Abstract
The segment polarity gene family, and its gene regulatory network, is at the basis of Drosophila embryonic development. The network's capacity for generating and robustly maintaining a specific gene expression pattern has been investigated through mathematical modelling. The models have provided several useful insights by suggesting essential network links, or uncovering the importance of the relative time scales of different biological processes in the formation of the segment polarity genes' expression patterns. But the developmental pattern formation process raises many other questions. Two of these questions are analysed here: the dependence of the signalling protein sloppy paired on the segment polarity genes and the effect of cell division on the segment polarity genes' expression patterns. This study suggests that cell division increases the robustness of the segment polarity network with respect to perturbations in biological processes.
Keywords: genetic networks, segment polarity genes, robustness
1. Introduction
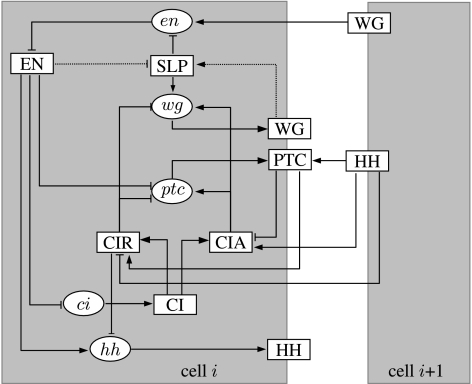
During the initial stages of development of the fruit fly Drosophila melanogaster, three families of genes are successively activated (Sanson 2001): the gap genes; the pair-rule genes; and the segment polarity genes. The resulting gene expression patterns contribute to gradually break the symmetry of the fertilized egg and accompany its transformation into a segmented embryo. Around stages 6 and 7 of embryonic development (i.e. approx. 3 hours after fertilization), the family of genes known as the segment polarity genes is activated: their expression patterns will define the position of the parasegmental grooves, the boundaries of the segments that form the body of the fruit fly. The segment polarity genes refine and maintain their expression through the network of intra- and intercellular regulatory interactions shown in figure 1. The stable expression pattern of these genes (specifically the expression of wingless and engrailed) defines and maintains the borders between different parasegments and contributes to subsequent developmental processes, including the formation of denticle patterns and of appendage primordia (Hooper & Scott 1992; Wolpert et al. 1998).
Figure 1.
Interactions for the segment polarity network. Squares represent proteins and ellipses represent mRNAs. Cell-to-cell communication is considered among nearest neighbours, through the wingless and hedgehog proteins. Only neighbouring cell i+1 is depicted, but both the cells i−1 and i+1 have a similar effect on cell i. This model was developed in Albert & Othmer (2003), except for the activation and inhibition links on protein SLP, which are introduced in this work. See main text for more details.
To try to understand and study this network and its properties, a first mathematical model was proposed by von Dassow et al. (2000). Some improvements to this model, including an alternative mathematical description (Albert & Othmer 2003) and analysis of its properties (Ingolia 2004; Chaves et al. 2006) have recently been presented. A common conclusion from these studies is that the network structure plays a fundamental role, that is, the interconnections among the genes and proteins that constitute the segment polarity network are the crucial factor for the robustness of the expression pattern with respect to biological (‘small’) perturbations.
Albert & Othmer (2003) proposed a Boolean version of the continuous model described in von Dassow et al. (2000). Boolean models provide a qualitative representation of a system, consisting of the nodes of the network (whose values are either 0=‘OFF’ or 1=‘ON’) and a set of logical rules to describe the regulatory links among them (activation or inhibition interactions). The choice of Boolean modelling in this context is very natural, as many genetic regulatory functions are known, but hardly any kinetic or binding parameters are available for the segment polarity network. Advantages of using Boolean rules include a clear modelling of the structure of interactions (i.e. the links among the system's nodes) and very intuitive qualitative representation of the system and its behaviour. In addition, various analytical methods can be used to study Boolean models (Glass & Kauffman 1973; Thomas 1973; Edwards & Glass 2000; Chaves et al. 2005).
The Boolean model proposed in Albert & Othmer (2003) improves on von Dassow et al. (2000) by adding an activating signal that initiates the expression of segment polarity genes. This is the protein sloppy paired (SLP) that is part of the pair-rule gene family. (Actually, there are two different proteins encoded by two genes sloppy paired, but they are known to have similar roles, and thus are referred to from now on as only one protein, SLP.) Further analysis of this model has provided many useful insights and contributed to better understanding the segment polarity network (Chaves et al. 2005, 2006); namely, it has shown the importance of considering different time scales for different biological processes (e.g. transcription and translation are generally slower than protein conformational changes; Papin et al. 2005) in the correct formation of the segment polarity genes' expression. It has also raised further questions, so, in this work, we have chosen to focus on two important issues. (i) The SLP-activating signal has been assumed constant; however, evidence shows (Alexandre & Vincent 2003) that it can also depend on the expression of some segment polarity genes. One of our goals is to introduce a Boolean rule to describe SLP, and thus obtain a more autonomous module. (ii) Most cited models of the segment polarity network assume four-cell-wide segments. However, evidence shows that there are rounds of cell division at stages 8 and 10 (Gonzalez et al. 1991; Hooper & Scott 1992). We will investigate the effect of segment width on the robustness properties of the segment polarity network.
2. Boolean models for genetic networks
In the model, each mRNA or protein is represented by a node of a network, and the interactions between them are encoded as directed edges (figure 1). The state of each node is 1 or 0, depending on whether the corresponding substance is present or not. The states of the nodes can change in time, and the next state of node ℓ is determined by a Boolean (logical) function of its state and the states of those nodes that have edges incident on it. In general, a Boolean or logical function is written as a statement acting on the inputs using the logical operators ‘AND’, ‘OR’ and ‘NOT’ and its output is 1 (0) if the statement is true (false).
The functions determining the state of each node are constructed from the interactions between nodes (such as displayed in figure 1) according to the following rules.
mRNAs/proteins are synthesized if their transcriptional activators/mRNAs are present.
The effect of transcriptional activators and inhibitors is never additive, but, rather, inhibitors are dominant.
mRNAs decay in the next updating step if not transcribed.
Transcription factors and proteins undergoing post-translational modification decay if their mRNA is not present.
Consider a regulatory network with N nodes, X1, …, XN. The expression of each node along time can be computed by iterating the Boolean rules, to obtain a discrete sequence
where denotes the expression of node ℓ at time instant kT, where T represents a (fixed) time unit. To compute , using X at the previous instants, several algorithms are available to iterate the Boolean rules. Some of these are briefly described next. The standard synchronous algorithm assumes that all N nodes are simultaneously updated, i.e.
Asynchronous algorithms allow different nodes to be updated at different times, for example according to a random order. Assuming that all nodes are updated exactly once during each time unit T, an asynchronous algorithm can be constructed by randomly assigning an updating order at each iteration. For example, if P=(P1, …, PN) is a permutation of {1, …, N}, let node ℓ be the th node to be updated in the kth iteration of the rules. Then
where τ1,k=k if and τ1,k=k+1 if , that is, use X1(k) if node 1 should be updated later than node ℓ, and use X1(k+1) if node 1 was updated already. The order of node updating may be different at each iteration k, since a new permutation Pk can be randomly generated once all nodes have been updated exactly k−1 times. Other asynchronous algorithms can be developed, for example by choosing the permutation Pk according to some criteria. In Chaves et al. (2005), one of the criteria consisted of choosing permutations where all protein nodes are updated first, and then all the mRNA nodes.
The steady states of a Boolean model () are fixed points of the vector function F, and consist of patterns that do not change with model updating. They can be found by solving the equations
It is not difficult to check that both synchronous and asynchronous updating schemes have the same steady states. However, note that different asynchronous algorithms may lead the system from the same initial condition to different steady states.
2.1 Analysis of Boolean networks
Useful techniques are available for the analysis of discrete logical models. In particular, Glass (1975) introduced a class of piecewise linear differential equations that combine logical rules for the synthesis of gene products with linear (free) decay by describing each node with two variables, one discrete () and one continuous (). For each node, a specific time scale () is also assigned. In a first approach, in equation (2.1), represents both degradation and synthesis rates, or a turnover rate. The model can be extended to allow distinct synthesis and degradation rates. From the set of Boolean rules , , a piecewise linear model can be obtained in the form
| (2.1) |
At each instant t, the discrete variable is defined as a function of the continuous variable according to a threshold value
| (2.2) |
where defines the fraction of ‘maximal concentration’ necessary for a protein or mRNA to regulate its successor nodes. As detailed in Chaves et al. (2006), the parameters represent different time scales for different biological processes (transcription, translation or post-translational modifications). Note that is also a scaling factor of the differential equation for . In fact, since solutions are piecewise increasing or decreasing exponentials, the evolution of is governed by the term . So, higher values of indicate that the variation rate of is higher. It is easy to see that the steady states of the piecewise linear equations (2.1) are still those of the Boolean model, since
independently of .
To study the effect of biological perturbations on the system, a natural way to proceed is to randomly assign values to time-scaling parameters , and numerically solve equations (2.1). Variations in represent perturbations in the relative time scales of each process: for instance, if αP1>αP2, then the total rate of change of protein P1 is faster than that of protein P2. Different combinations of {α1, …, αN} represent different scenarios for the segment polarity genes. By allowing to take values in an interval , it is possible to explore the space of biological fluctuations.
3. A Boolean model for the segment polarity network
As depicted in figure 1, the main segment polarity genes are wingless (wg), engrailed (en), hedgehog (hh), patched (ptc) and cubitus interruptus (ci) (e.g. Hooper & Scott 1989; Aza-Blanc et al. 1997; Sanson 2001). These code for their corresponding proteins (which will be respectively represented by the symbols WG, EN, HH, PTC and CI). The protein cubitus interruptus may be converted into a transcriptional activator (CIA), or may be cleaved to form a transcriptional repressor (CIR). The proteins EN, CIA and CIR are transcription factors, while WG and HH are secreted proteins and PTC is a transmembrane receptor protein.
The pair-rule gene product SLP activates wg transcription and represses en transcription. The WG protein is secreted from the cells that synthesize it (Hooper & Scott 1992; Pfeiffer & Vincent 1999) and initiates a signalling cascade leading to the transcription of en (Cadigan & Nusse 1997). EN promotes the transcription of the hh gene (Tabata et al. 1992) and represses the transcription of ci (Eaton & Kornberg 1990) and possibly ptc (Hidalgo & Ingham 1990; Taylor et al. 1993). The HH protein is also secreted, and binds to the HH receptor PTC on a neighbouring cell (Ingham & McMahon 2001). The intracellular domain of PTC forms a complex with SMO (van den Heuvel & Ingham 1996) in which SMO is inactivated by a post-translational conformation change (Ingham 1998). The binding of HH to PTC removes the inhibition of SMO and activates a pathway that results in the modification of CI (Ingham 1998). The CI protein can be converted into one of two transcription factors, depending on the activity of SMO. When SMO is inactive, CI is cleaved to form CIR, a transcriptional repressor that represses wg, ptc (Aza-Blanc & Kornberg 1999) and hh transcription (Ohlmeyer & Kalderon 1998; Méthot & Basler 1999). When SMO is active, CI is converted to a transcriptional activator, CIA, which promotes the transcription of wg and ptc (Jacinto et al. 1996; von Ohlen & Hooper 1997; Aza-Blanc & Kornberg 1999; Méthot & Basler 1999).
The expression pattern of these genes and proteins is repeated periodically along the embryo, and defines the parasegmental grooves. In wild-type embryos, the boundaries of the parasegments form between two consecutive cells, with one cell expressing wingless immediately anterior (to the left) to a cell expressing engrailed (Hooper & Scott 1992). At stages 6 and 7 of embryonic development, the parasegments are about four cells wide, and wingless mRNA is expressed in one of four cells, engrailed and hedgehog also in one of four cells, immediately posterior (to the right) to the cells expressing wg. Cubitus and patched mRNA are typically expressed in all cells but those expressing en. The corresponding proteins will later follow these patterns. Typically, CIA is present and CIR is absent in cells expressing wg, and either CIA is absent or CIR is present in cells not expressing wg.
3.1 Notation
Before proceeding, it is useful to introduce some notational conventions. Our model of the segment polarity network (and others mentioned) describes the evolution of a family of mRNAs and proteins in each cell of a parasegment of the embryo. The length of each parasegment is denoted by M. At stages 6 and 7 of development M=4, but, at later stages, parasegments with M≥4 cells will be considered. The notations for the model's variables encode the node name, a spatial coordinate (cell number) and a time instant. For example, wgi(k), i=1, …, M denotes the wingless mRNA concentration at time instant k in the ith cell of a parasegment; it is also convenient to write in short as wg(k), to denote the vector (wg1(k), …, wgM(k)). Similar notation is adopted for all the other mRNAs and proteins that form the segment polarity network (as listed above).
Periodic boundary conditions are assumed, meaning that: nodeM+1=node1 and node1−1=node M.
3.2 A new rule for SLP
The logical rule adopted for SLP in Albert & Othmer (2003) summarizes, in a simple but effective way, experimental observations on the regulatory activity of sloppy paired protein in the segment polarity network:
| (3.1) |
One other possible rule for SLP was studied in Chaves et al. (2006), to include recent evidence of engrailed protein inhibiting sloppy paired transcription (Alexandre & Vincent 2003). Additional regulation (again in the form of a constant input, RX) was represented by a combination of several possible effects from the pair-rule genes, namely runt, opa and factor X (Swantek & Gergen 2004) and of slp autoregulation
| (3.2) |
for i=1, …, 4. It is clear that both (3.1) and (3.2) impose somewhat strong conditions on the network, by assuming constant values on SLP or RX. To alleviate this constraint and incorporate the feedback of the segment polarity network on the sloppy paired protein, a reasonable hypothesis seems to include activation by wingless, as well as the observed inhibition by engrailed. This hypothesis is supported by some of the results reported in Bhat et al. (2000) and Lee & Frasch (2000). A similar modelling approach was already considered in von Dassow & Odell (2002) and Ingolia (2004). However, solely assuming activation by wingless and inhibition by engrailed does not explain why the domain of expression of SLP is wider than the domain of wingless and narrower than the domain where engrailed is absent, thus it is necessary to include the possibility of maintaining an initial pre-pattern. Thus, the following rule for SLP is proposed:
| (3.3) |
To test the feasibility of this rule, the resulting segment polarity model will be analysed, and predictions for several scenarios will be given, to be compared with biological observations. The Boolean rules for the other nodes are unchanged from those in Albert & Othmer (2003), and represent the network of interactions described above. A graphical representation of the model is given in figure 1, and the equations are summarized in table 1. Note that SMO does not appear in the equations. But its expression is given by Albert & Othmer (2003)
and substituted directly into the Cubitus activator and repressor proteins
The model now represents an autonomous network, with no external signals apart from the initial condition, which represents the known expression pattern at initiation of the segment polarity genes. It is now possible to investigate the mechanisms leading to the final segment polarity genes expression pattern (as observed for stages 8–10).
Table 1.
Regulatory functions governing the states of segment polarity gene products in the model. (Each node is labelled by its biochemical symbol and subscripts signify cell number in a segment. The dynamics of the system is evaluated according to X(k+1)=F(X(k)), for k=0, 1, …. For a system with M cells, there are thus N=13M variables, and X is a vector in {0, 1}N.)
| node | Boolean updating function |
|---|---|
| SLPi | SLPi(k+1)=(WGi(k) and not ENi(k)) or SLPi(k) |
| wgi | wgi(k+1)=(CIAi(k)) and SLPi(k) and not CIRi(k)) or [wgi(k) and (CIAi(k) or SLPi(k)) and not CIRi(k)] |
| WGi | WGi(k+1)=wgi(k) |
| eni | eni(k+1)=(WGi−1(k) or WGi+1(k)) and not SLPi(k) |
| ENi | ENi(k+1)=eni(k) |
| hhi | hhi(k+1)=ENi(k) and not CIRi(k) |
| HHi | HHi(k+1)=hhi(k) |
| ptci | ptci(k+1)=CIAi(k) and not ENi(k) and not CIRi(k) |
| PTCi | PTCi(k+1)=ptci(k) or (PTCi(k) and not HHi−1(k) and not HHi+1(k)) |
| cii | cii(k+1)= not ENi(k) |
| CIi | CIi(k+1)=cii(k) |
| CIAi | CIAi(k+1)=CIi(k) and [not PTCi(k) or HHi−1(k) or HHi+1(k) or hhi−1(k) or hhi+1(k)] |
| CIRi | CIRi(k+1)=CIi(k) and PTCi(k) and not HHi−1(k) and not HHi+1(k) and not hhi−1(k) and not hhi+1(k) |
The initial wild-type pattern (stage 7) is shown in table 2. Starting from this initial condition, the final wild-type pattern achieved by the segment polarity genes (stages 8–10) is of the form WT in table 3, which is indeed a steady state of the Boolean model, that is, this pattern satisfies X=F(X). The mathematical model admits several other steady states, some of which can be identified with mutant patterns (these are characterized and illustrated in Appendix A; see also Albert & Othmer 2003; Chaves et al. 2006). For instance, there is a non-segmented pattern (NS, en mutants), a state with a broad wg stripe (BS, ptc mutants), or an ectopic state (EC), where the boundary of the parasegment is displaced and inverted.
Table 2.
The initial wild-type pattern (stage 7). Nodes that are not indicated are set to 0.
| node | initial pattern |
|---|---|
| SLP(0) | 0011 |
| wg(0) | 0001 |
| en(0) | 1000 |
| hh(0) | 1000 |
| ptc(0) | 0111 |
| ci(0) | 0111 |
Table 3.
Mathematical steady-state patterns for the Boolean model with M=4.
| node | WT | NS | BS | EC |
|---|---|---|---|---|
| SLP(∞) | 0011 | 0011 | 0011 | 0011 |
| wg(∞)=WG(∞) | 0001 | 0000 | 0011 | 0010 |
| en(∞)=EN(∞) | 1000 | 0000 | 1100 | 0100 |
| hh(∞)=HH(∞) | 1000 | 0000 | 1100 | 0100 |
| ptc(∞) | 0101 | 0000 | 0011 | 1010 |
| PTC(∞) | 0111 | 1111 | 0011 | 1011 |
| ci(∞)=CI(∞) | 0111 | 1111 | 0011 | 1011 |
| CIA(∞) | 0101 | 0000 | 0011 | 1010 |
| CIR(∞) | 0010 | 1111 | 0000 | 0001 |
The challenge is then to study robustness of the convergence of the system X(k+1)=F(X(k)) to the desired wild-type state pattern WT, in table 3. For the system with the simpler SLP rule (3.1), Albert & Othmer (2003) verified that synchronous updating rules starting from the initial condition in table 2 always lead to convergence of the state of the system to WT (table 3). However, introducing totally asynchronous updating rules (which can be viewed as random perturbations to the time scales of the system), we have shown in Chaves et al. (2005) that there is a significant probability (approx. 40%) that the final state of the system is one of the ‘mutant’ states depicted in table 3. But if the asynchronous updating rules satisfy some ordering criteria, for instance in each round of iterations all proteins are updated first and then the mRNAs (corresponding to a separation of time scales of the various biological processes, with the protein binding and other kinetic events happening faster than transcription or translation), the convergence to the correct wild-type pattern is always higher than 87.5%. Therefore, the system robustly generates and maintains the segment polarity pattern, for a large range of biological perturbations. In §§5 and 6, similar results are stated for the new extended network, with SLP rule (3.3).
4. Rounds of cell division
The four-cell initial condition in table 2 is representative for up to the beginning of stage 8. There is evidence showing that several rounds of cell division occur during stages 8–10, summarized in table 4 (for more details, see Albert & Othmer 2003 and references therein). During stage 8 (3 h 10 min to 3 h 40 min after fertilization) there is an asynchronous round of cell division, and a second round of cell division in stage 10 (4 h 20 min to 5 h 20 min after fertilization), according to Hooper & Scott (1992). Slightly different observations by Gonzalez et al. (1991) indicate that the parasegment is approximately six cells wide at stages 8 and 9, and approximately eight cells wide at stage 10.
Table 4.
Approximate time frame for segment polarity genes' activation.
| developmental stage | hours from fertilization | cells per parasegment | expression pattern | references |
|---|---|---|---|---|
| 7 | 3 h 00 min | 4 | initial WT, table 2 | Hooper & Scott (1992) |
| 8 | 3 h 10 min–3 h 40 min | 4–5 | — | Hooper & Scott (1992) |
| 9 | 3 h 40 min–4 h 20 min | 5–6 | final WT, table 6 | Gonzalez et al. (1991) |
| 10 | 4 h 20 min–5 h 20 min | 6–8 | final WT, table 6 | Gonzalez et al. (1991), Hooper & Scott (1992) |
These observations lead us to investigate the model in the case when the parasegment is not four, but five, six or eight cells wide. We will not consider the case of a growing parasegment, which would require the introduction of spatial variables in the model, and hence other mathematical tools. Instead, we will take advantage of the fact that the rules governing the segment polarity model (depicted in table 1) are valid for any number of cells in a parasegment, as long as that number is fixed along time. Thus, we study time trajectories, from an initial condition to a steady state, for a fixed number of cells. We explore different initial conditions that represent the parasegment at different stages of cell division. Our goal is to check whether an analogous pattern is generated, independently of the fixed number of cells in a parasegment. If analogous patterns are generated, then one can check their robustness as a function of the number of cells. For instance, we will try to answer the question of whether a parasegment with six or eight cells is more or less vulnerable to environmental fluctuations than a parasegment with four cells.
The first question to be addressed is the initial condition: how to represent the initial stripes (the pattern in table 2) in a wider parasegment? As a first approach, either of the four cells may divide in the first round and it may be expected that the new daughter cell retains the expression levels of its mother cell. There are thus four possible initial conditions to consider, as shown in table 5, depending on whether the first, second, third or fourth cell divides from the state in table 2. One should also consider that, when the first round of cell division starts, the final pattern may be already (partly or fully) established (table 4). Thus, another possibility is to start from pattern WT in table 3, and again study the four cases arising from division of each cell. Using the analysis method described in §2.1, the results for these two limiting situations are shown in figure 2.
Table 5.
Possible initial conditions after the first round of cell division.
| dividing cell | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| SLP(0) | 00011 | 00011 | 00111 | 00111 |
| wg(0) | 00001 | 00001 | 00001 | 00011 |
| en(0) | 11000 | 10000 | 10000 | 10000 |
| hh(0) | 11000 | 10000 | 10000 | 10000 |
| ptc(0) | 00111 | 01111 | 01111 | 01111 |
| ci(0) | 00111 | 01111 | 01111 | 01111 |
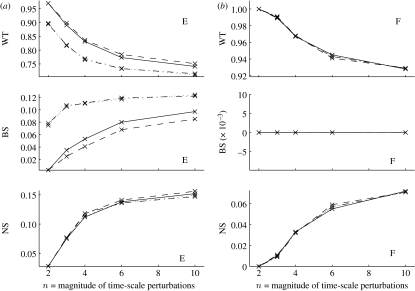
Figure 2.
Rate of convergence to wild-type (WT) and ‘mutant’ patterns, broad-striped (BS) or non-segmented (NS), after one round of cell division. The parasegment is now five cells wide, resulting from division of the first (solid lines), second (dashed lines), third (dot-dashed lines) or fourth (dotted lines) cell. The x-axis lists the magnitude of the time-scale perturbations, such that . (a) Cases (E) correspond to cell division at an early time during stage 8 (initial conditions are as in table 5), while in (b) cases (F) correspond to cell division after the full pattern is established. For cases (E), the curves corresponding to the division of the third and fourth cells have very similar values for both WT and BS, and thus overlap (dot-dashed and dotted lines). For cases (F), all curves are very similar.
For establishing the final pattern, some indications can be found in the literature. According to Cadigan et al. (1994), during stages 8–10, the SLP stripe is adjacent and anterior to en, overlapping wg and extending anterior. At stage 10 (when the parasegment may be approx. eight cells), the SLP stripe is three or four cells wide. This suggests that SLP is expressed at least in the last two cells and at most in the last half of the parasegment. According to the embryo stains shown in Alexandre & Vincent (2003), at stage 11 (when presumably there are more than 8 and up to 16 cells per parasegment), the ratio of ‘expressing’ to ‘not expressing’ cells for slp is 1 : 1.5. Similar ratios for other segment polarity genes include en and hh at 1 : 4, wg at 1 : 6 and ci at 3 : 1.
Putting together all these observations, it is reasonable to consider (in a five-cell segment) single-cell bands for wg, en and hh. For SLP, one may consider two alternatives: either a two- or a three-cell band. Indeed, for both a two- and a three-cell SLP band, there are steady-state solutions of the system depicted in table 1 with i=1, …, 5 (i.e. patterns satisfying X=F(X)), which correspond to wild-type expression patterns. All the mathematical steady states for the Boolean model, which are reachable from initial conditions with SLP=0⋯011 or SLP=0⋯0111, for any M≥4 are characterized in appendix A. For M=5, the wild-type patterns are depicted as WT (I) and (II) in table 6.
Table 6.
Some of the mathematical steady-state patterns for the Boolean model with M=5.
| node | WT (I) | NS (I) | BS (I) | WT (II) | NS (II) | BS (II) |
|---|---|---|---|---|---|---|
| SLP(∞) | 00011 | 00011 | 00011 | 00111 | 00111 | 00111 |
| wg(∞)=WG(∞) | 00001 | 00000 | 00011 | 00001 | 00000 | 00101 |
| en(∞)=EN(∞) | 10000 | 00000 | 10100 | 10000 | 00000 | 11000 |
| hh(∞)=HH(∞) | 10000 | 00000 | 10100 | 10000 | 00000 | 11000 |
| ptc(∞) | 01001 | 00000 | 01011 | 01001 | 00000 | 00101 |
| PTC(∞) | 01111 | 11111 | 01011 | 01111 | 11111 | 00111 |
| ci(∞)=CI(∞) | 01111 | 11111 | 01011 | 01111 | 11111 | 00111 |
| CIA(∞) | 01001 | 00000 | 01011 | 01001 | 00000 | 00101 |
| CIR(∞) | 00110 | 11111 | 00000 | 00110 | 11111 | 00010 |
Other patterns compatible with the steady states of the four-cell-wide model (compare tables 3 and 6) include five-cell versions of the non-segmented state (NS), the state with a broad wg stripe (BS), or the ectopic state (EC; not shown). Again, note that there are two alternative forms for each pattern, depending on the width of the SLP band (marked I and II, respectively). Observe that, while the WT and NS patterns differ only in the SLP expression, the effect on BS is more complex, with the en and wg bands forming in different ways, and thus also affecting expression of the other genes and proteins.
It is interesting to note that comparison of the two (mathematical) BS steady-state patterns with experimental data suggests that the patterns with a shorter SLP band (two cells) are biologically more relevant. In fact, the BS pattern is typically seen in ptc mutant embryos, with the obvious difference that ptc and PTC are not expressed in ptc mutants, while they are in the BS pattern. As observed in the work of Martinez Arias et al. (1988), in the ptc mutant embryos wingless is expressed in a broad domain that occupies half the parasegment. After cell division, engrailed is observed to form an extra stripe just anterior to wingless. This is further supported by the work on ptc mutants by Ingham et al. (1991), who report that the expression domain of wg broadens after gastrulation and that en transcription is induced in the cells anterior to the wg domain. This experimental evidence points to the state of the form I as the more plausible BS pattern. Hence, more generally, states with SLP=00011 represent the biological system more faithfully.
The patterns in table 6 extend (in an analogous form) as steady states of an M-cell-wide parasegment model (see appendix A). As in the five-cell model, we only consider two- or three-cell SLP bands.
5. Results
The method described in §2.1 was used to analyse the evolution of the segment polarity pattern, after the first and second rounds of cell division during stages 8–10 of embryonic development. The model described in table 1 was solved numerically with M cells (and i=1, …, M), for M=4, 5, 6, 8. For each biological case studied, a given initial condition was chosen (as indicated in the figure captions). To answer robustness questions, the time-scale constants were randomly chosen from a uniform distribution in intervals of the form
where the case n=1 is equivalent to the synchronous Boolean algorithm, and larger values n=2, 3, 4, 6, 10, 20 (as indicated in the x-axis of figures 2–4) represent different orders of magnitude of the perturbations. Therefore, n=2 represents a system where the relative time scales of different processes all have values between half and twice a normalized time unit (here 1), leading to small fluctuations. By contrast, n=10 represents a system where very large fluctuations are allowed, between one-tenth and ten times the normalized unit.
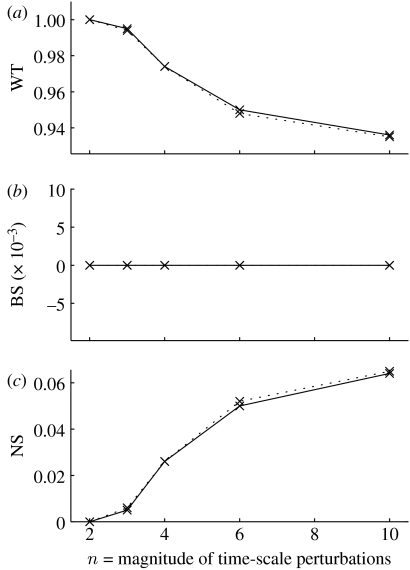
Figure 3.
Rate of convergence to (a) wild-type (WT) and (b) broad-striped (BS) or (c) non-segmented (NS) patterns, after two rounds of cell division. The parasegment is six cells wide, resulting from division of one of the five cells in the previous stage. The x-axis shows the magnitude of the time-scale perturbations, such that . The initial condition for the six-cell model is WT (I), as in table 6, with either of the five cells doubled (corresponding to cell division after the full pattern is established). The results are very similar for all the five possible initial conditions, all curves overlapping. Shown here are the curves for doubling the first (solid lines) and fourth (dotted lines) cells.
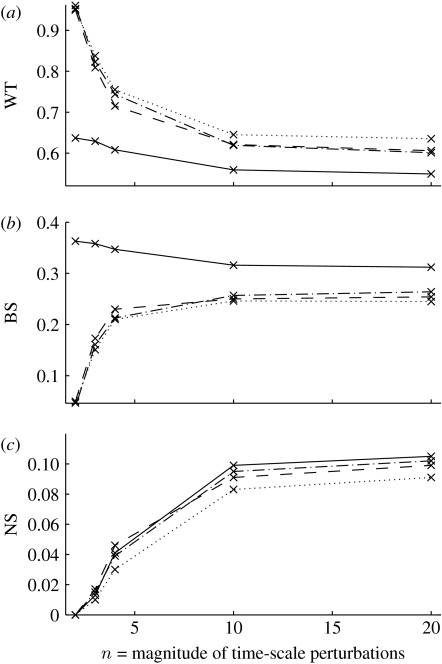
Figure 4.
Rate of convergence to (a) wild-type (WT) and ‘mutant’ patterns (b) broad-striped (BS) or (c) non-segmented (NS), starting from minimal patterning (5.1). The x-axis shows the magnitude of the time-scale perturbations, such that . Each of the four cases corresponds to four (solid lines)-, five (dashed lines)-, six (dot-dashed lines)- or eight (dotted lines)-cell-wide parasegments.
For each case studied (i.e. for each M and each n), j=1, …, 1000 numerical experiments were performed. For each numerical experiment j, a set of time scales was chosen from a uniform distribution on the interval [1/n,n], the threshold values were set to for all ℓ, and system (2.1) was numerically solved. The solution was observed to reach a steady state (from those indicated in appendix A), and this steady state was registered (either of the form WT, BS, NS or EC). Each experiment j represents a certain biological ‘scenario’: the values of indicate which nodes evolve at faster rates than others (see discussion in §2.1). The distribution of can be viewed as the result of biological perturbations due to, for instance, temperature changes or other stresses, which may induce delays in expression of some genes, or prompt faster signalling processes. Since the values are randomly chosen, no specific scenario is modelled, but instead all possible relative changes are explored. The goal is to measure the response of the system to ‘worst-case’ disruptions, by verifying how frequently the final steady state deviates from the correct wild-type pattern.
Results shown in the figures represent the probability of the system reaching a given steady state, that is, the frequency of each steady state over the 1000 replicate experiments
where if , and otherwise (here WT is either of form I or II). Similar expressions were used to compute the probabilities of the other patterns.
The results for the first round of cell division are shown in figure 2. As described in §4, two limiting cases were studied: either cell division occurs early, and the system starts from one of initial conditions in table 5, or the final pattern is already established, and the system starts from one of the four possible conditions arising by doubling each cell in the pattern WT (table 3).
It is clear that the expression pattern is not greatly disrupted by cell division, if it was already established with only four cells. It is interesting to observe that no broad-striped (BS) pattern is formed at this stage, that is, any perturbation either has no effect on groove formation (with probability higher than 90%) or it completely prevents groove formation (probability less than 10%). By contrast, if the pattern was not yet established with four cells, then there is still a significant possibility that strong perturbations to the system cause pattern disruption. In particular, division of the SLP-expressing cells (the last two) is more vulnerable, as the outcome may be the broad-striped pattern (corresponding to ptc mutants) with probability of approximately 10%, or the non-segmented pattern (corresponding to en mutants) with probability between 10 and 20%.
The second round of cell division most likely occurs at stages 9 and 10 of the embryonic development, when the wild-type pattern of the segment polarity network is already fully established in segments composed of five cells (table 4). Thus, as a starting condition for the six-cell parasegment model, we take the WT (I) pattern in table 6 and double each cell in turn, thus generating five possible initial conditions. Figure 3 shows that the network appears quite robust at this stage, in the sense that the pattern extends correctly into six cells with a probability of more than 95%. Moreover, no cell position is more vulnerable than another (in contrast to the extension from four to five cells).
Finally, to compare the robustness of four-, five-, six- or eight-cell-wide parasegments, under similar conditions, we have analysed the evolution of the system starting from a minimal pre-pattern
| (5.1) |
that is, only en (first cell), wg (last cell) and SLP (last two cells) are expressed. Taking these initial conditions, the capacity of M-cell-wide parasegments to generate the segment polarity pattern was verified, again considering several magnitudes of time-scale perturbations. The results are shown in figure 4. Interestingly, robustness to perturbations increases with the length of the parasegment, with a very significant increase from four to five cells. This indicates that a first round of cell division during stage 8 of embryonic development could be a very desirable event. Moreover, if the dividing cell is either the first or the second in the parasegment (see the discussion of figure 2), cell division contributes to an increased robustness of the segment polarity network.
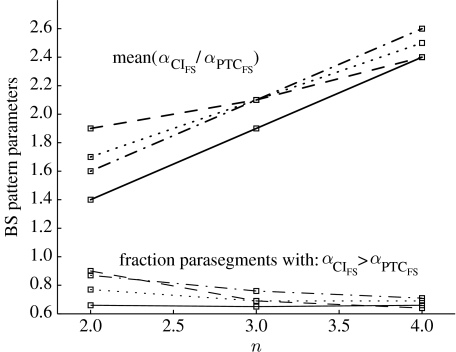
To further understand the mechanism responsible for higher robustness in wider parasegments, we explored the relative time scales for patched and cubitus interruptus proteins in the first cell expressing SLP, i.e. and . It was shown in Chaves et al. (2006) that (M=4, FS=3) is a necessary condition to obtain 100% convergence to the wild-type pattern (this result is now generalized in §6). Indeed, we verify that, among all parameter combinations leading to the BS pattern (say NBS), more than 65% break that condition, satisfying instead (figure 5).
Figure 5.
The average ratios () and percentage of (BS) parasegments satisfying the given condition (), as a function of the magnitude of the time-scale perturbations, n (x-axis; such that ). Curves correspond to four (solid lines)-, five (dashed lines)-, six (dot-dashed lines)- or eight (dotted lines)-cell-wide parasegments.
In addition, computing the average
we see that qFS>1 (figure 5), and is clearly lower for parasegments with four cells. In other words, a sharper difference between and is needed to disrupt normal pattern development in wider parasegments (presumably a less likely situation).
6. Generating the wild-type pattern with time-scale separation
As in Chaves et al. (2006), we will analyse the behaviours of trajectories of systems of the form (2.1), assuming that trajectories are well defined. Since the right-hand side of equations of this type are discontinuous, it is very difficult to give general existence and uniqueness theorems for solutions of initial-value problems. One must impose additional assumptions, ensuring that only a finite number of switches can take place on any finite time interval, and often tools from the theory of differential inclusions must be applied (see Gedeon 2003; Casey et al. 2006 for more discussion).
For a segment of M cells, SLP is typically expressed in the posterior part of the parasegment, in a group of adjacent cells (e.g. in the last two or three cells of a five-cell-wide parasegment). Then wingless is expressed in the last cell expressing SLP, while engrailed and hedgehog are expressed in the first cell of the parasegment. Thus, we will adopt the notation FS (respectively, LS) to denote the first cell (respectively, last cell) expressing SLP, and consider the following ‘generic’ initial condition:
| (6.1) |
where, typically, one expects to see LS=M and FS>M/2.
It was shown in Chaves et al. (2006) that a time-scale separation assumption guaranteed convergence of the piecewise linear system (2.1) to the wild-type pattern WT (table 3), for a four-cell-wide parasegment. We next state that a similar result holds for wider parasegments. Let AmRNA and AProt denote intervals for the scaling factors () of the system's mRNAs or proteins (respectively). Assume that AmRNA and AProt do not overlap, and satisfy
For all a∈AmRNA and b∈AProt: 0<2a<b.
Assume also that
θi=θ, for all i, and θ≤1/2; and
.
Such hypotheses seem quite reasonable from the biological point of view, as (A1) reflects the fact that post-translational modifications (such as protein conformational changes that happen on a scale of thousandths of a second) are usually faster than transcription or translation (which happen on a scale of hundreds of seconds; see Papin et al. 2005 and references therein). Hypothesis (A2) states that concentrations of protein (or mRNA) corresponding to less than 50% of its maximal value are sufficient to initiate an activation or inhibition by that protein. Hypothesis (A3) stems from observation of the numerical simulations (see discussion of ratio qFS>1 and figure 5), but the following theoretical results show that indeed it guarantees convergence to the wild-type steady state. It is thus a prediction of our model. Note that the following theorems are stated for any length M≥4 of the parasegment.
Theorem 6.1
Consider system (2.1) with initial condition (6.1). Suppose that assumptions (A1)–(A3) are satisfied. Then wgFS(t)=0 for all t.
▪
This shows that the steady state representing the broad-striped pattern (BS I or II) cannot ever be reached in system (2.1) from the initial condition (6.1), under assumptions (A1)–(A3).
Theorem 6.2
Consider system (2.1) with initial condition (6.1). Suppose that assumptions (A1)–(A3) are satisfied. Then wgLS(t)=1 and PTC1(t)=0 for all t.
▪
This shows that the steady states represented by the no segmentation or ectopic patterns (see tables 3 and 6) cannot ever be reached in system (2.1) from the initial condition (6.1).
From theorems 6.1 and 6.2, we conclude that, under the time-scale separation assumption (and for appropriate θ values), the piecewise linear model (2.1) can never converge to its steady states corresponding to mutant patterns such as BS (I or II), NS or EC, for any length of the parasegment. Thus, since the only steady states reachable from initial condition (6.1) are WT, BS, NS or EC (see appendix A) the wild-type pattern can be expected when starting from the initial condition (6.1).
The proofs are very similar to those in Chaves et al. (2006). One needs to consider first the modifications due to the dynamic rule for SLP, but then the arguments are analogous. For the four-cell-wide parasegment model, the proofs are based on the initial conditions and Boolean rules for the third and fourth cells, i.e. the ‘first and last cells expressing SLP’. The results can be easily adapted to deal more generally with SLP expressed in a group of adjacent cells, labelled FS–LS: indeed, to prove theorem 6.1, follow the proof given in Chaves et al. (2006), but substituting ‘node3’ by ‘nodeFS’ and ‘node2’ by ‘nodeFS–1’; to prove theorem 6.2, simply replace ‘node4’ by ‘nodeLS’. For completeness, the proofs are included in the electronic supplementary material.
We will now analyse the effect of the SLP rule on the evolution of the network.
Proposition 6.3
Consider system (2.1) with initial condition (6.1). Then
(6.2) and also
(6.3)
Proof
Since (at t=0) SLPi(0)=wgi(0)=WGi(0)=0, for all i=1, …, FS–1, to argue by contradiction, suppose that t1>0 is the first instant for which SLPi(t1)=1. First, observe that (since wgi starts at zero) wgi can only get activated after SLPi becomes 1. So, it holds that wgi(t)=0 for all t≤t1, for all i=1, …, FS–1. For any i=1, …, FS–1, note that SLPi(t1)=1 implies (looking at the initial conditions and the SLP rule)
And then WGi(ta)=1 implies
Now this implies (looking at the wgi rule)
But, finally, observe that this is impossible, a contradiction to our assumption, since both SLPi and wgi must be zero at tc<t1. This proves statement (6.2). The proof of (6.3) follows by direct application of the Boolean rules for SLP, en, EN, hh and HH.
▪
Note that this result holds for every trajectory, even when none of the hypotheses (A1)–(A3) are satisfied. In fact, this proposition guarantees that the anterior/posterior polarity in the parasegment is maintained throughout time, one of the first and crucial steps towards generating the wild-type pattern. Indeed, note that the absence of SLP strictly prevents wingless expression, while the presence of SLP strictly prevents engrailed expression. It is known that parasegment boundaries are defined between a cell expressing engrailed and another expressing wingless. In addition, SLP is expressed in bands of two or more cells (e.g. 0⋯011 or 0⋯0111). Therefore, parasegment boundaries can only form on the first (FS) or last (LS) cell expressing sloppy paired. This result is interesting for several reasons. First, it indicates that not all cells of the parasegment are important, and incorrect expression in several of the cells can be corrected during development. Second, if given the information that two cells are essential, most Drosophila scientists would expect that those two key cells are the first and last cells of the parasegment, or the (first) cell expressing engrailed and the (last) cell expressing wingless. Only one of the two cells that we find coincides with this expectation: the last cell of sloppy paired, that is also the (last) cell expressing wingless. The first cell expressing sloppy paired does not coincide with engrailed expression; on the contrary, it determines the boundary of engrailed expression. This result suggests that the pattern of the segment polarity genes, and specifically the wingless–engrailed boundary, depends critically on the sloppy paired expression. As we can see from appendix A, wild-type or mutant expression patterns evolve essentially around cells FS and LS. In wild-type, sloppy paired protein counteracts the symmetry of the en→wg positive feedback loop to generate a single asymmetric wingless–engrailed boundary around LS. In mutants, an extra wingless–engrailed boundary may also form around FS.
7. Conclusion
Two questions were discussed in this paper, and possible answers proposed, leading to improvements in modelling and understanding the properties of Drosophila segment polarity network. The new dynamical rule for regulation of sloppy paired is composed of activation by wingless and inhibition by engrailed proteins, together with a maintenance term. Thus, the network operates as an autonomous module with no ‘strong’ external inputs. Starting from an initial expression pattern corresponding to development stage 7, the network can robustly generate and maintain an expression pattern that faithfully reproduces the segment polarity genes pattern at stages 9 and 10. Moreover, with the natural hypothesis of time-scale separation, it is shown that the system cannot converge to any of the model's steady states corresponding to some mutant patterns. This result further shows that generating the wild-type segment polarity expression pattern depends essentially on the dynamics of the network at the first and last cells expressing sloppy paired protein. Rounds of cell division (and hence wider parasegments) will not disrupt the pattern: in fact, our results suggest that, as the number of cells in each parasegment increases, the system becomes more robust to perturbations in biological processes. One possible interpretation is that new cells in a parasegment strengthen the regulatory conditions defining the boundaries between parasegments, in such a way that those boundaries become much more difficult to break. The boundaries between parasegments are defined mainly by the relative positions of en and wg. According to the model, en can only be expressed in a cell immediately posterior or anterior to a cell expressing SLP. The results show that adding new cells between the first cell of the parasegment and the first cell expressing SLP leads to lower probability of formation of an extra en stripe just anterior to SLP. Hence, a system with higher number of cells per parasegment is more likely to generate the desired wild-type pattern.
Acknowledgments
Part of this study was done while both authors were visiting the Kavli Institute for Theoretical Physics, Santa Barbara, CA, USA, during July 2007. The authors acknowledge fruitful discussions with Boris Shraiman. Boolean modelling in R.A.'s group is funded by NSF grants MCB-0618402 and CCF-0643529 (CAREER), NIH grants 1R55AI065507-01A2 and 1 R01 GM083113-01, HFSP grant RGP20/2007 and USDA grant 2006-35100-17254.
Appendix A
A.1 Steady states of the mathematical model
The steady states of the Boolean model depicted in table 1 can be computed by solving the equation X=F(X). It was shown that these steady states are also those of the piecewise continuous model (2.1).
In our analysis, we focused on initial conditions of the form (6.1), with FS=M−1 or FS=M−2. Initial conditions of this form imply certain restrictions on the dynamics of both the Boolean and piecewise continuous model, namely that expression of SLP will not vary throughout time. This is established in proposition 6.1, and follows roughly from the fact that, in each cell i, SLPi can become expressed only if wgi has been expressed at some previous instant but, conversely, wgi can become expressed only if SLPi was expressed at some previous time. Thus, if both SLPi and wgi are absent at time 0, neither can become expressed at later times.
Thus, we compute only the steady states that can possibly be reached if the system starts from initial condition (6.1) (recall that different asynchronous updating strategies may lead the system from the same initial condition to different steady states). These are given in tables 7 and 8 (case FS=M−1), and tables 9 and 10 (case FS=M−2). All steady states can be derived from the expression of wingless in the first and last cells expressing SLP. Thus, there are four possible distinct steady states, associated with the four distinct combinations of the wingless values in the cells FS and LS as Wa and Wb, respectively.
Table 7.
Steady states for initial conditions of the form (6.1), FS=M−1 for M=4, 5. (The patterns for M=5 are represented in the five consecutive cells labelled 1, 2, 3, M−1 and M, while the patterns for M=4 are represented in the four consecutive cells M−1, M, 1 and 2. The symbols Wa, Wb and d represent digits in {0, 1}. Wa and Wb denote wingless expression in the FS and LS=M cells, respectively. The functions f and g are defined as and .)
| node | cell | ||||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | M−1 | M | 1 | 2 | |
| SLP | 0 | 0 | 0 | 1 | 1 | 0 | 0 |
| WG=wg | 0 | 0 | 0 | Wa | Wb | 0 | 0 |
| EN=en | Wb | 0 | Wa | 0 | 0 | Wb | Wa |
| HH=hh | Wb | 0 | Wa | 0 | 0 | Wb | Wa |
| ptc | 0 | 1−g(Wa, Wb) | 0 | Wa | Wb | (1−Wb)Wa | (1−Wa)Wb |
| PTC | 1−dWb | 1 | 1−dWa | 1 | 1 | f(Wa, Wb, d) | f(Wb, Wa, d) |
| CI=ci | 1−Wb | 1 | 1−Wa | 1 | 1 | 1−Wb | 1−Wa |
| CIA | 0 | 1−g(Wa, Wb) | 0 | Wa | Wb | (1−Wb)Wa | (1−Wa)Wb |
| CIR | 1−Wb | g(Wa, Wb) | 1−Wa | 1−Wa | 1−Wb | g(Wa, Wb) | g(Wa, Wb) |
Table 8.
Steady states for initial conditions of the form (6.1), FS=M−1 for M≥6 (the patterns for M=6 are obtained from the columns 1, 2, M−3, M−2, M−1 and M). (The symbols Wa, Wb and d represent digits in {0, 1}. Wa and Wa denote wingless expression in the FS and LS=M cells, respectively.)
| node | cell | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | … | M−5 | M−4 | M−3 | M−2 | M−1 | M | |
| SLP | 0 | 0 | 0 | … | 0 | 0 | 0 | 0 | 1 | 1 |
| WG=wg | 0 | 0 | 0 | … | 0 | 0 | 0 | 0 | Wa | Wb |
| EN=en | Wb | 0 | 0 | … | 0 | 0 | 0 | Wa | 0 | 0 |
| HH=hh | Wb | 0 | 0 | … | 0 | 0 | 0 | Wa | 0 | 0 |
| ptc | 0 | Wb | 0 | … | 0 | 0 | Wa | 0 | Wa | Wb |
| PTC | 1−dWb | 1 | 1 | … | 1 | 1 | 1 | 1−dWa | 1 | 1 |
| CI=ci | 1−Wb | 1 | 1 | … | 1 | 1 | 1 | 1−Wa | 1 | 1 |
| CIA | 0 | Wb | 0 | … | 0 | 0 | Wa | 0 | Wa | Wb |
| CIR | 1−Wb | 1−Wb | 1 | … | 1 | 1 | 1−Wa | 1−Wa | 1−Wa | 1−Wb |
Table 9.
Steady states for initial conditions of the form (6.1), FS=M−2 for M=5, 6. (The patterns for M=6 are represented in the six consecutive cells labelled 1, 2, 3, M−2, M−1 and M, while the patterns for M=5 are represented in the five consecutive cells M−2, M−1, M, 1 and 2. The symbols Wa, Wb and d represent digits in {0, 1}. Wa and Wb denote wingless expression in the FS and LS=M cells, respectively. The functions f and g are defined as and .)
| node | cell | |||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | M−2 | M−1 | M | 1 | 2 | |
| SLP | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 |
| WG=wg | 0 | 0 | 0 | Wa | 0 | Wb | 0 | 0 |
| EN=en | Wb | 0 | Wa | 0 | 0 | 0 | Wb | Wa |
| HH=hh | Wb | 0 | Wa | 0 | 0 | 0 | Wb | Wa |
| ptc | 0 | 1−g(Wa, Wb) | 0 | Wa | 0 | Wb | (1−Wb)Wa | (1−Wa)Wb |
| PTC | 1−dWb | 1 | 1−dWa | 1 | 1 | 1 | f(Wa, Wb, d) | f(Wb, Wa, d) |
| CI=ci | 1−Wb | 1 | 1−Wa | 1 | 1 | 1 | 1−Wb | 1−Wa |
| CIA | 0 | 1−g(Wa, Wb) | 0 | Wa | 0 | Wb | (1−Wb)Wa | (1−Wa)Wb |
| CIR | 1−Wb | g(Wa, Wb) | 1−Wa | 1−Wa | 1 | 1−Wb | g(Wa, Wb) | g(Wa, Wb) |
Table 10.
Steady states for initial conditions of the form (6.1), FS=M−2 for M≥7 (the patterns for M=7 are obtained from the columns 1, 2, M−4, M−3, M−2, M−1 and M). (The symbols Wa, Wb and d represent digits in {0, 1}. Wa and Wb denote wingless expression in the FS and LS=M cells, respectively.)
| node | cell | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | … | M−5 | M−4 | M−3 | M−2 | M−1 | M | |
| SLP | 0 | 0 | 0 | … | 0 | 0 | 0 | 1 | 1 | 1 |
| WG=wg | 0 | 0 | 0 | … | 0 | 0 | 0 | Wa | 0 | Wb |
| EN=en | Wb | 0 | 0 | … | 0 | 0 | Wa | 0 | 0 | 0 |
| HH=hh | Wb | 0 | 0 | … | 0 | 0 | Wa | 0 | 0 | 0 |
| ptc | 0 | Wb | 0 | … | 0 | Wa | 0 | Wa | 0 | Wb |
| PTC | 1−dWb | 1 | 1 | … | 1 | 1 | 1−dWa | 1 | 1 | 1 |
| CI=ci | 1−Wb | 1 | 1 | … | 1 | 1 | 1−Wa | 1 | 1 | 1 |
| CIA | 0 | Wb | 0 | … | 0 | Wa | 0 | Wa | 0 | Wb |
| CIR | 1−Wb | 1−Wb | 1 | … | 1 | 1−Wa | 1−Wa | 1−Wa | 1 | 1−Wb |
The steady state corresponding to the case (Wa,Wb)=(0,1) represents the wild-type (WT) expression pattern of the segment polarity genes. There are two (very similar) variants for this pattern, differing only on the value of PTC1, which can be either 1−0×1=1 or 1−1×1=0.
The steady state corresponding to the case (Wa, Wb)=(1,1) represents the broad-striped (BS) expression pattern, observed in ptc mutants (Tabata et al. 1992). These mutants express wingless in a stripe that is broader than that observed in wild type. For the case FS=M−1, the BS pattern expresses wg in two consecutive cells. For the case FS=M−2, wg is expressed in the first and last cells expressing SLP, but not in the middle cell. For this reason, we believe that initial conditions with SLP=0⋯011 represent the biological system more faithfully.
The steady state corresponding to the case (Wa, Wb)=(0, 0) represents the non-segmented (NS) expression pattern, observed in en mutants (Tabata et al. 1992). Here, patched and cubitus repressor proteins are ubiquituously expressed, and neither wingless nor engrailed are expressed.
Finally, the steady state corresponding to the case (Wa, Wb)=(1, 0) represents an ectopic (EC) expression pattern that, to our knowledge, has not been observed experimentally. The relative positions of en and wg are inverted, that is a segment could be formed, but with the boundary defined by a cell expressing engrailed to the left of a cell expressing wingless. In previous studies (Chaves et al. 2005, 2006), only a very small fraction of perturbations (less than 1%) to the mathematical model lead the system to this ectopic steady state. Here again there are two possible variants, differing only on the value of PTCFS–1, which can be either 1−0×1=1 or 1−1×1=0.
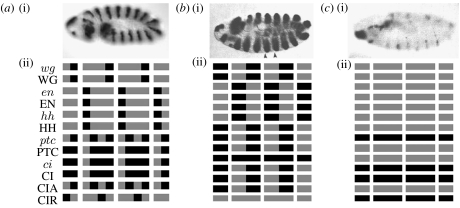
Some of these patterns are illustrated in figure 6 (this figure can also be found in Chaves et al. 2005). The wild-type broad-striped and no-segmentation mathematical patterns are shown for a four-cell-wide parasegment, and compared with the corresponding wild-type or mutant embryos.
Figure 6.
(a)(i) Illustration of the gene expression pattern of wingless on a gastrulating (stage 9) embryo. The parasegmental furrows form at the posterior border of the wg-expressing cells (Wolpert et al. 1998). (ii) Wild-type expression pattern of the Boolean model. Left corresponds to anterior and right to posterior in each parasegment. Horizontal rows correspond to the pattern of individual nodes, specified at the left side of the row, over two full and two partial parasegments. Each parasegment is assumed to be four cells wide. A black (grey) box denotes a node that is (is not) expressed. (b)(i) Wingless expression pattern in a patched knock-out mutant embryo at stage 11 (Tabata et al. 1992). The wingless stripes broaden, and secondary furrows appear at the middle of the parasegment, indicating a new en–wg boundary. (ii) Broad-striped steady state of the Boolean model. This state is obtained when wg, en or hh are initiated in every cell. A variant of this state is obtained when patched is kept off; the difference is in the fact that ptc and PTC are not expressed in the mutant state. This steady state agrees with all experimental observations on ptc mutants and heat-shocked genes (DiNardo et al. 1988; Martinez 1988; Ingham et al. 1991; Tabata et al. 1992; Bejsovec & Wieschaus 1993; Schwartz et al. 1995; Gallet et al. 2000). (c)(i) wingless expression pattern in an engrailed knock-out mutant embryo at stage 11 (Tabata et al. 1992). The initial periodic pattern is disappearing, and gives rise to a non-segmented embryonic lethal phenotype. (ii) Non-segmented steady state of the Boolean model. This steady state agrees with all experimental observations on wg, en and hh mutants (DiNardo et al. 1988; Hidalgo & Ingham 1990; Tabata et al. 1992; Schwartz et al. 1995; Gallet et al. 2000). Gene expression images can be obtained from http://www.fruitfly.org for (a) and Tabata et al. (1992) for (b and c).
Footnotes
One contribution of 10 to a Theme Supplement ‘Biological switches and clocks’.
Supplementary Material
Proofs of theorems 1 and 2
References
- Albert R., Othmer H.G. The topology of the regulatory interactions predicts the expression pattern of the Drosophila segment polarity genes. J. Theor. Biol. 2003;223:1–18. doi: 10.1016/s0022-5193(03)00035-3. doi:10.1016/S0022-5193(03)00035-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandre C., Vincent J.P. Requirements for transcriptional repression and activation by engrailed in Drosophila embryos. Development. 2003;130:729–739. doi: 10.1242/dev.00286. doi:10.1242/dev.00286 [DOI] [PubMed] [Google Scholar]
- Aza-Blanc P., Kornberg T.B. Ci a complex transducer of the hedgehog signal. Trends Genet. 1999;15:458–462. doi: 10.1016/s0168-9525(99)01869-7. doi:10.1016/S0168-9525(99)01869-7 [DOI] [PubMed] [Google Scholar]
- Aza-Blanc P., Ramirez-Weber F.A., Leget M.-P., Schwartz C., Kornberg T.B. Proteolysis that is inhibited by hedgehog targets Cubitus interruptus proteins to the nucleus and converts it to a repressor. Cell. 1997;89:1043–1053. doi: 10.1016/s0092-8674(00)80292-5. doi:10.1016/S0092-8674(00)80292-5 [DOI] [PubMed] [Google Scholar]
- Bejsovec A., Wieschaus E. Segment polarity gene interactions modulate epidermal patterning in Drosophila embryos. Development. 1993;119:501–517. doi: 10.1242/dev.119.2.501. [DOI] [PubMed] [Google Scholar]
- Bhat K.M., van Beers E.H., Bhat P. Sloppy paired acts as the downstream target of Wingless in the Drosophila CNS and interaction between sloppy paired and gooseberry inhibits sloppy paired during neurogenesis. Development. 2000;127:655–665. doi: 10.1242/dev.127.3.655. [DOI] [PubMed] [Google Scholar]
- Cadigan K.M., Nusse R. Wnt signaling: a common theme in animal development. Genes Dev. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. doi:10.1101/gad.11.24.3286 [DOI] [PubMed] [Google Scholar]
- Cadigan K.M., Grossniklaus U., Gehring W.J. Localized expression of sloppy paired protein maintains the polarity of Drosophila parasegments. Genes Dev. 1994;8:899–913. doi: 10.1101/gad.8.8.899. doi:10.1101/gad.8.8.899 [DOI] [PubMed] [Google Scholar]
- Casey R., de Jong H., Gouzé J.L. Piecewise linear models of genetic regulatory networks: equilibria and their stability. J. Math. Biol. 2006;52:27–56. doi: 10.1007/s00285-005-0338-2. doi:10.1007/s00285-005-0338-2 [DOI] [PubMed] [Google Scholar]
- Chaves M., Albert R., Sontag E.D. Robustness and fragility of boolean models for genetic regulatory networks. J. Theor. Biol. 2005;235:431–449. doi: 10.1016/j.jtbi.2005.01.023. doi:10.1016/j.jtbi.2005.01.023 [DOI] [PubMed] [Google Scholar]
- Chaves M., Sontag E.D., Albert R. Methods of robustness analysis for boolean models of gene control networks. IEE Proc. Syst. Biol. 2006;153:154–167. doi: 10.1049/ip-syb:20050079. doi:10.1049/ip-syb:20050079 [DOI] [PubMed] [Google Scholar]
- DiNardo S., Sher E., Heemskerk-Jongens J., Kassis J.A., O'Farrell P.H. Two-tiered regulation of spatially patterned engrailed gene expression during Drosophila embryogenesis. Nature. 1988;332:45–53. doi: 10.1038/332604a0. doi:10.1038/332604a0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton S., Kornberg T.B. Repression of ci–d in posterior compartments of Drosophila by engrailed. Genes Dev. 1990;4:1068–1077. doi: 10.1101/gad.4.6.1068. doi:10.1101/gad.4.6.1068 [DOI] [PubMed] [Google Scholar]
- Edwards R., Glass L. Combinatorial explosion in model gene networks. Chaos. 2000;10:691–704. doi: 10.1063/1.1286997. doi:10.1063/1.1286997 [DOI] [PubMed] [Google Scholar]
- Gallet A., Angelats C., Kerridge S., Thérond P.P. Cubitus interruptus-independent transduction of the hedgehog signal in Drosophila. Development. 2000;127:5509–5522. doi: 10.1242/dev.127.24.5509. [DOI] [PubMed] [Google Scholar]
- Gedeon T. Attractors in continuous-time switching networks. Commun. Pure Appl. Anal. 2003;2:187–209. [Google Scholar]
- Glass L. Classification of biological networks by their qualitative dynamics. J. Theor. Biol. 1975;54:85–107. doi: 10.1016/s0022-5193(75)80056-7. doi:10.1016/S0022-5193(75)80056-7 [DOI] [PubMed] [Google Scholar]
- Glass L., Kauffman S.A. The logical analysis of continuous, nonlinear biochemical control networks. J. Theor. Biol. 1973;39:103–129. doi: 10.1016/0022-5193(73)90208-7. doi:10.1016/0022-5193(73)90208-7 [DOI] [PubMed] [Google Scholar]
- Gonzalez F., Swales L.S., Bejsovec A., Skaer H., Martinez Arias A. Secretion and movement of wingless protein in the epidermis of the Drosophila embryo. Mech. Dev. 1991;35:43–54. doi: 10.1016/0925-4773(91)90040-d. doi:10.1016/0925-4773(91)90040-D [DOI] [PubMed] [Google Scholar]
- Hidalgo A., Ingham P. Cell patterning in the Drosophila segment: spatial regulation of the segment polarity gene patched. Development. 1990;110:291–301. doi: 10.1242/dev.110.1.291. [DOI] [PubMed] [Google Scholar]
- Hooper J.E., Scott M.P. The Drosophila patched gene encodes a putative membrane protein required for segmental patterning. Cell. 1989;59:751–765. doi: 10.1016/0092-8674(89)90021-4. doi:10.1016/0092-8674(89)90021-4 [DOI] [PubMed] [Google Scholar]
- Hooper J.E., Scott M.P. The molecular genetic basis of positional information in insect segments. In: Hennig W., editor. Early embryonic development of animals. Springer; Berlin, Germany: 1992. pp. 1–49. [DOI] [PubMed] [Google Scholar]
- Ingham P.W. Transducing hedgehog: the story so far. EMBO J. 1998;17:3505–3511. doi: 10.1093/emboj/17.13.3505. doi:10.1093/emboj/17.13.3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham P.W., McMahon A.P. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 2001;15:3059–3087. doi: 10.1101/gad.938601. doi:10.1101/gad.938601 [DOI] [PubMed] [Google Scholar]
- Ingham P.W., Taylor A.M., Nakano Y. Role of Drosophila patched gene in positional signaling. Nature. 1991;353:184–187. doi: 10.1038/353184a0. doi:10.1038/353184a0 [DOI] [PubMed] [Google Scholar]
- Ingolia N.T. Topology and robustness in the Drosophila segment polarity network. PLoS Biol. 2004;2:0805–0815. doi: 10.1371/journal.pbio.0020123. doi:10.1371/journal.pbio.0020123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto A., Alexandre C., Ingham P.W. Transcriptional activation of hedgehog target genes in Drosophila is mediated directly by the Cubitus interruptus protein, a member of the GLI family of zinc finger DNA-binding proteins. Genes Dev. 1996;10:2003–2013. doi: 10.1101/gad.10.16.2003. doi:10.1101/gad.10.16.2003 [DOI] [PubMed] [Google Scholar]
- Lee H.-H., Frasch M. Wingless effects mesoderm patterning and ectoderm segmentation events via induction of its downstream target sloppy paired. Development. 2000;127:5497–5508. doi: 10.1242/dev.127.24.5497. [DOI] [PubMed] [Google Scholar]
- Martinez Arias A., Baker N., Ingham P.W. Role of segment polarity genes in the definition and maintenance of cell states in the Drosophila embryo. Development. 1988;103:157–170. doi: 10.1242/dev.103.1.157. [DOI] [PubMed] [Google Scholar]
- Méthot N., Basler K. Hedgehog controls limb development by regulating the activities of distinct transcriptional activator and repressor forms of Cubitus interruptus. Cell. 1999;96:819–831. doi: 10.1016/s0092-8674(00)80592-9. doi:10.1016/S0092-8674(00)80592-9 [DOI] [PubMed] [Google Scholar]
- Ohlmeyer J.T., Kalderon D. Hedgehog stimulates maturation of Cubitus interruptus into a labile transcriptional activator. Nature. 1998;396:749–753. doi: 10.1038/25533. doi:10.1038/25533 [DOI] [PubMed] [Google Scholar]
- Papin J.A., Hunter T., Palsson B.O., Subramaniam S. Reconstruction of cellular signalling networks and analysis of their properties. Nat. Rev. Mol. Cell Biol. 2005;6:99–111. doi: 10.1038/nrm1570. doi:10.1038/nrm1570 [DOI] [PubMed] [Google Scholar]
- Pfeiffer S., Vincent J.-P. Signaling at a distance: transport of wingless in the embryonic epidermis of Drosophila. Cell Dev. Biol. 1999;10:303–309. doi: 10.1006/scdb.1999.0306. doi:10.1006/scdb.1999.0306 [DOI] [PubMed] [Google Scholar]
- Sanson B. Generating patterns from fields of cells. Examples from Drosophila segmentation. EMBO Rep. 2001;21:1083–1088. doi: 10.1093/embo-reports/kve255. doi:10.1093/embo-reports/kve255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz C., Locke J., Nishida C., Kornberg T.B. Analysis of cubitus interruptus regulation in Drosophila embryos and imaginal disks. Development. 1995;121:1625–1635. doi: 10.1242/dev.121.6.1625. [DOI] [PubMed] [Google Scholar]
- Swantek D., Gergen J.P. Ftz modulates runt-dependent activation and repression of segment—polarity gene transcription. Development. 2004;131:2281–2290. doi: 10.1242/dev.01109. doi:10.1242/dev.01109 [DOI] [PubMed] [Google Scholar]
- Tabata T., Eaton S., Kornberg T.B. The Drosophila hedgehog gene is expressed specifically in posterior compartment cells and is a target of engrailed regulation. Genes Dev. 1992;6:2635–2645. doi: 10.1101/gad.6.12b.2635. doi:10.1101/gad.6.12b.2635 [DOI] [PubMed] [Google Scholar]
- Taylor A.M., Nakano Y., Mohler J., Ingham P.W. Contrasting distributions of patched and hedgehog proteins in the Drosophila embryo. Mech. Dev. 1993;42:89–96. doi: 10.1016/0925-4773(93)90101-3. doi:10.1016/0925-4773(93)90101-3 [DOI] [PubMed] [Google Scholar]
- Thomas R. Boolean formalization of genetic control circuits. J. Theor. Biol. 1973;42:563–585. doi: 10.1016/0022-5193(73)90247-6. doi:10.1016/0022-5193(73)90247-6 [DOI] [PubMed] [Google Scholar]
- van den Heuvel M., Ingham P.W. Smoothened encodes a receptor-like serpentine protein required for hedgehog signaling. Nature. 1996;382:547–551. doi: 10.1038/382547a0. doi:10.1038/382547a0 [DOI] [PubMed] [Google Scholar]
- von Dassow G., Odell G.M. Design and constraints of the Drosophila segment polarity module: robust spatial patterning emerges from intertwined cell state switches. J. Exp. Zool. (Mol. Dev. Evol.) 2002;294:179–215. doi: 10.1002/jez.10144. doi:10.1002/jez.10144 [DOI] [PubMed] [Google Scholar]
- von Dassow G., Meir E., Munro E.M., Odell G.M. The segment polarity network is a robust developmental module. Nature. 2000;406:188–192. doi: 10.1038/35018085. doi:10.1038/35018085 [DOI] [PubMed] [Google Scholar]
- von Ohlen T., Hooper J.E. Hedgehog signaling regulates transcription through Gli/Ci binding sites in the wingless enhancer. Mech. Dev. 1997;68:149–156. doi: 10.1016/s0925-4773(97)00150-0. doi:10.1016/S0925-4773(97)00150-0 [DOI] [PubMed] [Google Scholar]
- Wolpert L., Beddington R., Brockes J., Jessell T., Lawrence P., Meyerowitz E. Principles of development. Current Biology Press; London, UK: 1998. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Proofs of theorems 1 and 2