Abstract
Primary astrocyte cultures were used to investigate the modulation of DNA repair as a tool for sensitizing astrocytes to genotoxic agents. Base excision repair (BER) is the principal mechanism by which mammalian cells repair alkylation damage to DNA and involves the processing of relatively nontoxic DNA adducts through a series of cytotoxic intermediates during the course of restoring normal DNA integrity. An adenoviral expression system was employed to target high levels of the BER pathway initiator, N-methylpurine glycosylase (MPG), to either the mitochondria or nucleus of primary astrocytes to test the hypothesis that an alteration in BER results in increased alkylation sensitivity. Increasing MPG activity significantly increased BER kinetics in both the mitochondria and nuclei. Although modulating MPG activity in mitochondria appeared to have little effect on alkylation sensitivity, increased nuclear MPG activity resulted in cell death in astrocyte cultures treated with methylnitrosourea (MNU). Caspase-3 cleavage was not detected, thus indicating that these alkylation sensitive astrocytes do not undergo a typical programmed cell death in response to MNU. Astrocytes were found to express relatively high levels of antiapoptotic Bcl-2 and Bcl-XL and very low levels of proapoptotic Bad and Bid suggesting that the mitochondrial pathway of apoptosis may be blocked making astrocytes less vulnerable to proapoptotic stimuli compared with other cell types. Consequently, this unique characteristic of astrocytes may be responsible, in part, for resistance of astrocytomas to chemotherapeutic agents.
Keywords: DNA repair, astrocytes, astrocytoma, apoptosis, mitochondria
INTRODUCTION
The grade IV astrocytoma is the most common primary brain tumor in adults. The infiltrative nature of these neoplasms, as well as their unusual resistance to radiation and chemotherapy, severely limit the efficacy of all currently available treatment options. In contrast with tumors of astrocytic origin, primary brain tumors of nonastrocytic origin, such as medulloblastomas and oligodendrogliomas, tend to respond better to DNA damaging anticancer therapy. As a consequence of this tumor type-specific response to cancer therapy, malignant astrocytomas cause a disproportionate number of cancer-related deaths and disability.
Nitrosoureas are widely used chemotherapeutic agents. These alkylating agents introduce DNA damage in both the nuclear and mitochondrial genomes of rapidly dividing tumor cells, which can lead to errors in transcription, mutations, recombinations and rearrangements. The capacity of a cell to resist these transformations depends heavily upon a variety of enzymatic pathways for repairing lesions in DNA of both the nucleus and mitochondria. While much attention has focused on DNA repair in the nucleus, differential repair of mitochondrial DNA (mtDNA) damage in CNS tissues shows an inverse correlation between mtDNA repair capacity and induction of apoptosis in these cell types (Harrison et al., 2005; Hollensworth et al., 2000; Ledoux et al., 1998). Specifically, astrocytes exhibit a greater capacity to repair mtDNA with no induction of apoptosis following treatment with DNA damaging agents compared with oligodendrocytes, microglia and neuronal cell types, all of which exhibit decreased mtDNA repair and increased induction of apoptosis. These findings suggest that, among the various CNS cell types, inherent cell-specific differences in mtDNA repair capacity may directly impact susceptibility to DNA damaging agents.
The base excision repair (BER) pathway is the principal mechanism by which mammalian cells repair alkylation damage in DNA (Krokan et al., 2000) and appears to be the predominant DNA repair system in mitochondria. BER of alkylation damage involves the processing of relatively nontoxic DNA adducts through a series of transient cytotoxic intermediates, which include apur-inic/apyrimidinic (AP) sites and single strand breaks (SSB), during the course of restoring DNA integrity. N-methylpurine DNA glycosylase (MPG) is an initiator glycosylase in BER that excises a wide variety of damaged bases such as N3-methyladenine, N7-methylguanine (7-meG), N3-methylguanine (Bonanno et al., 2002; Pendlebury et al., 1994), hypoxanthine (Saparbaev and Laval, 1994), 1,N6-ethenoadenine (Dosanjh et al., 1994), and 8-oxoguanine (8oxoG) (Bessho et al., 1993). Experimentally increasing the MPG to polymerase ratio in the nucleus as well as the mitochondria has been shown to affect the efficiency of BER and increase the sensitivity of both mouse and human cell lines to DNA damaging agents (Fishel et al., 2003; Rinne et al., 2004; Sobol et al., 2003). These findings along with others (Harrison et al., 2005) have given rise to the ‘imbalanced repair hypothesis’, which contends that increased MPG activity overwhelms subsequent pathway enzyme activities in cells treated with alkylating agents and consequently produces high levels of cytotoxic BER pathway intermediates in cellular DNA.
It is likely that unique characteristics of astrocytes play a role in the chemoresistence that is observed in astrocytic tumors. Therefore, the purpose of this study was to provide a better understanding of the relationship between DNA repair and chemoresistance in astrocytes. Primary cultures of astrocytes were used to investigate whether MPG modulation in mitochondria or the nucleus is an effective means for sensitizing astrocytic tumors to genotoxic anticancer agents. An adenoviral-based transient expression system was employed to target high levels of MPG to either the nucleus or mitochondria in astrocytes to test the hypothesis that a buildup of cytotoxic pathway intermediates due to altered BER causes increased sensitivity of astrocytes to DNA alkylating agents. The results from this study indicated that targeting MPG to the nucleus significantly sensitizes astrocytes to MNU and that a unique adaptation in the astrocyte apoptotic machinery may play a role in astrocyte resistance to chemotherapeutic agents.
MATERIALS AND METHODS
Cell Cultures
Primary rat astrocytes and cerebellar granule cells were cultured as previously described (Cohen and Wilkin, 1995; LeDoux et al., 1998). Briefly, for astrocytes, cerebral hemispheres from 1- to 4-day-old Sprague Dawley rats were aseptically removed, minced, dissociated into single-cell suspensions, and seeded. After 7 days, cultures were subjected to rotary shearing for 24 h. The cells that remained attached to the flasks were passaged and, after 1 day of recovery, pulse-treated with 1 × 10−5 M cytosine arabinoside. The resulting subcultures contained >95% astrocytes as determined by immunofluorescence using glial fibrillary acid protein (GFAP) as a marker.
For cerebellar granule cells, the cerebella of 7-day-old Sprague Dawley rats were aseptically removed, minced, dissociated into single cell suspensions, and plated. To prevent growth of non-neuronal cells, 1-day-old cultures were treated with the mitotic inhibitor fluorodeoxyuridine (80 µM). After 1–2 days, granule cell cultures were determined to be 95% pure using immunofluorescence staining with an antineurofilament antibody.
Vector Construction
Vector construction for the four viruses used in this study has been previously described (Rinne et al., 2004, 2005). The MPG, MTS-MPG and the E125A-MTS-MPG were ligated into the modified adenoviral shuttle vector pacAd5. The pacAd5 shuttle vector from the University of Iowa Gene Transfer Vector Core Laboratory was altered to contain an internal ribosomal entry site (IRES)-driven Enhanced Green Fluorescent Protein (EGFP) coding sequence (IRES2-EGFP; BD Biosciences Clontech, Palo Alto, CA). MPG, MTS-MPG, and E125A-MTS-MPG-containing adenoviral shuttle vectors along with an empty vector control (pacAd5 IRES2EGFP) were sent to the University of Iowa Gene Transfer Vector Core Laboratory for production and titering of replication- defective recombinant adenovirus. Titers ranged from 3 × 1010 to 1 × 1011 plaque forming units (pfu)/mL. Resultant adenoviruses (empty vector, MitoMPG, MutMitoMPG, and NucMPG) were aliquoted and stored at −80°C until use.
Adenoviral Transduction
Media (1% FBS) containing purified adenovirus at a multiplicity of infection (MOI) of 15 was placed on the cultures for 24 h at 37°C. Virus containing media were removed after 24 h and fresh media (10% FBS) was replaced for an additional 24 h. Transduction efficiencies were measured by EGFP fluorescence intensities of 104 cells using a FACS Calibur fluorescence-activated cell sorter (Becton Dickinson, San Jose, CA) and the results were analyzed with CellQuest 3.3 software (Becton Dickinson). To ensure comparable expression of MPG in the different astrocyte cultures FACS analysis was performed and only cells exhibiting similar high levels of viral transduction were used in the subsequent experiments.
Western Blot Analysis
Western blots were performed using either whole cell, mitochondrial or nuclear protein extracts from primary CNS cultures as previously described (Hollensworth et al., 2000). Subcellular localization of MPG was assessed using mouse anticytochrome c monoclonal antibody (Pharmingen), antiactin antibody (Sigma), or a monoclonal MPG antibody (kindly provided by Dr. Rabindra Roy, Georgetown University, Washington, DC). Pro- and antiapoptotic protein levels were assessed using mouse mono-clonal antibodies Bcl-2 (C-2), Bcl-XL (H-5), Bad (C-7), and Bax (B-9) as well as rabbit polyclonal antibodies Bak (G-23) and Bid (FL-195) (Santa Cruz). Caspase-3 cleavage was assessed using a rabbit polyclonal antibody to both full length and cleaved Caspase-3 (Cell Signaling). Membranes were incubated 1 h with antimouse or antirabbit IgG secondary antibodies (Jackson ImmunoResearch) at room temperature. Protein bands were visualized with chemiluminescence (SuperSignal, Pierce).
Enzyme Activity Assays
The oligonucleotide cleavage assay was performed using a 26-mer oligonucleotide containing a 1,N6-ethenoadenine adduct (EthenodA oligo) at the 15th position (Midland Cert. Reagent Co.). The EthenodA oligo was radiolabeled and allowed to anneal with a complementary oligonucleotide to form duplex DNA as previously described (Dobson et al., 2000). Activity assays contained 6 pmol of EthenodA labeled duplex oligonucleotide, 5 µL of 10 × REC Buffer 9 (Trevigen) and equal amounts of protein from either nuclear or mitochondrial extracts for a final concentration of 0.1 µg/µL in a total volume of 50 µL. Ten microliter aliquots were removed at 0, 5, 20, 40, and 60 min time points during the 37°C incubation and placed directly into 5 µL of stop buffer (80% formamide, 0.2% bromophenol blue dye) and held at 95°C for 5 min. One unit of purified recombinant mouse Aag (Trevigen) and 1 unit of purified human APE (Trevigen) were used as positive controls. The reaction contents were resolved on a 15% TBE-Urea Ready Gel (Bio-Rad) and visualized by autoradiography.
MNU Treatment
Methylnitrosourea (MNU) was dissolved in citrate buffer (pH 4.2) and diluted in HBSS to yield the desired concentration(s) while control cells were treated only with HBSS under the same conditions, 1 h in a 5% CO2, 37°C incubated environment. In one set of experiments, the Poly-ADP Ribose (PARP) inhibitor 3-amino-benzamide (3-AB, Sigma) at a concentration of 10 mM was added 2 h prior to the MNU exposure. The 3-AB remained in the media not only during the MNU exposure but was also in the media until the cells were analyzed.
DNA Repair Analysis
mtDNA damage and repair was detected using a quantitative Southern blot (QSB) technique in conjunction with a mitochondrial specific PCR-generated probe (Driggers et al., 1993; LeDoux et al., 1992). High molecular weight DNA was extracted from control and treated cells and digested to completion with BamHI. After heating, alkali treatment, and alkaline gel electrophoresis, QSB was performed using mtDNA specific probes and the data were analyzed by quantitative densitometry (LeDoux et al., 1992). Nuclear DNA repair capacity was assessed using qualitative alkaline gel electrophoresis (QAGE) analysis. QAGE analysis (Wassermann et al., 1990) is an adaptation of the QSB procedure where, following alkaline electrophoresis, gels are stained with ethidium bromide and total cellular DNA integrity is assessed using densitometric software (BioRad GS-250 molecular imager). For mtDNA repair experiments, BF is calculated densitometrically using a Poisson expression, BF = −ln(OD treated samples/OD control samples), and expressed as percent repair. QAGE analysis, involves a qualitative densitometric assessment to determine the average BamHI restriction fragment size (bp) of each DNA sample using the Lambda DNA/Hind III marker (Promega) as a standard. The fragment sizes are then used to calculate BF with the Poisson expression above and % repair at x-time is calculated: 100 × BF (0-time) – BF(x-time)/BF(0-time). The time following treatment at which 50% of DNA integrity was restored is represented as the average life of a DNA adduct.
Viability Assays
As previously described (Hollensworth et al., 2000), cell viability was determined using trypan blue dye exclusion in astrocyte cultures following MNU treatment. Percent viable = 100 × the number of cells that exclude trypan blue dye/total cell number. A minimum of 200 cells per data point were counted. Additionally, cultures were analyzed using a fluorescence activated cell sorter (FACS) and only cells exhibiting similar high levels of viral transduction were collected for MTS viability assays (Celltiter 96 Aqueous One kit available from Promega). Cell viability was expressed as a percent relative to matched untreated cultures.
Data Analyses
Statistical analysis was performed using two-way analysis of variance (ANOVA) with post hoc comparison by Tukey–Kramer multiple comparisons test. Inter-group differences with a P-value of < 0.05 were considered significant.
RESULTS
An Adenoviral-Based Transient Expression System Targets MPG in Primary Astrocytes
Transient overexpression and subcellular targeting of MPG was accomplished in primary astrocytes using a bicistronic adenoviral vector constructed to overexpress MTS-MPG as well as an IRES2-EGFP reporter (MitoMPG), thus facilitating the determination of transduction efficiencies in these cultures via fluorescence analysis. Additional control bicistronic adenoviral vectors also were constructed: IRES2-EGFP (empty vector) to control for the effects of viral transduction, nuclear localized MPG (NucMPG) to control for the effects of mitochondrial targeting, and mutant E125A MTS-MPG (MutMitoMPG) to control for the effects of increased MPG protein in mitochondria.
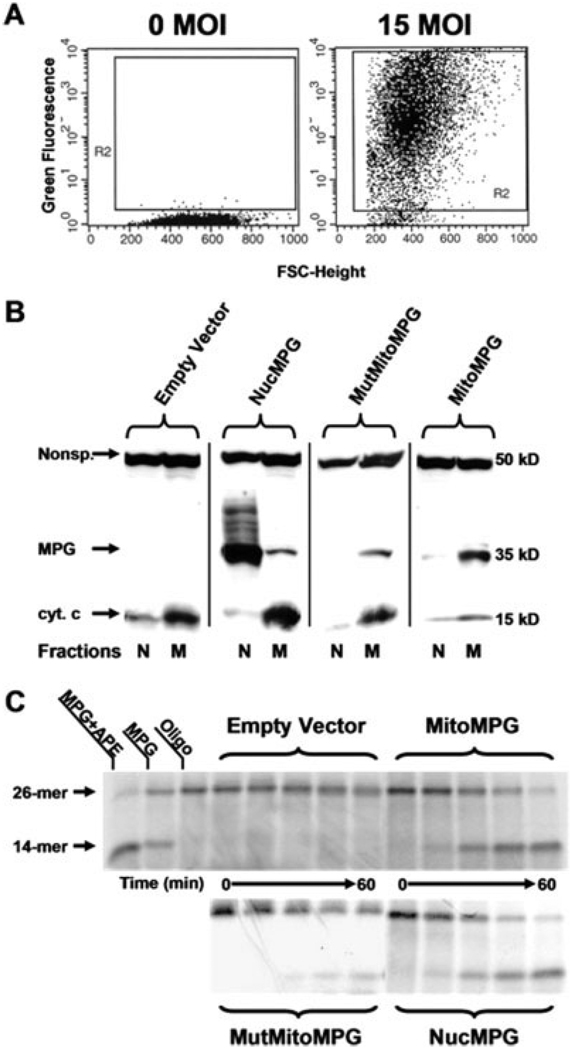
These recombinant nonreplicating adenoviruses were used to infect astrocytes in primary culture. An MOI, or virus-to-cell ratio, of 15 was found to transduce greater than 95% of the cells in these cultures within 24 h as monitored by EGFP fluorescence (Fig. 1A). Using Western blot analysis, it was determined that MPG transduced astrocytes expressed appreciable levels of MPG with appropriate subcellular localization (Fig. 1B). Although contamination from either fraction was detected in all of the samples, anticytochrome c antibody was used to show that the cell fractions were either mitochondrial or nuclear protein enriched. Oligonucleotide cleavage assays were performed to determine whether the expressed recombinant MPG possessed glycosylase activity. In this assay, lysates from transduced astrocytes were incubated with an oligonucleotide substrate containing an ethenoadenine adduct that is excised by MPG producing an AP site intermediate. The depurinated oligo substrate is then cleaved at the AP site by endogenous APE to generate a 14-mer product (Fig. 1C). These assays revealed that lysates taken from MitoMPG and NucMPG transduced astrocytes exhibit increased MPG activity relative to empty vector controls (Fig. 1C). Although the E125A MPG has been shown to possess minimal glycosylase activity (Lau et al., 2000), the in vitro assays also revealed a reduced level of MPG activity in mitochondrial lysates taken from MutMitoMPG astrocytes. These findings indicate that adenoviral delivery is an effective means of modulating MPG activity in either the nucleus or mitochondria of astrocytes maintained in primary culture.
Fig. 1.
Efficient transduction of the MPG gene in primary astrocyte cultures using an adenoviral expression system. A: FACS analysis of EGFP expression in samples of 10,000 cells taken from nontransduced (0 MOI) and transduced (15 MOI) cultures of primary astrocytes. B: Western blot analysis confirms appropriate subcellular localization of human MPG to either the nucleus (N) or mitochondria (M) in transduced rat astrocyte cultures. Anticytochrome c antibody was used to show that the cell fractions were either mitochondrial or nuclear protein enriched. Additionally, a nonspecific band that immunoreacts with the cyt. c antibody was included to show comparable protein loading between fractions. The human MPG antibody used in this study does not immunoreact with rat MPG. C: The oligonucleotide cleavage assays were performed using a 26-mer oligonucleotide containing a 1,N6-ethenoadenine adduct. The radiolabeled oligo was incubated at 37°C with either nuclear lysates (NucMPG) or mitochondrial lysates (empty vector, MitoMPG and MutMitoMPG) for 0, 5, 20, 40, and 60 min. The reaction products were resolved on a 15% TBE-urea gel and autoradiography was performed. One unit of purified recombinant mouse Aag (MPG) was used as a positive control as well as 1 unit of purified human APE.
Heightened MPG Activity Significantly Increases BER Kinetics in Transduced Astrocyte Cultures
MNU was used in these studies to generate DNA alkylation damage in transduced and nontransduced astrocytes. The majority of DNA adducts generated by MNU are N7-methylguanine (70%) and N3-methyladenine (8%) (Pegg, 1977), both of which serve as substrates for MPG. To determine whether modulating MPG activity affects mitochondrial BER in astrocytes treated with MNU, mtDNA repair capacity was assessed in transduced and nontransduced cultures using QSB analysis. QSB is a DNA repair assay that can detect the production and persistence of damaged sugars and alkylated bases as well as BER pathway intermediates, which include AP sites and SSB (LeDoux et al., 1993). The decreased intensity of the 10.8 kb BamHI restriction band indicates increased levels of mtDNA damage (Fig. 2A). Densitometric analysis of these data revealed that treatment with 0.5 mM MNU generated ~1.2 breaks per restriction fragment in all transduced astrocyte cultures, which is sufficient damage to conduct repair studies using QSB. No nuclear DNA damage was detected at these concentrations of MNU using either QSB or quantitative extended length PCR (data not shown).
Fig. 2.
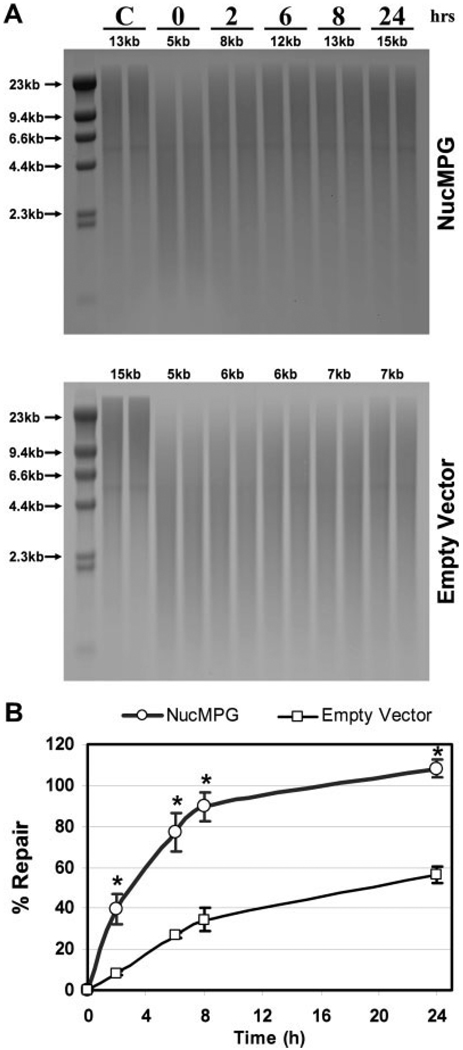
QSB analysis of mitochondrial BER reveals differential repair kinetics among the transduced astrocyte cultures treated with MNU. A: High molecular weight DNA from transduced cultures of astrocytes was isolated immediately (0) and at 2, 6, 8, and 24 h following a 0.5 mM MNU treatment and restricted to completion with BamHI. Control cultures (C) were incubated in drug diluent only. Duplicate samples were exposed to 0.1 N NaOH prior to alkaline electrophoresis and QSB analysis. Additional MitoMPG astrocyte DNA samples were processed in parallel with those used for QSB and transferred to membrane without electrophoresis (Slot Blot). Membranes were hybridized with a PCR generated mitochondrial probe and visualized by autoradiography. A decreased intensity of the 10.8 kb restriction band indicates increased levels of mtDNA damage. B: Autoradiographic bands from QSB were densitometrically analyzed to calculate mtDNA BF in astrocytes treated with 0.5 mM MNU. The mean + S.E. from three or more separate experiments is shown. An asterisk (*) indicates a significant difference (P < 0.05) compared with matched empty vector samples.
The mtDNA repair capacity of cultured astrocytes was determined by either harvesting cells immediately after a 1 h MNU treatment or allowing time for repair. Control cells were treated with HBSS without MNU for 1 h and then harvested. Compared with control samples, 0 h samples showed substantially fewer 10.8 kb fragments as indicated by a decrease in the intensity of the corresponding restriction band (Fig. 2A). Targeting MPG to the nucleus as well as empty vector transduction, as expected, had no significant impact on mtDNA repair. Control (nontransduced), empty vector and NucMPG astrocytes all repaired approximately 40 and 70% of the initial MNU induced mtDNA damage at 8 and 24 h, respectively (Fig. 2B). However, targeting the inactivated mutant E125A MPG to mitochondria significantly depressed mtDNA repair after both 8 and 24 h. Interestingly, MitoMPG transduced cultures exhibited a significant increase in mtDNA repair capacity that exceeded 100% at 8 h. To ensure that the increase in intensity observed at 8 and 24 h was not the result of either mtDNA replication or uneven loading, DNA samples processed in parallel with those used for QSB were transferred to a nylon support membrane without electrophoresis and hybridized with a mitochondrial probe (indicated as Slot Blot in Fig. 2A). These experiments showed comparable amounts of DNA in all samples. Therefore, the increasing restriction band densities observed in MitoMPG astrocyte mtDNA repair experiments reflect increasing mtDNA integrity over time following treatment with MNU, not an increase in replication of mtDNA or uneven loading.
Nuclear DNA repair capacity was assessed in NucMPG astrocytes using qualitative alkaline gel electrophoresis (QAGE) analysis to determine whether targeting the recombinant MPG to the nucleus caused alterations in nuclear BER kinetics. QAGE analysis is an adaptation of the QSB procedure where, following alkaline electrophoresis, gels are stained with ethidium bromide and total cellular DNA integrity is assessed using densitometric software. Because mtDNA is more sensitive to alkylation damage, higher doses of MNU are required to achieve similar, measurable levels of damage in chromosomal DNA. Empty vector and NucMPG transduced astrocytes were treated with 5 mM MNU for 1 h to generate BamHI restricted chromosomal DNA fragments averaging 5 kb in length (Fig. 3A), which is equivalent to the break frequency (BF) measured in mtDNA extracted from astrocytes treated with 0.5 mM MNU. Nuclear DNA repair capacity was determined by either harvesting cells immediately after a 1 h MNU treatment or allowing time for repair. At 8 and 24 h following treatment with MNU, DNA extracted from astrocytes with elevated nuclear MPG activity contained restriction fragment sizes of approximately 13 and 15 kb, respectively, compared with 7 kb in empty vector transduced astrocytes. Percent repair of initial nuclear DNA alkylation damage was significantly greater for NucMPG astrocytes at all time points measured (Fig. 3B). Similar to the mtDNA repair observed in MitoMPG astrocytes, nuclear BER in NucMPG astrocytes also was significantly enhanced 24 h following treatment.
Fig. 3.
QAGE analysis of nuclear BER reveals differential repair kinetics in empty vector and NucMPG transduced astrocyte cultures treated with MNU. A: High molecular weight DNA from transduced cultures of astrocytes was isolated immediately (0) and at 2, 6, 8, and 24 h following a 5 mM MNU treatment and restricted to completion with BamHI. Control cultures (C) were incubated in drug diluent only. Duplicate samples were exposed to 0.1 N NaOH and then subjected to alkaline electrophoresis. Gels were stained evenly with ethidium bromide and scanned in a UV imager. Nuclear DNA integrity in each sample was analyzed with densitometry software using the Lambda DNA/Hind III marker as a standard. Average BamHI restricted chromosomal DNA fragment sizes are indicated for each sample. A decrease in DNA fragment sizes indicates increased levels of chromosomal DNA damage. B: Average BamHI restricted chromosomal DNA fragment sizes determined by QAGE analysis were used to calculate nuclear DNA BF in astrocytes treated with 5 mM MNU. The mean + S.E. from three or more separate experiments is shown. An asterisk (*) indicates a significant difference (P < 0.05) compared with matched empty vector samples.
Overexpression of Nuclear MPG Significantly Increases the Sensitivity of Astrocytes to MNU
The sensitivity of astrocytes following treatment with MNU was evaluated using two different viability methods, trypan blue dye exclusion and an MTS assay. Trypan blue dye exclusion assays revealed no significant increase in sensitivity to MNU in astrocytes expressing MPG in the mitochondria (Fig. 4B). In fact, only cell cultures with heightened MPG activity in the nucleus exhibited a significant decrease in viability.
Fig. 4.
Astrocytes expressing a nuclear localized human MPG exhibit increased sensitivity to MNU. Transduced and nontransduced astrocytes were exposed to 5 mM MNU for 1 h, and viability was assessed at 24, 48, and 72 h following treatment. A: Phase contrast microscopy 24 h following MNU treatment. B: Trypan blue dye exclusion assays indicate a significant increase in sensitivity to MNU in NucMPG transduced cultures. C: Transduced as well as nontransduced (control) astrocytes were treated with 0, 0.5, 2.5, and 5 mM MNU for 1 h and then FACS analyzed. Cells exhibiting similar levels of viral transduction along with control astrocytes were sorted into 96-well plates. MTS assays were performed 24 h following MNU treatment. D: Empty vector (V), NucMPG (N) and nontransduced (C) astrocytes were exposed to 5 mM MNU for 1 h. At 6 and 24 h following treatment, cytosolic proteins were extracted. Protein (20 µg) from each sample was separated on 4–15% polyacrylamide gel. Caspase-3 cleavage was assessed using a rabbit polyclonal antibody to both full length and cleaved Caspase-3. Jurkat cells were treated with the broad-spectrum protein kinase inhibitor, staurosporine, to produce positive control lysates. Antiactin antibody was used to show equal loading. The mean + S.E. from three or more separate experiments is shown. An asterisk (*) indicates a significant difference (P < 0.05) when compared with matched empty vector.
Western blot analyses revealed particularly high levels of MPG in lysates taken from NucMPG transduced astrocytes (Fig. 1B). This relatively large amount of MPG is likely due to minor differences in nuclear and mitochondrial fractionation techniques, which may affect protein concentrations in these enriched lysates. However, the possibility exists that the NucMPG virus is more efficient than the MitoMPG virus. If the NucMPG transduced astrocytes express higher levels of MPG, then viral transduction efficiency might affect alkylation sensitivity. Therefore, to better assess MNU sensitivity, cultures were FACS analyzed and only cells exhibiting similar levels of viral transduction (i.e. similar levels of MPG expression) were collected for MTS viability assays. The results from the MTS experiments were consistent with the initial trypan blue assays; NucMPG transduced astrocytes were significantly more sensitive to 2.5 and 5 mM MNU compared with empty vector controls (Fig. 4C).
Exposure of cells to alkylating agents such as MNU results in the formation of SSB which can activate Poly-ADP-Ribose Polymerase (PARP) and cause cytotoxicity by depleting cells of NAD. To determine whether activation of PARP might play a role in the observed decrease in viability of NucMPG transduced astrocytes, a series of viability experiments were conducted using 3-amino-benzamide, a specific inhibitor of PARP. As can be seen in Table 1 treatment with 3-AB provided some protection suggesting that activation of PARP plays a role in the enhanced sensitivity seen in the NucMPG transduced astrocytes.
TABLE 1.
3-Aminobenzomide Increased Viability of MNU-Treated NuMPG Transduced Astrocytes
| MNU dose | Percentage of protection (%) |
|---|---|
| 2.5 mM | 2.1 ± 0.5 |
| 5 mM | 15 ± 3.3 |
A Lethal Exposure to MNU Does Not Induce Apoptosis in Astrocytes Overexpressing MPG
To further evaluate the heightened cell death in NucMPG transduced astrocytes, transduced as well as nontransduced cultures were assayed for apoptosis by detection of caspase-3 cleavage. Caspase-3 is a key executioner of apoptosis when proteolytically cleaved into its activated fragments (Fernandes-Alnemri et al., 1994). Western blot analysis detected no differences in levels of either full length or cleaved caspase-3 between empty vector and NucMPG transduced astrocytes at 6 and 24 h following treatment with 5 mM MNU (Fig. 4D). Additionally, TUNEL and Annexin-V assays revealed no MNU dose dependent changes in levels of apoptosis among NucMPG transduced, empty vector and nontransduced cultures (data not shown).
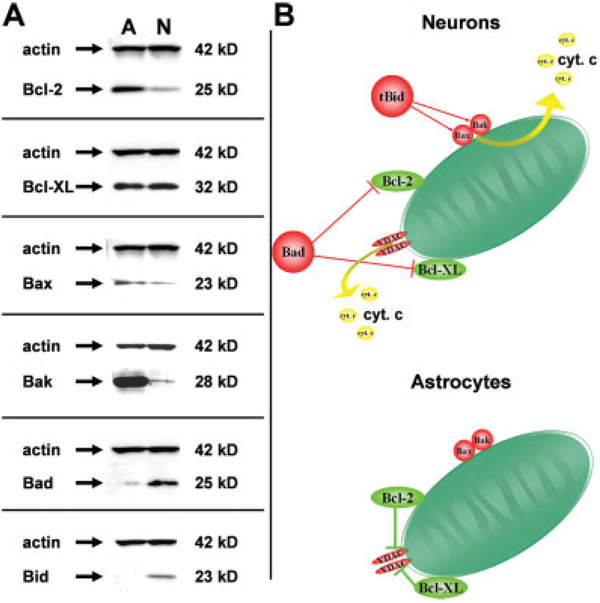
Previous studies from our laboratory have shown that, with the exception of astrocytes, most other CNS cell types readily undergo mitochondrial initiated apoptosis in response to treatment with DNA damaging agents (Harrison et al., 2005; Hollensworth et al., 2000; Ledoux et al., 1998). Therefore, we posed the question; are there intrinsic differences in the apoptotic machinery that might account for the inability of astrocytes to undergo programmed cell death following a lethal exposure to DNA damaging agents? Steady-state expression levels of both pro and antiapoptotic proteins were analyzed and comparisons were made with lysates taken from primary cultures of astrocytes and cerebellar granule cells, a neuronal cell type known to undergo mitochondrial initiated apoptosis following treatment with DNA damaging agents (Harrison et al., 2005). Western blot analysis revealed that astrocytes express high levels of the antiapoptotic proteins Bcl-2 and Bcl-XL (Fig. 5A). Astrocytes also were found to express relatively high levels of the pro-death proteins Bax and Bak; however, expression of the proapoptotic Bad and Bid appeared to be nearly absent. These adaptations suggest that the astrocyte apoptotic machinery may respond differently to DNA damage compared with other cell types in the CNS.
Fig. 5.
Intrinsic differences in apoptotic machinery might account for the inability of astrocytes to undergo programmed cell death following a lethal exposure to DNA damaging agents. Steady-state expression levels of both pro and antiapoptotic proteins were analyzed and comparisons were made with lysates taken from primary cultures of astrocytes (A) and cerebellar granule cells (N). A: Western blot analysis revealed differential expression of both proapoptotic (Bax, Bak, Bad, and Bid) and antiapoptotic (Bcl-2 and Bcl-XL) proteins in these cell types. Antiactin antibody was used to show equal loading. B: Because astrocytes express much higher steady-state levels of Bcl-2 and much lower levels of Bad and Bid, it is likely that the astrocyte apoptotic machinery is less amenable to triggering cell death in response to DNA damage.
DISCUSSION
Tumor type-specific responses to chemotherapy are known to affect the prognosis for various neoplasms of the CNS; for example, brain tumors of astrocytic origin are remarkably more resistant to radiation and chemotherapy compared with many CNS tumors arising from other lineages (Behin et al., 2003; Lefranc et al., 2005; Prados and Levin, 2000). Strong correlations have been observed among certain cellular responses unique to astrocytes and their ability to resist treatment with DNA damaging agents (Harrison et al., 2005). Therefore, these studies utilized primary cultures of astrocytes as a model to investigate DNA repair modulation as a potential tool for sensitizing astrocytic tumors to genotoxic anticancer agents.
Many chemotherapeutic agents modify DNA either through oxidation or alkylation reactions (Chabner and Longo, 2001) to produce adducts that are processed primarily by the BER pathway (Krokan et al., 2000). Interestingly, the intermediate AP sites and SSB generated in this pathway are more cytotoxic in many cases than the modified base lesions themselves (Chaudhry and Weinfeld, 1997; Lockhart et al., 1982; Loeb and Preston, 1986; Sagher and Strauss, 1983; Schaaper et al., 1983; Whitehouse et al., 2001). MNU was used in the present studies to generate alkylation damage. As with most other methylating agents, the vast majority of DNA adducts generated by MNU are N7-methylguanine (7-meG), which does not alter the DNA duplex structure and has the same coding properties as unmodified guanine (Pieper et al., 1989; Rinne et al., 2005). In fact, 7-meG is neither mutagenic nor cytotoxic and appears to be well tolerated by cells for generations (Karran et al., 1980; Lawley and Warren, 1976).
QSB and QAGE analyses of DNA repair capacities in transduced cultures clearly show that increasing MPG activity dramatically increases BER kinetics in both mitochondria and nuclei (Fig. 2 and Fig 3). This data suggests that, in astrocytes, glycosylase activity is likely the rate-limiting factor in both mitochondrial and nuclear BER of alkylation damage in DNA. It is interesting that the E125A mutant MPG significantly decreased mtDNA repair in astrocytes (Fig. 2). This mutation conserves the DNA binding ability of MPG, but glycosylase activity is eliminated. Thus, in vivo, the mutant MPG is likely binding to the mtDNA adducts and competitively inhibiting endogenous BER factors that would otherwise process the lesions.
The repair kinetics measured in control astrocytes that were treated with MNU indicate an average life of 8 h for DNA adducts in both nuclear and mitochondrial BER pathways. If glycosylase activity is indeed rate-limiting, then these adducts are most probably nontoxic 7-meGs. In MPG transduced astrocytes, glycosylase activity is in abundance, thus shifting the rate-limiting step down-stream in the pathway and increasing overall repair. The repair kinetics for treated MPG transduced cultures indicated an average life of 2 h for DNA adducts.
The increased rates of BER resulting from the shift to a downstream rate-limiting component was accompanied by a change in the nature of the DNA damage from less cytotoxic alkylated bases to more cytotoxic pathway intermediates, including SSB. Modulating MPG activity in the mitochondria appeared to have little effect on alkylation sensitivity in primary astrocytes (Fig. 4). However, the increased presence of these pathway intermediates in nuclear DNA resulting from increased MPG activity proved to be cytotoxic in astrocytes treated with MNU. One possible mechanism that could be responsible for the increase in cell killing is the activation of Poly-ADP Ribose Polymerase (PARP) with the subsequent depletion of NAD within the cells (Ying et al., 2005). PARP is abundant in the nucleus and is activated by SSB, which are generated at a 4-fold greater rate in NucMPG astrocytes. It is possible that MNU treatment may cause extensive activation of this pathway leading to a critical depletion of NAD+ pools and cell death without caspase-3 activation. Therefore, experiments were conducted which revealed that the use of 3-amino-benzamide, a specific inhibitor of PARP, provided some protection against cell killing, suggesting the activation of PARP plays a role in the enhanced sensitivity observed in the NucMPG transduced astrocytes.
Caspase-3 cleavage was not detected in either empty vector or NucMPG transduced astrocytes following MNU treatment. Astrocytes express much higher steady-state levels of antiapoptotic Bcl-2 and much lower levels of proapoptotic Bad and Bid, it is likely that the astrocyte apoptotic machinery, compared with that of neuronal cell types, is less amenable to triggering cell death in response to DNA damage. Therefore, when exposed to lethal levels of alkylating agents in the presence of increased nuclear MPG activity, astrocytes do not undergo typical programmed cell death.
The above mentioned adaptation in astrocyte apoptotic machinery makes sense in the context of CNS development where, in contrast to astrocytes, large numbers of neurons must undergo apoptosis to ensure the proper development of the brain. Bad is an important proapoptotic factor, which astrocytes appear to lack (Fig. 5). Recent studies have shown that targeting Bad to tumor cells is an effective means of restoring the apoptotic pathway and inducing programmed cell death (Antignani and Youle, 2005). However, further investigation is needed to determine whether modulating apoptotic machinery is a viable strategy for sensitizing astrocytes to DNA damaging agents.
Another interesting observation in these MPG overexpressing systems is the apparent enhancement of DNA repair at 24 h in the nucleus and, to a much greater extent, mitochondria. Similar DNA repair enhancements were previously revealed in oxidation repair studies of primary rat astrocytes as well as a rat insulinoma cell line (Driggers et al., 1993; Hollensworth et al., 2000) and were attributed to a treatment-induced mtDNA repair adaptation that repairs not only 100% of the experimentally induced damage, but also preexisting endogenous damage. Relatively high levels of endogenous oxidative damage are present in mtDNA in large part because of its close proximity to the cell’s main source of free radicals, the mitochondrial respiratory chain (Backer and Weinstein, 1980; Levy and Brabec, 1984; Niranjan et al., 1982; Rushmore et al., 1984; Wunderlich et al., 1970). MPG is known to excise various oxidative lesions (Dosanjh et al., 1994; Saparbaev and Laval, 1994); therefore, it is reasonable to conclude that heightened MPG activity increases the rate and extent to which both MNU generated as well as some preexisting endogenous DNA damages are repaired. This mtDNA repair adaptation is emphasized in MitoMPG astrocytes treated with sub-lethal levels of MNU. At 24 h following treatment with 0.5 mM MNU, mtDNA bands from MitoMPG transduced astrocytes were 60% greater than control, untreated MitoMPG astrocytes (Fig. 2). It is interesting that a greater percent of 0.5 mM MNU treated MitoMPG astrocytes survived flow cytometry and sorting compared with all other controls (Fig. 4C). This enhancement phenomenon is even more fascinating in NucMPG transduced astrocytes treated with a lethal 5 mM dose of MNU (Fig. 3 and Fig 4). These findings strongly implicate MPG-mediated rapid conversion of nontoxic 7-meG chromosomal adducts to toxic repair pathway intermediates as the mechanism of alkylation sensitivity in these cultures.
In conclusion, altering BER in the nucleus using MPG appears to be a more effective strategy for sensitizing astrocytes to alkylating agents than altering BER in the mitochondria. In spite of the increased repair rate, astrocytes overexpressing a nuclear localized MPG were significantly sensitized to treatment with MNU. Similar increases in sensitivity to MNU also were observed in a rat glioma cell line that was transduced with the NucMPG virus, though to a much lesser extent (data not shown). Using the present vector system, transduction efficiency in these cells was nearly 100-fold less than that of astrocytes in primary culture. However, it is likely that significant cellular toxicity could be obtained using a more appropriate vector. Future studies will be directed towards identifying an effective transduction system for these tumor cells.
Acknowledgments
Grant sponsor: National Institutes of Health; Grant numbers: ES03456, ES05865, NS047208, CA100045, AG19602.
REFERENCES
- Antignani A, Youle RJ. A chimeric protein induces tumor cell apoptosis by delivering the human Bcl-2 family BH3-only protein Bad. Biochemistry. 2005;44:4074–4082. doi: 10.1021/bi0477687. [DOI] [PubMed] [Google Scholar]
- Backer JM, Weinstein IB. Mitochondrial DNA is a major cellular target for a dihydrodiol-epoxide derivative of benzo[a]pyrene. Science. 1980;209:297–299. doi: 10.1126/science.6770466. [DOI] [PubMed] [Google Scholar]
- Behin A, Hoang-Xuan K, Carpentier AF, Delattre JY. Primary brain tumours in adults. Lancet. 2003;361:323–331. doi: 10.1016/S0140-6736(03)12328-8. [DOI] [PubMed] [Google Scholar]
- Bessho T, Roy R, Yamamoto K, Kasai H, Nishimura S, Tano K, Mitra S. Repair of 8-hydroxyguanine in DNA by mammalian N-methylpurine-DNA glycosylase. Proc Natl Acad Sci USA. 1993;90:8901–8904. doi: 10.1073/pnas.90.19.8901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonanno K, Wyrzykowski J, Chong W, Matijasevic Z, Volkert MR. Alkylation resistance of E. coli cells expressing different isoforms of human alkyladenine DNA glycosylase (hAAG) DNA Repair (Amst) 2002;1:507–516. doi: 10.1016/s1568-7864(02)00051-4. [DOI] [PubMed] [Google Scholar]
- Chabner BA, Longo DL. Cancer chemotherapy and biotherapy: Principles and practice. Philadelphia: Lippincott Williams & Wilkins; 2001. [Google Scholar]
- Chaudhry MA, Weinfeld M. Reactivity of human apurinic/apyrimidinic endonuclease and Escherichia coli exonuclease III with bistranded abasic sites in DNA. J Biol Chem. 1997;272:15650–15655. doi: 10.1074/jbc.272.25.15650. [DOI] [PubMed] [Google Scholar]
- Cohen J, Wilkin P. Neural cell culture: A practical approach. Oxford: IRL Press; 1995. [Google Scholar]
- Dobson AW, Xu Y, Kelley MR, LeDoux SP, Wilson GL. Enhanced mitochondrial DNA repair and cellular survival after oxidative stress by targeting the human 8-oxoguanine glycosylase repair enzyme to mitochondria. J Biol Chem. 2000;275:37518–37523. doi: 10.1074/jbc.M000831200. [DOI] [PubMed] [Google Scholar]
- Dosanjh MK, Roy R, Mitra S, Singer B. 1,N6-ethenoadenine is preferred over 3-methyladenine as substrate by a cloned human N-methylpurine-DNA glycosylase (3-methyladenine-DNA glycosylase) Biochemistry. 1994;33:1624–1628. doi: 10.1021/bi00173a002. [DOI] [PubMed] [Google Scholar]
- Driggers WJ, LeDoux SP, Wilson GL. Repair of oxidative damage within the mitochondrial DNA of RINr 38 cells. J Biol Chem. 1993;268:22042–22045. [PubMed] [Google Scholar]
- Fernandes-Alnemri T, Litwack G, Alnemri ES. CPP32, a novel human apoptotic protein with homology to Caenorhabditis elegans cell death protein Ced-3 and mammalian interleukin-1 beta-converting enzyme. J Biol Chem. 1994;269:30761–30764. [PubMed] [Google Scholar]
- Fishel ML, Seo YR, Smith ML, Kelley MR. Imbalancing the DNA base excision repair pathway in the mitochondria; targeting and over-expressing N-methylpurine DNA glycosylase in mitochondria leads to enhanced cell killing. Cancer Res. 2003;63:608–615. [PubMed] [Google Scholar]
- Harrison JF, Hollensworth SB, Spitz DR, Copeland WC, Wilson GL, LeDoux SP. Oxidative stress-induced apoptosis in neurons correlates with mitochondrial DNA base excision repair pathway imbalance. Nucleic Acids Res. 2005;33:4660–4671. doi: 10.1093/nar/gki759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollensworth SB, Shen C, Sim JE, Spitz DR, Wilson GL, LeDoux SP. Glial cell type-specific responses to menadione-induced oxidative stress. Free Radic Biol Med. 2000;28:1161–1174. doi: 10.1016/s0891-5849(00)00214-8. [DOI] [PubMed] [Google Scholar]
- Karran P, Lindahl T, Ofsteng I, Evensen GB, Seeberg E. Escherichia coli mutants deficient in 3-methyladenine-DNA glycosylase. J Mol Biol. 1980;140:101–127. doi: 10.1016/0022-2836(80)90358-7. [DOI] [PubMed] [Google Scholar]
- Krokan HE, Nilsen H, Skorpen F, Otterlei M, Slupphaug G. Base excision repair of DNA in mammalian cells. FEBS Lett. 2000;476:73–77. doi: 10.1016/s0014-5793(00)01674-4. [DOI] [PubMed] [Google Scholar]
- Lau AY, Wyatt MD, Glassner BJ, Samson LD, Ellenberger T. Molecular basis for discriminating between normal and damaged bases by the human alkyladenine glycosylase, AAG. Proc Natl Acad Sci USA. 2000;97:13573–13578. doi: 10.1073/pnas.97.25.13573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley PD, Warren W. Removal of minor methylation products 7-methyladenine and 3-methylguanine from DNA of Escherichia coli treated with dimethyl sulphate. Chem Biol Interact. 1976;12:211–220. doi: 10.1016/0009-2797(76)90100-9. [DOI] [PubMed] [Google Scholar]
- Ledoux SP, Shen CC, Grishko VI, Fields PA, Gard AL, Wilson GL. Glial cell-specific differences in response to alkylation damage. Glia. 1998;24:304–312. [PubMed] [Google Scholar]
- LeDoux SP, Wilson GL, Beecham EJ, Stevnsner T, Wassermann K, Bohr VA. Repair of mitochondrial DNA after various types of DNA damage in Chinese hamster ovary cells. Carcinogenesis. 1992;13:1967–1973. doi: 10.1093/carcin/13.11.1967. [DOI] [PubMed] [Google Scholar]
- LeDoux SP, Wilson GL, Bohr VA. Mitochondrial DNA repair and cell injury. San Diego: Academic Press; 1993. pp. 461–476. [Google Scholar]
- Lefranc F, Brotchi J, Kiss R. Possible future issues in the treatment of glioblastomas: Special emphasis on cell migration and the resistance of migrating glioblastoma cells to apoptosis. J Clin Oncol. 2005;23:2411–2422. doi: 10.1200/JCO.2005.03.089. [DOI] [PubMed] [Google Scholar]
- Levy GN, Brabec MJ. Binding of carbon tetrachloride metabolites to rat hepatic mitochondrial DNA. Toxicol Lett. 1984;22:229–234. doi: 10.1016/0378-4274(84)90071-7. [DOI] [PubMed] [Google Scholar]
- Lockhart ML, Deutsch JF, Yamaura I, Cavalieri LF, Rosenberg BH. Termination of DNA synthesis in vitro at apurinic sites but not at ethyl adducts on the template. Chem Biol Interact. 1982;42:85–95. doi: 10.1016/0009-2797(82)90144-2. [DOI] [PubMed] [Google Scholar]
- Loeb LA, Preston BD. Mutagenesis by apurinic/apyrimidinic sites. Annu Rev Genet. 1986;20:201–230. doi: 10.1146/annurev.ge.20.120186.001221. [DOI] [PubMed] [Google Scholar]
- Luo X, Budihardjo I, Zou H, Slaughter C, Wang X. Bid, a Bcl2 inter-acting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell. 1998;94:481–490. doi: 10.1016/s0092-8674(00)81589-5. [DOI] [PubMed] [Google Scholar]
- Niranjan BG, Bhat NK, Avadhani NG. Preferential attack of mitochondrial DNA by aflatoxin B1 during hepatocarcinogenesis. Science. 1982;215:73–75. doi: 10.1126/science.6797067. [DOI] [PubMed] [Google Scholar]
- Pegg AE. Formation and metabolism of alkylated nucleosides: Possible role in carcinogenesis by nitroso compounds and alkylating agents. Adv Cancer Res. 1977;25:195–269. doi: 10.1016/s0065-230x(08)60635-1. [DOI] [PubMed] [Google Scholar]
- Pendlebury A, Frayling IM, Santibanez Koref MF, Margison GP, Rafferty JA. Evidence for the simultaneous expression of alternatively spliced alkylpurine N-glycosylase transcripts in human tissues and cells. Carcinogenesis. 1994;15:2957–2960. doi: 10.1093/carcin/15.12.2957. [DOI] [PubMed] [Google Scholar]
- Pieper RO, Futscher BW, Erickson LC. Transcription-terminating lesions induced by bifunctional alkylating agents in vitro. Carcinogenesis. 1989;10:1307–1314. doi: 10.1093/carcin/10.7.1307. [DOI] [PubMed] [Google Scholar]
- Prados MD, Levin V. Biology and treatment of malignant glioma. Semin Oncol. 2000;27(3) Suppl 6:1–10. [PubMed] [Google Scholar]
- Rinne M, Caldwell D, Kelley MR. Transient adenoviral N-methylpurine DNA glycosylase overexpression imparts chemotherapeutic sensitivity to human breast cancer cells. Mol Cancer Ther. 2004;3:955–967. [PubMed] [Google Scholar]
- Rinne ML, He Y, Pachkowski BF, Nakamura J, Kelley MR. N-methylpurine DNA glycosylase overexpression increases alkylation sensitivity by rapidly removing non-toxic 7-methylguanine adducts. Nucleic Acids Res. 2005;33:2859–2867. doi: 10.1093/nar/gki601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushmore T, Snyder R, Kalf G. Covalent binding of benzene and its metabolites to DNA in rabbit bone marrow mitochondria in vitro. Chem Biol Interact. 1984;49:133–154. doi: 10.1016/0009-2797(84)90057-7. [DOI] [PubMed] [Google Scholar]
- Sagher D, Strauss B. Insertion of nucleotides opposite apurinic/apyrimidinic sites in deoxyribonucleic acid during in vitro synthesis: Uniqueness of adenine nucleotides. Biochemistry. 1983;22:4518–4526. doi: 10.1021/bi00288a026. [DOI] [PubMed] [Google Scholar]
- Saparbaev M, Laval J. Excision of hypoxanthine from DNA containing dIMP residues by the Escherichia coli, yeast, rat, and human alkylpurine DNA glycosylases. Proc Natl Acad Sci USA. 1994;91:5873–5877. doi: 10.1073/pnas.91.13.5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaper RM, Kunkel TA, Loeb LA. Infidelity of DNA synthesis associated with bypass of apurinic sites. Proc Natl Acad Sci USA. 1983;80:487–491. doi: 10.1073/pnas.80.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobol RW, Kartalou M, Almeida KH, Joyce DF, Engelward BP, Horton JK, Prasad R, Samson LD, Wilson SH. Base excision repair intermediates induce p53-independent cytotoxic and genotoxic responses. J Biol Chem. 2003;278:39951–39959. doi: 10.1074/jbc.M306592200. [DOI] [PubMed] [Google Scholar]
- Wassermann K, Kohn KW, Bohr VA. Heterogeneity of nitrogen mustard-induced DNA damage and repair at the level of the gene in Chinese hamster ovary cells. J Biol Chem. 1990;265:13906–13913. [PubMed] [Google Scholar]
- Whitehouse CJ, Taylor RM, Thistlethwaite A, Zhang H, Karimi-Busheri F, Lasko DD, Weinfeld M, Caldecott KW. XRCC1 stimulates human polynucleotide kinase activity at damaged DNA termini and accelerates DNA single-strand break repair. Cell. 2001;104:107–117. doi: 10.1016/s0092-8674(01)00195-7. [DOI] [PubMed] [Google Scholar]
- Wunderlich V, Schutt M, Bottger M, Graffi A. Preferential alkylation of mitochondrial deoxyribonucleic acid by N-methyl-N-nitrosourea. Biochem J. 1970;118:99–109. doi: 10.1042/bj1180099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying W, Alano CC, Garnier P, Swanson RA. NAD1 as a metabolic link between DNA damage and cell death. J Neurosci Res. 2005;79:216–223. doi: 10.1002/jnr.20289. [DOI] [PubMed] [Google Scholar]