Abstract
DNA methylation is a major epigenetic factor regulating genome reprogramming, cell differentiation and developmental gene expression. To understand the role of DNA methylation in central nervous system (CNS) neurons, we generated conditional Dnmt1 mutant mice that possess ∼90% hypomethylated cortical and hippocampal cells in the dorsal forebrain from E13.5 on. The mutant mice were viable with a normal lifespan, but displayed severe neuronal cell death between E14.5 and three weeks postnatally. Accompanied with the striking cortical and hippocampal degeneration, adult mutant mice exhibited neurobehavioral defects in learning and memory in adulthood. Unexpectedly, a fraction of Dnmt1−/− cortical neurons survived throughout postnatal development, so that the residual cortex in mutant mice contained 20–30% of hypomethylated neurons across the lifespan. Hypomethylated excitatory neurons exhibited multiple defects in postnatal maturation including abnormal dendritic arborization and impaired neuronal excitability. The mutant phenotypes are coupled with deregulation of those genes involved in neuronal layer-specification, cell death and the function of ion channels. Our results suggest that DNA methylation, through its role in modulating neuronal gene expression, plays multiple roles in regulating cell survival and neuronal maturation in the CNS.
INTRODUCTION
Neural development is composed of a cascade of genetic programs that precisely control stage-specific gene activities required for neural patterning, cell migration and neuronal connectivity. Recent studies indicate that the temporally- and spatially-controlled gene expression in the nervous system is not only regulated by the transcriptional machinery, but also subject to modulation by epigenetic mechanisms such as DNA methylation and chromatin remodeling. DNA methylation is catalyzed by a family of DNA methyltransferases (Dnmts) that include de novo (Dnmt3a and Dnmt3b) and maintenance methyltransferases (Dnmt1) (1–3). All three enzymes are expressed in the central nervous system (CNS) and are dynamically regulated during development and differentiation (4,5). Mechanistically, DNA methylation inhibits gene expression by either directly interfering with transcription factor binding to DNA (6) or recruitment of methyl CpG binding proteins (MBDs) which complex with co-repressor(s) and histone modification enzymes such as histotone deacetylases and methyltransferases to transform chromatin to a repressive state (reviewed in 5 and 7).
The important role for DNA methylation in CNS development and function is first implicated by the identification of MeCP2 mutations in mental retardation disorder Rett syndrome (8). As a prototype of the MBD proteins, MeCP2 is highly expressed in post-mitotic neurons and is involved in regulating neuronal gene expression including neurotrophin BDNF and transcription factor Dlx5/6 (9–11). MeCP2-deficient mice exhibit many typical Rett phenotypes including neuronal atrophy and stereotypic symptom such as irregular breathing pattern (12,13). To directly examine the role of DNA methylation in the CNS, we have previously created a mouse model with DNA hypomethylation in the entire developing CNS (14). By conditional deletion of Dnmt1 in neural precursor cells, we demonstrated that DNA hypomethylation in the CNS disrupts neural control of breathing at birth, leading to neonatal lethality of mutant mice (14). Furthermore, DNA hypomethylation in neural precursor/stem cells triggers precocious glial differentiation by activating the JAK-STAT pathway and the expression of glial marker genes such as glial fibrillary acidic protein (Gfap) (15–17). Thus, DNA methylation serves as a key epigenetic mechanism in the temporal control of neural stem cell differentiation.
In the current study, we attempted to address the role of DNA methylation during cortical neuronal maturation at the perinatal stages. We have crossed the Emx1-cre transgene (18) with the Dnmt1 conditional allele (Dnmt12lox) (14) to generate a strain of Dnmt1 mutant mice that are viable in adulthood but exhibit DNA hypomethylation exclusively in the dorsal forebrain. Although mutant mice have a normal lifespan, they develop cortical degeneration during early postnatal development and show obvious behavioral defects such as hyperactivity and impaired learning and memory. Surprisingly, a significant portion of hypomethylated cortical neurons was retained in the mutant brain, allowing us to address the effect of DNA hypomethylation on neural gene expression and the maturation of cortical neurons postnatally.
RESULTS
Mutant mice are viable with conditional Dnmt1 gene deletion in the dorsal forebrain
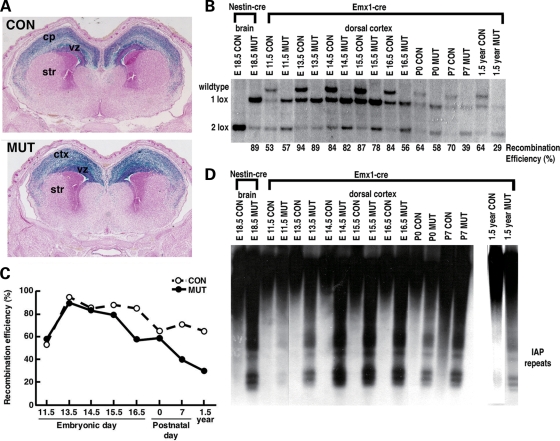
We used Emx1-cre to trigger in vivo deletion of Dnmt1 exclusively in telencephalic precursors, inducing DNA hypomethylation in excitatory neurons and astroglia of the cortex and hippocampus but not in inhibitory interneurons (18,19). We confirmed the gene deletion pattern in Emx1-cre; Dnmt12lox mice by crossing onto the R26R-lacZ reporter strain (20). The lacZ-staining pattern is restricted to the dorsal cortex and hippocampus in both E16.5 Emx1-cre; Dnmt12lox/+ heterozygous controls (designated as control group) and Emx1-cre; Dnmt12lox/2lox mutants (mutant group hereafter), indicating the pattern of Dnmt1 gene deletion in the CNS (Fig. 1A). It has been reported that Cre-mediated recombination in Emx-cre mice was initiated between E8.5 and E10.5 as characterized before (18,19). Southern blot analysis confirmed that significant Dnmt1 gene deletion was observed in E11.5, and the efficiency in mutant cortices peaks at 89% (E13.5) and declines to 58% at P0, down to 39% at P7, and maintains at ∼30% during adulthood (Fig. 1B and C). The decrease in Dnmt1 gene deletion efficiency in mutant brains when compared with controls is likely caused by degeneration of hypomethylated cells (see below). In parallel to the time course of Dnmt1 gene deletion, we found significant demethylation in mutant cortical cells as assessed by Southern blot analysis of the normally heavily methylated Intracisternal-A-Particle (IAP) retroviral element (Fig. 1D). These results suggest hypomethylated neurons are present in dorsal forebrain throughout the mutant animal's lifespan, but loss of hypomethylated neurons seems to occur at both pre- and post-natal stages.
Figure 1.
Hypomethylated cortical cells are detected in Emx1-cre; Dnmt12lox/2lox mutants throughout the normal life span (A) LacZ histology pattern shows brain regions with Dnmt1 gene deletion via the R26R reporter (ref. 20). (B) The recombination efficiency was analyzed by Southern blotting analysis. The percentages of Dnmt1 gene deletion are derived from the ratio of recombined null allele (1lox) over the sum of functional Dnmt12lox (2lox) and 1lox alleles. (C) The graph view of the efficiency of Dnmt1 gene deletion in control and mutant mice from E11.5 to adulthood at 1.5 years old. (D) Southern blot analysis of DNA methylation in dorsal cortex. DNA was digested with methyl-sensitive enzyme HpaII (CCGG) and was hybridized to an IAP probe. CON, Emx1-cre; Dnmt1210x/+ control; MUT, Emx1-cre; Dnmt1210x/210x mutant; Ctx, cortex; str, striatum.
Emx1-cre; Dnmt1 mutant mice exhibit severe cortical degeneration and neurobehavioral defects in learning and memory
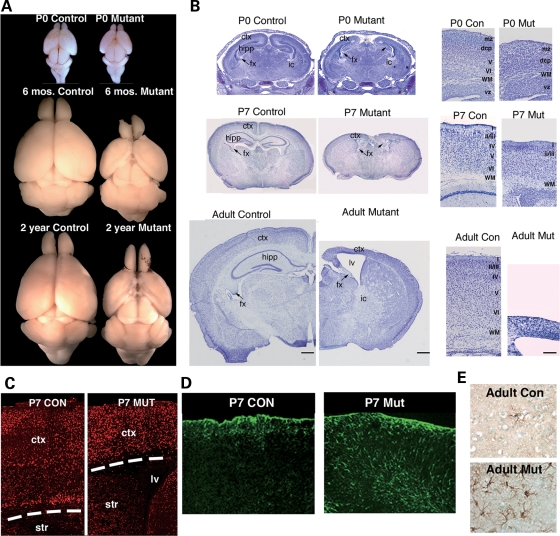
After conducting a careful histological examination of the brain morphology of control and mutant littermates, we found striking cortical degeneration in mutant brains at perinatal stages (Fig. 2A). Only a moderate decrease in cortical thickness was observed in mutant brains at P0; however, a severe loss of cortical and hippocampal volume was found during early postnatal stages (Fig. 2B). Indeed, quantitative measurements of cortical thickness indicate, as a percentage of control, a reduction of 36.7 ± 1.9% in P7 mutants and further diminishment by 68.3 ± 1.3% in adult mutants (Fig. 2B). Immunostaining for the post-mitotic neuronal marker NeuN confirms that the mutant cortex contains fewer neurons than heterozygous littermate controls (Fig. 2C). An increase of GFAP-positive reactive astroglia was accompanied with neuronal loss in the cortical layers, indicative of gliosis due to postnatal neuronal cell death (Fig. 2D and E).
Figure 2.
Cortical degeneration in dorsal forebrain of Emx1-cre; Dnmt12lox/2lox mutants (A and B) Microscopic images of cresyl-violet staining in control and mutant brain sections at newborn, early postnatal P7, and adult stages. (C) NeuN immunohistochemistry at P7 reveals a decrease in the density of neurons in the cortex of the mutant. (D and E). Immunostaining for GFAP in control and mutant cortex. Ctx, cortex; vz, ventricular zone; str, striatum; hipp, hippocampus; lv, lateral ventricle; fx, fornix; ic, internal capsule; I–IV, cortical layers; wm, white matter; mz, marginal zone; dcp, dense cortical plate. Con, Emx1-cre; Dnmt12lox/+ control; mut, Emx1-cre; Dnmt12lox/2lox mutant.
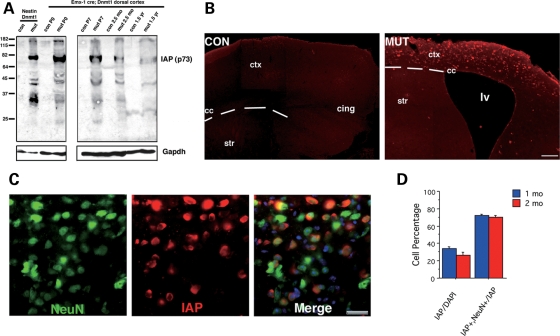
To visualize the distribution of hypomethylated neurons in mutant brains, we employed immunohistochemical analysis against the major core protein of IAP retroelements (21). IAP expression is undetected in methylation proficient cells, but is induced specifically in hypomethylated somatic cells (Fig. 3A) (22,23). In the cortex of one-month-old mutant mice, IAP immunoreactivity is restricted to the dorsal cortical zone, precisely corresponding to the area of Dnmt1 gene deletion (Fig. 3B). Co-localization of IAP immunoreactivity and NeuN marker showed that the majority of IAP+ cells are neurons. In one-month-old mutant brain, among the total 32.3% IAP+ cells, 70.7% are NeuN+ neurons (Fig. 3C and D). The remaining IAP+ cells (29.3%) are GFAP+ astrocytes that were also derived from Emx1-positive precursor cells. Similar results were observed in the two-month-old mutant mice (Fig. 3D). Thus, both demethylated neurons and GFAP+ astrocytes can survive in the adult mutant brain. In contrast, methylation proficient parvalbumin- and calbindin-positive GABAergic interneurons were negative for IAP immunoreactivity (data not shown). Inhibitory GABAergic interneurons originate from the medial ganglionic eminence during embryonic development and they migrate into dorsal cortex (24). Thus, interneurons are DNA methylation proficient from the ontogeny and are spared from Emx1-cre-mediated Dnmt1 gene deletion.
Figure 3.
Localization of demethylated cells in mutant brains by IAP immunostaining. (A) Western blotting of IAP protein in the dorsal cortex of wild type (con) and Emx1-cre; Dnmt12lox/2lox mutants mice (mut) and Nestin-cre; Dnmt12lox/2lox mutant mice (Nestin Dnmt1, see ref. 20). (B) Immunohistochemical detection of IAP in the dorsal cortex at 1 month of age in coronal sections. (scale bar = 100 microns) (C) Co-staining with NeuN (a pan-neuronal marker) reveals an IAP-positive staining pattern in mutant cortex only. (scale bar = 25 microns) (D) Cell counts show that approximately 28–32% of the total cells are IAP-positive in 1 or 2 months (mo) old mutant mice. Among those IAP+ cells, 70.67 ± 1.8% are NeuN+ neurons.
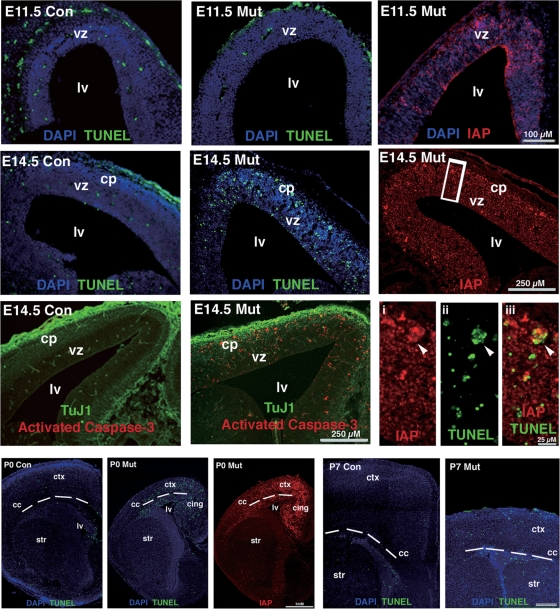
To determine whether hypomethylated neurons in mutant mice undergo apoptotic cell death during the pre- and post-natal development, we performed TUNEL assays in brain sections at ages E11.5, E14.5, P0 and P7 (Fig. 4). At E11.5, IAP staining showed the presence of hypomethylated neural precursor cells in mutant cortex; however, no difference in the number of TUNEL-positive cells was found between controls and mutants. In contrast, by E14.5 at the peak of neurogenesis, TUNEL-positive cells were significantly increased in the mutant cortical plate and ventricular zone, which co-localized with many IAP-positive cells (arrowhead in the insets of Fig. 4). Our results indicated that apoptotic cell death of hypomethylated cortical neurons and precursor cells commences at E14.5. Immunostaining with an antibody against activated caspase-3 confirmed that hypomethylated cells undergo a caspase-activated apoptosis at E14.5 (Fig. 4). Apoptotic cell death was also obvious in the dorsal cortex of mutant mice between P0 and P7, during this period when we observed a most dramatic reduction of cortical volume (Fig. 4). Taken together, our results indicated that severe cortical degeneration in Emx1-cre; Dnmt1 mutant mice is at least in part mediated by hypomethylation-induced apoptotic cell death at perinatal stages.
Figure 4.
Hypomethylated cells in the dorsal cortex undergo apoptotic cell death perinatally TUNEL immunohistochemistry were performed with E11.5, E14.5, P0 and P7 brain samples. No difference in levels of apoptotic index at E11.5 even though demethylated cells IAP-positive at this time point. By E14.5, there is a dramatic increase in apoptosis in the ventricular zone and the emerging cortical plate of the mutant embryo. This region is positive for strong IAP staining and activated caspase-3 immunoreactivity. The neurotubulin marker TUJ1 (green) is used to show the cortical plate (cp) boundary from the ventricular zone (vz). Note high magnification images from boxed region shows co-labeling of TUNEL-positive cells with IAP reactivity (arrowhead). Apoptotic signals are also easily detected in mutant cortex at P0 and P7. con, Emx1-cre; Dnmt12lox/+ control; mut, Emx1-cre; Dnmt12lox/2lox mutant; Lv, lateral ventricle; str, striatum; cc, corpus callosum; ctx, cortex.
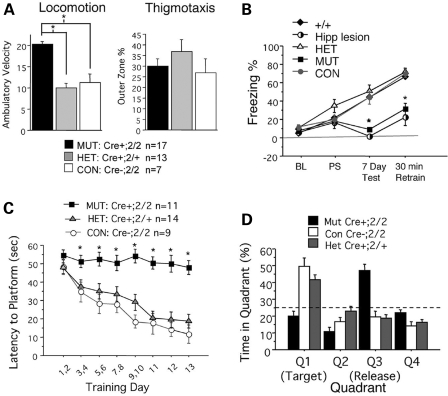
Given the observation of severe cortical degeneration in mutants, we next assessed the neurobehavioral defects using a battery of motor skill tasks and learning and memory trials on a cohort of age-matched, sex-matched mutant and control mice (age 6–8 months). We found that while mutant mice can perform just as well as controls in motor learning and coordination (Supplementary Material, Fig. S1), they exhibited severe deficits in learning and memory behaviors in both paradigms of fear conditioning and Morris hidden platform water maze (Fig. 5). In fact, the neurobehavioral defect in Emx1-cre; Dnmt1 mice resembles the deficit observed in hippocampal lesioned animals, which also show severely impaired learning and memory (Fig. 5B). Taken together, our data demonstrated that Dnmt1 deficiency in the developing dorsal forebrain leads to cortical and hippocampal degeneration in adult mutants, which exhibit behavioral defects in learning and memory.
Figure 5.
Locomotion, thigmotaxis, and Morris Water Maze behavioral tests of control and mutant mice. (A) Open field test assesses animal exploratory behavior. Mutants exhibit hyperactivity as a function of increased locomotor activity; however, mutants do not appear to have altered anxiety levels as measured by thigmotatic behavior. (B) Hippocampal dependent contextual fear tasks show a severe reduction of long term memory in the mutants, consistent with the loss of the hippocampal structure in mutants. Re-exposure to the fear conditioning chamber (7 day) elicited no fear response in mutant animals, akin to the lack of freezing seen in hippocampal lesioned animals. (C) Morris water maze testing the ability of mice to recall spatial navigation is a hippocampal specific learning and memory task. Over continuous learning trial days, mutants do not improve in finding the hidden platform, nor do they exhibit a learning curve for the task of locating the platform. (D) Conditional mutants score below chance (25%) in probe trials after a 14-day learning protocol, indicative of poor spatial memory in the mutants. For each category of mice, more than 11 adult mice/genotype around the age of 6 months were used in behavioral tests.
Deregulation of neuronal gene expression in hypomethylated forebrain
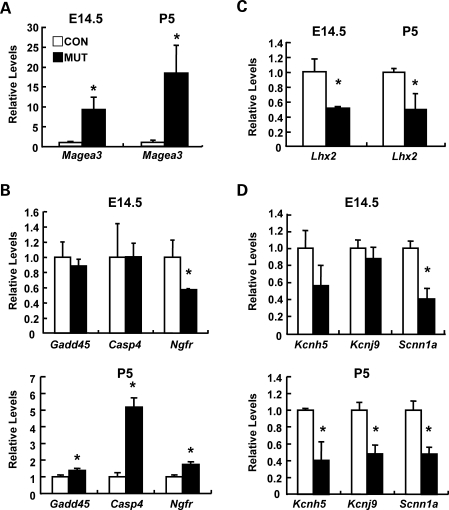
The generation of viable mutant mice containing hypomethylated cortical neurons allows us to profile the potential alteration of neuronal gene expression and further investigate cellular defects in dorsal cortex of Emx1-cre; Dnmt1 mutant mice. We choose to analyze potential alteration of neuronal gene expression in both prenatal E14.5 and early postnatal P5 when mutant neurons are still abundant in the dorsal cortex. At E14.5, the peak stage of neurogenesis, the cortical structure is by and large intact and demethylated neurons account for a majority of cells (82%). Thus, any gene deregulation at E14.5 mutant cortex reflects the effect of DNA hypomethylatoin on immature CNS neurons. On the other hand, P5 is a critical stage of neuronal maturation when proper regulation of genes involved in neural plasticity is established. Furthermore, a comparison of the altered gene expression profiles in these two stages also allows us to determine whether expression changes caused by DNA hypomethylation are stage-specific. Genome-wide gene expression profiling showed that in the P5 dorsal cortex of mutant mice, 1047 genes (6.1%) are up-regulated and 444 (2.6%) genes that are downregulated with a statistically significant 2-fold threshold (Supplementary Material, Tables S1 and S2). Using PANTHER classification categories (http://www.pantherdb.org/tools/compareToRefListForm.jsp), gene ontology analysis indicated that up-regulated genes exhibit an overrepresentation of genes involved in apoptosis and developmental process, consistent with the observed phenotype (Tables 1 and 2 and Supplementary Material, Fig. S2). Among those up-regulated genes, we confirmed the up-regulation of the Magea3 transcript in both E14.5 and P5 mutant cortex (Fig. 6A). This result is consistent with the demethylation state of mutant cortical cells because Magea3, an X-linked gene family member, is known to be repressed by DNA methylation in both somatic cells and embryonic stem cells (25) (Table 1). We further confirmed that the up-regulation of apoptosis-related genes such as Casp4, Gadd45a and Ngfr in P5 mutant cortex but not in E14.5 by real-time RT–PCR, consistent with a dramatic cortical degeneration in early postnatal stages but a relatively minor defect of neuronal cell death in embryonic development (Fig. 6B). Interestingly, deregulated gene list contained several transcriptional factors including Lhx2 (Fig. 6C) that regulates cortical layer specification and neuronal migration, suggesting a potential defect in cortical architecture in the dorsal cortex of mutant mice (24).
Table 1.
Up-regulation of genes involving in cortical layer-specification,cell death and other functions in dorsal cortex in P5 Emx-Cre; Dnmt1 mutant mice
| Gene symbol | Genebank | Gene name | Fold change |
|---|---|---|---|
| Layer-specific or area-specific expression genes | |||
| Lhx5 | NM_008499 | LIM homeobox protein 5 | 3.52 |
| Dlx5 | NM_010056 | distal-less homeobox 5 (Dlx5), transcript variant 1 | 2.47 |
| Nkx6-2 | NM_183248 | NK6 transcription factor related, locus 2 | 4.40 |
| Cart1 | NM_172553 | cartilage homeo protein 1 | 2.90 |
| Olig2 | NM_016967 | oligodendrocyte transcription factor 2 | 2.38 |
| Dlx2 | NM_010054 | distal-less homeobox 2 | 2.07 |
| Apoptosis- or survival-related genes | |||
| Ngfr | NM_033217 | nerve growth factor receptor (TNFR superfamily, member 16) | 2.17 |
| Casp4 | NM_007609 | caspase 4, apoptosis-related cysteine peptidase | 3.62 |
| Gadd45a | NM_007836 | growth arrest and DNA-damage-inducible 45 alpha | 2.20 |
| Fa s | NM_007987 | Fas (TNF receptor superfamily member) | 2.42 |
| MHC Antigen | |||
| H2-M3 | NM_013819 | histocompatibility 2, M region locus 3 | 2.50 |
| H2-Ab1 | NM_207105 | histocompatibility 2, class II antigen A, beta 1 | 2.93 |
| X-linked genes | |||
| Rhox2 | NM_029203 | reproductive homeobox 2 | 14.81 |
| Xlr3b | NM_011727 | X-linked lymphocyte-regulated 3B | 17.92 |
| Xlr | NM_011725 | X-linked lymphocyte-regulated complex | 6.11 |
| Xlr4b | NM_021365 | X-linked lymphocyte-regulated 4B | 31.92 |
| Mael | NM_175296 | maelstrom homolog (Drosophila) | 17.24 |
| Magea3 | NM_020017 | melanoma antigen, family A, 3 | 4.05 |
| Magea8 | NM_020020 | melanoma antigen, family A, 8 | 2.50 |
Table 2.
Down-regulation of genes involving in cortical layer-specification, ion channels, and cell survival in dorsal cortex in P5 Emx-Cre; Dnmt1 mutant mice
| Gene symbol | Genebank | Gene name | Fold change |
|---|---|---|---|
| Layer-specific or area-specific expression genes | |||
| Bhlhb5 | NM_021560 | basic helix-loop-helix domain containing, class B5 | 0.45 |
| Lhx2 | NM_010710 | LIM homeobox protein 2 | 0.46 |
| Ldb2 | NM_010698 | LIM domain binding 2 | 0.46 |
| Lmo4 | NM_010723 | LIM domain only 4 | 0.49 |
| Bcl11a | NM_016707 | B-cell CLL/lymphoma 11A (zinc finger protein) | 0.45 |
| Ion channel or neuronal activity | |||
| Kcnh3 | NM_010601 | potassium voltage-gated channel | 0.34 |
| Kcnj9 | NM_008429 | potassium inwardly-rectifying channel | 0.39 |
| Kcnq5 | NM_023872 | potassium voltage-gated channel | 0.28 |
| Kcnh5 | NM_172805 | potassium voltage-gated channel | 0.31 |
| Kcnk5 | NM_021542 | potassium channel | 0.35 |
| Kcnj6 | NM_010606 | potassium inwardly-rectifying channel | 0.41 |
| Trpa1 | NM_177781 | transient receptor potential cation channel | 0.36 |
| Scnn1a | NM_011324 | sodium channel | 0.50 |
| Neuronal development | |||
| Ntf3 | NM_008742 | neurotrophin3 | 0.43 |
| Bcl11a | NM_016707 | B-cell CLL/lymphoma 11A (zinc finger protein) | 0.45 |
Figure 6.
Hypomethylation alters neuronal gene expression in mutant cortex in both pre- and postnatal stages. (A and B) The expression of X-linked gene (Magea3, A) and apoptosis-related genes (Gadd45, Casp4, Ngfr, B) in the dorsal cortex of wild type (WT, open bar) or Emx1-cre; Dnmt1 mutant (mut, black bar) mice. (C and D) The expression of layer specific gene (Lhx2, C) and neuronal channels genes (Kcnh5, Kcnj9 and Scnn1a, D) in the dorsal cortex of wild type and mutant mice. Quantitative real-time PCR was performed in cDNAs derived from E14.5 and P5 dorsal cortex of each mouse by using the specific primers. Statistical significance was evaluated by the t-test. Mean ± SEM (n = 3). *P < 0.05.
We also found that a number of genes implicated in regulating neural activity and neurogenesis were down-regulated (Fig. 6C). For example, the neurotrophin gene Nt3 as well as many channel genes (Scnn1a, Kcnh5, Kcnj9) were significantly down-regulated, particularly in P5 mutant brain, which is a critical stage of neuronal maturation (Fig. 6D). These results suggest that mutant mice likely have defects in electrophysiological properties and neural cell differentiation.
Hypomethylation leads to defects in neuronal morphology and excitability
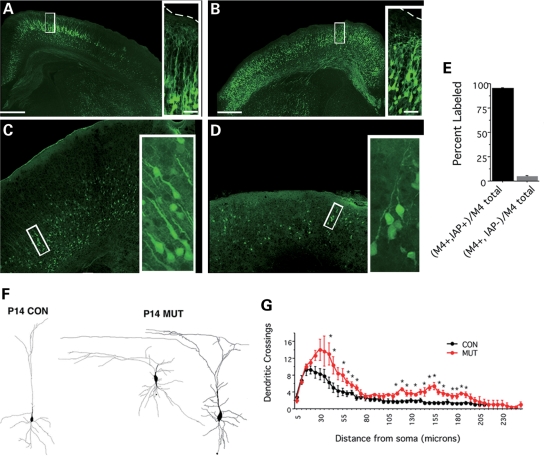
Given the above results, we subsequently examined the defects in morphology and neuronal excitability in mutant mice. Although hypomethylated neurons were retained in mutant brains at postnatal ages, cresyl violet histology denoted the failure of distinct cortical layer development in the mutant (Fig. 2B). To further analyze the morphological and physiological properties of a subset of cortical excitatory neurons, we crossed Dnmt1 mutants with BAC transgenic mice (M4-eGFP) expressing green fluorescent protein in a subset of cortical excitatory neurons of layers 2/3 and 5, as well as a subset of striatal neurons (Fig. 7A–D) (26). ∼95% of the cortical M4-eGFP neuronal population were IAP-positive in the mutant cortex (Fig. 7E), indicating that mutant neurons expressing M4-eGFP were indeed a subpopulation of hypomethylated excitatory neurons. Confocal images of M4+-EGFP-positive neurons demonstrated that mutant neurons do not maintain proper layer specificity and display abnormal branching (Fig. 7F). Consistent with the observed cell death in the cortex of one-week-old mutants, the number of demethylated eGFP-positive neurons decreased between P0 and P7, and the remaining neurons exhibited dystrophic neurites with loss of proper radial projections to the pial surface (Fig. 7F). Camera Lucida drawings of biocytin filled M4-eGFP+ control and mutant neurons revealed a significantly larger soma area in mutant neurons (con = 84.12 ± 10.88 µm2, mut = 140.4 ± 15.48 µm2; P < 0.01) and an increase in both basal and apical dendritic branching in demethylated neurons (Fig. 7G). These data suggest DNA methylation is important for the morphological maturation of neurons.
Figure 7.
Hypomethylated cortical neurons exhibit defects in dendritic arborization (A–D). Morphology of M4-egfp-positive neurons in control and mutant cortices at P0 (A and B) and P7 (C and D). (E) A high percentage of M4-egfp+ cells colocalize with IAP reactivity in mutant cortices (94.98 ± 0.85%). (F) Camera lucida drawings of individual M4-egfp-positive neurons filled with biocytin from either control or mutant cortex. (G) Sholl analysis to analyze dendritic branching reveals an increase in basal and apical branching patterns (P < 0.01, student t-tests).
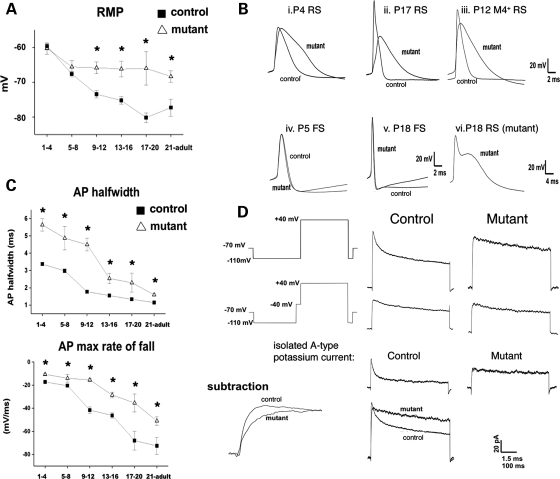
To directly examine whether genomic demethylation in excitatory neurons alters electrophysiological parameters, we performed whole-cell current clamp recordings in somatosensory cortical slices from mutant (age P3–P53) and control mice (age P1–P75). Excitatory mutant neurons possessed more depolarized resting membrane potentials after P9 and exhibited significantly broadened action potential (AP) half-widths when compared with controls at most ages examined (Fig. 8A–C). To rule out the possibility that the differences in electrophysiological parameters are caused by recording from different types of neurons, whole-cell recordings were also performed in the same subset of M4-EGFP-positive neurons in control and mutant mice. At P11–P12, the M4-eGFP+ mutant neurons possessed significantly increased AP half-widths [Value = mean ± SEM: con = 1.8 ± 0.1 ms (n = 6), mut = 2.9 ± 0.9 ms (n = 15); P < 0.05], demonstrating that prolonged APs are characteristic of hypomethylated neurons (Fig. 8B and C). The AP prolongation in mutant neurons occurred via decreased rate of maximal rise and, more prominently, a reduction of the maximal rate of fall, suggesting perturbations to both sodium and potassium channel function (Fig. 8D). Voltage-gated potassium currents recorded in somatic outside-out patches of excitatory neurons indicated similar voltage-dependence of activation and inactivation when compared with control (P9–P28). However, potassium currents in somatic patches from mutant neurons showed an absence of fast activation and inactivation, signifying the absence of the rapidly inactivating potassium current (IA) (Fig. 8D).
Figure 8.
Hypomethylated neurons exhibit defects in resting membrane and action potentials. (A) Significant changes in resting membrane potential (RMP) were graphed (*P < 0.05, student t-test). (B) Action potential wave forms in control and mutant cells including M4-eGFP-positive neurons. (C) Action potential half-width and maximum rate of repolarization in P1–4, 5–8, 9–12, 13–16, 17–20 and P21–adult regular spiking cortical neurons in control and mutant slices. Note the increased half-width resulting from decrease in the maximal rate of repolarization at all ages. (D) Voltage schematic and leak subtracted currents elicited by stepping from −110 mV to +40 mV, with or without a 50 ms pre-step to −40 mV in outside out patches pulled from control and mutant RS cortical neurons. Figures are averages of currents obtained from 23 control patches and 8 mutant patches. Subtraction of pulses with the pre-step from pulses without the pre-step result in isolation of the rapidly inactivating potassium conductance (IA) in control neurons but not mutant neurons. Note that potassium currents in mutant somatic outside out patches inactivate much more slowly than those obtained from control neurons.
Alterations of potassium channel Kv4.2 protein expression and mRNA expression of Kchips in hypomethylated neurons
A variety of studies suggest that the A-type fast inactivating potassium current is mediated by the Kv4.x channel family (27), and Kv4.2 is present both in the dendrites and in the neuronal soma of excitatory neurons (28). However, real-time RT–PCR assays did not find any significant differences in mRNA expression of Kv4.2 in mutant cortex (Supplementary Material, Fig. S3A). To determine whether there is a potential discordance between mRNA and protein expression of Kv4.2 potassium channel in hypomethylated neurons, we performed immunoblots for Kv4.2 using whole-cell cortical lysate from both control and mutant dorsal cortex region. We saw a decrease of Kv4.2 protein levels at both 2 weeks and 3 weeks of age, with an ∼30% reduction at P12 and 60% reduction at P21 (Supplementary Material, Fig. S3B). To verify the decrease seen in the immunoblots, we also examined the overall levels of Kv4.2 immunostaining in control and mutant cortex. We also see a decrease in Kv4.2 immunoreactivity in the mutant cortex with a loss of laminar-specific patterning seen in the control cortex (Supplementary Material, Fig. S3C). Because the mRNA levels of Kv4.2 are similar between control and mutant cortices, the reduced expression of Kv4.2 protein in Dnmt1−/− neurons suggests that an indirect mechanism, such as translational inhibition or protein instability may mediate the effect of DNA hypomethylation on the reduction of Kv4.2 protein expression. It is worth noting that the expression levels of many regulatory proteins for potassium channel such as Kchip1, Kchip2 and Kchip 4a were also altered in mutant cortex (Supplementary Material, Fig. S4). We suggest that the combined defects in the expression of Kv4.2 and Kchips as well as other types of potassium and sodium ion channels (Fig. 6) may underlie the defect in neuronal excitability in hypomethylated neurons postnatally.
DISCUSSION
As one of the major epigenetic modifications in the mammalian genome, DNA methylation is known to play multiple roles in embryogenesis including developmental gene regulation, genomic imprinting and X-inactivation. During the development of CNS, we have demonstrated that DNA methylation is not only essential for the vital postnatal CNS functions, but also controls the timing of astrogliogenesis (14,16). However, specific roles for DNA methylation in postnatal neuronal maturation are largely unknown. In this line of Emx1-cre; Dnmt1 mice, we demonstrated that a wave of cell death occurs in the developing CNS between E14.5 to three weeks postnatally, consistent with the previous findings that substantial DNA demethylation in somatic cells, but not in embryonic stem cells, leads to apoptotic cell death (14,29,31). Furthermore, our data showed that DNA hypomethylation affects neuronal excitability and dendritic growth. Our results may offer an explanation for the phenotypes in Nestin-cre; Dnmt1 mutant mice, where DNA hypomethylation in the CNS disrupts neural control of respiration and causes neonatal lethality (14). Perhaps defects in either neuronal excitability or connectivity in the hypomethylated CNS could perturb the generation and/or relay of respiratory drive, thus causing respiratory failure in neonatal mutant mice.
Multiple physiological defects in hypomethylated forebrain
In this study, we showed that Dnmt1 deletion in mutant cortices peaks at 89% on E13.5, declines thereafter, and finally stays at ∼30% in adulthood. Because we observed severe neuronal cell loss in mutant brain (Figs 3, 4 and 7), the decrease of Dnmt1 gene deletion certainly reflects on the degeneration of hypomethylated neurons. It is worth noting that methylation proficient GABAergic interneurons are present in postnatal mutant brains. Therefore, we cannot rule out the possibility that the preferential survival of GABAergic interneurons over hypomethylated excitatory neurons and astrocytes in postnatal CNS may also contributes to the observed decrease in the efficiency of Dnmt1 gene deletion.
One of the surprising observations in this study is that 20–30% of hypomethylated excitatory cortical neurons survive in Emx1-cre; Dnmt1 mutant mice throughout their lifespan, suggesting that a subset of hypomethylated neurons escape selection pressure during postnatal maturation. In previous models of Dnmt1/DNMT1 deletion in mouse fibroblasts and human colon cancer cells, apoptosis was partly triggered by a p53-dependent pathway (29,31). It remains to be determined whether loss of hypomethylated neurons in Emx1-cre; Dnmt1 mice is also mediated by p53-dependent pathway. Furthermore, it will be of interests to determine whether the survival of a subset of excitory neurons in Emx1-cre; Dnmt1 mutant mice is triggered by the cell autonomous inhibition of p53 or via a novel cell survival pathway.
Although we found massive neural degeneration and behavioral defects in learning and memory in mutant mice, these mice have a normal lifespan in a laboratory environment. Another group has also reported that mutant mice with the loss of dorsal forebrain can survive into adulthood, even though they display behavioral abnormality (32). These observations suggest that degeneration of the forebrain is not directly incompatible with viability of mutant mice in a laboratory environment.
Deregulation of multiple neuronal genes in hypomethylated neurons
Previous studies have demonstrated that activity-dependent regulation of the neurotrophin gene Bdnf in cortical neurons involves a DNA methylation-related chromatin remodeling mechanism (9,10). However, misregulation of Bdnf alone cannot not explain the pleiotropic phenotypes observed in this Emx1-cre; Dnmt1 model. It is likely that multiple genes are deregulated in the absence of Dnmt1 in the developing CNS. The availability of hypomethylated cortical neurons in Emx1-cre; Dnmt1 mutant mice at all developmental stages provides a unique opportunity to identify critical genes that are subjected to regulation by DNA methylation in a stage- and cell type-specific manner. Our microarray study showed that ∼1500 genes (8.5% of all annotated genes) that are either up-regulated or down-regulated in P5 mutant cortex postnatally. We further showed that the altered expression of a subset of neuronal genes commences in E14.5 mutant cortex, which including genes involved in neuronal survival, cell patterning as well as neuronal excitability. It is worth noting that not all deregulated genes are directly caused by DNA demethylation. First, while demethylation is compatible with gene up-regulation, the observed down-regulation of neuronal genes is most likely via a secondary effect. Second, when loss of excitatory neurons is severe in the dorsal cortex postnatally, we cannot rule out the possibility that the up-regulation of a subset of genes, such as those genes highly expressed in the inhibitory interneurons, is attributed to the shift of cell population towards the enrichment of interneurons.
In demethylated embryonic stem cells, we recently showed that ∼3% of genes are deregulated and most of up-deregulated genes are related to lineage-specific differentiation (25). Thus, the number of deregulated genes in hypomethylated and differentiated cells are significantly higher than that in undifferentiated embryonic stem cells. It is likely that the profiles of gene deregulation in different types of somatic cells are different. In demethylated mouse fibroblasts, ∼10% of genes are deregulated (29). In hypomethylated CNS neurons, up-regulated genes are over-represented by homeobox genes, other DNA-binding proteins, receptors and signal molecules for developmental processes. In the category of down-regulated genes, which are presumably through indirect mechanisms in hypomethylated cells, ion channel genes and cytoskeleton genes are over-represented. Our data support the notion that DNA methylation contributes to the tissue- and cell type-specific gene expression in the CNS. Recently, it has been reported that neuronal activity regulates gene expression through the induction of DNA demethylation (33,34), and these activity-dependent demethylation modulates excitatory neurotransmission and adult neurogenesis. Thus, our genetic model of Dnmt1 gene deletion in cortical excitatory neurons will be very useful to analyze the effect of DNA demethylation on activity-dependent neuronal gene regulation.
Deregulation of ion channel expression underlies defects in electrophysiological parameters in hypomethylated neurons
The survival of hypomethylated neurons in postnatal CNS allows us to probe electriphysiological defects in these cells. We have previously recorded thalmo-cortical projection pathways in this line of Emx1-cre; Dnmt1 mutant mice (35). We showed that hypomethylated neurons are unable to form somatosensory barrel maps in the cortex despite the ability for hypomethylated neurons to receive and elicit excitatory post-synaptic current upon thalamocortical projection stimulation. Nevertheless, recordings from hypomethylated cortical neurons revealed severely reduced long-term potentiation upon thalamocortical pathway stimulation. Our current study indicated that post-synaptic cortical neurons exhibit defects in neuronal excitability, which may underlie the observed defect in thalmo-cortical long-term potentiation.
Changes in ion channel expression, specifically Kv4.2, may result from direct cell-autonomous effects of demethylation or indirectly result from failure of maturation of intrinsic ionic currents because of degeneration of cortical neurons in the demethylated cortex. We favor a direct effect because electrophysiological studies in animals with severely disturbed cortical lamination do not show changes in AP repolarization (36,37). One explanation for the decrease in Kv4.2 protein without the concomitant decrease in mRNA levels is post-translational decay mediated by microRNA pathways. However, increased degradation, or perturbation of channel protein integration into cell membrane could also leads to a decrease in protein without alternation of mRNA level. Further molecular characterization of the expression of ion channels will help elucidate molecular mechanisms underlying the aberrant electrophysiological profiles of Dnmt1−/− neurons.
It has been known that the maturation of interneuronal gene expression depends on neuronal activity and trophic support (38,39). Moreover, the effect of excitatory neurons on interneurons is not only through neurotransmitters but only via neurotrophic factors, such as BDNF (40). Thus, although interneurons are present in the mutant brain, their physiological functions may be impaired as a consequence of the loss of demethylated excitatory neurons.
DNA methylation in neurodegeneration and aging
Recently, improper DNA methylation has been indirectly linked to neurodegenerative diseases. A pathological hallmark of these diseases is targeted neuronal degeneration leading to cell death. Apoptotic cell death pathways appear to be activated in neurodegenerative disorders (41). It is conceivable that genes activated during apoptosis are preferentially silenced in healthy cells. However, upon injury or insult de-repression of pro-apoptotic genes is necessary to trigger programmed cell death. Consistent with this hypothesis, inducing hypomethylation via depletion of the methionine pool in cultured primary neurons results in apoptosis (42). Interestingly, it has been postulated that folate deficiency, and by extension DNA hypomethylation, may enhance neural vulnerability to excitotoxic insults and oxidative stress in neurodegenerative diseases such as Alzheimer's disease and Parkinson's disease (42,43). Thus, biochemical evidence suggests a putative link between hypomethylation-induced gene de-regulation and apoptosis in neurodegenerative disorders.
The generation of Emx1-cre; Dnmt1 mutant mice creates a model system of severe cortical and hippocampal degeneration during early postnatal stages in mutant mice. Thus, our animal model will be valuable to study both behavioral and physiological parameters in association with cortical and hippcampal functions. Moreover, the severe disruption of cortical and hippocampal structures will provide a system to test the ability of in vitro generated neural precursor cells to replenish these degenerated structures via transplant therapy. While Emx1-cre; Dnmt1 mutant cortex is degenerated, the thalamus and striatum structures are still present (Fig. 2). We propose that Emx1-cre; Dnmt1 mutant mice is also a useful model system to examine the effects of severe cortical degeneration on target structures in thalmo-cortical and cortico-striatal pathways.
MATERIALS AND METHODS
Generation of Emx1-cre; Dnmt1 conditional mutants and southern analysis
The use of transgenic mice is approved by UCLA Institutional Animal Research Committee. Using the cre-loxP binary gene deletion strategy, we crossed female mice homozygous for the Dnmt1 conditional allele Dnmt12lox (14) with male mice carrying the Emx1-cre transgene (kindly provided by Dr S. Itohara at RIKEN Brain Research Institute, Japan). Control heterozygous and mutant offspring were obtained in expected Mendelian ratios. To assess the cre-recombination efficiency, we performed Southern hybridization using genomic DNA samples procured from dorsal cortex at multiple ages. We also used the IAP repeat probe to detect global methylation levels in the same DNA samples. Detailed explanation of probes used and methods were previously described (14). This conditional line was crossed to two reporter strains, the R26R ßgeo line (20) and the M4-egfp bac transgenic (26) for further analysis.
Histology, immunohistochemistry, TUNEL analysis and immunoblotting
Histological examination was performed on mouse brain sections collected from samples fixed with 2 or 4% PFA via cardiac perfusion (postnatal samples) or overnight immersion fixation (embryonic samples), cryoprotected in two changes of 30% sucrose, and cut in the coronal orientation using a cryostat (Leica CM1900). Sections were stored at −80°C until use. After thawing and hydration, sections were stained using 0.1% cresyl violet, dehydrated through alcohols and xylenes, and mounted with Permount (Fisher). LacZ detection was performed as previously described (5). Immunohistochemistry was performed from sections collected in the manner described earlier.
For TUNEL analysis, sections fixed with 4% PFA/PBS and stained using the Apoptag Fluoroscein In Situ Apoptosis Detection Kit (Chemicon) following the supplied manufacturer's protocol. For double labeling experiments, sections were first incubated with tDT enzyme as per kit instructions, then washed and incubated in 10% NGS in PBS to block nonspecific interactions for 1 h. Primary antibodies were added for overnight incubation at room temperature, then secondary antibodies were added in the dilution buffer supplied in the Apoptag kit.
Immunoblotting was performed as previously described (16). Antibodies and dilutions used were rabbit anti-Dnmt1 (PATH52, a gift from Dr T. Bestor, Columbia University), and mouse anti-GAPDH (Abcam, 1:5000).
Behavioral analysis
Spatial learning was assessed with the hidden platform version of the Morris water maze (44), as described previously (45). Briefly, animals were trained with two trials per day for 10 days at which time a probe trial with the platform removed was administered. As learning in all groups was low (data not shown), an additional 4 days of four trials per day was given at which time the final probe trial was administered. Acquisition data are presented as the group averages, in which escape latencies across four trials were averaged for each animal. On the day following the final training/probe trial, animals received three additional trials, in which the platform was clearly marked (visible platform test). Escape latencies for all trials during the visible-platform test are averaged for each animal. Statistical comparisons were made between groups (% time in training quadrant) with Student's t-tests. In addition, the percentage of time spent in the training quadrant was compared with that of a chance performance (25%) with a single group comparison. A selective search strategy was indicated if animals performed significantly above chance. The fear conditioning behavioral test follows up the procedure as described in Costa et al. (30).
Electrophysiology
Detailed methods on patch-clamp recordings from neurons in brain slices have been reported in the co-authors previous publications (46,47). P1–P73 control and mutant animals were anesthetized with halothane. Cortical slices were cut at the coronal angle or thalamocortical angle and incubated in ACSF at 35°C for 30 min and then at room temperature. Electrodes for somatic recordings were pulled from borosilicate glass capillaries to a final resistance of 3–6 mega Ohms (Warner Instruments). Biocytin (3 mg/ml) was also added to a potassium gluconate-based internal solution during most recordings. Somatic whole-cell recordings were performed with an Optopatch patch-clamp amplifier (Cairn Research, Kent, UK) in the current clamp configuration, from layer IV neurons in control slices. As cortical lamination was disrupted in mutant cortex, an attempt was made to record from neurons at approximately similar proportional distances from the pial surface, as in control cortical neurons. M4+ EGFP neurons were located in layer V in control cortex and in the inferior half of the cortical mantle in mutant cortex. Voltage-gated potassium currents were recorded in the outside out patch configuration in the presence of tetrodotoxin (500 nm). Current and voltage clamp traces were analyzed using custom made programs with Igor analysis software (Wavemetrics, Lake Oswego, OR, USA). Biocytin and tetrodotoxin were purchased from Sigma (St Louis, MO, USA). APs were elicited after hyperpolarizing all cells to −70 mV to reduce the effect of differing resting membrane potential on AP morphology.
Biocytin immunocytochemistry was performed as outlined (48). Cells were reconstructed at ×100 using camera lucida techniques. Sholl analysis was performed by superimposing concentric circles of onto reconstructed neurons and counting the number of dendritic intersections. Soma area was calculated using NIH Image software.
Microarray hybridization
Gene expression microarrays were done with Agilent Whole-Genome microarrays (G4122A) using the suggested protocol. Briefly, we ran the isolated RNA through a Qiagen RNeasy minElute column (Qiagen) and tested the quality of the RNA on a NanoChip (Agilent). We converted the RNA into cDNA and then the cDNA into cRNA using the Agilent Low RNA Input Linear Amplification Kit (Agilent). We using a Nanodrop (Nanodrop) to quantify the labeled cRNA and used 0.75 µg of each sample for hybridization. Probes were fragmented in a mix of labeled probes, 10× blocking reagent (Agilent) and 25× Fragmentation buffer (Agilent). Reaction was stopped with the addition of 2× Hybridization buffer (Agilent). We used Agilent Whole-Genome microarrays for expression studies. Slides were hybridized at 65° for 17 h at 4 revolutions per minute and then washed once in Agilent Gene Expression wash buffer 1 and once in Agilent Gene Expression wash buffer 2 before a quick wash in acetonitrile. Slides were scanned immediately after washing to prevent ozone degradation. Arrays were performed in triplicate. Microarray data described in this paper can be accessed with GEO number GSE14216.
Quantitative reverse transcription polymerase chain reaction
RNA was DNase I treated (Invitrogen) and then quantified again. cDNA conversion was done using the iScript kit (Bio-Rad). Quantitative PCR was done on an MyIQ Thermocycler (Bio-Rad) using the Sybr Green Supermix (Bio-Rad).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
FUNDING
This work was supported by National Institutes of Health (RO1 NS051411 to G.F.). L.K.H. was supported by NRSA 5F31NS051005 from NINDS and the ARCS Foundation. G.F. is a Carol Moss Spivak Scholar in Neuroscience.
ACKNOWLEDGEMENTS
We thank David Savin and Justin Sharim for processing and analyzing the camera lucida drawings.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Bestor T.H. The DNA methyltransferases of mammals. Hum. Mol. Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 2.Robertson K.D., Wolffe A.P. DNA methylation in health and disease. Nat. Rev. Genet. 2000;1:11–19. doi: 10.1038/35049533. [DOI] [PubMed] [Google Scholar]
- 3.Suzuki M.M., Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 4.Goto K., Numata M., Komura J.I., Ono T., Bestor T.H., Kondo H. Expression of DNA methyltransferase gene in mature and immature neurons as well as proliferating cells in mice. Differentiation. 1994;56:39–44. doi: 10.1046/j.1432-0436.1994.56120039.x. [DOI] [PubMed] [Google Scholar]
- 5.Feng J., Fouse S.D., Fan G. Epigenetic regulation of neural gene expression and neuronal function. Pediatr. Res. 2007;61:58R–63R. doi: 10.1203/pdr.0b013e3180457635. [DOI] [PubMed] [Google Scholar]
- 6.Watt F., Molloy P.L. Cytosine methylation prevents binding to DNA of a HeLa cell transcription factor required for optimal expression of the adenovirus major late promoter. Genes Dev. 1988;2:1136–1143. doi: 10.1101/gad.2.9.1136. [DOI] [PubMed] [Google Scholar]
- 7.Klose R.J., Bird A.P. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 8.Amir R.E., Van den Veyver I.B., Wan M., Tran C.Q., Francke U., Zoghbi H.Y. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 9.Martinowich K., Hattori D., Wu H., Fouse S., He F., Hu Y., Fan G., Sun Y.E. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302:890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- 10.Chen W.G., Chang Q., Lin Y., Meissner A., West A.E., Griffith E.C., Jaenisch R., Greenberg M.E. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003;302:885–889. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- 11.Horike S., Cai S., Miyano M., Cheng J.F., Kohwi-Shigematsu T. Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat. Genet. 2005;37:31–40. doi: 10.1038/ng1491. [DOI] [PubMed] [Google Scholar]
- 12.Chen R.Z., Akbarian S., Tudor M., Jaenisch R. Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat. Genet. 2001;27:327–331. doi: 10.1038/85906. [DOI] [PubMed] [Google Scholar]
- 13.Guy J., Hendrich B., Holmes M., Martin J.E., Bird A. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat. Genet. 2001;27:322–326. doi: 10.1038/85899. [DOI] [PubMed] [Google Scholar]
- 14.Fan G., Beard C., Chen R.Z., Csankovszki G., Sun Y., Siniaia M., Biniszkiewicz D., Bates B., Lee P.P., Kuhn R., et al. DNA hypomethylation perturbs the function and survival of CNS neurons in postnatal animals. J. Neurosci. 2001;21:788–797. doi: 10.1523/JNEUROSCI.21-03-00788.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takizawa T., Nakashima K., Namihira M., Ochiai W., Uemura A., Yanagisawa M., Fujita N., Nakao M., Taga T. DNA methylation is a critical cell-intrinsic determinant of astrocyte differentiation in the fetal brain. Dev. Cell. 2001;1:749–758. doi: 10.1016/s1534-5807(01)00101-0. [DOI] [PubMed] [Google Scholar]
- 16.Fan G., Martinowich K., Chin M.H., He F., Fouse S.D., Hutnick L., Hattori D., Ge W., Shen Y., Wu H., et al. DNA methylation controls the timing of astrogliogenesis through regulation of JAK-STAT signaling. Development. 2005;132:3345–3356. doi: 10.1242/dev.01912. [DOI] [PubMed] [Google Scholar]
- 17.Shimozaki K., Namihira M., Nakashima K., Taga T. Stage- and site-specific DNA demethylation during neural cell development from embryonic stem cells. J. Neurochem. 2005;93:432–439. doi: 10.1111/j.1471-4159.2005.03031.x. [DOI] [PubMed] [Google Scholar]
- 18.Iwasato T., Datwani A., Wolf A.M., Nishiyama H., Taguchi Y., Tonegawa S., Knopfel T., Erzurumlu R.S., Itohara S. Cortex-restricted disruption of NMDAR1 impairs neuronal patterns in the barrel cortex. Nature. 2000;406:726–731. doi: 10.1038/35021059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gorski J.A., Talley T., Qiu M., Puelles L., Rubenstein J.L., Jones K.R. Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J. Neurosci. 2002;22:6309–6314. doi: 10.1523/JNEUROSCI.22-15-06309.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 21.Kuff E.L., Fewell J.W. Intracisternal A-particle gene expression in normal mouse thymus tissue: gene products and strain-related variability. Mol. Cell. Biol. 1985;5:474–483. doi: 10.1128/mcb.5.3.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walsh C.P., Bestor T.H. Cytosine methylation and mammalian development. Genes Dev. 1999;13:26–34. doi: 10.1101/gad.13.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaudet F., Rideout W.M., 3rd, Meissner A., Dausman J., Leonhardt H., Jaenisch R. Dnmt1 expression in pre- and postimplantation embryogenesis and the maintenance of IAP silencing. Mol. Cell. Biol. 2004;24:1640–1648. doi: 10.1128/MCB.24.4.1640-1648.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Molyneaux B.J., Arlotta P., Menezes J.R., Macklis J.D. Neuronal subtype specification in the cerebral cortex. Nat. Rev. Neurosci. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- 25.Fouse S.D., Shen Y., Pellegrini M., Cole S., Meissner A., Van Neste L., Jaenisch R., Fan G. Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation. Cell Stem Cell. 2008;2:160–169. doi: 10.1016/j.stem.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gong S., Zheng C., Doughty M.L., Losos K., Didkovsky N., Schambra U.B., Nowak N.J., Joyner A., Leblanc G., Hatten M.E., et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- 27.Birnbaum S.G., Varga A.W., Yuan L.L., Anderson A.E., Sweatt J.D., Schrader L.A. Structure and function of Kv4-family transient potassium channels. Physiol. Rev. 2004;84:803–833. doi: 10.1152/physrev.00039.2003. [DOI] [PubMed] [Google Scholar]
- 28.Rhodes K.J., Carroll K.I., Sung M.A., Doliveira L.C., Monaghan M.M., Burke S.L., Strassle B.W., Buchwalder L., Menegola M., Cao J., et al. KChIPs and Kv4 alpha subunits as integral components of A-type potassium channels in mammalian brain. J. Neurosci. 2004;24:7903–7915. doi: 10.1523/JNEUROSCI.0776-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jackson-Grusby L., Beard C., Possemato R., Tudor M., Fambrough D., Csankovszki G., Dausman J., Lee P., Wilson C., Lander E., et al. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat. Genet. 2001;27:31–39. doi: 10.1038/83730. [DOI] [PubMed] [Google Scholar]
- 30.Costa R.M., Honjo T., Silva A.J. Learning and memory deficits in Notch mutant mice. Curr. Biol. 2003;13:1348–1354. doi: 10.1016/s0960-9822(03)00492-5. [DOI] [PubMed] [Google Scholar]
- 31.Chen T., Hevi S., Gay F., Tsujimoto N., He T., Zhang B., Ueda Y., Li E. Complete inactivation of DNMT1 leads to mitotic catastrophe in human cancer cells. Nat. Genet. 2007;39:391–396. doi: 10.1038/ng1982. [DOI] [PubMed] [Google Scholar]
- 32.Li H.S., Wang D., Shen Q., Schonemann M.D., Gorski J.A., Jones K.R., Temple S., Jan L.Y., Jan Y.N. Inactivation of Numb and Numblike in embryonic dorsal forebrain impairs neurogenesis and disrupts cortical morphogenesis. Neuron. 2003;40:1105–1118. doi: 10.1016/s0896-6273(03)00755-4. [DOI] [PubMed] [Google Scholar]
- 33.Nelson E.D., Kavalali E.T., Monteggia L.M. Activity-dependent suppression of miniature neurotransmission through the regulation of DNA methylation. J. Neurosci. 2008;28:395–406. doi: 10.1523/JNEUROSCI.3796-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma D.K., Jang M.H., Guo J.U., Kitabatake Y., Chang M.L., Pow-Anpongkul N., Flavell R.A., Lu B., Ming G.L., Song H. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323:1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Golshani P., Hutnick L., Schweizer F., Fan G. Conditional Dnmt1 deletion in dorsal forebrain disrupts development of somatosensory barrel cortex and thalamocortical long-term potentiation. Thalamus Relat. Syst. 2005;3:227–233. doi: 10.1017/S1472928807000222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luhmann H.J., Karpuk N., Qu M., Zilles K. Characterization of neuronal migration disorders in neocortical structures. II. Intracellular in vitro recordings. J. Neurophysiol. 1998;80:92–102. doi: 10.1152/jn.1998.80.1.92. [DOI] [PubMed] [Google Scholar]
- 37.Silva L.R., Gutnick M.J., Connors B.W. Laminar distribution of neuronal membrane properties in neocortex of normal and reeler mouse. J. Neurophysiol. 1991;66:2034–2040. doi: 10.1152/jn.1991.66.6.2034. [DOI] [PubMed] [Google Scholar]
- 38.Chattopadhyaya B., Di Cristo G., Higashiyama H., Knott G.W., Kuhlman S.J., Welker E., Huang Z.J. Experience and activity-dependent maturation of perisomatic GABAergic innervation in primary visual cortex during a postnatal critical period. J. Neurosci. 2004;24:9598–9611. doi: 10.1523/JNEUROSCI.1851-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang B., Kitamura A., Yasuda H., Sohya K., Maruyama A., Yanagawa Y., Obata K., Tsumoto T. Brain-derived neurotrophic factor acutely depresses excitatory synaptic transmission to GABAergic neurons in visual cortical slices. Eur. J. Neurosci. 2004;20:709–718. doi: 10.1111/j.1460-9568.2004.03523.x. [DOI] [PubMed] [Google Scholar]
- 40.Jin X., Hu H., Mathers P.H., Agmon A. Brain-derived neurotrophic factor mediates activity-dependent dendritic growth in nonpyramidal neocortical interneurons in developing organotypic cultures. J. Neurosci. 2003;23:5662–5673. doi: 10.1523/JNEUROSCI.23-13-05662.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mattson M.P., Kruman I.I., Duan W. Folic acid and homocysteine in age-related disease. Ageing Res. Rev. 2002;1:95–111. doi: 10.1016/s0047-6374(01)00365-7. [DOI] [PubMed] [Google Scholar]
- 42.Kruman I.I., Culmsee C., Chan S.L., Kruman Y., Guo Z., Penix L., Mattson M.P. Homocysteine elicits a DNA damage response in neurons that promotes apoptosis and hypersensitivity to excitotoxicity. J. Neurosci. 2000;20:6920–6926. doi: 10.1523/JNEUROSCI.20-18-06920.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duan W., Ladenheim B., Cutler R.G., Kruman I.I., Cadet J.L., Mattson M.P. Dietary folate deficiency and elevated homocysteine levels endanger dopaminergic neurons in models of Parkinson's disease. J. Neurochem. 2002;80:101–110. doi: 10.1046/j.0022-3042.2001.00676.x. [DOI] [PubMed] [Google Scholar]
- 44.Morris R.G.M. Spatial localization does not require the presence of local cues. Learning and Motivation. 1981;12:239–236. [Google Scholar]
- 45.Bourtchuladze R., Frenguelli B., Blendy J., Cioffi D., Schutz G., Silva A.J. Deficient long-term memory in mice with a targeted mutation of the cAMP-responsive element-binding protein. Cell. 1994;79:59–68. doi: 10.1016/0092-8674(94)90400-6. [DOI] [PubMed] [Google Scholar]
- 46.Golshani P., Jones E.G. Synchronized paroxysmal activity in the developing thalamocortical network mediated by corticothalamic projections and ‘silent’ synapses. J. Neurosci. 1999;19:2865–2875. doi: 10.1523/JNEUROSCI.19-08-02865.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Golshani P., Liu X.B., Jones E.G. Differences in quantal amplitude reflect GluR4- subunit number at corticothalamic synapses on two populations of thalamic neurons. Proc. Natl. Acad. Sci. USA. 2001;98:4172–4177. doi: 10.1073/pnas.061013698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hamam B.N., Kennedy T.E. Visualization of the dendritic arbor of neurons in intact 500 microm thick brain slices. J. Neurosci. Methods. 2003;123:61–67. doi: 10.1016/s0165-0270(02)00341-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.