Summary
HIV-1 intersubtype recombinants have an increasingly important role in shaping the AIDS pandemic. We sought to understand the molecular mechanisms that generate intersubtype HIV-1 recombinants. To approach this, we analyzed recombinants of HIV-1 subtypes B and C and identified their crossover junctions in the viral genome from the 5′ LTR to the end of pol. We identified 56 recombination events in 56 proviruses; the distribution of these events indicated an apparent recombination gradient: there were significantly more crossover junctions in the 3′ half than in the 5′ half of the analyzed region. HIV-1 subtypes B and C have different dimerization initiation signal (DIS). We hypothesized that the inability of subtype B and C RNAs to form perfect base-pairing of the DIS affects the dimeric RNA structure and causes a decrease in recombination events at the 5′ end of the viral genome. To test this hypothesis, we examined recombinants generated from a subtype C virus and a modified subtype B virus containing a subtype C DIS. In the 56 proviruses analyzed, we identified 96 recombination events, which are significantly more frequent than in the B/C recombinants. Furthermore, these crossover junctions were distributed evenly throughout the region analyzed, indicating that the recombination gradient was corrected by matching the DIS. Therefore, base-pairing at the DIS has an important function during HIV-1 reverse transcription, most likely in maintaining nucleic-acid structure in the complex. These findings not only reveal elements important to retroviral recombination but also provide insights into the generation of HIV-1 intersubtype recombinants that are important to AIDS epidemic.
Keywords: HIV-1, Subtype, Recombination, Dimerization initiation signal, RNA secondary structure
Introduction
The increasing prevalence of HIV-1 intersubtype recombinants establishes the importance of these variants in the AIDS epidemic 1; 2. These intersubtype recombinants continue to emerge and infect the human population 3; 4; 5; 6; 7; 8; 9; 10. Currently, there are at least 34 circulating recombinant forms that spread in various human populations and a large number of unique recombinant forms, each infecting an epidemiologically related population 2; 11. These intersubtype recombinants now contribute to almost 18% of new HIV-1 infections worldwide 1. The presence of such a high number of intersubtype recombinants in the current pandemic is caused by frequent recombination during HIV-1 replication 12; 13; 14; 15; 16; 17; 18.
High-frequency recombination is a common feature during retroviral replication 19; 20; 21; 22; 23. Like all orthoretroviruses, one HIV-1 virion contains two copies of the viral RNA genome that are in dimeric form, and each RNA molecule contains the complete genetic information required for viral replication 24; 25; 26; 27; 28. After entering the target cells, viral RNAs are reverse transcribed into DNA by the viral-encoded enzyme reverse transcriptase (RT). During reverse transcription, recombination can occur to generate DNA containing portions of genetic information from each copackaged RNA 19; 29; 30; 31. Although recombination can occur during reverse transcription of all viral particles, genotypically different recombinants (such as intersubtype recombinants) can only be generated from viruses containing two RNA molecules encoding different sequences (heterozygous virions) 22. Thus, the preference in selecting viral RNA for copackaging and the frequency of template switching during reverse transcription can influence the observed recombination frequencies 31; 32; 33; 34.
The dimerization initiation signal (DIS) is a 6-nt palindromic sequence located at the loop of the proposed stem-loop 1 (SL1) in the 5′ untranslated region of the HIV-1 genome. It was hypothesized that the DIS regions of the two RNA molecules first form base-pairing to initiate the dimerization process — hence, the term DIS 35; 36; 37; 38. Various strains of HIV-1 have two different DIS sequences. The DIS of subtypes B and D and some intersubtype recombinants contain a GCGCGC sequence, whereas the DIS of the other subtypes and recombinants contain a GUGCAC sequence 11; 39. We have shown that base-pairing of the DIS of two RNA molecules is a major determinant in HIV-1 copackaged RNA partner selection 34. Furthermore, we have shown that the identity of the DIS plays an important role in the copackaging of RNAs from different HIV-1 subtypes 32; 33. RNA genomes from variants with the same DIS regions are pacakged together more frequently than those with different DIS; therefore, the identity of the DIS can affect the potential of intersubtype HIV-1 recombination 32; 33.
Cis-acting elements that affect retroviral recombination have been a topic of interest for many years. It was hypothesized that the RNA dimerization site is also a recombination hot spot because the two RNA templates are close to each other, which facilitates RT template switching. The RNA dimerization site as a recombination hot spot was first proposed in the murine leukemia virus system 40. A similar hypothesis was also proposed in HIV-1 based on studies using in vitro transcribed RNAs 41; however, in vivo studies did not support this hypothesis 18. In addition to the DIS, other HIV-1 sequences have been proposed to have an effect on recombination 16; 42; 43; many of these sequences have predicted RNA secondary structures and are thought to stall reverse transcription, thereby increasing recombination. In most, if not all, of these experiments, the effects of the DIS or secondary structures are limited to a very short region. In this study, we describe an observed long-range recombination gradient that affects the recombination frequencies over a 2-kb region. Furthermore, this gradient can be corrected by changing only 3-nt sequence in the DIS region. These findings reveal that the DIS not only is important in selecting the copackaged RNA partners, but also plays a critical role in maintaining proper nucleic acid structures in the reverse transcription complex. These findings also provide insights into the generation of HIV-1 intersubtype recombinants.
Results
Strategy to Study Recombination Events within the Viral Sequences between a Subtype B and a Subtype C Virus
We sought to characterize the recombination events in the viral genome between HIV-1 subtypes B and C. We previously demonstrated that the difference in DIS sequences between two viruses reduces the formation of heterozygous viruses 32. Therefore, only a minority of the virions generated from cells coinfected with HIV-1 subtypes B and C are heterozygous, which can produce genotypically different recombinants. To avoid analyzing progeny viruses generated from homozygous virions, we first identified cell clones containing known recombinant proviruses by reconstitution of a functional green fluorescence protein gene (gfp) at the 3′ end of the viral genome. We then analyzed the viral sequences of the recombinants in the 5′ half of the genome. Recombination event in HIV-1 is not correlated 44; thus, we do not expect the recombination event in gfp would affect the crossover event in the HIV-1 genome.
We previously generated virus producer cell lines to measure the intersubtype recombination rates between HIV-1 subtypes B and C 32. These virus producer cells contain a subtype B-based and a subtype C-based HIV-1 vector, BT6 and CH0, respectively, which have similar genetic structures (Figure 1(a)). Both viruses have inactivating deletions in vif, vpr, vpu, and env; therefore, these cells can generate infectious viruses only after Env is supplemented. Additionally, each virus carries two marker genes: the subtype B virus carries a mouse thy-1.2 gene (thy) and a mutated gfp, whereas the subtype C virus carries a mouse heat-stable antigen gene (hsa) and a mutated gfp. The inactivating mutations in the subtype B and C viruses are located at the 3′ and 5′ ends of gfp, respectively. A recombination event between the mutations can reconstitute a functional gfp gene, the expression of which was used to identify recombinant proviruses.
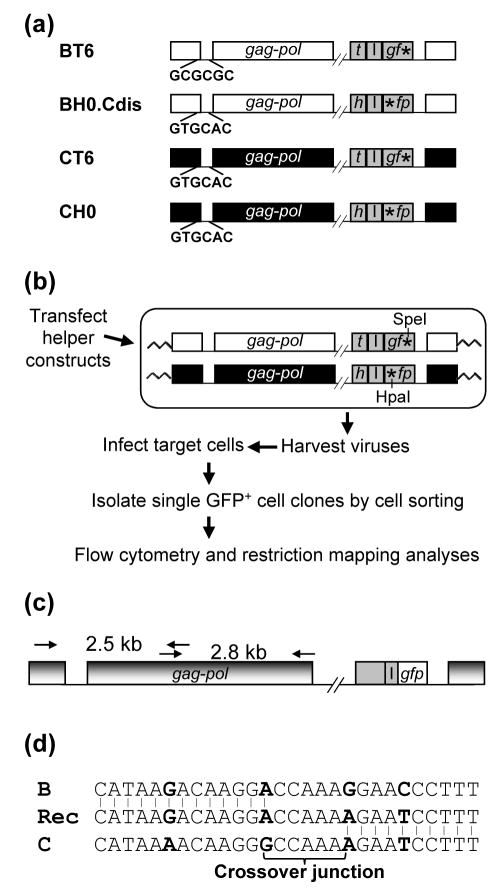
Figure 1.
System used to study the crossover junctions of HIV-1 recombinants. (a) Schematic structures of the HIV-1 vectors used to study intersubtype HIV-1 recombination. Subtype B and C proviruses are shown in white and black, respectively; reporter genes are shown in gray. h, hsa; t, thy; I, IRES; *, inactivating mutation in gfp. The DIS sequences of each vector are shown. Sequences between pol and the reporter genes are not shown (indicated by the double slash lines). All of the HIV-1 vectors also contained functional tat and rev. (b) Experimental approach for the generation of HIV-1 intersubtype recombinants. The restriction enzyme sites of the two gfp mutations are shown. (c) PCR amplification of GFP+ recombinant provirus. PCR primers that annealed to both HIV-1 subtype B and C proviruses were used to amplify a 2.5-kb and a 2.8-kb PCR product (from nt 113 to 2593, and from nt 2329 to 5095, respectively; NL4-3 nucleotide number designation). PCR products were sequenced directly. (d) Identification of the crossover junctions. A representative crossover junction is shown; Rec, recombinant. Nucleotides used as markers to identified crossover junctions are shown in boldface. The 5′ region of the recombinant sequence was derived from subtype B, whereas the 3′ region was derived from subtype C. Recombination occurred in the 5-nt region in the middle (crossover junction). All identified crossover junctions have two or more markers flanking each end.
The procedure we used to identify and isolate cell clones containing recombinant proviruses is described in Figure 1. Virus producer cells were transfected with two helper constructs, one expressing vesicular stomatitis virus G protein and the other expressing all the HIV-1 accessory genes (Figure 1(b)). Supernatants were harvested from the transfected cells 18 h posttransfection and were used to infect 293T target cells at a low multiplicity of infection (moi) (generally less than 0.1). Infected single GFP+ cell clones were isolated by cell sorting 48 h postinfection. These cell clones were expanded, and then characterized further by phenotypic and genotypic analyses. Although we infected the target cells at a low moi, double infection could occur. To ensure that each cell clone contained only a single recombinant provirus, we first performed flow cytometry on the cell clones and selected cell clones that were HSA+/GFP+ or Thy+/GFP+ but not positive for all three markers because each virus should only express hsa or thy, but not both. We then isolated genomic DNA from each cell clone and amplified a DNA fragment including the gfp gene by PCR. The 5′ and 3′ inactivating mutations in gfp also generated a HpaI and a SpeI site, respectively, whereas the wild-type gfp does not contain either restriction enzyme site. We mapped the amplified DNA fragment from each cell clone, and selected cell clones that only had a DNA fragment containing wild-type gfp. Using this protocol, we isolated 56 cell clones having the phenotype of HSA+/GFP+ or Thy+/GFP+ and containing a single GFP+ recombinant provirus.
Identification of Crossover Junctions in Recombinant Proviruses and Analyses of the Distribution of these Junctions in the Viral Genome
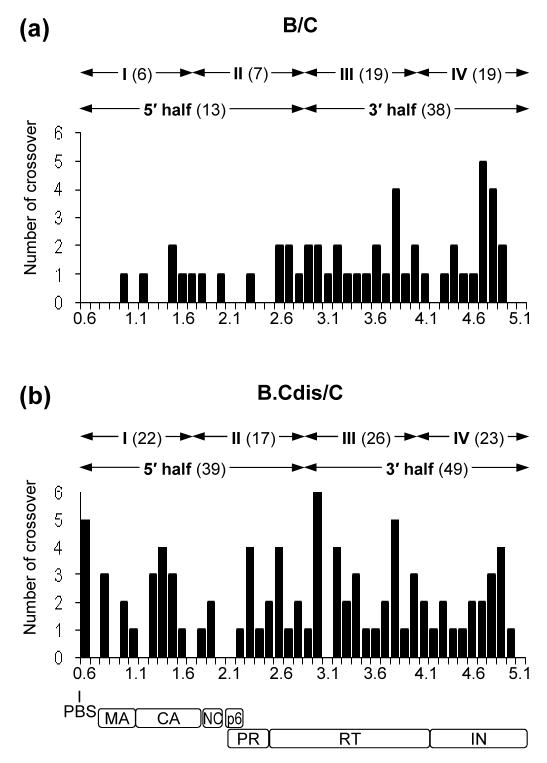
We PCR-amplified a 5-kb region of the proviral genome, comprising most of the 5′ LTR, 5′ untranslated region, and gag-pol, from each of the 56 GFP+ cell clones (Figure 1(c)). These PCR products were characterized by direct DNA sequencing, and all of the sequences were confirmed by two independent sequencing reactions. The sequences from each recombinant were compared with the parental subtype B and subtype C sequences, and crossover junctions were identified from the alignment (Figure 1(d)). Of the analyzed region in the 56 proviruses, we identified a total of 56 recombination events; 23 proviruses do not have recombination in the 5-kb region, 20 have one, and 13 have more than one recombination event. Among the viruses with more than one event, 6, 5, 1 and 1 proviruses have 2, 3, 4, and 5 recombination events, respectively (Figure 2(a)).
Figure 2.
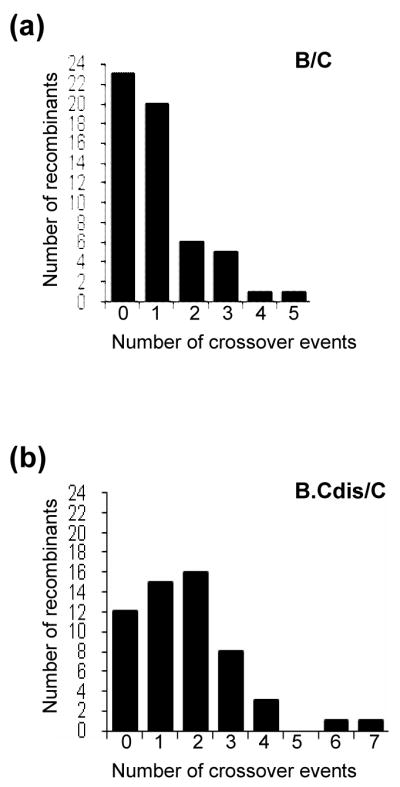
Number of crossover junctions in HIV-1 intersubtype recombinants. (a) Analysis of recombinants from a subtype B (BT6) and a subtype C (CH0) parent (marked as B/C). The DIS regions of these two parental viruses contain sequence differences. (b) Analysis of the recombinants from a subtype B parent with subtype C DIS (BH0.Cdis) and a subtype C (CT6) parent (marked as B.Cdis/C). The DIS sequences of these two parents are identical. The x axis represents the number of junctions in a recombinant; the y axis represents the number of recombinants observed.
We have analyzed the distribution of the 56 crossover junctions in the viral genome. Of the 56 crossovers, 5 are located at the 5′ LTR whereas 51 are located between the primer binding site (PBS) and the end of pol, which is a 4.4-kb region. The distribution of the 51 crossovers in this 4.4-kb region is uneven; only 13 are located at the 5′ 2.2-kb region (from the PBS to the 5′ end of the RT-coding region), whereas 38 junctions are located at the 3′ half [from 5′ end of RT- to the end of integrase (IN)-coding region] (Figure 3(a)). There are significantly fewer recombination events at the 5′ half compared with the 3′ half of the 4.4-kb region (p = 0.002, Wilcoxon Signed Ranks test). To eliminate the possibility that this uneven distribution was caused by one hot spot or cold spot for recombination in the viral genome, we divided the 4.4 kb region into four portions of equal lengths, designated I, II, III, and IV (Figure 3(a)). These portions have 6, 7, 19, and 19 crossover junctions respectively. Pairwise comparisons indicated that the numbers of crossovers are not significantly different between regions I and II, and between regions III and IV (for both comparisons, p = 1.000, Wilcoxon Signed Ranks test). However, the number of crossovers in either region I or II differs significantly from that in region III or IV (for comparison of I and III, p = 0.015; I and IV, p = 0.009; II and III, p = 0.034; II and IV, p = 0.034; Wilcoxon Signed Ranks test). Therefore, analyses of the location of the crossover jucntions suggest that there is a recombination gradient in intersubtype B and C HIV-1 recombination.
Figure 3.
Distribution of crossover junctions between the PBS to the end of pol in HIV-1 intersubtype recombinants. (a) Analysis of the recombinants from a subtype B and a subtype C parental virus that contains different DIS sequences. (b) Analysis of the recombinants from a modified subtype B virus with subtype C DIS and a subtype C parental virus. The x axis is shown in 0.1-kb increments and numbering is based on the NL4-3 viral DNA genome. The approximate locations of the PBS and viral genes are shown at the bottom. The y axis represents the number of crossover junctions observed in the 56 proviruses of each experimental group within the 0.1-kb regions. The total numbers of crossovers in the two halves and four quarters are shown above the plot. MA, matrix; CA, capsid, NC, nucleocapsid, PR, protease.
Elucidating the Cause of the Intersubtype Recombination Gradient
We envision two possible causes of the recombination gradient: uneven distribution of sequence identity and alteration of RNA structures. It is possible that the two parental viruses have variable levels of sequence identities among different regions of the genome, which affect the template-switching frequencies and result in a recombination gradient. Alternatively, it is possible that the two copackaged RNAs have different structures in various regions of the genome; the RNA structures at the 3′ region may be more accessible to intermolecular template switching than the ones at the 5′ region, causing the recombination gradient.
Analyses revealed that the nucleotide sequence identity of the two parental viruses in regions I, II, III, and IV are 87%, 86%, 87%, and 87%, respectively (data not shown). Therefore, it is unlikely that sequence identity among these regions caused the apparent recombination gradient. In contrast, the DIS regions of subtype B and subtype C viruses cannot form perfect base-pairing; furthermore, we have previously shown that the base-pairing of the DIS significantly affects the efficiency of RNA copackaging. Hence, we hypothesized that when subtype B and subtype C RNAs are copackaged, the imperfect base-pairing of the DIS between the two RNA molecules affects the RNA structures, thereby causing the reduced crossover events in the 5′ region of the viral genome compared with that in the 3′ region. To test our hypothesis, we examined the recombination events between a subtype C virus, CT6, and a subtype B virus with subtype C DIS, BH0.Cdis (Figure 1(a)) 32. These two viruses have the same nucleotide sequences in the 5-kb region as the aforementioned subtype B and C virus pair, except for a 3-nt sequence alteration in BH0.Cdis that makes the DIS sequences of CT6 and BH0.Cdis identical, thereby allowing the formation of perfect base-pairing of DIS sequences between the two RNAs. The DIS is located 75 nt downstream of the PBS and is between 0.6 and 0.7 in the x-axis of Figure 3.
Using a method similar to that described above, we harvested viruses generated from cells containing BH0.Cdis and CT6 proviruses, infected fresh target cells, and isolated and characterized 56 GFP+ target cell clones that harbored a single recombinant provirus with a reconstituted, functional gfp gene. The same 5-kb region in each provirus was characterized by DNA sequencing. Of the 56 proviruses, 12 did not have crossovers in the 5-kb region, whereas 15, 16, 8, 3, 1, and 1 provirus had 1, 2, 3, 4, 6, and 7 crossovers, respectively, in the analyzed region (Figure 2(b)), which yielded a total of 96 crossovers in these 56 proviruses. Therefore, when 3 nt of the 5-kb region was altered to allow base-pairing of the DIS regions of the two RNA, significantly more crossovers were observed (56 versus 96; p = 0.003, Mann-Whitney test).
Of the 96 recombination events, 8 occurred in the LTR region, whereas 88 occurred between the PBS and the end of pol. The positions of these 88 crossover junctions are shown in Figure 3(b); there are 39 junctions in the 5′ half and 49 junctions in the 3′ half region. Therefore, the numbers of crossover junctions in the two regions are not significantly different from each other (p = 0.401, Wilcoxon Signed Ranks Test). In addition, comparing the 5′ end of the analyzed region of the two groups of recombinants, there were significantly more junctions in the recombinants from viruses with matching DIS regions than those from viruses with different DIS regions (13 versus 39; p = 0.001, Mann-Whitney test). This difference is not seen in the 3′ end of the analyzed region of the two groups of recombinants (38 versus 49; p = 0.215, Mann-Whitney test). Additionally, we compared the recombination events in the four equal-distance portions: regions I, II, III, and IV of the 4.4-kb genome have 22, 17, 26, and 23 crossover junctions, respectively. There are no significant differences among any of the regions (p = 0.399, Friedman test). Hence, by changing 3-nt sequences and matching the DIS regions in subtype B and subtype C viruses, recombinants now have more crossover junctions and the junctions are more evenly distributed in the analyzed region of the genome.
Strand Transfer and Recombination in the LTR Region
We also analyzed DNA synthesis events that reconstituted the 5′ LTR region of the proviruses during reverse transcription. Of the 112 proviruses (56 from each group), initiation of reverse transcription occurred evenly between the subtype B and subtype C genomes (50 and 62, respectively; Table 1). Minus-strand DNA transfer occurred intra- and intermolecularly at similar frequencies (68 and 44, respectively). Furthermore, the minus-strand DNA initiation and transfer events were not different, regardless of whether the DIS regions of the two viruses were identical (p = 0.25 and p = 0.7, respectively, Chi-square test).
Table 1.
Initiation and transfer of minus-strand DNA
| Recombinant group | Minus-strand initiation |
Minus-strand transfer |
||
|---|---|---|---|---|
| B | C | Intraa | Intera | |
| BT6/CH0 | 22 | 34 | 33 | 23 |
| BH0.Cdis/CT6 | 28 | 28 | 35 | 21 |
|
| ||||
| Total | 50 | 62 | 68 | 44 |
intra and inter, intra- and intermolecular, respectively.
Among the 112 proviruses analyzed, only 3 had premature minus-strand DNA transfer, indicating that such events are not frequent. The TAR element dominates the R region; of the 97 nt of R, TAR occupies 59 nt. These three premature transfer events occurred at different locations: one occurred while copying the 5′ portion of the TAR region, one near the top of the TAR element, and the third within the 3′ end of TAR (Figure 4).
Figure 4.
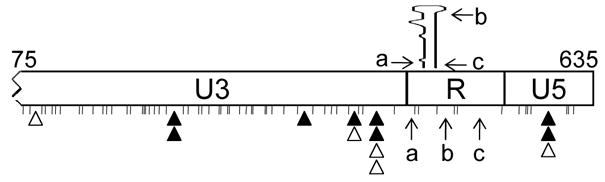
Distribution of crossover junctions and premature minus-strand DNA transfer in the 5′ LTR. The 5′ LTR from NL4-3 nt 75 – 635 is shown to scale. The nucleotide mismatches (markers) between subtype B and subtype C sequences are shown as short vertical lines. Open and solid triangles indicate the crossover junctions identified in the BT6/CH0 and BH0.Cdis/CT6 experimental groups, respectively. The structure of TAR is shown above the R region. Premature minus-strand DNA transfer events are indicated by arrows; event “a” was identified in the BT6/CH0 experimental group, whereas events “b” and “c” were identified in the BH0.Cdis/CT6 experimental group.
In addition to the strand transfer events required for the regeneration of the LTR, we also observed 13 recombination events: 3 occurred in U5 and 10 occurred in U3 (Figure 4). Therefore, recombination in the LTR appears to occur at similar rates when compared with the events observed between the PBS and the end of IN (for the LTR: 13 events in 58 kb analyzed; for PBS to IN: 140 events in 498 Kb analyzed).
Effects of RNA Secondary Structures on Recombination
It was proposed that secondary structures in the RNA template promote recombination. In addition to the TAR element, there are other well-documented RNA secondary structures: SL1 to SL4, and the gag-pol translational slippage site. Within the sequences that consritute SL1 to SL4, the two virus strains used in this study contain sequence variation between SL2 and SL3, but do not have variation between SL1 and SL2, or between SL3 and SL4. Therefore, we can only identify recombination events occurred within the SL1-SL2 region or within the SL3-SL4 region. We did not observe any recombination events in the SL1-SL2 region in the B/C experimental group, but observed five events in the B.Cdis/C experimental group (Figure 3). Based on the assumption of Poisson distribution, we calculated the probability of observing five events in the DIS region and our results suggest that DIS is a recombination hotspot when the two RNA molecules have the same sequence (p = 0.00001). In contrast, the SL3-SL4 region did not appear to experience more recombination; there was one crossover in the B.Cdis/C experimental group and none in the B/C group.
The translational slippage site sequence constitutes 44 nt (Figure 3, between 2.0 and 2.2 in the x axis); we observed one recombination event in this sequence in the 112 proviruses analyzed. Therefore, this secondary structure does not appear to experience more recombination events than the rest of the genome.
Fidelity of Recombination and Strand Transfer Events
Of the more than 556 kb of the viral genome analyzed, we observed 76 mutations in the viral genome, including 73 substitutions, one 1-nt deletion, and one 41-nt deletion. The overall mutation frequency is 1.4 × 10−4 per nucleotide analyzed; several events contribute to this frequency, including mutations accumulated from DNA transfection and two rounds of reverse transcription that first generate the provirus in the producer cells, then in the target cells.
We also analyzed mutations in the crossover junctions; this measurement was limited to events detected in one round of virus replication, because the two parental viruses were propagated and infected into the virus producer cells separately. Of the 152 recombination events detected in the viral genomes, 9 events contained mutations in the crossover junctions (Figure 5(a) to (c)). Therefore, most of the recombination events are accurate (143 of 152); only 6% of the events contained errors. Of the detected events that have mutations, five contained substitutions, two contained insertions caused by template misalignment, and two contained either a substitution or deletion that could be caused by misalignment. Considering that the total length of these 152 junctions is 3125 nt, the mutation frequency is 2.9 × 10−3/nt. Therefore, compared with the rest of the analyzed sequences, the crossover junctions have more errors (p = 0.002, Chi-square test).
Figure 5.
Mutations detected in crossover junctions and strand transfer events. (a) Substitutions in crossover junctions. Five of these events were observed. (b) Insertions caused by template misalignment in crossover junctions. Two of these events were observed; one has a 3-nt insertion and the other has a 1-nt insertion in the sequence. (c) Mutations that could be caused by template misalignment in crossover junctions. Two such events were observed. Recombinants in the alignments on the top and bottom have an insertion and a deletion, respectively. (d) Mutation (misalignment) occurred in a minus-strand DNA transfer event. This event corresponds to event “a” in Figure 4. Recombinant (Rec) sequences were aligned with subtype B and subtype C parental sequences. Markers used to identify crossover junctions are shown in uppercase boldface. Mutations are shown in lowercase boldface.
We also compared the accuracy between the obligatory minus-strand DNA transfer events and the recombination events in the genome. In the 112 minus-strand transfer events, we observe one error, which is a misalignment in a premature transfer event (Figure 5(d)). Therefore, the minus-strand DNA transfer events appear to be marginally more accurate than the recombination events (1 in 112 compared with 9 in 152; p = 0.05, Fisher’s Exact test). Although it was hypothesized that similar mechanisms are used for minus-strand DNA transfer and recombination, the accuracy of these events may be different, suggesting a mechanistic difference(s) between these two events.
Discussion
In this report, we describe studies that reveal an apparent recombination gradient between HIV-1 subtypes B and C that affects an approximately 2-kb region of the viral genome. Furthermore, this gradient is caused by improper base-pairing of the DIS sequences of the two RNAs, because this effect can be abolished by changing 3-nt sequences of the DIS region. To our knowledge, this is the first description of such a recombination gradient in HIV-1 and in the retroviral genome in general. This gradient reveals insights into viral RNA structures in the reverse transcription complex and also has important implications for the generation of intersubtype recombinants of HIV-1.
Base-pairing of DIS Sequences and the Apparent Recombination Gradient
Previously, we have shown that the DIS is largely responsible for the selection of the copackaged RNAs 32; 33; 34. In this study, we examined viruses that packaged two RNAs containing different DIS sequences and observed that the lack of perfect base-pairing between the two DIS regions caused an apparent recombination gradient with far fewer recombination events immediately downstream from the DIS than those in the pol region. Because changing the DIS can abolish the observed gradient, our data indicate that the long-range effect is caused by the DIS rather than by other local sequences. These results suggest that the two RNA molecules in the reverse transcription complex are organized in a particular structure(s) and that the base-pairing of the DIS sequences plays an important role in forming this structure. It is possible that the DIS serves as a nucleation point to allow proper arrangement of the dimeric RNA structures immediately downstream from it. Without this nucleation point, the 2-kb region immediately following the DIS is not in a suitable structure to allow recombination to occur. The effect of the DIS base-pairing diminishes after approximately 2 kb. Most of the pol regions experienced similar numbers of crossover events regardless of whether the DIS can form perfect base-pairing (Figure 3), which suggests that the rest of the RNA sequences are still in the proper dimer structure. This result is consistent with the conclusion generated by us and others that despite the importance of the DIS, base-pairing of the DIS sequences is not absolutely essential for the generation of virion RNA dimers 34; 36; 45; 46. Therefore, it is likely that there are other elements in the viral genome that also facilitate the formation of RNA dimers.
Fidelity of the Recombination and Strand Transfer Events
Whether HIV-1 recombination is an error-prone process has long been the subject of debate. Results from some in vitro recombination studies suggested that a large number of recombination events contain errors 47; 48, and it was proposed that misincorporation is the driving force behind high-frequency recombination 49. Analyses of a limited number of crossover junctions by in vivo assays suggested that recombination events may not be as error prone as some in vitro studies proposed 50. In our study, we analyzed a very large number of crossover junctions and found that most (94%) of the recombination events are accurate. Therefore, it is unlikely that misincorporation is the driving force of high-frequency HIV-1 recombination events. However, recombination events do have a higher error rate than the rest of the sequences that do not have apparent recombination; based on our analyses, the rate is likely to be one to two log higher. Within these few events of observed mutation in the crossover junctions, it is unclear whether misincorporation caused recombination or mistakes occurred during crossover to generate mutations in the junctions. Therefore, the cause and effect of mutations and recombination cannot be distinguished.
It has been widely accepted that the ability of the RT to switch template during minus-strand DNA transfer also allows recombination to occur during minus-strand DNA synthesis 29; 30; 31. However, our analyses indicated that minus-strand DNA transfer events may be more accurate than recombination events, suggesting that differences in the mechanistic details between these two events could cause the higher fidelity of the strand transfer events. One possible difference is the status of the template. Most of the minus-strand DNA transfer events occur at the end of the RNA template (109 of the 112 viruses studied or >97%), whereas the status of the template in recombination events is unclear. We compared the accuracy of the strong-stop minus-strand DNA transfer event (in which RT reached the end of the template before the transfer) with that of the recombination event; the accuracies of these two events are significantly different (0 in 109 compared with 9 in 152; p = 0.011, Fisher’s Exact test). We hypothesize that during the synthesis of the minus-strand strong stop DNA, RT reaches the end of the RNA template and pauses, allowing the degradation of the RNA template to expose sufficient nascent DNA to mediate accurate minus-strand DNA transfer. In contrast, the status of the templates and nascent DNA are unclear in recombination events. We propose that whether a transfer event occurs at the end of the template may affect the accuracy of the event.
The role of DIS in HIV-1 recombination
It has been suggested for many years that the retroviral RNA dimerization sites are recombination hot spots 40; 41. Although our study indicates that the DIS may indeed be a local hot spot, the most drastic effects of the DIS on HIV-1 recombination occurs when the DIS of the two HIV-1 strains do not form perfect base-pairing. Our results demonstrated that the DIS has two distinct roles in the generation of intersubtype HIV-1 recombinants. The sequences of the DIS alter the efficiencies of RNA copackaging: heterozygous virions form far less frequently than homozygous virions with RNAs from two HIV-1 subtypes containing different DIS sequences 32; 34. In this study, we demonstrated a second role of the DIS in HIV-1 intersubtype recombination. When the DIS sequences cannot form perfect base-pairing, recombination is affected during the reverse transcription of the rare heterozygous virion genome: there is a sharp decrease of recombination events that affect a 2-kb region immediately downstream of the DIS. These results have strong implications for two aspects of HIV-1 biology: first, the DIS plays a critical role in maintaining proper nucleic acid structures in the reverse transcription complex, and second, the sequence identities of the DIS affect the generation of intersubtype recombinants, which plays an important role in the AIDS pandemic.
Materials and Methods
HIV-1 Vectors, Virus Producer Cells, and the Generation of Single Cell Clones Containing Recombinant Proviruses
Subtype B-based BT6 and BH0.Cdis vectors, and subtype C-based CT6 and CH0 vectors have been described previously 32; 44. The human embryonic kidney cell line 293T was maintained in Dulbecco’s Modified Eagle’s Medium supplemented with 10% fetal calf serum, penicillin (50 U/ml), and streptomycin (50 μg/ml) at 37°C in 5% CO2. Virus producer cell lines containing two parent proviruses were generated as previously described 44. Single cell clones were isolated by cell sorting using a FACSVantage SE system with the FACSDiVi Digital option (BD Biosciences, Franklin Lakes, NJ). Phenotypes of cell clones were identified by staining cells with phycoerythrin-conjugated HSA antibody (BD Biosciences) and allophycocyanin-conjugated Thy-1.2 antibody (eBioscience, San Diego, CA), and then analyzed by flow cytometry. DNAs were isolated from the selected cell clones using QIAamp DNA Blood Mini Kit (Qiagen, Valencia, CA) following protocols recommended by the manufacturer.
Genotypic Analyses of a 5-kb Region in the Recombinant Proviruses
Proviral genome comprising the 5′ LTR, 5′ untranslated region, gag, and pol was amplified by PCR using the genomic DNA isolated from each GFP+ cell clone as a template. The PCR products were sequenced with overlapping primers and the resulting sequence contigs were assembled using the Gap4 program of the Staden Package 51. Every nucleotide was identified by at least two sequence contigs to ensure the accuracy of the DNA sequence. The assembled proviral sequences were aligned with the sequences of the two parental proviruses using ClustalX version 1.8.3 52. The crossover junctions, defined as stretches of homologous sequences flanked by mismatches between the two parental proviruses, were identified visually in the sequence alignment.
Acknowledgments
We thank Anne Arthur for expert editorial help; Vinay K. Pathak for intellectual input throughout the project and critical reading of the manuscript; Eric Freed, Michael Moore, Islam Mohammad for critical reading of the manuscript. This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Abbreviations
- HIV-1
human immunodeficiency virus type 1
- DIS
dimerization initiation signal
- SL
stem loop
- moi
multiplicity of infection
- PBS
primer binding site
- RT
reverse transcriptase
- IN
integrase
- gfp
green fluorescence protein gene
- thy
mouse thy1.2 gene
- hsa
mouse heat stable antigen gene
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hemelaar J, Gouws E, Ghys PD, Osmanov S. Global and regional distribution of HIV-1 genetic subtypes and recombinants in 2004. Aids. 2006;20:W13–23. doi: 10.1097/01.aids.0000247564.73009.bc. [DOI] [PubMed] [Google Scholar]
- 2.McCutchan FE. Global epidemiology of HIV. J Med Virol. 2006;78(Suppl 1):S7–S12. doi: 10.1002/jmv.20599. [DOI] [PubMed] [Google Scholar]
- 3.Arien KK, Vanham G, Arts EJ. Is HIV-1 evolving to a less virulent form in humans? Nat Rev Microbiol. 2007;5:141–51. [Google Scholar]
- 4.De Sa Filho DJ, Sucupira MC, Caseiro MM, Sabino EC, Diaz RS, Janini LM. Identification of two HIV type 1 circulating recombinant forms in Brazil. AIDS Res Hum Retroviruses. 2006;22:1–13. doi: 10.1089/aid.2006.22.1. [DOI] [PubMed] [Google Scholar]
- 5.Santos AF, Sousa TM, Soares EA, Sanabani S, Martinez AM, Sprinz E, Silveira J, Sabino EC, Tanuri A, Soares MA. Characterization of a new circulating recombinant form comprising HIV-1 subtypes C and B in southern Brazil. Aids. 2006;20:2011–2019. doi: 10.1097/01.aids.0000247573.95880.db. [DOI] [PubMed] [Google Scholar]
- 6.Tee KK, Li XJ, Nohtomi K, Ng KP, Kamarulzaman A, Takebe Y. Identification of a Novel Circulating Recombinant Form (CRF33_01B) Disseminating Widely Among Various Risk Populations in Kuala Lumpur, Malaysia. J Acquir Immune Defic Syndr. 2006 doi: 10.1097/01.qai.0000242451.74779.a7. [DOI] [PubMed] [Google Scholar]
- 7.Visawapoka U, Tovanabutra S, Currier JR, Cox JH, Mason CJ, Wasunna M, Ponglikitmongkol M, Dowling WE, Robb ML, Birx DL, McCutchan FE. Circulating and unique recombinant forms of HIV type 1 containing subsubtype A2. AIDS Res Hum Retroviruses. 2006;22:695–702. doi: 10.1089/aid.2006.22.695. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y, Lu L, Ba L, Liu L, Yang L, Jia M, Wang H, Fang Q, Shi Y, Yan W, Chang G, Zhang L, Ho DD, Chen Z. Dominance of HIV-1 subtype CRF01_AE in sexually acquired cases leads to a new epidemic in Yunnan province of China. PLoS Med. 2006;3:e443. doi: 10.1371/journal.pmed.0030443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sierra M, Thomson MM, Posada D, Perez L, Aragones C, Gonzalez Z, Perez J, Casado G, Najera R. Identification of 3 phylogenetically related HIV-1 BG intersubtype circulating recombinant forms in Cuba. J Acquir Immune Defic Syndr. 2007;45:151–60. doi: 10.1097/QAI.0b013e318046ea47. [DOI] [PubMed] [Google Scholar]
- 10.Wang B, Lau KA, Ong LY, Shah M, Steain MC, Foley B, Dwyer DE, Chew CB, Kamarulzaman A, Ng KP, Saksena NK. Complex patterns of the HIV-1 epidemic in Kuala Lumpur, Malaysia: Evidence for expansion of circulating recombinant form CRF33_01B and detection of multiple other recombinants. Virology. 2007 doi: 10.1016/j.virol.2007.05.033. [DOI] [PubMed] [Google Scholar]
- 11.Leitner T, Korber B, Daniels M, Calef C, Foley B. HIV-1 Subtype and Circulating Recombinant Form (CRF) Reference Sequences, 2005. In: Leitner T, Foley B, Hahn B, Marx P, McCutchan F, Mellors JW, Wolinsky S, Korber B, editors. The 2005 HIV Sequence Compendium. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory; Los Alamos, NM: 2005. [Google Scholar]
- 12.Clavel F, Hoggan MD, Willey RL, Strebel K, Martin MA, Repaske R. Genetic recombination of human immunodeficiency virus. J Virol. 1989;63:1455–9. doi: 10.1128/jvi.63.3.1455-1459.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jetzt AE, Yu H, Klarmann GJ, Ron Y, Preston BD, Dougherty JP. High rate of recombination throughout the human immunodeficiency virus type 1 genome. J Virol. 2000;74:1234–40. doi: 10.1128/jvi.74.3.1234-1240.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rhodes T, Wargo H, Hu WS. High rates of human immunodeficiency virus type 1 recombination: near-random segregation of markers one kilobase apart in one round of viral replication. J Virol. 2003;77:11193–200. doi: 10.1128/JVI.77.20.11193-11200.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Onafuwa A, An W, Robson ND, Telesnitsky A. Human immunodeficiency virus type 1 genetic recombination is more frequent than that of Moloney murine leukemia virus despite similar template switching rates. J Virol. 2003;77:4577–87. doi: 10.1128/JVI.77.8.4577-4587.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galetto R, Moumen A, Giacomoni V, Veron M, Charneau P, Negroni M. The structure of HIV-1 genomic RNA in the gp120 gene determines a recombination hot spot in vivo. J Biol Chem. 2004;279:36625–32. doi: 10.1074/jbc.M405476200. [DOI] [PubMed] [Google Scholar]
- 17.Levy DN, Aldrovandi GM, Kutsch O, Shaw GM. Dynamics of HIV-1 recombination in its natural target cells. Proc Natl Acad Sci U S A. 2004;101:4204–9. doi: 10.1073/pnas.0306764101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dykes C, Balakrishnan M, Planelles V, Zhu Y, Bambara RA, Demeter LM. Identification of a preferred region for recombination and mutation in HIV-1 gag. Virology. 2004;326:262–79. doi: 10.1016/j.virol.2004.02.033. [DOI] [PubMed] [Google Scholar]
- 19.Temin HM. Sex and recombination in retroviruses. Trends Genet. 1991;7:71–4. doi: 10.1016/0168-9525(91)90272-R. [DOI] [PubMed] [Google Scholar]
- 20.Hu WS, Rhodes T, Dang Q, Pathak V. Retroviral recombination: review of genetic analyses. Front Biosci. 2003;8:d143–55. doi: 10.2741/940. [DOI] [PubMed] [Google Scholar]
- 21.Negroni M, Buc H. Mechanisms of retroviral recombination. Annu Rev Genet. 2001;35:275–302. doi: 10.1146/annurev.genet.35.102401.090551. [DOI] [PubMed] [Google Scholar]
- 22.Hu WS, Temin HM. Genetic consequences of packaging two RNA genomes in one retroviral particle: pseudodiploidy and high rate of genetic recombination. Proc Natl Acad Sci U S A. 1990;87:1556–60. doi: 10.1073/pnas.87.4.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anderson JA, Bowman EH, Hu WS. Retroviral recombination rates do not increase linearly with marker distance and are limited by the size of the recombining subpopulation. J Virol. 1998;72:1195–202. doi: 10.1128/jvi.72.2.1195-1202.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duesberg PH. Physical properties of Rous Sarcoma Virus RNA. Proc Natl Acad Sci U S A. 1968;60:1511–8. doi: 10.1073/pnas.60.4.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kung HJ, Bailey JM, Davidson N, Nicolson MO, McAllister RM. Structure, subunit composition, and molecular weight of RD-114 RNA. J Virol. 1975;16:397–411. doi: 10.1128/jvi.16.2.397-411.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoglund S, Ohagen A, Goncalves J, Panganiban AT, Gabuzda D. Ultrastructure of HIV-1 genomic RNA. Virology. 1997;233:271–9. doi: 10.1006/viro.1997.8585. [DOI] [PubMed] [Google Scholar]
- 27.Fu W, Gorelick RJ, Rein A. Characterization of human immunodeficiency virus type 1 dimeric RNA from wild-type and protease-defective virions. J Virol. 1994;68:5013–8. doi: 10.1128/jvi.68.8.5013-5018.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fu W, Rein A. Maturation of dimeric viral RNA of Moloney murine leukemia virus. J Virol. 1993;67:5443–9. doi: 10.1128/jvi.67.9.5443-5449.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu WS, Temin HM. Retroviral recombination and reverse transcription. Science. 1990;250:1227–33. doi: 10.1126/science.1700865. [DOI] [PubMed] [Google Scholar]
- 30.Coffin JM. Structure, replication, and recombination of retrovirus genomes: some unifying hypotheses. J Gen Virol. 1979;42:1–26. doi: 10.1099/0022-1317-42-1-1. [DOI] [PubMed] [Google Scholar]
- 31.Hwang CK, Svarovskaia ES, Pathak VK. Dynamic copy choice: steady state between murine leukemia virus polymerase and polymerase-dependent RNase H activity determines frequency of in vivo template switching. Proc Natl Acad Sci U S A. 2001;98:12209–14. doi: 10.1073/pnas.221289898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chin MP, Rhodes TD, Chen J, Fu W, Hu WS. Identification of a major restriction in HIV-1 intersubtype recombination. Proc Natl Acad Sci U S A. 2005;102:9002–7. doi: 10.1073/pnas.0502522102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chin MP, Chen J, Nikolaitchik OA, Hu WS. Molecular determinants of HIV-1 intersubtype recombination potential. Virology. 2007;363:437–46. doi: 10.1016/j.virol.2007.01.034. [DOI] [PubMed] [Google Scholar]
- 34.Moore MD, Fu W, Nikolaitchik O, Chen J, Ptak RG, Hu WS. Dimer initiation signal of human immunodeficiency virus type 1: its role in partner selection during RNA copackaging and its effects on recombination. J Virol. 2007;81:4002–11. doi: 10.1128/JVI.02589-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clever JL, Wong ML, Parslow TG. Requirements for kissing-loop-mediated dimerization of human immunodeficiency virus RNA. J Virol. 1996;70:5902–8. doi: 10.1128/jvi.70.9.5902-5908.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muriaux D, Fosse P, Paoletti J. A kissing complex together with a stable dimer is involved in the HIV-1Lai RNA dimerization process in vitro. Biochemistry. 1996;35:5075–82. doi: 10.1021/bi952822s. [DOI] [PubMed] [Google Scholar]
- 37.Laughrea M, Jette L. A 19-nucleotide sequence upstream of the 5′ major splice donor is part of the dimerization domain of human immunodeficiency virus 1 genomic RNA. Biochemistry. 1994;33:13464–74. doi: 10.1021/bi00249a035. [DOI] [PubMed] [Google Scholar]
- 38.Skripkin E, Paillart JC, Marquet R, Ehresmann B, Ehresmann C. Identification of the primary site of the human immunodeficiency virus type 1 RNA dimerization in vitro. Proc Natl Acad Sci U S A. 1994;91:4945–9. doi: 10.1073/pnas.91.11.4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paillart JC, Shehu-Xhilaga M, Marquet R, Mak J. Dimerization of retroviral RNA genomes: an inseparable pair. Nat Rev Microbiol. 2004;2:461–72. doi: 10.1038/nrmicro903. [DOI] [PubMed] [Google Scholar]
- 40.Mikkelsen JG, Lund AH, Duch M, Pedersen FS. Recombination in the 5′ leader of murine leukemia virus is accurate and influenced by sequence identity with a strong bias toward the kissing-loop dimerization region. J Virol. 1998;72:6967–78. doi: 10.1128/jvi.72.9.6967-6978.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Balakrishnan M, Fay PJ, Bambara RA. The kissing hairpin sequence promotes recombination within the HIV-I 5′ leader region. J Biol Chem. 2001;276:36482–92. doi: 10.1074/jbc.M102860200. [DOI] [PubMed] [Google Scholar]
- 42.Derebail SS, DeStefano JJ. Mechanistic analysis of pause site-dependent and -independent recombinogenic strand transfer from structurally diverse regions of the HIV genome. J Biol Chem. 2004;279:47446–54. doi: 10.1074/jbc.M408927200. [DOI] [PubMed] [Google Scholar]
- 43.Moumen A, Polomack L, Roques B, Buc H, Negroni M. The HIV-1 repeated sequence R as a robust hot-spot for copy-choice recombination. Nucleic Acids Res. 2001;29:3814–21. doi: 10.1093/nar/29.18.3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rhodes TD, Nikolaitchik O, Chen J, Powell D, Hu WS. Genetic recombination of human immunodeficiency virus type 1 in one round of viral replication: effects of genetic distance, target cells, accessory genes, and lack of high negative interference in crossover events. J Virol. 2005;79:1666–77. doi: 10.1128/JVI.79.3.1666-1677.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berkhout B, van Wamel JL. Role of the DIS hairpin in replication of human immunodeficiency virus type 1. J Virol. 1996;70:6723–32. doi: 10.1128/jvi.70.10.6723-6732.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laughrea M, Jette L. HIV-1 genome dimerization: formation kinetics and thermal stability of dimeric HIV-1Lai RNAs are not improved by the 1–232 and 296–790 regions flanking the kissing-loop domain. Biochemistry. 1996;35:9366–74. doi: 10.1021/bi960395s. [DOI] [PubMed] [Google Scholar]
- 47.Wu W, Blumberg BM, Fay PJ, Bambara RA. Strand transfer mediated by human immunodeficiency virus reverse transcriptase in vitro is promoted by pausing and results in misincorporation. J Biol Chem. 1995;270:325–32. doi: 10.1074/jbc.270.1.325. [DOI] [PubMed] [Google Scholar]
- 48.Peliska JA, Benkovic SJ. Mechanism of DNA strand transfer reactions catalyzed by HIV-1 reverse transcriptase. Science. 1992;258:1112–8. doi: 10.1126/science.1279806. [DOI] [PubMed] [Google Scholar]
- 49.Palaniappan C, Wisniewski M, Wu W, Fay PJ, Bambara RA. Misincorporation by HIV-1 reverse transcriptase promotes recombination via strand transfer synthesis. J Biol Chem. 1996;271:22331–8. doi: 10.1074/jbc.271.37.22331. [DOI] [PubMed] [Google Scholar]
- 50.Zhuang J, Jetzt AE, Sun G, Yu H, Klarmann G, Ron Y, Preston BD, Dougherty JP. Human immunodeficiency virus type 1 recombination: rate, fidelity, and putative hot spots. J Virol. 2002;76:11273–82. doi: 10.1128/JVI.76.22.11273-11282.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Staden R. The Staden sequence analysis package. Mol Biotechnol. 1996;5:233–41. doi: 10.1007/BF02900361. [DOI] [PubMed] [Google Scholar]
- 52.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–82. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]