The purification, crystallization and preliminary X-ray crystallographic analyses of the AAV9 viral capsid are reported.
Keywords: adeno-associate virus serotype 9, viruses
Abstract
Adeno-associated virus (AAV) serotype 9, which is under development for gene-delivery applications, shows significantly enhanced capsid-associated transduction efficiency in muscle compared with other AAV serotypes. With the aim of characterizing the structural determinants of this property, the purification, crystallization and preliminary X-ray crystallographic analyses of the AAV9 viral capsid are reported. The crystals diffracted X-rays to 2.8 Å resolution using synchrotron radiation and belonged to the trigonal space group P32, with unit-cell parameters a = b = 251.0, c = 640.0 Å. There are three complete viral capsids in the crystal unit cell. The orientation and position of the asymmetric unit capsid have been determined by molecular-replacement methods and structure determination is in progress.
1. Introduction
Current approaches to corrective gene therapy include the utilization of viral vectors, such as adeno-associated viruses (AAVs), with the goal of achieving tissue/cell-specific delivery. The AAVs have been isolated from a variety of different hosts including humans and nonhuman primates and belong to the Dependovirus genus in the Parvoviridae family of ssDNA viruses (Muzyczka & Berns, 2001 ▶). They are nonpathogenic and hold great promise as recombinant vectors with the ability to transduce human tissue and infect both dividing and nondividing cells (Flotte & Carter, 1995 ▶; Kwon & Schaffer, 2008 ▶). The hundreds of genomic AAV species observed in human and nonhuman primates are grouped into different antigenic clades and clonal isolates represented by AAV1–AAV9, with AAV1 and AAV6 belonging to the same clade (Gao et al., 2004 ▶). The representative members exhibit varying cell-transduction efficiencies for different tissues mediated by their capsid viral protein (VP) amino-acid sequences (see, for example, Davidson et al., 2000 ▶; Kaludov et al., 2001 ▶; Gao et al., 2002 ▶; 2005 ▶; Rabinowitz et al., 2002 ▶; Burger et al., 2004 ▶; Mori et al., 2004 ▶; Walters et al., 2004 ▶; Pacak et al., 2006 ▶). For example, vectors generated from AAV9 isolated from human tissue have been reported to have the highest transduction efficiency (a twofold to 200-fold increase) in cardiac muscle and other tissues compared with representative serotypes in rodent models (Gao et al., 2004 ▶; Bish et al., 2008 ▶; Pacak et al., 2009 ▶). Thus, it is important to understand the capsid structure of the various serotypes in order to improve their efficacy as tissue/cell-specific targeting vectors.
AAV capsids consist of a total of 60 copies of three overlapping viral proteins, VP1–VP3, which combine to form a T = 1 icosahedral capsid. These proteins are translated from the same mRNA, are present in a predicted ratio of 1:1:10 in the AAV capsid and have calculated molecular weights of ∼81, 66 and 60 kDa, respectively. The entire sequence of VP3 is present within VP2, which contains an N-terminal extension, and is present within VP1 with a unique N-terminal sequence (VP1u). Molecular and biochemical studies show that the receptor attachment and transduction phenotypes of these viruses are dictated by sequences within the common VP3 region of the three VPs, while a phospholipase A2 function located within the VP1u is required for infection. The three-dimensional structures of three AAV serotypes (AAV2, AAV4 and AAV8) have been determined by X-ray crystallography and cryo-electron microscopy and image reconstruction (Xie et al., 2002 ▶; Walters et al., 2004 ▶; Padron et al., 2005 ▶; Govindasamy et al., 2006 ▶; Nam et al., 2007 ▶) and the crystallization of several others (AAV1, AAV3, AAV5, AAV6 and AAV7) have been reported (DiMattia et al., 2005 ▶; Lerch et al., 2009 ▶; Miller et al., 2006 ▶; Quesada et al., 2007 ▶; Xie et al., 2008 ▶). This paper reports the production, purification and preliminary X-ray crystallographic studies of AAV9. The long-term goal is to structurally identify the specific regions of the capsid that determine the differences in its tissue-transduction efficiency compared with other AAVs.
2. Materials and methods
2.1. Production and purification
Recombinant AAV9 capsids were produced using the Bac-to-Bac expression system (Gibco BRL) encoding the viral proteins VP1, VP2 and VP3 as previously described for AAV1 (Miller et al., 2006 ▶). Titred baculovirus stock was used to infect Sf9 insect cells in suspension culture. Suspended cells, cultured in Erlenmeyer flasks with Sf-900 II SFM media (Gibco/Invitrogen Corporation), were infected at a multiplicity of infection of 5.0 plaque-forming units per cell with the recombinant virus, followed by incubation at 300 K for 72 h. Prior to purification, virus capsids were released from the Sf9 cells by three freeze–thaw cycles in lysis buffer (50 mM Tris–HCl pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.2% Triton X-100), including the addition of benzonase (Merck KGaA, Germany) after the second cycle.
Purification involved the clarification of the cell lysate by centrifugation at 12 000g for 15 min at 277 K. The clarified cell lysate was pelleted through a 20%(w/v) sucrose cushion (in buffer containing 50 mM Tris–HCl pH 8.0, 300 mM NaCl, 5 mM EDTA, 0.625% Triton X-100) by ultracentrifugation at 149 000g for 3 h at 277 K. The pellet from the cushion was resuspended overnight with stirring at 277 K in buffer containing 25 mM Tris–HCl pH 8.0, 300 mM NaCl, 50 mM MgCl2 and 0.625% Triton X-100. The resuspended sample was subjected to several low-speed centrifugations at 500g to remove particulate material and further purified with a sucrose-step gradient [5–40%(w/v)] by ultracentrifugation at 150 000g for 3 h at 277 K. A visible blue fraction containing empty (no DNA) viral capsids, sedimenting at ∼25% sucrose, was extracted using a syringe and dialyzed against buffer containing 50 mM Tris–HCl pH 8.5, 350 mM NaCl and 2 mM MgCl2 at 277 K. The approximate concentration of the sample was calculated from optical density measurements at 280 nm, assuming an extinction coefficient of 1.7 for protein concentration in mg ml−1. Prior to crystallization, the purity and integrity of the viral capsids were monitored using SDS–PAGE and negative-stain electron microscopy, respectively.
2.2. Electron microscopy
Purified AAV9 capsids were viewed using a Joel JEM-100CX II electron microscope (EM). 4 µl purified virus at an estimated concentration of 1.0 mg ml−1 was spotted onto a 400 mesh carbon-coated copper grid (Ted Pella Inc., Redding, California, USA) for 1 min before blotting with filter paper (Whatman No. 5). The sample was negatively stained with 4 µl 2% uranyl acetate solution for 15 s, blotted dry and viewed by the EM.
2.3. Crystallization
Crystallization conditions were screened using the hanging-drop vapor-diffusion method (McPherson, 1982 ▶) in VDX 24-well plates with siliconized cover slips (Hampton Research, Laguna Niguel, California, USA). The screening was initially performed by varying the pH and the PEG 6000, glycerol and divalent-cation concentrations and was subsequently fine-tuned using an Additive Screening Kit (Hampton Research). Crystallization drops contained 1 µl purified virus solution and 1 µl precipitant solution mixed with an additive in a 4:1 ratio. Drops were equilibrated by vapor diffusion against 1 ml precipitant solution at room temperature. The precipitant solution contained constant concentrations of polyethylene glycol (PEG) 6000, glycerol, MgCl2 and NaCl in HEPES buffer and the additive solutions were varied.
2.4. Data collection and reduction
Prior to data collection, the crystals were flash-cooled to 100 K directly from the crystallization drops since they already contained glycerol, which served as a cryoprotectant. Diffraction images were collected on the X29 beamline at the National Synchrotron Light Source (NSLS, Brookhaven National Laboratory) at λ = 1.0809 Å using an ADSC Quantum Q315 CCD detector. A crystal-to-detector distance of 450 mm was used to record 0.3° oscillation images with an exposure time of 8 s. The measured diffraction intensities were indexed, integrated, scaled and merged using the HKL-2000 suite of programs (Otwinowski & Minor, 1997 ▶).
2.5. Molecular replacement: calculation of particle orientation and position
The orientations of the AAV9 virus capsids in the crystal unit cell were determined by the molecular-replacement method using the program AMoRe (Navaza, 2001 ▶). An AAV8 60-mer (PDB code 2qa0; Nam et al., 2007 ▶) generated from a VP3 monomer by icosahedral matrix multiplication using VIPERdb (Shepherd et al., 2006 ▶) was used as a search model. The cross-rotation and translation functions were performed using observed diffraction data in the 8.0–5.5 Å resolution range. The top solutions from the translation function were refined using a rigid-body refinement option (AMoRe, fitting) with data in the 8.0–4.0 Å resolution range.
To confirm the virus-capsid orientation, a self-rotation function was performed using the GLRF program (Tong & Rossmann, 1997 ▶). The self-rotation function was computed with ∼10% of the observed data between 10.0 and 5.0 Å resolution as large terms at κ = 72°, 120° and 180° to search for the fivefold, threefold and twofold noncrystallographic symmetry elements of the icosahedral virus capsid, respectively. A radius of integration of 120 Å was used for these searches.
3. Results and discussion
3.1. Crystallization
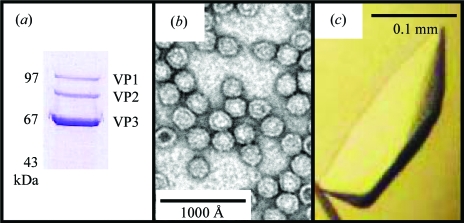
The AAV9 capsid sample purified through a sucrose gradient was greater than 95% pure as judged by SDS–PAGE (Fig. 1 ▶ a) and negative-stain electron microscopy (Fig. 1 ▶ b). From the crystallization trials, the best quality crystals (as judged by their diffraction quality, see §3.2) were obtained from crystallization conditions containing 5% PEG 6000, 100 mM NaCl, 30 mM HEPES pH 7.3, 30 mM MgCl2 and 25% glycerol with additives of 0.2 M ammonium sulfate or 20 mM EDTA. Both hexagonal and flat-shaped crystal habits were obtained in the drops containing ammonium sulfate, while elongated rod-shaped crystals (Fig. 1 ▶ c) grew in the drops containing EDTA. The crystals were observed after ∼1–2 weeks.
Figure 1.
Purification and crystallization of AAV9 viral capsids. (a) SDS–PAGE gel of the purified AAV9 sample, showing the presence of viral proteins VP1, VP2 and VP3. Note that on the SDS–PAGE gel the molecular weights of AAV9 VP1, VP2 and VP3 run slightly higher than the calculated molecular weights of 81, 66 and 60 kDa, respectively. (b) Negatively stained electron micrograph of AAV9 capsids. (c) Optical photograph of a rod-shaped AAV9 crystal of approximately 0.2 × 0.05 × 0.03 mm in size.
3.2. Data collection, processing, scaling and molecular replacement
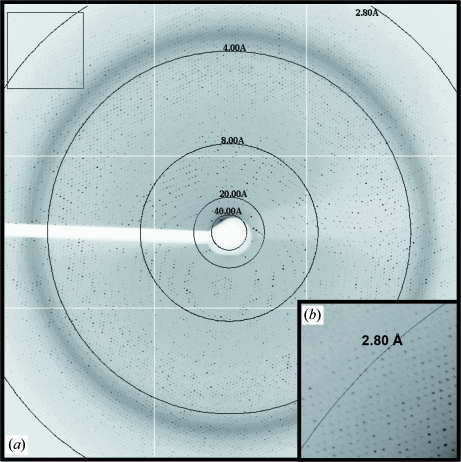
X-ray diffraction data were collected from crystals grown in the EDTA and ammonium sulfate additive conditions on the X29 beamline of NSLS, Brookhaven National Laboratory. The crystals were isomorphous and belonged to point group P3, with unit-cell parameters a = b = 251.0, c = 640.0 Å. Owing to the long c axis, the data were collected at a crystal-to-detector distance of 450 mm, which enabled resolution of the spots along this axis. Although the crystals diffracted to 2.5 Å resolution, at 450 mm data were collected to 2.8 Å resolution using an ADSC Quantum Q315 CCD detector (Fig. 2 ▶). Multiple data sets were collected and combined from a single crystal (grown in each condition) by translation to new positions once radiation damage became apparent. A more complete data set was collected from a crystal grown with the ammonium sulfate additive and the data statistics are shown in Table 1 ▶. Based on the unit-cell parameters and the P3 point group, packing considerations suggested the presence of three complete viral capsids in the unit cell and of one capsid in an asymmetric unit.
Figure 2.
X-ray diffraction image of an AAV9 crystal. (a) Image of a typical 0.3° oscillation photograph. The circles indicate resolution ranges of 40, 20, 8.0, 4.0 and 2.8 Å. (b) Close-up of the boxed region in (a) showing observed reflections to greater than 2.8 Å resolution.
Table 1. Crystal data-collection and processing statistics.
Values in parentheses are for the highest resolution shell.
| Wavelength (Å) | 1.0809 |
| Space group | P32 |
| Unit-cell parameters (Å) | a = b = 251.0, c = 640.0 |
| VM (Å3 Da−1) | 3.5 |
| No. of observations | 2183275 (173167) |
| No. of unique reflections | 960335 (72690) |
| Crystal mosaicity (°) | 0.3 |
| Resolution range (Å) | 40–2.8 (2.9–2.8) |
| Completeness (%) | 87.7 (66.4) |
| Rmerge† | 0.132 (0.361) |
| Redundancy | 2.3 (1.4) |
| I/σ(I) | 9.9 (1.7) |
R
merge = 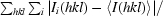
 , where Ii(hkl) is a single value of the measured intensity of the hkl reflection and 〈I(hkl)〉 is the mean of all measured values of the intensity of the hkl reflections.
, where Ii(hkl) is a single value of the measured intensity of the hkl reflection and 〈I(hkl)〉 is the mean of all measured values of the intensity of the hkl reflections.
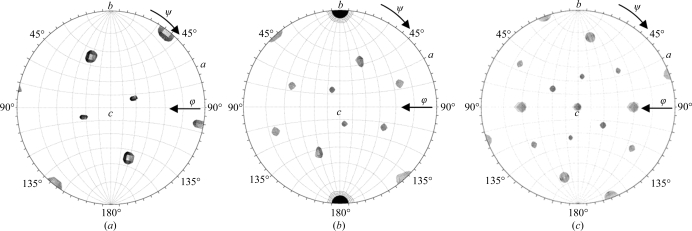
The molecular-replacement search procedures using the AAV8 capsid model in the program AMoRe (Navaza, 2001 ▶) yielded an unambiguous solution with a correlation coefficient of 0.41 and an R factor of 44% (10–4.0 Å) in space group P32, while the solution for space group P31 had a lower correlation coefficient of 0.21 and a higher R factor of 50% for data in the same resolution range. Thus, the P32 space group was used for further calculations. A self-rotation function search confirmed the orientation of the virus particle as determined by the molecular-replacement procedure (Fig. 3 ▶). Structure determination and refinement of AAV9 in space group P32 is in progress. It is anticipated that this structure of AAV9 will provide a molecular basis for the enhanced transduction efficiency of this serotype through comparison with the available AAV serotype structures and provide information that can be manipulated for improved gene-therapy applications.
Figure 3.
Stereographic projections showing self-rotation function search results for the X-ray diffraction data. The three viral capsids in the unit cell have the same orientation and the crystallographic threefold axis is parallel to the icosahedral threefold axis; thus, the self-rotation peaks from all three capsids overlap with each other. (a) The rotation function for κ = 72°, showing a search for fivefold icosahedral symmetry elements, using the observed data set in the 10–5.0 Å resolution range with a radius of integration of 120 Å. (b) The rotation function for κ = 120°, showing a search for threefold symmetry elements. (c) The rotation function for κ = 180°, showing a search for twofold symmetry elements.
Acknowledgments
The authors would like to thank the staff at the X29 beamline at the National Synchrotron Light Source (NSLS) Brookhaven National Laboratory. The NSLS is principally supported by the Offices of Biological and Environmental Research and of Basic Energy Sciences of the US Department of Energy and by the National Center for Research Resources of the National Institutes of Health (NIH). The research was funded by NIH project R01 GM082946 to RM, NM, SZ and MA-M.
References
- Bish, L. T., Morine, K., Sleeper, M. M., Sanmiguel, J., Wu, D., Gao, G., Wilson, J. M. & Sweeney, L. (2008). Hum. Gene Ther.19, 1359–1368. [DOI] [PMC free article] [PubMed]
- Burger, C., Gorbatyuk, O. S., Velardo, M. J., Peden, C. S., Williams, P., Zolotukhin, S., Reier, P. J., Mandel, R. J. & Muzyczka, N. (2004). Mol. Ther.10, 302–317. [DOI] [PubMed]
- Davidson, B. L., Stein, C. S., Heth, J. A., Martins, I., Kotin, R. M., Derksen, T. A., Zabner, J., Ghodsi, A. & Chiorini, J. A. (2000). Proc. Natl Acad. Sci. USA, 97, 3428–3432. [DOI] [PMC free article] [PubMed]
- DiMattia, M., Govindasamy, L., Levy, H. C., Gurda-Whitaker, B., Kalina, A., Kohlbrenner, E., Chiorini, J. A., McKenna, R., Muzyczka, N., Zolotukhin, S. & Agbandje-McKenna, M. (2005). Acta Cryst. F61, 917–921. [DOI] [PMC free article] [PubMed]
- Flotte, T. R. & Carter, B. J. (1995). Gene Ther.2, 357–362. [PubMed]
- Gao, G. P., Alvira, M. R., Wang, L., Calcedo, R., Johnston, J. & Wilson, J. M. (2002). Proc. Natl Acad. Sci. USA, 99, 11854–11859. [DOI] [PMC free article] [PubMed]
- Gao, G., Vandenberghe, L. H., Alvarez, R., Lu, Y., Calcedo, R., Zhou, X. & Wilson, J. M. (2004). J. Virol.78, 6381–6388. [DOI] [PMC free article] [PubMed]
- Gao, G., Vandenberghe, L. H. & Wilson, J. M. (2005). Curr. Gene Ther.5, 285–297. [DOI] [PubMed]
- Govindasamy, L., Padron, E., McKenna, R., Muzyczka, N., Kaludov, N., Chiorini, J. A. & Agbandje-McKenna, M. (2006). J. Virol.80, 11556–11570. [DOI] [PMC free article] [PubMed]
- Kaludov, N., Brown, K. E., Walters, R. W., Zabner, J. & Chiorini, J. A. (2001). J. Virol.75, 6884–6893. [DOI] [PMC free article] [PubMed]
- Kwon, I. & Schaffer, D. V. (2008). Pharm. Res.25, 489–499. [DOI] [PMC free article] [PubMed]
- Lerch, T. F., Xie, Q., Ongley, H. M., Hare, J. & Chapman, M. S. (2009). Acta Cryst. F65, 177–183. [DOI] [PMC free article] [PubMed]
- McPherson, A. (1982). Preparation and Analysis of Protein Crystals, 1st ed. New York: John Wiley & Sons.
- Miller, E. B., Gurda-Whitaker, B., Govindasamy, L., McKenna, R., Zolotukhin, S., Muzyczka, N. & Agbandje-McKenna, M. (2006). Acta Cryst. F62, 1271–1274. [DOI] [PMC free article] [PubMed]
- Mori, S., Wang, L., Takeuchi, T. & Kanda, T. (2004). Virology, 330, 375–383. [DOI] [PubMed]
- Muzyczka, N. & Berns, K. (2001). Parvoviridae: The Viruses and their Replication, 4th ed. New York: Lippincott Williams & Wilkins.
- Nam, H. J., Lane, M. D., Padron, E., Gurda, B., McKenna, R., Kohlbrenner, E., Aslanidi, G., Byrne, B., Muzyczka, N., Zolotukhin, S. & Agbandje-McKenna, M. (2007). J. Virol.81, 12260–12271. [DOI] [PMC free article] [PubMed]
- Navaza, J. (2001). Acta Cryst. D57, 1367–1372. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.296, 307–326. [DOI] [PubMed]
- Pacak, C. A., Mah, C. S., Thattaliyath, B. D., Conlon, T. J., Lewis, M. A., Cloutier, D. E., Zolotukhin, I., Tarantal, A. F. & Byrne, B. J. (2006). Circ. Res.99, e3–e9. [DOI] [PubMed]
- Pacak, C. A., Sakai, Y., Thattaliyath, B. D., Mah, C. S. & Byrne, B. J. (2009). Genet. Vaccines Ther.7, 3. [DOI] [PMC free article] [PubMed]
- Padron, E., Bowman, V., Kaludov, N., Govindasamy, L., Levy, H., Nick, P., McKenna, R., Muzyczka, N., Chiorini, J. A., Baker, T. S. & Agbandje-McKenna, M. (2005). J. Virol.79, 5047–5058. [DOI] [PMC free article] [PubMed]
- Quesada, O., Gurda, B., Govindasamy, L., McKenna, R., Kohlbrenner, E., Aslanidi, G., Zolotukhin, S., Muzyczka, N. & Agbandje-McKenna, M. (2007). Acta Cryst. F63, 1073–1076. [DOI] [PMC free article] [PubMed]
- Rabinowitz, J. E., Rolling, F., Li, C., Conrath, H., Xiao, W., Xiao, X. & Samulski, R. J. (2002). J. Virol.76, 791–801. [DOI] [PMC free article] [PubMed]
- Shepherd, C. M., Borelli, I. A., Lander, G., Natarajan, P., Siddavanahalli, V., Bajaj, C., Johnson, J. E., Brooks, C. L. III & Reddy, V. S. (2006). Nucleic Acids Res.34, D386–D389. [DOI] [PMC free article] [PubMed]
- Tong, L. & Rossmann, M. G. (1997). Methods Enzymol.276, 594–611. [PubMed]
- Walters, R. W., Agbandje-McKenna, M., Bowman, V. D., Moninger, T. O., Olson, N. H., Seiler, M., Chiorini, J. A., Baker, T. S. & Zabner, J. (2004). J. Virol.78, 3361–3371. [DOI] [PMC free article] [PubMed]
- Xie, Q., Bu, W., Bhatia, S., Hare, J., Somasundaram, T., Azzi, A. & Chapman, M. S. (2002). Proc. Natl Acad. Sci. USA, 99, 10405–10410. [DOI] [PMC free article] [PubMed]
- Xie, Q., Ongley, H. M., Hare, J. & Chapman, M. S. (2008). Acta Cryst. F64, 1074–1078. [DOI] [PMC free article] [PubMed]