Abstract
Quantifying the behavior of cells individually, and in clusters as part of a population, under a range of experimental conditions, is a challenging computational task with many biological applications. We propose a versatile algorithm for segmentation and tracking of multiple motile epithelial cells during wound healing using time-lapse video. The segmentation part of the proposed method relies on a level set-based active contour algorithm that robustly handles a large number of cells. The tracking part relies on a detection-based multiple-object tracking method with delayed decision enabled by multi-hypothesis testing. The combined method is robust to complex cell behavior including division and apoptosis, and to imaging artifacts such as illumination changes.
1. INTRODUCTION
Characterizing cell motility, cell deformation, and cell population dynamics are important parameters of interest in understanding basic biological processes such as growth [1], tissue repair [2], differentiation, metastatic potential [3], chemotaxis [4], or improving drug discovery. We focus on analyzing the behavior of single cells and the collective motion of the cell population, during in vitro wound healing after a scratch. Epithelial cells typically move together as a continuous sheet due to strong cell-to-cell adhesion. So wound closure is often quantified in terms of the change in wound gap area over time. However, this large scale measure of wound closure does not capture the cell layer reorganization that can occur hundreds of microns behind the wound edge [2, 3]. We investigate a combined approach of using level set-based active contour segmentation followed by multi-hypothesis tracking to accurately track individual epithelial cells from which cell-level and population-level statistics can be computed. The proposed algorithm is robust enough, that the epithelial cells imaged in phase contrast do not need to be marked or labeled in any way. Other advantages include the ability to track many cases of cell division and some cases of occlusions due to rounding on top of adjacent neighbors.
2. CELL SEGMENTATION USING ACTIVE CONTOURS
A level set-based active contour algorithm based on the Chan and Vese model [5] was used to automatically segment all of the cells in each frame of the image sequence. This algorithm does not require initialization or training and is robust to illumination shifts. A piecewise constant segmentation of a 2D gray scale image u(y) into two phases is obtained by minimizing the Mumford-Shah functional in Eq. 1 with respect to the scalar variables c1, c2 and φ; with φ(y) being the 2D level set function and u(y) being the intensity image.
| (1) |
In Eq. 1, c1 and c2 model the gray values of the two phases used to define the segmentation, with the boundary for the phases described by regularized Heaviside function H(φ) [5]. φ models the interface separating these two phases. The first two terms in the functional aim at maximizing the gray value homogeneity of the two phases, while the last term aims at minimizing the length of the boundary separating these two phases; μ1,μ2 and ν are adjustable constants associated with this functional. The Euler-Lagrange equation , to evolve the level set curve is:
| (2) |
The scalars c1 and c2 are updated at each level set iteration step using Eq. 3 for φ(y) > 0 and φ(y) < 0, respectively.
| (3) |
A regularized Heaviside function H2,∈ (φ(y)) is used to obtain a numerically stable Dirac delta function δ(φ) in Eq. 2 [5]. A suitable stopping criterion (such as a small change in energy, |φi+1 — φi| < kthresh where φi+1,φi are the level set functions at successive iterations) is used to terminate the evolving equation.
3. MULTIPLE CELL MOTION ANALYSIS USING GRAPHS
Motion analysis is used; to track cells over time, to compute kinematic parameters (e.g., velocity, acceleration etc.,) from trajectories, and to detect events such as cell division and cell apoptosis. The motion analysis module receives a cell mask from the segmentation module. Morphological operations (e.g., opening, closing) are applied to this mask to remove small noisy regions and to refine object boundaries. Connected component analysis is applied to the refined mask to identify distinct connected foreground regions. For each connected region, features such as bounding box, centroid, area, and support map are extracted. This information is arranged as a graph structure. Nodes of this graph represent the objects (cells) and their features. The correspondence analysis stage searches for potential object matches in frame(t) to objects in frame(t — 1). The graph is updated by linking nodes corresponding to objects in frame(t) with nodes of potential corresponding objects in frame(t — 1). The confidence value for each match is stored with each link. The segment generation module traces links on this graph to generate cell trajectories.
3.1. Multi-Hypothesis Cell Tracking
Cell tracking is useful to analyze the long term behavior of cells. Tracking is the process of establishing cell correspondences between consecutive frames. Typically the process involves a cycle of feature extraction, motion prediction, correspondence (gating, pruning, association), and update. Tracking algorithms typically rely upon feature correspondences or optical flow fields. However, these are ill-posed problems due to ambiguities arising from multiple matches, or zero matches. Prediction is used to resolve these ambiguities to some extent by providing a confidence region within which to search. Commonly used prediction methods include Kalman filters (and its variants like unscented Kalman filters etc.,) and particle filters [6]. Prediction is not used in the current version because the trajectories were short, but will be investigated in a future work. In the proposed system, cell-to-cell matching is done using a three level cell morphology overlap test. Three distance functions that depend upon size and shape similarity, and not just proximity, are used.
The proposed matching measures have several advantages over the often used Euclidean distance measure between object centroids. A particularly important case where the centroid measure fails is cell division, when epithelial cells often become elongated then split across the minor axis. This produces a big increase in the centroid distance that may result in discontinuities in the trajectory. However our overlap distance produces smaller increases and is more likely to find the right correspondence.
The feature correspondence distance measures developed for cell tracking are defined as:
| (4) |
| (5) |
| (6) |
where,
OA and OB represent distinct connected foreground regions corresponding to cells in two consecutive frames. Dolp (Eq. 4) measures the degree of overlap between two overlapping cell masks. Dmsk and Dbbx measure proximity between two non-overlapping masks. Dmsk is a simplified Hausdorff distance that measures minimum distance between two masks. It is computed in terms of the minimum number of dilations needed to overlap the two masks. It is defined in Eq. 5 where δk(msk(objA)) denote foreground mask for objectA dilated k times. Dbbx (Eq 6) measures the minimum distance between two bounding boxes. It is measured as the minimum corner to corner distance and is used for computational efficiency. Dmsk is only computed for cells with overlapping bounding boxes. Dbbx is used to measure proximity only for those cells with non-overlapping bounding boxes.
Using these distance measures tracking produces a match matrix Match and a confidence matrix ConfMatch for each frame(t). Match(i, j) indicates whether the ith object in frame(t) corresponds to the jth object in frame(t — 1). ConfMatch(i, j) denotes the confidence in Match(i, j).
The confidence in a match is computed based on both similarity and proximity of the associated objects, and the availability and quality of other possible matches. Two types of confidence values are considered: similarity confidence Confsim, and separation confidence Confsep. Similarity confidence for the matched objects is defined as,
| (7) |
where MAX_DIST is a user specified parameter. Separation confidence is a measure of how distinct the match is. It measures the competition between all possible matches for the current object. Separation confidence is defined as in Eq. 8,
| (8) |
where
and j* is the closest competitor in terms of distance. This measure favors matches without competitors, and matches with competitors having higher distances. Total similarity and total separation confidence values are computed as weighted sums of individual confidence values for all three distances. Similarity and separation confidence values are further combined into a single confidence value ConfMatch using a weighted sum.
For the final object to object association, a multi-hypothesis testing approach with delayed decision is used. Rather than picking a single best match or some subset of these matches with limited (two frame) information, many possible matches are kept and gradually pruned, as more information becomes available in the decision process. Besides one-to-one object matches, this scheme supports many-to-one, one-to-many, many-to-many, one-to-none, or none-to-one matches that may result from false detections, or false associations, segmentation errors, occlusion, entering, exiting, or dividing of cells.
3.2. Processing Segments and Forming Trajectories
The segment-extraction module analyzes match information by classifying the cells/objects into nine types; single, source, source-split, sink, inner, split, sink-merge, merge-split, merge, based on the number of parent and child objects. Trajectory segments (a linked list of inner objects starting with a source or split type object and ending with a merge or sink type object) are identified, and organized in a data structure called SegmentList. Extracted segments are labeled using a method similar to connected component labeling. Each segment without a parent is given a new label. Segments having parents inherit the labels of their parents. In case of multiple parents with inconsistent labels , the smaller label (older trajectory) is kept and a flag is set indicating the inconsistency. After labeling and pruning, trajectories are formed by linking segments sharing the same label.
3.3. Cell Trajectory Filtering
Filtering is performed at various levels of processing. At the image level small objects that may be due to noise, background clutter, or imaging and segmentation artifacts are removed by using morphological operators. At the correspondence level, gating eliminates unfeasible matches with distances above a threshold. At the confidence level, two types of pruning, absolute and relative, are performed. In absolute pruning, matches whose confidence values are below an absolute confidence threshold are eliminated from the possible match list. In relative pruning, first the most probable match (i.e., the match with highest confidence value) is determined. then matches whose confidences values are below a percentage of the highest confidence value are eliminated from the possible match list. At the segment level, very short segments that split from another segment and end unexpectedly or that start unexpectedly and merge to another segment are pruned. These segments are usually the result of temporal fragmentation in the segmentation stage.
4. EXPERIMENTAL RESULTS
The proposed approach was tested on a wound healing image sequence that consists of 46 frames of size 220 × 179 (29.33μm×23.86μm) cropped and subsampled from a larger sequence. The wound healing image sequence was obtained using a monolayer of cultured pig epithelial cells. The cells were handled by Salaycik et al. as described in [7]. Images were sampled uniformly over a 9:00:48 hour period and acquired using phase contrast microscopy with a 10× objective lens at a resolution of approximately 0.13μm per pixel.
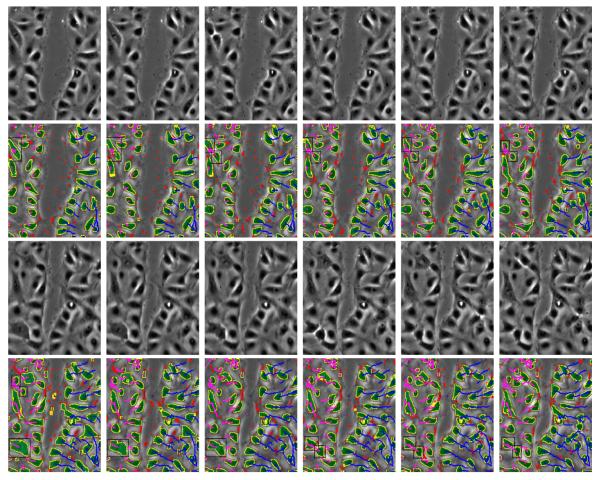
Some experimental results are shown in Figures 1, 2 and Table 1. Figure 1 shows the segmentation and tracking results for some selected images. Dividing cells (marked with black rectangles) are of particular interest in this sequence. Tracking correctly associates the cell trajectories before and after the division process. Figure 2 shows average velocities and directions per frame, for all the cells on one side of the wound and separately for those cells just along the wound margins. The cells on the right start moving faster (3.25 pixels/frame) compared to the cells on the left of the wound margin (1.5 pixels/frame). Table 1 shows some segmentation and tracking results compared to manual ground truth; most of the incorrect cell merges are due to incorrect segmentation around a lipid organelle. This also results in a series of false trajectory splits. The series of false splits caused by this lipid mass is counted as one false split in Table 1.
Fig. 1.
First row: Original frames 13-18. Second row: Segmentation and tracking results for frames 13-18. Third and fourth rows: Original frames 24-29 and results. Trajectories shown in rows 2 and 4 start from the first frame. Color key: yellow - post-processed contours, red - spurious contours, green - cell masks, blue and purple -trajectories, black - bounding box of the cell nuclei in the process of cell-division.
Fig. 2.
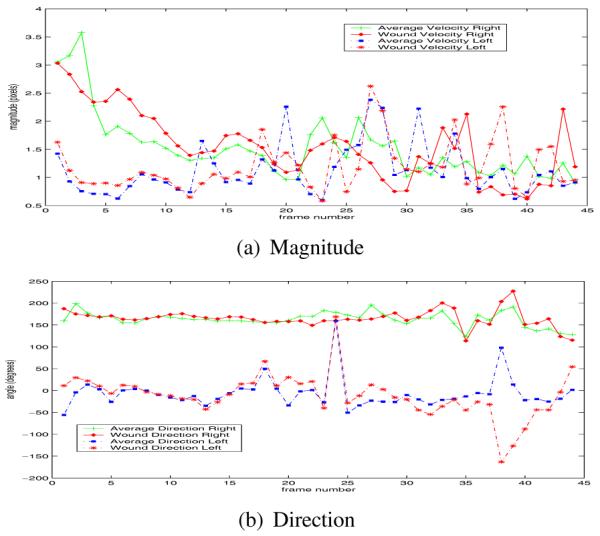
Components of average velocity. The four average velocity curves in (a) and (b) : average right motion (+), wound margin right motion (diamond), average left motion (square), wound margin left motion (*).
Table 1.
Detection and tracking performances for the wound closure dataset, N: Total nuber of cells/trajectories detected, FP: False positives, FN: False negatives, IM: Incorrect merges, IS: incorrect splits.
| N | FP | FN | IM | IS | |
|---|---|---|---|---|---|
| Cells | 2106 | 24 | 10 | 25 | 1 |
| Traj. | 64 | 0 | 0 | 0 | 2 |
5. CONCLUSION AND FUTURE WORK
We presented a level-set based active contour method for segmentation and a multi-hypothesis approach for robust tracking of cells. Filtering and pruning at different levels of processing enables the tracker to eliminate spurious objects and trajectories. Segmentation and tracking modules do not need initialization or training. The proposed confidence measures enable the system to delay the decisions until further levels of processing. The proposed approach will be evaluated with more diverse sequences.
Acknowledgments
This work is partially supported by a United States National Institutes of Health grant, NIBIB award R33 EB00573.
6. REFERENCES
- [1].Palaniappan K, Jiang HS, Baskin TI. CVPR - IEEE Workshop on Articulated and Nonrigid Motion. June. Vol. 1. Washington, DC: 2004. Non-rigid motion estimation using the robust tensor method; pp. 25–33. [Google Scholar]
- [2].Farooqui R, Fenteany G. Multiple rows of cells behind an epithelial wound edge extend cryptic lamellipodia to collectively drive cell-sheet movement. J. Cell Science. 2005 Jan;118(1):51–63. doi: 10.1242/jcs.01577. [DOI] [PubMed] [Google Scholar]
- [3].Ronot X, Doisy A, Tracqui P. Quantitative study of dynamic behavior of cell monolayers during in vitro wound healing by optical flow analysis. Cytometry. 2000;41(1):19–30. [PubMed] [Google Scholar]
- [4].Dufour A, Shinin V, Tajbakhsh S, Guillen-Aghion N, Olivo-Marin JC, Zimmer C. Segmenting and tracking fluorescent cells in dynamic 3-D microscopy with coupled active surfaces. IEEE Trans. Image Process. 2005 Sep;14(9):1396–1410. doi: 10.1109/tip.2005.852790. [DOI] [PubMed] [Google Scholar]
- [5].Chan T, Vese L. Active contours without edges. IEEE Trans. Image Process. 2001 Feb;10(2):266–277. doi: 10.1109/83.902291. [DOI] [PubMed] [Google Scholar]
- [6].Haykin S, deFreitas N. Special issue on sequential state estimation. Proc. IEEE. 2004 Mar;92(3):399–400. [Google Scholar]
- [7].Salaycik KJ, Fagerstrom CJ, Murthy K, Tulu US, Wadsworth P. Quantification of microtubule nucleation growth and dynamics in wound-edge cells. J. Cell Sci. 2005;118:4113–4122. doi: 10.1242/jcs.02531. [DOI] [PubMed] [Google Scholar]