Abstract
An important step towards understanding biological systems is to relate simple biochemical elements to dynamics. Here, we present the arguably simplest dynamical element in biochemical networks. It consists of a single protein with two states (active and inactive) and an external signal that catalyses the conversion between these two states. Further, there is steady synthesis and degradation of the inactive and active forms, respectively. As this element captures both structural and dynamical features of biochemical networks at the lowest level, we refer to it as a biochemical network unit (BioNetUnit). Using both simulations and mathematical analysis, we find that BioNetUnit shows perfect adaptation that leads to temporal responses to step changes in the incoming signal. Compared with a well-described adaptive system, which is found in bacterial chemotaxis, BioNetUnit has lower sensitivity and its adaptation time is less robust to the base signal levels. We show that these dynamical limitations lead to ‘once-and-only-once’ responses for certain signal sequences. These findings demonstrate that BioNetUnit is relevant in adaptive and cyclic processes. In particular, it could be seen as a generic representation for ligand-activated receptors that are desensitized upon continuous activation. The analysis of coupled BioNetUnits will show how the presented dynamics at single unit will change upon increased system complexity and how such systems would mediate biological functions.
Keywords: modularity, adaptive dynamics, computational modelling, chemotaxis, feed-forward loop, cell cycle
1. Introduction
All biological phenomena are the result of the underlying system dynamics. In the case of cellular responses, the dynamics result from the interactions of several proteins, constituting a pathway. Understanding such dynamics requires knowledge of the identity and the amount of proteins involved, their interactions and the kinetic rates governing those interactions. Alternatively, one could try to dissect universal structural and biochemical elements that always give rise to the same dynamics, so that pinpointing these elements in complex systems might lead to functional inference in a ‘bottom-up’ fashion. Put differently, such elements could be thought of as the modules or building blocks of cellular systems (Hartwell et al. 1999). There have been several attempts to link specific dynamics to key structural and biochemical elements. The general structural elements such as feed-forward loops and negative feedback are associated with response acceleration (Mangan & Alon 2003) and oscillations (Goldbeter 1996), respectively. These two elements (Behar et al. 2007) as well as a simple system, composed of a receptor and two proteins controlling its activity (Barkai & Leibler 1997), were proposed to underlie adaptation (i.e. transient response to sustained stimuli). Simple pathways of two or three proteins with their associated biochemistry are shown to underlie diverse sets of dynamics including bistability, amplification and oscillation (Tyson et al. 2003; Soyer et al. 2006). All these works concentrate on small systems of two or more interacting proteins as a proxy for a modular unit and analyse their dynamical capacities. It is also possible to apply such an approach at the lowest level possible, that of a single protein. Recently, it has been shown that the input–output relation in a two-component module could be highly robust to fluctuations in the levels of the components of the system (Shinar et al. 2007). Such findings and the obvious role of a single protein as the ‘building block’ of larger systems prompt us to consider here the dynamics of the simplest biochemical network module imaginable.
1.1 Introduction to BioNetUnit
From a biochemical perspective, the simplest building block of biological networks is a protein with all its associated reactions. Often proteins are found in two distinct states, where one state corresponds to an active form that can perform a downstream reaction. A protein is synthesized in the cell resulting in a constant flux into one of the states, while both states could be actively degraded. The conversion between the two states can be expected to occur with low base rates, or it could be actively catalysed by external signals (or enzymes). Many proteins found in the so-called two-component signalling pathways of bacteria are thought to function in this manner (Stock et al. 2000). Furthermore, a two-state model can be used as a first approximation to model more complex proteins with multiple internal states (see Nash et al. 2001). This generic system is arguably the simplest biochemical network unit (BioNetUnit) that could be thought of as the building block of biological pathways. Here, we concentrate on a special case of this system (figure 1a), where we assume that the synthesis results in the stable inactive form (P), the conversion of which to an unstable active form (Pa) is catalysed by an external signal (E). This scenario is true for many proteins that have to go through post-translational modifications to become active and/or unstable, i.e. phosphorylation can not only induce the activation of proteins but also enhance their degradation.
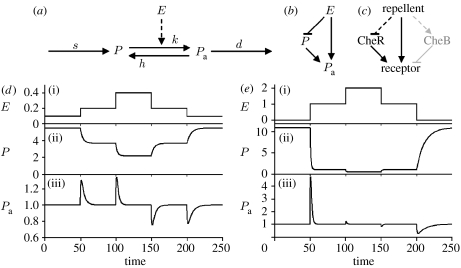
Figure 1.
(a) The biochemical and (b) abstract representation of BioNetUnit. Note that the latter corresponds to an incoherent feed-forward loop. (c) Direct (solid arrows) and indirect (dashed arrows) interactions at the receptor in the chemotaxis-regulating network of Escherichia coli. (d,e) The time course of input (E), inactive protein (P) and active protein (Pa) are shown in (i)–(iii), respectively. Note the difference in the magnitude of changes in E (small in (d) and large in (e)). Parameters used for BioNetUnit are s=1, k=0.1, h=0.1 and d=1.
Assuming simple mass-action kinetics, the resulting system dynamics can be easily written as a set of ordinary differential equations (to fit the dimensions, E should have a rate constant multiplier of 1 s−1, which we neglect here),
| (1.1) |
| (1.2) |
and solved for the steady-state concentrations of the two forms
| (1.3) |
and
| (1.4) |
As these equations show, the steady-state level of Pa is independent of the external signal level and is solely determined by the incoming (s) and outgoing (d) fluxes of the system. As a result, the active form of the protein would produce only transient responses to the incoming signal. In other words, the system would be perfectly adaptive. As shown in the electronic supplementary material, even if the protein activation is considered as an enzymatic reaction (i.e. proceeds through complex formation of P and E), the decoupling between the steady-state level of active protein and enzyme concentration remains.
While other simple structures have been proposed to lead to adaptive dynamics (Barkai & Leibler 1997; Behar et al. 2007), BioNetUnit is possibly the simplest adaptive biochemical system. As such, it is important to dissect the features in this module which render its dynamics adaptive. Structurally, we note that BioNetUnit contains a primitive integral feedback control, which has been shown previously to mediate adaptive dynamics (Yi et al. 2000; Behar et al. 2007). However, it is important to note that the independence of Pa steady state from E still holds if the inactivation reaction (h) is removed from the system, which means that BioNetUnit is adaptive even without any feedback. From a dynamical perspective, the distinctive feature of BioNetUnit is that it balances a zero-order reaction (the synthesis of P) against a first-order reaction (the degradation of Pa). In fact, this property of BioNetUnit would remain intact even if we assume multiple intermediary steps for the activation of P. Following a derivation for the dynamics of proteins with multiple phosphorylation sites (Gunawardena 2005), we can suppose that there is a number of intermediary steps going from P0 to Pn (corresponding to the active form), and with arbitrarily complex rate functions transforming Pi to Pi+1. The assumption that only P0 (Pn) is synthesized (degraded) and the flux at steady state from and to any species has to satisfy mass balance results in
| (1.5) |
where f and g stand for the flux from Pi to Pi+1 and vice versa, respectively. Hence, at steady state we would have Pn (protein active form) equal to s/d, resulting in an adaptive response to any incoming signals at the intermediary steps. Note that this formulation still corresponds to a two-state protein model, with all forms except Pn assumed to be inactive (or have only negligible activity).
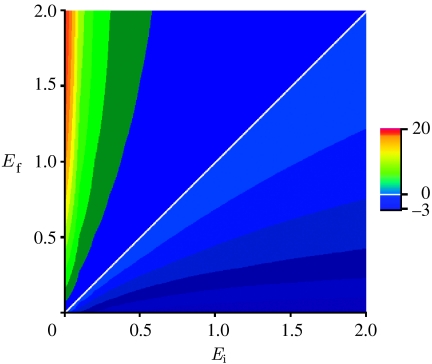
While the active form of the protein in BioNetUnit would always give transient responses to an incoming signal, it is important to understand how this transient response would be shaped. The increase in Pa results from the conversion of the inactive form when the signal level increases from a given initial (Ei) to a final value (Ef). As the system settles back to steady state, Pa reaches its original value (i.e. s/d), while P will reach a new steady state depending on Ef (see figure 1d and also figure 2a in the electronic supplementary material, which is derived from analytical solutions of the differential equations given above). The level of the initial increase in Pa will depend on both the base signal level (Ei) and the signal magnitude (i.e. ΔE). The former value would dictate the inactive protein (P) levels before a change in signal, while the latter would define the flux into the active form during signal change. For example, a high value of Ei leads to low levels of P prior to signal increase, and results in a low increase in Pa (figure 1e). The relation between the total Pa response and the different signal sequences is shown in figure 2.
Figure 2.
Heat map showing peak value of Pa after a step jump from Ei to Ef in the signal level. The signal change was done after the system reached steady state with Ei, and total Pa response (integral of the area between steady-state and perturbed Pa solutions) was registered.
The inverse relation between the steady-state level of P and E leads to a decrease in the former for every increase in the latter. The effects of this kind of ‘substrate depletion’ can be seen in both figures 1e and 2; the system cannot produce significant responses after responding to a large signal jump from a low to high value (see also figure 2 in the electronic supplementary material). This property of the system could be exploited to generate ‘once-and-only-once’ responses. Suppose that there was a threshold in the system relating the magnitude of the Pa peak to a physiological response. Further, imagine that the activation is driven by E, whose activity cycles between low and high values. Under this scenario, Pa would rise above the threshold only once during one cycle of E, resulting in a once-and-only-once physiological response during that cycle.
In summary, this arguably simplest biochemical system (BioNetUnit) consisting of a single protein (and its associated reactions) produces adaptive dynamics under biologically plausible assumptions. Further, it could lead to a once-and-only-once response under cyclic signals. Below we give examples of biological processes that are underlined by adaptive and once-and-only-once dynamics.
2. Biological relevance
2.1 Adaptation of membrane receptors
The description of BioNetUnit is highly reminiscent of several membrane receptors found in higher eukaryotes, such as G-protein-coupled receptors. These receptors appear on the cell surface in an inactive form, where they will be activated upon ligand binding. The continuous presence of ligand causes the active receptor form to be internalized for degradation resulting in an adaptive response (Sorkin & Von Zastrow 2002), with internalization being crucial for proper adaptation (Mashukova et al. 2006). We note that while this process could involve multiple steps governed by different time constants, their sequence follows the same scenario as presented in figure 1a. Given that the adaptive dynamics of BioNetUnit is independent of intermediary steps (see above), it could act as a very general description for adaptation observed at receptor level. Obviously, signalling pathways contain several regulatory interactions at various levels above the adaptation process at receptors, which can lead to additional properties (Shankaran et al. 2007).
2.2 Adaptation in chemotaxis
A complex system where such additional interactions are well described and the system dynamics is adaptive is the one underlying the chemotaxis behaviour of Escherichia coli (Berg & Tedesco 1975; Blair 1995). The input to the chemotaxis system is an external ligand concentration (i.e. attractants), while the system output is the phosphorylated form of the protein CheY. The external signals influence the activity of the membrane-bound receptors, which act as kinases to CheY. By binding to the motor protein, CheY effectively controls the swimming behaviour of the bacterium. There are several other proteins in the system that influences receptor and CheY activities, resulting in a complexity (Bray et al. 1993, 1998; Levin et al. 2002; Sourjik & Berg 2004; Endres & Wingreen 2006; Lipkow 2006) well beyond that of the BioNetUnit. Despite its increased complexity, the qualitative behaviour of the chemotaxis system is similar to the BioNetUnit; the response at CheY is perfectly adaptive with respect to the incoming signal at the receptor (Barkai & Leibler 1997; Sourjik & Berg 2002a).
What are the key features in the BioNetUnit or the chemotaxis system which underlie the qualitative similarities in their dynamics? As discussed above, the key features of the BioNetUnit are the presence of an incoherent feed-forward element and the balancing of a zero-order reaction against a first-order reaction. The former can be visualized better if we think of P and Pa as different species (figure 1b). In this case, the signal (E) acts as an inhibitor of the former and an activator of the latter, generating an incoherent feed-forward element (Mangan & Alon 2003). In the E. coli chemotaxis system, two proteins (CheR and CheB) control the activity level of the receptors through methylation and demethylation reactions. It is assumed that CheR and CheB act only on the active and inactive receptors, respectively (Barkai & Leibler 1997; Rao et al. 2004; Endres & Wingreen 2006). This assumption is crucial for proper adaptive dynamics and results in external signals having two separate effects. The first effect is the direct activation (deactivation) of the receptors, while the second, indirect, effect results from the assumption that the activation (deactivation) increases the rate of demethylation (methylation) and hence deactivation (activation). Thus, the external signals could be thought of as acting both on the receptor and on the CheB/CheR couple, creating an incoherent feed-forward loop (figure 1c). Interestingly, the assumption regarding the actions of CheR and CheB generates a system that balances a zero-order reaction against a first-order reaction (Alon 2006), as seen in the BioNetUnit. Both incoherent feed-forward (Yi et al. 2000; Tyson et al. 2003; Behar et al. 2007) and balancing a zero-order against a first-order reaction (Alon 2006; Ruoff et al. 2007) are suggested to involve adaptive dynamics. Incorporating both these proposed outlines in a primitive way, BioNetUnit stands out possibly as the simplest adaptive biochemical system.
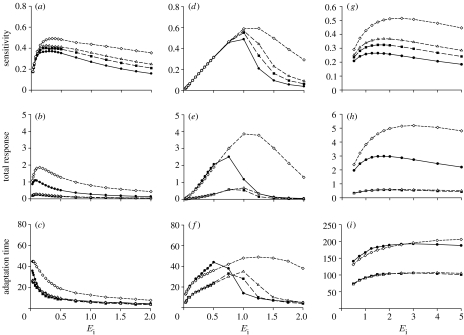
While the full chemotaxis system is similar to the BioNetUnit in qualitative terms, it is plausible to expect that quantitative properties of the dynamics obtained from BioNetUnit are not sufficient for chemotaxis with sensitivity and range as seen in E. coli (Segall et al. 1986; Bray 2002; Sourjik & Berg 2002b; Bray et al. 2007). Indeed, we find significant differences between the quantitative properties of the chemotaxis system (derived from the E. coli chemotaxis model presented in Rao et al. 2004) and BioNetUnit, independent of whether or not complex formation is taken into account. As shown in figure 3, the sensitivity (defined as percentage of Pa peak over percentage change in signal) in BioNetUnit remains high over a range of basal signal levels (Ei), qualitatively similar to that of the chemotaxis system (figure 3a,g). BioNetUnit with enzymatic kinetics (i.e. signal operates as an enzyme) shows a narrower range of sensitivity and its peak shifts to higher basal enzyme levels (figure 3d). Similarly, the total response (defined as the area under the Pa curve) varies widely with the varying basal signal level for the BioNetUnit model, but remains more or less constant for the detailed chemotaxis model (figure 3b,e,h). This difference is mainly due to the sensitivity of adaptation time (time to get back to steady-state Pa level). In BioNetUnit, stepwise increases from larger basal levels (Ei) produce responses that last for shorter periods (figure 3c,f, see also figure 1d(ii)). By contrast, the adaptation time in the chemotaxis system is robust to the changes in basal levels (figure 3i).
Figure 3.
Analysis of responses for 10 or 50% changes in ligand (E) level in BioNetUnit (a–c) without PE complex formation or (d–f) detailed enzymatic reactions and (g–i) in a detailed E. coli chemotaxis model of Rao et al. (2004). (a,d,g) Sensitivity of peak heights ((ΔPa/)/(ΔE/Ei)) depending on initial E level (Ei). (b,e,h) Integral of the area between steady-state and perturbed Pa solutions. (c,f,i) Time from Pa increase to settling back to steady state ±0.1%. For the chemotaxis model, the response is taken to be the total concentration of active receptors. For the enzymatic reactions of E on P we used k+1=100, k−1=9 and k2=1, resulting in a Michaelis constant of 0.1 (see equations in the electronic supplementary material). Circles, 50% up; squares, 10% up; triangles, 10% down; diamonds, 50% down.
It is plausible to expect that these quantitative differences between the BioNetUnit and the chemotaxis system relate to the increased complexity in the latter. Besides containing multiple proteins, the chemotaxis system contains a receptor that has multi-level (i.e. both ligand and other proteins control activity) and multi-step (i.e. multiple methylation sites) regulation. That the existence of multiple methylation sites provides a damping effect on the receptor activation and effectively improves the adaptation properties is shown in the detailed molecular models of chemotaxis (Endres & Wingreen 2006; Hansen et al. 2008). By contrast, for BioNetUnit the inactive form of the protein is easily depleted because the system has no possibility of damping the effects of E. As a consequence, the BioNetUnit produces much smaller responses (i.e. Pa peaks) for subsequent signals, resulting in weaker sensitivity and shorter adaptation times. This effect is somewhat compensated in the presence of enzymatic reactions (figure 3f) as complex formation results in saturation.
2.3 Role for BioNetUnit in cyclic processes
As discussed above, the dynamical limitations resulting from substrate depletion open up the possibility of once-and-only-once responses in BioNetUnit. This type of response plays an important role in cell-cycle regulation, where DNA replication and activation of cell division machinery have to occur only once in the presence of periodic signals that take the form of oscillations in cyclin-dependent kinase (Cdk) activity (Morgan 1995). In particular, increase in Cdk (bound by an S-phase cyclin) causes the induction of DNA replication (Piatti et al. 1996), which has to be initiated only once during one cell cycle. In other words, the replication complex (RC) has to be activated only once during that period. This can be achieved by setting up a system like BioNetUnit, where substrate depletion and adaptive dynamics are at work. Imagine for example that E and P correspond to Cdk activity and inactive RC, respectively, and that there is a threshold level of activated RC (Pa), which has to be surpassed to induce DNA replication. In such a case, high enough increases at Cdk level would cause RC activation above the threshold (resulting in replication induction), after which the latter would adapt (i.e. return to pre-stimulus levels). Owing to the associated substrate depletion, a further increase in Cdk level, say at entry into mitosis, cannot induce RC activation above threshold anymore and hence no DNA re-replication can be induced. Note that although BioNetUnit would provide perhaps the simplest implementation for this, other adaptive systems with substrate depletion should be able to provide the desired effect. For example, it has been proposed that an adaptive feed-forward loop regulates the septation initiation network, which induces cell division, through once-and-only-once dynamics (Csikász-Nagy et al. 2007).
3. Conclusion
In this paper we introduced what is arguably the simplest biological network unit imaginable, which is composed of a single protein and its associated reactions. Assuming two states of the protein, corresponding to the active and inactive forms, and the synthesis and the degradation acting preferentially on the inactive and active forms, respectively, we analyse the dynamical capacity of this small system. We find that under such conditions the steady-state level of the active protein would be independent of the signal that drives activation. In other words, the system provides perfect adaptation to incoming signals. This finding is robust against the assumptions regarding complex formation and the number of intermediary steps during the activation of the protein.
The biochemical system (BioNetUnit) presented here provides a description for dynamics of cell-membrane receptors, where activation occurs via ligand binding and the desensitization of active form is achieved via internalization. Similarly, several proteins where phosphorylation or other post-translational processes lead to activation and where the active form is more amenable to degradation are expected to follow the adaptive dynamics presented here.
We find that adaptation times in BioNetUnit are sensitive to basal signal levels, narrowing the range of its operability. Interestingly such limitations can lead to the usage of BioNetUnit as a once-and-only-once switch, where large signals can deplete the system and diminish subsequent responses. Such implementations would make any adaptive system, where substrate depletion and response threshold are in place, a possible regulator of cyclic processes such as the cell cycle. In metabolic networks, a mechanism similar to BioNetUnit could provide adaptation in a system where an enzyme converts a substrate that is steadily produced by an upstream reaction to another one which is used up in a subsequent reaction; in other words, the same structure as in figure 1a with P and Pa corresponding to substrates and with the hydrolysis rate set to zero. It is possible that such enzymatic reactions are involved in temperature compensation (Ruoff et al. 2007).
The concept of proteins and their associated biochemistry as simple building blocks of the cell could be highly useful to understand the physiological behaviour and help future efforts in systems and synthetic biology. As proteins are ubiquitous in cell biology, it should be straightforward to find the right experimental setting to test the results of this analysis. Conversely, BioNetUnit could be built in a controlled environment using the tools of synthetic biology and could be further used as a true building block of cellular systems. Finally, we note that further computational and mathematical analysis of interacting BioNetUnits might provide better understanding of more complex biological process with a wider selection of dynamical features such as oscillation and bistability.
Acknowledgments
We thank John J. Tyson and Bela Novak for their fruitful discussions, and Guy Shinar, Eszter Sipos Szabó and an anonymous reviewer for important comments and suggestions. We acknowledge the financial support from the Italian Ministry of University and Research Project FIRB (RBPR0523C3). A.C.N. acknowledges the support from the Hungarian Science Foundation Project OTKA (F-60414) and is also thankful to all the members of the Biological Switches and Clocks Meeting (organized by KITP with support by the National Science Foundation Project PHY05-51164).
Footnotes
One contribution of 10 to a Theme Supplement ‘Biological switches and clocks’.
Supplementary Material
Detailed elementary model of BioNetUnit; analytic solution of the mass action version of BioNetUnit
References
- Alon U. Chapman and Hall; New York, NY: 2006. An introduction to systems biology: design principles of biological circuits. [Google Scholar]
- Barkai N., Leibler S. Robustness in simple biochemical networks. Nature. 1997;387:913–917. doi: 10.1038/43199. doi:10.1038/43199 [DOI] [PubMed] [Google Scholar]
- Behar M., Hao N., Dohlman H.G., Elston T.C. Mathematical and computational analysis of adaptation via feedback inhibition in signal transduction pathways. Biophys. J. 2007;93:806–821. doi: 10.1529/biophysj.107.107516. doi:10.1529/biophysj.107.107516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg H.C., Tedesco P.M. Transient response to chemotactic stimuli in Escherichia coli. Proc. Natl Acad. Sci. USA. 1975;72:3235–3239. doi: 10.1073/pnas.72.8.3235. doi:10.1073/pnas.72.8.3235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair D.F. How bacteria sense and swim. Annu. Rev. Microbiol. 1995;49:489–522. doi: 10.1146/annurev.mi.49.100195.002421. doi:10.1146/annurev.mi.49.100195.002421 [DOI] [PubMed] [Google Scholar]
- Bray D. Bacterial chemotaxis and the question of gain. Proc. Natl Acad. Sci. USA. 2002;99:7–9. doi: 10.1073/pnas.022641699. doi:10.1073/pnas.022641699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray D., Bourret R.B., Simon M.I. Computer simulation of the phosphorylation cascade controlling bacterial chemotaxis. Mol. Biol. Cell. 1993;4:469–482. doi: 10.1091/mbc.4.5.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray D., Levin M.D., Morton-Firth C.J. Receptor clustering as a cellular mechanism to control sensitivity. Nature. 1998;393:85–88. doi: 10.1038/30018. doi:10.1038/30018 [DOI] [PubMed] [Google Scholar]
- Bray D., Levin M.D., Lipkow K. The chemotactic behavior of computer-based surrogate bacteria. Curr. Biol. 2007;17:12–19. doi: 10.1016/j.cub.2006.11.027. doi:10.1016/j.cub.2006.11.027 [DOI] [PubMed] [Google Scholar]
- Csikász-Nagy A., Kapuy O., Gyorffy B., Tyson J.J., Novak B. Modeling the septation initiation network (SIN) in fission yeast cells. Curr. Genet. 2007;51:245–255. doi: 10.1007/s00294-007-0123-4. doi:10.1007/s00294-007-0123-4 [DOI] [PubMed] [Google Scholar]
- Endres R.G., Wingreen N.S. Precise adaptation in bacterial chemotaxis through “assistance neighborhoods”. Proc. Natl Acad. Sci. USA. 2006;103:13 040–13 044. doi: 10.1073/pnas.0603101103. doi:10.1073/pnas.0603101103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldbeter A. Cambridge University Press; Cambridge, UK: 1996. Biochemical oscillations and cellular rhythms. [Google Scholar]
- Gunawardena J. Multisite protein phosphorylation makes a good threshold but can be a poor switch. Proc. Natl Acad. Sci. USA. 2005;102:14 617–14 622. doi: 10.1073/pnas.0507322102. doi:10.1073/pnas.0507322102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen C.H., Endres R.G., Wingreen N.S. Chemotaxis in Escherichia coli: a molecular model for robust precise adaptation. PLoS Comput. Biol. 2008;4:e1. doi: 10.1371/journal.pcbi.0040001. doi:10.1371/journal.pcbi.0040001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L.H., Hopfield J.J., Leibler S., Murray A.W. From molecular to modular cell biology. Nature. 1999;402:C47–C52. doi: 10.1038/35011540. doi:10.1038/35011540 [DOI] [PubMed] [Google Scholar]
- Levin M.D., Shimizu T.S., Bray D. Binding and diffusion of CheR molecules within a cluster of membrane receptors. Biophys. J. 2002;82:1809–1817. doi: 10.1016/S0006-3495(02)75531-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipkow K. Changing cellular location of CheZ predicted by molecular simulations. PLoS Comput. Biol. 2006;2:e39. doi: 10.1371/journal.pcbi.0020039. doi:10.1371/journal.pcbi.0020039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan S., Alon U. Structure and function of the feed-forward loop network motif. Proc. Natl Acad. Sci. USA. 2003;100:11 980–11 985. doi: 10.1073/pnas.2133841100. doi:10.1073/pnas.2133841100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashukova A., Spehr M., Hatt H., Neuhaus E.M. Beta-arrestin2-mediated internalization of mammalian odorant receptors. J. Neurosci. 2006;26:9902–9912. doi: 10.1523/JNEUROSCI.2897-06.2006. doi:10.1523/JNEUROSCI.2897-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan D.O. Principles of CDK regulation. Nature. 1995;374:131–134. doi: 10.1038/374131a0. doi:10.1038/374131a0 [DOI] [PubMed] [Google Scholar]
- Nash P., Tang X., Orlicky S., Chen Q., Gertier F.B., Mendenhall M.D., Sicheri F., Pawson T., Tyers M. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature. 2001;414:514–521. doi: 10.1038/35107009. doi:10.1038/35107009 [DOI] [PubMed] [Google Scholar]
- Piatti S., Bohm T., Cocker J.H., Diffley J.F., Nasmyth K. Activation of S-phase-promoting CDKs in late G1 defines a “point of no return” after which Cdc6 synthesis cannot promote DNA replication in yeast. Genes Dev. 1996;10:1516–1531. doi: 10.1101/gad.10.12.1516. doi:10.1101/gad.10.12.1516 [DOI] [PubMed] [Google Scholar]
- Rao C.V., Kirby J.R., Arkin A.P. Design and diversity in bacterial chemotaxis: a comparative study in Escherichia coli and Bacillus subtilis. PLoS Biol. 2004;2:E49. doi: 10.1371/journal.pbio.0020049. doi:10.1371/journal.pbio.0020049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruoff P., Zakhartsev M., Westerhoff H.V. Temperature compensation through systems biology. FEBS J. 2007;274:940–950. doi: 10.1111/j.1742-4658.2007.05641.x. doi:10.1111/j.1742-4658.2007.05641.x [DOI] [PubMed] [Google Scholar]
- Segall J.E., Block S.M., Berg H.C. Temporal comparisons in bacterial chemotaxis. Proc. Natl Acad. Sci. USA. 1986;83:8987–8991. doi: 10.1073/pnas.83.23.8987. doi:10.1073/pnas.83.23.8987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankaran H., Wiley H.S., Resat H. Receptor downregulation and desensitization enhance the information processing ability of signalling receptors. BMC Syst. Biol. 2007;1:48. doi: 10.1186/1752-0509-1-48. doi:10.1186/1752-0509-1-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinar G., Milo R., Martinez M.R., Alon U. Input output robustness in simple bacterial signaling systems. Proc. Natl Acad. Sci. USA. 2007;104:19 931–19 935. doi: 10.1073/pnas.0706792104. doi:10.1073/pnas.0706792104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorkin A., Von Zastrow M. Signal transduction and endocytosis: close encounters of many kinds. Nat. Rev. Mol. Cell Biol. 2002;3:600–614. doi: 10.1038/nrm883. doi:10.1038/nrm883 [DOI] [PubMed] [Google Scholar]
- Sourjik V., Berg H.C. Binding of the Escherichia coli response regulator CheY to its target measured in vivo by fluorescence resonance energy transfer. Proc. Natl Acad. Sci. USA. 2002a;99:12 669–12 674. doi: 10.1073/pnas.192463199. doi:10.1073/pnas.192463199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourjik V., Berg H.C. Receptor sensitivity in bacterial chemotaxis. Proc. Natl Acad. Sci. USA. 2002b;99:123–127. doi: 10.1073/pnas.011589998. doi:10.1073/pnas.011589998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourjik V., Berg H.C. Functional interactions between receptors in bacterial chemotaxis. Nature. 2004;428:437–441. doi: 10.1038/nature02406. doi:10.1038/nature02406 [DOI] [PubMed] [Google Scholar]
- Soyer O.S., Salathe´ M., Bonhoeffer S. Signal transduction networks: topology, response and biochemical processes. J. Theor. Biol. 2006;238:416–425. doi: 10.1016/j.jtbi.2005.05.030. doi:10.1016/j.jtbi.2005.05.030 [DOI] [PubMed] [Google Scholar]
- Stock A.M., Robinson V.L., Goudreau P.N. Two-component signal transduction. Annu. Rev. Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. doi:10.1146/annurev.biochem.69.1.183 [DOI] [PubMed] [Google Scholar]
- Tyson J.J., Chen K.C., Novak B. Sniffers, buzzers, toggles and blinkers: dynamics of regulatory and signaling pathways in the cell. Curr. Opin. Cell Biol. 2003;15:221–231. doi: 10.1016/s0955-0674(03)00017-6. doi:10.1016/S0955-0674(03)00017-6 [DOI] [PubMed] [Google Scholar]
- Yi T.M., Huang Y., Simon M.I., Doyle J. Robust perfect adaptation in bacterial chemotaxis through integral feedback control. Proc. Natl Acad. Sci. USA. 2000;97:4649–4653. doi: 10.1073/pnas.97.9.4649. doi:10.1073/pnas.97.9.4649 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Detailed elementary model of BioNetUnit; analytic solution of the mass action version of BioNetUnit