Abstract
Illuminating the molecular identity and regulation of early progenitor cells in the olfactory sensory epithelium represents an important challenge in the field of neural development. We show in both mouse and zebrafish that the winged helix transcription factor Foxg1 is expressed in an early progenitor population of the olfactory placode. In the mouse, Foxg1 is first expressed throughout the olfactory placode but later becomes restricted to the ventrolateral olfactory epithelium. The essential role of Foxg1 in olfactory development is demonstrated by the strikingly severe phenotype of Foxg1 knock-out mice: older embryos have no recognizable olfactory structures, including epithelium, bulb, or vomeronasal organs. Initially, a small number of olfactory progenitors are specified but show defects in both proliferation and differentiation. Similarly, antisense RNA knockdown of Foxg1 expression in the zebrafish shows a reduction in the number of neurons and mitotic cells in olfactory rosettes, mirroring the phenotype seen in the mouse Foxg1 null mutant. Using mosaic analysis in the zebrafish, we show that Foxg1 is required cell-autonomously for the production of mature olfactory receptor neurons. Therefore, we identified an evolutionarily conserved requirement for Foxg1 in the development of the vertebrate olfactory system.
Keywords: olfaction, Foxg1, olfactory placode, progenitor, olfactory epithelium zones, zebrafish
Introduction
The sensory epithelium of the vertebrate nose arises from the olfactory placode during embryogenesis. Placodes form as thickenings at the junction of the neural and superficial ectoderm that give rise to peripheral neurons and sensory structures. All placodal tissue is thought to arise from a common primordium of cells that undergo migrations, shape changes such as invagination or delamination, and proliferation to form the nascent sensory structures (Schlosser, 2005). Within the olfactory placode, the order and progression of proneural genes regulating olfactory neurogenesis have been illuminated by genetic analysis of basic helix-loop-helix (bHLH) family proteins (Guillemot et al., 1993; Cau et al., 1997, 2002). This has led to the current model in which a pool of rapidly proliferating transit amplifying cells expressing Ascl1 (formerly Mash1) give rise to Ngn1- and NeuroD-positive immediate neuronal precursors. However, little is known about the cells that give rise to the Ascl1 committed neuronal progenitors.
Forkhead family transcription factors comprise a large group of winged helix molecules with diverse roles in development, cancer, aging, and metabolism (Jonsson and Peng, 2005; Wijchers et al., 2006). One forkhead family member, Foxg1, affects proliferation, differentiation, patterning, and cell fate determination of telencephalic progenitor cells (Xuan et al., 1995; Hanashima et al., 2002, 2004, 2007; Martynoga et al., 2005; Muzio and Mallamaci, 2005). Specifically, Foxg1 affects the capacity and timing of cortical neuron production by resetting the progressive restriction in cortical progenitor potential to produce earlier-born Cajal-Retzius cells (Hanashima et al., 2004, 2007; Shen et al., 2006). Loss of Foxg1 also leads to dorsoventral patterning defects in the eye (Huh et al., 1999) in addition to aberrant projections of retinal ganglion cell axons at the optic chiasm (Pratt et al., 2004). Peripherally, Foxg1 is a marker of migrating ectodermal placodal cells (Hatini et al., 1999) and, in the otic placode, Foxg1 is required for the generation of crista neurons (Pauley et al., 2006). We were therefore interested in determining whether Foxg1 is required for the development of the olfactory placode.
In the present study, we show that Foxg1 is expressed in progenitor cells throughout the olfactory placode at early embryonic stages. In mice harboring a targeted mutation of the Foxg1 gene (Hebert and McConnell, 2000), we observe initial specification of some olfactory placode cells but a failure of those cells to proliferate and differentiate into mature olfactory neurons. Mice lacking Foxg1 ultimately fail to form all olfactory structures, an effect we ascribe to defects in the early olfactory placode, with a subsequent increase in apoptosis. In zebrafish, Foxg1 hypomorphs show a reduction in the number of proliferating cells as well as olfactory neurons. Using mosaic analysis in zebrafish embryos, we demonstrate that Foxg1 is required cell-autonomously within the olfactory lineage to produce mature olfactory receptor neurons (ORNs). Together, these results demonstrate a conserved role of Foxg1 in the proliferation and differentiation of cells comprising the olfactory placode, providing insight into the early steps of olfactory development.
Materials and Methods
Mice.
Homozygous Foxg1-Cre mice described by Hebert and McConnell (2000) resulted in a null mutant phenotype for Foxg1. Foxg1-Cre heterozygotes were maintained on a mixed 129/C57BL/6 background resulting from mating with other strains such as Gt(ROSA)26Sortm2Sho (The Jackson Laboratory, Bar Harbor, ME) (Mao et al., 2001). Ascl1 heterozygous mice were described previously (Guillemot et al., 1993). Observation of a vaginal plug was considered embryonic day 0.5 (E0.5). Genotyping primers used were as follows: Foxg1-F GACCCCTGATTTTGATGTGTG, Foxg1-R GGGCAACAACCACTCCTTCTC, CRE-F GCGGTCTGGCAGTAAAAACTATC, CRE-R GTGAAACAGCATTGCTGTCACTT, Ascl1wt CTCTTAGCCCAGAGGAAC, Mashmut1 GCAGCGCATCGCCTTCTATC, Mashmut2 CCAGGACTCAATACGCAGGG.
Zebrafish.
Tübingen longfin Danio rerio were maintained at 28.5°C and bred naturally using a timed light cycle. Morpholino (MO) oligonucleotides were injected in 1× Danieau buffer [58 mm NaCl, 0.7 mm KCl, 0.4 mm MgSO4, 0.6 mm Ca (NO3)2, 5 mm HEPES, pH 7.6] with 0.1% phenol red into the yolk of one-cell stage embryos. Morpholino sequence directed against the transcriptional start site of zebrafish Foxg1 was designed by Gene Tools (Philomath, OR): 5′-TCCCATATCCAACATCACAAGTAAG (Foxg1 MO) and mismatch (MM) 5′-TCCGATATGCAAGATCAGAACTAAG. Using 9.2 ng of Foxg1 MO per injection, we observed defects consistent with those found in mouse such as reduced telencephalic regions and aberrant projections in the optic chiasm (Xuan et al., 1995; Pratt et al., 2004). Antibody-labeled zebrafish were mounted in 1.2% low-melting point agarose and imaged on a Nikon (Tokyo, Japan) confocal microscope. Optical sections were acquired at 3 μm intervals and collapsed into a maximum projection image for analysis of phosphohistone H3 (PH3) and terminal deoxynucleotidyl transferase-mediated biotinylated UTP nick end labeling (TUNEL)-positive cells.
Transplantation experiments were performed using standard techniques (Ho and Kane, 1990). Donor embryos were labeled with 10 kDa lysine-fixable rhodamine dextran at the one-cell stage. Embryos in which no transplanted cells contributed to the olfactory rosettes were discarded. Mosaic embryos used in the analysis had an average of nine transplanted cells per rosette at 48 h postfertilization (hpf; the time of analysis), which represents a small fraction of the entire cellular composition. Morpholino injection of donor and host embryos was identical to that described above. Mosaic embryos were processed for calretinin immunohistochemistry at 48 hpf and confocal scanned as described. “Find Edges” was applied to the green channel in ImageJ to outline calretinin-positive cells. Cell counts were made examining confocal slices throughout rosettes and were comparable between two independent observers.
RNA in situ hybridization.
In situ hybridization of digoxygenin-labeled probes was performed as described previously (Lin et al., 2004). Antisense RNA probes were generated using the following sequences: Foxg1 (bp 1710–2321 and 1590–1940 of NM_008241), Sox2 (bp 1882–2260 of X94127), Ascl1 and Ngn1 (gift from M. Prasol, University of California, Berkeley, CA), Fgf8 (full length; gift from R. Fletcher and R. Harland, University of California, Berkeley, CA), Sox10 (bp 1253–1588 of AF047043), Nqo1 (bp 720–1224 of Av074850), and zebrafish Foxg1 (bp 502–855 of NM_131067), Cxcr4b (bp 648–1001 of NM_131834), olfactory marker protein (OMP) (bp 582–893 of NM_173281), and Fgf8a (gift from S. Amacher, University of California, Berkeley, CA).
Histological analyses.
Immunohistochemistry was performed on 4% PFA fixed whole zebrafish embryos or mouse tissue cryosectioned at 16 or 20 μm. The following antibodies were used: rabbit anti-green fluorescent protein (GFP; 1:200; Invitrogen, San Diego, CA), chicken anti-GFP (1:200; Abcam, Cambridge, MA), rabbit anti-PH3 (1:1000, or 1:250 when used with TUNEL; Millipore, Billerica, MA), mouse anti-calretinin (1:1000; Swant, Bellinzona, Switzerland), rabbit anti-Foxg1 (1:25, 2–3 d; Abcam), guinea pig anti-Ascl1 (1:10,000), rabbit anti-OMACS (olfactory-specific medium-chain acyl CoA synthetase; 1:250), and guinea pig anti-OlfCc1 directed against the peptide sequence [C]-SLVSTDSNTTDPPESVD-[amide]; antibody specificity for OlfCc1 was demonstrated by preadsorption with free peptide (data not shown). Hoechst (1:5000; Invitrogen) and BOBO3 (1:5000; Invitrogen) were used to label nuclei. Secondary antibodies labeled with Alexa Fluor 488 or 568 (Invitrogen) were used at 1:200. TUNEL staining was performed according to protocol (In Situ Cell Death Detection Kit; Roche Diagnostics, Indianapolis, IN), except that slides were microwave heated for 1 min in 0.1 m sodium citrate, pH 6, before treatment. Skeleton staining followed the method of Otto et al. (1997).
Cell quantitation.
The olfactory epithelial area was defined morphologically based on nuclear counterstain to identify borders in an unbiased manner. For mice, all PH3 and TUNEL counts were made focusing through 16 μm sections at 40× on a Nikon epifluorescence microscope. Placodes forming left and right nostrils were counted separately, because the right nostril often appeared more severely hypoplastic. However, PH3 and TUNEL did not show a left/right bias at early stages, so counts from each placode were treated separately toward the final n. Whole placode PH3 values were calculated from four placodes of two heterozygous mice and three placodes of two knock-out mice. Per section counts were calculated from averaging representative sections throughout the dorsoventral axis of the animal. PH3 and TUNEL data were taken from six wild-type and five mutant animals at E10.5 and five wild-type and seven knock-outs at E11.5. For zebrafish PH3 and TUNEL cell counts, values from the left and right nostrils were averaged, and n represents the number of fish analyzed. All values are presented as the mean ± SEM and were analyzed using a Student's t test.
Results
Foxg1 is expressed in early precursors of the olfactory lineage
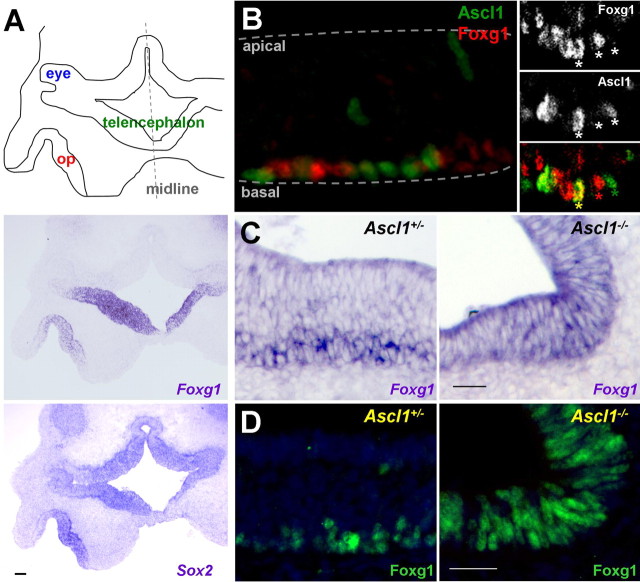
We used multiple approaches to investigate the expression of Foxg1 and placed it in the lineage of the developing olfactory epithelium. First, we determined the expression pattern of Foxg1 mRNA during early mouse development using in situ hybridization. At E10.5, Foxg1 is expressed throughout the invaginating olfactory placode in a pattern similar to a putative stem cell marker, Sox2 (Fig. 1A) (Donner et al., 2007). We next localized Foxg1 protein by immunohistochemistry, which revealed two important findings. First, we observed that from midgestation onward, Foxg1-positive cells are found basally within the olfactory epithelium (Figs. 1B–D, 2A), which is the site of transit amplifying progenitors that are responsible for regenerating olfactory neurons throughout the life of the animal (Cuschieri and Bannister, 1975; Graziadei et al., 1978). Second, Foxg1 protein is transiently coexpressed with the bHLH transcription factor Ascl1 at E16.5 (Fig. 1B). To date, Ascl1 is the earliest molecular marker in the olfactory neuron lineage and is expressed in a proliferative transit amplifying cell population (Guillemot et al., 1993; Cau et al., 1997). A null mutation in the Ascl1 gene results in the absence of more mature cells in this lineage, as highlighted by the loss of markers for the immediate neuronal precursors, immature neurons, and mature neurons (Cau et al., 1997, 2002).
Figure 1.
Foxg1-positive cells are early olfactory lineage precursors. In situ hybridization (A, C) and immunohistochemistry (B, D) were performed to localize Foxg1 to developing olfactory structures in the embryonic mouse. A, Schematic of transverse section from an E10.5 mouse embryo. op, Olfactory placode. The right side is out of the field of view, and anterior is toward the bottom. Localization of Foxg1 mRNA shows a similar expression pattern in the olfactory placode as that of a probe for Sox2, a putative olfactory progenitor marker. B, Foxg1 protein localizes to the basal cell layer of E16.5 olfactory epithelium (red), adjacent to and sometimes overlapping with the proneural bHLH transcription factor Ascl1 (green). The inset shows a cell from a different tissue section expressing both Foxg1 and Ascl1 (yellow asterisk). C, D, In E16.5 Ascl1+/− mice, Foxg1 mRNA (C) and protein (D) are expressed in the basal cell layers of the olfactory epithelium. The presence of Foxg1 in Ascl1−/− mice places Foxg1 upstream of Ascl1. Scale bars: A, 100 μm; C, D, 50 μm.
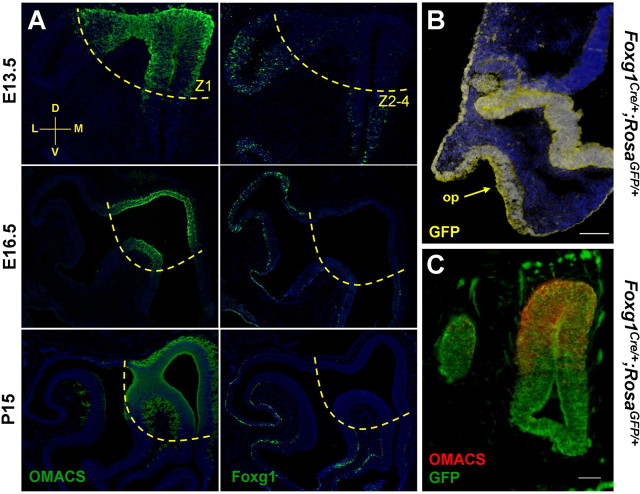
Figure 2.
Foxg1 becomes restricted to ventrolateral regions of the olfactory epithelium during development. A, Time course of Foxg1 and OMACS immunohistochemistry. In coronal sections through the olfactory epithelium, there is complementary expression of the dorsomedial (Z1) marker OMACS and Foxg1 in the ventrolateral zones (Z2–4). B, Transverse section of an E10.5 Foxg1-Cre;Rosa-GFP mouse demonstrates that cells of the olfactory placode (arrow), telencephalon, nasal retina, lens, and surface ectoderm are derived from Foxg1-expressing cells. C, Using the same lineage tracing strategy coupled with an anti-OMACS antibody, it is clear that although Foxg1 becomes downregulated dorsally (A), the dorsal tissue did express Foxg1 at an earlier time in development. A coronal section at E13.5 with dorsal to the top and medial to the right is shown. Scale bars: B, 100 μm; C, 50 μm.
We reasoned that if Foxg1 can be detected in Ascl1 mutant embryos, it must be present in a cell type at the same or previous developmental commitment as Ascl1-expressing cells. At E16.5 in the wild-type olfactory epithelium, Foxg1 mRNA and protein are restricted to the basal cell layer (Fig. 1C,D). Foxg1 expression is maintained in the olfactory epithelium of E16.5 Ascl1 mutants (Fig. 1C,D), suggesting that Foxg1 is upstream of Ascl1. As expected, the Ascl1 mutant epithelium is thinner (caused by the loss of neurons and more mature neuronal precursors) and has lost its laminar structure (Fig. 1C,D) (Murray et al., 2003). Foxg1 mRNA and protein were also present at E13.5 in olfactory epithelium devoid of Ascl1 (data not shown). From these experiments, we conclude that Foxg1 is expressed before Ascl1, the earliest proneural transcription factor identified within the olfactory neuron lineage in the mouse.
Zonal restriction of Foxg1 expression in the olfactory epithelium
The murine olfactory epithelium is patterned along the dorsomedial–ventrolateral axis. This organization plays an essential role in odorant receptor choice as well as in guiding topographic projections of ORN axons to the olfactory bulb (Miyamichi et al., 2005; Imai and Sakano, 2007). However, the mechanisms that generate and maintain this regional identity are not understood. In general, zones are defined by the expression of odorant receptors confined within one of four nonoverlapping regions (Ressler et al., 1993; Vassar et al., 1993; Miyamichi et al., 2005). Several other molecules show zonal restriction patterns in addition to the odorant receptors. For example, cytoprotective enzymes are found in the dorsomedial-most zone (zone 1), whereas the guidance molecule Npn2 and transcription factors Msx1 and Eya2 increase in a gradient ventrolaterally through zones 2–4 (Norlin et al., 2001; Oka et al., 2003; Tietjen et al., 2003; Gussing and Bohm, 2004).
Whereas Foxg1 expression is widespread in the early olfactory placode (E10.5) (Fig. 1A), at E11.5, cells expressing Foxg1 become restricted to ventral regions, distinct from cells expressing OMACS, a marker for cells of the dorsomedial olfactory epithelium (data not shown) (Oka et al., 2003). As shown in Figure 2A, a complementary expression pattern between Foxg1 and OMACS is evident at E13.5. At E16.5, Foxg1 is localized specifically to zones 2–4 in the basal cell layers of the olfactory epithelium (Fig. 2A). This pattern persists, as shown by immunohistochemistry of postnatal day 15 coronal sections (Fig. 2A). Therefore, Foxg1 represents the earliest transcription factor exhibiting a zonally restricted pattern both during development and maintained in a progenitor population at later stages.
Is Foxg1 expressed early in all cells fated to give rise to olfactory structures, or do dorsal ORNs arise from a Foxg1-independent progenitor population? To address this question, we used lineage tracing to ask whether cells of the dorsal olfactory epithelium arose from Foxg1-expressing precursors. Knock-in mice expressing Cre recombinase under the control of the Foxg1 promoter (Foxg1-Cre) (Hebert and McConnell, 2000) were crossed to mice containing a lox-STOP-lox-GFP transgene at the Rosa26 locus (Rosa-GFP) (Mao et al., 2001). GFP-labeled descendents of Foxg1-expressing cells are found in the olfactory placode, surface ectoderm, developing telencephalon (but not diencephalon), lens, and nasal retina at E10.5 (Fig. 2B), as expected from published expression patterns (Tao and Lai, 1992; Hatini et al., 1994; Pratt et al., 2004). At E13.5, GFP is expressed not only in the ventral but also in the dorsal olfactory epithelium, as shown by colocalization with the zone 1 marker OMACS (Fig. 2C). Together with the results of the Ascl1 epistasis experiment (Fig. 1C,D), we conclude that Foxg1 is expressed in one of the earliest cell types fated to give rise to olfactory epithelium in mouse but later exhibits a zonally restricted expression pattern.
Foxg1 is required for establishing the olfactory neuronal lineage
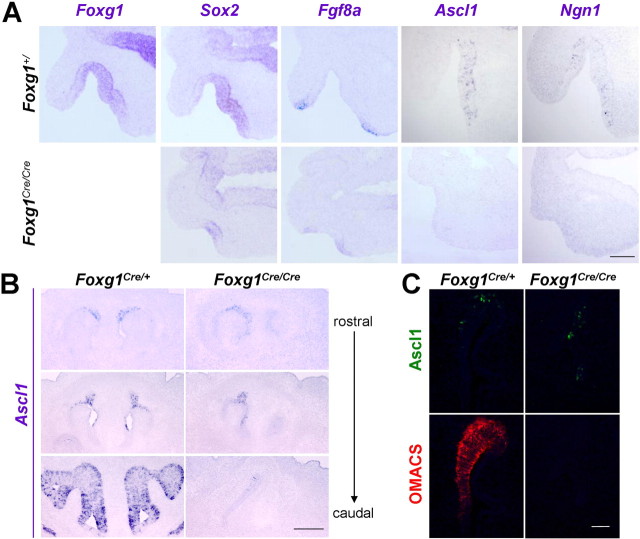
To investigate the role of Foxg1 during development of the olfactory system, we examined knock-out mice homozygous for an insertion of Cre recombinase into the Foxg1 coding region (Hebert and McConnell, 2000). In the developing neocortex, premature differentiation has been demonstrated to deplete the neural progenitor pool in the Foxg1 mutant (Xuan et al., 1995; Hanashima et al., 2002). We were therefore interested in whether an early overproduction of olfactory neurons would result from the Foxg1 null mutation. In wild-type mice at E10.5, the putative stem cell marker Sox2 labels a large portion of cells in the olfactory placode, whereas the proneural markers Ascl1 and Ngn1 are scattered throughout the epithelium (Fig. 3A). Fgf8, which has been shown to induce an olfactory fate from undifferentiated ectoderm (Bailey et al., 2006), is found at the tips of invaginating olfactory placodes. Foxg1 mutant tissue displays a severe reduction in the area comprising Sox2-positive cells (Fig. 3A), but their presence indicates that at least some cells are in fact specified as olfactory tissue. Few Ascl1- or Ngn1-positive cells (indicative of differentiation within the neuronal lineage) can be detected at E10.5, whereas Fgf8 expression in the peripheral ectoderm remains intact. Similar in situ hybridizations performed at E15.5 show the absence of all olfactory markers, arguing against a delay in neuronal development (Fig. S1, available at www.jneurosci.org as supplemental material). These findings contrast with a significant increase in neurons in the dorsal telencephalon of Foxg1 mutant mice (Xuan et al., 1995; Hanashima et al., 2002), suggesting that accelerated differentiation, with a concomitant depletion of a progenitor pool, does not account for the observed effects of the Foxg1 mutation on olfactory development. Instead, in the olfactory placode, we observe impaired rather than accelerated differentiation.
Figure 3.
Diminished expression of neuronal lineage markers in Foxg1Cre/Cre olfactory placode. A, RNA in situ hybridizations of the neuronal lineage markers Sox2, Ascl1, and Ngn1 show little expression in Foxg1 null mice at E10.5. However, Fgf8 expression in adjacent non-neural ectoderm persists. B, In the Foxg1 mutant, some Ascl1-positive cells are present in the rostral dorsomedial nasal cavity at E13.5. Conversely, Ascl1-positive cells were not found in the ventrolateral region of olfactory epithelium in the mutant, where Foxg1 is normally most highly expressed. C, To test whether the Ascl1-positive cells remaining in Foxg1 mutants represent a dorsal zone fate, we colabeled tissue sections with antibodies directed against Ascl1 and the dorsal marker OMACS. Of nine mutants, all were negative for OMACS expression, as shown here in a representative section at E13.5. Scale bars: A, 100 μm; B, 200 μm; C, 50 μm.
Defects in Foxg1 mutant olfactory epithelia are present throughout the dorsoventral axis
Because Foxg1 expression begins to show a dorsoventral bias by E11.5, we asked whether the phenotype of Foxg1 mutant animals might result from a differential requirement for Foxg1 in the olfactory epithelium zones. An intriguing possibility, one that would parallel the greater severity of the Foxg1 mutation on ventral versus dorsal telencephalon (Xuan et al., 1995; Hanashima et al., 2002; Martynoga et al., 2005; Muzio and Mallamaci, 2005), is that the Sox2-positive cells we observe in the E10.5 Foxg1 mutants comprise cells destined to adopt the zone 1 (dorsomedial) fate, whereas ventrolaterally fated cells are not specified or fail to proliferate altogether. Consistent with this idea, in E12.5–E13.5 mutants, a small number of cells expressing Ascl1 and Gap43 (a marker of immature neurons) were found in a dorsomedial location (Fig. 3B) (data not shown). To test whether this remaining tissue represents cells with a zone 1 identity (vs nonzone 1-fated cells that were simply displaced to a dorsomedial location), we performed double-label immunohistochemistry for Ascl1 and the zone 1 marker OMACS (Oka et al., 2003) (Fig. 2A). Of nine mutants showing Ascl1-positive cells in the olfactory epithelium, none displayed detectable OMACS expression (Fig. 3C). Similar results were obtained with another zone 1 marker, Nqo1 (data not shown). Therefore, the few remaining cells showing expression of the proneural gene Ascl1 do not appear to represent Foxg1-independent, zone 1 cells. These results suggest that, although zone 1 eventually ceases to express Foxg1 later in embryogenesis, Foxg1 is required for the earlier development of cells giving rise to all zones along the dorsoventral axis of the olfactory epithelium.
Foxg1 mutant mice eventually fail to develop olfactory structures and exhibit craniofacial defects
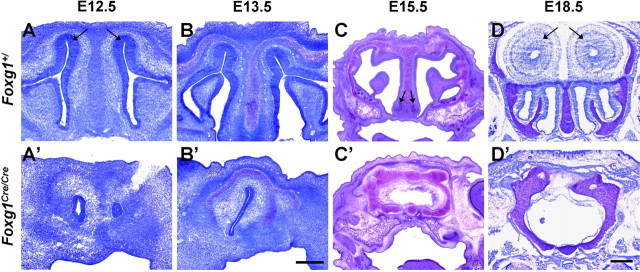
We next wanted to determine the consequences of the Foxg1 null mutation on the formation of olfactory structures as development progresses. In addition to abnormal eye and cortical structures reported previously (Xuan et al., 1995; Pratt et al., 2004), the absence of Foxg1 results in a dramatic disruption not only of the olfactory sensory epithelium but to the overall structure of the nasal cavity and other craniofacial structures (Fig. 4) (Fig. S2, available at www.jneurosci.org as supplemental material). By E12.5, Foxg1 mutant mice show rostral airways developing asymmetrically and no stratified olfactory epithelium (Fig. 4A,A′). The right side was often found to be more severely underdeveloped at this stage, similar to the phenotype observed in a Dlx5 null mutant (Acampora et al., 1999; Depew et al., 1999). Although the olfactory epithelium, vomeronasal organs, and olfactory bulbs are essentially absent in Foxg1 mutants, rostral structures appear more highly developed at E13.5. (Fig. 4B,B′). To determine whether these defects simply reflect a developmental delay, we also examined E15.5 and E18.5 embryos. At these later stages, cartilage and bone normally found in the septum, turbinates, and cribriform plate are disorganized in the mutant with increasing severity caudally, resulting in an empty cavity devoid of all markers of olfactory epithelium (Fig. 4C,C′,D,D′) (Fig. S1, available at www.jneurosci.org as supplemental material) (data not shown). These observations reveal a critical role for Foxg1 in the development of the peripheral olfactory system and craniofacial structures more generally. However, the severity of the Foxg1 null phenotype at late embryonic stages precludes a straightforward interpretation of the cellular mechanisms underlying these defects. Therefore, understanding the origin of the observed phenotype necessitates a more detailed analysis earlier in embryogenesis.
Figure 4.
Foxg1Cre/Cre mice fail to develop olfactory structures. Coronal sections of wild-type (A–D) and Foxg1Cre/Cre (A′–D′) mice at the indicated embryonic stages were stained with toluidine blue. Foxg1 mutant mice show asymmetrically developing airways rostrally and no stratified olfactory epithelium (A, arrows), vomeronasal organs (C, arrows), or olfactory bulbs (D, arrows). No difference was observed between Foxg1+/+ and Foxg1Cre/+ mice. The right nostril of the animal is in the right half of the image. Scale bars: A–B′, 200 μm; C–D′, 400 μm.
Olfactory placodes have fewer proliferating progenitors in Foxg1 mutants
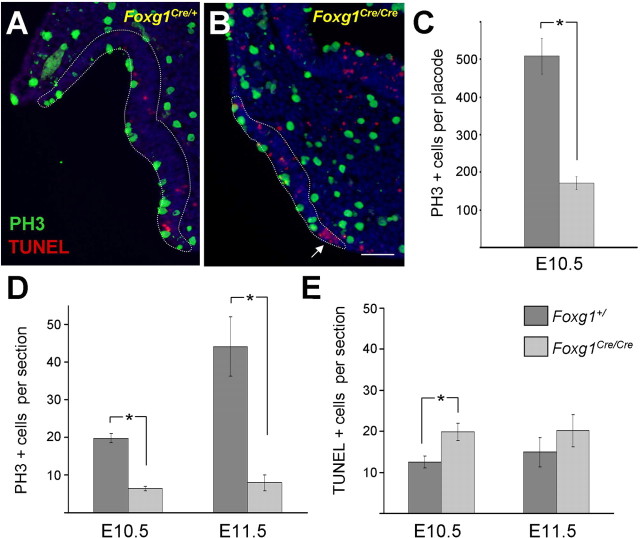
Foxg1 has been shown to function in proliferation, differentiation, and cell fate in cortical progenitors as well as cell death in the neural tube (Xuan et al., 1995; Hanashima et al., 2002, 2004, 2007; Ahlgren et al., 2003). To examine more closely the basis of the defects observed in olfactory development, we quantitated the number of proliferating cells in olfactory placodes from Foxg1 mutant mice. Using an antibody directed against PH3 to detect mitotic cells, at E10.5, we found one-third the number of proliferating cells per olfactory placode of Foxg1 mutants versus wild-type animals (171 ± 17 vs 509 ± 47; p < 0.001 in two-tailed t test) (Fig. 5C). To extend these findings, tissue sections were prepared from E10.5 and E11.5 mice, and the numbers of PH3-positive cells were counted and normalized per section. Confirming the results based on counting proliferative cells per whole placode, we found a similar decrease in PH3-positive cells per section in E10.5 mutants compared with control (6 ± 1 vs 20 ± 1; p = 5 × 10−6) (Fig. 5A,B,D). At E11.5, proliferation increases dramatically in wild-type placodes (44 ± 8 PH3-positive cells per section vs 20 ± 1 at E10.5). This developmental increase in proliferative cells was not observed in the Foxg1 mutant placodes, resulting in fivefold fewer PH3-positive cells in mutant versus control placodes at E11.5 (8 ± 2 vs 44 ± 8; p = 0.0004) (Fig. 5D). Consistent with these results, a decrease in bromodeoxyuridine-positive cells was also observed in the Foxg1 null mutant (data not shown) (Hatini et al., 1999).
Figure 5.
Temporal analysis of proliferation and apoptosis in the mouse olfactory placode. PH3 (green) and TUNEL (red) were used to identify proliferating and apoptotic cells, respectively. Representative transverse sections are shown from a control E10.5 embryo (A) and Foxg1Cre/Cre (B). Olfactory epithelium borders are indicated by dashed white lines. C, Wild-type mice have threefold more proliferating cells per placode than Foxg1 mutants (p = 0.001, Student's t test; n = 4 wild-type placodes, n = 3 mutant placodes). D, Quantitation of PH3-positive cells per section shows that mitotic cells increase with time in wild-type mice, whereas mice lacking functional Foxg1 fail to expand the pool of proliferative cells. There is a threefold difference in the number of proliferating cells at E10.5 (p = 5 × 10−6; n = 6 wild-type, 5 mutant mice) and 5.5-fold difference at E11.5 (p = 4 × 10−4; n = 5 wild-type, 7 mutant mice). E, Foxg1 homozygous null mice show a small but significant increase in cell death at E10.5 (p = 0.009; n = 6 wild-type, 5 mutant mice), although the increase at E11.5 is not significant (p = 0.4; n = 5 wild-type, 7 mutant mice). A subsequent wave of apoptosis was observed at E12.5 (Fig. S2, available at www.jneurosci.org as supplemental material). Asterisks indicate statistical significance as noted above. Scale bar, 100 μm.
The Foxg1 null mutation causes a secondary increase in cell death in the olfactory placode
To assess whether cell death might also be playing a role in the reduction of olfactory neurons, we undertook a temporal analysis of apoptosis within the frontonasal structures of mouse embryos. Tissue sections were prepared from E10.5 and E11.5 mice, and the numbers of TUNEL-positive cells were counted and normalized per section. Using this assay, we measured a modest but statistically significant increase in cell death in the olfactory placode of Foxg1 null compared with wild-type mice at E10.5 (20 ± 2 vs 13 ± 1; p = 0.009) (Fig. 5E). Similarly, there was a slight increase in apoptosis found in E11.5 mutants compared with wild-type controls; however, this latter increase was not statistically significant (20 ± 4 vs. 15 ± 4; p = 0.4). By E12.5, we observed a dramatic increase in apoptotic cell death extending beyond the olfactory epithelial area in the mutant (Fig. S3, available at www.jneurosci.org as supplemental material). The widespread cell death observed at this later stage probably underlies the severe loss of both olfactory and craniofacial structures observed in the Foxg1 mutant mouse (Fig. 4).
Notably, we observed an increase in apoptosis in the medial and lateral tips of the E10.5 mouse olfactory pits (Fig. 5B, arrow) (Fig. S4, available at www.jneurosci.org as supplemental material). Although it is difficult to ascertain whether cells in this region of the mutant are truly olfactory (vs neighboring non-neural ectoderm), these observations suggest that Foxg1 may play a role in regulating programmed cell death early in a restricted region of the olfactory placode.
Conserved pattern of Foxg1 expression in zebrafish embryos
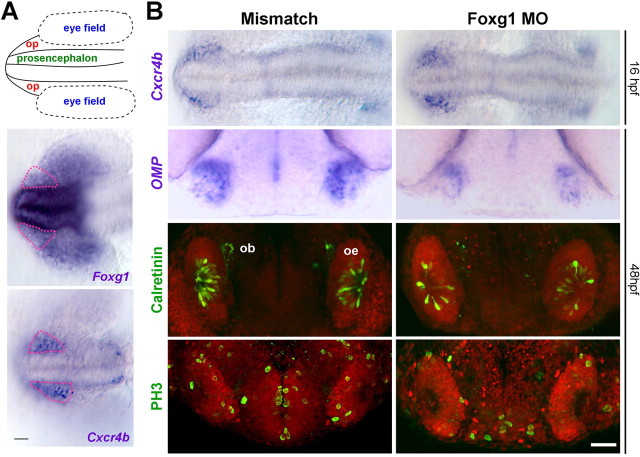
Previous reports of Foxg1 expression in forebrain structures of amphioxus, Xenopus, and medaka (Bourguignon et al., 1998; Toresson et al., 1998; Kitagawa et al., 2004) suggest an evolutionarily conserved role for Foxg1. We therefore asked whether Foxg1 is expressed in the early zebrafish olfactory placode by localizing Foxg1 expression using RNA in situ hybridization. In the zebrafish, Cxcr4b serves as a marker for the olfactory placode field (Miyasaka et al., 2007). In embryos at 16 hpf, Cxcr4b-positive cells migrate anteriorly toward the presumptive olfactory rosettes (Whitlock and Westerfield, 2000). As shown in Figure 6A, there is prominent expression of Foxg1 in the developing prosencephalon (the presumptive telencephalon), with a lower level of expression in the adjacent olfactory placode (pink outline) and nasal retina. Foxg1 transcript is also detected in the otic placodes, which are out of the field of view (data not shown). Therefore, the expression of Foxg1 in the embryonic zebrafish is similar to its expression in the developing mammalian nervous system (Tao and Lai, 1992; Hatini et al., 1999).
Figure 6.
Morpholino knockdown of Foxg1 in zebrafish results in fewer proliferative cells and olfactory receptor neurons. A, Schematic of rostral 16 hpf zebrafish embryo. op, Olfactory placode; dorsal view, anterior to the left. Foxg1 mRNA is predominantly expressed in the prosencephalon with lighter staining in the olfactory placode (pink dashed outline) and nasal retina. Cxcr4b indicates the position of the olfactory placode field. B, Embryos were analyzed with multiple markers after injection of Foxg1 morpholino or a five-base mismatch sequence. In situ hybridization of Cxcr4b mRNA shows that olfactory progenitors are specified in 16 hpf Foxg1 morpholino-injected fish. However, at 48 hpf, Foxg1 morphant embryos display a significant reduction of mature ORN markers (OMP in ciliated neurons and calretinin in microvillous neurons). PH3 labels 1.5-fold fewer mitotic cells in Foxg1 morphant placodes. Note the reduction of PH3-positive cells in the prosencephalon. Data are summarized in Tables 1 and 2. Fluorescence images represent a maximum projection of confocal stacks from a coronal view. ob, Olfactory bulb; oe, olfactory epithelium. Scale bar, 50 μm.
Foxg1 knockdown in zebrafish mirrors defects in Foxg1-deficient mice
To determine whether the requirement for Foxg1 in olfactory development is conserved across vertebrates, we studied the effects of reducing Foxg1 expression in the developing zebrafish embryo. Foxg1 hypomorphs (morphants) were created by injecting zebrafish embryos with an antisense morpholino oligonucleotide directed against the start site of the zebrafish Foxg1 gene (MO) or an MM morpholino oligonucleotide as a control. The effects of these injections were then assessed at various stages of development using multiple molecular markers.
Similar to the phenotype observed in Foxg1 mutant mice, at 12 and 16 hpf, there was no effect of Foxg1 knockdown on the expression of Fgf8 (Fig. S5, available at www.jneurosci.org as supplemental material). Olfactory placode is specified in morphant embryos as judged by the number of cells expressing Cxcr4b in MO and MM morpholino-injected fish (Fig. 6B). These results suggest that, if Foxg1 is required for the development of the olfactory epithelium, it is acting at a stage later than the initial specification of olfactory placodal cells. We therefore proceeded to investigate the effect of Foxg1 knockdown on cell proliferation and differentiation in the olfactory epithelium.
Teleost olfactory rosettes contain two major classes of ORNs: the ciliated and microvillous cells (Hansen and Zielinski, 2005). Ciliated olfactory neurons were monitored using in situ hybridization of OMP mRNA at 48 hpf (Celik et al., 2002; Sato et al., 2005). Although the olfactory placodes in Foxg1 morpholino-injected fish appear mostly intact, we nonetheless observed a decrease in OMP expression in Foxg1 morphants (Fig. 6B). Using an anti-calretinin antibody to identify microvillous neurons (Miyasaka et al., 2005), we observed 1.6-fold fewer calretinin-positive cells in the MO- versus MM-injected embryos at 48 hpf (Fig. 6B, Table 1). We also observed a similar reduction of microvillous neurons expressing OlfCc1, a C family G-protein-coupled receptor that is expressed in a large fraction of microvillous ORNs (Speca et al., 1999; Alioto and Ngai, 2006) (Fig. S5, available at www.jneurosci.org as supplemental material). Together, these observations indicate that knockdown of Foxg1 expression causes a decrease in both ciliated and microvillous olfactory neurons in the developing zebrafish olfactory epithelium.
Table 1.
Summary of calretinin-positive cells in the olfactory epithelium of 48 hpf zebrafish embryos injected with Foxg1 and mismatch morpholino oligonucleotides
| Experiment | Morpholino | Average calretinin+ cells/epithelium | n | p |
|---|---|---|---|---|
| 1 | Mismatch | 31 | 7 | 3.6 × 10−7 |
| Foxg1 | 18 | 24 | ||
| 2 | Mismatch | 31 | 8 | 0.014 |
| Foxg1 | 23 | 7 | ||
| 3 | Mismatch | 30 | 12 | 3.5 × 10−6 |
| Foxg1 | 16 | 20 |
n, Number of fish assayed; p, p value from two-tailed Student's t test.
We also analyzed cell proliferation in 48 hpf zebrafish olfactory epithelia, which are rapidly expanding at this stage. There was a 1.5-fold reduction in the number of proliferating cells in olfactory rosettes of Foxg1 morphants (Fig. 6B, Table 2). As expected from the known role of Foxg1 in cell proliferation in the developing mouse telencephalon (Xuan et al., 1995), in Foxg1 morphants, we also observed a 1.8-fold reduction of PH3-positive nuclei in the rostral telencephalic region from which the olfactory bulb neurons arise (Fig. 6B, Table 2). Therefore, in both zebrafish and mouse, reduction or elimination of Foxg1 causes a significant decrease in the number of proliferating cells and mature neurons in the regions that ultimately give rise to olfactory structures.
Table 2.
Summary of PH3-positive cells in the olfactory epithelium and telencephalon of 48 hpf zebrafish embryos injected with Foxg1 and mismatch morpholino oligonucleotides
| Experiment | Morpholino | Average PH3+ cells/epithelium | n | p | PH3+ cells/ telencephalon | p |
|---|---|---|---|---|---|---|
| 1 | Mismatch | 8.5 | 9 | 0.017 | 12 | 0.022 |
| Foxg1 | 5.6 | 14 | 7.9 | |||
| 2 | Mismatch | 11 | 24 | 2.0 × 10−7 | 17 | 2.6 × 10−6 |
| Foxg1 | 7.3 | 28 | 9.9 | |||
| 3 | Mismatch | 12 | 15 | 0.007 | 17 | 1.3 × 10−5 |
| Foxg1 | 7.7 | 11 | 8.6 |
n, Number of fish assayed; p, p value from two-tailed Student's t test.
We also asked whether apoptosis is altered in zebrafish Foxg1 morphants. Zebrafish embryos injected with Foxg1 MO or MM morpholinos were processed for TUNEL staining at 24, 36, and 48 hpf. However, this analysis did not reveal a difference in the number of TUNEL-positive cells in the olfactory placodes at any of these stages (data not shown). Together with the analysis of proliferation, these observations suggest that the loss of mature olfactory neurons in zebrafish Foxg1 morphants is primarily attributable to a requirement of Foxg1 for proliferation and/or differentiation of olfactory progenitor cells.
Mosaic analysis in zebrafish demonstrates a cell-autonomous requirement for Foxg1 in olfactory development
The results presented thus far demonstrate a role of Foxg1 in the development of both mouse and zebrafish olfactory epithelium. Is Foxg1 required cell-autonomously by cells in the olfactory placode, or does it exert its effects noncell-autonomously through the interaction with other cells in the developing embryo? For example, because the telencephalon and peripheral olfactory system develop in close apposition, it is possible that the defects present in Foxg1 mutant brain structures are responsible for the aberrant olfactory development described above. We therefore pursued a mosaic analysis in the zebrafish to determine whether the requirement for Foxg1 is cell-autonomous within the olfactory lineage.
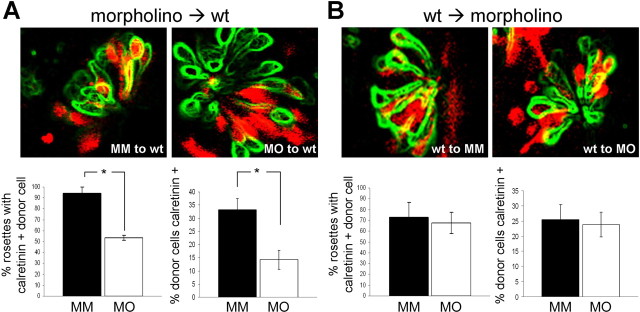
We transplanted sphere stage cells from zebrafish embryos injected with a lineage tracer and Foxg1 MO or MM morpholino to the animal pole of wild-type embryos. Mosaic animals were analyzed for expression of the ORN marker calretinin by transplanted donor cells within the host olfactory rosette at 48 hpf. Representative rosettes are shown in Figure 7A. The differentiation of donor cells into mature calretinin-positive neurons was quantitated using two different calculations. First, we determined the percentage of olfactory placodes containing at least one calretinin-positive donor cell, a stringent metric for assessing the capacity of the morphant donor cells to differentiate into mature ORNs. In MM morpholino controls, 94% of olfactory rosettes contained at least one transplanted calretinin-positive donor cell (31 of 34 rosettes over three different transplantation experiments). In contrast, only 53% (18 of 34) of rosettes contained calretinin-positive cells from Foxg1 MO-injected blastomeres (p = 0.009). Second, we determined the percentage of transplanted cells within the olfactory rosettes that differentiated into calretinin-positive mature ORNs. With MM donor cells, we found that 33% of the mosaic cells expressed calretinin (n = 28 rosettes). However, in Foxg1 MO donors, only 14% of the cells colabeled with calretinin (n = 34 rosettes; p = 0.001). Therefore, using both methods of analysis, we found a statistically significant reduction in mature olfactory neurons in Foxg1 MO versus MM donor cells (Fig. 7A). We conclude that in the presence of wild-type structures, Foxg1 is required cell-autonomously for the production of mature ORNs.
Figure 7.
Mosaic analysis in zebrafish demonstrates cell-autonomous requirement for Foxg1 in production of mature olfactory receptor neurons. A, Confocal optical section of olfactory rosettes from 48 hpf mosaic zebrafish embryos processed for calretinin immunohistochemistry (green outline; see Materials and Methods) and rhodamine dextran lineage tracer (red), indicating the presence of transplanted cells. Differentiation of donor cells into mature calretinin-positive neurons was quantitated based on the percentage of olfactory placodes containing at least one calretinin-positive donor cell (bottom left) or the percentage of donor cells that are also calretinin positive (bottom right). Using either method of quantitation, there is a statistically significant difference between the MM (black bars) and MO (white bars) conditions (asterisks; p = 0.009 for percentage of placodes containing at least one calretinin-positive donor cell; p = 0.001 for percentage of calretinin-positive donor cells). B, The converse experiment was performed in which wild-type (wt) cells were transplanted to morpholino-injected host zebrafish embryos. There is no apparent difference in the ability of wild-type cells to differentiate into mature ORNs in host embryos injected with MM or MO (black and white bars, respectively; p = 0.46 for percentage of placodes containing at least one calretinin-positive donor cell; p = 0.78 for percentage of calretinin-positive donor cells).
To test whether there is also a noncell-autonomous contribution, we performed the converse experiment in which wild-type cells were transplanted to morphant embryos. However, in these experiments, we observed no difference in the ability of transplanted cells to differentiate into mature olfactory neurons by both of the measures described above (Fig. 7B). A Foxg1 defective embryonic environment, therefore, does not impair development of the peripheral olfactory system. Rather, our transplantation analysis demonstrates that Foxg1 is required cell-autonomously within the olfactory lineage for production of mature ORNs.
Discussion
Development of the olfactory system requires the careful orchestration of a sequence of events, including initial specification, proliferation, and differentiation. Errors in any of these processes can lead to aberrant formation of olfactory tissue. Here, we show that Foxg1 functions in both mouse and zebrafish to ensure proper proliferation and differentiation of olfactory placode progenitors.
Role of Foxg1 in proliferation and differentiation of olfactory neuron progenitors
Previous studies have shown that a null mutation in the Foxg1 gene causes a marked decrease in volume of the cerebral hemispheres (Xuan et al., 1995; Hanashima et al., 2002, 2004; Martynoga et al., 2005; Muzio and Mallamaci, 2005). This morphological defect is caused by the premature onset of neuronal differentiation in the dorsal telencephalon, coupled with a decrease in cell proliferation in both dorsal and ventral structures (Xuan et al., 1995; Martynoga et al., 2005). In contrast to the effects of the Foxg1 mutation on dorsal cortical development, in the olfactory placode, we found no evidence for accelerated differentiation of olfactory progenitor cells; markers for the olfactory lineage were found to be reduced in Foxg1 null animals and morphant zebrafish embryos.
We show in both species that olfactory progenitors are specified but have defects in proliferation and differentiation when Foxg1 protein is reduced or absent. Moreover, this effect is cell-autonomous within the olfactory neuron lineage as assayed by zebrafish mosaic analysis. We attribute the highly dysmorphic frontonasal structures in mouse to be the result of apoptotic cell death following the primary defects described above. Increased cell death was not observed in zebrafish embryos, and we note that zebrafish olfactory rosettes maintained their morphological integrity compared with olfactory structures in mouse. It is likely that the hypomorphic nature of morpholino knockdown results in a less severe phenotype than that caused by genetic mutation.
Through what signaling pathways might Foxg1 exert its effects on the proliferation of olfactory progenitors? Previous studies have shown that TGFβ promotes p21Cip1-induced cell cycle arrest, a process negatively regulated by Foxg1 (Dou et al., 2000; Seoane et al., 2004; Siegenthaler et al., 2007). Because TGFβ signaling is prevalent in the developing olfactory epithelium, an increase in p21Cip1-mediated G1 cell cycle arrest may be a proximate effect of the Foxg1 mutation.
Placing Foxg1 in the context of other olfactory placode development genes
What genes and signaling pathways are required for specification and progression through the olfactory neuron lineage? A study in the chick suggests that Fgf8 expressed in the anterior neural ridge specifies olfactory tissue expressing Foxg1 (Bailey et al., 2006). It is unclear whether neighboring non-neural ectoderm secreting Fgf8 at the olfactory placode stage continues to specify new placodal cells later in development (Kawauchi et al., 2005), or whether all existing Foxg1-positive cells are specified at the neural plate stage and instead rely on Fgf8 for survival in the olfactory placode (Song et al., 2004). Whatever the case, from the present data, we propose that Foxg1 is required to establish a population of proliferating progenitor cells in the invaginating olfactory pit. These early placode progenitors, likely regulated by Foxg1 together with multiple other transcription factor genes (e.g., Pax, Sox, Dlx, Hes), generate a second population of proliferating cells committed to the neuronal lineage, the Ascl1-positive proneural transit amplifying cells, which ultimately differentiate into mature ORNs (Cau et al., 1997, 2002). Our present data demonstrate that Foxg1 plays an important and evolutionarily conserved role in the proliferation and differentiation of early olfactory progenitor cells. Future studies should help identify the transcription factor combinatorials underlying olfactory neurogenesis.
Regional identity in the olfactory epithelium
The mammalian olfactory epithelium displays a striking spatial organization, such that odorant receptors and other molecular markers are restricted to circumscribed zones that radiate along the dorsomedial–ventrolateral axis (Ressler et al., 1993; Vassar et al., 1993; Alenius and Bohm, 1997; Yoshihara et al., 1997; Norlin et al., 2001; Oka et al., 2003; Gussing and Bohm, 2004; Miyamichi et al., 2005; Tietjen et al., 2005). Dorsoventral organization within the olfactory epithelium plays an essential role in odorant receptor choice as well as in guiding topographic projections of ORN axons to the olfactory bulb (Miyamichi et al., 2005; Imai and Sakano, 2007). At a functional level, olfactory neurons of the dorsomedial epithelium mediate innate responses to odors, whereas the ventrolateral epithelium appears to subserve learned, perceptual functions (Imai and Sakano, 2007; Kobayakawa et al., 2007). However, the mechanisms that establish regionalization within the olfactory epithelium are mostly unknown.
Here, we show that Foxg1 becomes progressively more restricted to ventrolateral regions of the olfactory epithelium during development. Indeed, Foxg1 is the earliest identified transcription factor exhibiting zonal specificity. In one scenario, Foxg1 is required only for the specification and/or proliferation of cells fated to occupy the ventrolateral (but not dorsomedial) zones of the olfactory epithelium. Alternatively, Foxg1 could be required at a time before zonal specification (e.g., for proliferation of the initial pool of olfactory precursors). Arguing against the former model, we found that the residual cells in the Foxg1 mutant epithelium do not express OMACS, a molecular marker of zone 1 cells. Together with the overall severity of the Foxg1 null mutation, these observations strongly suggest that Foxg1 is required for the development of the olfactory placode, before the appearance of zonal restrictions within the developing olfactory epithelium. At later stages of embryogenesis (as well as through postnatal life), Foxg1 is expressed ventrolaterally and in the basal cell layer of the olfactory epithelium, the site of the transit amplifying olfactory progenitor cells. This pattern of expression implies that at these later stages, Foxg1 is required for the production and regeneration of olfactory neurons in the ventrolateral, but not dorsomedial, olfactory epithelium. Future experiments using conditional inactivation of Foxg1 should clarify the role of this transcription factor in ventrolateral olfactory progenitor cells.
Footnotes
This work was supported by a grant from the National Institute on Deafness and Other Communication Disorders (J.N.). We thank S. McConnell for Foxg1-Cre mice, J. Johnson for Ascl1 mutant mice and Ascl1 antibody, and K. Kobayakawa and H. Sakano for OMACS antibody. We thank S. Amacher, M. Prasol, and R. Fletcher for additional reagents; K. Vranizan and H. Janovjak for help with analyzing transplantation data; and J. Scolnick for helpful discussions throughout the course of this study.
References
- Acampora D, Merlo GR, Paleari L, Zerega B, Postiglione MP, Mantero S, Bober E, Barbieri O, Simeone A, Levi G. Craniofacial, vestibular and bone defects in mice lacking the Distal-less-related gene Dlx5. Development. 1999;126:3795–3809. doi: 10.1242/dev.126.17.3795. [DOI] [PubMed] [Google Scholar]
- Ahlgren S, Vogt P, Bronner-Fraser M. Excess FoxG1 causes overgrowth of the neural tube. J Neurobiol. 2003;57:337–349. doi: 10.1002/neu.10287. [DOI] [PubMed] [Google Scholar]
- Alenius M, Bohm S. Identification of a novel neural cell adhesion molecule-related gene with a potential role in selective axonal projection. J Biol Chem. 1997;272:26083–26086. doi: 10.1074/jbc.272.42.26083. [DOI] [PubMed] [Google Scholar]
- Alioto TS, Ngai J. The repertoire of olfactory C family G protein-coupled receptors in zebrafish: candidate chemosensory receptors for amino acids. BMC Genomics. 2006;7:309. doi: 10.1186/1471-2164-7-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey AP, Bhattacharyya S, Bronner-Fraser M, Streit A. Lens specification is the ground state of all sensory placodes, from which FGF promotes olfactory identity. Dev Cell. 2006;11:505–517. doi: 10.1016/j.devcel.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Bourguignon C, Li J, Papalopulu N. XBF-1, a winged helix transcription factor with dual activity, has a role in positioning neurogenesis in Xenopus competent ectoderm. Development. 1998;125:4889–4900. doi: 10.1242/dev.125.24.4889. [DOI] [PubMed] [Google Scholar]
- Cau E, Gradwohl G, Fode C, Guillemot F. Mash1 activates a cascade of bHLH regulators in olfactory neuron progenitors. Development. 1997;124:1611–1621. doi: 10.1242/dev.124.8.1611. [DOI] [PubMed] [Google Scholar]
- Cau E, Casarosa S, Guillemot F. Mash1 and Ngn1 control distinct steps of determination and differentiation in the olfactory sensory neuron lineage. Development. 2002;129:1871–1880. doi: 10.1242/dev.129.8.1871. [DOI] [PubMed] [Google Scholar]
- Celik A, Fuss SH, Korsching SI. Selective targeting of zebrafish olfactory receptor neurons by the endogenous OMP promoter. Eur J Neurosci. 2002;15:798–806. doi: 10.1046/j.1460-9568.2002.01913.x. [DOI] [PubMed] [Google Scholar]
- Cuschieri A, Bannister LH. The development of the olfactory mucosa in the mouse: light microscopy. J Anat. 1975;119:277–286. [PMC free article] [PubMed] [Google Scholar]
- Depew MJ, Liu JK, Long JE, Presley R, Meneses JJ, Pedersen RA, Rubenstein JL. Dlx5 regulates regional development of the branchial arches and sensory capsules. Development. 1999;126:3831–3846. doi: 10.1242/dev.126.17.3831. [DOI] [PubMed] [Google Scholar]
- Donner AL, Episkopou V, Maas RL. Sox2 and Pou2f1 interact to control lens and olfactory placode development. Dev Biol. 2007;303:784–799. doi: 10.1016/j.ydbio.2006.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou C, Lee J, Liu B, Liu F, Massague J, Xuan S, Lai E. BF-1 interferes with transforming growth factor beta signaling by associating with Smad partners. Mol Cell Biol. 2000;20:6201–6211. doi: 10.1128/mcb.20.17.6201-6211.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziadei PP, Levine RR, Graziadei GA. Regeneration of olfactory axons and synapse formation in the forebrain after bulbectomy in neonatal mice. Proc Natl Acad Sci USA. 1978;75:5230–5234. doi: 10.1073/pnas.75.10.5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemot F, Lo LC, Johnson JE, Auerbach A, Anderson DJ, Joyner AL. Mammalian achaete-scute homolog 1 is required for the early development of olfactory and autonomic neurons. Cell. 1993;75:463–476. doi: 10.1016/0092-8674(93)90381-y. [DOI] [PubMed] [Google Scholar]
- Gussing F, Bohm S. NQO1 activity in the main and the accessory olfactory systems correlates with the zonal topography of projection maps. Eur J Neurosci. 2004;19:2511–2518. doi: 10.1111/j.0953-816X.2004.03331.x. [DOI] [PubMed] [Google Scholar]
- Hanashima C, Shen L, Li SC, Lai E. Brain factor-1 controls the proliferation and differentiation of neocortical progenitor cells through independent mechanisms. J Neurosci. 2002;22:6526–6536. doi: 10.1523/JNEUROSCI.22-15-06526.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanashima C, Li SC, Shen L, Lai E, Fishell G. Foxg1 suppresses early cortical cell fate. Science. 2004;303:56–59. doi: 10.1126/science.1090674. [DOI] [PubMed] [Google Scholar]
- Hanashima C, Fernandes M, Hebert JM, Fishell G. The role of Foxg1 and dorsal midline signaling in the generation of Cajal-Retzius subtypes. J Neurosci. 2007;27:11103–11111. doi: 10.1523/JNEUROSCI.1066-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen A, Zielinski BS. Diversity in the olfactory epithelium of bony fishes: development, lamellar arrangement, sensory neuron cell types and transduction components. J Neurocytol. 2005;34:183–208. doi: 10.1007/s11068-005-8353-1. [DOI] [PubMed] [Google Scholar]
- Hatini V, Tao W, Lai E. Expression of winged helix genes, BF-1 and BF-2, define adjacent domains within the developing forebrain and retina. J Neurobiol. 1994;25:1293–1309. doi: 10.1002/neu.480251010. [DOI] [PubMed] [Google Scholar]
- Hatini V, Ye X, Balas G, Lai E. Dynamics of placodal lineage development revealed by targeted transgene expression. Dev Dyn. 1999;215:332–343. doi: 10.1002/(SICI)1097-0177(199908)215:4<332::AID-AJA5>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Hebert JM, McConnell SK. Targeting of cre to the Foxg1 (BF-1) locus mediates loxP recombination in the telencephalon and other developing head structures. Dev Biol. 2000;222:296–306. doi: 10.1006/dbio.2000.9732. [DOI] [PubMed] [Google Scholar]
- Ho RK, Kane DA. Cell-autonomous action of zebrafish spt-1 mutation in specific mesodermal precursors. Nature. 1990;348:728–730. doi: 10.1038/348728a0. [DOI] [PubMed] [Google Scholar]
- Huh S, Hatini V, Marcus RC, Li SC, Lai E. Dorsal-ventral patterning defects in the eye of BF-1-deficient mice associated with a restricted loss of shh expression. Dev Biol. 1999;211:53–63. doi: 10.1006/dbio.1999.9303. [DOI] [PubMed] [Google Scholar]
- Imai T, Sakano H. Roles of odorant receptors in projecting axons in the mouse olfactory system. Curr Opin Neurobiol. 2007;17:507–515. doi: 10.1016/j.conb.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Jonsson H, Peng SL. Forkhead transcription factors in immunology. Cell Mol Life Sci. 2005;62:397–409. doi: 10.1007/s00018-004-4365-8. [DOI] [PubMed] [Google Scholar]
- Kawauchi S, Shou J, Santos R, Hebert JM, McConnell SK, Mason I, Calof AL. Fgf8 expression defines a morphogenetic center required for olfactory neurogenesis and nasal cavity development in the mouse. Development. 2005;132:5211–5223. doi: 10.1242/dev.02143. [DOI] [PubMed] [Google Scholar]
- Kitagawa D, Watanabe T, Saito K, Asaka S, Sasado T, Morinaga C, Suwa H, Niwa K, Yasuoka A, Deguchi T, Yoda H, Hirose Y, Henrich T, Iwanami N, Kunimatsu S, Osakada M, Winkler C, Elmasri H, Wittbrodt J, Loosli F. Genetic dissection of the formation of the forebrain in Medaka, Oryzias latipes. Mech Dev. 2004;121:673–685. doi: 10.1016/j.mod.2004.03.010. [DOI] [PubMed] [Google Scholar]
- Kobayakawa K, Kobayakawa R, Matsumoto H, Oka Y, Imai T, Ikawa M, Okabe M, Ikeda T, Itohara S, Kikusui T, Mori K, Sakano H. Innate versus learned odour processing in the mouse olfactory bulb. Nature. 2007;450:503–508. doi: 10.1038/nature06281. [DOI] [PubMed] [Google Scholar]
- Lin DM, Yang YH, Scolnick JA, Brunet LJ, Marsh H, Peng V, Okazaki Y, Hayashizaki Y, Speed TP, Ngai J. Spatial patterns of gene expression in the olfactory bulb. Proc Natl Acad Sci USA. 2004;101:12718–12723. doi: 10.1073/pnas.0404872101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X, Fujiwara Y, Chapdelaine A, Yang H, Orkin SH. Activation of EGFP expression by Cre-mediated excision in a new ROSA26 reporter mouse strain. Blood. 2001;97:324–326. doi: 10.1182/blood.v97.1.324. [DOI] [PubMed] [Google Scholar]
- Martynoga B, Morrison H, Price DJ, Mason JO. Foxg1 is required for specification of ventral telencephalon and region-specific regulation of dorsal telencephalic precursor proliferation and apoptosis. Dev Biol. 2005;283:113–127. doi: 10.1016/j.ydbio.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Miyamichi K, Serizawa S, Kimura HM, Sakano H. Continuous and overlapping expression domains of odorant receptor genes in the olfactory epithelium determine the dorsal/ventral positioning of glomeruli in the olfactory bulb. J Neurosci. 2005;25:3586–3592. doi: 10.1523/JNEUROSCI.0324-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyasaka N, Sato Y, Yeo SY, Hutson LD, Chien CB, Okamoto H, Yoshihara Y. Robo2 is required for establishment of a precise glomerular map in the zebrafish olfactory system. Development. 2005;132:1283–1293. doi: 10.1242/dev.01698. [DOI] [PubMed] [Google Scholar]
- Miyasaka N, Knaut H, Yoshihara Y. Cxcl12/Cxcr4 chemokine signaling is required for placode assembly and sensory axon pathfinding in the zebrafish olfactory system. Development. 2007;134:2459–2468. doi: 10.1242/dev.001958. [DOI] [PubMed] [Google Scholar]
- Murray RC, Navi D, Fesenko J, Lander AD, Calof AL. Widespread defects in the primary olfactory pathway caused by loss of Mash1 function. J Neurosci. 2003;23:1769–1780. doi: 10.1523/JNEUROSCI.23-05-01769.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzio L, Mallamaci A. Foxg1 confines Cajal-Retzius neuronogenesis and hippocampal morphogenesis to the dorsomedial pallium. J Neurosci. 2005;25:4435–4441. doi: 10.1523/JNEUROSCI.4804-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norlin EM, Alenius M, Gussing F, Hagglund M, Vedin V, Bohm S. Evidence for gradients of gene expression correlating with zonal topography of the olfactory sensory map. Mol Cell Neurosci. 2001;18:283–295. doi: 10.1006/mcne.2001.1019. [DOI] [PubMed] [Google Scholar]
- Oka Y, Kobayakawa K, Nishizumi H, Miyamichi K, Hirose S, Tsuboi A, Sakano H. O-MACS, a novel member of the medium-chain acyl-CoA synthetase family, specifically expressed in the olfactory epithelium in a zone-specific manner. Eur J Biochem. 2003;270:1995–2004. doi: 10.1046/j.1432-1033.2003.03571.x. [DOI] [PubMed] [Google Scholar]
- Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, Selby PB, Owen MJ. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- Pauley S, Lai E, Fritzsch B. Foxg1 is required for morphogenesis and histogenesis of the mammalian inner ear. Dev Dyn. 2006;235:2470–2482. doi: 10.1002/dvdy.20839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt T, Tian NM, Simpson TI, Mason JO, Price DJ. The winged helix transcription factor Foxg1 facilitates retinal ganglion cell axon crossing of the ventral midline in the mouse. Development. 2004;131:3773–3784. doi: 10.1242/dev.01246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ressler KJ, Sullivan SL, Buck LB. A zonal organization of odorant receptor gene expression in the olfactory epithelium. Cell. 1993;73:597–609. doi: 10.1016/0092-8674(93)90145-g. [DOI] [PubMed] [Google Scholar]
- Sato Y, Miyasaka N, Yoshihara Y. Mutually exclusive glomerular innervation by two distinct types of olfactory sensory neurons revealed in transgenic zebrafish. J Neurosci. 2005;25:4889–4897. doi: 10.1523/JNEUROSCI.0679-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlosser G. Evolutionary origins of vertebrate placodes: insights from developmental studies and from comparisons with other deuterostomes. J Exp Zoolog B Mol Dev Evol. 2005;304:347–399. doi: 10.1002/jez.b.21055. [DOI] [PubMed] [Google Scholar]
- Seoane J, Le HV, Shen L, Anderson SA, Massague J. Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell. 2004;117:211–223. doi: 10.1016/s0092-8674(04)00298-3. [DOI] [PubMed] [Google Scholar]
- Shen Q, Wang Y, Dimos JT, Fasano CA, Phoenix TN, Lemischka IR, Ivanova NB, Stifani S, Morrisey EE, Temple S. The timing of cortical neurogenesis is encoded within lineages of individual progenitor cells. Nat Neurosci. 2006;9:743–751. doi: 10.1038/nn1694. [DOI] [PubMed] [Google Scholar]
- Siegenthaler JA, Tremper-Wells BA, Miller MW. Foxg1 haploinsufficiency reduces the population of cortical intermediate progenitor cells: effect of increased p21 expression. Cereb Cortex. 2007 doi: 10.1093/cercor/bhm209. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Hui JN, Fu KK, Richman JM. Control of retinoic acid synthesis and FGF expression in the nasal pit is required to pattern the craniofacial skeleton. Dev Biol. 2004;276:313–329. doi: 10.1016/j.ydbio.2004.08.035. [DOI] [PubMed] [Google Scholar]
- Speca DJ, Lin DM, Sorensen PW, Isacoff EY, Ngai J, Dittman AH. Functional identification of a goldfish odorant receptor. Neuron. 1999;23:487–498. doi: 10.1016/s0896-6273(00)80802-8. [DOI] [PubMed] [Google Scholar]
- Tao W, Lai E. Telencephalon-restricted expression of BF-1, a new member of the HNF-3/fork head gene family, in the developing rat brain. Neuron. 1992;8:957–966. doi: 10.1016/0896-6273(92)90210-5. [DOI] [PubMed] [Google Scholar]
- Tietjen I, Rihel JM, Cao Y, Koentges G, Zakhary L, Dulac C. Single-cell transcriptional analysis of neuronal progenitors. Neuron. 2003;38:161–175. doi: 10.1016/s0896-6273(03)00229-0. [DOI] [PubMed] [Google Scholar]
- Tietjen I, Rihel J, Dulac CG. Single-cell transcriptional profiles and spatial patterning of the mammalian olfactory epithelium. Int J Dev Biol. 2005;49:201–207. doi: 10.1387/ijdb.041939it. [DOI] [PubMed] [Google Scholar]
- Toresson H, Martinez-Barbera JP, Bardsley A, Caubit X, Krauss S. Conservation of BF-1 expression in amphioxus and zebrafish suggests evolutionary ancestry of anterior cell types that contribute to the vertebrate telencephalon. Dev Genes Evol. 1998;208:431–439. doi: 10.1007/s004270050200. [DOI] [PubMed] [Google Scholar]
- Vassar R, Ngai J, Axel R. Spatial segregation of odorant receptor expression in the mammalian olfactory epithelium. Cell. 1993;74:309–318. doi: 10.1016/0092-8674(93)90422-m. [DOI] [PubMed] [Google Scholar]
- Whitlock KE, Westerfield M. The olfactory placodes of the zebrafish form by convergence of cellular fields at the edge of the neural plate. Development. 2000;127:3645–3653. doi: 10.1242/dev.127.17.3645. [DOI] [PubMed] [Google Scholar]
- Wijchers PJ, Burbach JP, Smidt MP. In control of biology: of mice, men and Foxes. Biochem J. 2006;397:233–246. doi: 10.1042/BJ20060387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan S, Baptista CA, Balas G, Tao W, Soares VC, Lai E. Winged helix transcription factor BF-1 is essential for the development of the cerebral hemispheres. Neuron. 1995;14:1141–1152. doi: 10.1016/0896-6273(95)90262-7. [DOI] [PubMed] [Google Scholar]
- Yoshihara Y, Kawasaki M, Tamada A, Fujita H, Hayashi H, Kagamiyama H, Mori K. OCAM: a new member of the neural cell adhesion molecule family related to zone-to-zone projection of olfactory and vomeronasal axons. J Neurosci. 1997;17:5830–5842. doi: 10.1523/JNEUROSCI.17-15-05830.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]