Abstract
The Triticeae Full-Length CDS Database (TriFLDB) contains available information regarding full-length coding sequences (CDSs) of the Triticeae crops wheat (Triticum aestivum) and barley (Hordeum vulgare) and includes functional annotations and comparative genomics features. TriFLDB provides a search interface using keywords for gene function and related Gene Ontology terms and a similarity search for DNA and deduced translated amino acid sequences to access annotations of Triticeae full-length CDS (TriFLCDS) entries. Annotations consist of similarity search results against several sequence databases and domain structure predictions by InterProScan. The deduced amino acid sequences in TriFLDB are grouped with the proteome datasets for Arabidopsis (Arabidopsis thaliana), rice (Oryza sativa), and sorghum (Sorghum bicolor) by hierarchical clustering in stepwise thresholds of sequence identity, providing hierarchical clustering results based on full-length protein sequences. The database also provides sequence similarity results based on comparative mapping of TriFLCDSs onto the rice and sorghum genome sequences, which together with current annotations can be used to predict gene structures for TriFLCDS entries. To provide the possible genetic locations of full-length CDSs, TriFLCDS entries are also assigned to the genetically mapped cDNA sequences of barley and diploid wheat, which are currently accommodated in the Triticeae Mapped EST Database. These relational data are searchable from the search interfaces of both databases. The current TriFLDB contains 15,871 full-length CDSs from barley and wheat and includes putative full-length cDNAs for barley and wheat, which are publicly accessible. This informative content provides an informatics gateway for Triticeae genomics and grass comparative genomics. TriFLDB is publicly available at http://TriFLDB.psc.riken.jp/.
The recent accumulation of nucleotide sequences for agricultural species, including crops and domestic animals, now permits the application of genome-wide comparative analyses of model organisms with the goal of identifying key genes involved in phenotypic characteristics (Cogburn et al., 2007; Flicek et al., 2008; Paterson, 2008; Tanaka et al., 2008). The integration of genomic resources derived from various related species, such as large-scale collections of cDNAs and data from whole-genome sequencing projects, with various types of knowledge bases permits the sharing of information about gene function between models and applied organisms.
Integrative databases that house the sequences of systematically collected full-length cDNA clones have become fundamental initial resources for the bold promotion of the study of genomics in various organisms (Hayashizaki, 2003; Imanishi et al., 2004; Maeda et al., 2006; Tanaka et al., 2008; Yamasaki et al., 2008). In plants, full-length cDNA sequence resources are being used for a variety of purposes. For example, they are being used to create accurate genome annotations, for comparative analyses that link the genomic information of model and applied species, as sequence resources for protein sequence-based comparative analyses, and as sequence datasets for identifying proteins corresponding to peptides that have been detected using proteomics (Itoh et al., 2007; Ralph et al., 2008; Alexandrov et al., 2009; Seki and Shinozaki, 2009). Furthermore, collections of full-length cDNA clones have been used for systematic screening of characteristic gene functions by phenotyping overexpressor pools of full-length cDNA libraries. This technique was established recently in the Arabidopsis (Arabidopsis thaliana) “FOX hunting” (for Full-length cDNA Over-eXpressor gene hunting) system. Full-length cDNAs are key resources that provide a link between the genome, the transcriptome, and the proteome (Tochitani and Hayashizaki, 2007). Thus, full-length cDNAs and their annotations provide an initial gateway to various omics data (Sakurai et al., 2005; Maeda et al., 2006). To respond to the increasing amount of plant genome data, user interfaces that provide a seamless integration of comparative functions will be required to perform knowledge mining not only within community databases for specified organisms, but also across datasets integrated with multiple plant species (Spannagl et al., 2007; Ware, 2007). Full-length cDNA resources and modeled proteomes should be integrated with various types of representative sequence resources not only in the same species, but also in related species, thereby making it possible to exchange genomic knowledge and gain insight from comparative genomics (Dong et al., 2005). Furthermore, comparative allocation between transcripts from related species and the genome sequences of model organisms should be informative for both species, allowing for predictions and comparisons of gene structure and splicing patterns and assisting in the detection of genes and flanking sequences of possible counterparts (Mitchell et al., 2007; Zhu and Buell, 2007). The protein domain structure of modeled full-length protein sequences should also be compiled so that comparative sequence family analyses can be performed (Horan et al., 2005; Conte et al., 2008). Transfer of the genomic and genetic information of model organisms to related applied plant species should be promoted, particularly in the case of plant families that include both sequenced organisms and crops. This transfer should include full-length transcript-related datasets that are enriched with annotations, such as orthologous gene sequences and DNA markers (Sato and Tabata, 2006; Sato et al., 2007).
The Poaceae are a plant family that includes four major food staple crop species: wheat (Triticum aestivum), maize (Zea mays), rice (Oryza sativa), and barley (Hordeum vulgare). cDNA and/or genome sequence data for crops of the Poaceae have recently been accumulating in the public domain. Completion of the whole-genome sequencing of rice and its curated annotation using full-length cDNA data have benefited comparative plant genomics by increasing our understanding of genome-wide features and accelerating practical cereal breeding (International Rice Genome Sequencing Project, 2005; Itoh et al., 2007). The draft genome sequence of sorghum (Sorghum bicolor; in the Panicoideae subfamily) has been released and used for genomic comparisons with maize (Paterson et al., 2009). Large-scale EST and full-length cDNA collections of maize have also been used as resources to aid ongoing genome sequencing (Lai et al., 2004; Jia et al., 2006). ESTs of common wheat and barley (in the Pooideae subfamily) have been collected on a large scale to establish a comprehensive sequence resource for gene discovery and a reliable database of gene expression (Zhang et al., 2004; Mochida et al., 2006). Progress has now been made in both the barley and wheat genome sequencing projects, and full-length cDNA-related databases are expected to be key resources for genome annotation in these crops (Paux et al., 2008; Schulte et al., 2009). Sequence information from the large-scale collection of full-length cDNA clones of wheat and barley has been released to the public domain (Sato et al., 2009; K. Kawaura, K. Mochida, A. Enju, T. Totoki, A. Toyoda, Y. Sakaki, C. Kai, J. Kawai, Y. Hayashizaki, M. Seki, K. Shinozaki, and Y. Ogihara, unpublished data). As a result of these comprehensive efforts, there are now more than 15,000 nucleotide sequence entries available for the Triticeae (a tribe in the Pooideae), each of which potentially covers full-length coding sequences (CDSs).
To integrate our genomic knowledge of plants and facilitate further discoveries, many public databases that contain important plant genomics resources and that have effective interfaces have been established (Supplemental Fig. S1). PlantGDB, The Institute for Genomic Research (TIGR) Gene Indices, TIGR Plant Transcript Assemblies, and HarvEST provide clustered and representative transcript sequences resulting from advances in large-scale EST compilation. Each of these databases is useful not only for the provision of comprehensive transcripts, but also for comparisons among plant species (Liang et al., 2000; Lee and Quackenbush, 2003; Childs et al., 2007; Close et al., 2007; Duvick et al., 2008). The integration of genetic markers with corresponding genomic and/or transcriptomic sequences is facilitating genome-wide genetic approaches. Gramene is a database established for plant comparative genomics that provides genetic maps of various plant species (Jaiswal et al., 2006; Ware, 2007; Liang et al., 2008). GrainGenes is a popular site for information regarding genetic markers in Triticeae species (Carollo et al., 2005). We also recently released a new database (Triticeae Mapped EST Database [TriMEDB], http://TriMEDB.psc.riken.jp/) that focuses on genetically mapped cDNA markers of barley and diploid wheat. TriMEDB allows researchers to perform cDNA-based genetic knowledge comparisons among Triticeae species and syntenic regions of the rice genome (Mochida et al., 2008). Furthermore, genome annotations and modeled proteome datasets from the sequenced plant species (i.e. Arabidopsis and rice) can be used effectively for genome-wide comparative studies, such as comprehensive gene family constructions. Such studies can themselves yield databases that are useful for further phylogenetic studies (Horan et al., 2005; Conte et al., 2008; Wall et al., 2008). However, there are no databases that assemble the modeled proteome-based data of Triticeae species together with annotations based on comparative genomics that have effective links to knowledge databases of other plant species. The integration of resources for full-length CDSs of the Triticeae species will be important for comparative studies among Gramineae species, as well as a wide range of other species.
Therefore, to fill the gap in our knowledge of full-length CDSs of the Triticeae and, thus, to facilitate comparative grass genomics, we gathered the relational annotations of full-length CDSs of wheat and barley into a new database with the following specific properties. The first property was to provide predicted domain structures as well as other protein domain-oriented annotations of entire amino acid sequences that have been deduced from full-length CDSs and from CDSs clustered with proteome datasets of other plant species. The second was to provide seamless cross references to previously released sequence data resources, which was accomplished by annotating each of the database entries with possible identical sequences and/or counterparts in various transcripts and also by annotating the modeled proteome data resources of plant species, all with related reference links. The aim of this was to integrate knowledge and thus increase our understanding of gene annotations. Third, each of the entries in the database was related to the genetically mapped cDNAs of barley and diploid wheat, which in turn were bidirectionally integrated with TriMEDB. This yields a synergistic data relationship and extends the application of these resources to provide potential genetic positions of full-length transcripts on linkage maps of Triticeae in silico.
Here we describe our novel database. The Triticeae Full-Length CDS Database (TriFLDB) integrates knowledge of full-length CDSs of Triticeae crops with insights into comparative grass genomics. Currently, TriFLDB consists of 8,530 wheat and 7,341 barley putative full-length CDSs and related information. TriFLDB can be accessed via the Web interface at http://TriFLDB.psc.riken.jp/.
RESULTS AND DISCUSSION
Dataset, Design, and Search Interface of TriFLDB
The dataset integrated into the initial version of TriFLDB is summarized in Table I. Full-length CDSs were predicted using the full-length open reading frame (ORF) methods employed in the japonica rice full-length cDNA project (Kikuchi et al., 2003). As supporting information, DECODER (Fukunishi and Hayashizaki, 2001), which was originally used for full-length CDS prediction in the functional annotation of the mouse transcriptome 3 (FANTOM3), was also applied to full-length CDS prediction (Furuno et al., 2003). From the results of both methods, we predicted that 87.8% of the full-length barley cDNAs and 87.0% of wheat cDNAs contained putative full-length CDSs (Table II).
Table I.
Data sources for TriFLCDS
| Organisms | DNA | CDS/Protein | Source |
|---|---|---|---|
| Barley | 2,348 | 2,348 | Genpept/GenBank |
| 5,006 | 4,993 | BarleyDB | |
| Barley total | 7,354 | 7,341 | |
| Wheat | 2,393 | 2,393 | Genpept/GenBank |
| 6,158 | 6,137 | RIKEN/NBRP Komugi | |
| Wheat total | 8,551 | 8,530 | |
| Total | 15,905 | 15,871 |
Table II.
CDS predictions for barley and wheat full-length cDNAs using longest-frame prediction and DECODER
| BarleyDB Barley Full-Length cDNA | RIKEN Wheat Full-Length cDNA | |
|---|---|---|
| No. sequences examined | 5,006 | 6,158 |
| Longest frames predicted | 5,002 | 6,142 |
| DECODER CDS frames predicted | 4,576 | 5,562 |
| DECODER fragments predicted | 430 | 596 |
| Consistent frames predicted | 4,397 | 5,360 |
| % of predicted full CDS-containing clones | 87.8 | 87.0 |
We integrated full-length CDS data for wheat and barley with various annotations into a database capable of providing insights for comparative genomics. First, we retrieved full-length cDNA data and protein data deduced from full-length CDSs and analyzed it bioinformatically. This yielded sequence annotations, hierarchical protein clustering, and sequence similarity-based mapping of Triticeae full-length CDSs compared to the rice and sorghum genomes (Fig. 1).
Figure 1.
Schematic representation of the informatics workflow used to generate TriFLCDS entries and related annotations. The user can access the three types of TriFLDB content using sequence similarity and keyword searches or the genome browser Gbrowse to access data on homology mapping between TriFLCDSs and the rice and sorghum genomes (bottom).
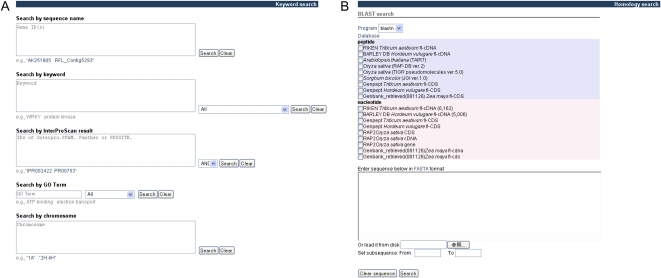
To access housed full-length CDS entries, TriFLDB provides a Web-based search interface enabling keyword and sequence similarity searches (Fig. 2). It is possible to search with keyword strings from BLAST definitions as well as with identifiers from databases such as PFAM, Prosite, and Panther. Gene Ontology (GO) terms assigned in the InterProScan results can also be used, as predicted chromosomal locations from TriMEDB (Fig. 2A). National Center for Biotechnology Information (NCBI) BLAST has also been implemented on the TriFLDB Web site. The BLAST service allows users to perform a homology search against multiple-sequence datasets. The database for this BLAST service consists of wheat and barley full-length cDNAs and their transcribed amino acid sequences, as well as the Arabidopsis proteome dataset from The Arabidopsis Information Resource (TAIR) and the Rice Annotation Project Database (RAP-DB) and TIGR rice databases (Fig. 2B). These search interfaces provide users with effective access to Triticeae full-length CDS (TriFLCDS) entries by using various types of queries that are also used in the databases for other plant species. For wheat and barley, this approach permits knowledge of model organisms, such as rice and Arabidopsis, which could be used for gene discovery and crop improvement (Bellgard et al., 2004; Varshney et al., 2006).
Figure 2.
Search interfaces for accessing TriFLDB content. The user can search TriFLCDS not only with sequence identifiers, but also by using various types of strings, such as keywords in the descriptions of BLAST hit sequences, identifiers of conserved protein domains and related GO terms found by InterProScan searches, and the predicted allocated chromosome name (A). The user can also access sequence similarity searches for TriFLCDS entries and for sequence sets of other plant species (B). [See online article for color version of this figure.]
Annotation of Triticeae Full-Length CDSs
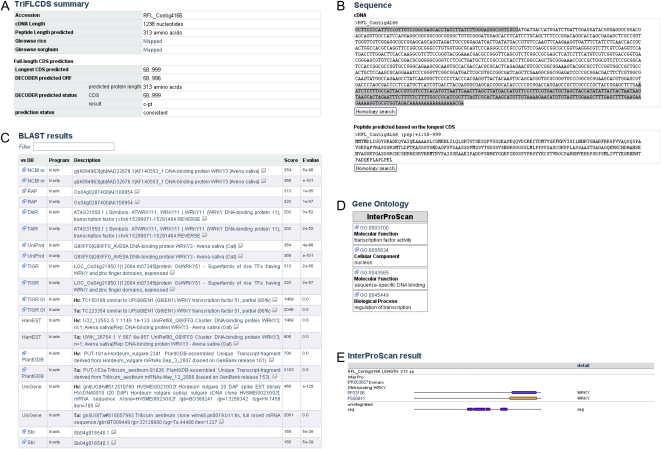
The Web interface displays information on TriFLCDSs that includes the results of CDS predictions and the nucleotide and deduced protein sequences (Fig. 3, A B). To provide annotations based on sequence similarity, nucleotide sequences of TriFLCDS entries were used as the query to search against the sequence sets provided in various public data resources. Because assignment of full-length CDSs with clustered representative transcript sequences makes it possible to use complete ORFs, which facilitates the molecular elucidation of CDS function and gene structure, TriFLCDS entries were assigned to clustered, representative transcript sequences of wheat and barley using separate BLASTN searches against the NCBI UniGene, Plant GDB, TIGR Gene Index, and HarvEST databases. In total, 7,030 (95.8%) full-length CDSs from barley and 7,719 (90.5%) from wheat were assigned to at least one representative transcript derived from these clustered cDNA sequence datasets (Supplemental Fig. S2A).
Figure 3.
Typical example of the detailed annotation of TriFLCDS entries. A, Summary table for a TriFLCDS entry, including the results of full-length CDS predictions. B, Nucleotide sequence and deduced protein sequence. C, Results of a similarity search against various sequence resources. D, List of the GO terms associated with the TriFLCDS entry. E, Domain structure predicted by InterProScan. [See online article for color version of this figure.]
To obtain clues about gene function, TriFLCDS entries were also searched against the annotated protein datasets of Arabidopsis, rice (RAP-DB and TIGR), and sorghum, as well as against representative nonredundant protein data repositories (nr of NCBI and UniProt of the European Bioinformatics Institute [EBI]). We found hits with significant similarity to more than 80% of the TriFLCDS entries in Arabidopsis and to at least 87% in rice and sorghum (Supplemental Fig. S2B). The results of the similarity searches for each of the TriFLCDS entries are shown on the Web interface, and, whenever possible, links to the original data for each hit are provided so as to enable browsing of additional related information (Fig. 3C). For domain-based functional annotation, the deduced protein data were subjected to a domain search using InterProScan. In total, 13,162 (82.9%) entries were assigned to at least one identifier of the database used in InterPro. Using the Web interface, the user can browse each of the results of the domain search, along with the predicted GO classification (Fig. 3, D and E). A synopsis of the results of the similarity search against various sequence resources is shown on the Web interface, and this should allow researchers to determine the annotation status of the searched entries and the predicted annotation of the most likely counterparts in other databases. This should help users to build hypotheses that are related to gene function.
To construct a dataset that relates the proteins predicted in TriFLDB to those of other plant species, we grouped TriFLCDSs hierarchically into homologous clusters with the protein datasets for Arabidopsis, rice, and sorghum. Clustering with a 90% identity threshold produced 10,639 clusters containing one or more protein sequences derived from wheat or barley full-length CDSs. This indicates that the current version of TriFLDB contains putative full-length CDSs that correspond to more than 10,000 nonredundant genes (Supplemental Table S1).
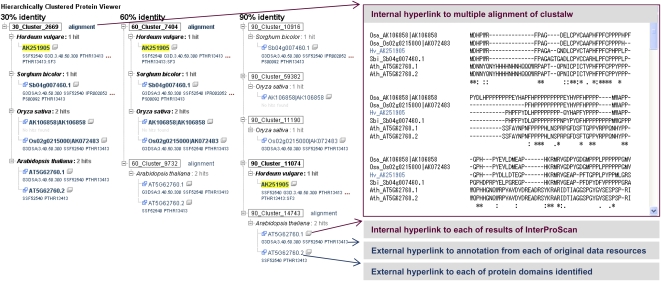
Hierarchically clustered data have been added to TriFLDB and are presented together with information on the domain structure predicted for each protein sequence. This information can be browsed via a Web-based hierarchical structure, which is a viewing interface that contains annotated domain data as well as hyperlinks to the reference databases (Fig. 4). The interface provides the structure and relationships of the modeled proteomes of Arabidopsis, rice, and sorghum, and includes TriFLDB entries that are clustered according to global amino acid identities. Since all of the TriFLCDS entries in the viewer are reciprocally related on each annotation page, the user can navigate to the detailed annotation pages of other TriFLCDS entries classified in the same cluster. To provide clues for sequence comparison among clustered proteins, the interface provides four kinds of hyperlinks to internal and external data resources. The user can confirm a multiple alignment of each clustered dataset, and these can be captured in the clustalw format and shown in a subwindow opened from the alignment hyperlink in each cluster. The protein domain search results from InterProScan for all clustered entries can be browsed, and hyperlinks to the original protein domain knowledge resources are also provided. The domain identifiers listed for each of the sequence entries should allow a clear assessment of the sharing status of the domain structure among the clustered sequences. Hyperlinks to referenced annotations in the modeled proteome datasets of Arabidopsis, rice, and sorghum are also provided to permit comparisons of domain structures among clustered genes with seamless browsing.
Figure 4.
Example of a hierarchically clustered protein sequence TriFLDB entry containing sequences from Arabidopsis, rice, and sorghum, presented with the hyperlinked destination for each of the referenced annotations. The Web interface employs a tree-form viewer to provide the results of clustering with global amino acid sequence identity thresholds of 30%, 60%, and 90%. The viewer also provides identifiers for clustered sequences via four kinds of hyperlinks: multiple alignment, InterProScan domain search results, the annotation page of the original data resources, and each data resource for the protein domain families identified. [See online article for color version of this figure.]
The detailed annotations of each of the TriFLCDS entries that have been inferred via sequence similarity as well as predicted protein domains should facilitate the prediction of possible gene functions, as well as the configuration of further functional analyses and/or the narrowing down of candidate genes in Triticeae.
Integration of Full-Length CDS Data and Genetically Allocated cDNA Markers of Triticeae
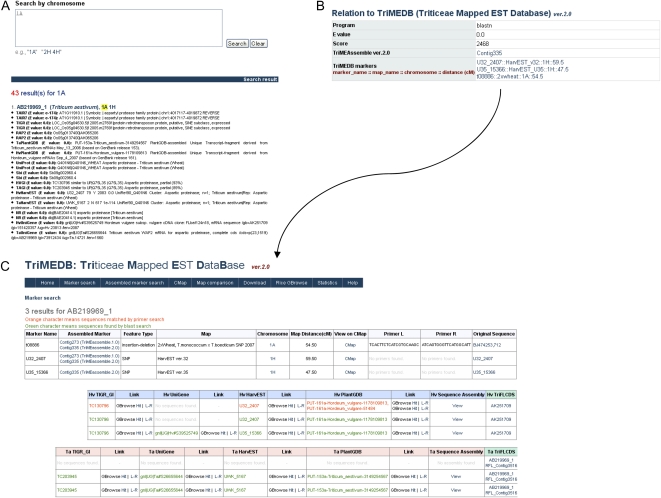
Genetic localization of full-length CDSs will greatly facilitate the positional cloning of targeted genes in wheat and barley. We related mapped EST markers to full-length cDNA sequences and to CDSs of barley and wheat to generate a table showing the map locations of full-length transcripts in Triticeae. Out of 3,605 mapped cDNAs, 2,182 (60.5%) demonstrated significant similarity to full-length CDSs of either barley or wheat (Table III). TriFLDB entries assigned to mapped wheat and barley cDNA markers can be searched using wheat and barley chromosome names, and relational links are provided on the Web interface together with additional annotations (Fig. 5, A and B). The user can browse information on corresponding cDNA markers at the TriMEDB interface (Fig. 5C) and can search for cDNA markers related to full-length CDSs via the TriMEDB search interface (http://TriMEDB.psc.riken.jp/cgi-bin/TriMEDB/marker_search.pl). The integration of mapped ESTs with full-length CDSs can provide valuable information, especially when accompanied by annotations, such as predictions of whole-gene structure. This information can be used to coordinate nucleotide polymorphism discoveries with marker development. Moreover, genome-scale genotyping will facilitate forward genetic approaches, such as QTL analyses and association studies (Varshney et al., 2006).
Table III.
Assignment of nonredundant EST markers of TriMEDB v. 2.0 (3,605 marker groups) to full-length CDS entries in TriFLDB
| Organism | No. of Entries of TriFLDB Used for Similarity Search | No. of Marker Groups of TriMEDB Assigned to TriFLCDS (%) |
|---|---|---|
| Barley | 7,341 | 1,486 (41.2) |
| Wheat | 8,530 | 1,457 (40.4) |
| Total | 15,871 | 2,182 (60.5) |
Figure 5.
Example of the interdatabase relationship between TriFLDB and TriMEDB. The user can search TriFLCDS entries using chromosome names as a result of the connection to genetically mapped cDNA markers in TriMEDB. A, The annotation page for each of the TriFLCDS entries provides a relational link to the assigned cDNA marker information. B, The user can browse detailed information related to the corresponding cDNA markers. The TriMEDB interface provides a link to TriFLDB for browsing corresponding full-length CDSs (C). [See online article for color version of this figure.]
Assignment and Assembly of Wheat and Barley ESTs into TriFLCDSs
Full-length CDSs are useful for obtaining accurate sequence clusters and for the assembly of cDNA sequences. To determine the relationships between TriFLCDSs and the released ESTs of wheat and barley, we conducted BLAST similarity searches with the ESTs against TriFLCDS entries. Each query EST demonstrating ≥80% nucleotide identity to TriFLCD and with a high-scoring segment pair (HSP) alignment of at least 100 bp was included. Their distributions and identities are summarized in Figure 6A. The results indicate that 238,142 (49.3%) of the released barley ESTs were assigned to the barley full-length CDS entries in TriFLDB and that 481,804 (47.5%) of the released wheat ESTs were assigned to and the wheat full-length CDS entries in TriFLDB, which enabled the identification of the corresponding full-length CDSs. Using the wheat and barley ESTs that were assigned to respective TriFLCDS entries, we assembled each sequence group into contigs to facilitate transcript assembly using the full-length CDSs. As an example of the interface of the database, the cDNA assemblies of the ESTs that were assigned to entries in TriFLDB are shown in Figure 6B. We found that 311,429 (64.5%) barley ESTs and 621,688 (61.3%) wheat ESTs could be assigned to at least one of the barley or wheat full-length CDS entries of TriFLDB. These assignments are most likely between pairs of identical or possibly homologous sequences. These data should provide for a more accurate assembly of cDNAs and better predictions of the structure of the wheat and barley transcripts. The assembly of ESTs into the best possible transcripts is helpful not only for gaining representative transcripts, but also, in many cases, for using them as initial sequence resources for the design of genomic tools, such as probes for oligo arrays and sequence-tagged site primers (Mitchell et al., 2007). The full-length CDS-based assembly of ESTs can also provide reference alignments for the identification of polymorphisms among released transcript sequences. Such identifications are also useful for designing transcript sequence-based markers. Given the expected growth in the number of transcript sequences that will be released, the integration of additional full-length CDSs and ESTs of Triticeae should become an important data resource that enhances genomic infrastructures in Triticeae.
Figure 6.
Summarized results of EST assignment to TriFLCDSs on the basis of similarity searches, and a typical example of clustered ESTs assembled together with full-length CDSs to form a contig sequence. Pairwise alignments between ESTs and possible full-length counterpart TriFLCDSs with at least 80% identity and a HSP of at least 100 bp were plotted under each of the search conditions. The BLAST-searched combinations of queried ESTs and full-length CDSs indicated by asterisks (*) in the graph were used for sequence assembly against the wheat and barley entries provided in TriFLDB. TaEST and HvEST refer to the EST datasets of wheat and barley, respectively, that were used as query data in the BLAST search. TaFLCDS and HvFLCDS refer to the TriFLCDS entries for wheat and barley, respectively, that were used as the database in the BLAST search (A). Barley and wheat ESTs assigned to the TriFLCDSs of barley and wheat, respectively, were assembled, and each of these is shown in TriFLDB. A barley entry, AK251905, is shown as an example of the assembly of barley ESTs and TriFLCDS (B). [See online article for color version of this figure.]
Comparative Mapping of TriFLCDSs onto the Rice and Sorghum Genomes
To visualize predicted exon-intron structures and the comparative genomic features of Triticeae transcripts in rice and sorghum, sequence similarity-based mapping of TriFLCDSs onto the rice and sorghum genomes was performed. The Generic Genome Browser (Gbrowse; Donlin, 2007) was used to visualize the predicted gene structure of each full-length CDS (Fig. 7). Because the database for Gbrowse contains current rice and sorghum gene annotations, the user can compare predicted gene structures of TriFLCDSs with their possible counterparts in rice and sorghum. A total of 12,486 full-length CDSs were mapped onto the rice genome, and 12,242 onto the sorghum genome under the set threshold (Supplemental Table S2). About 80% of the TriFLCDS entries were assigned to the sequenced genomes, yielding predictions of gene structures, such as exon boundaries that should be useful for designing probes and/or sequence-tagged site primers for gene mapping in barley and wheat. Although most of the entries were allocated to the annotated gene regions of rice and sorghum, some were allocated to the nonannotated regions, as summarized in Supplemental Table S2. Comparative mapping of full-length CDSs and cDNAs can provide evidence of gene structure and splicing patterns among homologous genes. Because the genomic browser in TriFLDB for the rice genome is completely interrelated with that in TriMEDB, users can also compare and confirm knowledge of cDNAs and full-length mapped sequences in the rice genome. This feature is useful for designing markers and for browsing syntenic regions of the rice genome. Recent progress has been realized in the genome sequencing of Brachypodium distachyon as a model grass species (http://www.brachypodium.org/). In the future, TriFLDB will include the Brachypodium genome draft as a third reference genome, which will permit further integration and facilitate the comparative genomics of grasses. The resulting comprehensive full-length CDS resource should be useful for annotating the Brachypodium genome and for comparative studies of Triticeae species (Bossolini et al., 2007; Faris et al., 2008; Ozdemir et al., 2008).
Figure 7.
Typical example of the results of comparative mapping of TriFLCDSs onto the rice (A) and sorghum (B) genomes, shown here in the Gbrowse interface. The interface provides allocated TriFLCDS entries together with annotated gene information from the original data resources. [See online article for color version of this figure.]
The database structure of the current version of TriFLDB and its relationship with TriMEDB are depicted in a schematic diagram showing the data handling and generated relational datasets with corresponding Web interfaces (Supplemental Fig. S3). The genome resources related to Triticeae species will continue to accumulate (Schulte et al., 2009). Therefore, it is important that comparable datasets from related organisms be accessible and cross referenced (Childs, 2009). Because they feature segmentalized data tables and various types of interfaces for browsing, the database structures of TriFLDB and TriMEDB should be able to respond to such expected increases in genomic resources in Triticeae, as well as in other model plant species. The Poaceae are a good example of how genomic knowledge of crops, such as wheat, barley, and maize, is facilitated by comparative genomics with a model organism, such as rice (Paterson et al., 2005). Integration of the information present in our database, in which the modeled proteome data of applied crops is related to functional annotations of model species, increases the ability to cross reference between these species, and thereby facilitates knowledge exchange and application of databases to comparative crop genomics.
CONCLUSION
This integrative Web-based database interface provides information on putative full-length CDSs of wheat and barley that will facilitate the comparative genomics of grasses. The database should meet the broad demands of researchers who need to search for information related to Triticeae genes with the goal of a greater understanding of Gramineae species. The database should accelerate progress in Triticeae genomics and plant comparative genomics, as well as facilitate molecular breeding programs.
MATERIALS AND METHODS
Prediction and Retrieval of Full-Length CDSs
We retrieved cDNA sequences of completely sequenced wheat (Triticum aestivum) full-length cDNAs using a primer walking method with the Phred/Phrap package (Ewing and Green, 1998) to generate the assembly at RIKEN and the Kihara Institute for Biological Research, Yokohama City University, Japan (K. Kawaura, K. Mochida, A. Enju, T. Totoki, A. Toyoda, Y. Sakaki, C. Kai, J. Kawai, Y. Hayashizaki, M. Seki, K. Shinozaki, and Y. Ogihara, unpublished data). We also retrieved barley (Hordeum vulgare) sequences corresponding to the proper accession IDs that were reported by Sato et al. (2009) from the BarleyDB database (http://www.shigen.nig.ac.jp/barley/) of the Research Institute for Bioresources at Okayama University in Japan.
The sequences were first checked for sequence contamination and extensive simple repeats using the SeqClean script (http://compbio.dfci.harvard.edu/tgi/software/). Vector sequences were then trimmed using the univec_core db of NCBI (http://www.ncbi.nlm.nih.gov/VecScreen/UniVec.html) with the cross_match utility of the Phred/Phrap package. Contamination was identified via BLASTN sequence similarity searches against both the Escherichia coli K12 genome (U00096) and the bacteriophage phi_X174 (J02482) genome sequences. Sequences with a threshold e value less than 1e-100 were removed.
CDS prediction was performed based on the longest ORF using those sequences that had passed through the sequence cleaning step. As supporting information, we used the results for full-length CDS prediction from DECODER (Fukunishi and Hayashizaki, 2001). The nucleotide and deduced amino acid sequences corresponding to the predicted full-length ORFs were used to generate further annotations in TriFLDB. The deduced protein sequences and corresponding CDSs of GenPept entries (GenPept Release 165.0, ftp://ftp.ncifcrf.gov/) were also retrieved using the full-length CDS feature; 2,348 barley sequences and 2,393 wheat sequences were retrieved. Predicted CDSs less than 30 bp in length as well as disproportionately short CDSs (CDS/cDNA < 10%) were then removed. A total of 15,871 CDSs of barley and wheat were entered into TriFLDB (Table I).
Sequence Annotations
To annotate the CDSs of TriFLDB with predicted gene functions, we searched the sequence data against the following protein and nucleotide datasets using the BLAST algorithm (Altschul et al., 1997): the nr protein database of NCBI (ftp://ftp.ncbi.nih.gov/blast/db); UniProt/trembl of EBI (http://www.uniprot.org/downloads); the protein data of RAP-DB v. 2 (http://rapdb.dna.affrc.go.jp/); the TIGR Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/); the protein data for predicted genes of the sorghum (Sorghum bicolor) genome in JGI v. 1.4 (http://genome.jgi-psf.org/Sorbi1/Sorbi1.home.html); the protein data present in TAIR release 7 (ftp://ftp.arabidopsis.org/home/tair/Sequences/blast_datasets/); the cDNA sequences of barley and wheat in UniGene (ftp://ftp.ncbi.nih.gov/repository/UniGene/); the TIGR Plant Transcript Assemblies (http://plantta.jcvi.org/); Plant GDB (http://www.plantgdb.org/); and HarvEST (http://harvest.ucr.edu/). All of the similarity searches using BLASTN were performed with threshold e values of less than 1e-200 for same-species combinations and 1e-100 for cross-species combinations among wheat and barley, and the top scoring hit for each query was applied. All similarity searches with BLASTX and BLASTP against protein datasets to find possible functional descriptions were performed with a threshold e-value of less than 1e-5, and the top scoring hit for each query was applied. The definition strings used for the similarity searches of each database have been assembled as a keyword database to allow users to specify queries with keywords to retrieve relevant gene information from TriFLDB.
Conserved domains in the deduced protein sequence of each TriFLCDS were identified with InterProScan and the InterPro database (http://www.ebi.ac.uk/interpro/). The domain data were also used to assign GO terms to each TriFLCDS, which are also available as search query terms for the TriFLCDSs. Links to each of the original datasets interrelated with the TriFLCDS entries are provided on the TriFLDB Web interface.
Hierarchical Clustering of Deduced Protein Sequences with Plant Proteome Data
The nonredundant set of TriFLCDSs and groupings with other plant proteins on the basis of sequence similarity assists in the identification of the unique genes of Triticeae plants, as well as in acquiring proteins with sequence similarity to those in other plants. Through the use of the CD-HIT package (Li and Godzik, 2006), the TriFLCDSs were hierarchically organized into protein clusters with the protein datasets from Arabidopsis (Arabidopsis thaliana), rice (Oryza sativa), and sorghum using global amino acid sequence identity thresholds of 100% to 30% in 10% decrements. The hierarchically clustered data were imported into a Web-based hierarchical structure-viewing interface.
Clustering of Wheat and Barley ESTs with TriFLCDSs
As of April 15, 2008, the dbEST database of NCBI (NCBI-GenBank Flat File Release 165.0) contained more than 0.5 million entries for barley and more than 1 million for wheat. These sequences were retrieved from GenBank and were cleaned up as follows. First, low-complexity and/or repetitive sequences were removed using SeqClean with the default parameter settings. Repetitive sequence regions of the remaining sequences were identified and masked with RepeatMasker (http://www.repeatmasker.org/), with optional use of the nonredundant Gramineae repeat-sequence dataset derived from TIGR as the target database (Ouyang and Buell, 2004). Vector sequences were then masked using the cross_match utility in the Phred/Phrap package (Ewing and Green, 1998) and the UniVec dataset of NCBI. A similarity search using cross_match against wheat mitochondrial and chloroplast genome sequences was also performed to eliminate contaminant organelle sequences. Finally, ESTs (482,904 for barley and 1,014,305 for wheat) containing ≥100 bp of unmasked sequences were clustered and assembled. Sequence similarity searches between the Triticeae ESTs and full-length CDS data were used to create potential realistic assemblies based on the full-length CDSs of barley and wheat using BLASTN with an e value of ≤1e-20. The ESTs grouped with the TriFLCDS were assembled into contigs using CAP3 (Huang and Madan, 1999) with the default parameter settings. Each ace format file for CAP3 output corresponding to TriFLCDS was applied to retrieve positional information for contig alignment of the assembled ESTs.
Comparative Mapping onto the Rice and Sorghum Genomes
To provide comparative sequence mapping information for the TriFLCDS entries that were allocated to the genome sequences of rice and sorghum, we mapped the nucleotide sequences of TriFLCDSs onto the genome sequences of rice (International Rice Genome Sequencing Project v. 4, http://rgp.dna.affrc.go.jp/IRGSP/download.html) and sorghum (JGI v. 1.4) based on nucleotide sequence similarity. A combination of BLASTN and SIM4 (Pidoux et al., 2003) was used to reduce any inconsistencies in the map positions. To ascertain the most similar regions of the rice and sorghum genomes, a BLASTN similarity search with a threshold e value of less than 1e-10 was conducted to find the highest hit. Then, all other HSPs that were found in a 10-kb window upstream and downstream of the endpoints of the top HSP obtained in the BLAST hit were collected along with the genome sequence. Finally, a region that encompassed a 5-kb window upstream and downstream of the endpoints of the collected HSPs was retrieved to generate pairwise alignments between the retrieved genomic sequence and TriFLCDS.
Pairwise alignment using SIM4 with default parameter settings was then performed to predict the genomic structure in the comparative alignment between the two sequences that were used as input. The comparative genome mapping results have been implemented in Gbrowse with the gene annotations for rice and sorghum provided by RAP-DB and JGI, respectively. To map TriFLCDSs onto the nonannotated regions of each genome, the TriFLCDSs homologous to the plant organelle sequences that were filtered out were mapped onto both genomes and compared with the mapped region using the genome annotations RAP-DB v. 2 and Sbi 1.4. The wheat and barley chloroplast genomes (AB042240, EF115541) and the wheat mitochondrial genome (AP008982) were searched using BLASTN with a threshold e value of less than 1e-20 to subtract possible FLCDSs derived from the organelle genomes.
Data Integration with the Triticeae Mapped EST Database
To assign genetically mapped ESTs to the full-length transcripts of the TriFLDB entries, we searched the dataset of 15,871 TriFLCDS nucleotide sequences with the mapped EST markers housed in TriMEDB (http://TriMEDB.psc.riken.jp/) using BLASTN with a threshold e value of less than 1e-130. The table of relationships between the mapped ESTs and the full-length transcripts generated by this homology search was imported into TriMEDB as a database for Cmap (http://gmod.org/wiki/Cmap) to visualize linkage map images. The comparative data from the mapping of cDNA markers of TriMEDB onto the rice genome were also integrated into the Gbrowse interface of TriFLDB. Cross referencing between the Web interfaces of TriMEDB and TriFLDB was also implemented.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Schematic overview of representative databases covering genome informatics areas for wheat and barley, together with those for rice and Arabidopsis.
Supplemental Figure S2. Summarized results of similarity-search-based annotation of TriFLCDSs against various sequence resources provided in public domains.
Supplemental Figure S3. A schematic diagram of the database structure of TriFLDB together with that of TriMEDB.
Supplemental Table S1. Distribution of deduced peptide sequences of TriFLCDS with proteome data of Arabidopsis, rice, and sorghum hierarchically clustered by threshold of amino acid identity.
Supplemental Table S2. Summarized results of mapping of similarity between TriFLCDS and the rice and sorghum genome sequences.
Supplementary Material
Acknowledgments
The authors thank Dr. K. Sato of Okayama University, Japan, for permitting the integration of released data into TriFLDB. The authors also thank Dr. T. Close of the University of California for permitting integration of the released data from HarvEST barley v. 1.68 to update TriMEDB. We also thank Dr. Y. Hayashizaki of the RIKEN Omics Science Center for DECODER.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Kazuo Shinozaki (sinozaki@rtc.riken.jp).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open access articles can be viewed online without a subscription.
References
- Alexandrov NN, Brover VV, Freidin S, Troukhan ME, Tatarinova TV, Zhang H, Swaller TJ, Lu YP, Bouck J, Flavell RB, et al (2009) Insights into corn genes derived from large-scale cDNA sequencing. Plant Mol Biol 69 179–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellgard M, Ye J, Gojobori T, Appels R (2004) The bioinformatics challenges in comparative analysis of cereal genomes: an overview. Funct Integr Genomics 4 1–11 [DOI] [PubMed] [Google Scholar]
- Bossolini E, Wicker T, Knobel PA, Keller B (2007) Comparison of orthologous loci from small grass genomes Brachypodium and rice: implications for wheat genomics and grass genome annotation. Plant J 49 704–717 [DOI] [PubMed] [Google Scholar]
- Carollo V, Matthews DE, Lazo GR, Blake TK, Hummel DD, Lui N, Hane DL, Anderson OD (2005) GrainGenes 2.0. An improved resource for the small-grains community. Plant Physiol 139 643–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs KL (2009) Genomic and genetic database resources for the grasses. Plant Physiol 149 132–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs KL, Hamilton JP, Zhu W, Ly E, Cheung F, Wu H, Rabinowicz PD, Town CD, Buell CR, Chan AP (2007) The TIGR Plant Transcript Assemblies database. Nucleic Acids Res 35 D846–D851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close TJ, Wanamaker S, Roose ML, Lyon M (2007) HarvEST: an EST database and viewing software. Methods Mol Biol 406 161–178 [DOI] [PubMed] [Google Scholar]
- Cogburn LA, Porter TE, Duclos MJ, Simon J, Burgess SC, Zhu JJ, Cheng HH, Dodgson JB, Burnside J (2007) Functional genomics of the chicken—a model organism. Poult Sci 86 2059–2094 [DOI] [PubMed] [Google Scholar]
- Conte MG, Gaillard S, Lanau N, Rouard M, Perin C (2008) GreenPhylDB: a database for plant comparative genomics. Nucleic Acids Res 36 D991–D998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Q, Kroiss L, Oakley FD, Wang BB, Brendel V (2005) Comparative EST analyses in plant systems. Methods Enzymol 395 400–418 [DOI] [PubMed] [Google Scholar]
- Donlin MJ (2007) Using the Generic Genome Browser (GBrowse). Curr Protoc Bioinformatics Chapter 9: Unit 9.9 [DOI] [PubMed]
- Duvick J, Fu A, Muppirala U, Sabharwal M, Wilkerson MD, Lawrence CJ, Lushbough C, Brendel V (2008) PlantGDB: a resource for comparative plant genomics. Nucleic Acids Res 36 D959–D965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B, Green P (1998) Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 8 186–194 [PubMed] [Google Scholar]
- Faris JD, Zhang Z, Fellers JP, Gill BS (2008) Micro-colinearity between rice, Brachypodium, and Triticum monococcum at the wheat domestication locus Q. Funct Integr Genomics 8 149–164 [DOI] [PubMed] [Google Scholar]
- Flicek P, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, et al (2008) Ensembl 2008. Nucleic Acids Res 36 D707–D714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukunishi Y, Hayashizaki Y (2001) Amino acid translation program for full-length cDNA sequences with frameshift errors. Physiol Genomics 5 81–87 [DOI] [PubMed] [Google Scholar]
- Furuno M, Kasukawa T, Saito R, Adachi J, Suzuki H, Baldarelli R, Hayashizaki Y, Okazaki Y (2003) CDS annotation in full-length cDNA sequence. Genome Res 13 1478–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashizaki Y (2003) RIKEN mouse genome encyclopedia. Mech Ageing Dev 124 93–102 [DOI] [PubMed] [Google Scholar]
- Horan K, Lauricha J, Bailey-Serres J, Raikhel N, Girke T (2005) Genome cluster database: a sequence family analysis platform for Arabidopsis and rice. Plant Physiol 138 47–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Madan A (1999) CAP3: a DNA sequence assembly program. Genome Res 9 868–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanishi T, Itoh T, Suzuki Y, O'Donovan C, Fukuchi S, Koyanagi KO, Barrero RA, Tamura T, Yamaguchi-Kabata Y, Tanino M, et al (2004) Integrative annotation of 21,037 human genes validated by full-length cDNA clones. PLoS Biol 2 e162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436 793–800 [DOI] [PubMed] [Google Scholar]
- Itoh T, Tanaka T, Barrero RA, Yamasaki C, Fujii Y, Hilton PB, Antonio BA, Aono H, Apweiler R, Bruskiewich R, et al (2007) Curated genome annotation of Oryza sativa ssp. japonica and comparative genome analysis with Arabidopsis thaliana. Genome Res 17 175–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal P, Ni J, Yap I, Ware D, Spooner W, Youens-Clark K, Ren L, Liang C, Zhao W, Ratnapu K, et al (2006) Gramene: a bird's eye view of cereal genomes. Nucleic Acids Res 34 D717–D723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia J, Fu J, Zheng J, Zhou X, Huai J, Wang J, Wang M, Zhang Y, Chen X, Zhang J, et al (2006) Annotation and expression profile analysis of 2073 full-length cDNAs from stress-induced maize (Zea mays L.) seedlings. Plant J 48 710–727 [DOI] [PubMed] [Google Scholar]
- Kikuchi S, Satoh K, Nagata T, Kawagashira N, Doi K, Kishimoto N, Yazaki J, Ishikawa M, Yamada H, Ooka H, et al (2003) Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science 301 376–379 [DOI] [PubMed] [Google Scholar]
- Lai J, Dey N, Kim CS, Bharti AK, Rudd S, Mayer KF, Larkins BA, Becraft P, Messing J (2004) Characterization of the maize endosperm transcriptome and its comparison to the rice genome. Genome Res 14 1932–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Quackenbush J (2003) Using the TIGR gene index databases for biological discovery. Curr Protoc Bioinformatics Chapter 1: Unit 1.6 [DOI] [PubMed]
- Li W, Godzik A (2006) Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22 1658–1659 [DOI] [PubMed] [Google Scholar]
- Liang C, Jaiswal P, Hebbard C, Avraham S, Buckler ES, Casstevens T, Hurwitz B, McCouch S, Ni J, Pujar A, et al (2008) Gramene: a growing plant comparative genomics resource. Nucleic Acids Res 36 D947–D953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang F, Holt I, Pertea G, Karamycheva S, Salzberg SL, Quackenbush J (2000) An optimized protocol for analysis of EST sequences. Nucleic Acids Res 28 3657–3665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda N, Kasukawa T, Oyama R, Gough J, Frith M, Engstrom PG, Lenhard B, Aturaliya RN, Batalov S, Beisel KW, et al (2006) Transcript annotation in FANTOM3: mouse gene catalog based on physical cDNAs. PLoS Genet 2 e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell RA, Castells-Brooke N, Taubert J, Verrier PJ, Leader DJ, Rawlings CJ (2007) Wheat Estimated Transcript Server (WhETS): a tool to provide best estimate of hexaploid wheat transcript sequence. Nucleic Acids Res 35 W148–W151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida K, Kawaura K, Shimosaka E, Kawakami N, Shin IT, Kohara Y, Yamazaki Y, Ogihara Y (2006) Tissue expression map of a large number of expressed sequence tags and its application to in silico screening of stress response genes in common wheat. Mol Genet Genomics 276 304–312 [DOI] [PubMed] [Google Scholar]
- Mochida K, Saisho D, Yoshida T, Sakurai T, Shinozaki K (2008) TriMEDB: a database to integrate transcribed markers and facilitate genetic studies of the tribe Triticeae. BMC Plant Biol 8 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang S, Buell CR (2004) The TIGR Plant Repeat Databases: a collective resource for the identification of repetitive sequences in plants. Nucleic Acids Res 32 D360–D363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozdemir BS, Hernandez P, Filiz E, Budak H (2008) Brachypodium genomics. Int J Plant Genomics 2008 536104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH (2008) Genomics of sorghum. Int J Plant Genomics 2008 362451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, et al (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457 551–556 [DOI] [PubMed] [Google Scholar]
- Paterson AH, Freeling M, Sasaki T (2005) Grains of knowledge: genomics of model cereals. Genome Res 15 1643–1650 [DOI] [PubMed] [Google Scholar]
- Paux E, Sourdille P, Salse J, Saintenac C, Choulet F, Leroy P, Korol A, Michalak M, Kianian S, Spielmeyer W, et al (2008) A physical map of the 1-gigabase bread wheat chromosome 3B. Science 322 101–104 [DOI] [PubMed] [Google Scholar]
- Pidoux AL, Richardson W, Allshire RC (2003) Sim4: a novel fission yeast kinetochore protein required for centromeric silencing and chromosome segregation. J Cell Biol 161 295–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph SG, Chun HJ, Kolosova N, Cooper D, Oddy C, Ritland CE, Kirkpatrick R, Moore R, Barber S, Holt RA, et al (2008) A conifer genomics resource of 200,000 spruce (Picea spp.) ESTs and 6,464 high-quality, sequence-finished full-length cDNAs for Sitka spruce (Picea sitchensis). BMC Genomics 9 484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai T, Satou M, Akiyama K, Iida K, Seki M, Kuromori T, Ito T, Konagaya A, Toyoda T, Shinozaki K (2005) RARGE: a large-scale database of RIKEN Arabidopsis resources ranging from transcriptome to phenome. Nucleic Acids Res 33 D647–D650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Nakamura Y, Asamizu E, Isobe S, Tabata S (2007) Genome sequencing and genome resources in model legumes. Plant Physiol 144 588–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Shin IT, Seki M, Shinozaki K, Yoshida H, Takeda K, Yamazaki Y, Conte M, Kohara Y (2009) Development of 5006 full-length CDNAs in barley: a tool for accessing cereal genomics resources. DNA Res 16: 81–89 [DOI] [PMC free article] [PubMed]
- Sato S, Tabata S (2006) Lotus japonicus as a platform for legume research. Curr Opin Plant Biol 9 128–132 [DOI] [PubMed] [Google Scholar]
- Schulte D, Close TJ, Graner A, Langridge P, Matsumoto T, Muehlbauer G, Sato K, Schulman AH, Waugh R, Wise RP, et al (2009) The international barley sequencing consortium—at the threshold of efficient access to the barley genome. Plant Physiol 149 142–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Shinozaki K (2009) Functional genomics using RIKEN Arabidopsis thaliana full-length cDNAs. J Plant Res (in press) [DOI] [PubMed]
- Spannagl M, Noubibou O, Haase D, Yang L, Gundlach H, Hindemitt T, Klee K, Haberer G, Schoof H, Mayer KF (2007) MIPSPlantsDB—plant database resource for integrative and comparative plant genome research. Nucleic Acids Res 35 D834–D840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Antonio BA, Kikuchi S, Matsumoto T, Nagamura Y, Numa H, Sakai H, Wu J, Itoh T, Sasaki T, et al (2008) The Rice Annotation Project Database (RAP-DB): 2008 update. Nucleic Acids Res 36 D1028–D1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tochitani S, Hayashizaki Y (2007) Functional screening revisited in the postgenomic era. Mol Biosyst 3 195–207 [DOI] [PubMed] [Google Scholar]
- Varshney RK, Hoisington DA, Tyagi AK (2006) Advances in cereal genomics and applications in crop breeding. Trends Biotechnol 24 490–499 [DOI] [PubMed] [Google Scholar]
- Wall PK, Leebens-Mack J, Muller KF, Field D, Altman NS, dePamphilis CW (2008) PlantTribes: a gene and gene family resource for comparative genomics in plants. Nucleic Acids Res 36 D970–D976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware D (2007) Gramene: a resource for comparative grass genomics. Methods Mol Biol 406 315–33018287700 [Google Scholar]
- Yamasaki C, Murakami K, Fujii Y, Sato Y, Harada E, Takeda J, Taniya T, Sakate R, Kikugawa S, Shimada M, et al (2008) The H-Invitational Database (H-InvDB), a comprehensive annotation resource for human genes and transcripts. Nucleic Acids Res 36 D793–D799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Sreenivasulu N, Weschke W, Stein N, Rudd S, Radchuk V, Potokina E, Scholz U, Schweizer P, Zierold U, et al (2004) Large-scale analysis of the barley transcriptome based on expressed sequence tags. Plant J 40 276–290 [DOI] [PubMed] [Google Scholar]
- Zhu W, Buell CR (2007) Improvement of whole-genome annotation of cereals through comparative analyses. Genome Res 17 299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.