Abstract
Killer cell immunoglobulin-like receptors (KIR) regulate natural killer cell response against infection and malignancy. KIR genes are variable in the number and type, thereby discriminating individuals and populations. Herein, we analyzed the KIR gene content diversity in four native populations of Iran. The KIR genomic diversity was comparable between Bakhtiari and Persian and displayed a balance of A and B KIR haplotypes, a trend reported in Caucasian and African populations. The KIR gene content profiles of Arab and Azeri were comparable and displayed a preponderance of B haplotypes, a scenario reported in the natives of America, India, and Australia. A majority of the B haplotype carriers of Azeri and Arab had a centromeric gene-cluster (KIR2DS2-2DL2-2DS3-2DL5). Remarkably, this cluster was totally absent from the American natives but occurred at highest frequencies in the natives of India and Australia in combination with another gene cluster at the telomeric region (KIR3DS1-2DL5-2DS5-2DS1). Therefore, despite having similar frequencies of B haplotypes, the occurrence of B haplotype-specific KIR genes, such as 2DL2, 2DL5, 3DS1, 2DS1, 2DS2, 2DS3, and 2DS5 in Azeri and Arab were substantially different from the natives of America, India, and Australia. In conclusion, each Iranian population exhibits distinct KIR gene content diversity, and the Indo-European KIR genetic signatures of the Iranians concur with geographic proximity, linguistic affinity, and human migrations.
Electronic supplementary material
The online version of this article (doi:10.1007/s00251-009-0378-7) contains supplementary material, which is available to authorized users.
Keywords: NK cells, KIR genes, Immunity-related genes, Polymorphism, Iranian populations, Persian
Introduction
Natural killer (NK) cells are fast-acting lymphocytes that provide the first line of defense against infection and tumor transformation (Trinchieri 1989). Human NK cells largely use killer cell immunoglobulin-like receptors (KIR) to distinguish the unhealthy targets from the healthy self (Lanier 2005). A family of 16 homologous genes clustered at the leukocyte receptor complex on chromosome 19q13.4 encodes KIR receptors (Vilches and Parham 2002; Wilson et al. 2000). Fourteen of them encode receptors that trigger either inhibition (3DL1-3, 2DL1-3, and 2DL5) or activation (3DS1, 2DS1-2DS5) or both (2DL4) and two pseudogenes (2DP1 and 3DP1) that do not encode a cell-surface receptor. The inhibitory KIRs recognize distinct motifs of HLA class I molecules and trigger signals that stop NK cell function, while the ligands for the activating KIRs are unknown. Epidemiological studies suggest that the activating KIRs may recognize pathogen-derived or pathogen-induced cell surface determinants and trigger NK response against the unhealthy targets, which may yield immunopathology or resistance to infection (Khakoo and Carrington 2006).
The number and type of KIR genes vary substantially between haplotypes and display sequence polymorphism (Hou et al. 2008; Hsu et al. 2002; Martin et al. 2004; Middleton et al. 2007; Shilling et al. 2002; Whang et al. 2005; Wilson et al. 2000). On the basis of gene content, KIR haplotypes are broadly classified into two groups (Uhrberg et al. 1997). Group A haplotypes have a fixed gene content comprising KIR3DL3-2DL3-2DP1-2DL1-3DP1-2DL4-3DL1-2DS4-3DL2 but are diversified through allelic polymorphism of the individual genes. In contrast, group B haplotypes have variable gene content comprising several genes and alleles that are not part of the A haplotype. Particularly, KIR2DS1, 2DS2, 2DS3, 2DS5, 2DL2, 2DL5, and 3DS1 are associated only with group B haplotypes, and thus B haplotypes generally encode more activating KIR receptors than the A haplotype that encodes a single activating receptor, KIR2DS4. Four framework genes (KIR3DL3, 3DP1, 2DL4, and 3DL2) are conserved on both A and B haplotypes and therefore ubiquitously present in all individuals.
All human populations have both group A and B haplotypes, but their frequencies vary considerably (Parham 2005; Single et al. 2007; Yawata et al. 2002a). In Africans and Caucasians, the A and B haplotypes are equally distributed, suggestive of a balancing selection. Conversely, the A haplotype is overrepresented in Northeast Asians (Chinese, Japanese and Koreans), while the B haplotype occurred most frequently in the natives of India, Australia, and America (Ewerton et al. 2007; Flores et al. 2007; Gendzekhadze et al. 2006; Jiang et al. 2005; Norman et al. 2002; Rajalingam et al. 2002; Toneva et al. 2001; Whang et al. 2005; Yawata et al. 2002b). Herein, we investigated the KIR gene content diversity in four native populations of Iran, a country with central geographic location which served as a gateway of human movements during the past 60,000 years.
Materials and methods
Study subjects and DNA extraction
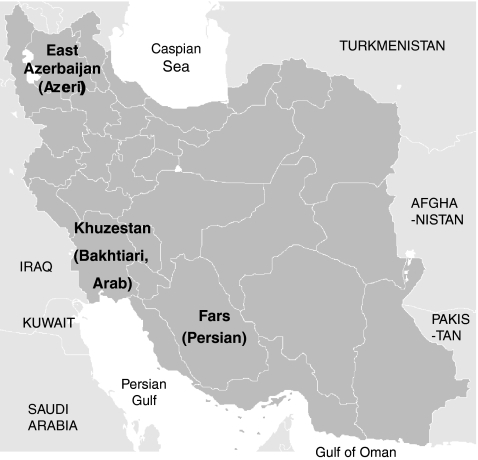
A total of 504 unrelated individuals belonging to four native populations of Iran were included in this study (Fig. 1). Persian people (n = 248), who speak Persian as their primary language, live in Fars province. Azeri people (n = 84) live in East Azerbaijan province and speak Turkic language. Bakhtiari people (n = 96) live in Khuzestan province and speak Luri, a dialect of Persian language. Arab people (n = 76) live in Khuzestan province and speak Arabic. The study was reviewed and approved by the appropriate Institutional Review Boards of human research protection. Genomic DNA was extracted from peripheral blood samples using either standard salting out method or by QIAamp blood kit (Qiagen, Hilden, Germany). The quality and quantity of DNA samples were determined by UV spectrophotometry, and the concentration was adjusted to 100 ng/μL.
Fig. 1.
Map of Iran showing the provinces of four study populations. DNA samples of Persian population were collected from Fars province, Azeri were collected from East Azerbaijan province, and Bakhtiari and Arab were collected from Khuzestan province
KIR genotyping
The presence and absence of 16 distinct KIR genes was determined using our recently developed duplex sequence-specific priming-based polymerase chain reaction (SSP-PCR) typing system (Ashouri et al. 2009). The unique and unusual KIR genotypes were further confirmed by re-typing using our alternative SSP-PCR typing method (Du et al. 2007). Both of the typing methods were validated extensively using the UCLA International KIR exchange reference DNA samples, which provided identical KIR genotyping results, indicating that the specificity and sensitivity of the two methods were comparable (Ashouri et al. 2009).
The KIR genotyping data of world populations used for comparison in this study was extracted from the following publications: Han Chinese (Jiang et al. 2005), Korean (Whang et al. 2005), Japanese (Yawata et al. 2002b), Vietnamese, Australian Aborigine (Toneva et al. 2001), Thai, British Caucasian, Palestinian Arab (Norman et al. 2001), Warao, Bari, Yucpa (Gendzekhadze et al. 2006), Australian Caucasian (Witt et al. 1999), New York Caucasian (Hsu et al. 2002), Finnish, French Caucasian, Senegal African, Guadeloupe Caribbean, and Reunion, a population from Indian Ocean origin (Denis et al. 2005), American Caucasian, Hispanic, African American (Du et al. 2007), Greek (Niokou et al. 2003), Afro-Caribbean, Trinidad Asian, Pakistani (Norman et al. 2002), North Indian (Rajalingam et al. 2002), Chinese, Malay and Indian migrants in Singapore (Lee et al. 2008), Parsi and Maharastrian of India (Kulkarni et al. 2008), Paravar, Kanikar, Mollukurumba (Rajalingam et al. 2008), Basque population (Santin et al. 2006), Cook Island, Samoan, Tokelau, Tongan (Velickovic et al. 2006), Mestizo, Huichol, Purepecha, Tarahumara (Gutierrez-Rodriguez et al. 2006), Northern Irish (Middleton et al. 2007), Wichis and Chiriguanos (Flores et al. 2007), and Amazonian Amerindian (Ewerton et al. 2007).
Prediction of haplogroups from genotypes
KIR gene content of a given individual is conventionally called “KIR genotype,” which is variable among individuals. The KIR gene content was used to infer group A and B KIR haplotypes and to assign each person to one of three genotypes: AA, BB, and AB. Individuals having only genes of the group A KIR haplotypes (KIR3DL3-2DL3-2DL1-2DP1-3DP1-2DL4-3DL1-2DS4-3DL2) were considered to be homozygous for the A haplotype and assigned the KIR genotype AA. Individuals lacking any of the four A haplotype associated genes (KIR2DL1, 2DL3, 3DL1, and 2DS4) that have a known function and vary among individuals in their existence were regarded to be homozygous for group B haplotypes and assigned the KIR genotype BB. All other individuals were regarded to be heterozygous for A and B haplotypes and assigned the KIR genotype AB. The individuals with AB genotypes had all nine genes present on the A haplotype, as well as one or more B haplotype specific genes (2DL2, 2DL5, 2DS1, 2DS2, 2DS3, 2DS5, and 3DS1). The AB and BB genotypes were previously referred together as KIR genotype Bx (McQueen et al. 2007).
Classification of genotypes on the basis of centromeric and telomeric gene clusters
Based on the linkage disequilibrium, we recognized two frequently occurring gene clusters (Du et al. 2008). One cluster comprises KIR2DS2-2DL2-2DS3-2DL5 genes and is located at the centromeric half of the KIR gene complex, while another cluster comprises KIR3DS1-2DL5-2DS1-2DS5 genes and is located at the telomeric half of the complex. For simplicity, we call these clusters C4 and T4, in which “C” represents centromeric, “T” represents telomeric, and “4” indicates the number of genes. On the basis of the presence and absence of C4 and T4 clusters, the Bx genotypes were further divided into the following four subsets: C4Tx (presence of C4 and absence of T4), CxT4 (absence of C4 and presence of T4), C4T4 (presence of both C4 and T4), and CxTx (absence of both C4 and T4). These Bx subsets were substantially variable in activating KIR gene content.
Data analysis and statistical methods
The percentage of individuals carrying each KIR gene in four population groups was determined by direct counting (individuals positive for the gene divided by the individuals tested per population × 100). Frequencies of A and B haplotypes were calculated using the following formula: group A = 2nAA + nAB/2N and group-B = 2nBB + nAB/2N, where nAA, nAB, and nBB were the numbers of AA, AB, and BB genotypes and N was the total number of individuals tested. Differences between populations in the frequencies of individuals carrying each KIR gene and genotype were estimated by two-tailed Fisher exact probability (p) test, and p < 0.05 was considered to be statistically significant. The principal components analysis (PCA) of carrier frequency of KIR genes was carried out using the Minitab statistical software.
Results
Frequency of activating KIR gene carriers differ among Iranian populations
All 16 known KIR genes were detected in each of the native Iranian populations analyzed in this study, and the frequencies of individuals carrying each KIR gene were compared in Table 1. Four framework KIR genes (2DL4, 3DL2, 3DL3 and 3DP1) were detected in all 504 individuals analyzed in this study. Overall, the A haplotype associated KIR genes occurred more frequently than the B haplotype associated KIR genes, and their frequencies were comparable between populations. Arab and Azeri revealed similar carrier frequency of each KIR gene but differed considerably from those of Persian and Bakhtiari populations (Table 1). Particularly, the carriers of KIR2DL5, 2DS2, and 2DS3 were more in Arab and Azeri populations compared to the Persian and Bakhtiari populations. The carriers of KIR3DS1 and 2DS5 occurred most frequently in Bakhtiari, and the difference was statistically significant in comparison with Persian population.
Table 1.
Comaprison of carrier frequency of KIR genes in four Iranian populations
| KIR | Persian(Per) | Bakhtiari (Bak) | Arab (Arb) | Azeri (Aze) | p values | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| n = 248 | n = 96 | n = 76 | n = 84 | Per vs. Bak | Per vs. Arb | Per vs. Aze | Bak vs. Arb | Bak vs. Aze | Arb vs. Aze | |
| %F (N) | %F (N) | %F (N) | %F (N) | |||||||
| A haplotype associated KIR genes | ||||||||||
| 2DL1 | 98.0 (244) | 94.8 (91) | 100 (76) | 98.0 (83) | ||||||
| 2DL3 | 91.0 (226) | 89.6 (86) | 89.5 (68) | 89.2 (75) | ||||||
| 3DL1 | 94.0 (238) | 95.8 (92) | 85.5 (65) | 90.5 (76) | ||||||
| 2DS4 | 96.0 (239) | 97.9 (94) | 98.7 (75) | 98.8 (83) | ||||||
| B haplotype associated KIR genes | ||||||||||
| 2DL2 | 56.8 (141) | 54.1 (52) | 63.1 (48) | 67.9 (57) | ||||||
| 2DL5 | 58.0 (144) | 54.1 (52) | 67.1 (51) | 73.8 (62) | 0.013 | 0.0083 | ||||
| 3DS1 | 33.0 (83) | 45.8 (44) | 42.1 (32) | 38.0 (32) | 0.035 | |||||
| 2DS1 | 35.0 (89) | 42.7 (41) | 44.7 (34) | 39.2 (33) | ||||||
| 2DS2 | 54.0 (135) | 49.0 (47) | 56.3 (49) | 70.2 (59) | 0.015 | 0.046 | 0.0041 | |||
| 2DS3 | 38.3 (95) | 27.1 (26) | 50.0 (38) | 53.5 (45) | 0.015 | 0.0025 | 0.0004 | |||
| 2DS5 | 25.4 (63) | 39.6 (38) | 35.5 (27) | 34.5 (29) | 0.012 | |||||
| Framework genes/pseudogenes | ||||||||||
| 2DL4 | 100 (248) | 100 (96) | 100 (76) | 100 (84) | ||||||
| 3DL2 | 100 (248) | 100 (96) | 100 (76) | 100 (84) | ||||||
| 3DL3 | 100 (248) | 100 (96) | 100 (76) | 100 (84) | ||||||
| 2DP1 | 98.0 (243) | 96.9 (93) | 98.7 (75) | 100 (84) | ||||||
| 3DP1 | 100 (248) | 100 (96) | 100 (76) | 100 (84) | ||||||
Frequency (%F) of carriers of each gene is expressed as a percentage and defined as the number of individuals carrying the gene (N) divided by the number of individuals studied (n) in the given population group. The p values are given only for those pairwise comparisons indicating significant (<0.05) differences
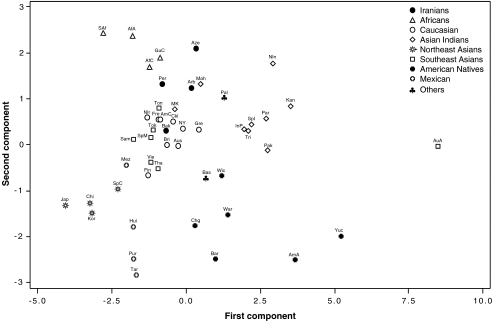
The carrier frequency of variably occurring KIR genes in four Iranian populations were compared with those reported in other ethnic populations using the PCA analysis (Fig. 2). Apart from a few outliers, distinct geographic clusters of Africans, Northeast Asians, Mexicans, American Natives, Asian Indians, and Caucasians were noticed on the PCA plot (Fig. 2). Four Iranian populations studied in this paper were mapped somewhat close to each other but considerably isolated from the Northeast Asians, Mexicans, and American Natives. Bakhtiari was plotted within the Caucasian group, while Persian was mapped between Caucasian and African groups. Iranian Arab was more closely clustered to the Maharashtrians, a Western Indian population than the Palestine Arab. Azeri stood isolated and revealed some affinity to the Iranian Arab.
Fig. 2.
Principal component analysis (PCA) of carrier frequency of nine variable KIR genes. The PCA graph built upon the frequencies of individuals carrying nine variably occurring KIR genes (2DL1-3, 2DS1-4, 3DL1, and 3DS1) shows a global view relationship between the four Iranian populations studied in this paper and other previously reported world populations. The other seven genes (2DL5, 2DS5, 2DL4, 3DL2, 3DL3, 2DP1, and 3DP1) were excluded from the analysis because they were either invariably present in all individuals or not typed in some populations. Jap Japanese, Chi Han Chinese, Kor Korean, SpC Singapore Chinese, Hui Huichol, Pur Purepecha, Tar Tarahumara, Chg Chiriguanos, Wic Wichis, War Warao, Bar Bari, AmA Amazonian Amerindian, Yuc Yucpa, Vie Vietnamese, Tha Thai, Reu Reunion, Gre Greek, CkI Cook Island, Sam Samoan, Tok Tokelau, Ton Tongan, Mez Mestizo, Bri British Caucasian, Aus Australian Caucasian, NIr Northern Irish, NY New York Caucasian, Fin Finnish, Fre French Caucasian, AmC American Caucasian, SpM Singapore Malay, His Hispanic, MK Mollukurumba, SAf Senegal African, GuC Guadeloupe Caribbean, AfA African American, AfC Afro-Caribbean, Per Persian, Bak Bakhtiari, Arb Iranian Arab, Aze Azeri, Pal Palestinian Arab, Mah Maharashtrian, Tri Trinidad Asian, Pak Pakistani, NIn North Indian, Par Paravar, Kan Kanikar, InP Indian Parsi, Bas Basque population, SpI Singapore Indians, AuA Australian Aborigine
Each Iranian population displays distinct KIR gene content diversity
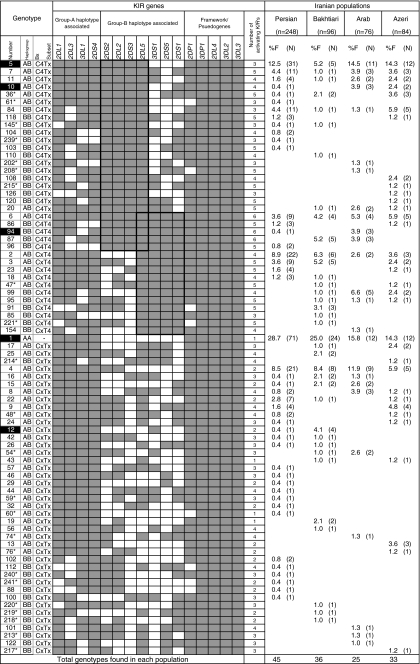
Within the study panel of 504 unrelated Iranians, we found 78 distinct KIR gene content profiles (genotypes) carrying a different number and combination of 16 KIR genes (Fig. 3). Only 10% of these genotypes (1, 2, 4–7, 11, and 84) were observed in all four Iranian populations, but their combined frequencies were 72.6% in Persian, 52.1% in Bakhtiari, 57.9% in Arab, and 55.9% in Azeri. Comparison with other ethnic populations revealed that only Caucasian populations carried all of these eight KIR genotypes, and their combined frequencies were comparable to those observed in Iranian populations (Supplement Figure 1a, b). Genotype 1, the homozygous combination of A haplotypes, occurred most frequently in Persian (28.7%), Bakhtiari (25%), and Arab (15.8%; Fig. 3). This is the only genotype found in all ethnic populations investigated to date, and its highest occurrence was reported in the Northeast Asians (Jiang et al. 2005; Whang et al. 2005; Yawata et al. 2002b) (Supplement Figures 1a, b). In Azeri, genotypes 1 and 5 occur at similar frequencies, 14.3% each. Genotype 5 was the second most common profile in Persian (12.5%) and Arab (14.5%), while it occurred only 5.2% in Bakhtiari populations.
Fig. 3.
KIR gene content diversity of Iranian populations. Within 504 unrelated individuals representing four linguistic Iranian populations, 78 genotypes that differed by the presence (shaded box) and absence (white box) of 16 KIR genes were observed. The frequency of each genotype is presented in percentage frequency (%F) and defined as the number of individuals carrying the genotype (N) divided by the number of individuals studied (n) in the given population. Genotypes with identical gene content listed in this figure and in the Supplementary Figure 1a, b are marked with the same number. Unique genotypes that were not reported from other ethnic populations are identified by asterisk. Based on the gene content, genotypes were grouped as we described in the text. The genotypes that significantly differed (p < 0.05) among Iranian populations are marked by dark boxes: genotypes 1 (Persian vs. Arab, p = 0.035, Persian vs. Azeri, p = 0.013), 5 (Bakhtiari vs. Azeri, p = 0.044), 10 (Persian vs. Arab, p = 0.041), 12 (Persian vs. Bakhtiari, p = 0.041), 94 (Persian vs. Arab, p = 0.041)
The majority of the genotypes characterized in this study (43/78, 55%) were unique to one of the four Iranian populations, and their combined frequency in the entire study panel was 11.9% (Fig. 3). Eighteen of these 43 unique genotypes (59–61, 74, 76, 202, 208, 213–215, 217–221, and 239–241) were not reported in other ethnic populations studied previously (Supplement Figures 1a, b). Five genotypes (36, 47, 48, 54, and 145) that occurred in more than one Iranian population were also not reported in other ethnic populations. Therefore, a set of 23 KIR genotypes appeared to be unique to the Iranians and are yet to be detected in other populations (Fig. 3 and Supplement Figures 1a, b).
Nearly 75% of the Iranian Arab and Azeri carry a B KIR haplotype
Of the 78 observed genotypes, 40 were predicted to be the heterozygous combination of A and B haplotypes (AB genotypes), 37 were predicted to be the homozygous combination of B haplotypes (BB genotypes), and one was the homozygous combination of A haplotype (AA genotypes; Fig. 3). Over 55% of each Iranian population carried AB genotypes with the highest prevalence in Azeri (65.5%; Table 2). The predicted A haplotypes occurred more frequently than the B haplotypes in Persian and Bakhtiari, and the extreme of this scenario was reported in Northeast Asians (Jiang et al. 2005; Whang et al. 2005; Yawata et al. 2002b; Supplement Figures 1a, b). In contrast, predicted B haplotypes occurred more frequently in Arab and Azeri compared to the A haplotypes (Table 2). This scenario was previously observed in the natives of America, India, and Australia (Gendzekhadze et al. 2006; Norman et al. 2002; Rajalingam et al. 2008; Toneva et al. 2001).
Table 2.
Comparison of genotypes, haplotypes and linkage groups in four Iranian populations
| Types | Persian (Per) | Bakhtiari (Bak) | Arab (Arb) | Azeri (Aze) | Per vs. Bak | Per vs. Arb | Per vs. Aze | Bak vs. Arb | Bak vs. Aze | Arb vs. Aze |
|---|---|---|---|---|---|---|---|---|---|---|
| n = 248 | n = 96 | n = 76 | n = 84 | |||||||
| %F (N) | %F (N) | %F (N) | %F (N) | |||||||
| AA genotype | 28.7 (71) | 25.0 (24) | 15.8 (12) | 14.3 (12) | 0.035 | 0.013 | ||||
| BB genotypes | 12.8 (32) | 18.3 (18) | 24.6 (19) | 20.2 (17) | 0.0132 | |||||
| AB genotypes | 58.4 (145) | 55.8 (54) | 59.1 (45) | 65.5 (55) | ||||||
| C4Tx genotypes | 27.4 (68) | 13.5 (13) | 31.6 (24) | 40.5 (34) | 0.0068 | 0.028 | 0.000043 | |||
| CxT4 genotypes | 15.3 (38) | 20.8 (20) | 11.8 (9) | 11.9 (10) | ||||||
| C4T4 genotypes | 6.0 (15) | 9.3 (9) | 13.1 (10) | 7.1 (6) | ||||||
| CxTx genotypes | 22.5 (56) | 31.2 (30) | 27.3 (21) | 26.1 (22) | ||||||
| A haplogroups | 57.6 (286) | 53.1 (102) | 44.7 (68) | 46.4 (78) | 0.0053 | 0.012 | ||||
| B haplogroups | 42.3 (210) | 46.8 (90) | 55.2 (84) | 53.5 (90) | 0.0053 | 0.012 | ||||
| C4 gene-cluster | 33.5 (83) | 23.0 (22) | 44.7 (34) | 47.6 (40) | 0.026 | 0.0031 | 0.00056 | |||
| T4 gene-cluster | 21.4 (53) | 30.2 (29) | 25.0 (19) | 19.0 (16) |
The haplotype A and B were determined by using the following formula: group A = 2NAA + NAB/2n and group B = 2NBB + NAB/2n, where NAA, NAB, and NBB are the numbers of AA, AB, and BB genotypes, n = total number of individual.
Nearly half of the Iranian Arab and Azeri populations carried KIR2DS2-2DL2-2DS3-2DL5 gene cluster
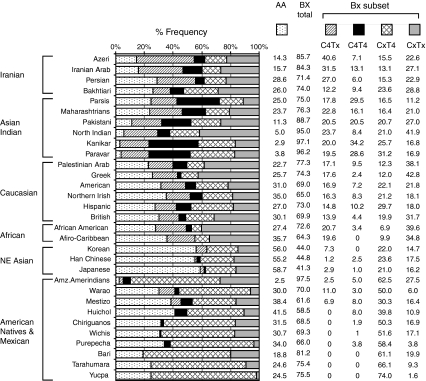
All four Iranian populations investigated in these study carried all four subsets of Bx genotypes (CxT4, C4Tx, C4T4, and CxTx), while some ethnic populations studied previously lacked one or more of these subsets (Fig. 4). Within our study panel of 504 Iranians, individuals displaying C4Tx genotypes carried three to five activating KIR genes (61.9% carried three activating KIRs), while individuals displaying CxT4 genotypes carried three to five activating KIR genes (59.0% carried four activating KIRs), individuals displaying C4T4 genotypes carried five to six activating KIR genes (75.5% carried six activating KIRs), and individuals displaying CxTx carried one to four activating KIRs (60.2% carried two activating KIRs). The Azeri (40.5%), Arab (31.6%), and Persian (27.4%) comprised the higher frequencies of Bx genotypes with C4Tx configuration compared to any other population studied thus far (Fig. 4). Interestingly, the C4Tx subset was virtually absent from American natives, in whom CxT4 occurred predominantly at a frequency of 70%. In Bakhtiari, the Bx genotypes missing C4 and T4 gene clusters (CxTx) were more common compared to the other three Iranian populations. Overall, the constellation of Bx genotypes of Iranians was comparable to Asian Indians and Caucasians but differed substantially from the American natives, Northeast Asians, and Africans (Fig. 4).
Fig. 4.
Subsets of KIR gene content profiles and their frequency in populations. The frequencies of distinct KIR genotype subsets in four Iranian populations are shown in comparison with other previously studied populations. The individuals carrying KIR3DL3-2DL3-2DL1-2DP1-3DP1-2DL4-3DL1-2DS4-3DL2, a fixed gene content characteristic of A haplotypes, were considered to have AA genotypes (two copies of A haplotypes). Remainders carried Bx genotypes, which comprised either one copy of A haplotype and one copy of B haplotype (AB genotypes) or two copies of B haplotypes (BB genotypes). Based on the presence and absence of two distinct gene clusters (C4, KIR2DS2-2DL2-2DS3-2DL5; T4, KIR3DS1-2DL5-2DS1-2DS5), the Bx genotype carries were divided into four subsets: C4Tx (presence of C4 and absence of T4), CxT4 (absence of C4 and presence of T4), C4T4 (presence of both C4 and T4), CxTx (absence of both C4 and T4)
Discussion
Persian and Bakhtiari populations were the descendants of ancient Elamites and Aryans, who arrived in parts of Greater Iran from central Asia in the second millennium bc. Genetic studies describing mitochondrial DNA (mtDNA) sequence variation, Y chromosome SNP, and HLA gene polymorphism revealed close affinities among Persian, Bakhtiari, and European Caucasian populations (Farjadian et al. 2009; Nasidze et al. 2008). Consistent with the ethnic ancestry and genetic homology, the KIR gene content diversity determined in Bakhtiari and Persian populations were comparable to those reported in Caucasian populations and revealed a balance of A and B KIR haplotypes. Compared to the Bakhtiari and Caucasian populations, the Persian displayed a higher frequency of genotype 5, which lacks T4 gene cluster (KIR3DS1, 2DS5 and 2DS1 genes). The genotype 5 occurred most frequently in African populations (Denis et al. 2005; Du et al. 2007; Norman et al. 2002) but is completely absent in American natives (Ewerton et al. 2007; Flores et al. 2007; Gendzekhadze et al. 2006). Unlike the Bakhtiari, a nomadic pastoralist tribal group that straddles the central Zagros Mountains in the province of Khuzistan, the Persian had several historical admixtures, which presumably is the source for the high incidence of genotype 5.
Compared to the geographic neighbors, a greater variability was recently observed at the KIR locus in Indian Parsi, a descendent of Iranian Zoroastrians who emigrated to Western India in the seventh century ad (Kulkarni et al. 2008). Notably, the Indian Parsi had a significantly higher frequency of KIR3DS1 than did the Northern Indian populations, and possible evolutionary pressure was suggested to retain such high frequency of KIR3DS1 in Indian Parsi. The Y chromosome SNP data showed that the Parsis of India resembled Iranian populations rather than their Indian neighbors (Qamar et al. 2002). KIR gene content diversity data of native Iranian populations was not available at that time for comparison. Herein, we compared the Indian Parsi data with those of other world populations, including four native Iranian populations characterized in this study and three Southern Indian tribal populations studied recently (Rajalingam et al. 2008). The analyses revealed that the KIR gene content profiles of Indian Parsi were more comparable to the Indian populations than the native Iranians. Specifically, 29.5% of Parsis of India were the carriers of C4T4 Bx genotypes, while only 6% of the Persian displayed this constellation. Consequently, the Parsis of India comprised more activating KIR genes than the Iranian Persian. It is intriguing to postulate that the Parsis of India gained the C4 (KIR2DS2-2DL2-2DS3-2DL5) and T4 (KIR3DS1-2DL5-2DS5-2DS1) gene clusters by accumulating the activating KIR genes during their migration to India or following their settlement in India through population bottlenecks and episodes of selection by infectious disease.
The Iranian Arab population mainly occupies the Khuzestan province of Iran. Historical evidence indicates that their ancestors, Arab tribes such as the Bakr bin Wael and Bani Tamim, entered Iran in the seventh century ad, although there may have been an earlier Arabic presence in Iran (Morony 2006). The mtDNA HV1 sequence polymorphisms and Y chromosome bi-allelic diversity showed that the North African Arabs were far more distant genetically from the Iranian Arab (Nasidze et al. 2008). Moreover, the Iranian Arab shared close relatedness to the neighboring geographic groups. Haplogroups J2 and G were especially intriguing because they were found in very high frequencies in Bakhtiari and Iranian Arab (Nasidze et al. 2008). Despite the genetic affinity and geographic proximity, the Iranian Arab carried the highest frequency of B haplotypes among the four Iranian populations, while the Bakhtiari displayed a balance of A and B haplotypes. Intriguingly, the KIR genomic diversity of Iranian Arab was more similar to the Indian Maharashtrians and Iranian Azeri than the Palestinian Arab. Particularly, the Iranian Arab, Azeri, and Indian Maharashtrians displayed a comparable constellation of Bx genotypes with high frequencies of the C4 gene cluster (KIR2DS2-2DL2-2DS3-2DL5) compared to the Palestinian Arab (Supplement Figure 1b).
Comparative analyses of KIR data from world populations revealed a link between the prehistoric human migrations and the evolution of two groups of KIR haplotypes distinguished by their content of activating KIR genes (Rajalingam et al. 2008). The natives of America, India, and Australia, who had extensive prehistoric migrations, carried high frequencies of B haplotypes, and presumably, they acquired these activating KIR enriched haplotypes to survive different environmental challenges on their journey. The natives of India and Australia were considered to be the most ancient human dispersal out of Africa, which happened about 60,000 years ago, most likely via the tropical coast of the Arabian peninsula, India, Southeast Asia, and Australia. An increasing frequency cline of B haplotypes from Arabs (51.8% in Palestinian Arab and 55.3% in Iranian Arab) toward Pakistani (60.5%), Indian tribes (~70%), and Australian aborigines (>70%) suggests that the expansion of B haplotypes in the most ancient migratory group began in Arabian peninsula that bridges Africa and Asia.
Azeri, one of the major Iranian ethnic populations that speaks an Indo-European language, has mixed genetic (mtDNA and Y chromosome) and cultural elements of Persian, Caucasian, and Turkic people (Ashrafian-Bonab et al. 2007; Nasidze et al. 2003; Quintana-Murci et al. 2004). However, by displaying a dominance in B KIR haplotypes that carry C4 gene cluster (KIR2DS2-2DL2-2DS3-2DL5), the Azeri population was more related to Iranian and Indian populations. It is not clear whether a local specific selection or a racial admixture is responsible for the dominance of B haplotypes in Azeri. In conclusion, the genetic pool supplied by the waves of prehistoric migration, subsequent racial admixture, and local-specific selection appeared to be the force that determined the KIR diversity in modern Iranians.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Comparison of Iranian KIR genotypes with those reported in global populations. Distinct KIR genotypes and their frequencies in Iranian populations were compared with those characterized previously in other populations. A total of 242 distinct KIR genotypes in the combined pool of 4,638 unrelated individuals were observed, which differ from each other by the presence (shaded box) and absence (white box) of 16 KIR genes. The 'a' identifies populations not tested for 2DL5, 'b' identifies populations not tested for 3DL3, 'c' identifies populations not tested for 2DP1 and 3DP1, 'd' identifies populations not tested for 2DL4, 'e' identifies populations that were typed 2DS5 using a primer set recognizing an unreal substitution (resulting in false-negative results), 'f' identifies populations for which only the frequently occurring KIR genotypes were published, and therefore, the KIR genotype data for 35% Vietnamese, 11% Finnish, 18% French, 20% Senegal African, 30% Guadeloupe Caribbean, 24% Reunion, and 37% Australian Aborigine are not available for this analysis, 'g' identifies populations not tested for 2DP1. Frequencies of genotypes that occur in over 7% in the populations are highlighted with gray shade. 'nt' Not tested for the given KIR gene, 'nd' not determined for the given population because the complete gene profile data is not available, 'n' number of individuals studied in each population. (XLS 272 kb)
(XLS 266 kb)
Acknowledgments
This work was supported by the start-up funds from the UCLA Department of Pathology and Laboratory Medicine (to R.R.) and by Shiraz Institute for Cancer Research (grant number 85-3073 to A.G.). Elham Ashouri was supported by a fellowship from the Ministry of Health and Medical Education, The Islamic Republic of Iran. The authors thank Dr. Samadi, Dr. Shalbafzadeh, Dr. Afrasiabi, Dr. Dehbozorgian, Miss Chenari, Mr. Saki, Mr. Naeimi, Mr. Aliparsti, and Mr. Malekzadeh for their help in sample collection.
Open Access This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s00251-009-0378-7) contains supplementary material, which is available to authorized users.
Contributor Information
Abbas Ghaderi, Phone: +98-711-2303687, FAX: +98-711-2304952, Email: ghaderia@sums.ac.ir.
Raja Rajalingam, Phone: +98-310-8251467, FAX: +98-310-2063216, Email: rrajalingam@mednet.ucla.edu.
References
- Ashouri E, Ghaderi A, Reed EF, Rajalingam R (2009) A novel duplex SSP-PCR typing method for KIR gene profiling. Tissue Antigens 73. doi:10.1111/j.1399-0039.2009.01259.x [DOI] [PubMed]
- Ashrafian-Bonab M, Lawson Handley LJ, Balloux F (2007) Is urbanization scrambling the genetic structure of human populations? A case study. Heredity 98:151–156. doi:10.1038/sj.hdy.6800918 [DOI] [PMC free article] [PubMed]
- Denis L, Sivula J, Gourraud PA, Kerdudou N, Chout R, Ricard C, Moisan JP, Gagne K, Partanen J, Bignon JD (2005) Genetic diversity of KIR natural killer cell markers in populations from France, Guadeloupe, Finland, Senegal and Reunion. Tissue Antigens 66:267–276. doi:10.1111/j.1399-0039.2005.00473.x [DOI] [PubMed]
- Du Z, Gjertson DW, Reed EF, Rajalingam R (2007) Receptor-ligand analyses define minimal killer cell Ig-like receptor (KIR) in humans. Immunogenetics 59:1–15. doi:10.1007/s00251-006-0168-4 [DOI] [PubMed]
- Du Z, Sharma SK, Spellman S, Reed EF, Rajalingam R (2008) KIR2DL5 alleles mark certain combination of activating KIR genes. Genes Immun 9:470–480. doi:10.1038/gene.2008.39 [DOI] [PubMed]
- Ewerton PD, Leite Mde M, Magalhaes M, Sena L, Melo dos Santos EJ (2007) Amazonian Amerindians exhibit high variability of KIR profiles. Immunogenetics 59:625–630. doi:10.1007/s00251-007-0229-3 [DOI] [PubMed]
- Farjadian S, Ota M, Inoko H, Ghaderi A (2009) The genetic relationship among Iranian ethnic groups: an anthropological view based on HLA class II gene polymorphism. Mol Biol Rep. doi:10.1007/s11033-008-9403-4 [DOI] [PubMed]
- Flores AC, Marcos CY, Paladino N, Capucchio M, Theiler G, Arruvito L, Pardo R, Habegger A, Williams F, Middleton D, Fainboim L (2007) KIR genes polymorphism in Argentinean Caucasoid and Amerindian populations. Tissue Antigens 69:568–576. doi:10.1111/j.1399-0039.2007.00824.x [DOI] [PubMed]
- Gendzekhadze K, Norman PJ, Abi-Rached L, Layrisse Z, Parham P (2006) High KIR diversity in Amerindians is maintained using few gene-content haplotypes. Immunogenetics 58:474–480. doi:10.1007/s00251-006-0108-3 [DOI] [PubMed]
- Gutierrez-Rodriguez ME, Sandoval-Ramirez L, Diaz-Flores M, Marsh SG, Valladares-Salgado A, Madrigal JA, Mejia-Arangure JM, Garcia CA, Huerta-Zepeda A, Ibarra-Cortes B, Ortega-Camarillo C, Cruz M (2006) KIR gene in ethnic and Mestizo populations from Mexico. Hum Immunol 67:85–93. doi:10.1016/j.humimm.2005.11.007 [DOI] [PubMed]
- Hou L, Steiner NK, Chen M, Belle I, Kubit AL, Ng J, Hurley CK (2008) Limited allelic diversity of stimulatory two-domain killer cell immunoglobulin-like receptors. Hum Immunol 69:174–178. doi:10.1016/j.humimm.2008.01.009 [DOI] [PubMed]
- Hsu KC, Liu XR, Selvakumar A, Mickelson E, O’Reilly RJ, Dupont B (2002) Killer Ig-like receptor haplotype analysis by gene content: evidence for genomic diversity with a minimum of six basic framework haplotypes, each with multiple subsets. J Immunol 169:5118–5129 [DOI] [PubMed]
- Jiang K, Zhu FM, Lv QF, Yan LX (2005) Distribution of killer cell immunoglobulin-like receptor genes in the Chinese Han population. Tissue Antigens 65:556–563. doi:10.1111/j.1399-0039.2005.00412.x [DOI] [PubMed]
- Khakoo SI, Carrington M (2006) KIR and disease: a model system or system of models? Immunol Rev 214:186–201. doi:10.1111/j.1600-065X.2006.00459.x [DOI] [PubMed]
- Kulkarni S, Single RM, Martin MP, Rajalingam R, Badwe R, Joshi N, Carrington M (2008) Comparison of the rapidly evolving KIR locus in Parsis and natives of India. Immunogenetics 60:121–129. doi:10.1007/s00251-008-0279-1 [DOI] [PubMed]
- Lanier LL (2005) NK cell recognition. Annu Rev Immunol 23:225–274. doi:10.1146/annurev.immunol.23.021704.115526 [DOI] [PubMed]
- Lee YC, Chan SH, Ren EC (2008) Asian population frequencies and haplotype distribution of killer cell immunoglobulin-like receptor (KIR) genes among Chinese, Malay, and Indian in Singapore. Immunogenetics 60:645–654. doi:10.1007/s00251-008-0321-3 [DOI] [PubMed]
- Martin AM, Kulski JK, Gaudieri S, Witt CS, Freitas EM, Trowsdale J, Christiansen FT (2004) Comparative genomic analysis, diversity and evolution of two KIR haplotypes A and B. Gene 335:121–131. doi:10.1016/j.gene.2004.03.018 [DOI] [PubMed]
- McQueen KL, Dorighi KM, Guethlein LA, Wong R, Sanjanwala B, Parham P (2007) Donor-recipient combinations of group A and B KIR haplotypes and HLA class I ligand affect the outcome of HLA-matched, sibling donor hematopoietic cell transplantation. Hum Immunol 68:309–323. doi:10.1016/j.humimm.2007.01.019 [DOI] [PMC free article] [PubMed]
- Middleton D, Meenagh A, Gourraud PA (2007) KIR haplotype content at the allele level in 77 Northern Irish families. Immunogenetics 59:145–158. doi:10.1007/s00251-006-0181-7 [DOI] [PubMed]
- Morony M (2006) Arab II: Arab conquest of Iran. The encyclopaedia Iranica. Center for Iranian Studies, Columbia University, New York
- Nasidze I, Quinque D, Rahmani M, Alemohamad SA, Stoneking M (2008) Close genetic relationship between Semitic-speaking and Indo-European-speaking groups in Iran. Ann Hum Genet 72:241–252. doi:10.1111/j.1469-1809.2007.00413.x [DOI] [PubMed]
- Nasidze I, Sarkisian T, Kerimov A, Stoneking M (2003) Testing hypotheses of language replacement in the Caucasus: evidence from the Y-chromosome. Hum Genet 112:255–261 [DOI] [PubMed]
- Niokou D, Spyropoulou-Vlachou M, Darlamitsou A, Stavropoulos-Giokas C (2003) Distribution of killer cell immunoglobulin-like receptors in the Greek population. Hum Immunol 64:1167–1176. doi:10.1016/j.humimm.2003.08.100 [DOI] [PubMed]
- Norman PJ, Carrington CV, Byng M, Maxwell LD, Curran MD, Stephens HA, Chandanayingyong D, Verity DH, Hameed K, Ramdath DD, Vaughan RW (2002) Natural killer cell immunoglobulin-like receptor (KIR) locus profiles in African and South Asian populations. Genes Immun 3:86–95. doi:10.1038/sj.gene.6363836 [DOI] [PubMed]
- Norman PJ, Stephens HA, Verity DH, Chandanayingyong D, Vaughan RW (2001) Distribution of natural killer cell immunoglobulin-like receptor sequences in three ethnic groups. Immunogenetics 52:195–205. doi:10.1007/s002510000281 [DOI] [PubMed]
- Parham P (2005) MHC class I molecules and KIRs in human history, health and survival. Nat Rev Immunol 5:201–214. doi:10.1038/nri1570 [DOI] [PubMed]
- Qamar R, Ayub Q, Mohyuddin A, Helgason A, Mazhar K, Mansoor A, Zerjal T, Tyler-Smith C, Mehdi SQ (2002) Y-chromosomal DNA variation in Pakistan. Am J Hum Genet 70:1107–1124. doi:10.1086/339929 [DOI] [PMC free article] [PubMed]
- Quintana-Murci L, Chaix R, Wells RS, Behar DM, Sayar H, Scozzari R, Rengo C, Al-Zahery N, Semino O, Santachiara-Benerecetti AS, Coppa A, Ayub Q, Mohyuddin A, Tyler-Smith C, Qasim Mehdi S, Torroni A, McElreavey K (2004) Where west meets east: the complex mtDNA landscape of the southwest and Central Asian corridor. Am J Hum Genet 74:827–845. doi:10.1086/383236 [DOI] [PMC free article] [PubMed]
- Rajalingam R, Du Z, Meenagh A, Luo L, Kavitha VJ, Pavithra-Arulvani R, Vidhyalakshmi A, Sharma SK, Balazs I, Reed EF, Pitchappan RM, Middleton D (2008) Distinct diversity of KIR genes in three southern Indian populations: comparison with world populations revealed a link between KIR gene content and pre-historic human migrations. Immunogenetics 60:207–217. doi:10.1007/s00251-008-0286-2 [DOI] [PubMed]
- Rajalingam R, Krausa P, Shilling HG, Stein JB, Balamurugan A, McGinnis MD, Cheng NW, Mehra NK, Parham P (2002) Distinctive KIR and HLA diversity in a panel of north Indian Hindus. Immunogenetics 53:1009–1019. doi:10.1007/s00251-001-0425-5 [DOI] [PubMed]
- Santin I, de Nanclares GP, Calvo B, Gaafar A, Castano L, Bilbao JR (2006) Killer cell immunoglobulin-like receptor (KIR) genes in the Basque population: association study of KIR gene contents with type 1 diabetes mellitus. Hum Immunol 67:118–124. doi:10.1016/j.humimm.2006.02.036 [DOI] [PubMed]
- Shilling HG, Guethlein LA, Cheng NW, Gardiner CM, Rodriguez R, Tyan D, Parham P (2002) Allelic polymorphism synergizes with variable gene content to individualize human KIR genotype. J Immunol 168:2307–2315 [DOI] [PubMed]
- Single RM, Martin MP, Gao X, Meyer D, Yeager M, Kidd JR, Kidd KK, Carrington M (2007) Global diversity and evidence for coevolution of KIR and HLA. Nat Genet 39:1114–1119. doi:10.1038/ng2077 [DOI] [PubMed]
- Toneva M, Lepage V, Lafay G, Dulphy N, Busson M, Lester S, Vu-Trien A, Michaylova A, Naumova E, McCluskey J, Charron D (2001) Genomic diversity of natural killer cell receptor genes in three populations. Tissue Antigens 57:358–362. doi:10.1034/j.1399-0039.2001.057004358.x [DOI] [PubMed]
- Trinchieri G (1989) Biology of natural killer cells. Adv Immunol 47:187–376. doi:10.1016/S0065-2776(08)60664-1 [DOI] [PMC free article] [PubMed]
- Uhrberg M, Valiante NM, Shum BP, Shilling HG, Lienert-Weidenbach K, Corliss B, Tyan D, Lanier LL, Parham P (1997) Human diversity in killer cell inhibitory receptor genes. Immunity 7:753–763. doi:10.1016/S1074-7613(00)80394-5 [DOI] [PubMed]
- Velickovic M, Velickovic Z, Dunckley H (2006) Diversity of killer cell immunoglobulin-like receptor genes in Pacific Islands populations. Immunogenetics 58:523–532. doi:10.1007/s00251-006-0124-3 [DOI] [PubMed]
- Vilches C, Parham P (2002) KIR: diverse, rapidly evolving receptors of innate and adaptive immunity. Annu Rev Immunol 20:217–251. doi:10.1146/annurev.immunol.20.092501.134942 [DOI] [PubMed]
- Whang DH, Park H, Yoon JA, Park MH (2005) Haplotype analysis of killer cell immunoglobulin-like receptor genes in 77 Korean families. Hum Immunol 66:146–154. doi:10.1016/j.humimm.2004.10.013 [DOI] [PubMed]
- Wilson MJ, Torkar M, Haude A, Milne S, Jones T, Sheer D, Beck S, Trowsdale J (2000) Plasticity in the organization and sequences of human KIR/ILT gene families. Proc Natl Acad Sci U S A 97:4778–4783. doi:10.1073/pnas.080588597 [DOI] [PMC free article] [PubMed]
- Witt CS, Dewing C, Sayer DC, Uhrberg M, Parham P, Christiansen FT (1999) Population frequencies and putative haplotypes of the killer cell immunoglobulin-like receptor sequences and evidence for recombination. Transplantation 68:1784–1789. doi:10.1097/00007890-199912150-00024 [DOI] [PubMed]
- Yawata M, Yawata N, Abi-Rached L, Parham P (2002a) Variation within the human killer cell immunoglobulin-like receptor (KIR) gene family. Crit Rev Immunol 22:463–482 [PubMed]
- Yawata M, Yawata N, McQueen KL, Cheng NW, Guethlein LA, Rajalingam R, Shilling HG, Parham P (2002b) Predominance of group A KIR haplotypes in Japanese associated with diverse NK cell repertoires of KIR expression. Immunogenetics 54:543–550. doi:10.1007/s00251-002-0497-x [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Below is the link to the electronic supplementary material.
Comparison of Iranian KIR genotypes with those reported in global populations. Distinct KIR genotypes and their frequencies in Iranian populations were compared with those characterized previously in other populations. A total of 242 distinct KIR genotypes in the combined pool of 4,638 unrelated individuals were observed, which differ from each other by the presence (shaded box) and absence (white box) of 16 KIR genes. The 'a' identifies populations not tested for 2DL5, 'b' identifies populations not tested for 3DL3, 'c' identifies populations not tested for 2DP1 and 3DP1, 'd' identifies populations not tested for 2DL4, 'e' identifies populations that were typed 2DS5 using a primer set recognizing an unreal substitution (resulting in false-negative results), 'f' identifies populations for which only the frequently occurring KIR genotypes were published, and therefore, the KIR genotype data for 35% Vietnamese, 11% Finnish, 18% French, 20% Senegal African, 30% Guadeloupe Caribbean, 24% Reunion, and 37% Australian Aborigine are not available for this analysis, 'g' identifies populations not tested for 2DP1. Frequencies of genotypes that occur in over 7% in the populations are highlighted with gray shade. 'nt' Not tested for the given KIR gene, 'nd' not determined for the given population because the complete gene profile data is not available, 'n' number of individuals studied in each population. (XLS 272 kb)
(XLS 266 kb)