Abstract
Outside the pseudoautosomal regions, the mammalian sex chromosomes are thought to have been genetically isolated for up to 350 million years. However, in humans pathogenic XY translocations occur in XY-homologous (gametologous) regions, causing sex-reversal and infertility. Gene conversion might accompany recombination intermediates that resolve without translocation and persist in the population. We resequenced X and Y copies of a translocation hotspot adjacent to the PRKX and PRKY genes and found evidence of historical exchange between the male-specific region of the human Y and the X in patchy flanking gene-conversion tracts on both chromosomes. The rate of X-to-Y conversion (per base per generation) is four to five orders of magnitude more rapid than the rate of Y-chromosomal base-substitution mutation, and given assumptions about the recombination history of the X locus, tract lengths have an overall average length of ∼100 bp. Sequence exchange outside the pseudoautosomal regions could play a role in protecting the Y-linked copies of gametologous genes from degeneration.
Main Text
The human sex chromosomes derive from an ancestral pair of autosomes;1 the Y chromosome has degenerated so that normal crossover with the X is now restricted to the specialized XY-homologous pseudoautosomal regions (Figure 1A). Other XY-homologous loci (“gametologs”2) are regarded as evolutionarily isolated and include 25 genes with functional copies on both sex chromosomes.3 Although gene conversion (nonreciprocal transfer of sequence without crossover) is frequent4–6 among sequences on the male-specific region of the Y chromosome (MSY7), the highly resolved MSY binary-marker phylogeny8 reveals no signal of recombination with other chromosomes. The only evidence to date for inter-gametolog exchange in mammals is the observation of two gene-conversion events in the ZFX (MIM 314980)/ZFY (MIM 490000) gene pair during 10–15 million years of evolution within the Felidae.9
Figure 1.
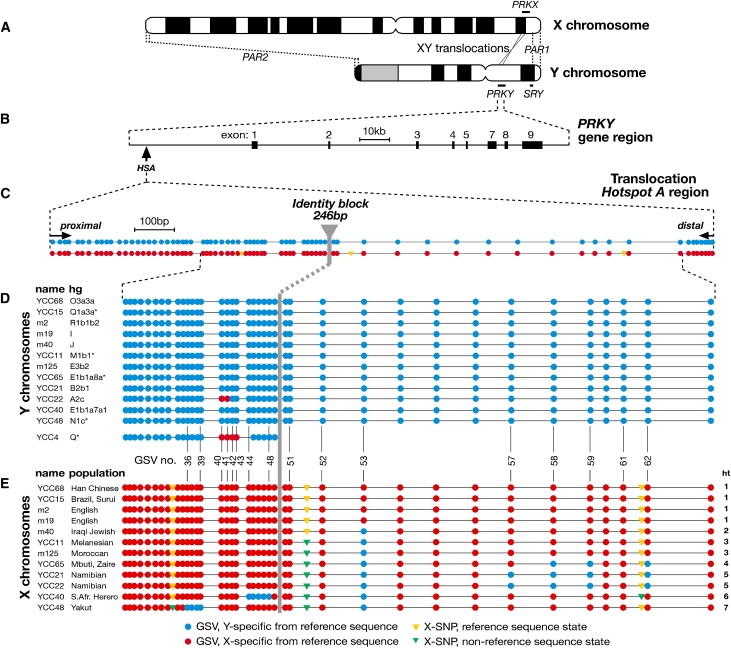
XY Translocations between PRKX and PRKY, the PRKY Gene Region, and Gametologous Sequence Variants and Conversions around Translocation Hotspot A
(A) Idiograms of the sex chromosomes (not to scale) showing the pseudoautosomal regions (PAR1, PAR2), the SRY gene, and the PRKX and PRKY gene pair.
(B) The PRKY gene region, showing positions of exons and HSA. Exon numbering is based on PRKX13, so exon 6 is absent. Gene orientation varies as a result of a polymorphic Yp inversion14, and the orientation shown is that permissive for XY translocation15 (opposite to the Y reference sequence7). There is no reason to expect orientation to influence the probability of conversion.
(C) Sequence states of GSVs in the Y- and X-chromosomal reference sequences of an ∼1.9 kb region encompassing HSA. The positions of 66 surveyed gametologous sequence variants (GSVs) are indicated by circles (see key below figure), and some are numbered sequentially. Y- and X-specific primers (arrows) cover the proximal- and distal-most four GSVs.
(D) Sequence states of GSVs in Y-chromosomal copies of part of the region. Names of DNA samples and their Y-chromosomal haplogroups8 are given to the left. Samples with the prefix “YCC” are from the Y Chromosome Consortium, and haplogroup information is as described.8,21 Other human samples of known haplogroup are from collections of the authors. Below is shown the shorter region surveyed in the Y chromosome of sample YCC4.
(E) Sequence states of GSVs in X-chromosomal copies; DNA sample names and their populations of origin are given to the left. Haplotypes (ht) are numbered to the right.
Translocations between the short arms of the human sex chromosomes can transfer the sex-determining SRY gene (MIM 480000), causing sex reversal10 (46,XX males [MIM 278850] and 46,XY females [MIM 233420]), demonstrating that recombination intermediates can indeed form between gametologs (Figure 1A). In the best-understood class of such translocations, nonallelic homologous recombination (NAHR) occurs within an approximately 140 kb region encompassing the PRKX (MIM 300083) and PRKY (MIM 400008) gametologous gene pair11,12 (Figure 1B). Overall sequence similarity between these regions, lying in stratum 5 of the X chromosome and recombinationally isolated 29–32 million years ago3, is 94%. Most breakpoints cluster in one of two hotspots (hotspot A [HSA]: 12/30 cases13); exchange in this hotspot has occurred within a 246 bp segment of sequence identity.
Because of their associated infertility, PRKX/Y recombination intermediates resolving with translocation yield products that are lost from the population. However, resolution of intermediates can also occur without translocation, and we hypothesized that branch migration, heteroduplex formation, and repair prior to resolution could lead to heritable genetic exchange (gene conversions) between the sex chromosomes. To seek evidence of this, we resequenced the X and Y copies of the HSA region in normal males; we reasoned that past gene conversion events would be indicated by switching of GSVs (gametologous sequence variants) from the sequence states usually observed for one sex chromosome to those of the other.
We chose twelve males, all sampled with informed consent in accordance with appropriate institutional and national ethical approval, for resequencing to maximize the chance of observing informative variants: their populations of origin were diverse (thereby maximizing X diversity), and their Y chromosomes belonged to a wide range of haplogroups (hgs) in the Y phylogeny. We designed primers in regions of high X-Y divergence to amplify ∼1.9 kb X- and Y-specific segments (Table S1 in the Supplemental Data available online), within which a comparison of the X- and Y-chromosomal reference sequences (Figure S1) revealed a total of 66 GSVs centered on the 246 bp identity block (Figure 1C).
Eleven of the 12 Y-chromosomal sequences were identical to the reference sequence. However, in one hgA2c Y chromosome (YCC22) carried by a Namibian male, two GSVs lying 4 bp apart differed (Figure 1D). In principle, differences could be due either to mutation or to gene conversion from the X gametolog. Here, gene conversion is the only plausible explanation because the variant GSVs have both switched to X-chromosomal sequence states and lie adjacent to each other, such that they represent a single gene conversion tract that is between 4 and 138 bp in length (the maximum length is the distance between the two GSVs flanking the conversion tract); neither GSV corresponds to a highly mutable CpG dinucleotide. This represents the first evidence, to our knowledge, of heritable genetic exchange between the human sex chromosomes outside the pseudoautosomal regions. We surveyed the same subregion in a sample of 23 additional diverse Y chromosomes (Table S2) and found the same tract in two additional hgA2 Y chromosomes, suggesting common ancestry for the event. Surprisingly, we also found an overlapping but longer tract of conversion, involving four GSVs, in a chromosome belonging to hgQ. Survey of a further 32 diverse hgQ chromosomes from the Americas, Central Asia, and Europe failed to provide any other examples, so this independent conversion event remains a singleton.
Given a figure for the number of generations encompassed within the phylogeny relating all sampled chromosomes, we can estimate a likely range of rates for X-to-Y conversion in this region by basing our approach on that of Repping et al.14 In their study, they resequenced ∼80 kb of DNA in each of 47 Y chromosomes covering most of the major branches of the Y phylogeny to ascertain nucleotide divergence without bias; this revealed a total of 95 base substitutions. Assuming a TMRCA of 118,000 years and a 25 year generation time, they estimated that 47 chromosomes encompassed 52,000 generations.14 The 67 Y chromosomes we surveyed also included most of the haplogroups in the tree, but they also contained multiple representatives for individual haplogroups, and the number of additional mutations these would contribute is uncertain. We estimated a likely lower and upper bound on the time encompassed in the tree, as follows: for the lower bound we assumed that each additional chromosome in a given major haplogroup contributed no additional mutations in excess of the haplogroup-specific branch length given by Repping et al.14, and for the upper bound we assumed that each additional chromosome contributed an additional number of mutations equivalent to the number it would have contributed had it descended from the root of the tree independently. We also assumed that hgG, unsampled by Repping et al.,14 contributed nine mutations (close to the average height of the tree). This led to a range of total mutations of 177–557, corresponding to 96,535–302,080 generations. The observation of two gene-conversion events within this time range corresponds to a rate of ∼6.6 × 10−6 to ∼2.1 × 10−5 events per generation.
To compare this conversion rate meaningfully with that of mutation by base substitution, we can consider it as an effective rate of change per base per generation by multiplying the rate for conversion events by the number of converted base pairs. Given the distributions of GSVs, the two events we observe convert between 22 and 325 bp of converted DNA in total. This yields a range for the effective conversion rate per base per generation of ∼1.45 × 10−4 to ∼6.82 × 10−3—four to five orders of magnitude more rapid than the rate of Y-chromosomal base-substitution mutation (2.3 × 10−8 per base per generation14). The rate is of a similar order to the rate calculated for Y-Y conversion (2.2 × 10−4) between the arms of palindromes;4 these are >99.8% similar in sequence due to conversion because the arms share a single evolutionary history, and conversion eliminates variants as they arise. The X and Y copies of HSA, by contrast, have independent evolutionary histories, so independent conversions can maintain diversity among sequences. Note that the per-base-per-generation rate we estimate is for all bases, regardless of whether they are GSVs detectable in conversion. Given that the overall average similarity between the X and Y gametologs in the studied 1890 bp region is ∼96.5%, only 3.5% of converted bases on average will be detectable as switched GSVs.
How does the conversion rate compare with the rate of XY translocation at HSA? The population occurrence of PRKX/PRKY translocation XX males has been estimated as 1/80,00015, and we assume that this represents the rate at which XY translocations give rise to derived X chromosomes. The reciprocal translocation product is a derived Y chromosome, which should arise in sperm at the same rate, although it is observed in the population (in XY females) much more rarely.15 The likely overall rate of PRKX/PRKY translocation in sperm is therefore ∼1/40,000. Of such translocations, 12/30 have been observed to be due to translocation at HSA,13 yielding a rate of ∼1 × 10−5 per generation, which is of a similar order to the conversion rate that we observe.
The 12 X chromosomes analyzed showed much greater diversity than the Y chromosomes: seven distinct haplotypes involved 17 variable sites (Figure 1E). We know that three sites are not GSVs because the human reference sequences do not differ, and orthologous sequence states in chimpanzee X and Y chromosomes indicate that they are most probably SNPs arising on the human X chromosome. The remaining 14 sites represent switches to the states seen in the Y reference sequence, and only one of them lies at a CpG dinucleotide: they are therefore probably gene conversions. The X haplotypes include four examples where more than one adjacent GSV is switched, and two involving isolated GSVs. Interpretation of patterns on the X is more complex than that on the Y because recombination among X chromosomes in female meiosis could divide or unify individual conversion tracts. However, if we assume that each set of adjacent GSVs represents a single conversion tract, then six gene conversion events is the minimum required to explain the observed haplotypes, and tract lengths lie in a range between 1 and 269 bp. If we include Y-chromosomal events, the average length of gene conversion tracts (estimated according to Wolf et al.16) is ∼100 bp, around 3-fold higher than the average length estimated for Y-chromosomal inter-paralog conversion.5
The lack of a temporal phylogenetic framework means that a conversion rate for the X chromosome cannot be estimated as was done for the Y. However, although more X- than Y-chromosomal conversions have been observed here, this does not necessarily imply a true rate difference because three times as many observed events are expected on the X as a result of its larger effective population size. None of the observed conversion events are contiguous with the identity block at the heart of HSA, suggesting patchy conversion within extended or migrated heteroduplexes. A single perfect consensus motif17 associated with recombination and genome instability exists in the X-chromosomal copy of the sequenced region (it is disrupted by a GSV on the Y), but its significance is hard to judge because it lies several hundred base pairs away from the identity block and the observed conversion events. Notably, 10/66 GSVs in the studied region are also annotated as X-chromosomal SNPs in dbSNP, and in each case the two alleles correspond to the two GSV states. This could represent either further evidence for gene conversion or misannotation of GSVs as SNPs: all except one are unvalidated and were discovered in a panel of five DNA donors of unspecified sex. This underscores the need for careful chromosome-specific analysis in the search for conversion events.
Comparing the human sequences with orthologs from great apes allowed us to ask whether conversion in this region has a deeper evolutionary history. We extracted the chimpanzee X- and Y-linked orthologs and the gorilla X-linked ortholog from sequence databases and aligned them (Figure S2). In the absence of X-Y exchange, the phylogeny should be dominated by the X-Y divergence (because this is ∼5 times more ancient than the divergence of the most recent common ancestor of the three species), bifurcating, and lacking in reticulations. However, for HSA the phylogeny is strongly reticulated (Figure 2), suggesting a history of conversion. Eighteen examples of converted sites are evident in the sequence alignments (Figure S2) and, like the conversions observed among the human sequences, cluster around the location of the human identity block as well as more distally. Three of the sites demonstrate Y-to-X conversions, but the opposite direction of conversion cannot be reliably identified because there is no gorilla Y sequence in the comparisons.
Figure 2.
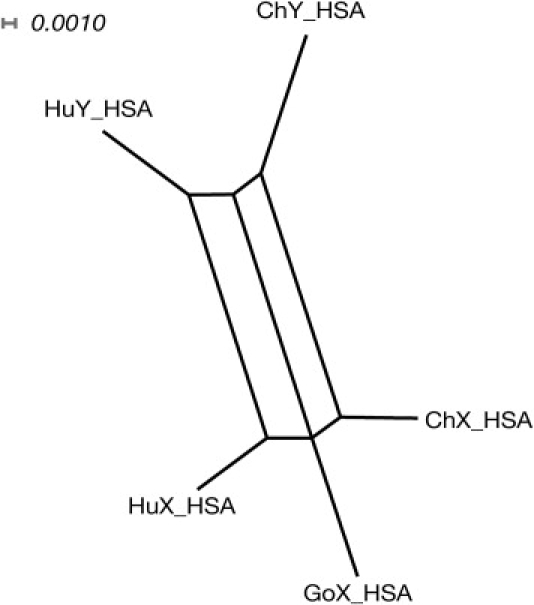
Phylogenetic Network of HSA Sequences in Humans and Great Apes
Phylogenetic split network (based on alignment shown in Figure S2) showing the relationships between HSA segments, produced with SplitsTree422 via the “NeighborNet” method and the “uncorrectedP” distance. Lengths of edges represent the proportion of sites at which sequences differ, as indicated by the scale-bar. The reticulated structure indicates a history of gene conversion. Abbreviations are as follows: Hu, human; Ch, chimpanzee; and Go, gorilla.
Our findings suggest that the traditional view of genetically isolated human sex chromosomes requires revision because sequence variants can be exchanged outside the pseudoautosomal regions. Other known hotspots for recurrent translocations, such as those involving the regions around the steroid sulfatase gene (STS [MIM 300747]) and its Y-linked pseudogene18, as well as less well-characterized examples such as those causing XYXq syndrome19, may also be sites of conversion activity. However, although it is likely that translocations in some regions would be lethal and/or would yield acentric or dicentric chromosomes, the same would not be true of conversions, so gametologous exchange could be more general. It could play a key role in the molecular evolution of the ∼8.6 Mb3 of gametologous sequences on the X and Y chromosomes and counteract the degeneration20 of the Y-linked copies of gametologous genes.
Acknowledgments
We thank Raymond Dalgleish for assistance and Celia May for helpful comments. Z.H.R. and P.B. were supported by the Wellcome Trust, and M.A.J. was supported by a Wellcome Trust Senior Fellowship in Basic Biomedical Science (grant no. 057559).
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim
SplitsTree4, http://www.splitstree.org/
Accession Numbers
The GenBank accession numbers for the variant Y-chromosomal sequences around HSA reported in this paper are GQ281274 and GQ281275, and those for the variant X-chromosomal sequences around HSA are GQ281268–GQ281273.
References
- 1.Ohno S. Springer-Verlag; Berlin: 1967. Sex chromosomes and sex-linked genes. [Google Scholar]
- 2.Garcia-Moreno J., Mindell D.P. Rooting a phylogeny with homologous genes on opposite sex chromosomes (gametologs): A case study using avian CHD. Mol. Biol. Evol. 2000;17:1826–1832. doi: 10.1093/oxfordjournals.molbev.a026283. [DOI] [PubMed] [Google Scholar]
- 3.Ross M.T., Grafham D.V., Coffey A.J., Scherer S., McLay K., Muzny D., Platzer M., Howell G.R., Burrows C., Bird C.P. The DNA sequence of the human X chromosome. Nature. 2005;434:325–337. doi: 10.1038/nature03440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rozen S., Skaletsky H., Marszalek J.D., Minx P.J., Cordum H.S., Waterston R.H., Wilson R.K., Page D.C. Abundant gene conversion between arms of massive palindromes in human and ape Y chromosomes. Nature. 2003;423:873–876. doi: 10.1038/nature01723. [DOI] [PubMed] [Google Scholar]
- 5.Bosch E., Hurles M.E., Navarro A., Jobling M.A. Dynamics of a human interparalog gene conversion hotspot. Genome Res. 2004;14:835–844. doi: 10.1101/gr.2177404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adams S.M., King T.E., Bosch E., Jobling M.A. The case of the unreliable SNP: Recurrent back-mutation of Y-chromosomal marker P25 through gene conversion. Forensic Sci. Int. 2006;159:14–20. doi: 10.1016/j.forsciint.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 7.Skaletsky H., Kuroda-Kawaguchi T., Minx P.J., Cordum H.S., Hillier L., Brown L.G., Repping S., Pyntikova R., Ali J., Bieri T. The male-specific region of the human Y chromosome: A mosaic of discrete sequence classes. Nature. 2003;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- 8.Karafet T.M., Mendez F.L., Meilerman M., Underhill P.A., Zegura S.L., Hammer M.F. New binary polymorphisms reshape and increase resolution of the human Y-chromosomal haplogroup tree. Genome Res. 2008;18:830–838. doi: 10.1101/gr.7172008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pecon-Slattery J., Sanner-Wachter L., O'Brien S.J. Novel gene conversion between X-Y homologues located in the nonrecombining region of the Y chromosome in Felidae (Mammalia) Proc. Natl. Acad. Sci. USA. 2000;97:5307–5312. doi: 10.1073/pnas.97.10.5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferguson-Smith M.A. Karyotype-phenotype correlations in gonadal dysgenesis and their bearing on the pathogenesis of malformations. J. Med. Genet. 1965;39:142–155. doi: 10.1136/jmg.2.2.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weil D., Wang I., Dietrich A., Poustka A., Weissenbach J., Petit C. Highly homologous loci on the X and Y chromosomes are hot-spots for ectopic recombinations leading to XX maleness. Nat. Genet. 1994;7:414–419. doi: 10.1038/ng0794-414. [DOI] [PubMed] [Google Scholar]
- 12.Klink A., Schiebel K., Winkelmann M., Rao E., Horsthemke B., Lüdecke H.-J., Claussen U., Scherer G., Rappold G. The human protein kinase gene PKX1 on Xp22.3 displays Xp/Yp homology and is a site of chromosomal instability. Hum. Mol. Genet. 1995;4:869–878. doi: 10.1093/hmg/4.5.869. [DOI] [PubMed] [Google Scholar]
- 13.Schiebel K., Winkelmann M., Mertz A., Xu X., Page D.C., Weil D., Petit C., Rappold G.A. Abnormal XY interchange between a novel isolated protein kinase gene, PRKY, and its homologue, PRKX, accounts for one third of all (Y+)XX males and (Y-)XY females. Hum. Mol. Genet. 1997;6:1985–1989. doi: 10.1093/hmg/6.11.1985. [DOI] [PubMed] [Google Scholar]
- 14.Repping S., van Daalen S.K., Brown L.G., Korver C.M., Lange J., Marszalek J.D., Pyntikova T., van der Veen F., Skaletsky H., Page D.C. High mutation rates have driven extensive structural polymorphism among human Y chromosomes. Nat. Genet. 2006;38:463–467. doi: 10.1038/ng1754. [DOI] [PubMed] [Google Scholar]
- 15.Jobling M.A., Williams G., Schiebel K., Pandya A., McElreavey K., Salas L., Rappold G.A., Affara N.A., Tyler-Smith C. A selective difference between human Y-chromosomal DNA haplotypes. Curr. Biol. 1998;8:1391–1394. doi: 10.1016/s0960-9822(98)00020-7. [DOI] [PubMed] [Google Scholar]
- 16.Wolf A., Millar D.S., Caliebe A., Horan M., Newsway V., Kumpf D., Steinmann K., Chee I.S., Lee Y.H., Mutirangura A. A gene conversion hotspot in the human growth hormone (GH1) gene promoter. Hum. Mutat. 2009;30:239–247. doi: 10.1002/humu.20850. [DOI] [PubMed] [Google Scholar]
- 17.Myers S., Freeman C., Auton A., Donnelly P., McVean G. A common sequence motif associated with recombination hot spots and genome instability in humans. Nat. Genet. 2008;40:1124–1129. doi: 10.1038/ng.213. [DOI] [PubMed] [Google Scholar]
- 18.Ballabio A., Carrozzo R., Gil A., Gillard B., Affara N., Ferguson-Smith M.A., Fraser N., Craig I., Rocchi M., Romeo G. Molecular characterization of human X/Y translocations suggests their aetiology through aberrant exchange between homologous sequences on Xp and Yq. Ann. Hum. Genet. 1989;53:9–14. doi: 10.1111/j.1469-1809.1989.tb01117.x. [DOI] [PubMed] [Google Scholar]
- 19.Lahn B.T., Ma N., Breg W.R., Stratton R., Surti U., Page D.C. Xq-Yq interchange resulting in supernormal X-linked gene-expression in severely retarded males with 46,XYq karyotype. Nat. Genet. 1994;8:243–250. doi: 10.1038/ng1194-243. [DOI] [PubMed] [Google Scholar]
- 20.Graves J.A. Sex chromosome specialization and degeneration in mammals. Cell. 2006;124:901–914. doi: 10.1016/j.cell.2006.02.024. [DOI] [PubMed] [Google Scholar]
- 21.Y Chromosome Consortium A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res. 2002;12:339–348. doi: 10.1101/gr.217602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huson D.H., Bryant D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


