Abstract
The freshwater snail Biomphalaria glabrata is closely associated with the transmission of human schistosomiasis. An ecologically sound method has been proposed to control schistosomiasis using genetically modified snails to displace endemic, susceptible ones. To assess the viability of this form of biological control, studies towards understanding the molecular makeup of the snail relative to the presence of endogenous mobile genetic elements are being undertaken since they can be exploited for genetic transformation studies. We previously cloned a 1.95 Kb BamHI fragment in B. glabrata (BGR2) with sequence similarity to the human long interspersed nuclear element (LINE or L1). A contiguous, full-length sequence corresponding to BGR2, hereafter-named nimbus (BgI), has been identified from a B. glabrata bacterial artificial chromosome (BAC) library. Sequence analysis of the 65,764 bp BAC insert contained one full-length, complete nimbus (BgI) element (element I), two full-length elements (elements II and III) containing deletions and flanked by target site duplications and 10 truncated copies. The intact nimbus (BgI) contained two open reading frames (ORFs 1 and 2) encoding the characteristic hallmark domains found in non-long terminal repeat retrotransposons belonging to the I clade; a nucleic acid binding protein in ORF1 and an apurinic/apyrimidinic endonuclease, reverse transcriptase and RNase H in ORF2. Phylogenetic analysis revealed that nimbus (BgI) is closely related to Drosophila (I factor), mosquito Aedes aegypti (MosquI) and chordate ascidian Ciona intestinalis (CiI) retrotransposons. Nimbus (BgI) represents the first complete mobile element characterized from a mollusk that appears to be transcriptionally active and is widely distributed in snails of the neotropics and the Old World.
Keywords: Biomphlaria glabrata, Schistosoma mansoni, nimbus (BgI), Non-LTR retrotransposon, Mobile genetic element, Reverse transcriptase, I clade
1. Introduction
Transmission of the parasitic blood fluke, Schistosoma mansoni, one of the causative agents of the chronic debilitating disease, schistosomiasis, persists in the tropics despite efforts in reducing its prevalence. The snail Biomphalaria glabrata is the major intermediate host responsible for transmission of human schistosomiasis in the Western Hemisphere. With schistosomiasis still prevalent in 74 countries and with an estimated 600 million people at risk, the search for an effective vaccine continues (Pearce, 2003; Lebens et al., 2004; Tran et al., 2006), as does the development of novel tools that will target the intermediate snail stage of the parasite’s life cycle.
Variations in snail-host susceptibility and parasite infectivity may have helped shape the genomes of both these organisms (Cribb et al., 2001) and several laboratories are involved in identifying genes that govern the outcome of parasite infection in the snail (Raghavan et al., 2003; Lockyer et al., 2004; Zhang et al., 2004; Bouchut et al., 2007). Data from studies using expressed sequence tags (ESTs) showed that sequences related to retrotransposons occur in B. glabrata (Raghavan et al., 2003). By identifying those genes that render a snail resistant to infection, we can envision future genetic transformation of snails with such gene(s) using mobile genetic elements (MGEs). With the success of germ line transformation in insects (Coates et al., 1998; Jasinskiene et al., 1998; Kidwell and Wattam, 1998) and mollusks (Boulo et al., 2000) using MGEs, it is conceivable that this strategy can be extended to B. glabrata in a way that will make the genetic manipulation of this snail a practical possibility.
Transposable elements are ubiquitous in eukaryotes and have emerged as one of the key molecular evolutionary forces shaping the genome (Fedoroff, 1999). Eukaryotic MGEs are grouped into two major classes (I and II) according to their mechanism of transposition (Finnegan, 1989; Charlesworth et al., 1994). Class I elements transpose via an RNA intermediate, using reverse transcriptase (RT) to copy the sequence that is transposed, while Class II elements transpose directly from DNA to DNA. The Class I elements are further subdivided into sub-classes, namely those that possess long terminal repeats (LTRs) at their ends and those that do not (non-LTRs), with LTR retrotransposons being more efficiently transposed (Eickbush, 1992).
The non-LTR retrotransposable elements or long interspersed nuclear elements (LINE or L1-like elements) are the most abundant (Eickbush, 1994; Malik et al., 1999). They comprise 17% of the human genome and play a powerful role in modulating gene expression. The presence of the L1 retrotransposon was found to reduce gene expression in a mechanism that truncated mRNA (Han and Boeke, 2004) and a synthetic construct showed a 200-fold increase in transposition compared with the original sequence (Han et al., 2004). In addition, a human L1 harboring a retrotransposon cassette was shown to effect changes in the neural progenitor cells both in vitro and in the brains of transgenic mice in vivo (Muotri et al., 2005), demonstrating that synthetic retrotransposons have the potential to be practical tools for manipulating mammalian and perhaps other genomes.
We previously identified a 1.95 Kb BamHI repetitive sequence (BGR2) related to the human L1 and Drosophila melanogaster I factor in B. glabrata (Knight et al., 1992) whose open reading frame (ORF) was found to encode an RT. Subsequent gene discovery studies using ESTs identified a cDNA transcript (Bg37) with 97% sequence identity to the previously reported BGR2 element. In addition to the identification of this transcript, varying levels of RT enzyme activity were detected in the resistant and susceptible snails before and after parasite infection (Raghavan et al., 2003). Using the cDNA Bg37 and the genomic BGR2 as probes, the complete non-LTR element, from here on named nimbus (BgI), was isolated from a B. glabrata (BS-90) bacterial artificial chromosome (BAC) library.
Very few mobile elements have been characterized in mollusks. Aside from the truncated BGR2 element (Knight et al., 1992), pearl from the cupped oyster Crassostrea virginica (molluscan class Bivalvia) is the only other putative non-autonomous DNA transposon to be identified in the phylum Mollusca (Gaffney et al., 2003). Here, we provide the discovery and characterization of nimbus (BgI), an MGE of the freshwater snail B. glabrata and the first full-length, non-LTR retrotransposon from a mollusk.
2. Materials and methods
2.1. Snails and DNA isolation
Snails used in this study were either recent field isolates or laboratory maintained. Of the laboratory maintained snails, two different B. glabrata snail stocks were used. The BS-90 stock (wild type, pigmented) isolated in Salvador, Brazil (Paraense and Correa, 1963), has been maintained in the laboratory since its isolation and serves as a stable stock that displays the parasite-resistant phenotype at any age (either as an adult or juvenile). The M-line stock (albino) was bred for high susceptibility to infection (Newton, 1955) and although genotypic variability among and within these snails exists, this stock continues to be useful as one that displays the parasite susceptibility phenotype. Recent field-isolated species of Biomphalaria (Biomphalaria tenagophila, Biomphalaria straminea, Biomphalaria alexandrina, Biomphalaria pfeifferi, Biomphalaria sudanica, Biomphalaria schrammi) or species of different genera (Bulinus truncatus, Oncomelania hupensis hupensis, Planorbis sp.) that can either serve or not serve as intermediate hosts were collected from several countries in Africa (Old World) and South America (New World). Once in the laboratory, snails were maintained at ambient temperature in aerated water and fed romaine lettuce. Prior to DNA isolation, snails were treated with ampicillin (100 mg/ml) overnight at room temperature to avoid potential contamination from resident bacteria. DNA was isolated from whole individual snails and from adult schistosomes as previously described (Knight et al., 1998).
2.2. Haemocyte cDNA library and sequencing of ESTs
Haemocyte cDNA libraries were constructed from either normal or parasite-exposed BS-90 snails. The construction of the libraries and processing of individual clones to generate ESTs have previously been described (Raghavan et al., 2003).
2.3. Probe preparation and Southern hybridization
Probes ([32P]-labeled) used for Southern hybridization were prepared from purified PCR products (572 bp for BGR2 and 272 bp for Bg37) from recombinant plasmids containing either the BamH1 genomic L1- related element (BGR2; GenBank Acc. No. X60372) or the cDNA insert of nimbus (Bg37, GenBank Acc. No. EF413179) identified from the haemocyte ESTs (GenBank Acc. No. AW740305) as described previously (Knight et al., 1992; Raghavan et al., 2003). For the Southern blots, BamHI and PstI genomic DNA digests were separated on a 1% agarose gel, blotted on to Zeta Probe™ membrane (Bio-Rad Laboratories, Hercules, CA) and probed with either the ([32P]-labeled) BGR2 or Bg37 (1 x 105 cpm/ml each of acid precipitable counts). Hybridization was performed at 65°C for 18 h and membranes washed according to standard stringent conditions (Maniatis et al., 1982). The blots were stripped between hybridizations with the two different probes and tested for residual radioactivity by autoradiography prior to re-probing.
2.3. Copy number analysis
Copy number analysis of nimbus (BgI) was performed by slot blot analysis by comparing the hybridization signal of nimbus (BgI) with that of the single copy B. glabrata ferritin gene using the same blot. Briefly, nylon membrane (Hybond N+, Amersham) cut to fit the slot-blot manifold (BioRad) was equilibrated with deionized water and 6 X sodium chloride-sodium citrate (SSC). After placing the pre-wet membrane on three layers of Whatman 3MM paper (pre-equilibrated with 6 X SSC) in the slot-blot manifold, DNA (10-fold serial dilutions from 10 μg to 1 ng in a total volume of 150 μl tris-EDTA (TE) buffer, pH 8.0) was pipetted into microcentrifuge tubes containing 12 μl of 5 M NaOH and 3 μl 0.5 M EDTA. Samples were boiled (10 min), snap-chilled on ice and neutralized (equal volume of 2 M ammonium acetate) before dispensing into wells. Samples were pulled through the slots by gentle vacuum and wells washed with 2 X SSC. After u.v. cross-linking, blots were cut into strips before hybridizing individual strips with either [32P]-labeled ferritin or the nimbus (BgI) (RT domain) as probes under standard hybridization conditions (Maniatis et al., 1982). Membranes were washed at moderate stringency (1 X SSC, 0.5% SDS at 60°C) before subjecting to autoradiography on X-ray film with intensifying screens at −70°C for 7 days. Densitometric analyses of the autoradiograph were performed on a Macintosh computer using the public domain NIH Image program (developed at the U.S. National Institutes of Health and available on the Internet at http://rsb.info.nih.gov/nih-image/). Autoradiographs were captured digitally and analyzed using the NIH ImageJ program.
2.4. PCR analysis of genomic DNA
Genomic DNA (50 ng) from the various snail isolates was processed for PCR and amplified using Taq polymerase (Promega, WI). Amplified products were resolved by gel electrophoresis (1% agarose) in TBE (0.089 M Tris-borate pH 8.0, 0.089 M boric acid, 0.002 M EDTA) buffer and visualized by u.v. trans-illumination of the ethidium-stained gel. Gene specific primers (GSP) used in PCR were purchased from Sigma Genosys (St. Louis, MO) and designed using the Accelrys GCG program, “Prime” (Deveraux et al., 1984), for the following samples: haemocyte EST (Bg37), Forward primer (Bg37for)- 5′ GCTCCATTAAACCGAACAGAC 3′ and Reverse primer (Bg37rev)- 5′CCCCGTAGATCATTGCTAAC; Genomic LINE (BGR2) RT domain, Forward primer (BGR2-3)- 5′ ATCACCGACCTACTTGCACC 3′ and Reverse primer (BGR2-4)- 5′ GATTCGGCTTACTGCCTTCC 3′ with the following conditions; denaturing: 94°C for 1 min, annealing: 56°C for 1 min, extension: 72°C for 2 min, for 30 cycles.
2.5. RT-PCR
Total mRNA was isolated from whole snails and treated with RNase- free DNase (RQ1, Promega, WI) as described previously (Miller et al., 2001). First strand cDNA was synthesized using the treated mRNA in the presence of an oligo (dT) primer, either in the presence or absence (control reactions with no RT were performed to preclude any DNA contamination) of RT (SuperScript RT, Invitrogen Life Technologies, Carlsbad, CA) as previously described (Miller et al., 2001). Second strand amplifications of the templates were performed using the same GSPs of the haemocyte EST cDNA(Bg37) as described above. As a positive control, the constitutively expressed B. glabrata myoglobin gene (Dewilde et al., 1998) was amplified in parallel using the same conditions as described previously (Miller et al., 2001) and the following primer sets: myo5′primer- 5′ GATGTTCGCCAATGTTCCC 3′ and myo3′primer- 5′ AGCGATCAAGTTTCCCCAG 3′. Another control included a no-cDNA template reaction, for both sets of amplification reactions (Bg37 and myoglobin) to rule out any contamination of the PCR reagents used. The PCR products were analyzed on a 1% agarose gel and the bands visualized by u.v. transillumination of ethidium bromide-stained gel.
2.6. Biomphalaria glabrata (BS90) BAC library and isolation of nimbus (BgI)
High molecular weight DNA was made from the head foot region of the BS90 (resistant) snail (six adults, 10 mm in diameter) after incubating overnight at room temperature in ampicillin (100 μg/ml). Dissected tissue was plunged directly into liquid nitrogen, crushed into a fine powder at −70°C before resuspending in 3 vol. of 50 mM EDTA (pH 8.0). After the addition of molten agarose at 70°C (InCert, FMC BioProducts, ME) made up in 125 mM EDTA (pH 7.5), the mixture was dispensed into moulds (500 μl) and allowed to solidify for 20 min at 4°C. DNA-impregnated agarose plugs were removed from the moulds and incubated in CTAB (2% w/v cetyl trimethyl ammonium bromide, 1.4 M NaCl, 0.2% 2-mercaptoethanol, 20 mM EDTA, 100 mM Tris/HCl pH8.0) proteinase K (2 mg/ml) lysis buffer for 24 h at 55°C. Digestion was allowed to proceed for a second day with a single change of buffer before washing the plugs at 10 min intervals (six to eight times) at 55°C in 50 mM EDTA (pH 8.0) followed by washing (five to six times) in TE buffer (10 mM Tris.HCl, 0.1mM EDTA pH 8.0) at 37°C and four times at room temperature. Washed plugs were incubated for 2 h at 37°C in TE buffer containing 1.0 mM phenylmethylsulphonyl fluoride (PMSF). After one change of buffer, incubation in protease inhibitor was continued overnight at 37°C before repeating the series of washes as described above. Washed plugs were stored indefinitely in TE buffer at 4°C and digested with restriction enzyme HindIII before ligation. Fragments between 100-50 Kb were resolved by transverse alternating field electrophoresis (TAFE; Stewart et al., 1988) before ligation into the HindIII site of the BAC vector (pBeloBAC11), performed in collaboration with Molecular Genetics (Huntsville, AL). From three separate ligation reactions 21,952 colonies were obtained with a size range of inserts between 60–88 Kb which translates approximately into two-fold coverage of the 931 Mb estimated genome size of the B. glabrata snail. Individual colonies were picked into 384 well plates (stored at −70°C in 50% glycerol) and gridded onto Luria-agar chloramphenicol (12.5 μg/μl) plates which were overlaid with nylon membranes (Zetaprobe) and probed using the [32P]-labeled PCR products from BGR2 (572 bp) or the transcript Bg37 (272 bp). DNA lifted from colonies (384-well format, high-density gridded membranes) in duplicate were processed for hybridization and washed under high stringency conditions as described in Maniatis et al. (1982). Clones that hybridized to both probes were processed for further sequencing only if signals were present in duplicate lifts and after verification by nested PCR to check for the expected size product using either BGR2 or Bg37 GSPs and DNA from the positive BAC clones as a template.
2.7. Sequence and phylogenetic analysis
DNA sequences were analyzed using the Accelrys GCG program (Deveraux et al., 1984), EMBOSS (Rice et al., 2000) and comparisons made with sequences in the protein and nucleic acid public databases using the BLAST algorithm (Altschul et al., 1990). All sequences have been submitted to GenBank with accession numbers (EF413179, nimbus cDNA Bg37; EF413180, nimbus BgI element and EF418587, complete BAC sequence BRIBAC72bg_line5). In addition, all sequences mentioned in this paper are designated with BRI numbers and clone identification numbers.
For phylogenetic analyses, the B. glabrata nimbus (BgI) RT sequence was added to previously reported alignments (Permanyer et al., 2003, 2006). The B. glabrata BGR element (that was a partial sequence of nimbus, BgI) was replaced with the complete nimbus (BgI) RT sequence (GenBank Acc. No. EF413180), the Aedes aegypti mosquI element was added to the alignment and sequences with very large gaps were eliminated from the alignment. The multiple sequences were then clustered using CLUSTAL X 1.83 (Thompson et al., 1997) with pair-wise gap penalties. Phylogenetic analysis was performed using the neighbor-joining method with corrections for multiple substitutions, excluding positions with gaps and an unrooted tree. The tree was drawn using NJ plot (Perrière and Gouy, 1996). Confidence in each node was assessed by 1,000 bootstrap replicates.
3. Results
3.1. Bg37 transcript hybridizes to the same B. glabrata Bam H1 fragment as does the L1 related clone BGR2
Knight et al. (1992) demonstrated that a major repetitive element (BGR2) in the genome of B. glabrata that is present as a 1.95Kb BamHI fragment was homologous to the human L1-like retrotransposon (Fanning and Singer, 1987). Attempts at finding the corresponding transcript for BGR2 (indicative of its active transcription) by conventional Northern blots were unsuccessful. During gene discovery studies involving haemocytes (immune effector cells), an EST (GenBank Acc. No. AW740305) corresponding to this BamH1 repetitive element was identified (Raghavan et al., 2003), offering evidence that this L1-related retrotransposon might be active in the snail. This 476 bp cDNA transcript, named Bg37, was fully sequenced and contained additional sequence extending the 5′ end than was previously available in the original BGR2 sequence. The sequence of this transcript has been submitted to GenBank with the accession number EF413179.
Because of the significant match between the Bg37 transcript and BGR2 (97% sequence similarity) we used Southern hybridization to re-examine whether the transcribed version was organized as the same BamH1 repetitive element (BGR2) (Knight et al., 1992) in the snail genome. DNA isolated from either resistant (BS-90) or susceptible (M-line) snails and the parasite (S. mansoni) after digestion (BamH1 and Pst1), was blotted and probed as described in Materials and methods. Blots were probed either with the [32P]-labeled 1.95 Kb genomic L1-like element BGR2 (Fig. 1A) or the radiolabeled 476 bp nimbus (BgI) transcript Bg37 (Fig. 1B) after stripping and re-probing. As expected, results indicated that both probes recognize the same 2 Kb BamH1 repetitive fragment within the snail genome as previously described (Knight et al., 1992). Likewise, both probes hybridized to the same repetitive ~ 3.5Kb Pst1 restricted band in the genomes of either the resistant (BS-90) or susceptible (M-line) snails. No signal was detected with these probes against S. mansoni DNA, indicating the absence of this sequence in the parasite.
Fig. 1.
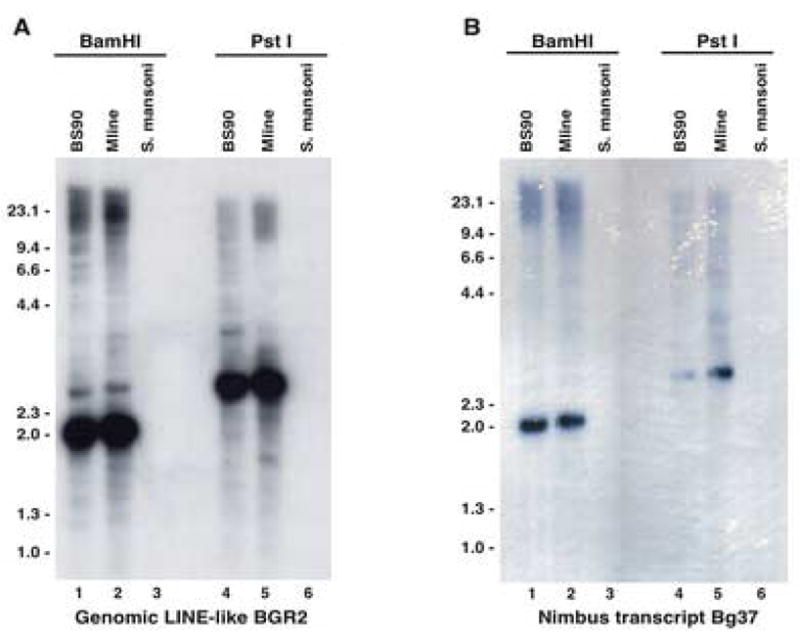
Southern hybridization of Biomphalaria glabrata. Five μg of genomic DNA from B. glabrata-resistant (BS-90) and -susceptible (M-line) snails and the parasite (Schistosoma mansoni) DNA were digested with either BamH1 or Pst1 and probed using either the [32P]-labeled 1.95 Kb insert from clone BGR2 (GenBank X60372 and PIR PC1123; Panel A) or the [32P]-labeled 476 bp nimbus (BgI) transcript Bg37 (GenBank EF413179 and EST GenBank Acc. No. AW740305; Panel B).
3.2. The presence of an active transcript corresponding to the L1 related sequence is revealed by RT-PCR
Nested GSPs based on the 476 bp haemocyte nimbus (BgI) transcript Bg37 were used to amplify a 272 bp fragment by RT-PCR using DNase -treated RNA prepared from parasite-resistant and -susceptible adult and juvenile snails (Fig. 2A lanes 2–5). Results showed that a transcript corresponding to this retrotransposon is actively expressed in both adult and juvenile snails. Myoglobin was used as the housekeeping gene (352 bp) to indicate that equivalent amounts of mRNA were used during the RT-PCR reaction (Fig. 2B lanes 2–5). Since the PCR reactions were performed to saturation, no quantitative basis could be attributed to age-related differential regulation of the L1 related Bg37 transcript. In addition, in both Figs. 2A and 2B lanes 7–10 reflect PCRs performed using Bg37 and myoglobin primers, respectively, on the four snail samples in the absence of RT to rule out DNA contamination. The lack of product from these samples (aside from primer dimers) indicates the absence of DNA contamination in the RNA samples. Lanes 6 and 11 in both Figs. 2A and 2B show the “no template” negative control to rule out any contamination from the PCR reagents.
Fig. 2.
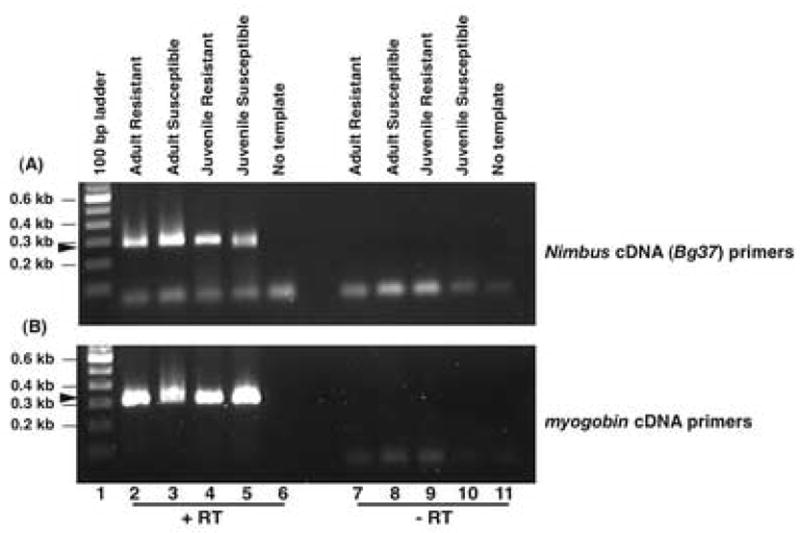
RT-PCR analysis of resistant and susceptible adult and juvenile Biomphalaria glabrata. Snail RNA was converted to cDNA by reverse transcription (RT) using nimbus (BgI) cDNA (Bg37) primers (panel A) or the housekeeping gene myoglobin (panel B). Lane 1, 100 bp DNA ladder; lane 2, adult resistant snail cDNA; lane 3, adult susceptible snail cDNA; lane 4, juvenile resistant snail cDNA; lane 5, juvenile susceptible snail cDNA and lane 6, no template control. To rule out DNA contamination in the snail RNA samples cDNA prepared without RT in the first strand synthesis was used to perform PCR using either Bg37 primers (Panel A, lanes 7–11) or myglobin primers (Panel B, lanes 7–11). Lane 7, adult resistant snail cDNA; lane 8, adult susceptible snail cDNA; lane 9, juvenile resistant snail cDNA; lane 10, juvenile susceptible snail cDNA and lane 11, no template control. Amplified products were analyzed on a 1% agarose gel. Arrows indicate the position of the PCR products for (A) nimbus (BgI) cDNA (Bg37, 272 bp) and (B) myoglobin (352 bp).
3.3. Copy number analysis of the retrotransposable elements
Copy number analysis for the retrotransposon nimbus (BgI) was deduced by slot blot analysis compared with hybridization of the same blots with the single copy ferritin gene (Adema et al., 2006). Ten-fold serial dilutions of denatured and neutralized B. glabrata BS-90 DNA from 10 μg-1 ng were spotted. The blots were cut into strips before hybridizing individual strips with either the labeled, ferritin gene or nimbus (BgI) RT as probes (Figs. 3A and B, respectively). The probe for nimbus (BgI) detected a signal in as little as 10 ng of B. glabrata DNA, while the single copy ferritin probe detected a signal at the higher concentration (1 μg) of genomic DNA. Nimbus (BgI) was detected at a 100-fold lower concentration of genomic DNA compared with the single-copy ferritin gene. This result indicated that at least 100 copies (including partial copies) were present throughout the B. glabrata genome.
Fig. 3.
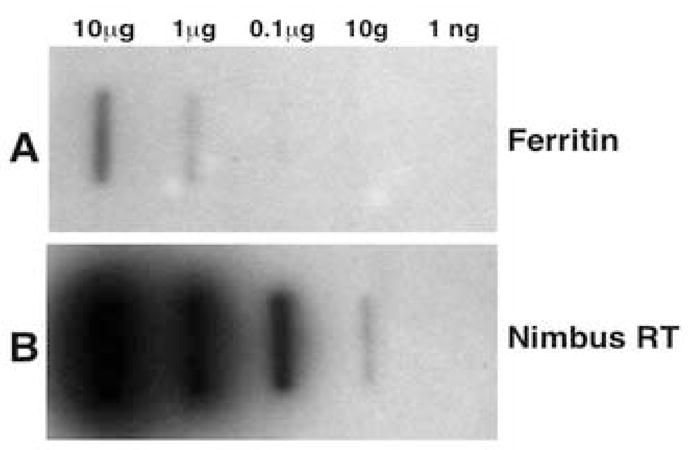
Relative distribution of the nimbus (BgI) retrotransposable element in the diploid genome of Biomphalaria glabrata (BS-90). Single copy gene (ferritin) was used for the copy number estimation of nimbus (BgI). Serial 10-fold dilutions of B. glabrata genomic DNA (10 μg-1 ng) was probed with either 100 ng of ferritin (429 bp panel A) or nimbus (BgI) RT domain (572 bp, panel B)
3.4. The L1 related retrotransposon is widely distributed among snails of different species and geographical origins
Because these studies were conducted with laboratory maintained snails, our concern was that the nimbus (BgI) retrotransposon represents an MGE unique to these snails and not to those in their natural environment. We were, therefore, interested in analyzing the distribution of this element in both field isolates of the same species and snails of other genera from different geographical regions (Old and New World). In Fig. 4, the distribution of the nimbus (BgI) retrotransposon in the genomes of 11 different snails was examined by PCR using GSPs designed from the corresponding nimbus (BgI) cDNA Bg37. Of the Biomphalaria snails examined, all are successful hosts for the transmission of S. mansoni. Geographically, B. sudanica, B. alexandrina and B. pfeifferi (lanes 9–11) are of African (Old World) origin, whereas the others are neotropical (lanes 3–8). DNA samples from snails that transmit schistosomes of other species e.g. B. truncatus (host for Schistosoma haematobium, lane 12) and O. hupensis (host for Schistosoma japonicum, lane 13) were also included in the analysis, as was DNA from Planorbis (lane 8), an organism that represents a pulmonate gastropod that is not an intermediate host for human schistosomes. As shown in Fig. 4, the PCR product (272 bp) corresponding to the expected size of the nimbus (BgI) cDNA transcript (Bg37) product was amplified from DNA from all the snails examined. The uniform distribution of this retrotransposon in all these snails, including phylogenetically distant Pomatiopsidae Oncomelania and more recent Basommatophora snails (Biomphalaria and Bulinus), provides evidence (Dejong et al., 2003) of an ancient colonization of the genomes of these snails by this L1-like retrotransposon.
Fig. 4.
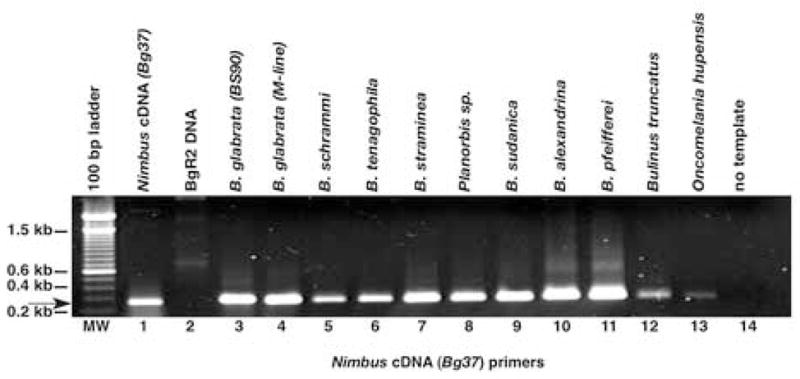
Distribution of nimbus (BgI) retrotransposon in the Old World and New World snail DNAs using cDNA (Bg37) primers. DNA was extracted from 10 different snails originating from the Old and New World. Fifty ng of DNA was used in each amplification reaction primed using nimbus (BgI) cDNA gene-specific primers (GSPs). Ten μl of each PCR product was run on a 1% agarose gel and the bands visualized using ethidium bromide. Lanes: MW, 100 bp ladder; 1, nimbus (BgI) cDNA +ve control; 2, LINE-BgR2 DNA; 3, Biomphalaria glabrata (resistant) BS-90 DNA; 4, susceptible M-line DNA; 5, Biomphalaria schrammi; 6, Biomphalaria tenagophila; 7, Biomphalaria straminea; 8, Planorbis sp.; 9, Biomphalaria sudanica; 10, Biomphalaria alexandrina; 11, Biomphalaria pfeifferi; 12, Bulinus truncatus; 13, Oncomelania hupensis; 14, No DNA –ve control. Arrow denotes 272 bp product amplified using the Bg37 GSPs.
3.5. Identification and sequence analysis of nimbus (BgI), a novel family of non-LTR retrotransposon in B. glabrata
A genomic BAC library of B. glabrata BS-90 was screened either with the [32P]-labeled nimbus (BgI) transcript Bg37 (272 bp) or the original BGR2 RT region (572 bp) amplified RT-PCR products as probes. Of the 10 BAC clones that cross-hybridized with both probes, five were subjected to preliminary BAC sequencing. Of these, one clone named BRIBAC72bg_line5 was selected for complete sequencing and closure due to the presence of a large number of RT domains. The entire BAC insert (65,764 bp, GenBank Acc. No. EF418587, Fig. 5) contained one complete, full-length nimbus (BgI) element (element I), two full-length elements containing deletions and substitutions (elements II and III) and 10 truncated copies (elements A, B and C), all highlighted in dark grey. In addition, a truncated copy of an unrelated non-LTR retrotransposon CR1 (shown in black), one truncated copy of another non-LTR retrotransposon, RTE (in the reverse orientation, shown in black) and two truncated copies of the LTR retrotransposon gypsy (highlighted in black) were found in this clone. The full-length nimbus (BgI) elements (I, II and III) displayed the characteristic hallmarks of Class 1 non-LTR retrotransposons,, ORF1 (encoding a nucleic acid binding protein) and ORF2 encoding the apurinic/apyrimidinic endonuclease (APE), RT and RNase H (RNH) domains. While element I was complete, elements II and III showed multiple deletions in ORFs 1 and 2. The other ten truncated copies of nimbus (BgI) in the BAC clone contained no ORF1 and only a partial ORF2.
Fig. 5.
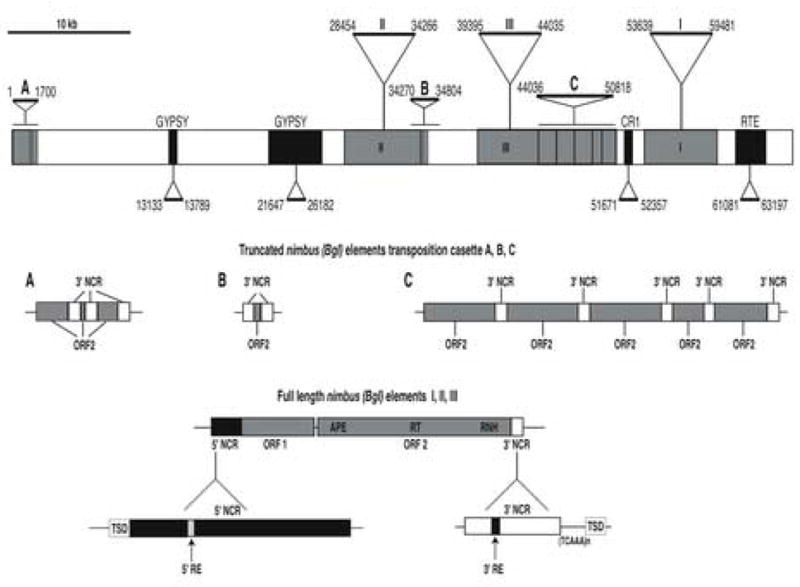
A schematic representation of the Biomphalaria glabrata bacterial artificial chromosome (BAC) bg-line 5 (GenBank Acc. No. EF418587) containing multiple copies of the transposable element nimbus (BgI). The 65,764 bp BAC insert contained one full-length-complete nimbus (BgI) element (Element I), two full-length but incomplete nimbus (BgI) elements (Elements II and III) and 10 truncated copies that are present in multiple transposition cassettes (Elements A, B, and C). All the nimbus sequences are shown in dark grey. Truncated copies of other non-long terminal repeat (LTR) retrotransposons CR1 and RTE (black) and the LTR retrotransposon gypsy (black) were also present in the BAC sequence. A 10 Kb bar scale line indicates the relative length of the BAC clone. The beginning and end position of each of the transposable elements within the BAC clone is indicated. The detailed organization of the full-length elements (I, II and III) is shown at the bottom of this Fig. Each element contains two open reading frames (ORF1 and ORF2) separated by a small intervening sequence. ORF2 contains three3 domains (apurinic/apyrimidinic endonuclease (APE), reverse transcriptase (RT) and RNase H (RNH)). Preceding ORF1 is the 5′ non-coding region (NCR) whose expanded schematic is shown in black directly below it. This region consists of a terminal 5′ target site duplication (TSD) (white), the conserved sequences of the 5′ NCR (black) with a 16 bp 5′ repeat element (RE) (white). Following ORF2 is the 3′NCR whose expanded schematic is shown in white directly below it. This region consists of a terminal 3′ TSD (white), the conserved sequence of the 3′ NCR (white) with its 16 bp 3′ RE (shown in black) and separated from the 3′ TSD by the variable tandem TCAAn or CAAn repeats. The truncated copies of nimbus are present as tranposition cassettes (A: three copies; B: two copies and C: five copies). The detailed organization of these truncated transposition cassettes are shown in panels A, B and C (grey boxes). Nucleotide sequences of the 5′ and 3′ NCRs are provided in Supplementary Fig. 1B.
The nucleotide and amino acid sequences of the full-length nimbus (BgI) retrotransposon (GenBank Acc. No. EF413179) is shown in Supplementary Fig. 1A. Blastx (Altschul et al., 1990) analysis of the coding regions (ORF1 and 2) showed significant sequence similarities (40–50%) to other non-LTR retrotransposons from Aedes aegypti (Tu and Hill, 1999), the fruit fly Drosophila melanogaster (Berezikov et al., 2000) and the sea squirt Ciona intestinalis (Permanyer et al., 2003), with E values of > e−20.
Analysis of ORF1 in the 5,869 bp complete nimbus (BgI) retrotransposon (Fig. 5, element I and Supplementary Fig. 1A) revealed the presence of zinc finger domains. ORF1, also known as the nucleic acid binding domain (NABD, defined by horizontal arrows labeled nucleocapsid), had three zinc finger domains of the CCHC type mainly found in the NABD of retroviruses. Two of the nimbus (BgI) NABD domains had the invariant sequence type CX2CX4HX4C and the other, the variant sequence CX2CX4HX6C (Supplementary Fig. 1A, underlined amino acids). Putative promoter sequences TATA and CAAT (Supplementary Fig. 1A, underlined in bold italics) were present at nucleotide positions −112 and −164, respectively, upstream of the start codon for ORF1. ORF1 was separated from ORF2 by a short stretch of 20 nucleotides before the start of ORF2. ORF2 encoded the APE (domains defined by horizontal arrows labeled endonuclease), RT (domains defined by horizontal arrows labeled reverse transcriptase) and RNH (domains defined by horizontal arrows labeled RNase H). The non-coding regions (NCRs) 5′NCR (565 bp) were present prior to the start of ORF1 and the 3′ NCR (234 bp) at the end of ORF2.
The 5′ and 3′ NCRs of all nimbus (BgI) elements were complex (Supplementary Fig. 1B). The 5′ NCR contains a highlighted 16 bp repeat element (RE) at the 5′ end (5′RE: 5′-TTTAAATTCTCACTAC-3′) that has a direct counterpart (3′RE: 5′-TTTAAATTTTTACTAC-3′) in the 3′ NCR. While we have been unable to identify a function for these REs, they are intriguing since they differ from each other with the subsitution of two “T” residues in place of “C” residues and are invariant in their positioning relative to the nimbus (BgI) element. The 3′ end of the NCR is characterized by the presence of a variable tandem repeats (TCAAn or CAAn). These features are different from the fruit fly (Fawcett et al., 1986) or mosquito (Tu and Hill, 1999) I elements that have either poly (A) or “TAA” tandem repeats. These nimbus (BgI) variable tandem repeats are AT rich (75%), and define the junction of the 3′ target site duplication (TSD) at the end of each element in cis.
Putative TSDs define the 5′ and 3′ ends of the full-length nimbus (BgI) elements. The TSDs also vary in sequence and size (Supplementary Fig. 1B) in the three full-length elements (Element I TSD: 5′-ACCATTCGACCC-3′; Element II TSD: 5′-ACATG-3′ and Element III TSD: 5′-TGACAGAGA-3′). Multiple insertions of the truncated nimbus (BgI) elements (Fig. 5, regions A, B and C) in close juxtaposition in the BAC clone made the identification of the TSDs on either ends of these elements difficult (Supplementary Fig. 1B). However, both the 5′ and 3′ ends of these truncated elements resemble the 3′ NCR of the full-length elements.
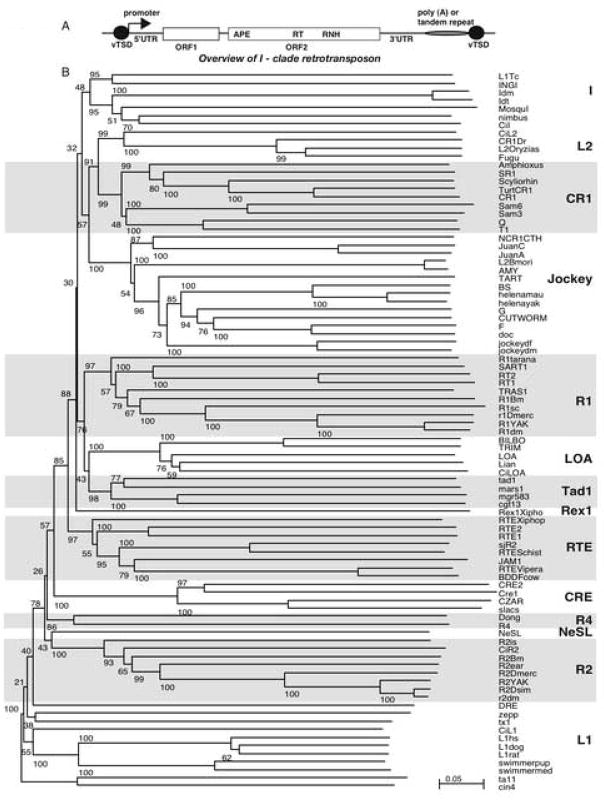
3.6. Phylogenetic analysis of the non-LTR retrotransposon nimbus (BgI)
A schematic diagram of a reprentative retrotransposable element belonging to the I-clade is shown in Fig. 6A. Phylogenetic analysis was performed using multiple sequence alignments of the RT domain (conserved regions 0–5) comprising of 440 amino acids of the nimbus (BgI) element and 88 other known elements of the 14 non-LTR clades. As shown in Fig. 6B, all clades were supported with significant bootstrap values (> 50%) using the neighbour-joining method used by the program NJ plot (Perrière and Gouy, 1996). Based on this analysis we found that nimbus (BgI) belongs to clade I (Malik et al., 1999), along with the CiI element of C. intenstinalis (Permanyer, et al., 2003), mosquI from the Aedes aegypti (Tu and Hill 1999), I factor from two different species of Drosophila (Fawcett et al., 1986), and the trypanosome L1 and INGI elements (Kimmel et al., 1987; Martin et al., 1995). Since this is an unrooted tree, the relationship between the major groups cannot be assigned definitively.
Fig. 6.
A) A schematic diagram of a representative retrotransposon of the I-clade. The general organization of an I-clade element is represented schematically showing the hallmark domains, open reading frame 1 (ORF1), coding for the nucleic acid binding domain protein, ORF2, coding for the apurinic/apyrimidinic endonuclease (APE), reverse transcriptase (RT) and RNase H (RNH). The 5′ and 3′ untranslated regions (5′ UTR and 3′ UTR), tandem repeats (regions containing either a tandem repeat or poly(A) tail) and variable target site duplication sites (vTSDs) which are direct repeats at the 5′ and 3′ ends of the element. B) Phylogenetic tree of the reverse transcriptase (RT) domain of non-long terminal repeat (LTR) elements. The elements that share an evolutionary relationship with nimbus (BgI) belong to clade I. The name of each non-LTR element is shown at the end of each branch and the clade into which they group is boxed and the clade name indicated in bold text. The multiple sequences were aligned using CLUSTAL W 1.83.1 software (gap opening=10, gap extension=0.2, gap separation distance=4) using the GONNET matrix for proteins. Four hundred and forty amino acids from the RT domain of nimbus (BgI) were used in the alignment. The initial sequences used in this alignment were obtained from Permanyer et al. (2003) (http://genomebiology.com/2003/4/11/R73). The original snail BGR sequence (GenBank Acc. No. X60372, PIR Acc. No. PC1123) has been substituted with the RT domain from the complete nimbus (BgI) (GenBank Acc. No. EF413180) element and in addition, the mosquito sequence mosquI (Acc. No. AF134900) has been added to the alignment. The phylogenetic tree shown here is unrooted, with all characters considered of equal weight and unordered, and constructed using the neighbor-joining method. Confidence in each node was assessed by 1,000 bootstrap replicates and the values are indicated next to each branch.
4. Discussion
MGEs, or transposable elements, are major components of eukaryotic genomes but are poorly understood (Craig, 2002). They are one of the principal driving forces in eukaryotic evolution (Charlesworth et al., 1994) and can disrupt genes, induce genomic rearrangements, influence gene expression and mobilize various types of non-autonomous sequences. While non-LTRs had been identified in a large number of metazoans, the lack of a large pool of sequences has hampered the identification of MGEs in mollusks. This paucity will be addressed by the sequencing of the 931 Mb snail genome (Gregory 2003) which is underway (Raghavan and Knight, 2006) and should provide us with more information on the genomic organization, copy number and distribution of this and other MGEs in the snail genome.
It was serendipitous that aa L1-like element BGR2, with an ORF encoding RT, showing similarity to human L1 (Scott et al., 1987) and drosophila I factor (Fawcett et al., 1986) was discovered in the snail B. glabrata, when genomic DNA analysis involving festriction fragment length polymorphism (RFLP) studies were being conducted (Knight et al., 1992). With the availability of snail ESTs and BAC libraries, more of these non-LTR sequences were identified and a functional but variable RT enzyme activity was also demonstrated in the snail (Raghavan et al., 2003). Using the B. glabrata EST transcript (Bg37) and the original genomic BGR2 as probes, we have now identified from a B. glabrata (BS-90) BAC library the corresponding contiguous snail non-LTR retrotransposon that we have named nimbus (BgI) to denote its potential for mobility in the snail genome. Since nimbus clusters with I factor (group V) of non-LTRs (Finnegan, 1989; Busseau et al., 1994; Berezikov et al., 2000), the designation BgI has also been included in the naming of the snail element to signify which clade it belongs to (clade I).
Southern hybridization results (Fig. 1) showing that both genomic (BGR2) and transcript (Bg37) probes hybridize to the same discrete fragments (2.0, Kb BamH1 and 3.5 Kb, PstI) demonstrates the conserved organization of both active and inactive elements in the genome. The copy number of the nimbus (BgI) element indicated the presence of ~ 100 copies distributed throughout the B. glabrata genome. While the copy number of nimbus (BgI) compared with other non-LTR elements is lower than expected, variation between copy numbers of non-LTR elements belonging to the various clades have been reported (Permanyer et al., 2006). MosquI element from A. aegypti, to which nimbus (BgI) is related, is present in only14 copies.
Nucleotide sequence analysis of the entire insert (65,764 bp) of a BAC clone that satisfied the screening strategy we adopted revealed the presence of several non-LTR sequences in this single clone. The BS-90 BAC clone contained three full-length nimbus (BgI) elements (Fig. 5, elements I, II and III) and 10 truncated copies (Fig. 5, elements A, B and C). Truncated copies of other non-LTR retrotransposons (CR1 and RTE) and the LTR retrotransposon (gypsy) were also present in the BAC sequence. It is therefore possible that this clone represents a transposition hot spot in the B. glabrata genome, a phenomenon that has been observed with the mosquito mosquI element (Tu and Hill, 1999). Transposition activity of non-LTR retrotransposons frequently generates 5′ truncated copies that are unable to transpose because they lack the coding and promoter sequences that are essential for transposition (Luan et al. 1993) and consequently evolve as pseudogenes.
Phylogenetic relationship of the various non-LTR retrotransposons can only be assigned via the common RT domain of all the elements, since this domain is absolutely required to achieve active transposition. The RT domain was used to establish the phylogeny of nimbus (BgI) and the 14 reported clades of the non-LTR retrotransposable elements. In the neighbour-joining tree (Fig. 6B), while most of the clades were supported with significant bootstrap values (≥50%), clade I showed the lowest bootstrap value (70%), in agreement with previous analyses (Malik et al., 1999; Tu and Hill, 1999; Permanyer et al., 2003, 2006). However, we believe our results are the first reported from a gastropod for the full-length sequence of a non-LTR retrotransposon that may be functional and may provide some insight into the evolutionary timeline of the colonization of invertebrate genomes by these MGEs.
The full-length nimbus (BgI) elements contained two ORFs, ORF1 and ORF2. ORF1 revealed the presence of three zinc finger motifs of the CCHC type that are characteristic of this domain ((Dawson et al., 1997; Apweiler et al., 2001). This region, usually composed of 14 amino acids and resembling the ‘zinc fingers’ of transcription factors, is the only highly conserved sequence element among the retroviral proteins (Katz and Jentoft, 1989) and is required for viral genome packaging and the early infection process. ORF2 codes for an APE, RT and RNH (Fawcett et al., 1986, Feng et al., 1998). Like other active non-LTR retrotransposons, the two ORFs from nimbus (BgI) are flanked by 5′ and 3′ NCRs and a direct repeat indicating possible TSD sites. While this site by itself is variable in both size (six to 12 nucleotides) and composition (see Results section 3.5), they are present within a larger sequence of 150 bp or more (Finnegan, 1997). The variable TSDs may be generated during the asymmetric cleavage of the target DNA sequence by the nimbus (BgI) endonuclease (APE). While most non-LTR retrotransposons are terminated at the 3′ end either by an A-rich sequence or by tandem TAAn repeats as in the fruit fly and mosquito I elements, we found variable tandem-repeats (TCAAn or CAAn) immediately preceding the 3′ TSD in both the truncated and full-length copies of nimbus (BgI). Several features about this snail element are unique among other known elements of the I-clade, These include the complex nature of the 5′ and 3′ NCRs that are highly conserved, different types of TSDs, the invariant 5′ and 3′ 16 bp REs, and the variable tandem repeats at the 3′ ends. These novel features are interesting enough to warrant further investigation. Intriguingly, the nimbus 5′ and 3′ NCRs have been discovered to be highly conserved in the genome of S. mansoni when screened against the S. mansoni whole genome shotgun database available either at the Sanger Center (using BLASTN) or NCBI (using discontiguous Mega BLAST). Significant nucleotide matches (approximately 80% identity, data not shown) between the nimbus 5′ and 3′ NCRs and parasite might be an indication of possible horizontal transfer of this sequence between the snail host and its parasite. Although several MGEs have been characterized from the schistsosomes (S. mansoni and S. japonicum; Brindley et al., 2003), none have so far been identified as belonging to the I-clade. In conclusion, nimbus (BgI) is a complete, mobile element of the freshwater snail B. glabrata and the first full-length MGE from a mollusk, a discovery that may provide insights into the snail/parasite relationship and, at a future date, be useful for genetic transformation studies aimed towards reducing snail-borne diseases.
Supplementary Material
A). The 5,869 sequence of the complete nimbus (BgI) element (GenBank EF413180). Capital letters denote coding sequences and small letters, the non-coding sequences. Position 1 in the numbering scheme is the first nucleotide of the start codon ATG (indicated by bold text) that begins the open reading frame 1 (ORF1; nucleic acid binding protein) coding region. A short, non-coding region of 20 bp separates ORF1 and ORF2. The ORF2 sequence encodes the apurinic/apyrimidinic endonuclease (APE), reverse transcriptase (RT) and RNase H (RNH) and horizontal arrows delineate these three hallmark domains. The amino acids of the various ORFs are shown in single letter codes and also labeled from 1-461 for ORF1 and 1-1,222 for ORF2. The three zinc finger regions composed of the sequence CX2CX4HX4C and the variant CX2CX4HX6C are underlined. The nimbus(BgI) coding region is flanked at either end by the 5′ and 3′ non-coding regions (NCRs) that are represented in small letters. The 3′ NCR consists of a conserved sequence containing a 16 bp repeat element (RE; bold, underlined region and Supplementary Fig. 1B) at the end of which is a stretch of “TCAAn or CAA” variable tandem repeats (highlighted in bold) that occur prior to the 3′ TSD (capital letters, boxed region) and defines the end of one element and the beginning of the next sequence. The 5′ NCR also begins with a 5′ target site duplication (TSD) (capital letters, boxed region), and contains a 16 bp 5′ RE (bold, underlined region and Supplementary Fig. 1B) and putative promoter sequences (CAAT and TATA, italics) upstream of the initiation codon of ORF1. B) Multiple alignment of the 5′ non-coding regions (NCRs) and 3′ NCRs of the nimbus (BgI) elements. The top panel shows the alignment of the 3′ NCRs of all nimbus (BgI) elements in the order they are arranged in the bacterial artificial chromosome (BAC) clone. The full-length elements are labeled I, II and III and the truncated transposition cassettes as A, B and C (the individual elements within each cassette is designated A1, A2 and A3; B1 and B2; C1, C2, C3. C4 and C5). The 3′ NCR panel is defined by the name of the element, the beginning bp, the 3′ NCR conserved domains (shown in red and blue), the 16 bp 3′ repeat element (RE) (shown in green), the variable tandem repeat (TCAAn or CAAn) region shown in orange and the start of the next region that may or may not contain the target site duplications (TSDs) (the 3′ TSDs shown in black are identifiable in full-length elements only) and the end bp position. The bottom panel shows the alignment of the three 5′ NCRs of the full-length nimbus (BgI) elements in the sequential order in which they appear 5′-3′ in the BAC clone. The full-length elements are labeled as I, II and III. The 5′ NCR panel is defined by the name of the element, the beginning bp position, the end of the previous region that contain the 5′ TSDs (black), the 5′ NCR conserved domains (blue and red) containing the 16 bp 5′ RE (shown in green), and the end bp position.
Acknowledgments
We wish to acknowledge the Biowulf PC/Linux cluster at the NIH, Bethesda, MD (http://biowulf.nih.gov) used for the sequence analysis and Dr. Peter FitzGerald for his help and support with the bioinformatics undertaken for this work. We also thank Maria Ragland for her help in the BAC library construction, Fred Lewis for his support and, Mitzi Sereno for her help in editing this manuscript. Thanks goes to the following for sending us field-isolated snails used in this study, Drs. C.P. de Souza, D. Minchella, E.S Loker and to Dr. H. Shizuya for sending us the pBeloBAC11 vector. This work was funded by; USF-Sandler Foundation Grant, 9000005939 and NIH-NIAID Grant, AI-063480-01.
Footnotes
Note: Supplementary data associated with this article (Supplementary Fig. 1A, sequence of nimbus and 1B, alignment of the 5′ and 3′ non-coding regions of nimbus).
Note: Nucleotide sequence data reported in this paper have been submitted to GenBank with the following accession numbers: EF413179 (nimbus cDNA Bg37), EF413180 (nimbus BgI element) and EF418587 (complete sequence of the BAC clone BRIBAC72bg_line5)
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adema CM, Luo M, Hanelt B, Hertel LA, Marshall JJ, Zhang S, DeJong RJ, Kim H, Kudrna D, Wing RW, Soderlund C, Knight M, Lewis FA, Caldeira RL, Jannotti-Passos LK, Carvalho OS, Loker ES. A bacterial artificial chromosome library for Biomphalaria glabrata, intermediate snail host of Schistosoma mansoni. Mem Inst Oswaldo Cruz. 2006;101(Suppl I):167–177. doi: 10.1590/s0074-02762006000900027. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Apweiler R, Attwood TK, Bairoch A, Bateman A, Birney E, Biswas M, Bucher P, Cerutti L, Corpet F, Croning MD, Durbin R, Falquet L, Fleischmann W, Gouzy J, Hermjakob H, Hulo N, Jonassen I, Kahn D, Kanapin A, Karavidopoulou Y, Lopez R, Marx B, Mulder NJ, Oinn TM, Pagni M, Servant F, Sigrist CJ, Zdobnov E. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 2001;29:37–40. doi: 10.1093/nar/29.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E, Bucheton A, Busseau I. A search for reverse transcriptase-coding sequences reveals new non-LTR retrotransposons in the genome of Drosophila melanogaster. Genome Biol. 2000;1:RESEARCH0012. doi: 10.1186/gb-2000-1-6-research0012. Epub 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchut A, Coustau C, Gourbal B, Mitta G. Compatibility in the Biomphalaria glabrata/Echinostoma caproni model: new candidate genes evidenced by a suppressive subtractive hybridization approach. Parasitology. 2007;134:575–88. doi: 10.1017/S0031182006001673. Epub 2006. [DOI] [PubMed] [Google Scholar]
- Boulo V, Cadoret JP, Shike H, Shimizu C, Miyanohara A, Burns JC. Infection of cultured embryo cells of the pacific oyster, Crassostrea gigas, by pantropic retroviral vectors. In Vitro Cell Dev Biol Anim. 2000;36:395–399. doi: 10.1290/1071-2690(2000)036<0395:IOCECO>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Brindley PJ, Laha T, McManus DP. Loukas A. Mobile genetic elements colonizing the genomes of metazoan parasites. Trends Parasitol. 2003;19:79–87. doi: 10.1016/s1471-4922(02)00061-2. Review. [DOI] [PubMed] [Google Scholar]
- Busseau I, Chaboissier MC, Pelisson A, Bucheton A. I factors in Drosophila melanogaster: transposition under control. Genetica. 1994;93:101–16. doi: 10.1007/BF01435243. Review. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Sniegowski P, Stephan W. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature. 1994;371:215–20. doi: 10.1038/371215a0. [DOI] [PubMed] [Google Scholar]
- Coates CJ, Jasinskiene N, Miyashiro L, James AA. Mariner transposition and transformation of the yellow fever mosquito, Aedes aegypti. Proc Natl Acad Sci USA. 1998;95:3748–3751. doi: 10.1073/pnas.95.7.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig NL. In: Mobile DNA II. Craig NL, Craigie R, Gellert M, Lambowitz A, editors. Washington DC: ASM press publishers; 2002. pp. 3–11. [Google Scholar]
- Cribb TH, Bray RA, Littlewood DT. The nature and evolution of the association among digeneans, molluscs and fishes. Int J Parasitol. 2001;31:997–1011. doi: 10.1016/s0020-7519(01)00204-1. [DOI] [PubMed] [Google Scholar]
- Dawson A, Hartswood E, Paterson T, Finnegan DJ. A LINE-like transposable element in Drosophila, the I factor, encodes a protein with properties similar to those of retroviral nucleocapsids. EMBO J. 1997;16:4448–4455. doi: 10.1093/emboj/16.14.4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejong RJ, Morgan JA, Wilson WD, Al-Jaser MH, Appleton CC, Coulibaly G, D’Andrea PS, Doenhoff MJ, Haas W, Idris MA, Magalhaes LA, Mone H, Mouahid G, Mubila L, Pointier JP, Webster JP, Zanotti-Magalhaes EM, Paraense WL, Mkoji GM, Loker ES. Phylogeography of Biomphalaria glabrata and B. pfeifferi, important intermediate hosts of Schistosoma mansoni in the New and Old World tropics. Mol Ecol. 2003;12:3041–56. doi: 10.1046/j.1365-294x.2003.01977.x. [DOI] [PubMed] [Google Scholar]
- Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–95. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewilde S, Winnepenninckx B, Arndt MH, Nascimento DG, Santoro MM, Knight M, Miller AN, Kerlavage AR, Geoghagen N, Van Marck E, Liu LX, Weber RE, Moens L. Characterization of the myoglobin and its coding gene of the mollusc Biomphalaria glabrata. J Biol Chem. 1998;273:13583–92. doi: 10.1074/jbc.273.22.13583. [DOI] [PubMed] [Google Scholar]
- Eickbush TH. Transposing without ends: the non-LTR retrotransposable elements. New Biol. 1992;4:430–40. Review. [PubMed] [Google Scholar]
- Eickbush TH. Origin and evolutionary relationship of retroelements. In: Morse SS, editor. Evolutionary Biology of Viruses. Raven Press; 1994. pp. 121–157. [Google Scholar]
- Fanning TG, Singer MF. LINE-1: a mammalian transposable element. Biochim Biophys Acta. 1987;910:203–12. doi: 10.1016/0167-4781(87)90112-6. Review. [DOI] [PubMed] [Google Scholar]
- Fawcett DH, Lister CK, Kellett E, Finnegan DJ. Transposable elements controlling I-R hybrid dysgenesis in D. melanogaster are similar to mammalian LINEs. Cell. 1986;47:1007–1015. doi: 10.1016/0092-8674(86)90815-9. [DOI] [PubMed] [Google Scholar]
- Fedoroff NV. Transposable elements as a molecular evolutionary force. Ann NY Acad Sci. 1999;870:251–64. doi: 10.1111/j.1749-6632.1999.tb08886.x. Review. [DOI] [PubMed] [Google Scholar]
- Feng Q, Schumann G, Boeke JD. Retrotransposon R1Bm endonuclease cleaves the target sequence. Proc Natl Acad Sci USA. 1998;95:2083–2088. doi: 10.1073/pnas.95.5.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan DJ. Eukaryotic transposable elements and genome evolution. Trends Genet. 1989;5:103–107. doi: 10.1016/0168-9525(89)90039-5. Review. [DOI] [PubMed] [Google Scholar]
- Finnegan DJ. Transposable elements: how non-LTR retrotransposons do it. Curr Biol. 1997;7:R245–8. doi: 10.1016/s0960-9822(06)00112-6. Review. [DOI] [PubMed] [Google Scholar]
- Gaffney PM, Pierce JC, Mackinley AG, Titchen DA, Glenn WK. Pearl, a novel family of putative transposable elements in bivalve mollusks. J Mol Evol. 2003;56:308–316. doi: 10.1007/s00239-002-2402-5. [DOI] [PubMed] [Google Scholar]
- Gregory TR. Genome size estimates for two important freshwater molluscs, the zebra mussel (Dreissena polymorpha) and the schistosomiasis vector snail (Biomphalaria glabrata) Genome. 2003;46:841–844. doi: 10.1139/g03-069. [DOI] [PubMed] [Google Scholar]
- Han JS, Boeke JD. A highly active synthetic mammalian retrotransposon. Nature. 2004;429:314–318. doi: 10.1038/nature02535. [DOI] [PubMed] [Google Scholar]
- Han JS, Szak ST, Boeke JD. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature. 2004;429:268–274. doi: 10.1038/nature02536. [DOI] [PubMed] [Google Scholar]
- Jasinskiene N, Coates CJ, Benedict MQ, Cornel AJ, Rafferty CS, James AA, Collins FH. Stable transformation of the yellow fever mosquito, Aedes aegypti, with the Hermes element from the housefly. Proc Natl Acad Sci USA. 1998;95:3743–3747. doi: 10.1073/pnas.95.7.3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz RA, Jentoft JE. What is the role of the cys-his motif in retroviral nucleocapsid (NC) proteins? Bioessays. 1989;11:176–181. doi: 10.1002/bies.950110605. [DOI] [PubMed] [Google Scholar]
- Kidwell MG, Wattam AR. An important step forward in the genetic manipulation of mosquito vectors of human disease. Proc Natl Acad Sci USA. 1998;95:3349–3350. doi: 10.1073/pnas.95.7.3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel BE, ole-MoiYoi OK, Young JR. Ingi, a 5.2-kb dispersed sequence element from Trypanosoma brucei that carries half of a smaller mobile element at either end and has homology with mammalian LINEs. Mol Cell Biol. 1987;7:1465–1475. doi: 10.1128/mcb.7.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight M, Miller A, Raghavan N, Richards C, Lewis F. Identification of a repetitive element in the snail Biomphalaria glabrata: relationship to the reverse transcriptase-encoding sequence in LINE-1 transposons. Gene. 1992;118:181–187. doi: 10.1016/0378-1119(92)90187-t. [DOI] [PubMed] [Google Scholar]
- Knight M, Miller AN, Geoghagen NSM, Lewis FA, Kerlavage AR. Expressed sequence tags (ESTs) of Biomphalaria glabrata, an intermediate snail host of Schistosoma mansoni: use in the identification of RFLP markers. Malacologia. 1998;39:175–182. [Google Scholar]
- Lebens M, Sun JB, Czerkinsky C, Holmgren J. Current status and future prospects for a vaccine against schistosomiasis. Expert Rev Vaccines. 2004;3:315–328. doi: 10.1586/14760584.3.3.315. Review. [DOI] [PubMed] [Google Scholar]
- Lockyer AE, Noble LR, Rollinson D, Jones CS. Schistosoma mansoni: resistant specific infection-induced gene expression in Biomphalaria glabrata identified by fluorescent-based differential display. Exp Parasitol. 2004;107:97–104. doi: 10.1016/j.exppara.2004.04.004. [DOI] [PubMed] [Google Scholar]
- Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- Malik HS, Burke WD, Eickbush TH. The age and evolution of non-LTR retrotransposable elements. Mol Biol Evol. 1999;16:793–805. doi: 10.1093/oxfordjournals.molbev.a026164. [DOI] [PubMed] [Google Scholar]
- Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning, A Laboratory Manual. Cold Spring Harbor Laboratory; Cold Spring Harbor, New York: 1982. [Google Scholar]
- Martin F, Maranon C, Olivares M, Alonso C, Lopez MC. Characterization of a non-long terminal repeat retrotransposon cDNA (L1Tc) from Trypanosoma cruzi: homology of the first ORF with the ape family of DNA repair enzymes. J Mol Biol. 1995;247:49–59. doi: 10.1006/jmbi.1994.0121. [DOI] [PubMed] [Google Scholar]
- Miller AN, Raghavan N, FitzGerald PC, Lewis FA, Knight M. Differential gene expression in haemocytes of the snail Biomphalaria glabrata: effects of Schistosoma mansoni infection. Int J Parasitol. 2001;31:687–696. doi: 10.1016/s0020-7519(01)00133-3. [DOI] [PubMed] [Google Scholar]
- Muotri AR, Chu VT, Marchetto MC, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- Newton WL. The establishment of a strain of Australorbis glabratus which combines albinism and high susceptibility to infection with Schistosoma mansoni. J Parasitol. 1955;41:526–528. [PubMed] [Google Scholar]
- Paraense W, Correa L. Variation in susceptibility of populations of Australorbis glabratus to a strain of Schistosoma mansoni. Rev Inst Med Trop Sao Paulo. 1963;5:15–22. [PubMed] [Google Scholar]
- Pearce EJ. Progress towards a vaccine for schistosomiasis. Acta Trop. 2003;86:309–313. doi: 10.1016/s0001-706x(03)00062-7. Review. [DOI] [PubMed] [Google Scholar]
- Permanyer J, Gonzàlez-Duarte R, Albalat R. The non-LTR retrotransposons in Ciona intestinalis: new insights into the evolution of chordate genomes. Genome Biol. 2003;4:R73. doi: 10.1186/gb-2003-4-11-r73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Permanyer J, Albalat R, Gonzàlez-Duarte R. Getting closer to a pre-vertebrate genome: the non-LTR retrotransposons of Branchiostoma floridae. Int J Biol Sci. 2006;2:48–53. doi: 10.7150/ijbs.2.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrière G, Gouy M. WWW-Query: An on-line retrieval system for biological sequence banks. Biochimie. 1996;78:364–369. doi: 10.1016/0300-9084(96)84768-7. [DOI] [PubMed] [Google Scholar]
- Raghavan N, Miller AN, Gardner M, FitzGerald PC, Kerlavage AR, Johnston DA, Lewis FA, Knight M. Comparative gene analysis of Biomphalaria glabrata haemocytes pre- and post-exposure to miracidia of Schistosoma mansoni. Mol Biochem Parasitol. 2003;126:181–191. doi: 10.1016/s0166-6851(02)00272-4. [DOI] [PubMed] [Google Scholar]
- Raghavan N, Knight M. The snail (Biomphalaria glabrata) genome project. Trends Parasitol. 2006;22:148–151. doi: 10.1016/j.pt.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends in Genetics. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Rowling JK. Harry Potter and the Sorcerer’s Stone. Scholastic, Arthur A. Levine books; USA: 1998. [Google Scholar]
- Scott AF, Schmeckpeper BJ, Abdelrazik M, Comey CT, O’Hara B, Rossiter JP, Cooley T, Heath P, Smith KD, Margolet L. Origin of the human L1 elements: proposed progenitor genes deduced from a consensus DNA sequence. Genomics. 1987;1:113–125. doi: 10.1016/0888-7543(87)90003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart G, Furst A, Avdalovic N. Transverse alternating field electrophoresis (TAFE) Biotechniques. 1988;6:68–73. [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran MH, Pearson MS, Bethony JM, Smyth DJ, Jones MK, Duke M, Don TA, McManus DP, Correa-Oliveira R, Loukas A. Tetraspanins on the surface of Schistosoma mansoni are protective antigens against schistosomiasis. Nat Med. 2006;12:835–40. doi: 10.1038/nm1430. Epub. 2006, Jun 18. [DOI] [PubMed] [Google Scholar]
- Tu Z, Hill JJ. MosquI, a novel family of mosquito retrotransposons distantly related to the Drosophila I factors, may consist of elements of more than one origin. Mol Biol Evol. 1999;16:1675–1686. doi: 10.1093/oxfordjournals.molbev.a026081. [DOI] [PubMed] [Google Scholar]
- Zhang SM, Adema CM, Kepler TB, Loker ES. Diversification of Ig superfamily genes in an invertebrate. Science. 2004;305:251–254. doi: 10.1126/science.1088069. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A). The 5,869 sequence of the complete nimbus (BgI) element (GenBank EF413180). Capital letters denote coding sequences and small letters, the non-coding sequences. Position 1 in the numbering scheme is the first nucleotide of the start codon ATG (indicated by bold text) that begins the open reading frame 1 (ORF1; nucleic acid binding protein) coding region. A short, non-coding region of 20 bp separates ORF1 and ORF2. The ORF2 sequence encodes the apurinic/apyrimidinic endonuclease (APE), reverse transcriptase (RT) and RNase H (RNH) and horizontal arrows delineate these three hallmark domains. The amino acids of the various ORFs are shown in single letter codes and also labeled from 1-461 for ORF1 and 1-1,222 for ORF2. The three zinc finger regions composed of the sequence CX2CX4HX4C and the variant CX2CX4HX6C are underlined. The nimbus(BgI) coding region is flanked at either end by the 5′ and 3′ non-coding regions (NCRs) that are represented in small letters. The 3′ NCR consists of a conserved sequence containing a 16 bp repeat element (RE; bold, underlined region and Supplementary Fig. 1B) at the end of which is a stretch of “TCAAn or CAA” variable tandem repeats (highlighted in bold) that occur prior to the 3′ TSD (capital letters, boxed region) and defines the end of one element and the beginning of the next sequence. The 5′ NCR also begins with a 5′ target site duplication (TSD) (capital letters, boxed region), and contains a 16 bp 5′ RE (bold, underlined region and Supplementary Fig. 1B) and putative promoter sequences (CAAT and TATA, italics) upstream of the initiation codon of ORF1. B) Multiple alignment of the 5′ non-coding regions (NCRs) and 3′ NCRs of the nimbus (BgI) elements. The top panel shows the alignment of the 3′ NCRs of all nimbus (BgI) elements in the order they are arranged in the bacterial artificial chromosome (BAC) clone. The full-length elements are labeled I, II and III and the truncated transposition cassettes as A, B and C (the individual elements within each cassette is designated A1, A2 and A3; B1 and B2; C1, C2, C3. C4 and C5). The 3′ NCR panel is defined by the name of the element, the beginning bp, the 3′ NCR conserved domains (shown in red and blue), the 16 bp 3′ repeat element (RE) (shown in green), the variable tandem repeat (TCAAn or CAAn) region shown in orange and the start of the next region that may or may not contain the target site duplications (TSDs) (the 3′ TSDs shown in black are identifiable in full-length elements only) and the end bp position. The bottom panel shows the alignment of the three 5′ NCRs of the full-length nimbus (BgI) elements in the sequential order in which they appear 5′-3′ in the BAC clone. The full-length elements are labeled as I, II and III. The 5′ NCR panel is defined by the name of the element, the beginning bp position, the end of the previous region that contain the 5′ TSDs (black), the 5′ NCR conserved domains (blue and red) containing the 16 bp 5′ RE (shown in green), and the end bp position.