPS, 200 μM T16 acceptor peptide (Table II), and enzyme protein. Time or temperature was varied as indicated. UDP-[U-14C]GlcNAc (288 mCi/mmol, Dupont NEN) or UDP-[3H]GalNAc (60 Ci/mmol; American Radiochemical) was substituted for UDP-[3H]GlcNAc in some trials. Other variations included use of pH 8.0 instead of 8.5 and alternate peptide substrates (Table II). For the pH optimization study, the buffer was 100 mM Tris-maleate. The reaction was started by the addition of protein and stopped by dilution with 900 μL 50 mM Na2EDTA (pH 8.0). After centrifugation at 15,000 g × 15 min at 22°C, or 100,000 g × 60 min for microsomes, the reaction supernatant was loaded onto a Waters 1 mL C18 Sep-Pak, and bound radioactivity was determined after elution with MeOH and scintillation counting in BioSafe-II (Research Products International Corp.) as described (Ercan and West 2005).
One milliliter of the C18 Sep-Pak flow-through/wash pool was applied to a 0.8 mL Dowex-1 (Cl− form) and eluted under positive pressure, and an additional 1 mL H2O was applied and collected in the same way. Breakdown activity was based on the sum of the dpm values of the two fractions.
Characterization of reaction products
For identification of the incorporated sugar, the 3H-labeled glycopeptide (T16) was eluted from a C18 Sep-Pak, dried by vacuum centrifugation, hydrolyzed in 6 N HCl, and analyzed by HPAEC on a CarboPac PA-1 column (Dionex) as previously described (Teng-umnuay et al. 1998). The 3H-glycopeptide was subjected to β-elimination by dissolving in 0.5 mL freshly prepared 1 M NaBH4 in 0.1 M NaOH and incubated for 18 h at 45°C. The reaction was stopped by the slow addition of 29 μL glacial acetic acid. The sample was loaded onto 2 mL of Dowex 50 W (X4-400; H+-form), eluted 3× with 4 mL of water and passed through a 1-mL C18 Sep-Pak. The column was washed with 1.5 mL of water, and all 13.5 mL were pooled and freeze-dried, suspended in 1 mL MeOH and dried by vacuum centrifugation. The addition of MeOH and drying were repeated 6× to eliminate borate. Recovery of radioactivity was 69%. Samples were suspended in water, mixed with 1–10 nmol/10 μL of GlcNAcitol and GalNAcitol (prepared from GlcNAc and GalNAc by Na cyanoborohydride reduction), and separated by HPAEC at 0.4 mL/min on a CarboPac MA-1 column using a gradient of NaOH (60–660 mM) on a Dionex DX-600 HPLC (Corradini et al. 1997). Standards were detected by pulsed amperometry, and fractions were collected and counted in a liquid scintillation counter to detect radioactivity.
The UDP-GlcNAc breakdown product, collected as the Dowex-1 flow-through fraction from a reaction containing 70 μM UDP-[14C]GlcNAc (1 μCi) and mono-Q-purified enzyme, was dried, redissolved in water with 1 nmol of GlcNAc, passed through a 1 mL Dowex-50 (H+-form), and analyzed on a CarboPac PA-1 column as mentioned above.
Antibody preparation
For bacterial expression, TcOGNT2cat DNA was PCR amplified using primers E5S3 and E5AS3 (supplementary Table I) and cloned into pCR4TOPO as mentioned above. DNA was excised with NdeI and BamHI and cloned into pET15-TEVi (van der Wel et al. 2005) predigested with the same enzymes. The resulting expression plasmid, pET15TEVTcOGNT2cat, was transformed into E. coli strain ER2566 (New England Biolabs), and clonal transfectants were induced with 0.5 mM isopropyl 1-thio-β-d-galactopyranoside (RPI) for 6 h at 22°C. E. coli was incubated in lysozyme, lysed in a French pressure cell in the presence of protease inhibitors, and incubated in nucleases according to standard procedures. TcOGNT2 was found exclusively in inclusion bodies which were washed with 2% (v/v) Triton X-100, 2 M urea, 100 mM Tris-HCl (pH 8.0), 5 mM EDTA, and 5 mM DTT. Because of insolubility, TcOGNT2 was solubilized by probe sonication in 6 M guanidine isothiocyanate in 50 mM Tris-HCl (pH 8.8), 1 mM DTT. After centrifugation, the supernatant was diluted 10-fold in 6 M urea, 50 mM Tris-HCl (pH 6.8), 0. 5 M NaCl, 5 mM imidazole, loaded onto a Ni+2-primed His-Trap HP column (Pharmacia Biotech), and eluted in 0.1–0.25 M imidazole in 0.5 M NaCl. The EtOH-precipitated protein was electrophoresed on a gradient polyacrylamide slab gel, and the major Coomassie blue-stained band at Mr 49,000 was excised, minced, and used to immunize New Zealand White female rabbits using complete Freund's adjuvant for the initial injection and incomplete Freund's for booster injections. Preimmune serum and anti-TcOGNT2cat antiserum from bleeds 5–7 were used at 1:100–500.
GenBank accession numbers
The TcOGNT2 haplotype nucleotide sequences reported in this paper have been submitted to the GenBankTM/EBI Data Bank with accession numbers EU599317 and EU599318.
Supplementary data
Supplementary data for this article is available online at http://glycob.oxfordjournals.org/.
Funding
The Oklahoma Center for Advancement of Science and Technology (Oklahoma Health Research Program HR04-141 (20041406) to C.M.W.), the National Institutes of Health (R01 GM37539 to C.M.W.), the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) (research grant and postdoctoral fellowship to N.H.), and the Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ) (to N.H.).
Acknowledgments
We thank Stephen M. Beverley (Washington University) for helpful suggestions. We are grateful to Wendy Ives of the Oklahoma Center for Medical Glycobiology Core Laboratory for assistance with the HPAEC analyses.
Conflict of interest statement
None.
Abbreviations
- FBS
fetal bovine serum
- HPAEC
high-performance (or pH) anion exchange chromatography
- pp-αGlcNAcT
UDP-GlcNAc:polypeptide N-α-acetylglucosminyltransferase
References
- Acosta-Serrano A, Almeida IC, Freitas-Junior L, Yoshida N, Schenkman S. The mucin-like glycoprotein super-family of Trypanosoma cruzi: Structure and biological roles. Mol Biochem Parasitol. 2001;114:143–150. doi: 10.1016/s0166-6851(01)00245-6. [DOI] [PubMed] [Google Scholar]
- Alcaide P, Fresno M. The Trypanosoma cruzi membrane mucin AgC10 inhibits T cell activation and IL-2 transcription through l-selectin. Int Immunol. 2004;16:1365–1375. doi: 10.1093/intimm/dxh138. [DOI] [PubMed] [Google Scholar]
- Almeida IC, Ferguson MAJ, Schenkman S, Travassos LR. Lytic anti-α-galactosyl antibodies from patients with chronic Chagas disease recognize novel O-linked oligosaccharides on mucin-like glycosylphosphatidylinositol anchored proteins of Trypanosoma cruzi. Biochem J. 1994;304:793–802. doi: 10.1042/bj3040793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argibay PF, Di Noia JM, Hidalgo A, Mocetti E, Barbich M, Lorenti AS, Bustos D, Tambutti M, Hyon SH, Frasch ACC, et al. Trypanosoma cruzi surface mucin TcMuc-e2 expressed on higher eukaryotic cells induces human T cell anergy, which is reversible. Glycobiology. 2002;12:25–32. doi: 10.1093/glycob/12.1.25. [DOI] [PubMed] [Google Scholar]
- Breitling R, Klingner S, Callewaert N, Pietrucha R, Geyer A, Ehrlich G, Hartung R, Muller A, Contreras R, Beverley SM, et al. Non-pathogenic trypanosomatid protozoa as a platform for protein research and production. Protein Expr Purif. 2002;25:209–218. doi: 10.1016/s1046-5928(02)00001-3. [DOI] [PubMed] [Google Scholar]
- Brener Z. Biology of Trypanosoma cruzi. Annu Rev Microbiol. 1973;27:347–482. doi: 10.1146/annurev.mi.27.100173.002023. [DOI] [PubMed] [Google Scholar]
- Buscaglia CA, Campo VA, Di Noia JM, Torrecilhas ACT, De Marchi CR, Ferguson MAJ, Frasch ACC, Almeida IC. The surface coat of the mammal-dwelling infective trypomastigote stage of Trypanosoma cruzi is formed by highly diverse immunogenic mucins. J Biol Chem. 2004;279:15860–15869. doi: 10.1074/jbc.M314051200. [DOI] [PubMed] [Google Scholar]
- Buscaglia CA, Campo VA, Frasch ACC, Di Noia JM. Trypanosoma cruzi surface mucins: Host-dependent coat diversity. Nat Rev Microbiol. 2006;4:229–236. doi: 10.1038/nrmicro1351. [DOI] [PubMed] [Google Scholar]
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The carbohydrate-active enZymes database (CAZy): An expert resource for glycogenomics. Nucleic Acids Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SK, Yeh JC, Cummings RD. Secretion of alpha1,3-galactosyltransferase by cultured cells and presence of enzyme in animal sera. Glycoconj J. 1997;14:809–819. doi: 10.1023/a:1018533804015. [DOI] [PubMed] [Google Scholar]
- Corradini C, Canali G, Cogliandro E, Nicoletti I. Separation of alditols of interest in food products by high-performance anion-exchange chromatography with pulsed amperometric detection. J Chromatogr A. 1997;791:343–349. doi: 10.1016/s0021-9673(97)00827-3. [DOI] [PubMed] [Google Scholar]
- Coura JR. Chagas disease: What is known and what is needed – A background article. Mem Inst Oswaldo Cruz. 2007;102(Suppl 1):113–122. doi: 10.1590/s0074-02762007000900018. [DOI] [PubMed] [Google Scholar]
- Hellman U, Wernstedt C, Góñez J, Heldin CH. Improvement of an “In-Gel” digestion procedure for the micropreparation of internal protein fragments for amino acid sequencing. Anal Biochem. 1995;224:451–455. doi: 10.1006/abio.1995.1070. [DOI] [PubMed] [Google Scholar]
- El-Sayed NM, Myler PJ, Bartholomeu DC, Nilsson D, Aggarwal G, Tran AN, Ghedin E, Worthey EA, Delcher AL, Blandin G, et al. The genome sequence of Trypanosoma cruzi, ethiologic agent of Chagas disease. Science. 2005;309:409–415. doi: 10.1126/science.1112631. [DOI] [PubMed] [Google Scholar]
- Ercan A, West CM. Kinetic analysis of a Golgi UDP-GlcNAc:polypeptide-Thr/Ser N-acetyl-alpha-glucosaminyltransferase from Dictyostelium. Glycobiology. 2005;15:489–500. doi: 10.1093/glycob/cwi034. [DOI] [PubMed] [Google Scholar]
- Frasch ACC. Functional diversity in the trans-sialidase and mucin families in Trypanosoma cruzi. Parasitol Today. 2000;16:282–286. doi: 10.1016/s0169-4758(00)01698-7. [DOI] [PubMed] [Google Scholar]
- Freire T, Robello C, Soulé S, Ferreira F, Osinaga E. Sialyl-Tn antigen expression and O-linked GalNAc-Thr synthesis by Trypanosoma cruzi. Biochem Biophys Res Commun. 2003;312:1309–1316. doi: 10.1016/j.bbrc.2003.11.060. [DOI] [PubMed] [Google Scholar]
- Fritz TA, Raman J, Tabak LA. Dynamic association between the catalytic and lectin domains of human UDP-GalNAc:polypeptide alpha-N-acetylgalactosaminyltransferase-2. J Biol Chem. 2006;281:8613–8619. doi: 10.1074/jbc.M513590200. [DOI] [PubMed] [Google Scholar]
- Gerken TA, Zhang J, Levine J, Elhammer A. Mucin core O-glycosylation is modulated by neighboring residue glycosylation status. Kinetic modeling of the site-specific glycosylation of the apo-porcine submaxillary mucin tandem repeat by UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferases T1 and T2. J Biol Chem. 2002;277:49850–49862. doi: 10.1074/jbc.M205851200. [DOI] [PubMed] [Google Scholar]
- Guérardel Y, Balanzino L, Maes E, Leroy Y, Coddeville B, Oriol R, Strecker G. The nematode Caenorhabditis elegans synthesizes unusual O-linked glycans: Identification of glucose-substituted mucin-type O-glycans and short chondroitin-like oligosaccharides. Biochem J. 2001;357:167–182. doi: 10.1042/0264-6021:3570167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hagen FK, Nehrke K. cDNA cloning and expression of a family of UDP-N-acetyl-d-galactosamine:polypeptide N-acetylgalactosaminyltransferase sequence homologs from Caenorhabditis elegans. J Biol Chem. 1998;273:8268–8277. doi: 10.1074/jbc.273.14.8268. [DOI] [PubMed] [Google Scholar]
- Hang HC, Bertozzi CR. The chemistry and biology of mucin-type O-linked glycosylation. Bioorg Med Chem. 2005;13:5021–5034. doi: 10.1016/j.bmc.2005.04.085. [DOI] [PubMed] [Google Scholar]
- He CY, Ho HH, Malsam J, Chalouni C, West CM, Ullu E, Toomre D, Warren G. Golgi duplication in Trypanosoma brucei. J Cell Biol. 2004;165:313–321. doi: 10.1083/jcb.200311076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heise N, Raper J, Buxbaum L, Peranovich TMS, Cardoso de Almeida ML. Identification of complete precursors of glycosylphosphatidylinositol protein anchors of Trypanosoma cruzi. J Biol Chem. 1996;271:16877–16887. doi: 10.1074/jbc.271.28.16877. [DOI] [PubMed] [Google Scholar]
- Hoft DF, Farrar PL, Kratz-Owens K, Shaffer D. Gastric invasion by Trypanosoma cruzi and induction of protective mucosal immune responses. Infect Immun. 1996;64:3800–3810. doi: 10.1128/iai.64.9.3800-3810.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C, Todeschini AR, Agrellos OA, Previato JO, Mendonça-Previato L. Heterogeneity in the biosynthesis of mucin O-glycans from Trypanosoma cruzi tulahuen strain with the expression of novel galactofuranosyl-containing oligosaccharides. Biochemistry. 2004;43:11889–11897. doi: 10.1021/bi048942u. [DOI] [PubMed] [Google Scholar]
- Ju T, Cummings RD. A unique molecular chaperone Cosmc required for activity of the mammalian core 1 beta 3-galactosyltransferase. Proc Natl Acad Sci USA. 2002;99:16613–16618. doi: 10.1073/pnas.262438199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K, Jeanneau C, Tarp MA, Benet-Pagès A, Lorenz-Depiereux B, Bennett EP, Mandel U, Strom TM, Clausen H. Polypeptide GalNAc-transferase T3 and familial tumoral calcinosis. Secretion of fibroblast growth factor 23 requires O-glycosylation. J Biol Chem. 2006;281:18370–18377. doi: 10.1074/jbc.M602469200. [DOI] [PubMed] [Google Scholar]
- Keane TM, Creevey CJ, Pentony MM, Naughton TJ, McInerney JO. Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol Biol. 2006;6:29. doi: 10.1186/1471-2148-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConville MJ, Mullin KA, Ilgoutz SC, Teasdale RD. Secretory pathway of trypanosomatid parasites. Microbiol Mol Biol Rev. 2002;66:122–154. doi: 10.1128/MMBR.66.1.122-154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendonça-Previato L, Todeschini AR, Heise N, Previato JO. Protozoan parasite-specific carbohydrate structures. Curr Opin Struct Biol. 2005;15:499–505. doi: 10.1016/j.sbi.2005.08.011. [DOI] [PubMed] [Google Scholar]
- Morgado-Diaz JA, Nakamura CV, Agrellos OA, Dias WB, Previato JO, Mendonça-Previato L, de Souza W. Isolation and characterization of the Golgi complex of the protozoan Trypanosoma cruzi. Parasitol. 2001;123:33–43. doi: 10.1017/s0031182001007946. [DOI] [PubMed] [Google Scholar]
- Mortara RA, Silva S, Araguth MF, Blanco SA, Yoshida N. Polymorphism of the 35- and 50-kilodalton surface glycoconjugates of Trypanosoma cruzi metacyclic trypomastigotes. Infect Immun. 1992;60:4673–4678. doi: 10.1128/iai.60.11.4673-4678.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nepomuceno-Silva JL, Yokoyama K, de Mello LD, Mendonca SM, Paixão JC, Baron R, Faye JC, Buckner FS, Van Voorhis WC, Gelb MH, et al. TcRho1, a farnesylated Rho family homologue from Trypanosoma cruzi: Cloning, trans-splicing, and prenylation studies. J Biol Chem. 2001;276:29711–29718. doi: 10.1074/jbc.M102920200. [DOI] [PubMed] [Google Scholar]
- Previato JO, Andrade AFB, Pessolani MCV, Mendonca-Previato L. Incorporation of sialic acid into Trypanosoma cruzi macromolecules. A proposal for a new metabolic route. Mol Biochem Parasitol. 1985;16:85–96. doi: 10.1016/0166-6851(85)90051-9. [DOI] [PubMed] [Google Scholar]
- Previato JO, Jones C, Gonçalves LPB, Wait R, Travassos LR, Mendonça-Previato L. O-Glycosidically linked N-acetylglucosamine-bound oligosaccharides from glycoproteins of Trypanosoma cruzi. Biochem J. 1994;301:151–159. doi: 10.1042/bj3010151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Previato JO, Sola-Penna M, Agrellos OA, Jones C, Oeltmann T, Travassos LR, Mendonça-Previato L. Biosynthesis of O-N-acetylglucosamine-linked glycans in Trypanosoma cruzi. Characterization of the novel uridine diphospho-N-acetylglucosamine:polypeptide N-acetyl glucosaminyltransferase-catalyzing formation of N-acetylglucosamine-α1-O-threonine. J Biol Chem. 1998;273:14982–14988. doi: 10.1074/jbc.273.24.14982. [DOI] [PubMed] [Google Scholar]
- Previato JO, Wait R, Jones C, DosReis GA, Todeschini AR, Heise N, Mendonça-Previato L. Glyconositolphospholipids from Trypanosoma cruzi: Structure, biosynthesis and immunobiology. Adv Parasitol. 2004;56:1–41. doi: 10.1016/s0065-308x(03)56001-8. [DOI] [PubMed] [Google Scholar]
- Ruiz RC, Rigoni VL, Gonzalez J, Yoshida N. The 35/50 kDa surface antigen of Trypanosoma cruzi metacyclic trypomastigotes, an adhesion molecule involved in host cell invasion. Parasite Immunol. 1993;15:121–125. doi: 10.1111/j.1365-3024.1993.tb00591.x. [DOI] [PubMed] [Google Scholar]
- Schenkman S, Ferguson MAJ, Heise N, de Almeida ML, Mortara RA, Yoshida N. Mucin-like glycoproteins linked to the membrane by glycosylphosphatidylinositol anchor are the major acceptors of sialic acid in a reaction catalyzed by trans-sialidase in metacyclic forms of Trypanosoma cruzi. Mol Biochem Parasitol. 1993;59:293–304. doi: 10.1016/0166-6851(93)90227-o. [DOI] [PubMed] [Google Scholar]
- Schenkman S, Jiang MS, Hart GW, Nussenzweig V. A novel cell surface trans-sialidase of Trypanosoma cruzi generates a stage-specific epitope required for invasion of mammalian cells. Cell. 1991;65:1117–1125. doi: 10.1016/0092-8674(91)90008-m. [DOI] [PubMed] [Google Scholar]
- Tarleton RL, Reithinger R, Urbina JA, Kitron U, Gürtler RE. The challenges of Chagas’ disease – Grim outlook or glimmer of hope. PLoS Med. 2007;4:e332. doi: 10.1371/journal.pmed.0040332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ten Hagen KG, Fritz TA, Tabak LA. All in the family: The UDP-GalNAc:polypeptide N-acetylgalactosaminultransferases. Glycobiology. 2003;13:1R–16R. doi: 10.1093/glycob/cwg007. [DOI] [PubMed] [Google Scholar]
- Teng-umnuay P, Morris HR, Dell A, Panico M, Paxton T, West CM. The cytoplasmic F-box binding protein Skp1 contains a novel pentasaccharide linked to hydroxyproline in Dictyostelium. J Biol Chem. 1998;273:18242–18249. doi: 10.1074/jbc.273.29.18242. [DOI] [PubMed] [Google Scholar]
- Teng-umnuay P, Van Der Wel H, West CM. Identification of a UDP-GlcNAc:Skp1-hydroxyproline GlcNAc-transferase in the cytoplasm of Dictyostelium. J Biol Chem. 1999;274:36392–36402. doi: 10.1074/jbc.274.51.36392. [DOI] [PubMed] [Google Scholar]
- Tian E, Ten Hagen KG. A UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase is required for epithelial tube formation. J Biol Chem. 2007;282:606–614. doi: 10.1074/jbc.M606268200. [DOI] [PubMed] [Google Scholar]
- Todeschini AR, da Silveira EX, Jones C, Wait R, Previato JO, Mendonça-Previato L. Structure of O-glycosidically linked oligosaccharides from glycoproteins of Trypanosoma cruzi CL-Brener strain: Evidence for the presence of O-linked sialyl-oligosaccharides. Glycobiology. 2001;11:47–55. doi: 10.1093/glycob/11.1.47. [DOI] [PubMed] [Google Scholar]
- Tomlinson S, Raper J. Natural immunity to trypanosomes. Parasitol Today. 1998;14:354–359. doi: 10.1016/s0169-4758(98)01295-2. [DOI] [PubMed] [Google Scholar]
- Trasar-Cepeda C, Gil-Sotres F, Leirós MC. Thermodynamic parameters of enzymes in grassland soils from Galicia, NW Spain. Soil Biol Biochem. 2007;39:311–319. [Google Scholar]
- Van Der Wel H, Ercan A, West CM. The Skp1 prolyl hydroxylase from Dictyostelium is related to the hypoxia-inducible factor-alpha class of animal prolyl 4-hydroxylases. J Biol Chem. 2005;280:14645–14655. doi: 10.1074/jbc.M500600200. [DOI] [PubMed] [Google Scholar]
- Van Der Wel H, Morris H, Panico M, Paxton T, Dell A, Kaplan L, West CM. Molecular cloning and expression of a UDP-GlcNAc:hydroxyproline polypeptide GlcNAc-transferase that modifies Skp1 in the cytoplasm of Dictyostelium. J Biol Chem. 2002;277:46328–46337. doi: 10.1074/jbc.M208024200. [DOI] [PubMed] [Google Scholar]
- Wang F, Metcalf T, Van Der Wel H, West CM. Initiation of mucin-type O-glycosylation in Dictyostelium is homologous to the corresponding step in animals and is important for spore coat function. J Biol Chem. 2003;278:51395–51407. doi: 10.1074/jbc.M308756200. [DOI] [PubMed] [Google Scholar]
- West CM, Van Der Wel H, Sassi S, Gaucher E. Cytoplasmic glycosylation of protein-hydroxyproline and its relationship to other glycosylation pathways. Biochim Biophys Acta. 2004;1673:29–44. doi: 10.1016/j.bbagen.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- Wojczyk BS, Stwora-Wojczyk MM, Hagen FK, Striepen B, Hang HC, Bertozzi CR, Roos DS, Spitalnik SL. cDNA cloning and expression of UDP-N-acetyl-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase T1 from Toxoplasma gondii. Mol Biochem Parasitol. 2003;131:93–107. doi: 10.1016/s0166-6851(03)00196-8. [DOI] [PubMed] [Google Scholar]
- Yoshida N. Molecular basis of mammalian cell invasion by Trypanosoma cruzi. An Acad Bras Cienc. 2006;78:87–111. doi: 10.1590/s0001-37652006000100010. [DOI] [PubMed] [Google Scholar]
- Yoshida N, Mortara RA, Araguth MF, Gonzalez JC, Russo M. Metacyclic neutralizing effect of monoclonal antibody 10D8 directed to the 35- and 50-kilodalton surface glycoconjugates of Trypanosoma cruzi. Infect Immun. 1989;57:1663–1667. doi: 10.1128/iai.57.6.1663-1667.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachara NE, Packer NH, Temple MD, Slade MB, Jardine DR, Karuso P, Moss CJ, Mabbutt BC, Curmi PMG, Williams KL, et al. Recombinant prespore-specific antigen from Dictyostelium discoideum is a beta-sheet glycoprotein with a spacer peptide modified by O-linked N-acetylglucosamine. Eur J Biochem. 1996;238:511–518. doi: 10.1111/j.1432-1033.1996.0511z.x. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhang P, West CM. A linking function for the cellulose-binding protein SP85 in the spore coat of Dictyostelium discoideum. J Cell Sci. 1999;112:4367–4377. doi: 10.1242/jcs.112.23.4367. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.