Abstract
The phytohormone abscisic acid (ABA) is a key regulator of plant growth and development as well as plant responses to situations of decreased water availability. Protein phosphatases type 2C (PP2Cs) from group A, which includes the ABI1/HAB1 and PP2CA branches, are key negative regulators of ABA signaling. Specifically, HAB1, ABI1, ABI2, and PP2CA have been shown to affect both seed and vegetative responses to ABA. To further understand their contribution to ABA signaling and to unravel possible genetic interactions and functional redundancy among them, we have generated different combinations of double and triple mutants impaired in these PP2Cs. Interestingly, hab1-1pp2ca-1 and abi1-2pp2ca-1 double mutants showed reduced water loss and enhanced resistance to drought stress, which further supports the role of PP2CA in vegetative responses to ABA. Two triple hab1-1abi1-2abi2-2 and hab1-1abi1-2pp2ca-1 mutants were generated, which showed an extreme response to exogenous ABA, impaired growth, and partial constitutive response to endogenous ABA. Thus, transcriptomic analysis revealed a partial up-regulation/down-regulation of a subset of ABA-responsive genes in both triple mutants in the absence of exogenous ABA. Comparison of ABA responses in the different pp2c mutants showed that a progressive increase in ABA sensitivity could be obtained through combined inactivation of these PP2Cs. These results indicate that ABA response is finely tuned by the integrated action of these genes, which is required to prevent a constitutive response to endogenous ABA that might have a deleterious effect on growth and development in the absence of environmental stress.
Protein phosphorylation/dephosphorylation events in abscisic acid (ABA) signaling involve several known protein kinases and phosphatases (Finkelstein et al., 2002; Hirayama and Shinozaki, 2007; Verslues and Zhu, 2007). For instance, the guard cell-specific protein kinase AAPK from Vicia faba and the orthologous OST1/SnRK2.6 regulate ABA-induced stomatal closure (Li et al., 2000; Mustilli et al., 2002; Yoshida et al., 2002). The Arabidopsis (Arabidopsis thaliana) genome contains 10 SnRK2s; among them, SnRK2.2, SnRK2.3, and SnRK2.6/OST1 are key regulators of ABA signaling. Both SnRK2.2 and SnRK2.3 regulate ABA responses in germination, growth, and gene expression, whereas SnRK2.6/OST1 specifically regulates stomatal aperture (Mustilli et al., 2002; Fujii et al., 2007). Another protein kinase involved in ABA signaling is PKABA1, which is induced by ABA and suppresses GA-inducible gene expression in barley (Hordeum vulgare) aleurone layers (Gomez Cadenas et al., 1999). In addition to the above-described calcium-independent protein kinases, several calcium-dependent protein kinases that belong either to the calcium-dependent protein kinases or to the SnRK3/CIPK family mediate ABA signaling (Sheen, 1996; Guo et al., 2002; Kim et al., 2003; Mori et al., 2006; Cheong et al., 2007).
On the other hand, protein phosphatases type 2A (PP2As) and type 2C (PP2Cs) are involved in the regulation of ABA signaling. Disruption of the PP2A regulatory subunit RCN1 confers ABA insensitivity in Arabidopsis, which suggests that RCN1 functions as a positive transducer of ABA signaling (Kwak et al., 2002). Recent results have reported that a catalytic subunit of PP2A functions as a negative regulator of ABA signaling (Pernas et al., 2007). According to the classification of Schweighofer et al. (2004), 76 Arabidopsis genes qualify as members of the big PP2C family. Among them, group A contains most of the PP2Cs that are associated with ABA signaling, which are separated in two subgroups (i.e. the ABI1 and PP2CA branches). The pioneering evidence for the involvement of PP2Cs in the regulation of ABA signaling process was provided by the identification of the ABA-insensitive abi1-1 and abi2-1 mutants and the cloning of the corresponding loci (Koornneef et al., 1984; Leung et al., 1994, 1997; Meyer et al., 1994; Rodriguez, et al., 1998). Currently, genetic evidence indicates that at least six Arabidopsis PP2Cs, namely, ABI1, ABI2, PP2CA/AHG3, AHG1, HAB1, and HAB2, act as negative regulators of ABA signaling (Gosti et al., 1999; Merlot et al., 2001; Tahtiharju and Palva, 2001; Gonzalez-Garcia et al., 2003; Leonhardt et al., 2004; Saez et al., 2004, 2006; Kuhn et al., 2006; Yoshida et al., 2006; Nishimura et al., 2007). AHG1 and PP2CA/AHG3 appear to play an essential role for ABA signaling in germination and postgermination growth (Kuhn et al., 2006; Yoshida et al., 2006; Nishimura et al., 2007), but the ahg1-1 mutant has no ABA-related phenotype in adult plants (Nishimura et al., 2007). The analysis of a double ahg1-1ahg3-1 mutant suggests that AHG1 has specific functions in seed development and germination, shared partially with PP2CA/AHG3 (Nishimura et al., 2007). Kuhn et al. (2006) showed that PP2CA/AHG3 plays an essential role for ABA signaling both in seed and vegetative tissue, as the pp2ca mutant alleles showed ABA hypersensitivity in germination, growth, and stomatal closure assays. Conversely, 35S:PP2CA expression caused ABA insensitivity in seed germination and ABA-induced stomatal closure assays (Kuhn et al., 2006). Previous work of Sheen (1998) showed that PP2CA can block ABA-mediated gene induction when transiently overexpressed in protoplasts. These results suggest that PP2CA function might be related to other PP2Cs from the ABI1 branch; however, genetic interactions among both branches have not been investigated.
Single reduction/loss-of-function alleles from ABI1, ABI2, and HAB1 produced phenotypic effects on ABA signaling to a different extent and it was apparent from double mutant analyses that some functional redundancy occurs among them (Merlot et al., 2001; Saez et al., 2006). For instance, inactivation of both HAB1 and ABI1 led to a stronger response to ABA than that found in either hab1-1 or abi1-2 monogenic mutants. A similar trend was obtained by combination of the recessive abi1-1R4 and abi2-1R1 alleles (Merlot et al., 2001). Combined inactivation of close members of a gene family is usually required to unravel possible functional genetic redundancy and to establish a functional hierarchy among them. This fact has been particularly evident in hormonal signaling pathways (Hua and Meyerowitz, 1998; Kwak et al., 2003; Higuchi et al., 2004; Achard et al., 2006; Iuchi et al., 2007). Alternatively, gain-of-function approaches can circumvent genetic redundancy, as deduced from the global ABA-insensitive phenotype found in the dominant mutants abi1-1D and abi2-1D, as well as the transgenic lines 35S:HAB1, 35S:PP2CA, and 35S:hab1Gly-246Asp (Koornneef et al., 1984; Saez et al., 2004; Kuhn et al., 2006; Robert et al., 2006).
In this article, we have combined loss-of-function mutations in the ABI1, ABI2, HAB1, and PP2CA genes to unravel their contributions to ABA signaling. The hab1-1, abi1-2, and pp2ca-1 alleles have been described previously (Leonhardt et al., 2004; Saez et al., 2004, 2006; Kuhn et al., 2006) and in this article we have identified a loss-of-function allele for ABI2, named abi2-2. Different combinations of mutations were generated (Table I) and, as a result, two triple mutants, hab1-1abi1-2abi2-2 and hab1-1abi1-2pp2ca-1, were generated, which showed an extreme response to exogenous ABA and a partial constitutive response to endogenous ABA. Additionally, hab1-1pp2ca-1 and abi1-2 pp2ca-1 double mutants showed stronger responses to ABA than single parental mutants, which reveals a genetic interaction between both branches of PP2Cs for the regulation of ABA signaling.
Table I.
List of T-DNA mutants used in this work
The hab1-1, abi1-2, and pp2ca-1 single and double hab1-1abi1-2 mutants have been described previously (Saez et al., 2004, 2006; Kuhn et al., 2006). The abi2-2 mutant was obtained from the NASC. Different crosses were performed to generate the double and triple mutants mentioned in the table.
| Single Mutants | Double Mutants | Triple Mutants |
|---|---|---|
| hab1-1 SALK2104 | hab1-1abi1-2 | hab1-1abi1-2pp2ca-1 |
| abi1-2 SALK72009 | abi1-2abi2-2 | hab1-1abi1-2abi2-2 |
| pp2ca-1 SALK28132 | hab1-1pp2ca-1 | |
| abi2-2 SALK15166 | abi1-2pp2ca-1 |
RESULTS
Generation and Analysis of Double hab1-1pp2ca-1 and abi1-2pp2ca-1 Mutants
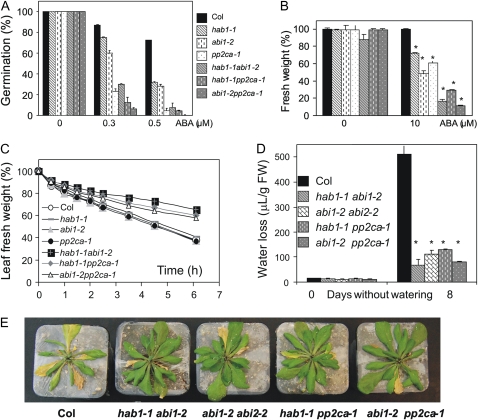
Seeds of pp2ca-1 are strongly ABA-hypersensitive, whereas pp2ca-1 seedlings show only a moderated enhanced sensitivity to ABA-mediated inhibition of root growth and loss of fresh weight of detached rosette leaves in pp2ca-1 was similar to wild type (Kuhn et al., 2006). We wondered whether combination of the pp2ca-1 mutation with either hab1-1 or abi1-2 might reinforce ABA responses as it was described previously for the double hab1-1abi1-2 mutant (Saez et al., 2006). Therefore, the corresponding double mutants were generated and their ABA response analyzed. ABA-mediated inhibition of seed germination was stronger in the pp2ca-1 single mutant than in hab1-1 and abi1-2 single mutants (Fig. 1A), which is in agreement with previous results from Yoshida et al. (2006). Interestingly, both double hab1-1pp2ca-1 and abi1-2pp2ca-1 mutants showed enhanced sensitivity to ABA-mediated inhibition of seed germination and growth inhibition than single parental mutants (Fig. 1, A and B). Likewise, short-term water loss assays showed that combined inactivation of either PP2CA and HAB1 or PP2CA and ABI1 resulted in a phenotype of reduced water loss compared to Columbia wild type (Col) or single parental mutants (Fig. 1C). Additionally, water-loss data were obtained under greenhouse conditions after exposing 21-d-old plants to drought stress by stopping irrigation and minimizing soil evaporation. Figure 1D shows that, after 8 d, the double mutants showed reduced water loss as compared to wild type. In agreement with these data, the double mutants did not show symptoms of wilting and they had turgid green rosette leaves, whereas wild-type plants wilted and rosette leaves yellowed (Fig. 1E).
Figure 1.
ABA-hypersensitive germination and growth inhibition of hab1-1, abi1-2, pp2ca-1, and double hab1-1abi1-2, hab1-1pp2ca-1, abi1-2pp2ca-1 mutants compared to wild type. A, Percentage of seeds that germinated and developed green cotyledons in the presence of the indicated concentrations of ABA. Approximately 200 seeds of each genotype were sowed on each plate and scored 10 d later. The assays were done in the period encompassed between 1 to 2 months after seed harvesting. Values are averages ± sd for three independent experiments (not indicated in the figure); P < 0.05 (Student's t test) when comparing data from each genotype and wild type in ABA medium. B, Percentage of fresh weight from the different mutants as compared to wild type. The percentage was calculated with respect to the fresh weight of wild type in Murashige and Skoog medium either lacking or containing 10 μm ABA. Fresh weight of wild type was reduced by 39% in plates supplemented with ABA as compared to medium lacking ABA. Values are averages from three independent experiments ± sd (n = 15). * indicates P < 0.01 (Student's t test) when comparing data from each genotype and wild type in the same assay conditions. C, Reduced water loss measured in detached leaves of double hab1-1abi1-2, hab1-1pp2ca-1, abi1-2pp2ca-1 mutants as compared to wild type or single parental mutants. Values are averages from two independent experiments (n = 5). sd values were lower than 8%. D, Quantification of water loss in 30-d-old plants after 8 d without watering. Values are averages from two independent experiments ± sd (n = 10). * as in B. FW, Fresh weight. E, Enhanced drought resistance of the indicated double mutants as compared to wild type. Photograph was taken 10 d after withholding water. Shoot was cut to better show the effect of drought on rosette leaves.
Generation and Analysis of Double abi1-2abi2-2 Mutants
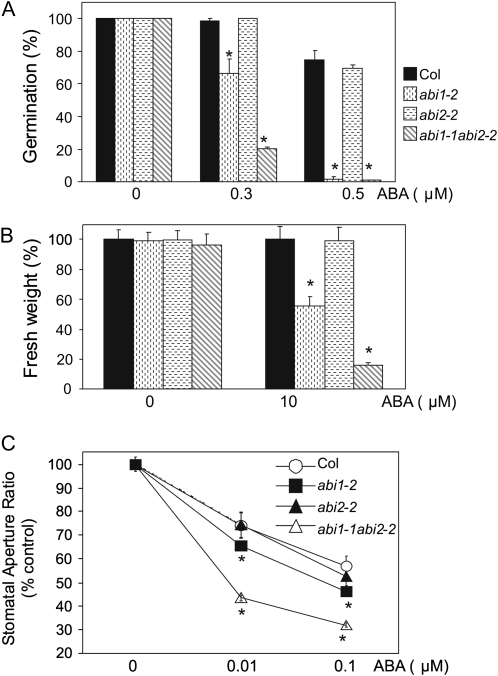
A T-DNA mutant of ABI2 was identified in the Salk collection, corresponding to donor stock number SALK_015166, and it was named abi2-2. Sequencing of the T-DNA flanking region in abi2-2 showed that the insertion was localized 19 nucleotides upstream of the TGA stop codon. The T-DNA insertion severely impaired ABI2 expression, based on quantitative real-time (qRT)-PCR analyses (see Fig. 3); however, the abi2-2 allele showed wild-type responses in ABA-mediated inhibition of seed germination and growth assays, as well as ABA-induced stomatal closing (Fig. 2). In contrast, the double abi1-2abi2-2 mutant showed higher sensitivity to ABA-mediated inhibition of germination and growth by ABA than single parental mutants (Fig. 2, A and B). Additionally, it showed enhanced ABA-induced stomatal closing (Fig. 2C), reduced water loss (Fig. 1D), and enhanced resistance to drought stress (Fig. 1E) as compared to wild-type plants.
Figure 3.
Growth impairment and ABA-hypersensitive growth inhibition of triple hab1-1abi1-2abi2-2 (triple abi2-2) and hab1-1abi1-2pp2ca-1 (triple pp2ca-1) mutants as compared to wild type. A, qRT-PCR analysis of HAB1, PP2CA, ABI1, and ABI2 transcript levels in wild type and triple hab1-1abi1-2abi2-2 and hab1-1abi1-2pp2ca-1 mutants. Gene expression was analyzed in mRNAs extracted from 14-d-old seedlings treated with 10 μm ABA for 3 h. Data are averages ± sd from three independent experiments. B, Reduced growth in both Murashige and Skoog (MS) medium and soil of triple mutants compared to wild type. The photographs were taken after 3 weeks of plant growth in Murashige and Skoog medium or soil. The graphics show the quantification of fresh weight (n = 30) and leaf area (n = 10) in triple mutants with respect to wild type. Data are averages ± sd from two independent experiments. *, P < 0.01 (Student's t test) when comparing data from the mutant and wild type in the same assay conditions. C, ABA-hypersensitive root growth inhibition of double and triple mutants compared to wild type and single parental mutants. Representative seedlings were selected and a photograph was taken after 7 d of growth in medium lacking or supplemented with 10 μm ABA. Data are averages ± sd from three independent experiments (n = 15). *, P < 0.01 (Student's t test) when comparing data from the mutant and wild type in the same assay conditions. [See online article for color version of this figure.]
Figure 2.
ABA-hypersensitive response of double abi1-2abi2-2 mutant compared to wild type and single parental mutants. A, Percentage of seeds that germinated and developed green cotyledons in the presence of the indicated concentrations of ABA. Approximately 200 seeds of each genotype were sowed on each plate and scored 10 d later. The assays were done in the period encompassed between 1 to 2 months after seed harvesting. Values are averages ± sd for three independent experiments; *, P < 0.05 (Student's t test) when comparing data from each genotype and wild type in the same assay conditions. B, Percentage of fresh weight from the different mutants as compared to wild type. The percentage was calculated with respect to the fresh weight of wild type in Murashige and Skoog medium either lacking or containing 10 μm ABA. Fresh weight of wild type was reduced by 22% in plates supplemented with ABA as compared to medium lacking ABA. Values are averages from three independent experiments ± sd (n = 15); *, P < 0.01 (Student's t test) when comparing data from each genotype and wild type in the same assay conditions. C, ABA-hypersensitive stomatal closing in double abi1-2abi2-2 mutant. Stomatal apertures were measured 2 h and 30 min after addition of 0.01 or 0.1 μm ABA. Data represent average aperture ratio (width/length) of three independent experiments ± se (n = 30–40 stomata per experiment); *, P < 0.01 (Student's t test) when comparing data from the mutant and wild type in the same assay conditions.
Generation and Analysis of ABA Response in Triple hab1-1abi1-2pp2ca-1 and hab1-1abi1-2abi2-2 Mutants
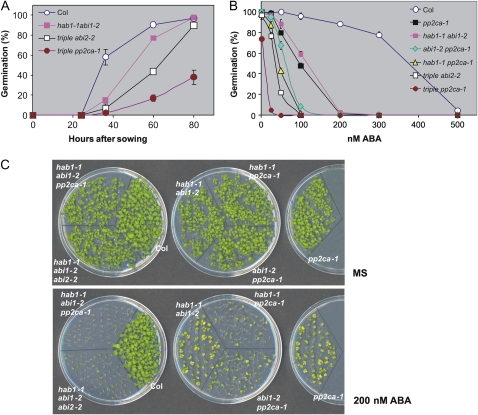
To further study the effect on ABA signaling of the combined inactivation of negative regulators of the pathway, we generated two triple mutants impaired in three of the above-described PP2Cs. Thus, hab1-1abi1-2pp2ca-1 and hab1-1abi1-2abi2-2 triple mutants were generated and qRT-PCR analyses confirmed that expression of HAB1, ABI1, and either PP2CA or ABI2, respectively, was severely impaired (Fig. 3A). The phenotype of the triple mutants in the absence and presence of exogenous ABA was analyzed. Interestingly, both mutants showed impaired growth in the absence of exogenous ABA, either under in vitro conditions or when plants were grown in soil under greenhouse conditions (Fig. 3B). The impaired growth of these mutants suggests that combined inactivation of three negative regulators of ABA signaling might lead to a partial constitutive response to endogenous ABA levels. For instance, roots of these mutants were shorter than those of wild type and root growth was extremely hypersensitive to ABA-mediated inhibition of growth (Fig. 3C). Indeed, root growth of the triple mutants in the absence of exogenous ABA was approximately 60% to 65% of wild type. This decrease was very similar to that obtained in wild-type seedlings grown in 10 μm ABA (Fig. 3C). Additionally, a comparison of root growth for different ABA-hypersensitive mutants reveals a progressive increase in their sensitivity to ABA-mediated inhibition of growth (Fig. 3C), which suggests that PP2Cs act in concert to modulate root growth sensitivity to ABA.
Germination was slower in triple mutants than in wild type (Fig. 4A) and, indeed, the germination percentage of triple hab1-1abi1-2pp2ca-1 mutants was 40% of wild type 80 h after sowing (Fig. 4A). Seeds of the triple mutants were hypersensitive to very low concentrations (nm) of ABA in germination assays (Fig. 4B). Thus, the inhibitory ABA concentration to achieve 50% inhibition (ABA-IC50) of seed germination was 10 and 40 nm for the triple hab1-1abi1-2pp2ca-1 and hab1-1abi1-2abi2-2 mutants, respectively, whereas it was 380 nm for wild-type seeds. Interestingly, the germination percentage of the triple hab1-1abi1-2pp2ca-1 mutant in the absence of exogenous ABA was similar to that found in wild-type seeds germinated in 300 nm ABA. Double mutants that contain the pp2ca-1 allele were more sensitive to ABA-mediated inhibition of germination than hab1-1abi1-2. Thus, the double hab1-1abi1-2 mutant had an ABA-IC50 of 120 nm, whereas the ABA-IC50 for the double hab1-1pp2ca-1 and abi1-2pp2ca-1 mutants was 50 and 70 nm, respectively. Early seedling growth was more inhibited by ABA in the triple mutants than in the double mutants, and double mutants containing the pp2ca-1 allele were more ABA-hypersensitive than hab1-1abi1-2 or single pp2ca-1 mutants (Fig. 4C; Supplemental Fig. S1). Therefore, taking into account the sensitivity to ABA-mediated inhibition of germination and a dose-response (25–500 nm ABA) analysis of ABA-mediated inhibition of early growth (Supplemental Fig. S1), the following hierarchy of ABA hypersensitivity can be established: hab1-1abi1-2pp2ca-1 > hab1-1abi1-2abi2-2 > hab1-1pp2ca-1 > abi1-2pp2ca-1 > pp2ca-1 ∼ hab1-1abi1-2.
Figure 4.
ABA-hypersensitive germination and early seedling growth inhibition of different mutants compared to wild type. A, Seed germination time course of triple hab1-1abi1-2abi2-2 (triple abi2-2) and hab1-1abi1-2pp2ca-1 (triple pp2ca-1) mutants compared to wild type. Data are averages ± sd from three independent experiments. B, Percentage of seeds that germinated and developed green cotyledons in the presence of the indicated concentrations of ABA. Approximately 200 seeds of each genotype were sowed on each plate and scored 5 d later. The assays were done in the period encompassed between 1 to 2 months after seed harvesting. Data are averages ± sd from three independent experiments. C, ABA-hypersensitive early seedling growth inhibition of different mutants compared to wild type. Approximately 80 seeds of each genotype were sowed on each plate and the photograph was taken 12 d later. MS, Murashige and Skoog medium.
Finally, measurements of both stomatal aperture and aperture ratio (width/length) reveal that stomata of the triple hab1-1abi1-2abi2-2 mutant were more closed than wild type in the absence of exogenous ABA (Fig. 5, A and B). Stomatal aperture measurements also showed that triple hab1-1abi1-2abi2-2 mutant stomata were extremely hypersensitive to ABA-induced stomatal closing in the range of 10 to 100 nm ABA (Fig. 5). Interestingly, stomatal apertures in the triple hab1-1abi1-2abi2-2 mutant in the absence of exogenous ABA were similar to those found in wild type treated with 10 nm ABA. Additionally, we measured stomatal conductance in the absence of exogenous ABA for both triple mutants. As a result, we found decreased stomatal conductance for water vapor (gs) and decreased transpiration (E) in both triple mutants as compared to wild type (Fig. 5, C and D). This result indicates that, in the absence of exogenous ABA, stomata of triple mutants have a lower aperture than wild type.
Figure 5.
Reduced stomatal aperture in the absence of exogenous ABA for triple hab1-1abi1-2abi2-2 (triple abi2-2) and hab1-1abi1-2pp2ca-1 (triple pp2ca-1) mutants. A and B, ABA-hypersensitive stomatal closing in triple hab1-1abi1-2abi2-2 mutant as compared to wild type. Plants were kept overnight in high humidity and then leaves were preincubated for 2 h in opening solution. Stomatal apertures were measured 2 h and 30 min after addition of 0, 0.01, or 0.1 μm ABA. Data represent the average of three independent experiments ± sem (n = 30–40 stomata per experiment). C and D, Leaf gas-exchange measurements reveal both reduced stomatal conductance and transpiration in triple pp2ca-1 and triple abi2-2 mutants as compared to wild type. Data represent the average of two independent experiments ± sem (n = 10 plants/experiment). *, P < 0.01 (Student's t test) when comparing data from each genotype and Col in the same assay conditions.
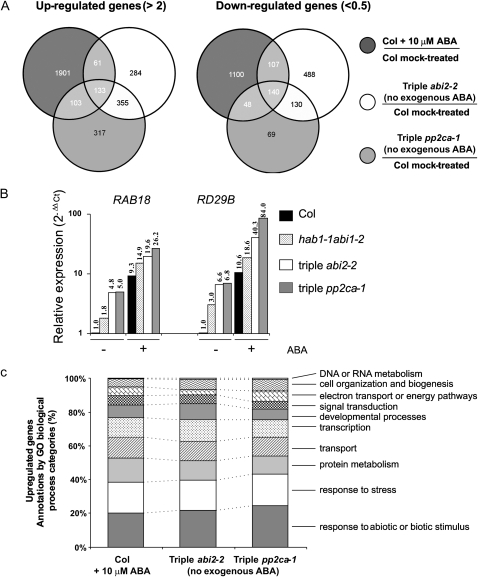
Transcriptomic Profiling Suggests That Triple Mutants Show a Constitutive Response to Endogenous ABA
The above data suggested that the phenotype of triple hab1-1abi1-2pp2ca-1 and hab1-1abi1-2abi2-2 mutants might reflect a partial constitutive response to endogenous ABA levels. To challenge this hypothesis, we have compared transcriptomic profiles of wild type and triple mutants using whole-genome long-oligonucleotide microarrays. First, we have identified the total number of genes up-regulated (>2-fold) and down-regulated (<0.5) in wild type by treatment with 10 μm ABA for 3 h (false discovery rate P < 0.05), which qualify as ABA-responsive genes according to the ratio of expression Col + 10 μm ABA/Col no exogenous ABA (Fig. 6A). In addition, we have compared whole-genome expression of triple mutants with respect to wild type, in the absence of exogenous ABA (ratio of expression for triple no exogenous ABA/Col no exogenous ABA; Fig. 6A). The number of genes that (1) showed enhanced or diminished expression in the triple mutants with respect to the wild type, in the absence of exogenous ABA and (2) were ABA-responsive are indicated in Figure 6A and a complete list of these genes is provided as Supplemental Table S1. Thus, 194 or 266 ABA-responsive genes were up-regulated in the triple abi2-2 or pp2ca-1 mutants, respectively, in the absence of exogenous ABA. Conversely, 247 and 188 ABA-responsive genes were down-regulated in the triple abi2-2 or pp2ca-1 mutants, respectively, in the absence of exogenous ABA. Interestingly, 133 or 140 ABA-responsive genes were up-regulated or down-regulated, respectively, both in triple abi2-2 and pp2ca-1 mutants in the absence of exogenous ABA with respect to wild type. Independent confirmation of these results was obtained by qRT-PCR for two genes, RAB18 and RD29B, which are up-regulated by ABA treatment (Fig. 6B). In the absence of exogenous ABA, these gene markers showed a 5- to 7-fold higher expression in the triple mutants than in wild type. Interestingly, expression of these genes in the triple mutants (in the absence of exogenous ABA) was approximately 50% to 60% of that found in wild type treated with 10 μm ABA. Finally, both triple mutants showed enhanced up-regulation by ABA of RAB18 and RD29B, as their expression level upon ABA treatment was between 2- and 8-fold higher than in wild type (Fig. 6B).
Figure 6.
Triple hab1-1abi1-2abi2-2 (triple abi2-2) and hab1-1abi1-2pp2ca-1 (triple pp2ca-1) mutants show partial constitutive up-regulation and down-regulation of ABA-responsive genes in the absence of exogenous ABA. A, Venn diagrams. Number of ABA-responsive genes that show up-regulation and down-regulation in triple mutants in the absence of exogenous ABA. B, Expression of RAB18 and RD29B in triple mutants with respect to wild type. qRT-PCR analyses were made in triplicate on RNA samples of 2-week-old seedlings that were either mock or 10 μm ABA-treated for 3 h. Numbers indicate the expression level of the RAB18 and RD29B genes under mock (−) or ABA treatment (+) in each mutant genotype with respect to the wild type (value 1). C, Comparison of GO categories for ABA-responsive genes in ABA-treated wild type versus triple mutants in the absence of exogenous ABA.
In summary, a partial constitutive up-regulation and down-regulation of ABA-responsive genes was found in both triple mutants in the absence of exogenous ABA (Supplemental Table S1). Furthermore, gene ontology (GO) analysis performed at The Arabidopsis Information Resource (TAIR; http://www.Arabidopsis.org/tools/bulk/go/index.jsp) reveals that these genes belong to the same categories that are overrepresented in ABA-treated wild-type seedlings, as, for instance, genes related to plant response to abiotic/biotic stimulus and stress response (oxidative stress, osmotic, salt, heat shock, and cold; Fig. 6C). Additionally, we identified ABA-responsive genes that were differentially up-regulated/down-regulated (>2-fold) between both triple mutants when compared with each other (Supplemental Table S2). For instance, genes encoding ABA-responsive seed storage proteins and AAA-type ATPases showed enhanced up-regulation in triple abi2-2, whereas genes encoding defensin-like proteins showed enhanced down-regulation compared to triple pp2ca-1 (Supplemental Table S2). Interestingly, some genes encoding late embryogenesis-abundant proteins showed enhanced up-regulation in triple pp2ca-1 compared to triple abi2-2 (Supplemental Table S2).
Finally, to clarify whether the phenotypes described for the triple mutants reflect either enhanced synthesis of ABA or enhanced signaling, we have measured endogenous ABA levels in 12-d-old seedlings (Supplemental Fig. S2). Interestingly, endogenous ABA levels did not significantly differ when compared with data measured in triple mutants with respect to wild type (Supplemental Fig. S2). Moreover, after osmotic stress treatment (350 mm mannitol), both triple mutants did not produce more ABA than wild type (Supplemental Fig. S2). Therefore, these results indicate that the phenotypes observed in both triple mutants reflect a genuine effect of enhanced ABA signaling.
DISCUSSION
In this article, we show that different degrees of ABA hypersensitivity, ranging from mild to extreme, can be engineered through progressive inactivation of PP2Cs from group A. Thus, a fine tuning of ABA signaling can be accomplished in Arabidopsis through genetic inactivation of these genes, which might be of biotechnological interest once the corresponding orthologous genes are identified in crops. For instance, from this article and previous results of Saez et al. (2006), we conclude that inactivation of two major PP2Cs (HAB1/ABI1, ABI1/ABI2, ABI1/PP2CA, HAB1/PP2CA) is enough in Arabidopsis to generate drought-avoidant plants. Taking into account the important advance in RNA-mediated gene-silencing technology, either through the use of hairpin RNAi or artificial microRNAs (Ossowski et al., 2008), it is reasonable to suggest that combined inactivation of these PP2Cs might be attained in crop plants where genome information is available. However, this work also shows that overactivation of ABA signaling is detrimental for plant growth in the absence of water stress because triple mutants that show a partial constitutive response to ABA are impaired in growth. Therefore, the multiplicity of PP2Cs that regulate ABA signaling appears to be a versatile mechanism to adequately control ABA response both in the absence or presence or stress.
The analysis of combined mutations in PP2Cs from group A has revealed a genetic interaction between the ABI1/HAB1 and PP2CA branches, which extends our knowledge on these PP2Cs. For instance, although it was known that PP2CA plays a major role in regulating ABA signaling in seeds, the phenotypes of double mutants that combine the pp2ca-1 allele with either hab1-1 or abi1-2 reveal that PP2CA also has a key role in regulating water loss in vegetative tissue. This evidence could not be obtained from the analysis of single pp2ca mutants, which show similar water loss than wild type due to a slight ABA-hypersensitive response, as discussed by Kuhn et al. (2006). Additionally, enhanced sensitivity to ABA-mediated inhibition of seed germination was found in the double hab1-1pp2ca-1 and abi1-2pp2ca-1 mutants, whose ABA-IC50 was 50 and 70 nm, respectively, compared to pp2ca-1 (ABA-IC50 = 110 nm). These double mutants were also more sensitive to ABA-mediated inhibition of root growth than pp2ca-1. Therefore, taken together, these results confirm that PP2CA, as shown previously for HAB1 and ABI1, is a key regulator of ABA signaling both in seeds and vegetative tissues (Kuhn et al., 2006). This conclusion is consistent with gene expression data found at public databases (Supplemental Fig. S3). In addition, the phenotypes of the different mutants hereby reported suggest that PP2Cs provide a threshold of negative regulation required for a normal response to ABA. If ABI1/HAB1 and PP2CA branches regulate independently ABA signaling, the loss of one branch would be enough to overcome this threshold and, therefore, lead to ABA hypersensitivity. For instance, even though HAB1 and ABI1 are expressed in pp2ca-1, this mutant shows a strong ABA-hypersensitive phenotype in seeds. Alternatively, it is possible that ABA signaling is negatively regulated through a mechanism that involves an accurate PP2C dosage effect. For instance, such a mechanism is supported for the progressive increase in root growth sensitivity to ABA observed in single, double, and triple pp2c mutants (Fig. 4C).
Finally, we show that triple pp2c mutants are extremely sensitive to exogenous ABA. Particularly noticeable is the nanomolar sensitivity of the triple mutants for ABA-mediated inhibition of seed germination, which is particularly appealing, taking into account that other PP2Cs that regulate ABA signaling in seed are still active in such mutants. For instance, the triple hab1-1abi1-2abi2-2 mutant shows an ABA-IC50 of 40 nm, even though two PP2Cs (PP2CA/AHG3 and AHG1) that are essential components of ABA signaling in seed are active. This fact suggests the existence of at least two branches of PP2Cs to negatively regulate ABA signaling in seed. Additionally, triple mutants show a partial constitutive response to ABA in different assays. For instance, root growth of triple mutants in the absence of exogenous ABA was very similar to wild type in the presence of 10 μm ABA. Partial stomatal closure was found in triple mutants as compared to wild type and, interestingly, in the absence of exogenous ABA the triple mutants showed a stomatal aperture only slightly higher than wild type treated with 10 nm ABA. This partial constitutive response to ABA was also found by gene expression analysis. Thus, expression of RAB18 and RD29B in triple mutants, in the absence of exogenous ABA, was only 50% to 60% of that found in wild type treated with 10 μm ABA. Whole transcriptome analysis reveals that approximately 15% or approximately 25% of genes up-regulated or down-regulated by ABA in wild type, respectively, were constitutively up-regulated or down-regulated in the triple mutants in the absence of exogenous ABA. Even though this effect represents only a fraction of the whole set of ABA-responsive genes, it leads to impaired plant growth. This phenotype is reminiscent of that found in plants that overexpress DRE-binding protein DREB1A under the control of a 35S promoter, which show constitutive expression of many stress tolerance genes, but also show severe growth retardation under normal growing conditions (Kasuga et al., 1999). Similarly, constitutive expression of ABRE-binding factors ABF3 and ABF4 also led to constitutive expression of ABA-responsive genes and growth retardation (Kang et al., 2002). Therefore, PP2Cs play a major role in regulating ABA signaling both under stress as well as normal growth conditions. Additionally, the phenotype of triple pp2c mutants serves to illustrate the importance of ABA in stress response as well as growth regulation.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants were routinely grown under greenhouse conditions in pots containing a 1:3 vermiculite:soil mixture. For in vitro culture, seeds were surface sterilized by treatment with 70% ethanol for 20 min, followed by commercial bleach (2.5% sodium hypochlorite) containing 0.05% Triton X-100 for 10 min and, finally, four washes with sterile distilled water. Stratification of the seeds was conducted in the dark at 4°C for 3 d. Then seeds were sowed on plates composed of Murashige and Skoog basal salts, 0.1% MES acid, 1% agar, and 1% Suc. The pH was adjusted to 5.7 with KOH before autoclaving. Plates were sealed and incubated in a controlled-environment growth chamber at 22°C under a 16-h-light/8-h-dark photoperiod at 80 to 100 μE m−2 s−1.
Mutant Identification by PCR Screening
The hab1-1, abi1-2, and pp2ca-1 alleles have been described previously (Leonhardt et al., 2004; Saez et al., 2004, 2006; Kuhn et al., 2006). A line containing a single T-DNA insertion in ABI2 was identified in the SALK T-DNA collection (Alonso et al., 2003), SALK_15166, and obtained from the Nottingham Arabidopsis Stock Centre (NASC; http://nasc.nott.ac.uk). To identify individuals homozygous for the T-DNA insertion, genomic DNA was obtained from kanamycin-resistant seedlings and submitted to PCR genotyping using the following ABI2 primers: forward 5′-AGTGACTTCAGTGCGGCGAGT and reverse 5′-CCTTCTTTTTCAATTCAAGGAT. As a T-DNA left-border primer of the pROK2 vector, we used LBpROK2: 5′-GCCGATTTCGGAACCACCATC.
To generate the abi1-2abi2-2, abi1-2pp2ca-1, and hab1-1pp2ca-1 double mutants, we transferred pollen of either abi2-2 or pp2ca-1 to the stigmas of emasculated flowers of abi1-2 and hab1-1. To generate the triple hab1-1abi1-2abi2-2 and hab1-1abi1-2pp2ca-1 mutants, we crossed either abi1-2abi2-2 or abi1-2pp2ca-1 double mutants with hab1-1abi1-2 (Saez et al., 2006; Rodrigues et al., 2009), respectively. The resulting F2 individuals were genotyped by PCR for the presence of the double or triple mutants.
Germination, Growth, and Stomatal Aperture Assays
To measure sensitivity to ABA-mediated inhibition of germination, seeds (approximately 200 seeds/experiment) were plated on solid medium composed of Murashige and Skoog basal salts, 1% Suc, and increasing concentrations of ABA. To score seed germination, either radicle emergence or the percentage of seeds that had germinated and developed fully green expanded cotyledons was determined. The ABA-resistant growth was scored as described by Saez et al. (2006). Data were obtained for three independent experiments, each done with 15 plants. To quantify root growth inhibition, 5-d-old seedlings grown vertically onto Murashige and Skoog plates were transferred to either Murashige and Skoog plates or Murashige and Skoog plates supplemented with 10 μm ABA. After 7 d, the plates were scanned on a flatbed scanner to produce image files suitable for quantitative analysis using National Institutes of Health Image software (ImageJ version 1.37). Data were obtained for three independent experiments, each done with 15 plants. Assays of ABA-induced stomatal closing were performed as described by Saez et al. (2006). Data were expressed as the average of three experiments where 30 to 40 stomata were measured for each one.
Leaf Gas-Exchange Measurements
For leaf gas-exchange measurements, plants were grown in hydroponics (Hoagland solution at 50% in 250-mL nontransparent pots) inside a growth chamber (12 h at 150 μmol photons m−2 s−1, approximately 60% humidity, and 26°C day/20°C night air temperature). Nutrition solution was changed every 2 to 3 d to prevent nutrition and water limitations. From preliminary light response curves, photosynthesis was proved to be saturating at photon flux densities above 500 μmol photons m−2 s−1. Light-saturating net photosynthesis (AN), leaf conductance for water vapor (gs), transpiration rate (E), and substomatal CO2 concentration (Ci) were therefore measured under steady-state conditions at 800 μmol photons m−2 s−1 with an open gas-exchange infrared gas analyzer (Li-6400; LI-COR) on the youngest fully expanded leaf of 38- to 45-d-old plants for Col and triple mutants. CO2 concentration in the leaf chamber was maintained at 400 μmol CO2 mol−1 air, whereas humidity and vapor pressure deficit were kept around 45% and 2 kPa, respectively.
Drought Stress and Water Loss Assays
Two different water-loss assays were performed. Short-term assays were performed basically as described by Saez et al. (2006) in detached leaves at the same developmental stage and size from 21-d-old plants. Four samples of three leaves per genotype were excised and fresh weight was determined by submitting the leaves to the drying atmosphere of a flow laminar hood for 6 h. Data are averages ± se from three independent experiments (n = 5). Long-term assays were performed after withholding water in plants maintained under greenhouse conditions basically as described by Saez et al. (2006). Eight plants of each genotype (two independent experiments) were grown under normal watering conditions for 21 d and then subjected to drought stress for 8 d by completely terminating irrigation and minimizing soil evaporation by covering pots with plastic wrap film. Eight leaves from each plant were removed, weighed, incubated in demineralized water for 3 h, and weighed again. The difference in weight was considered as water loss.
RNA Analyses
These assays were performed as described by Saez et al. (2006). Briefly, plants were grown on Murashige and Skoog plates supplemented with 1% Suc. After 10 d, approximately 30 to 40 seedlings were either mock or 10 μm ABA treated. After 3 h, plant material was collected and frozen in liquid nitrogen. qRT-PCR amplifications and measurements were performed using an ABI PRISM 7000 sequence detection system (Perkin-Elmer Applied Biosystems). The sequences of the primers used for PCR amplifications were the following ones: for HAB1 (At1g72770), forward 5′-AACTGCTGTTGTTGCCTTG and reverse 5′-GGTTCTGGTCTTGAACTTTCT; for ABI1 (At4g26080), forward 5′-ATGATCAGCAGAACAGAGAGT and reverse 5′-TCAGTTCAAGGGTTTGCT; for ABI2 (At5g57050), forward 5′-AGTGACTTCAGTGCGGCGAGT and reverse 5′-CCTTCTTTTTCAATTCAAGGAT; for PP2CA (At3g11410), forward 5′-CTTTGTCGTAACGGTGTAGC and reverse 5′-TTGCTCTAGACATGGCAAGA; for RAB18 (At5g66400), forward 5′-ATGGCGTCTTACCAGAACCGT and reverse 5′-CCAGATCCGGAGCGGTGAAGC; for RD29B (At5g52300), forward 5′-ATGGAGTCACAGTTGACACGTCC and reverse 5′-GAGATAGTCATCTTCACCACCAGG; and for β-actin-8 (At1g49420), forward 5′-AGTGGTCGTACAACCGGTATTGT and reverse 5′-GAGGATAGCATGTGGAAGTGAGAA.
qRT-PCR amplifications were monitored using Eva-Green fluorescent stain (Biotium). Relative quantification of gene expression data was carried out using the 2−ΔΔCT or comparative CT method (Livak and Schmittgen, 2001). Expression levels were normalized using the CT values obtained for the β-actin-8 gene. The presence of a single PCR product was further verified by dissociation analysis in all amplifications. All quantifications were made in triplicate on RNA samples obtained from three independent biological replicas.
RNA Amplification, Labeling, and Microarray Hybridization
These assays were performed as described by Rodrigues et al. (2009), according to MIAME guidelines (Brazma et al., 2001).
Identification of Differentially Expressed Genes and GO Analysis
Significance analysis of microarrays (Tusher et al., 2001) was performed on the three normalized datasets to identify differentially expressed genes. The parameters for significance analysis of microarrays were adjusted so that the false discovery rate probability for every experiment was below 0.05. A 2-fold expression cutoff was considered. A functional category analysis of the genes simultaneously up-regulated or down-regulated in the three genotypes (Col, triple abi2-2, and triple pp2ca-1) was carried out by the Munich Information Center for Protein Sequences (MIPS; http://mips.gsf.de/proj/funcatDB/search_main_frame.html). Only overrepresented categories with a P value smaller than 0.05 were further considered. Functional categorization of up-regulated genes in each of the genotypes, based on the high-level terms in the GO hierarchy, was conducted using TAIR (http://www.Arabidopsis.org/tools/bulk/go/index.jsp). Venn diagrams were generated to illustrate differences in expression.
ABA Content Measurement
Three independent biological samples of 12-d-old seedlings grown in Murashige and Skoog plates were used for ABA content determination. Seedlings were either mock- or 350 mm mannitol treated for 8 h. After measuring fresh weight, plants were ground in liquid nitrogen and then extracted with 1 mL extraction buffer (80% methanol, 100 mg/L butylated hydroxytoluene, and 0.5 g/L citric acid monohydrate) overnight at 4°C (Xiong et al., 2001). After centrifugation at 1,000g for 20 min at 4°C, supernatants were collected and dried. Samples were resuspended in buffer containing 10% methanol, 25 mm Tris, pH 7.5, 100 mm NaCl, and 1 mm MgCl2, and they were subjected to ABA measurement using the Phytodetek ABA test kit (Agdia).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. ABA-hypersensitive early seedling growth inhibition of different mutants compared to wild type at different concentrations of ABA ranging from 25 to 500 nm ABA.
Supplemental Figure S2. Comparison of the endogenous ABA levels in wild type and triple mutants under nonstressed conditions or after treatment with 350 mm mannitol for 8 h.
Supplemental Figure S3. Cladogram and gene expression data from clade A PP2Cs.
Supplemental Table S1. ABA-responsive genes constitutively up-regulated (threshold of 2-fold, ratio >2) or down-regulated (threshold of 2-fold, ratio <0.5) in the triple mutants in the absence of exogenous ABA.
Supplemental Table S2. Differential expression of ABA-responsive genes constitutively up-regulated or down-regulated in the triple abi2-2 and pp2ca-1 mutants compared each other (genes selected previously from Supplemental Table S1).
Supplementary Material
Acknowledgments
We thank Joseph Ecker and the Salk Institute Genomic Analysis Laboratory for providing the sequence-indexed Arabidopsis T-DNA insertion mutants, and the Arabidopsis Biological Resource Center/Nottingham Arabidopsis Stock Centre for distributing these seeds.
This work was supported by the Ministerio de Educación y Ciencia and Fondo Europeo de Desarrollo Regional (grant nos. BIO2005–01760 and BIO2008–00221 to P.L.R., and grant no. BFU2008–01072/BFI to J.F.), Consejo Superior de Investigaciones Científicas (fellowship to S.R. and J.S.), and the National Science Foundation and the National Institutes of Health (grant nos. MCB0417118 and GM060396 to J.I.S.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Pedro L. Rodriguez (prodriguez@ibmcp.upv.es).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open access articles can be viewed online without a subscription.
References
- Achard P, Cheng H, De Grauwe L, Decat J, Schoutteten H, Moritz T, Van Der SD, Peng J, Harberd NP (2006) Integration of plant responses to environmentally activated phytohormonal signals. Science 311 91–94 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, et al (2001) Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet 29 365–371 [DOI] [PubMed] [Google Scholar]
- Cheong YH, Pandey GK, Grant JJ, Batistic O, Li L, Kim BG, Lee SC, Kudla J, Luan S (2007) Two calcineurin B-like calcium sensors, interacting with protein kinase CIPK23, regulate leaf transpiration and root potassium uptake in Arabidopsis. Plant J 52 223–239 [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala SS, Rock CD (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell (Suppl) 14 S15–S45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Verslues PE, Zhu JK (2007) Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 19 485–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Cadenas A, Verhey SD, Holappa LD, Shen Q, Ho TH, Walker-Simmons MK (1999) An abscisic acid-induced protein kinase, PKABA1, mediates abscisic acid-suppressed gene expression in barley aleurone layers. Proc Natl Acad Sci USA 96 1767–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Garcia MP, Rodriguez D, Nicolas C, Rodriguez PL, Nicolas G, Lorenzo O (2003) Negative regulation of abscisic acid signaling by the Fagus sylvatica FsPP2C1 plays a role in seed dormancy regulation and promotion of seed germination. Plant Physiol 133 135–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosti F, Beaudoin N, Serizet C, Webb AA, Vartanian N, Giraudat J (1999) ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell 11 1897–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Xiong L, Song CP, Gong D, Halfter U, Zhu JK (2002) A calcium sensor and its interacting protein kinase are global regulators of abscisic acid signaling in Arabidopsis. Dev Cell 3 233–244 [DOI] [PubMed] [Google Scholar]
- Higuchi M, Pischke MS, Mahonen AP, Miyawaki K, Hashimoto Y, Seki M, Kobayashi M, Shinozaki K, Kato T, Tabata S, et al (2004) In planta functions of the Arabidopsis cytokinin receptor family. Proc Natl Acad Sci USA 101 8821–8826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama T, Shinozaki K (2007) Perception and transduction of abscisic acid signals: keys to the function of the versatile plant hormone ABA. Trends Plant Sci 12 343–351 [DOI] [PubMed] [Google Scholar]
- Hua J, Meyerowitz EM (1998) Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell 94 261–271 [DOI] [PubMed] [Google Scholar]
- Iuchi S, Suzuki H, Kim YC, Iuchi A, Kuromori T, Ueguchi-Tanaka M, Asami T, Yamaguchi I, Matsuoka M, Kobayashi M, et al (2007) Multiple loss-of-function of Arabidopsis gibberellin receptor AtGID1s completely shuts down a gibberellin signal. Plant J 50 958–966 [DOI] [PubMed] [Google Scholar]
- Kang JY, Choi HI, Im MY, Kim SY (2002) Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14 343–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1999) Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotechnol 17 287–291 [DOI] [PubMed] [Google Scholar]
- Kim KN, Cheong YH, Grant JJ, Pandey GK, Luan S (2003) CIPK3, a calcium sensor-associated protein kinase that regulates abscisic acid and cold signal transduction in Arabidopsis. Plant Cell 15 411–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Reuling G, Karssen CM (1984) The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Physiol Plant 61 377–383 [Google Scholar]
- Kuhn JM, Boisson-Dernier A, Dizon MB, Maktabi MH, Schroeder JI (2006) The protein phosphatase AtPP2CA negatively regulates abscisic acid signal transduction in Arabidopsis, and effects of abh1 on AtPP2CA mRNA. Plant Physiol 140 127–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak JM, Moon JH, Murata Y, Kuchitsu K, Leonhardt N, DeLong A, Schroeder JI (2002) Disruption of a guard cell-expressed protein phosphatase 2A regulatory subunit, RCN1, confers abscisic acid insensitivity in Arabidopsis. Plant Cell 14 2849–2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak JM, Mori IC, Pei ZM, Leonhardt N, Torres MA, Dangl JL, Bloom RE, Bodde S, Jones JD, Schroeder JI (2003) NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J 22 2623–2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt N, Kwak JM, Robert N, Waner D, Leonhardt G, Schroeder JI (2004) Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant. Plant Cell 16 596–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J, Bouvier-Durand M, Morris PC, Guerrier D, Chefdor F, Giraudat J (1994) Arabidopsis ABA response gene ABI1: features of a calcium-modulated protein phosphatase. Science 264 1448–1452 [DOI] [PubMed] [Google Scholar]
- Leung J, Merlot S, Giraudat J (1997) The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell 9 759–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang XQ, Watson MB, Assmann SM (2000) Regulation of abscisic acid-induced stomatal closure and anion channels by guard cell AAPK kinase. Science 287 300–303 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25 402–408 [DOI] [PubMed] [Google Scholar]
- Merlot S, Gosti F, Guerrier D, Vavasseur A, Giraudat J (2001) The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J 25 295–303 [DOI] [PubMed] [Google Scholar]
- Meyer K, Leube MP, Grill E (1994) A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science 264 1452–1455 [DOI] [PubMed] [Google Scholar]
- Mori IC, Murata Y, Yang Y, Munemasa S, Wang YF, Andreoli S, Tiriac H, Alonso JM, Harper JF, Ecker JR, et al (2006) CDPKs CPK6 and CPK3 function in ABA regulation of guard cell S-type anion- and Ca(2+)-permeable channels and stomatal closure. PLoS Biol 4 e327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustilli AC, Merlot S, Vavasseur A, Fenzi F, Giraudat J (2002) Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14 3089–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N, Yoshida T, Kitahata N, Asami T, Shinozaki K, Hirayama T (2007) ABA-Hypersensitive Germination1 encodes a protein phosphatase 2C, an essential component of abscisic acid signaling in Arabidopsis seed. Plant J 50 935–949 [DOI] [PubMed] [Google Scholar]
- Ossowski S, Schwab R, Weigel D (2008) Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J 53 674–690 [DOI] [PubMed] [Google Scholar]
- Pernas M, Garcia-Casado G, Rojo E, Solano R, Sanchez-Serrano JJ (2007) A protein phosphatase 2A catalytic subunit is a negative regulator of abscisic acid signalling. Plant J 51 763–778 [DOI] [PubMed] [Google Scholar]
- Robert N, Merlot S, N'guyen V, Boisson-Dernier A, Schroeder JI (2006) A hypermorphic mutation in the protein phosphatase 2C HAB1 strongly affects ABA signaling in Arabidopsis. FEBS Lett 580 4691–4696 [DOI] [PubMed] [Google Scholar]
- Rodrigues A, Santiago J, Rubio S, Saez A, Osmont KS, Gadea J, Hardtke CS, Rodriguez PL (2009) The short-rooted phenotype of the brevis radix mutant partly reflects root ABA hypersensitivity. Plant Physiol 149 1917–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez PL, Benning G, Grill E (1998) ABI2, a second protein phosphatase 2C involved in abscisic acid signal transduction in Arabidopsis. FEBS Lett 421 185–190 [DOI] [PubMed] [Google Scholar]
- Saez A, Apostolova N, Gonzalez-Guzman M, Gonzalez-Garcia MP, Nicolas C, Lorenzo O, Rodriguez PL (2004) Gain-of-function and loss-of-function phenotypes of the protein phosphatase 2C HAB1 reveal its role as a negative regulator of abscisic acid signalling. Plant J 37 354–369 [DOI] [PubMed] [Google Scholar]
- Saez A, Robert N, Maktabi MH, Schroeder JI, Serrano R, Rodriguez PL (2006) Enhancement of abscisic acid sensitivity and reduction of water consumption in Arabidopsis by combined inactivation of the protein phosphatases type 2C ABI1 and HAB1. Plant Physiol 141 1389–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweighofer A, Hirt H, Meskiene I (2004) Plant PP2C phosphatases: emerging functions in stress signaling. Trends Plant Sci 9 236–243 [DOI] [PubMed] [Google Scholar]
- Sheen J (1996) Ca2+-dependent protein kinases and stress signal transduction in plants. Science 274 1900–1902 [DOI] [PubMed] [Google Scholar]
- Sheen J (1998) Mutational analysis of protein phosphatase 2C involved in abscisic acid signal transduction in higher plants. Proc Natl Acad Sci USA 95 975–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahtiharju S, Palva T (2001) Antisense inhibition of protein phosphatase 2C accelerates cold acclimation in Arabidopsis thaliana. Plant J 26 461–470 [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 98 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verslues PE, Zhu JK (2007) New developments in abscisic acid perception and metabolism. Curr Opin Plant Biol 10 447–452 [DOI] [PubMed] [Google Scholar]
- Xiong L, Ishitani M, Lee H, Zhu JK (2001) The Arabidopsis LOS5/ABA3 locus encodes a molybdenum cofactor sulfurase and modulates cold stress- and osmotic stress-responsive gene expression. Plant Cell 13 2063–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida R, Hobo T, Ichimura K, Mizoguchi T, Takahashi F, Aronso J, Ecker JR, Shinozaki K (2002) ABA-activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis. Plant Cell Physiol 43 1473–1483 [DOI] [PubMed] [Google Scholar]
- Yoshida T, Nishimura N, Kitahata N, Kuromori T, Ito T, Asami T, Shinozaki K, Hirayama T (2006) ABA-hypersensitive germination3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiol 140 115–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.