Abstract
It is commonly accepted that proteins have evolutionarily conserved 3-dimensional structures, uniquely defined by their amino acid sequence. Here, we question the direct association of structure to sequence by comparing multiple models of identical proteins. Rapidly growing structural databases contain models of proteins determined independently multiple times. We have collected these models in the database of the redundant sets of protein structures and then derived their conformational states by clustering the models with low root-mean-square deviations (RMSDs). The distribution of conformational states represented in these sets is wider than commonly believed, in fact exceeding the possible range of structure determination errors, by at least an order of magnitude. We argue that differences among the models represent the natural distribution of conformational states. Our results suggest that we should change the common notion of a protein structure by augmenting a single 3-dimensional model by the width of the ensemble distribution. This width must become an indispensible attribute of the protein description. We show that every protein contains regions of high rigidity (solid-like) and regions of high mobility (liquid-like) in different and characteristic contribution. We also show that the extent of local flexibility is correlated with the functional class of the protein. This study suggests that the protein-folding problem has no unique solution and should be limited to defining the folding class of the solid-like fragments even though they may constitute only a small part of the protein. These results limit the capability of modeling protein structures with multiple conformational states.
Keywords: conformational ensemble, conformational states, protein folding
Our basic understanding of the structure of a protein has been radically changing with time (1, 2). The initial notion that proteins are basically unstructured has been replaced by the notion of uniquely and beautifully folded rigid structures (3, 4). Currently, even this notion is being gradually modified to include elements of mobility necessary for protein function (5, 6). Understanding the protein function at the molecular level is an extremely daunting problem in biology (7, 8).
Structural studies of proteins (rigid models) provided important results leading to a better understanding of many biological processes. However, proteins must undergo significant energy and volume fluctuations (9). Despite the mounting evidence that changes in protein structures are necessary to produce a desired function (10), the dominant paradigm of protein structure-function relationship is still based on the concept of a rigid protein with a unique structure. Pictures of solid, apparently rigid structures dominate textbooks, scientific magazines, and even the popular press. Although crystallographers at large are well aware of conformational variability of proteins, a change of a paradigm is gradual as expressed in a series of recent papers (11, 12). Moreover, recent discoveries of intrinsically disordered proteins have increased our awareness of the flexibility of protein structures (13).
One of the main reasons proteins are represented by solid, rigid bodies is that the dominating experimental technique of structural biology is X-ray crystallography. This technique produces a single structure. Here, we argue that because of the rapid growth of the Protein Data Bank (PDB) (14), we now have a valuable and unanticipated window into the wide range of protein conformational flexibility. A systematic review of the accumulated protein structures has allowed us to gain insight into the structural differences between independently obtained models of identical proteins.
Availability of multiple models of the same or very closely related proteins was studied earlier to establish the principles of structure/sequence co-conservation. In a classical paper, Chothia and Lesk (15) explored the divergence of structures with reduced homology. Brian Matthews (16) investigated the resilience of structure to sequence changes and Martin Karplus (17, 18) investigated conformational variability of the crystal structures of a single protein. Only recently the availability of multiple structures allowed us to explore this subject in a more quantitative manner (19). However, none of these studies addressed the question of the structural uniqueness and identity of the individual protein nor performed a comprehensive review of the available structures of identical proteins present in the PDB.
Although some experimental techniques, such as NMR, provide direct measures of the flexibility of a protein (20), X-ray crystallography provides only limited information about protein mobility. There is, however, an additional source of information about a protein's mobility that only recently has come into focus (21). When 2 (or more) models of the same protein are deposited into the PDB, they can vary by as much as 0.1–0.4 Å (14). When the conditions vary the changes can reach tens of angstroms (22). Such “redundant” depositions now dominate the PDB (≈50,000 depositions represents ≈15,000 proteins) (14).
This redundancy is largely ignored in most large-scale analyses of protein structures, and a given analysis is usually performed on non-redundant (or “culled”) sets of PDB proteins (23, 24). Non-redundant sets are very useful for some purposes, such as classifying proteins into fold groups but, as we argue here, not for other purposes, such as analyzing the conformational ensemble for a single protein (25, 26). To investigate the latter, we have prepared a redundant set of proteins by specifically selecting clusters of independently solved models of the same protein. This particular database will be the focus of most of our analyses, as described in the Research and Methods of this paper.
A recent paper explored this database of “redundant” protein structure and reported the discovery of dual-personality (DP) fragments that can be found in either an ordered or a disordered state among different members of the same cluster (21). The DP fragments have unique features that differentiate them from both regularly folded and intrinsically unstructured/disordered fragments. An analysis of these differences among redundant structures can provide insights into intrinsic proteins' variability and into how a protein structure reacts to small changes in its environment.
In this paper, we study the structural differences in “redundant” structures. For many types of analyses, a general description of an ensemble for a single protein would be needed (27). It is needed not only for assessing proteins' similarities, or in more general tasks such as classifying proteins or building databases of distantly homologous domains, but also in practical tasks like solving protein structures by molecular replacement. Finally, in anticipation of the next Critical Assessment of Structure Prediction (CASP) competition the description of the structural ensemble could serve as a valuable tool for properly assessing the modeling results (28).
Results
I. The Database of Redundant Protein Structures.
The database of redundant protein structures we used was generated from the collection of X-ray structure files deposited in the PDB before July 2007. The PDB is highly redundant because an average protein is represented more than 4 times. Different chains in one protein structure (the same PDB ID) were treated as separate entries, which further increased the redundancy. A total of 68,881 entries (independent protein chains) were collected for processing. After removing the data in accordance with the procedures specified in the Methods section (deposited before 1990; resolution < 2.5; R-value < 0.25), we reduced this number by 37% to 43,525 individual entries.
The clustering at the 100% level sequence identity resulted in 12,406 clusters (individual proteins). Out of these 7,206 (54%) were represented by more than 1 entry and were therefore included in our further analysis. Out of 7,206 multiple representative clusters about 50% had a single pair of structures to compare. The size of individual clusters varied from approximately 220 to 2 structures in the cluster. Approximately 600 clusters contained more than 10 structures in the cluster; the majority of the clusters in the database contained less than 10 structures in the cluster.
The total number of non-redundant pairs used in the analysis was 220,345. The size of the proteins varied from less than 100 residues to approximately 1,500 residues. The largest number of structures and the resulting pairs was in the range of 100–400 amino acids in length, representing 5,298 clusters (individual proteins) that constituted 73% of the clusters.
II. Distribution of RMSDs.
Our analyses were constructed using the most commonly accepted measure of similarity of protein structures: the root mean square deviations (RMSD) calculated between backbone atoms of selected pairs of structures. The results (Fig. 1 and S1) showed a large divergence in the RMSDs in individual clusters, as well as in the entire database. The 47,615 pairs, representing 3,720 clusters, had RMSDs larger than 1 Å. The largest divergence measured was 23.4 Å. An example of the RMSD frequency distribution in cluster 29 is presented in Fig. S1. The plot shows several maxima that correspond to different conformational substates, represented by 7 models in the cluster. The distribution of the entire set of RMSDs representing all of the clusters (Fig. 1) shows a large peak near 0.3 Å (compatible with crystallographic errors estimated to be ≈0.25 Å) but with a large shoulder that extends to 24 Å (Fig. 1). Different scales in the Insets show the details of the extended tail of the distribution with several smaller maxima around 3, 7 10, and 15 Å. These maxima originated from a relative overabundance of individual structures in clusters that have large conformational changes. The logarithmic scale accentuates the relationship, suggesting a hidden scaling principle of structural divergence.
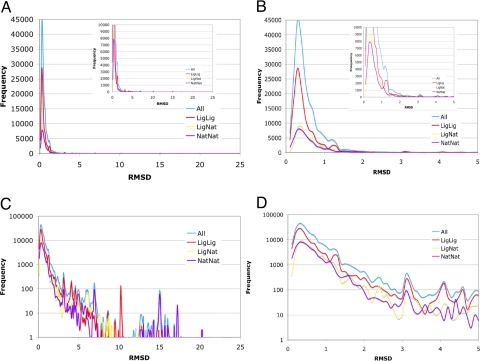
Fig. 1.
Global distributions of pair-wise RMSDs combined for all of the clusters. The distribution is wide and maximum reaches 23.7 Å. Panels A and C show the distributions within the maximal range of 24 Å and panels B and D within 5 Å RMSD. The blue line represents all-to-all RMSDs, red line ligand-ligand, yellow line ligand-native RMSDs, and dark-blue line native-native RMSDs distribution. Insets show the smaller frequency scale to show similarity of the different distributions regardless of the scale. Panels C and D show the same plots as in A and B with frequency in logarithmic scale.
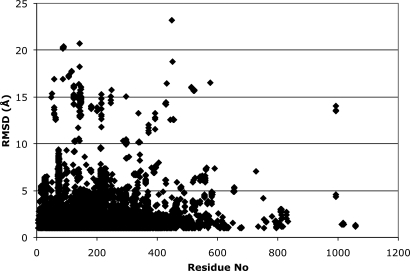
It is commonly believed that upon binding of a ligand the structures rigidify, and therefore, one would expect that the distribution calculated for proteins only containing a ligand should be different with a diminished contribution of large RMSDs. To investigate this possibility we identified the structures containing non-metal ligands. We subdivided the entire set of models into 2 subsets, liganded and unliganded (or native). Subsequently, we calculated the distributions within each of these states. In Fig. 1, we present the distributions for all RMSDs, ligand-containing structures, unliganded structures, and RMSDs between liganded and nonliganded. Contrary to our expectation, all those distributions appear to have the same shape on a linear as well as on a logarithmic scale, suggesting that the distribution has a universal character. Another naive expectation is that the RMSD should increase with the length of the protein. Again, contrary to this expectation, there is no clear dependence of the RMSD values versus the length of the proteins (Fig. 3). This fact testifies to the universal character of the obtained distributions as they describe the proteins' design principles versus the errors of experimental methodologies.
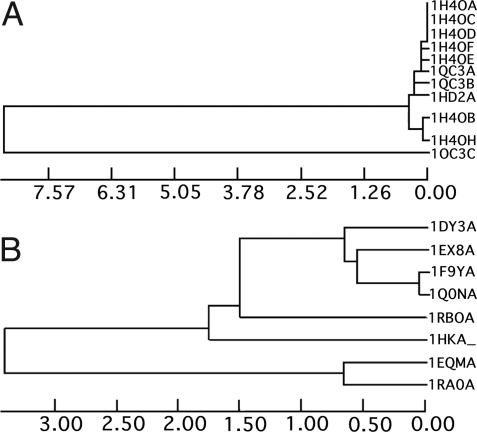
Fig. 3.
Example of trees of conformational states obtained by subclustering the RMSDs in individual cluster.
Fig. 2.
Plot of the RMSDs versus the length of the protein sequence.
III. Distribution of Conformational States.
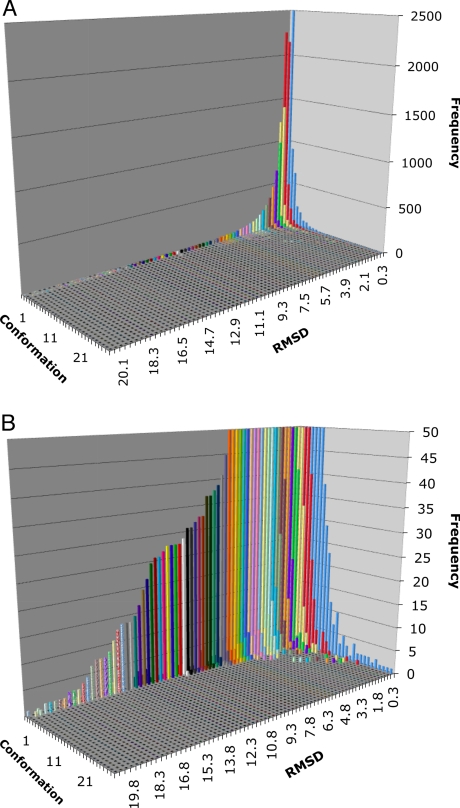
To avoid the weaknesses inherent in a direct interpretation of the RMSD distribution (overrepresentation of many similar structures and the influence of outliers), we designed a method to derive conformational states. The method relied on subclustering of models with similar RMSD values. This clustering procedure directly produced a tree of conformational states that represent individual subclusters as branches obtained at a particular level of RMSD. An example of “deep” and “shallow” trees is shown in Fig. 4. In Fig. 5, we show the global distribution of conformational states (subclusters or branches) present in all clusters in our database. The number of conformational states is derived by “cutting” the tree (represented in Fig. 4) at a particular RMSD value and counting the branches. This distribution shows a main peak centered below 1 Å RMSD and between 2–3 conformational states with a very wide shoulder. The main peak extends toward larger RMSDs and a higher number of conformational states. The main body of the peak extends smoothly up to 5 Å RMSDs and 10 conformational states. Beyond those limits the distribution levels off and decays at approximately 25Å and 50 conformational states.
Fig. 4.
Global distribution of the conformational states of all clusters obtained by subclustering the RMSDs in individual cluster. A single bar represents the number of clusters with the given number of subcluster (conformational states) in reference to the RMSD cutoff at which the number was obtained. (Δ) The global distribution in full scale. (R) The same distribution as in A with a scale of frquency limited to 50 models.
Fig. 5.
Global distribution of the conformational states of all clusters obtained by subclustering the RMSDs in individual cluster. A single bar represents the numberof clusters with the given number of subcluster (conformational states) in reference to the RMSD cutoff at which the number was obtained. (A) The full scale distribution of conformational states in the PDB, (B) The samedistribtion with frequency scale limited to 50.
IV. Sliding RMSD.
An individual crystal structure can be interpreted as a snapshot taken from a full ensemble of possible conformational states. This interpretation leads us to ask about the physical origin of the structural divergence. Is a protein structure deforming elastically or is it comprised off a small number of rigid components moving against each other? We designed a technique to address this question. We calculated the RMSD of a small fragment (for instance “window RMSD” of 25 amino acids) and then slid the superposition window by 10 residues along the sequence. If the fragments are relatively rigid, the window RMSD plotted along the sequence shows broad valleys with low RMSD. However if a hinge is encountered in the structure, a sudden increase in RMSD is observed. The method is therefore sensitive to sudden and localized changes between compared proteins. The sensitivity of the method depends on the size of the sliding fragment and the rate of sliding. Use of a larger fragment imparts a greater averaging effect on small changes and accentuates large conformational changes.
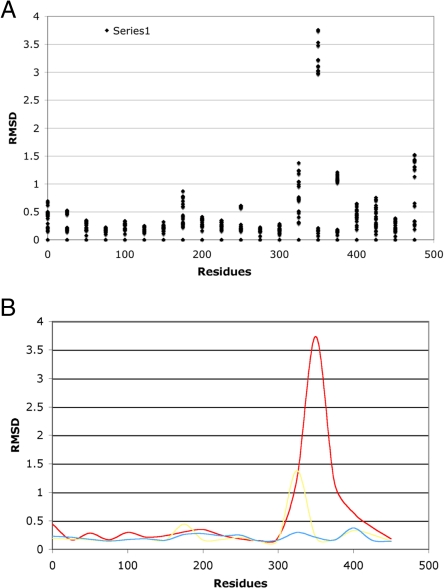
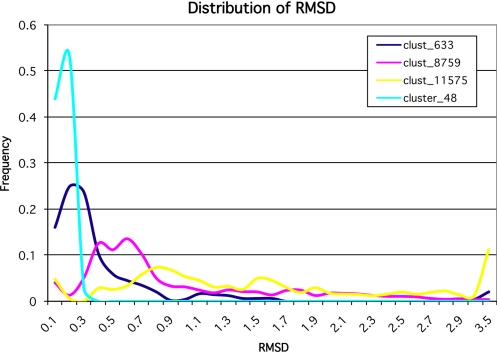
The method was applied to several clusters. The clusters were selected to show different type of a protein's behavior such as the size of the maximal RMSD and the number of conformational states. The results for clusters 48, 633, 8,791, and 11,575 are presented in Figs. 5 and S2. Cluster 48 represents a very large but rigid molecule (transhydroxylase) with a maximum RMSD of approximately 0.5 Å and only a single conformational state at the RMSD approximately 0.5 Å. Cluster 633 represents a large but less rigid molecule (diphtheria toxin) with 2 conformational states at the level of 1 Å. The structure has a single hinge motion. Cluster 8,791 represents calmodulin, which has a large number of conformational states and a 2-domain structure. Finally, cluster 11,575 represents a fragment of apolipoprotein A, a highly mobile structure with the largest number of conformational states detected in this study.
Fig. 5 and Fig. S2 shows 2 panels for each structure. Both show the sliding window RMSD as a function of a window position in the protein. The first panel shows a collection of all RMSDs for the particular sliding window position. A collection of dots represents clustering of conformational states and the magnitude of RMSD. The clustering of dots at the bottom of the figure describes a fragment of high rigidity. The clustering of dots at the top of the figure defines a hinge in the structure (represented by several models). The second panel shows the sliding RMSD for 3 pairs selected to illustrate a small, intermediate, and large structural divergence.
As expected from the maximal RMSD for the cluster 48, the divergence of RMSDs is small and the frequency of small RMSDS is very high as shown in Fig. 7, which displays the global frequency of the individual RMSDs. When the structure is rigid the distribution is narrow and the peak located in the region of small RMSD. For clusters 633, 8,791, and 11,575 the main peak gradually diminishes describing decreasing rigidity and the center of the distribution moves toward the higher RMSD values. The shift in the peak of the distribution is a direct measure of how rigid the molecule is. The distribution appears to be unique for every molecule tested and suggests that every molecule has its own optimization principle. This optimization leads to the characteristic balance between rigidity and mobility, for the particular structure. Our method provides another quantitative measure of this balance. The plots in Fig. 5 can be subdivided into regions of low and high RMSD. The number and identity of residues belonging to each category can be easily computed and the results provide the basis for a new classification regarding the contents of mobile fragments. This property appears to be directly correlated with the functional class of the protein as represented by 4 particular clusters.
Fig. 7.
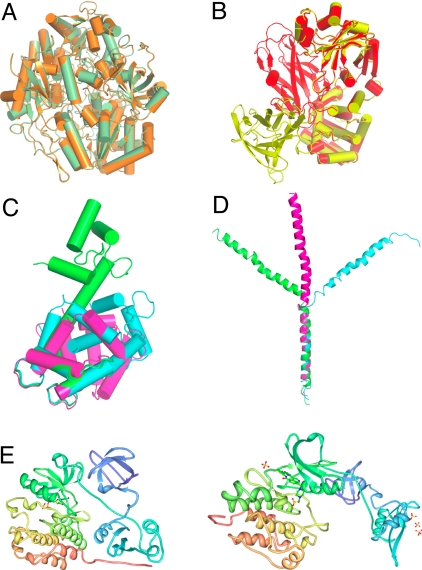
Examples of proteins representing different ‘mobility classes’ found in the PDB. (A) cluster 48 representing a large enzyme transhydroxylase (1,236 amino acids) with a single conformational state. (B) Cluster 633 representing diphtheria toxin with a single hinge that represents a movement of the entire domain shown in yellow and red models. (C) Cluster 8,791 representing calmodulin that has the entire family of different conformational states represented. Three of these states are depicted; green an open state, blue half closed state, and purple the completely closed state. (D) Cluster 11,575 representing a fragment of apolipoprotein A. Three conformational states are represented out of many available in 2 independent crystal structures. (E) The Src kinase representing the largest conformational change detected in the PDB models represented in our database that comprises 23.7 Å RMSD between models 2src and 1y57.
This classification can also be augmented by directly correlating the functional class with the number of conformational states. We have tested this idea by asking whether an individual keyword such as a “motor,” or a “transduction,” or an “enzyme” present in the PDB description record, can be directly associated with the average number of conformational states measured at a given RMSD level (for instance 0.6 Å). The preliminary results indicated that clusters having “motor” and “transduction” as keywords had a higher average number of conformational states than the remainder of the database. The keyword “enzyme” in contrast had a comparable average number of conformational states to the rest of the database.
Two caveats must be taken into account when interpreting our preliminary result: (i) the keywords in the PDB are not accurate descriptors of the proteins in individual clusters and (ii) an individual protein (cluster) can have several functional associations. For instance, the largest number of conformational states found in our database belongs to the cluster describing CDK2 kinase, an enzyme associated with signal transduction. Other proteins with a high number of conformational states were hemoglobin and insulin. Both proteins are difficult to classify in a single functional category. Nevertheless, we clearly demonstrated the utility of the above approach in classifying the proteins in the PDB. In the future, the statistical characteristics of the individual clusters relating to the number of conformational states can be used as an aid in assigning additional function to an individual protein.
V. Detailed Examples.
We studied, in detail, 4 clusters 48, 633, 8,791, and 11,575. We also focused on the SRC kinase that showed the largest structural divergence for a single protein. The clusters were selected to show the wide divergence of protein structure behavior and correlation of the functional class with the level of mobility in the individual cluster. Cluster 48 represents a large but very rigid molecule. It contains only 2 crystal structures but each has 6 independent chains so the resulting number of individual RMSDs is statistically significant. The number of sliding RMSDs is proportional to the length and number of chain pairs and therefore it is quite large. As shown in Figs. 5–7, the structure appears to be very rigid and shows only a single conformational state.
Fig. 6.
An example ofthe 25 amino acds sliding window RMSD distribution in cluster 633 (diphtheria toxin). (A) Dots represent all individual 25 a.a. RMSDs wherens B shows examples of 3 models with conformational states with small (yellow), intermediate (red), and large divergence (blue).
The second example illustrates a single conformational change. Cluster 633 represents the diphtheria toxin. The cluster is comprised of 7 structures with 3 of them having 2 independent chains. The total number of chains was comparable to that of cluster 48. The clustering procedure, as Fig. 5 clearly shows, produced 2 dominant conformational states with a hinge localized around residue 370. Fig. 5 clearly suggested by the increase in a local RMSD the presence of 2 potential hinges around residues 150 and 350. This very localized mobility created a substantial change in the sliding RMSD global distribution by shifting the maximum from approximately 0.2 Å to 0.4 Å and producing a much longer tail (Fig. 6) in the distribution.
The third example was cluster 8,791, which represents calmodulin. This very well-investigated signaling molecule is traditionally described by 2 conformational states (open and closed). The analysis of the global distribution of RMSDs as well as the sliding RMSDs provides an interesting picture. As seen in Fig. S2, calmodulin possesses a near continuum of conformational states that represent a wide range of bent angles. However, the commonly assumed stable structures of the C and N-terminal lobes appear to be much more flexible than commonly assumed, producing a significant shift in the maximum of the sliding RMSDs around 0.7 Å and with a much more pronounced shoulder. This result suggests that calmodulin has an intrinsic mobility designed into its helical segment and also into its relatively well-folded terminal domains.
The fourth example is the cluster 11,575 representing apolipoprotein A. This structure is one of the most mobile, and it does not have a defined tertiary structure. It is dominated by the helical arrangement that appears to change from molecule to molecule. This flexible design is most visible in the shift of the maximum in sliding RMSD distribution to approximately 0.9 Å, and a shoulder that represents a significant proportion of RMSDs in excess of 3.5 Å. This design strongly suggests a synergism with highly flexible structures of the phospholipids.
The largest conformational change we detected was in cluster 1,168 that represents the Src kinase. A change of almost 24 Å was detected between 2 crystal forms of the protein. The structure that can be subdivided into 4 domains undergoes a pronounced transformation with 2 pairs of domains twisting away. The changes are caused by the activation process, which releases the autoinhibitory fragment upon binding of an inhibitor. This conformational change was described in detail earlier (26).
Discussion
As indicated in the introduction, our understanding of the protein structure and its connection with the sequence has changed over the years (10). After an initial period accentuated with a common belief in unstructured proteins, a new paradigm has emerged. The new paradigm declared that the sequence uniquely determined the 3-dimensional structure of the protein. Two discoveries reinforcing this notion were rewarded with Nobel prizes. Dr. Pauling received it for predicting the secondary structure organization (29), and Dr. Anfinsen was rewarded for formulating the thermodynamic theory of protein folding (30). However, later developments in the field of protein folding, especially the Paracelsus challenge (31, 32) and the discovery of intrinsically unstructured proteins (13) in combination with the existence of prions (33, 34), substantially modified the view regarding the association of a sequence with its corresponding protein structure. Additionally, an example of a complete structural ambiguity was also published recently (35).
Recently, a new paradigm emerged based on the notion of the conformational ensemble. This view has been most prominently propagated in the works of the Frauenfelder, Freire, Nussinov, and Wolynes groups (6, 36–39). The experimental hints in support of this new paradigm, were provided over the years by a variety of different techniques (40, 41), in particular by NMR (19) and other spectroscopic methods (42). Recently, single-molecule studies allowed us to glimpse into the distribution itself (ion channel opening, kinesins walking on tubulin). This latest paradigm is clearly capable of describing a full spectrum of behavior of different proteins from very stable, self-folding proteins to intrinsically unstructured proteins. This new paradigm is reflected in a recent call by crystallographers to change the representation of protein models deposited in the PDB from a single model to a multimodel representation (11). A single X-ray data set cannot describe a full ensemble of conformational states, even if during the X-ray structure determination the molecular dynamics was used (41, 43). To provide such a description multiple structure determinations must be carried out.
This study showed that a majority of proteins in the PDB have multiple conformational states. The differences between the states are significantly larger than a possible crystallographic error, or an in-the-crystal structure variation (>≈0.3 Å). Actually, only about 25% of the models of high-resolution structures represent a single conformer in the PDB. The remaining 75% show at least 2 conformers with RMSD divergence greater than 0.6 Å, but some proteins apparently have as many as approximately 40 conformational states. Some of these changes could be associated with the environmental changes such as structural changes upon ligand binding. The results of this study suggest that they represent a natural conformational distribution. This view is confirmed by the results of the calculations performed for apo-structures as well as liganded structures that produced a very similar distribution of conformational states (Fig. 1). We have concluded, in agreement with previous results (11, 12) that any individual protein cannot be described by a single model, despite the fact that the original model was obtained by X-ray crystallography. This conclusion is true even when the structural picture is supplemented by the B-factors (quasi-dynamical information).
Protein structure is, in reality, a broad ensemble of individual models (conformers) sampling a wide conformational space (20). The individual proteins differ, sometimes significantly, in the width and shape of this ensemble as it relates to function (8). Independent crystallographic experiments sample the distribution at different points, providing a lower bound estimate of its size. The last point is especially important, because every additional experimental technique will only make the distribution broader (vide NMR), and cannot make it narrower. A rapid increase in size of the PDB can only expand and further emphasize the picture we present above. This view is further strengthened if lower resolution structures present in the PDB are included.
The experiments with sliding window RMSDs allowed us to study the internal mechanism of protein flexibility. Preliminary results suggest that every protein has a unique composition of rigid (solid) and mobile (liquid) components. The simplified view of the protein as individual folding units (rigid body elements) connected by flexible loops has to be replaced with an elastic medium model in which certain fragments of the protein are stiffer than the others and the remaining fragments differ in plasticity. This view supports the success of the Gaussian network model capable of explaining internal protein mobility (44).
A varying degree of protein flexibility indicates that the classical formulation of the protein folding problem might not have a unique solution and the coexistence of many conformers at the particular set of conditions may be the case. This fact was recently suggested as a mechanism for evolving new functions and most likely for protein evolution in general (35, 45). The sequences that do not code for a particular preference in the secondary structure formation play an important role in changes of the structure and the formation of new functions (35).
Observations presented here are the tip of an iceberg. A statistical analysis of the rapidly growing body of structural information on proteins is certain to provide greater insight into the nature of the protein structure. Many more structures that are added to the PDB each day, certainly will improve chances for a better template selection for modeling of an unknown structure. However, this work defines a limit on our ability to produce a reliable model in the absence of knowledge of the entire ensemble. This work also offers a clear delineation of possible limits on our predictability of protein structures in general and associated with it capability of inferring the function. One thing is certain, that a classical paradigm of a DNA sequence defining the protein structure, which in turn defines protein function, has to be reinterpreted to provide for a broader understanding of a protein structure and biology in general.
Methods
The methodology used in this paper follows closely the one described in our previous publication (21). More details can be found in the SI Text. We constructed the database of redundant protein structures deposited in the PDB on or before January, 2007. We used the structures that fulfilled the criteria: (i) deposited after 1990; (ii) resolution higher than 2.5 Å; (iii) R-value < 0.25.
We used “SEQRES” records of all of the PDB entries to identify identical proteins and to align them using “blast2seq” program in the National Center for Biotechnology Information (NCBI) toolkit. We removed artifacts such as Sel-Met by converting to Met and removed His-tags. Subsequently, we computed the RMSD in all clusters of the same sequence proteins in an integrated environment of BOS (v3.0) (www.helixgenomics.com). The numerically close RMSDs were used to cluster models with the UPGMA algorithm (unweighted pair group method with arithmetic mean). Clustered RMSDs were used to determine the nodes of conformational speciation. To study the local structural divergence, we calculated the RMSD for 25 amino acid structural pairs by sliding it along the structure.
Supplementary Material
Acknowledgments.
This work was supported by National Institutes of Health Grant R01 GM64881 (to B.S.) and P20 Grant GM076221 to the Joint Center for Molecular Modeling (Y.Z. and A.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812152106/DCSupplemental.
References
- 1.Tanford C, Reynolds J. Nature's Robots: A History of Proteins. New York: Oxford University Press; 2001. [Google Scholar]
- 2.Pauling L. How my interest in proteins developed. Protein Sci. 1993;2:1060–1063. doi: 10.1002/pro.5560020620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Branden C, Tooze J. Introduction to Protein Structure. London: Taylor and Francis; 1999. [Google Scholar]
- 4.Lezon TR, Banavar JR, Lesk AM, Maritan A. What determines the spectrum of protein native state structures? Proteins. 2006;63:273–277. doi: 10.1002/prot.20869. [DOI] [PubMed] [Google Scholar]
- 5.Caspar DL, Clarage J, Salunke DM, Clarage M. Liquid-like movements in crystalline insulin. Nature. 1988;332:659–662. doi: 10.1038/332659a0. [DOI] [PubMed] [Google Scholar]
- 6.Frauenfelder H, Fenimore PW, Young RD. Protein dynamics and function: Insights from the energy landscape and solvent slaving. IUBMB Life. 2007;59:506–512. doi: 10.1080/15216540701194113. [DOI] [PubMed] [Google Scholar]
- 7.Petsko GA, Ringe D. Protein Structure and Function. Oxford, UK: Blackwell Publishing; 2004. [Google Scholar]
- 8.Henzler-Wildman K, Kern D. Dynamic personalities of proteins. Nature. 2007;450:964–972. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- 9.Cooper A. Thermodynamic fluctuations in protein molecules. Proc Natl Acad Sci USA. 1976;73:2740–2741. doi: 10.1073/pnas.73.8.2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morange M. The protein side of the central dogma: Permanence and change. Hist Philos Life Sci. 2006;28:513–524. [PubMed] [Google Scholar]
- 11.Furnham N, Blundell TL, DePristo MA, Terwilliger TC. Is one solution good enough? Nat Struct Mol Biol. 2006;13:184–185. doi: 10.1038/nsmb0306-184. [DOI] [PubMed] [Google Scholar]
- 12.DePristo MA, de Bakker PIW, Blundell TL. Heterogeneity and inaccuracy in protein structures solved by X-ray crystallography. Structure. 2004;12:831–838. doi: 10.1016/j.str.2004.02.031. [DOI] [PubMed] [Google Scholar]
- 13.Dunker AK, et al. Intrinsically disordered protein. J Mol Graphics Model. 2001;19:26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- 14.Berman HM, et al. The Protein Data Bank. Nucl Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chothia C, Lesk AM. The relation between the divergence of sequence and structure in proteins. EMBO J. 1986;5:823–826. doi: 10.1002/j.1460-2075.1986.tb04288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matthews BW. Structural and genetic analysis of the folding and function of T4 lysozyme. FASEB J. 1996;10:35–41. doi: 10.1096/fasebj.10.1.8566545. [DOI] [PubMed] [Google Scholar]
- 17.Kuriyan J, Osapay K, Burley SK, Brünger AT, Hendrickson WA, Karplus M. Exploration of disorder in protein structures by X-ray restrained molecular dynamics. Proteins. 1991;10:340–358. doi: 10.1002/prot.340100407. [DOI] [PubMed] [Google Scholar]
- 18.Zoete V, Michielin O, Karplus M. Relation between sequence and structure of HIV-1 protease inhibitor complexes: A model system for the analysis of protein flexibility. J Mol Biol. 2002;315:21–52. doi: 10.1006/jmbi.2001.5173. [DOI] [PubMed] [Google Scholar]
- 19.Kosloff M, Kolodny R. Sequence-similar, structure-dissimilar protein pairs in the PDB. Proteins. 2008;71:891–902. doi: 10.1002/prot.21770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindorff-Larsen K, Best RB, Depristo MA, Dobson CM, Vendruscolo M. Simultaneous determination of protein structure and dynamics. Nature. 2005;433:128–132. doi: 10.1038/nature03199. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, Stec B, Godzik A. Between order and disorder in protein structures: Analysis of “dual personality” fragments in proteins. Structure. 2007;15:1141–1147. doi: 10.1016/j.str.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mowbray SL, Helgstrand C, Sigrell JA, Cameron AD, Jones TA. Errors and reproducibility in electron-density map interpretation. Acta Crystallogr D. 1999;55:1309–1319. doi: 10.1107/s0907444999005211. [DOI] [PubMed] [Google Scholar]
- 23.Kallberg Y, Persson B. KIND-a non-redundant protein database. Bioinformatics. 1999;15:260–261. doi: 10.1093/bioinformatics/15.3.260. [DOI] [PubMed] [Google Scholar]
- 24.Wang G, Dunbrack RL., Jr PISCES: A protein sequence culling server. Bioinformatics. 2003;19:1589–1591. doi: 10.1093/bioinformatics/btg224. [DOI] [PubMed] [Google Scholar]
- 25.Hilser VJ. Modeling the native state ensemble. Methods Mol Biol. 2001;168:93–116. doi: 10.1385/1-59259-193-0:093. [DOI] [PubMed] [Google Scholar]
- 26.Schneider TR. A genetic algorithm for the identification of conformationally invariant regions in protein molecules. Acta Crystallogr D. 2002;58:195–208. doi: 10.1107/s0907444901019291. [DOI] [PubMed] [Google Scholar]
- 27.Lätzer J, Eastwood MP, Wolynes PG. Simulation studies of the fidelity of biomolecular structure ensemble recreation. J Chem Phys. 2006;125:214905. doi: 10.1063/1.2375121. [DOI] [PubMed] [Google Scholar]
- 28.Jauch R, Yeo HC, Kolatkar PR, Clarke ND. Assessment of CASP7 structure predictions for template free targets. Proteins Struct Funct Bioinf. 2007;69(Suppl 8):57–67. doi: 10.1002/prot.21771. [DOI] [PubMed] [Google Scholar]
- 29.Pauling L, Corey RB, Branson HR. The structure of proteins, two hydrogen-bonded helical configurations of the polypeptide chain. Proc Natl Acad Sci USA. 1951;37:205–511. doi: 10.1073/pnas.37.4.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 31.Rose GD. Protein folding and the Paracelsus challenge. Nat Struct Biol. 1997;4:512–514. doi: 10.1038/nsb0797-512. [DOI] [PubMed] [Google Scholar]
- 32.Dalal S, Balasubramanian S, Regan L. Protein alchemy: Changing beta-sheet into alpha-helix. Nat Struct Biol. 1997;4:548–552. doi: 10.1038/nsb0797-548. [DOI] [PubMed] [Google Scholar]
- 33.Prusiner SB. Prions. Proc Natl Acad Sci USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dobson CM. Structural biology: Prying into prions. Nature. 2005;435:747–749. doi: 10.1038/435747a. [DOI] [PubMed] [Google Scholar]
- 35.Stieglitz KA, Zhang W, Roberts MF, Stec B. Structure of the tetrameric IMPase (TM1415) from a hyperthermophile Thermotoga maritime. FEBS J. 2007;274:2461–9. doi: 10.1111/j.0014-2956.2007.05779.x. [DOI] [PubMed] [Google Scholar]
- 36.Hilser VJ, Dowdy D, Oas TG, Freire E. The structural distribution of cooperative interactions in proteins: Analysis of the native state ensemble. Proc Natl Acad Sci USA. 1998;95:9903–9908. doi: 10.1073/pnas.95.17.9903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsai CD, Ma B, Kumar S, Wolfson H, Nussinov R. Protein folding: Binding of conformationally fluctuating building blocks via population selection. Crit Rev Biochem Mol Biol. 2001;36:399–433. doi: 10.1080/20014091074228. [DOI] [PubMed] [Google Scholar]
- 38.Shoemaker BA, Wang J, Wolynes PG. Exploring structures in protein folding funnels with free energy functionals: The transition state ensemble. J Mol Biol. 1999;287:675–694. doi: 10.1006/jmbi.1999.2613. [DOI] [PubMed] [Google Scholar]
- 39.Onuchic JN, Luhey-Schulten Z, Wolynes PG. Theory of protein folding: The energy landscape perspective. Annu Rev Phys Chem. 1997;48:545–600. doi: 10.1146/annurev.physchem.48.1.545. [DOI] [PubMed] [Google Scholar]
- 40.Wilson MA, Brunger AT. The 1.0 Å crystal structure of Ca(2+)-bound calmodulin: an analysis of disorder and implications for functionally relevant plasticity. J Mol Biol. 2000;301:1237–1256. doi: 10.1006/jmbi.2000.4029. [DOI] [PubMed] [Google Scholar]
- 41.Brunger AT, Adams PD. Molecular dynamics applied to X-ray structure refinement. Acc Chem Res. 2002;35:404–412. doi: 10.1021/ar010034r. [DOI] [PubMed] [Google Scholar]
- 42.Balakrishnan G, Weeks CL, Ibrahim M, Soldatova AV, Spiro TG. Protein dynamics from time resolved UV Raman spectroscopy. Curr Opin Struct Biol. 2008;18:623–629. doi: 10.1016/j.sbi.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Levin EJ, Kondrashov DA, Wesenberg GE, Phillips GN., Jr Ensemble refinement of protein crystal structures: Validation and application. Structure. 2007;15:1040–1052. doi: 10.1016/j.str.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Erman B. The gaussian network model: Precise prediction of residue fluctuations and application to binding problems. Biophys J. 2006;91:3589–3599. doi: 10.1529/biophysj.106.090803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hilser VJ, Thompson EB. Intrinsic disorder as a mechanism to optimize allosteric coupling in proteins. Proc Natl Acad Sci USA. 2007;104:8311–835. doi: 10.1073/pnas.0700329104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.