Abstract
The origin recognition complex (ORC) is a 6-subunit complex required for the initiation of DNA replication in eukaryotic organisms. ORC is also involved in other cell functions. The smallest Drosophila ORC subunit, Orc6, is important for both DNA replication and cytokinesis. To study the role of Orc6 in vivo, the orc6 gene was deleted by imprecise excision of P element. Lethal alleles of orc6 are defective in DNA replication and also show abnormal chromosome condensation and segregation. The analysis of cells containing the orc6 deletion revealed that they arrest in both the G1 and mitotic stages of the cell cycle. Orc6 deletion can be rescued to viability by a full-length Orc6 transgene. The expression of mutant transgenes of Orc6 with deleted or mutated C-terminal domain results in a release of mutant cells from G1 arrest and restoration of DNA replication, indicating that the DNA replication function of Orc6 is associated with its N-terminal domain. However, these mutant cells accumulate at mitosis, suggesting that the C-terminal domain of Orc6 is important for the passage through the M phase. In a cross-species complementation experiment, the expression of human Orc6 in Drosophila Orc6 mutant cells rescued DNA replication, suggesting that this function of the protein is conserved among metazoans.
Keywords: DNA replication, ORC, Chromatin
The hexameric origin recognition complex (ORC) is an essential component for eukaryotic DNA replication. It was originally discovered in Saccharomyces cerevisiae, and subsequent studies both in yeast and in higher eukaryotes laid the foundation for understanding the functions of this important key initiation factor. ORC binds to origin sites in an ATP-dependent manner and directs the assembly of the prereplicative complex (pre-RC) at the origins (1, 2). ORC subunits and/or complete ORC complexes have also been identified in many metazoan species (1, 3), suggesting the existence of common mechanisms for the initiation of DNA replication in all eukaryotes. ORC genes are essential for cell survival. Mutational analysis of ORC-related genes in yeast and in higher eukaryotes reveals defects in DNA replication (reviewed in refs. 1 and 3). In other studies, immunodepletion experiments using either Xenopus or Drosophila replication-competent extracts indicate an absolute requirement for ORC to initiate DNA replication (4–6). Acute depletion of ORC gene expression in human cells by RNAi resulted in cell cycle arrest (7, 8). In addition to initiating DNA replication, ORC is involved in other functions described previously in detail (1, 3, 9).
The Orc6 protein is the least conserved of all ORC subunits. In S. cerevisiae, Orc6 is not important for DNA binding, but it is required for cell survival (10–12) and the maintenance of the pre-RC, specifically for recruitment of Cdt1 followed by MCM loading (13, 14). Schizosaccharomyces pombe and metazoan Orc6 proteins (6, 15, 16) are more homologous, similar in size, and considerably smaller than the S. cerevisiae Orc6. In Drosophila, Orc6 is essential for ORC-dependent DNA binding and DNA replication (17, 18). In Xenopus and human systems Orc6 is less tightly associated with the core complex, and some of the published data suggest that Orc6 may not be important for these activities (19–21). This apparent inconsistency may reflect the difference in affinity of Orc6 for the core ORC1-5 complex in distant metazoan species. Drosophila Orc6 and human Orc6 also have a function in cytokinesis (7, 22, 23). This function in Drosophila is attributed to the C-terminal domain of Orc6 (22). To study the Orc6 functions in a living organism, we generated and characterized the Orc6-deletion mutant in Drosophila. We have analyzed in detail the mutant phenotypes associated with a lethal allele of the Drosophila orc6 gene alone or with different versions of fly or human Orc6 rescue transgenes, gaining further insight into the roles Orc6 plays through the cell cycle in metazoan species.
Results
Drosophila Orc6 Accumulates on Chromosomes in Late Mitosis.
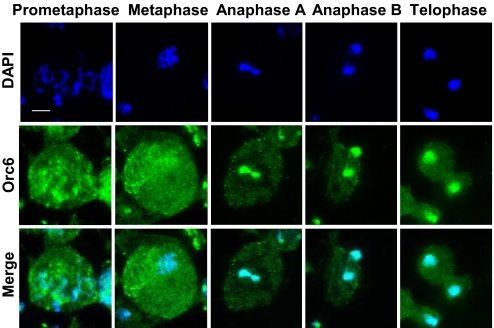
In Drosophila cells Orc6 colocalizes with other ORC subunits but also displays distinct cytoplasmic and membrane staining in both embryonic and tissue culture cells, reflecting its functions in both DNA replication and cytokinesis (17, 22). Analysis of mitotic stages in developing Drosophila neuroblasts revealed that at prometaphase and metaphase Orc6 was present in the nucleus but was weakly associated with the DNA (Fig. 1). However, beginning at anaphase, Orc6 staining of the segregating chromosomes became intense along the length of the chromatids and persisted further into telophase (Fig. 1). The observed pattern of Orc6 staining in this experiment is remarkably similar to those of both Drosophila Orc2 and Xenopus Orc1, which were also weakly associated with DNA at metaphase but present at the later stages of mitosis (4, 24, 25). Most likely, at these stages ORC is deposited onto the replication origins in preparation for the next cell cycle.
Fig. 1.
Drosophila Orc6 accumulates on chromosomes in anaphase through telophase. Immunofluorescence images of wild-type Drosophila neuroblasts stained with affinity-purified anti-Orc6 antibody (green) are shown in metaphase, anaphase, and telophase stages. DNA is stained with DAPI (blue). (Scale bar: 5 μm.)
Generation of an Orc6 Mutation in Drosophila.
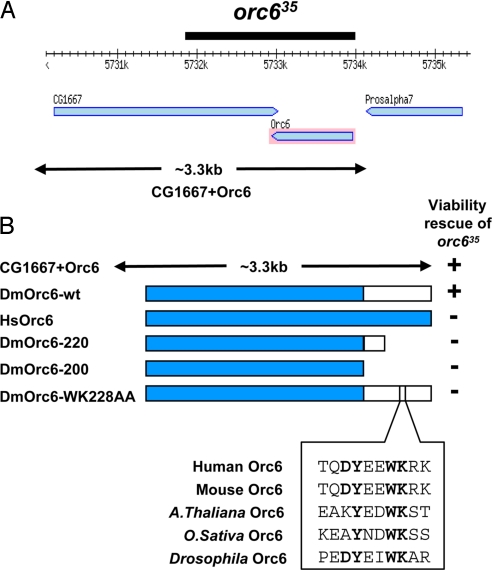
To study the functions of Orc6 in vivo in live animals, we generated a deletion of the orc6 gene in Drosophila by using the method of P element imprecise excision. Several lethal deletions of the orc6 genomic region were identified and their boundaries mapped by sequencing. Fig. 2A shows a map of the genomic region of the second chromosome containing the orc6 gene, and it also shows the boundaries of the obtained deletion used in the current study. This third-instar lethal deletion, called orc635, includes the whole orc6 gene and a part of overlapping CG1667, which has no apparent or predicted function.
Fig. 2.
Generation and rescue of Orc6 mutant. Fragment of genomic map from Drosophila database and limits of the orc635 deletion are shown (A). Drosophila wild-type (DmOrc6), human (HsOrc6), truncated C terminus mutants (DmOrc6-220, DmOrc6-200), and substitution mutant (DmOrc6-WK228AA), all fused with GFP, were used in the rescue experiments (B). The predicted cytokinesis domain of Orc6 is shown in white. Conservative tryptophan (228) and lysine (229) amino acid residues of Drosophila Orc6 were replaced with alanines to create DmOrc6-WK228AA clone. An alignment of corresponding Orc6 sequences between different species is shown in the box.
To rescue the orc635 deletion, we used a 3.3-kb genomic clone containing the wild-type orc6 gene together with whole CG1667. This genomic construct is depicted in Fig. 2A. In addition, full-length, GFP-fused Orc6 transgene under control of the native Orc6 promoter was used to rescue the lethality associated with the obtained deletion. Both constructs, the genomic clone containing the orc6 gene and CG1667, as well as the full-length GFP-Orc6 transgene alone—were successfully able to rescue the deletion mutant (Fig. 2B), demonstrating that the lethality is indeed associated with the deleted orc6 gene. CG1667 had no effect on viability in these experiments. Next, we tested the ability of the human orc6 gene to rescue the orc635 deletion. As shown in Fig. 2B, the expression of the full-length GFP-fused human Orc6 protein in Drosophila orc635 did not rescue lethal orc635 mutation, indicating that these 2 protein homologues cannot substitute each other in viability experiments.
Orc6 protein consists of 2 functional domains, which are important for DNA replication and cytokinesis functions (22). The larger, N-terminal domain is important for replication, whereas the smaller, C-terminal domain is required for the interaction with Pnut protein and cytokinesis function of Orc6, as demonstrated by our earlier in vitro and cell culture-based studies (18, 23). To learn more about the functions of the C-terminal domain in vivo, we designed GFP-Orc6 transgenes containing C-terminal deletions or point mutations and used them in rescue experiments. Two C-terminal deletion mutations, Orc6-200 and Orc6-220, were used. These mutations do not interfere with the replicative function of Orc6 and reconstituted ORC in vitro; however, they are defective for the interaction with Pnut protein and induce cytokinetic defects when expressed in Drosophila tissue culture cells (22). The Orc6-WK228AA point amino acid mutant was chosen based on sequence analysis of Orc6 homologues derived from different species of animals and plants. As shown in Fig. 2B, W228 and K229 are conserved amino acids in metazoan and plant organisms and are localized at the extreme C terminus of Orc6, just beyond the boundary of the Orc6-220 deletion. We hypothesized that these amino acids might be important for nonreplicative functions of Orc6. As expected, no rescue to viability was observed when C-terminal Orc6 mutants were used (Fig. 2B), suggesting that this domain of Orc6 is important for survival of the animals. The detailed analysis of the described rescue experiments with all Orc6 clones is presented in Table S1. All GFP-Orc6 transgenes used in this study were tested for expression in Drosophila tissues by Western blot analysis (Fig. S1). All transgenes expressed in vivo at similar levels.
Orc6 Mutant Is Defective in DNA Replication and Arrests in both G1 and Mitosis.
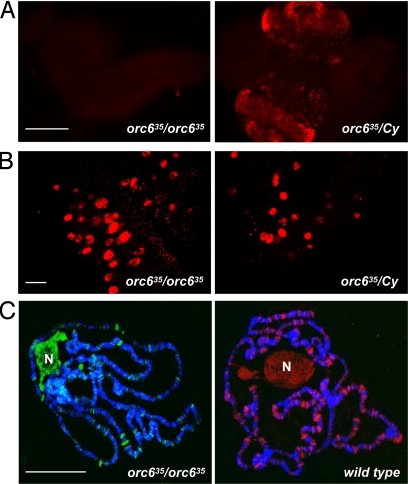
To investigate whether the Orc6 mutant obtained in the course of our studies is also defective for DNA synthesis, BrdU incorporation levels in larval neuroblasts were determined (Fig. 3A). The optic lobes of Orc6 mutant brains were significantly smaller in size compared with the wild-type or heterozygous animals, indicating that brain development and cell proliferation stop prematurely when the maternal supply of Orc6 protein is depleted. BrdU incorporation was also severely reduced in homozygous Orc6-deletion mutant larval brains compared with the large levels of BrdU incorporation in heterozygous brains (Fig. 3A), demonstrating the lack of DNA replication. Correspondingly, immunoblot analysis of Orc6-null mutant brain extracts did not show the presence of Orc6 protein compared with the immunoblots using heterozygous brain cell extracts (Fig. S2). No imaginal discs were detected in late third instar larvae of mutant animals. These phenotypes represent severe proliferation defects also observed in other replication initiation factor mutants (24, 26–30).
Fig. 3.
DNA replication in Orc6 mutant. BrdU incorporation level (red) in third-instar larval brains (A) and in salivary glands of orc635 mutant (B) compared with wild-type or orc635/Cy heterozygous animals. (C) Immunofluorescence experiments using affinity-purified anti-Orc6 antibody on polytene chromosomes isolated from wild-type animals (red) and from orc635 mutant (green) show the presence of Orc6 at specific loci along the chromosome length and in the nucleolus (N). (Scale bars: A and B, 100 μm; C, 50 μm.)
Recently, it was reported that Orc1 was not required for endoreplication in Drosophila salivary glands (30). We analyzed DNA replication in salivary glands isolated from Orc6 mutant animals. Salivary glands appeared normal morphologically but were reduced in size. BrdU incorporation in salivary glands was close to the levels of the wild-type or heterozygous animals (Fig. 3B), suggesting that DNA replication was not affected by the deletion of Orc6. We have also tested the salivary glands of Orc6-deletion mutant animals for the presence of Orc6 protein by immunostaining and Western blotting experiments. Immunoblotting experiments did not reveal detectable amounts of Orc6 in the protein extracts isolated from the salivary glands of mutant animals compared with heterozygous flies (Fig. S2). However, immunostaining experiments using several independently prepared batches of affinity-purified Orc6 antibody consistently revealed distinct Orc6-positive bands on salivary gland chromosomes isolated from Orc6-null homozygous flies (Fig. 3C). The number of Orc6-positive bands was reduced, but nevertheless always detectable on the polytene chromosomes prepared from the salivary glands of Orc6 mutant flies. These results suggest that low amounts of maternally deposited Orc6 protein are still present in salivary glands and may therefore contribute to the observed levels of DNA replication.
Our immunostaining experiments also revealed high amounts of Orc6 localized in the nucleolus of the salivary gland cells (Fig. 3C). We have observed similar staining patterns for other Drosophila ORC subunits, including Orc1 and Orc2. Moreover, ORC was found in the nucleolus in human cells (31, 32). These findings may indicate a potential interaction of ORC with components of the ribosome biogenesis pathway (31, 33).
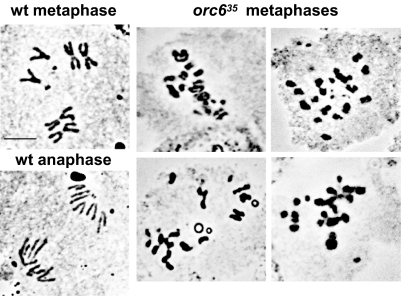
Earlier studies of Orc2 and Orc5 mutants in Drosophila confirmed that cells bearing these mutations arrest both in G1 and during mitosis (24, 29). We have also found that the Orc6 mutant has double arrest points during the cell cycle. First, determination of the ratio of cells with single centrosomes to those with duplicated centrosomes clearly confirmed that Drosophila Orc6-null cells were delayed in cell cycle progression (Table 1). Only a single centrosome is apparent when neuroblasts enter G1 phase. During S phase the centrosomes start separating, and 2 centrosomes are apparent in G2 phase. Further, during mitosis, the 2 poles of the spindle are also clearly marked by γ-tubulin antibody staining. By using this assay, the ratio of G1 cells was found to be increased by a factor of ≈7 in the Drosophila Orc6-deletion mutant, demonstrating the accumulation of G1 cells in the absence of Orc6 protein (Table 1, second row). Second, we identified mitotic cells by immunostaining of Drosophila neuroblasts with antibody against the mitotic marker phosphohistone H3. Overall, the number of mitotic cells observed in the Orc6-deletion mutant decreased compared with the wild-type or heterozygous tissues, probably because of a large number of cells arrested at G1 phase (Table 2, second row). At the next step, chromosome spreads were scored for the presence of chromosomes in each stage of mitosis. No cells at anaphase or telophase stages were found in cells carrying the orc635 deletion. Instead, a large number of metaphase-like figures were observed, in which highly condensed chromosomes were present with incomplete or disorganized alignment (Fig. 4). In addition, individual chromatids appeared aberrantly condensed and fragmented. Chromosome spreads were prepared from third instar larval brains to analyze the degree of chromosome condensation in orc635 homozygous cells. The representative data from wild-type and mutant cells are shown in Fig. 4. In general, the mitotic chromosomes in mutant neuroblasts appeared shorter and thicker, and sister chromatid cohesion was frequently lost (Fig. 4). Extensive chromosome fragmentation was also observed. Therefore, we conclude that Orc6-deletion mutant, in addition to the G1 arrest, also has a defect in establishing a true metaphase and progressing past this point.
Table 1.
Effect of Orc6 transgenes on G1 arrest of the orc635 neuroblasts
| Fly genotype | No. of cells |
G1:S/G2/M ratio | |
|---|---|---|---|
| G1,1 centrosome | S/G2/M,2 centrosomes | ||
| orc635/Cy | 154 | 119 | 1.3:1 |
| orc635/orc635 | 208 | 28 | 7.4:1 |
| orc635/Cy, GFP-orc6 | 286 | 250 | 1.1:1 |
| orc635/orc635, GFP-orc6 | 234 | 211 | 1.1:1 |
| orc635 /Cy, GFP-orc6-200 | 108 | 93 | 1.2:1 |
| orc635/orc635, GFP-orc6-200 | 324 | 33 | 9.8:1 |
| orc635/Cy, GFP-orc6-220 | 267 | 243 | 1.1:1 |
| orc635/orc635, GFP-orc6-220 | 235 | 187 | 1.2:1 |
| orc635/Cy, GFP-orc6-WK228AA | 176 | 144 | 1.2:1 |
| orc635/orc635,GFP-orc6-WK228AA | 256 | 236 | 1.1:1 |
| orc635/Cy, GFP-Hs-orc6 | 141 | 139 | 1.01:1 |
| orc635/orc635, GFP-Hs-orc6 | 317 | 221 | 1.4:1 |
Several independent fly stocks have been tested for each GFP-Orc6 transgene. In every case, tested stocks have shown similar G1 to S/G2/M ratios.
Table 2.
Effect of GFP-Orc6 transgenes on cell cycle progression of orc635 mutant neuroblasts
| Fly genotype | Mitotic to total cells ratio (%) |
|---|---|
| orc635/Cy | 79:3,677 (2.1) |
| orc635/orc635 | 18:1,425 (1.2) |
| orc635/Cy, GFP-orc6 | 45:2,384 (1.9) |
| orc635/orc635, GFP-orc6 | 25:1,488 (1.7) |
| orc635/Cy, GFP-orc6-200 | 64:2,495 (2.6) |
| orc635/orc635, GFP-orc6-200 | 11:1,777 (0.6) |
| orc635/Cy, GFP-orc6-220 | 23:1,712 (1.3) |
| orc635/orc635, GFP- orc6-220 | 31:764 (4) |
| orc635/Cy, GFP-orc6-WK228AA | 16:802 (1.9) |
| orc635/orc635, GFP-orc6-WK228AA | 180:2,541 (7) |
| orc635/Cy, GFP-Hs-orc6 | 60:3,200 (1.8) |
| orc635/orc635, GFP-Hs-orc6 | 64:3,738 (1.7) |
At least 3 independent fly stocks have been tested for each GFP-Orc6 transgene.
Fig. 4.
Aceto-Orcein squashes of orc635 brains. Metaphase-like chromosomes in orc635 mutant neuroblasts are abnormally condensed and fragmented compared with the wild-type chromosomes (wt). (Scale bar: 5 μm.)
Effect of Orc6 Transgenes on DNA Replication and Cell Cycle Progression in the Orc6-Deletion Mutant.
Next, we analyzed the effect of transgenes, described in Fig. 2, on DNA replication, cell cycle progression, and cell morphology. As shown in Fig. 2B and Table S1, only transgenes containing the full-length orc6 gene were able to rescue orc635 flies to the adult stage, confirming that full-length Orc6 protein is required for viability. Transgenic orc635 animals carrying constructs containing either human orc6 gene or Drosophila Orc6 with deletions or point mutations within the C-terminal domain of the protein did not survive beyond third larval instar stage. However, detailed analysis revealed that Orc6-null cells carrying these transgenes did not exhibit the G1 arrest characteristic for all described ORC mutants in Drosophila. Again, we quantified the ratio of cells containing 1 and 2 centrosomes for all transgenes used in this study. The results are summarized in Table 1. The number of cells in G1 stage is increased by a factor of ≈7 in the Drosophila orc635 mutant, demonstrating the accumulation of G1 cells in the absence of Orc6 protein (Table 1, second row). However, the expression of the Orc6-220-deletion mutant, Orc6-WK228AA mutant, or human Orc6 protein resulted in the release of orc635 mutant cells from G1 arrest (Table 1). In other words, the ratio of cells containing either 1 or 2 centrosomes was indistinguishable from cells obtained from heterozygous siblings for all of these transgenes, with the sole exception of Orc6-200, which did not differ from orc635 homozygous mutant.
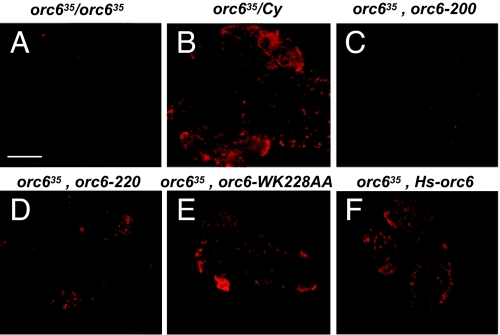
Next, we asked whether these orc635 mutant cells containing different Orc6 transgenic constructs would be able to pass through the S phase and replicate DNA. The results are present in Fig. 5. Again, the orc635 mutant carrying the Orc6-200 gene did not differ in that respect from orc635 mutant cells, because no BrdU incorporation was detected in brain neuroblasts. However, chromosomes from neuroblasts isolated from Orc6-null mutant carrying Orc6-220, Orc6-WK228AA, or human Orc6 transgenes were all able to incorporate BrdU, and therefore were able to replicate DNA (Fig. 5). Brains observed in these larvae were still significantly reduced in size compared with heterozygous or wild-type larval brains, suggesting severe defects in cell proliferation and larval brain development.
Fig. 5.
BrdU incorporation in Orc6 transgenic mutants. DNA replication in orc635 mutant neuroblasts (A) is rescued by the expression of Drosophila GFP-Orc6-220 (C terminus-truncated mutant; D), GFP-Orc6-WK228AA (amino acid substitution mutant; E), and human GFP-HsOrc6 (F), but not by the expression of Drosophila GFP-Orc6-200 (C). BrdU incorporation in heterozygous orc635/Cy neuroblasts is shown in B. (Scale bar: 100 μm.)
Further analysis revealed that neuroblasts carrying the Orc6-220 and Orc6-WK228AA transgenes accumulated at mitosis (Table 2). The number of mitotic neuroblasts derived from these transgenic flies increased 4- to 8-fold compared with orc635 mutant cells. The expression of the shorter Orc6-200 transgene did not have any effect, and the number of mitotic cells carrying this transgene was indistinguishable from the orc635 homozygous mutant neuroblasts and consistently lower than in wild-type or heterozygous tissues. The ratio of mitotic to total cells in orc635 neuroblasts expressing human Orc6 protein increased by the factor of ≈2 compared with Orc6-null cells and was close to the ratio of mitotic to total cells in heterozygous sibling brains.
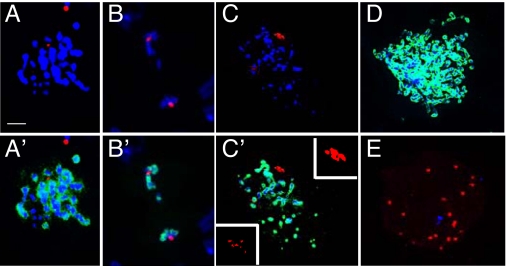
Again, a large number of metaphase-like figures were observed in cells expressing Orc6 C-terminal mutants (Fig. 6A), similarly to Orc6-null cells. However, we also observed cells at different mitotic stages, cells with multiple centrosomes, and polyploid cells when Orc6-220 and Orc6-WK228AA transgenes were expressed in orc635 mutant. For example, anaphase-like figures containing broken chromosomes were observed (Fig. 6B). These anaphase chromosomes contained mutant Orc6 proteins, as shown in Fig. S3 for Orc6-WK228AA protein. Polyploid cells with multiple centrosomes (Fig. 6 C and D) as well as cells containing multiple centrosomes with almost no DNA (Fig. 6E) were also detected.
Fig. 6.
Cell cycle defects in Orc6 transgenic mutants. orc635 neuroblasts expressing GFP-Orc6-220 or GFP-Orc6-WK228AA are arrested in metaphase-like stage (A), but they also display anaphase-like figures (B) and proceed further in the cell cycle to form cells with multiple centrosomes and polyploid cells (C–E). Two clusters of multicentrosome bodies (enlarged 5× in C′ Insets) are shown on metaphase-like figures in C. DNA is stained with DAPI (blue); centrosomes are stained with anti-γ-tubulin antibody (red); mitotic chromosomes are stained with antibody raised against phosphohistone H3 (green). Merged images are shown in A′, B′, C′, D, and E. (Scale bar: 10 μm.)
Discussion
In our studies, we have analyzed in detail the mutant phenotype associated with a lethal allele of the Drosophila orc6 gene, gaining further insight into Orc6 functions during the cell cycle in metazoan species. The Orc6 protein is the most evolutionary diverged and enigmatic among all ORC subunits because its functions in various eukaryotic organisms differ. Orc6 is dispensable for DNA binding in budding yeast (12) but is required for origin recognition in Drosophila (17, 18). Orc6 is tightly associated with core ORC subunits in yeast and Drosophila, and it is crucial for DNA replication in these species (1, 6, 12). In contrast, human Orc6 is loosely bound to the core complex (20), and its role in DNA replication has not yet been completely established. This controversy has partially resulted from the use of different experimental systems that generally do not permit an organismal view of the DNA replication process under physiological conditions.
The functional ORC complex begins to assemble on chromosomes during anaphase. We have found that during mitotic stages, Orc6 localization is remarkably similar to that of other ORC subunits, such as Drosophila Orc2 and Xenopus Orc1. All of these proteins were weakly associated with DNA at metaphase but present at the later stages of mitosis (4, 24, 25). This pattern of binding of Drosophila ORC to the chromatin depends on the cessation of mitotic cyclin activity (25). Most likely, at these stages functional 6-subunit ORC is deposited onto the replication origins in preparation for the next cell cycle.
The analysis of a mutant phenotype for the Orc6 gene in Drosophila during the course of our studies confirmed in a living animal the important role of Orc6 in cell functions. Similarly to the other reported ORC mutants (24, 27–30), we observed vastly reduced BrdU labeling in the proliferative neural tissue of larvae homozygous for the Orc6 mutation, consistent with a crucial role for ORC in the DNA replication. These data together with the increased occurrence of cells with a single centrosome indicate an early cell cycle delay, resulting in G1 arrest in Orc6-null mutant neuroblasts. The amount of cells arrested in G1 increased ≈7-fold compared with the wild-type or heterozygous cells, suggesting that the inactivation of Orc6 leads to defects in pre-RC assembly. Some of the orc635 mutant cells, however, accumulated at a stage with many characteristic features of metaphase. Metaphase arrest with abnormally condensed chromosomes has also been observed in other ORC and replication protein mutants (24, 29). It appears that in all of these cases, insufficient levels of replication proteins due to gradually depleted maternal deposits result in incomplete DNA replication and broken chromosomes that would lead to cell cycle arrest at the early stages of mitosis by a mechanism sensitive to chromosomal integrity.
Recent studies on an Orc1 mutant suggested that ORC may be dispensable for the endoreplication in Drosophila (30). Our analysis of the Orc6-deletion mutant similarly revealed that despite DNA replication defects observed in Drosophila brain cells, the development and DNA synthesis in salivary glands were similar to the wild-type or heterozygous cells. Western blotting experiments did not reveal detectable Orc6 protein in salivary glands isolated from homozygous mutant animals. It is possible that as maternal supplies of Orc6 are depleted, the Western blotting analysis may not be sensitive enough to detect the residual Orc6 protein. Immunofluorescence experiments, however, consistently showed Orc6-specific bands throughout polytene chromosomes at the same stage. The number of the Orc6-positive bands was ≈5- to 10-fold lower in polytene chromosomes derived from Orc6-deletion mutant cells than in chromosomes from control wild-type or heterozygous cells, but they were always detectable with many independent batches of affinity-purified rabbit polyclonal or mouse monoclonal antibody against Drosophila Orc6. Thus, we cannot at this point exclude the possibility that the residual maternal deposits of Orc6 protein are sufficient to support endoreplication in the salivary glands, augmented by a lower turnover of ORC in polyploid cells than in mitotically active cells.
From our earlier biochemical and cell culture studies of Orc6 activities, we concluded that Orc6 protein in Drosophila consisted of 2 functional domains important for DNA binding and replication (18), as well as for cytokinesis, through interaction with the septin protein Pnut (22, 23). In the current study, we wanted to address the functions of Drosophila Orc6 in a living organism. We found that only the constructs containing a full-length Drosophila Orc6 gene, but not its C-terminal mutants, were able to rescue orc6− animals to viability, indicating that both domains of Orc6 are important for animal survival. C-terminal mutants of Orc6 were active in replication and DNA binding in vitro and in cell culture assays (18, 22). Therefore, a priori we expected to observe at least partial rescue of DNA replication in developing larval tissues expressing these mutants. Indeed, constructs containing Drosophila Orc6-220 deletion mutant and Orc6-WK228AA, and also human Orc6, were able to rescue DNA replication in brain neuroblasts of orc635 larvae. Cells arrested previously at G1 stage were now able to pass G1 arrest block and undergo a round of DNA replication. However, brains in orc635 animals containing these transgenes were still significantly smaller and undeveloped compared with the wild-type or heterozygous flies. Imaginal discs were also not detected, suggesting that normal cell cycle progression and tissue development were not restored. Consistent with this observation, we found a significant increase in neuroblasts arrested during mitosis, indicating that the N-terminal domain of Orc6 is not sufficient to rescue the mitotic function of the protein. It is also interesting to note that the Orc6-200 mutant, which was active in DNA binding and replication in vitro, failed to rescue DNA replication in our current in vivo study. This shorter Orc6-200 protein was also less tightly associated with core ORC1-5 subunits during reconstitution and purification assays, suggesting that this particular deletion may extend deeper into the replicative domain of the protein, resulting in observed replication defects in live animals.
Orc6 is the least conserved ORC subunit; however, human Orc6 protein was able to rescue DNA replication in Drosophila larval brains lacking the endogenous Orc6. Orc6 in human cells is loosely bound to core ORC and also found to be dispensable for DNA binding and DNA replication on Orc2-depleted Xenopus egg extracts (21, 34). Other studies have shown that Orc6 is important for DNA replication, because depletion of the protein from human cells results in DNA replication defects (7). In our current studies the expression of human Orc6 in Drosophila orc6− cells relieved an observed G1 arrest in orc635 mutants, resulting in a rescue of DNA replication. The ability of human Orc6 protein to support DNA replication in Drosophila cells suggests that the 2 proteins are homologous in replication function and also provides an opportunity for further molecular dissection of human Orc6 in vivo, by using Drosophila as a model system.
Despite their ability to replicate DNA, the orc635 mutant cells carrying the Orc6-220, Orc6-WK228AA, and human Orc6 transgenes accumulated at the stage of mitosis with the significant increase in the number of mitotic cells, compared with orc6-null mutant. Many cells were arrested with metaphase-like figures with uncondensed and broken chromosomes, suggesting that DNA may be underreplicated in these mutants. Arrest at metaphase also suggests that C-terminal Orc6 mutants may be defective during ORC-dependent assembly of pre-RC at the final stages of mitosis. It is possible that the N terminus of Orc6 is necessary, but not wholly sufficient, to rescue all Orc6 functions in DNA replication. However, we also found cells at different mitotic stages, which suggests that some cells escape metaphase arrest. Furthermore, the appearance of multinucleated and polyploid cells as well as cells with multiple centrosomes indicates that cell cycle progression through mitosis and cytokinesis is defective in cells carrying C-terminal mutations of Orc6 protein. The role of ORC subunits in coordinating DNA replication and centrosome copy number in human cells has been reported recently (35).
Overall, our data provide evidence in a living organism that DNA replication function of Orc6 is associated mainly with its N-terminal domain, whereas the C-terminal domain is necessary for the passage through the M phase. The whole Orc6 protein, however, is required for the survival of Drosophila. DNA replication function of Orc6 can also be rescued by human Orc6 protein, suggesting the conservation of replicative function among metazoan species.
Materials and Methods
Generation of Orc6-Deletion Mutant.
Deletions of Orc6 gene have been generated by imperfect excision of P element-based transposon P{EPgy2}EY04071 (FBti0027288). This transposon is mapped 71 bp upstream of the Orc6 start codon, according to the sequence information released by the Gene Disruption Project. To initiate excision, the homozygous females y1w67c23; P{w+mCy+mDint2=EPgy2}Ey04071 (Bloomington stock 15714) were crossed to males of jump stock y1w1118; CyO, PBac{w+mC=Delta 2-3. Exel}2/amosTft, bearing transposase on a second chromosome, marked by Curly. F1 Curly male progeny y1w67c23; P{w+mCy+mDint2=EPgy2}Ey04071/CyO, PBac{w+mC=Delta 2-3. Exel}2 were collected and crossed to y1w1118; If/CyO females. The resulting F2 progeny were screened for white-eyed flies. White-eyed flies were crossed individually to y1w1118; If /CyO to set up stocks. Genomic DNA of these mutants was isolated and analyzed by PCR with primers mmup7: 5′-GAGTACCTCGACCATGTTGCC-3′ and mmdown1: 5′-AATGCCACGACGCTGTTTCTG-3′ or mmup2: 5′-CAGGCTATCCCGCTTAAACACC-3′ and ORC6-H-R: 5′- CTAAGCCTCGAGAAGCTGGC-3′. PCR fragments were cloned by using TOPO TA or Zero Blunt TOPO PCR (Invitrogen) cloning kits and sequenced.
Rescue of the Orc6 Mutant.
The orc6 native promoter was amplified by PCR from genomic DNA and inserted into the EcoRI site of the pUAS vector upstream of wild-type or mutant GFP-Orc6 genes. CG1667+Orc6 genomic clone was created by PCR of wild-type genomic DNA with primers 5′-CCACAAATATGGCACGATTGC-3′ and 5′-CGAAGACAGCGACAATGAGACG-3′. The PCR product was double-digested with EcoRI/XbaI and cloned into the pUAS vector. Obtained constructs were sequenced, purified, and injected into w1118 fly embryos. The expression of fused products was verified by Western blot analysis with anti-Orc6 antibody. In rescue experiments of the orc6 mutant, progeny from heterozygous orc6/Cy; GFP-Orc6/+ were analyzed for the presence of orc6/orc6; GFP-Orc6 flies. The percentage of rescued flies was calculated based on expected segregation.
Site-Directed Mutagenesis.
The 2-amino acid mutant Orc6-WK228AA was generated by replacing amino acids W228 and K229 with alanines following the Stratagene site-directed mutagenesis protocol (www.stratagene.com/manual/200516.pdf).
Third-Instar Larval Brain Squashes.
Brains of third-instar larvae were squashed in acetoorcein without colchicine treatment and hypotonic shock, following a general protocol described previously (37). This procedure allows observation of all phases of mitosis.
BrdU Labeling and Immunostaining of Third-Instar Larval Brains.
Larval brains were soaked in PBS with 1 μM BrdU for 30 min at 25 °C. BrdU incorporation was detected by using a monoclonal antibody (Becton Dickinson). Immunostaining protocols of larval brains and polytene chromosomes have been described previously (18, 36).
Supplementary Material
Acknowledgments.
We thank Kirill Popov and Guillermo Marques for helpful discussions and advice. This work is supported by National Institutes of Health Grant GM69681.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0902670106/DCSupplemental.
References
- 1.Bell SP. The origin recognition complex: From simple origins to complex functions. Genes Dev. 2002;16:659–672. doi: 10.1101/gad.969602. [DOI] [PubMed] [Google Scholar]
- 2.Machida YJ, Hamlin JL, Dutta A. Right place, right time, and only once: Replication initiation in metazoans. Cell. 2005;123:13–24. doi: 10.1016/j.cell.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 3.Chesnokov IN. Multiple functions of the origin recognition complex. Int Rev Cytol. 2007;256:69–109. doi: 10.1016/S0074-7696(07)56003-1. [DOI] [PubMed] [Google Scholar]
- 4.Romanowski P, Madine MA, Rowles A, Blow JJ, Laskey RA. The Xenopus origin recognition complex is essential for DNA replication and MCM binding to chromatin. Curr Biol. 1996;6:1416–1425. doi: 10.1016/s0960-9822(96)00746-4. [DOI] [PubMed] [Google Scholar]
- 5.Rowles A, et al. Interaction between the origin recognition complex and the replication licensing system in Xenopus. Cell. 1996;87:287–296. doi: 10.1016/s0092-8674(00)81346-x. [DOI] [PubMed] [Google Scholar]
- 6.Chesnokov I, Gossen M, Remus D, Botchan M. Assembly of functionally active Drosophila origin recognition complex from recombinant proteins. Genes Dev. 1999;13:1289–1296. doi: 10.1101/gad.13.10.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prasanth SG, Prasanth KV, Stillman B. Orc6 involved in DNA replication, chromosome segregation, and cytokinesis. Science. 2002;297:1026–1031. doi: 10.1126/science.1072802. [DOI] [PubMed] [Google Scholar]
- 8.Machida YJ, Teer JK, Dutta A. Acute reduction of an origin recognition complex (ORC) subunit in human cells reveals a requirement of ORC for Cdk2 activation. J Biol Chem. 2005;280:27624–27630. doi: 10.1074/jbc.M502615200. [DOI] [PubMed] [Google Scholar]
- 9.Sasaki T, Gilbert DM. The many faces of the origin recognition complex. Curr Opin Cell Biol. 2007;19:337–343. doi: 10.1016/j.ceb.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 10.Li JJ, Herskowitz I. Isolation of ORC6, a component of the yeast origin recognition complex by a one-hybrid system. Science. 1993;262:1870–1874. doi: 10.1126/science.8266075. [DOI] [PubMed] [Google Scholar]
- 11.Newlon CS. Two jobs for the origin replication complex. Science. 1993;262:1830–1831. doi: 10.1126/science.8266070. [DOI] [PubMed] [Google Scholar]
- 12.Lee DG, Bell SP. Architecture of the yeast origin recognition complex bound to origins of DNA replication. Mol Cell Biol. 1997;17:7159–7168. doi: 10.1128/mcb.17.12.7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Semple JW, et al. An essential role for Orc6 in DNA replication through maintenance of pre-replicative complexes. EMBO J. 2006;25:5150–5158. doi: 10.1038/sj.emboj.7601391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen S, de Vries MA, Bell SP. Orc6 is required for dynamic recruitment of Cdt1 during repeated Mcm2-7 loading. Genes Dev. 2007;21:2897–2907. doi: 10.1101/gad.1596807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moon KY, Kong D, Lee JK, Raychaudhuri S, Hurwitz J. Identification and reconstitution of the origin recognition complex from Schizosaccharomyces pombe. Proc Natl Acad Sci USA. 1999;96:12367–12372. doi: 10.1073/pnas.96.22.12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dhar SK, Dutta A. Identification and characterization of the human ORC6 homolog. J Biol Chem. 2000;275:34983–34988. doi: 10.1074/jbc.M006069200. [DOI] [PubMed] [Google Scholar]
- 17.Chesnokov I, Remus D, Botchan M. Functional analysis of mutant and wild-type Drosophila origin recognition complex. Proc Natl Acad Sci USA. 2001;98:11997–12002. doi: 10.1073/pnas.211342798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Balasov M, Huijbregts RP, Chesnokov I. Role of the Orc6 protein in origin recognition complex-dependent DNA binding and replication in Drosophila melanogaster. Mol Cell Biol. 2007;27:3143–3153. doi: 10.1128/MCB.02382-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gillespie PJ, Li A, Blow JJ. Reconstitution of licensed replication origins on Xenopus sperm nuclei using purified proteins. BMC Biochem. 2001;2:15. doi: 10.1186/1471-2091-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vashee S, Simancek P, Challberg MD, Kelly TJ. Assembly of the human origin recognition complex. J Biol Chem. 2001;276:26666–26673. doi: 10.1074/jbc.M102493200. [DOI] [PubMed] [Google Scholar]
- 21.Vashee S, et al. Sequence-independent DNA binding and replication initiation by the human origin recognition complex. Genes Dev. 2003;17:1894–1908. doi: 10.1101/gad.1084203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chesnokov IN, Chesnokova ON, Botchan M. A cytokinetic function of Drosophila ORC6 protein resides in a domain distinct from its replication activity. Proc Natl Acad Sci USA. 2003;100:9150–9155. doi: 10.1073/pnas.1633580100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huijbregts RP, Svitin A, Stinnett MW, Renfrow MB, Chesnokov I. Drosophila Orc6 facilitates GTPase activity and filament formation of the septin complex. Mol Biol Cell. 2009;20:270–281. doi: 10.1091/mbc.E08-07-0754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loupart ML, Krause SA, Heck MS. Aberrant replication timing induces defective chromosome condensation in Drosophila ORC2 mutants. Curr Biol. 2000;10:1547–1556. doi: 10.1016/s0960-9822(00)00844-7. [DOI] [PubMed] [Google Scholar]
- 25.Baldinger T, Gossen M. Binding of Drosophila ORC proteins to anaphase chromosomes requires cessation of mitotic cyclin-dependent kinase activity. Mol Cell Biol. 2009;29:140–149. doi: 10.1128/MCB.00981-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Landis G, Kelley R, Spradling AC, Tower J. The k43 gene, required for chorion gene amplification and diploid cell chromosome replication, encodes the Drosophila homolog of yeast origin recognition complex subunit 2. Proc Natl Acad Sci USA. 1997;94:3888–3892. doi: 10.1073/pnas.94.8.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pinto S, et al. latheo encodes a subunit of the origin recognition complex and disrupts neuronal proliferation and adult olfactory memory when mutant. Neuron. 1999;23:45–54. doi: 10.1016/s0896-6273(00)80752-7. comment 1–2. [DOI] [PubMed] [Google Scholar]
- 28.DiAntonia A. Learning something ORIGINal at the Drosophila neuromuscular junction. Neuron. 1999;23:1–2. doi: 10.1016/s0896-6273(00)80743-6. [DOI] [PubMed] [Google Scholar]
- 29.Pflumm MF, Botchan MR. Orc mutants arrest in metaphase with abnormally condensed chromosomes. Development. 2001;128:1697–16707. doi: 10.1242/dev.128.9.1697. [DOI] [PubMed] [Google Scholar]
- 30.Park SY, Asano M. The origin recognition complex is dispensable for endoreplication in Drosophila. Proc Natl Acad Sci USA. 2008;105:12343–12348. doi: 10.1073/pnas.0805189105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Killian A, et al. Inactivation of the RRB1-Pescadillo pathway involved in ribosome biogenesis induces chromosomal instability. Oncogene. 2004;23:8597–8602. doi: 10.1038/sj.onc.1207845. [DOI] [PubMed] [Google Scholar]
- 32.Thomae AW, et al. Interaction between HMGA1a and the origin recognition complex creates site-specific replication origins. Proc Natl Acad Sci USA. 2008;105:1692–1697. doi: 10.1073/pnas.0707260105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Du YC, Stillman B. Yph1p, an ORC-interacting protein: Potential links between cell proliferation control, DNA replication, and ribosome biogenesis. Cell. 2002;109:835–848. doi: 10.1016/s0092-8674(02)00773-0. [DOI] [PubMed] [Google Scholar]
- 34.Dhar SK, Delmolino L, Dutta A. Architecture of the human origin recognition complex. J Biol Chem. 2001;276:29067–29071. doi: 10.1074/jbc.M103078200. [DOI] [PubMed] [Google Scholar]
- 35.Hemerly AS, Prasanth SG, Siddiqui K, Stillman B. Orc1 controls centriole and centrosome copy number in human cells. Science. 2009;323:789–793. doi: 10.1126/science.1166745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sullivan W, Ashburner M, Hawley RS. Drosophila Protocols. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2000. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.