Abstract
Most human genes undergo alternative splicing, but aberrant splice forms are hallmarks of many cancers, usually resulting from mutations initiating abnormal exon skipping, intron retention, or the introduction of a new splice sites. We have identified a family of aberrant splice variants of HAS1 (the hyaluronan synthase 1 gene) in some B lineage cancers, characterized by exon skipping and/or partial intron retention events that occur either together or independently in different variants, apparently due to accumulation of inherited and acquired mutations. Cellular, biochemical, and oncogenic properties of full-length HAS1 (HAS1-FL) and HAS1 splice variants Va, Vb, and Vc (HAS1-Vs) are compared and characterized. When co-expressed, the properties of HAS1-Vs are dominant over those of HAS1-FL. HAS1-FL appears to be diffusely expressed in the cell, but HAS1-Vs are concentrated in the cytoplasm and/or Golgi apparatus. HAS1-Vs synthesize detectable de novo HA intracellularly. Each of the HAS1-Vs is able to relocalize HAS1-FL protein from diffuse cytoskeleton-anchored locations to deeper cytoplasmic spaces. This HAS1-Vs-mediated relocalization occurs through strong molecular interactions, which also serve to protect HAS1-FL from its otherwise high turnover kinetics. In co-transfected cells, HAS1-FL and HAS1-Vs interact with themselves and with each other to form heteromeric multiprotein assemblies. HAS1-Vc was found to be transforming in vitro and tumorigenic in vivo when introduced as a single oncogene to untransformed cells. The altered distribution and half-life of HAS1-FL, coupled with the characteristics of the HAS1-Vs suggest possible mechanisms whereby the aberrant splicing observed in human cancer may contribute to oncogenesis and disease progression.
About 70–80% of human genes undergo alternative splicing, contributing to proteomic diversity and regulatory complexities in normal development (1). About 10% of mutations listed so far in the Human Gene Mutation Database (HGMD) of “gene lesions responsible for human inherited disease” were found to be located within splice sites. Furthermore, it is becoming increasingly apparent that aberrant splice variants, generated mostly due to splicing defects, play a key role in cancer. Germ line or acquired genomic changes (mutations) in/around splicing elements (2–4) promote aberrant splicing and aberrant protein isoforms.
Hyaluronan (HA)3 is synthesized by three different plasma membrane-bound hyaluronan synthases (1, 2, and 3). HAS1 undergoes alternative and aberrant intronic splicing in multiple myeloma, producing truncated variants termed Va, Vb, and Vc (5, 6), which predicted for poor survival in a cohort of multiple myeloma patients (5). Our work suggests that this aberrant splicing arises due to inherited predispositions and acquired mutations in the HAS1 gene (7). Cancer-related, defective mRNA splicing caused by polymorphisms and/or mutations in splicing elements often results in inactivation of tumor suppressor activity (e.g. HRPT2 (8, 9), PTEN (10), MLHI (11–14), and ATR (15)) or generation of dominant negative inhibitors (e.g. CHEK2 (16) and VWOX (17)). In breast cancer, aberrantly spliced forms of progesterone and estrogen receptors are found (reviewed in Ref. 3). Intronic mutations inactivate p53 through aberrant splicing and intron retention (18). Somatic mutations with the potential to alter splicing are frequent in some cancers (19–25). Single nucleotide polymorphisms in the cyclin D1 proto-oncogene predispose to aberrant splicing and the cyclin D1b intronic splice variant (26–29). Cyclin D1b confers anchorage independence, is tumorogenic in vivo, and is detectable in human tumors (30), but as yet no clinical studies have confirmed an impact on outcome. On the other hand, aberrant splicing of HAS1 shows an association between aberrant splice variants and malignancy, suggesting that such variants may be potential therapeutic targets and diagnostic indicators (19, 31–33). Increased HA expression has been associated with malignant progression of multiple tumor types, including breast, prostate, colon, glioma, mesothelioma, and multiple myeloma (34). The three mammalian HA synthase (HAS) isoenzymes synthesize HA and are integral transmembrane proteins with a probable porelike structural assembly (35–39). Although in humans, the three HAS genes are located on different chromosomes (hCh19, hCh8, and hCh16, respectively) (40), they share a high degree of sequence homology (41, 42). HAS isoenzymes synthesize a different size range of HA molecules, which exhibit different functions (43, 44). HASs contribute to a variety of cancers (45–55). Overexpression of HASs promotes growth and/or metastatic development in fibrosarcoma, prostate, and mammary carcinoma, and the removal of the HA matrix from a migratory cell membrane inhibits cell movement (45, 53). HAS2 confers anchorage independence (56). Our work has shown aberrant HAS1 splicing in multiple myeloma (5) and Waldenstrom's macroglobulinemia (6). HAS1 is overexpressed in colon (57), ovarian (58), endometrial (59), mesothelioma (60), and bladder cancers (61). A HAS1 splice variant is detected in bladder cancer (61).
Here, we characterize molecular and biochemical characteristics of HAS1 variants (HAS1-Vs) (5), generated by aberrant splicing. Using transient transfectants and tagged HAS1 family constructs, we show that HAS1-Vs differ in cellular localization, de novo HA localization, and turnover kinetics, as compared with HAS1-FL, and dominantly influence HAS1-FL when co-expressed. HAS1-Vs proteins form intra- and intermolecular associations among themselves and with HAS1-FL, including covalent interactions and multimer formation. HAS1-Vc supports vigorous cellular transformation of NIH3T3 cells in vitro, and HAS1-Vc-transformed NIH3T3 cells are tumorogenic in vivo.
EXPERIMENTAL PROCEDURES
Cell Culture and Reagents
HeLa, HEK293, and KMS-12BM (multiple myeloma line) cells were originally obtained from the American Type Culture Collection (Manassas, VA). The cells were grown at 37 °C in 5% CO2 in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, streptomycin, and penicillin. Monoclonal antibodies against hemagglutinin epitopes, anti-mouse horseradish peroxidase, anti-rabbit horseradish peroxidase, and protein A-Sepharose were obtained from Sigma; protease inhibitor mixture was from Roche Applied Science; and transfection reagent Lipofectamine 2000, TRIzol reagent, reverse transcriptase III, and HiFi Taq were from Invitrogen. The GFP-fused murine Has3 (GFP-mHAS3) construct was a gift from Prof. Markku Tammi (Kuopio University, Finland).
Plasmid Construction
pCDNA3 were engineered to express different 10-amino acid epitope tags at the BamHI and EcoRI sites by ligation of annealed oligonucleotides, and cDNAs were inserted at the EcoRI site to express N-terminally tagged proteins. The 10 amino acid tags were hemagglutinin epitope (HE) and A2 (monoclonal antibody against A2 was a kind gift of Prof. Greg Matlashewski, McGill University, Quebec, Canada) (62). To clone HAS1-FL and HAS1-Vs, first-strand cDNAs were synthesized from KMS-12BM cells using Superscript-III from Invitrogen following the manufacturer's protocol with minor modifications as follows. 4 μg of total RNA were used for the reverse transcription reaction (20 μl) where oligo(dT)25, 10% DMSO, and 1 unit of Pfu (Fermentas, Burlington, Canada) were used. All RNA preparations were made DNA-free before subsequent experiments. PCRs were performed with 6% DMSO with HiFi Taq, where primers were made for two-step PCR cycles (94 and 68 °C annealing/extension) spanning the respective genes' start to stop codons. All of the tagged cDNAs were also subcloned in the HindIII and NotI site of pCEP4-puro, a vector derived from pCEP4 (Invitrogen), where the hygromycin resistance gene was replaced with a puromycin resistance gene. All plasmids were prepared with Qiagen (Mississauga, Canada) Maxiprep columns.
Enzyme Capture Assay of Active HAS1 and Its Variants
This procedure was designed and standardized based on a procedure described in Ref. 63. 2 × 106 HeLa cells were transfected for 40 h and incubated with fresh medium 2 h prior to harvesting cells and lysis in 1.2 ml of lysis buffer (25 mm HEPES, pH 7.4, 150 mm NaCl, 5 mm MgCl2, 0.5% Nonidet P-40, and protease inhibitor mixture) on the plate. Lysis was performed at room temperature for 30 min with mild vortexing every 5 min and microcentrifugation at 8,000 rpm for 8 min, and supernatants were collected and incubated with 0.6 μg/ml biotinylated HA-binding protein (bHABP; Seikagaku America, Cape Cod, MA) for 1 h at room temperature on a tube rotor. Each sample was divided into two equal parts, and the enzyme-elongating HA-bHABP complex was precipitated with streptavidin-Sepharose beads (GE Healthcare) at room temperature for 1 h and washed twice (3 min each) with lysis buffer, followed by two washes with phosphate-buffered saline (PBS). Samples were incubated either with 100 μl of hyaluronidase (HAase; type 4 from bovine testes; Sigma) (500 units/ml PBS) or with 100 μl of PBS for 1 h or overnight at 37 °C followed by three washes with PBS. The protein was extracted with 4× SDS-PAGE sample buffers, heated for 10 min at 85 °C, and immunoblotted with tag-specific antibody to detect proteins associated with elongating HA.
Immunofluorescence Microscopy
For immunofluorescence experiments, cells (105 in 1 ml of medium in 12-well plate) grown on coverglasses were transfected for 40 h, ringed with PBS twice, and fixed in 3.7% formalin in medium (without any supplements) for 20 min at room temperature. Cells were rinsed two times, for 5 min each, with PBS with 2% glycine (w/v). The cells were then permeabilized with PBS supplemented with 0.1% Triton X-100, blocked for 1–2 h at room temperature with 10% milk in PBS supplemented with 0.1% Triton X-100. Cells were incubated overnight with anti-epitope monoclonal antibody in blocking solution at 4 °C with gentle rocking. Following three washes with PBS supplemented with 0.1% Triton X-100, cells were incubated with secondary antibody and/or phalloidin conjugated to Alexa Fluor Dyes (Molecular Probes, Inc., Eugene, OR) in blocking solution for 1–2 h at room temperature. After staining with 4′,6-diamidino-2-phenylindole (0.5 μg/ml) in PBS supplemented with 0.1% Triton X-100 and three washes, coverglasses were mounted with PermaFluor aqueous mountant (Thermo Scientific via Fisher). The slides were viewed with a Zeiss 100M laser-scanning microscope.
For HA staining, HeLa cells were grown on collagen-coated (10 μg/cm2) 8-well chamber slides (Nalge Nunc). Cells transfected for 40 h were chilled on ice and stained for membrane with FM-4–64FX dye (Molecular Probes) according to the supplier's protocol followed by fixing with 3.7% formaldehyde in HBSS for 15 min on ice. Cells were treated without or with HAase (100 units/ml HBSS at 37 °C for 30 min) following extensive washes with HBSS before or after permeabilization with PBS supplemented with 1% BSA + 0.1% Triton X-100. Cells were blocked with 3% BSA in PBS and incubated with bHABP (4 μg/ml) and anti-tag antibodies in PBS supplemented with 1% BSA + 0.1% Triton X-100 overnight at 4 °C. Avidin-Alexa-633- and Alexa-594-conjugated secondary antibodies were incubated in PBS supplemented with 1% BSA + 0.1% Triton X-100 for 1–2 h at room temperature.
Relative Measurement of HA via a Competitive Enzyme-linked Immunosorbent Assay-like Method
2 × 106 HEK293 cells were transfected with pCEP4-puro constructs for 40 h, followed by incubation in 1 ml of serum-free medium for 6 h. Supernatants and cells (in 2 ml of fresh medium, manually lifted) were harvested and divided equally into two parts. Cells were incubated with or without HAase (1000 units/ml) in serum-free medium for 1 h at 37 °C with inverting tubes every 5 min, followed by washing three times with fresh medium and resuspension in 1 ml of PBS. Similarly, supernatants were treated. All of the samples were digested with protease K (final 50 μg/ml) (Invitrogen) at 60 °C for 2 h with mixing every 15 min. Cold (−20 °C) absolute ethanol was added into digested product to a final concentration of 77% ethanol, mixed vigorously, and incubated at −30 °C overnight following centrifugation at 14,000 rpm for 30 min at 4 °C. The precipitate was again washed in −20 °C absolute ethanol in a centrifuge for 15 min, air-dried and digested with DNase and RNase at 37 °C overnight in PBS. HA was precipitated again and dissolved in 100 and 50 μl of PBS for cell and supernatant compartments, respectively, making the final concentration 10 times the original volume of harvested materials. Control HA (10 μg/ml in PBS) was precipitated twice similarly. 5 μl of each extracted sample were assayed per well (total 35 μl in 6% BSA plus an equal volume of bHABP). The enzyme-linked immunosorbent assay-like method followed exactly a procedure described previously (64) with two changes; we used 0.5 μg/ml bHABP for preincubation and a 1:5000 dilution of avidin-horseradish peroxidase (Sigma). Relative abundance of HA in 10× samples was expressed as the percentage of saturating 10 μg/ml HA as follows, ({(APBS − Asample)/(APBS − A10 μg/ml HA)} × 100). Assays were performed in triplicate.
Western Blot (WB) and Immunoprecipitation Analysis
The transferred nitrocellulose membranes were blocked for 1–2 h in 10% milk in PBS plus 0.1% Tween 20 (PBS-T), treated with primary antibody overnight at 4 °C in the blocking solution, washed three times with PBS-T, and finally incubated with horseradish peroxidase-conjugated secondary antibodies. The image was developed using a luminol-based chemiluminescence reaction. For immunoprecipitations, cells (HeLa, 106 cells, 40 h post-transfection) were washed twice with cold (4 °C) PBS and lysed in 1 ml of RIPA buffer (25 mm Tris (pH 7.5), 150 mm NaCl, 1% Nonidet P-40, 1% deoxycholate, 0.1% SDS, and protease inhibitor mixture) on ice for 15–20 min. Cell debris was discarded after centrifugation at 16,000 × g for 10 min. Cleared lysate was incubated at 4 °C for 2 h to overnight with antibody, followed by protein A-Sepharose (GE Life Sciences) for 1 h. Immunoprecipitates were washed four times with cold RIPA buffer and subjected to PAGE followed by Western blot analysis.
Multimers and Intermolecular Bond Formation Assay
To detect multimer formation, HeLa cells (∼106 cells) were transfected with the indicated plasmids, divided into three aliquots at 18 h post-transfection, and allowed to grow for a further 12 h, followed by treatment with protein cross-linker disuccinimidyl suberate (DSS; Thermo Scientific via Fisher) according to the manufacturer's instructions. In brief, cells from two aliquots were lifted with PBS-EDTA, washed twice with PBS, and treated with or without 10 mm methyl-β-cyclodextrin (Sigma) in medium (without fetal bovine serum) at 37 °C for 1 h, washed three times with PBS, and subjected to the cross-linking reaction using ∼0.33 × 106 cells in 50 μl, with final concentration of 5 mm DSS (from freshly made 100 mm stock in DMSO) at room temperature for 30 min. The reaction was quenched by the addition of amino groups to the concentration of 50 mm Tris. For another aliquot, cells were first lysed in RIPA buffer (0.33 × 106 cells in 100 μl), and then cleared lysates were subjected to a cross-linking reaction as above. After quenching reactions, the cells or the lysate were mixed with 4× SDS loading dye (0.25 m Tris, pH 6.8, 6% SDS, 20% glycerol, 10% β-mercaptoethanol) following WB analysis. For detecting intermolecular bond (disulfide bond) formation, transfected HeLa cells were harvested directly from plates using extraction buffer (50 mm Tris, pH 7.5, 137.5 mm NaCl, 5 mm EDTA, 0.5% Triton X-100, 10% glycerol, protease inhibitor mixture) on ice, pipetting up and down many times, using 300 μl for 1 × 106 seeded cells followed by clearing with centrifugation at 2000 rpm for 10 min at 4 °C. The extracts were then mixed and boiled in 4× SDS loading dye with or without 10% β-mercaptoethanol, followed by native PAGE and WB analysis.
Subcellular Fractionation
For subcellular fractionation, the Qproteome Cell Compartment kit (Qiagen) according to the supplier's instructions, using a protocol scaled down to 106 transfected HeLa cells at 40 h post-transfection. The extracted proteins were acetone-precipitated according to the supplier's instructions, followed by WB analysis.
Anchorage-independent Transformation and in Vivo Tumorigenicity Assay
NIH3T3 cells were transfected with the indicated plasmids and selected with 2 mg/ml Geneticin (Invitrogen) for 3 weeks to generate selected colonies. Pools of selected colonies were maintained in the selection medium, periodically tested for the expression of the transfected gene products by reverse transcription-PCR, and evaluated in an in vitro soft agar transformation assay and an in vivo tumorigenicity assay in NOD-SCID mice. Early (3 weeks) and late passage (10 weeks) NIH3T3 transfectants were used to determine cellular growth in semisolid medium. 3% low melting agarose (Invitrogen) was autoclaved in water and kept at 40 °C in a water bath. Medium was preheated in the same bath. In a 6-well plate, 1.5 ml/well agarose/medium mixture (0.6% agarose final) was layered and left to solidify at room temperature for 40 min, followed by a layer of 2 ml/well cell/medium/agarose mixture (total of 105 cells in 0.3% agarose in medium) and kept at room temperature for 1 h. The semisolid medium was overlaid with 1.5 ml of selection medium and incubated at 37 °C for 3 weeks, changing overlaying medium every 4 days. Images of soft agar colonies were captured on day 22. For the tumorigenicity assay, female NOD-SCID (5–6 weeks old) mice (Charles River Laboratories, St. Constant, Canada) were subcutaneously injected with stable NIH3T3 transfectants at 1 × 106 cells/animal in a 50-μl volume in their right flanks. Tumor volume was measured by Vivarium staff.
RESULTS
Expression of Full-length HAS1 and Its Variants
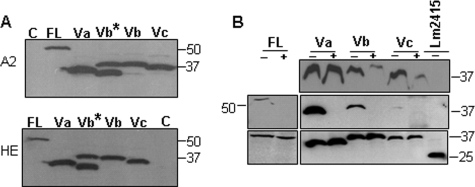
Human HAS1-FL and HAS1-Vs cDNAs were cloned from KMS-12BM (a multiple myeloma cell line) (Fig. 1), PCR-amplified, and subcloned into the mammalian expression vector pCDNA3, already engineered to express different small (10-amino acid) fusion tags at the N terminus of the expressed protein for differential detection and capture experiments using highly specific monoclonal antibodies. Fig. 2A shows a representative WB analysis of transfectants expressing HAS1-FL or each of the HAS1-Vs in HeLa cells. As seen in Fig. 2A, HAS1-Vb transfectants produce two protein bands, because the 59-base intronic sequence of HAS1-Vb (Fig. 1) was spliced out in almost half of the transcripts, confirmed by reverse transcription-PCR and subsequent sequencing (data not shown). This 59-base sequence of the retained partial intron resides between a strong pair of splice donor/acceptor sites (3′ of exon-3 and 5′ of exon-5). To obtain HAS1-Vb protein with the expected size (about ∼38 kDa), we replaced the last nucleotide of the 59-base intron (cag → caa, code Gln) to maintain the in frame coding for Gln while inhibiting the unwanted splicing. As expected, this silent point mutation prevented loss of the 59-base partial intron encoded in HAS1-Vb cDNA and produced a single HAS1-Vb protein of the expected size (Fig. 2A). This mutated construct was used for transfection of HAS1-Vb in all subsequent experiments.
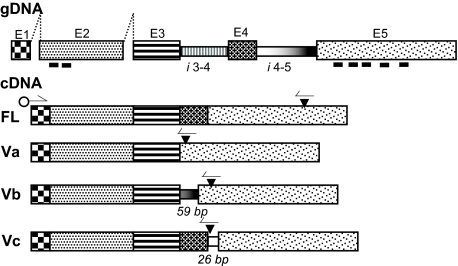
FIGURE 1.
HAS1-FL and HAS1-Vs constructs. This drawing illustrates the cDNA constructs of HAS1-FL and all the variants (HAS1-Va, HAS1-Vb, and HAS1-Vc) with genomic sequence (gDNA) (top bar) (5). E1–E5 are exons, and i3-4 and i4-5 are the introns joining E3 to E4 and E4 to E5, respectively. Thick black bars under E2 and E5 show positions coding transmembrane domains (not scaled). Filled triangles on cDNAs point to stop codons, half-arrows indicate PCR primer positions for reverse transcription-PCR, and open circles indicate the position of 10 amino acid tags when subcloned in the expression vector. HAS1-Va results from skipping of E4, which leads to formation of a frameshift generated a premature termination codon (fg-PTC) at 56 bp downstream of the deletion. HAS1-Vb results from skipping of E4 and partial retention of the last 59 bp of the 3′ part of i4-5 joining to the 5′-end of E5 and thus formation of an fg-PTC at 93 bp downstream of E5. HAS1-Vc results from retention of first 26 bp of the 5′ part of i4-5 joining to the 5′-end of E5 without any exon skipping and thus formation of an fg-PTC 1 bp downstream of the retained 26-bp partial intron.
FIGURE 2.
Aberrantly spliced human HAS1 variants are translated to protein and synthesize HA. A, WB analysis of N-terminally 10-amino acid tagged HAS1-FL and HAS1-Vs, 40 h post-transfection in HeLa cells. Lane C, control plasmid (pCDNA3); lane FL, HAS1-FL; lane Va, HAS1-Va; lane Vb, HAS1-Vb; lane Vc, HAS1-Vc. Lane Vb*, expression of original cDNA of HAS1-Vb showing two protein bands as the 59-base intron (Fig. 1A) was spliced out in almost half of the mRNAs; lane Vb, the reconstructed plasmid where one silent point mutation was introduced to prevent unwanted splicing, thereby producing the expected size single band. Upper and lower panels indicate expression individual proteins with N-terminally fused 10-amino acid tags A2 and HE, respectively, as indicated on the left, and protein molecular mass markers in kDa are marked on the right of each panel. B, enzyme capture assay of HAS1 and HAS1-Vs to determine enzymatic activity to detect de novo synthesis of HA associated with HeLa cell transfectants. The pulled down enzyme-HA-bHABP-streptavidin-Sepharose complex was treated with (+) or without (−) hyaluronidase followed by WB with tag-specific antibody as described under “Experimental Procedures.” An unrelated protozoan protein (Lm2415) with the same tag was used as control. The top and middle panels indicate hyaluronidase treatment for 1 and 16 h, respectively. The bottom panel represents straight WB of total cell lysate (0.1% input) in duplicates for the middle panel experiment. The results shown are representative of at least four independent experiments. Protein molecular weights are marked on both sides of the panel.
Different HASs differ in enzyme stability, kinetics, and length of HA produced (43). Thus, it is important to evaluate enzymatic activity of the HAS1-Vs and HAS1-FL cDNA constructs used. To examine this, HeLa cells were transfected with the indicated plasmids, and an enzyme capture assay was designed to determine enzymatically active HAS1s bound to elongating HA (Fig. 2B); an active HAS1 is expected to be bound to its polymerizing end of HA and thus can be captured with bHAPB (63), pulled down with streptavidin beads, and, as a control, treated with PBS or with HAase to remove bound enzyme from the bead complex. As demonstrated in Fig. 2B, HAS1-FL and HAS1-Vs were precipitated in association with elongating HA. Treatment with hyaluronidase prevented the precipitation of HAS1-FL and HAS1-Vs, indicating that the active enzymes continue to be bound on the elongating end of synthesizing HA. This demonstrates that all of the cDNA constructs (HAS1-FL and HAS1-Vs) synthesize de novo HA when expressed.
The Localizations of HAS1-Vs Differ from That of HAS1-FL
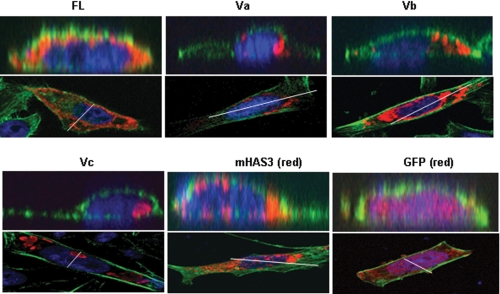
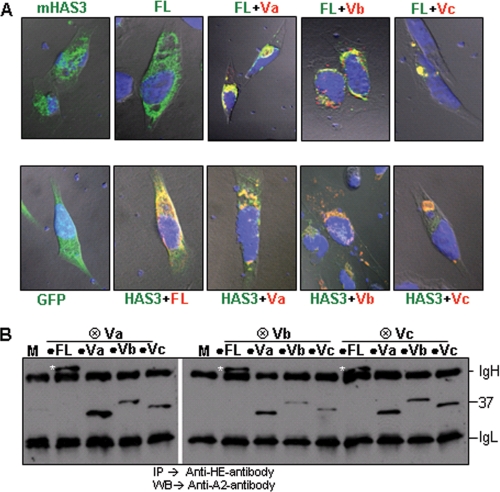
HASs are integral membrane proteins with an active site located on the cytoplasmic face and a predicted porelike structure for HA extrusion through the plasma membrane (65). HAS1-Vs retain only two transmembrane domains (TMDs) in comparison with HAS1-FLs, which contain seven TMDs (Fig. 1), and there is no signal sequence yet identified in the N-terminal part of HAS1-FL/Vs (see the GeneCards and UniProtKB sites on the World Wide Web). Using transfectants, we identified differential localization patterns for HAS1-FL and HAS1-Vs and compared them with that of mHAS3. HeLa cells were transiently transfected with tagged HAS1-FL or HAS-Vs, GFP-mHAS3, and GFP constructs and subjected to immunofluorescence with tag-specific monoclonal antibodies. Using analysis of confocal microscopic three-dimensional Z stacks (Fig. 3), HAS1-Vs were shown to localize predominantly as concentrated patches in cytoplasmic spaces in contrast to HAS1-FL, which has a diffuse cellular localization similar to that of mHAS3. The analysis of Fig. 3 lacks the resolution to allow determination of whether or not HAS1 is localized to the plasma membrane. Transient expression of HAS1-Va and HAS1-Vb was concentrated around the nucleus; HAS1-Vc localized to organelle-shaped concentric foci in cytoplasm. In KMS-12BM cells, transfected HAS1-Vs had a similar cytoplasmic localization (supplemental Fig. 1). To determine the organelle association(s) of HAS1-FL and each of the HAS1-Vs, HeLa and KMS-12BM cells were co-transfected with different organelle-specific markers fused with the fluorescent tag mCherry; most of the HAS1-Vs proteins and a part of HAS1-FL were shown to be expressed in association with the Golgi apparatus (supplemental Fig. 2). Different subcellular localization patterns of HAS1-Vs may result from the loss of five TMDs and different C-terminal peptide sequences acquired because of aberrant splicing (Fig. 1).
FIGURE 3.
Differential localization of HAS1 and its variants. Upper panels, a virtual vertical plane (position indicated as a white line on the lower panel) of cells obtained by three-dimensional Z-stacking confocal analysis showing localization pattern of HAS1-FL and HAS1-Vs in HeLa cells (colored red) as indicated on the panel. GFP and GFP-mHAS3 are pseudocolored red. Expressed N-terminally tagged proteins were tracked using tag-specific monoclonal antibody and Alexa594-conjugated secondary antibody. Actin filaments were stained with Alexa dye-conjugated phalloidin (green) to visualize the overall shape of cells, and the nucleus was stained with 4′,6-diamidino-2-phenylindole (blue). Lower panels represent the respective middle horizontal plane of cells of the Z stacks used in the upper panel.
The Localizations of de Novo HA Synthesized by HAS1-Vs Differ from That of HAS1-FL
Results of enzyme capture assay and localization of HAS1-Vs infer the intracellular localization of de novo HA synthesized by them. To directly determine relative abundance of intracellular and cell surface HA produced by these cDNA constructs, competitive enzyme-linked immunosorbent assay-like analysis (ELISA-LA) (Fig. 4A) and detailed fluorescence microscopy for HA staining (Fig. 4, B and C) were performed using bHAPB. This representative ELISA-LA result is a qualitative assessment for the relative abundance of HA present in a sample; the linear relationship of HA concentration determined by ELISA-LA is consistently valid for only a window of HA concentrations from 20 to 80 ng/ml (64). To detect the synthesis of extracellular HA by HAS1 transfectants, supernatants were collected over a 6-h period, followed by HA extraction and treatment with or without HAase, as shown in Fig. 4A (top). This analysis, although not quantitative, shows that HAS1-FL, GFP-mHAS3, and HAS1-Vs transfectants produce extracellular HA. The ELISA-LA results also indicate the presence of newly synthesized HA on the outer surface of the cell membrane for HAS1-FL and GFP-mHAS3 transfectants, as defined by its loss after digestion of intact cells with HAase. Cell surface HA was not detected on HAS1-Vs transfectants (Fig. 4A, bottom). HAS1-Vs-expressing cells appear to synthesize de novo HA inside cells, based on the observation that only minimal amounts of cellular HA were lost after digestion of intact cells with HAase (Fig. 4A, bottom).
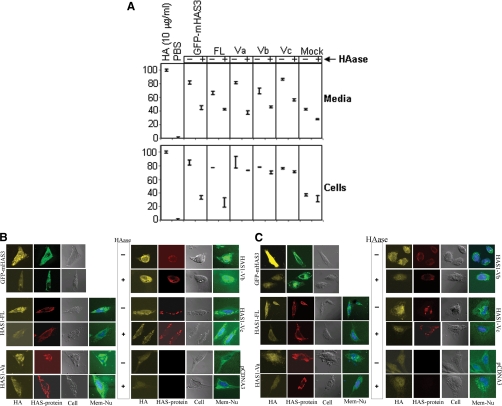
FIGURE 4.
Localization of de novo HA produced by each of the transfected cDNAs. A, competitive enzyme-linked immunosorbent assay-like analysis with bHABP. Top, HA extracted from supernatant harvested from a 6-h incubation of indicated HEK293 transfected cells were treated with (+) or without (−) HAase. 10 μg/ml HA and PBS were used as saturating (100) and zero concentration (0) against limited bHABP used, where free bHABP competes for coated HA on the plate. Bottom, live transfected HEK293 cells with the indicated plasmids were incubated in medium with (+) or without (−) HAase to remove cell surface-bound HA molecules followed by extraction of HA. HAase treatment for 1 h resulted in lysis of at least 8% cells, as determined by microscopic observations, which may reflect a diminished signal. This assay is a qualitative indicator for the presence of HA; it is not quantitative for the full range of HA concentrations. All of the samples were tested in triplicates, and S.E. values are indicated. B, fluorescence microscopy for HA staining. HeLa cells were transfected with the indicated plasmids (marked on the left and right side of the respective panels), fixed, and treated with (+) or without (−) HAase to remove cell surface-bound HA molecules (as marked in the space between the two panels) before permeabilizing and staining. Photographs presenting HA staining by bHABP, N-terminal tag immunostaining, phase-contrast, and membrane/nucleus staining (HA, HAS-protein, Cell, and Mem + Nu, respectively) are indicated at the bottom of each column of photographs. Staining for GFP-mHAS3 appears as green, whereas for other plasmids, membrane and nucleus staining appear as green and blue, respectively. C, similar HA staining was performed as above (B), except in this case, transfected cells were fixed and permeabilized first before treatment with (+) or without (−) HAase to remove both cell surface-bound and intracellular HA molecules.
To directly detect cell surface and intracellular HA, we used confocal microscopy and bHABP staining to confirm HA localization. HAS1 transfectants were stained with bHABP with or without HAase treatment to detect surface-bound HA (Fig. 4B). After washing to remove extracellular HA, HAS1-FL- and GFP-mHAS3-transfected cells were stained by bHABP, and this staining was lost when treated with HAase, indicating the presence of cell surface HA (Fig. 4B, top and middle left). In contrast, washed, intact HAS1-Vs transfectants remained brightly stained even after HAase treatment, indicating that intracellular HA was present (Fig. 4B, bottom left and right).
To further confirm the synthesis of de novo intracellular HA by HAS1-Vs transfectants, washed cells were permeabilized, followed by treatment with HAase, and stained with bHABP (Fig. 4C). This treatment will remove both surface and intracellular HA. In contrast to the HAase-resistant staining of “intact” HAS1-Vs transfectants shown in Fig. 4B, staining of permeabilized cells resulted in the loss of all bHABP binding by HAS1-Vs transfectants, indicating that the only de novo HA synthesized by HAs1-Vs is intracellular.
Fig. 4 shows that in both biochemical and microscopic analyses, intracellular HA remained detectable in all of the HAS1-Vs after removing cell-surface/extracellular HA by HAase treatment. In contrast, HA signals were significantly diminished by HAase in GFP-mHAS3- and HAS1-FL-transfected cells with intact membranes. These results confirm that newly synthesized HA in HAS1-FL, like GFP-mHAS3, was exposed on the outer surface of the cell membrane, and HAS1-Vs expressing cells appear to synthesize de novo HA primarily inside cells.
Plasma Membrane Translocation of HAS1-FL and mHAS3 Is Inhibited by HAS1-Vs
In patient cells, HAS1-Vs and HAS1-FL are almost always coexpressed in various combinations (5). Since localization patterns between HAS1-FL/mHAS3 and HAS1-Vs vary widely, we evaluated the influence, if any, of HAS1-Vs on HAS1-FL/mHAS3 or vice versa. HeLa and KMS-12BM cells were transiently co-transfected with HAS1-FL and each of the HAS1-Vs, followed by immunostaining and confocal analysis. As shown in Fig. 5A, in transfectants co-expressing any of the HAS1-Vs with either HAS1-FL (top) or mHAS3 (bottom), the latter were relocated to the inner cytoplasmic space. Both HAS1-FL and co-expressed HAS1-Vs proteins were closely associated (yellow staining) in the cytoplasm at locations characteristic of individual HAS1-Vs, in both HeLa and KMS-12BM cells (Fig. 5A and supplemental Fig. 3, respectively). Likewise, mHAS3 was also closely associated with co-expressed HAS1-Vs proteins, as shown in Fig. 5A (bottom).
FIGURE 5.
HAS1-Vs influence the localization of HAS1-FL and mHAS3. A, HAS1-Vs dominantly relocate HAS1-FL (top). As indicated in these confocal images, HeLa cells were transfected with GFP-mHAS3 or HAS1-FL alone or in combination with differentially tagged HAS1-Va, HAS1-Vb, and HAS1-Vc and stained. Green, HAS1-FL; red, all HAS1-Vs; blue, nucleus. Areas of coexpression appear as yellow. HAS1-Vs also dominantly relocate mHAS3 (lower panel). GFP alone and GFP-mHAS3 in combination with tagged HAS1-FL, HAS1-Va, HAS1-Vb, and HAS1-Vc were expressed and stained in red. B, biochemical association of HAS1-FL with each of the HAS1-Vs and among HAS1-Vs HE-tagged (⊗) HAS1-Vs co-transfected with A2-tagged (●) HAS1-FL or HAS1-Vs and mock transfection (M), as indicated in the panel. The samples were immunoprecipitated (IP) with anti-HE monoclonal antibody and immunoblotted (WB) with anti-A2 monoclonal antibody. The white asterisks represent the full-length protein on the blot. On the right of the panel, IgH and IgL represent heavy and light chain, respectively; the position of the 37-kDa size standard is indicated.
The dominant relocalization of HAS1-FL by HAS1-Vs may be governed by their intermolecular interactions. HeLa cells were co-transfected with differentially tagged HAS1-FL and each of the HAS1-Vs in different combinations, as shown in Fig. 5B. Cleared lysates were subjected to immunoprecipitation with monoclonal antibody specific to one tag followed by WB with the other tag-specific monoclonal antibody. Analysis of the RIPA-stable protein association pattern detected homomultimers of each of the HAS1-Vs with themselves (e.g. HE-Va with A2-Va) and heteromultimers consisting of HAS1-Vs with each other (e.g. HE-Va with A2-Vb or A2-Vc) and of HAS1-FL with each of the HAS1-Vs (Fig. 5B). This work indicates active biochemical/biophysical associations of expressed HAS1-Vs with each other and with HAS1-FL, remaining stable when lysed in RIPA buffer, which dissociates all but strong non-covalent or covalent intermolecular associations.
HAS1-Vs Form Multimers and Intermolecular Bonds
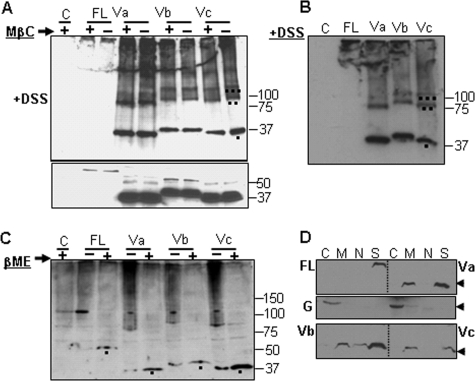
To further characterize biochemical/biophysical properties of the multimolecular associations among individual HAS1-FL and HAS1-Vs, transiently transfected HeLa cells were treated with protein cross-linker DSS, a homobifunctional, non-cleavable, and membrane-permeable linker that cross-links protein molecules associated within the 11.4-Å space arm distance. Cells were cross-linked as follows: 1) cross-linking performed on intact transfected cells followed by WB analysis (Fig. 6A) and 2) detergent-extracted (RIPA) lysates from transfected cells, followed by DSS cross-linking and WB analysis (Fig. 6B). Others have shown that murine Has3 localizes to lipid rafts in transfected MCF7 cells (67). To exclude the possibility that an artifactual association results from physical accumulation of these proteins in lipid rafts when overexpressed, transfected cells were treated with or without methyl-β-cyclodextrin prior to DSS cross-linking (Fig. 6A). Methyl-β-cyclodextrin treatment disrupts lipid rafts by removing cholesterol from the membrane and thus diffuses non-interacting lipid raft-associated monomers. HeLa cells were transfected with the indicated plasmids and divided into three aliquots. For two aliquots, cells were treated with or without methyl-β-cyclodextrin, followed by DSS cross-linking (Fig. 6A). The third aliquot was extracted with RIPA buffer, followed by DSS cross-linking (Fig. 6B). As shown in WB analysis (Fig. 6, A and B), cross-linking of either intact cells or RIPA-extracted lysate indicated strong interactions among expressed proteins (for each of HAS1-Vs) forming multimeric complexes, which were not influenced by mere physical accumulation of monomers in lipid rafts. HAS1-FL transfectants expressed less protein than did transfectants with HAS1-Vs. After cross-linking, most of the HAS1-FL protein and a portion of each HAS1-V were excluded from the resolving gel (Fig. 6, A and B), raising the possibility of their association with other higher order complexes.
FIGURE 6.
HAS1-Vs form homo- and heteromultimers and have differential cytoskeleton-anchoring patterns. A, top, WB analysis of HeLa transfections (with (+) or without (−) methyl-β-cyclodextrin (MβC) treatment on intact cells), which were subsequently DSS-cross-linked, run on 4–20% SDS-PAGE, and immunoblotted with tag-specific monoclonal antibody. Bands of monomer, dimer, and trimer are indicated as one, two, and three dots marked under the respective bands in the far right lane as an example; many higher order bands are also seen. Bottom, WB analysis of corresponding methyl-β-cyclodextrin-treated but non-cross-linked portion of cells, run on 10% SDS-PAGE. Lane C, vector DNA transfection. The numbers on the right mark protein molecular masses in kDa. B, WB analysis showing DSS cross-linking following RIPA lysis of transfected cells with the indicated plasmids, run on 4–20% SDS-PAGE and immunoblotted with tag-specific monoclonal antibody. Bands of monomer, dimer, and trimer are indicated as one, two, and three dots marked under the respective bands in the far right lane as an example. C, formation of cross-molecular disulfide bonds within individual HAS1-FL and HAS1-Vs Cell extracts treated with (+) or without (−) β-mercaptoethanol (βME) and run on 6% native PAGE followed by WB analysis with tag-specific antibody. The black dots under the bands indicate monomers released after disulfide bond-reduced protein when β-mercaptoethanol was added (+). Dark smears of higher protein assembly appear in (−)-lanes when disulfide bonds were not reduced. On the right, protein molecular masses in kDa are marked. D, association of HAS1-FL and HAS1-Vs with different subcellular fractionations. Transfected HeLa cells were fractionated to four portions: soluble cytoplasmic proteins (lane C); total solubilized non-nuclear membranes as well as lumen proteins of those organelles (lane M); total solubilized nuclear proteins (lane N), and insoluble cytoskeletal proteins (lane S). The fractions were subjected to WB analysis. Top, fractions from HAS1-FL and HAS1-Va. Middle, a separate WB analysis with anti-glyceraldehyde-3-phosphate dehydrogenase; glyceraldehyde-3-phosphate dehydrogenase is used as a representative soluble cytoplasmic protein. G, analysis on the same cellular fractions obtained from HAS1-FL- and HAS1-Va-transfected cells. Bottom, fractions from HAS1-Vb and HAS1-Vc. The arrowheads on the right mark the 37 kDa position.
To determine whether these HAS1 multiprotein associations were due to intermolecular disulfide bond formation, cellular extracts were treated with or without disulfide-reducing agent. As shown in Fig. 6C, post-transfection HeLa cell extracts were treated with or without 10% β-mercaptoethanol, followed by native PAGE and WB analysis. Results indicated that complex associations and multimeric patterns of expressed HAS1-FL and HAS1-Vs were disrupted when disulfide bonds were reduced. The smears in the untreated lanes represent multimers. Monomers were released upon treatment with β-mercaptoethanol, confirming HAS1-FL and each of the HAS1-Vs form intermolecular disulfide bonds. To authenticate reducing reagents, mouse IgG was reduced to separate heavy and light chains (supplemental Fig. 4) using the same reagents as in Fig. 6C. Although predicted by some, to our knowledge, this is the first demonstration that HAS1 family members multimerize, form heteromultimers, and are linked by intermolecular disulfide bonds.
Differential Cytoskeletal Association of HAS1-FL and HAS1-Vs
HA is abundant in extracellular matrix as well as being associated with intracellular structural elements. Although localized expression of mouse HASs on cellular protrusions was demonstrated in previous reports (67), underlying cytoskeleton association-driven remodeling was not investigated. To investigate possible cytoskeletal associations of human HAS1-FL and HAS1-Vs, cellular fractionation was performed on transiently transfected HeLa cells followed by WB analysis. The first fraction (Fig. 6D, lane C) consisted of soluble cytoplasmic proteins; the second fraction (lane M) consisted of solubilized plasma membrane, non-nuclear organelle membrane, and lumen proteins of those organelles; the third fraction (lane N) consisted of soluble, membrane-bound, and chromatin-bound nuclear proteins; and the last fraction (lane S) consisted of mainly insoluble cytoskeletal proteins. Membrane-bound (plasma and non-nuclear organelle) proteins were fractionated by solubilizing membranes, which liberates all but cytoskeleton-anchored membrane proteins. As demonstrated in Fig. 6D, HAS1-FL was found exclusively in the insoluble cytoskeleton fractions. In contrast, each of the HAS1-Vs distributes between cytoskeleton and non-cytoskeleton membrane-bound fractions, with HAS1-Vb showing much wider distributions (minor proportions were seen in nuclear and possibly in soluble cytoplasmic portions). This work supports the idea that human HAS1 is tightly associated with the cellular cytoskeletal network.
Differential Turnover Rates of HAS1-Vs Influence HAS1-FL
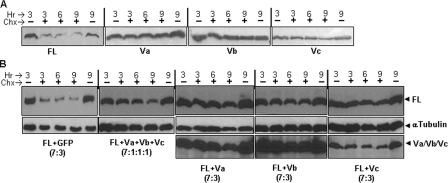
HA in extracellular matrix has a high turnover rate (half-life of <6 h), and plasma membrane-associated HASs are reported to have extremely short half-lives (68–71). Moreover, turnover kinetics has been found to alter when proteins and/or their variants relocate to different cell compartments (72, 73). Since each of the HAS1-Vs dominantly relocates HAS1-FL to the cytoplasm, thereby disrupting the normal trafficking of HAS1-FL, its turnover kinetics may be altered. To determine individual turnover kinetics, each HAS1-FL or HAS1-Vs plasmid was transfected into HeLa cells, followed by cycloheximide (50 μg/ml in medium) treatment for 3, 6, or 9 h and harvested in SDS-loading dye for WB analysis. Untreated cells at 3 and 9 h were used as untreated and natural degradation controls, respectively. Cycloheximide interferes with the translocation step in translation and thus stops de novo protein synthesis. As shown in Fig. 7A, HAS1-FL band intensity was reduced to less than half within 3 h of cycloheximide treatment, whereas each of the HAS1-Vs was found to be resistant to degradation, even if treated for as long as 9 h. This demonstrates that HAS1-Vs have much slower turnover rates than does HAS1-FL. Furthermore, it is important to analyze the influence of co-expression with HAS1-Vs on HAS1-FL, since HAS1-Vs associate with and dominantly relocate HAS1-FL (Fig. 5). As indicated in Fig. 7B, HeLa cells were co-transfected with HAS1-FL and each of the HAS1-Vs and later exposed to cycloheximide for the indicated period of time. Our experiments reproducibly demonstrated that co-expression of each of the HAS1-Vs individually or in a mixture (HAS1-Va, -Vb, and -Vc) protect HAS1-FL protein and dramatically prolong its cellular life as compared with control HAS1-FL protein co-expressed with GFP. Transcripts of HAS1-Vs and HAS1-FL are coexpressed in various combinations and ratios in clinical samples (5). It is thus important to examine the influence of HAS1-Vs and HAS1-FL on each other at different plasmid input combinations and ratios. Turnover kinetics were found to be similar in cotransfection experiments using either equal (1:1; data not shown) or lower ratios of HAS1-FL to HAS1-Vs (HAS1-FL/HAS1-Vs ratio of 7:3 or HAS1-FL/HAS1-Va/HAS1-Vb/HAS1-Vc ratio of 7:1:1:1) plasmid inputs, implying that HAS1-FL was protected in the presence of a very low relative abundance of HAS1-Vs, as shown in Fig. 7B. Slightly faster turnover kinetics of HAS1-Vc, alone or in combination with HAS1-FL, were observed in comparison with HAS1-Va or HAS1-Vb. Nevertheless, HAS1-Vc increased the stability of HAS1-FL when compared with HAS1-FL/GFP co-expressors. This indicates that co-expressed HAS1-Vs not only predominantly relocate HAS1-FL proteins to cytoplasm, thereby disrupting normal trafficking of HAS1-FL, but also dominantly alter turnover kinetics by protecting HAS1-FL from degradation. Relocalization and increased stability of HAS1-FL may have a significant influence on hyaluronan dynamics and cell cycle regulation.
FIGURE 7.
Differential turnover kinetics of HAS1-FL and HAS1-Vs singly and together. A, 106 HeLa cells were transfected and divided into five equal parts, which were exposed to medium with (+) or without (−) cycloheximide (Chx) for different time periods in hours (Hr), harvested in an equal volume of 4× SDS-loading dye, run on 10% SDS-PAGE, and immunoblotted with tag-specific monoclonal antibody. B, essentially same experimental design was followed as for A except for transfection, where mixtures of HAS1-FL with different plasmids (Vs) were used in different ratios and combinations, as indicated. Top, level of HAS1-FL proteins in the sample. The membrane was stripped and reblotted using α-tubulin antibody, as shown in the middle. Bottom, stripping and reblotting of the same membrane with tag-specific monoclonal antibody to show the level of either HAS1-Va, HAS1-Vb, or HAS1-Vc proteins.
HAS1-Vc Mediates Cellular Transformation and Tumorigenicity
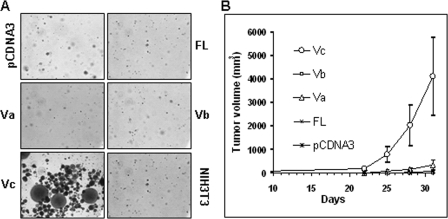
Overexpression of HASs has been associated with malignant transformation (45, 53, 56). Elevated expression of HAS genes accompanies oncogenic viral transformations (44) or inflammatory stimuli (74), and aberrant endogenous synthesis of HA or treatment with exogenous HA in vitro has been shown in multiple model systems to promote cancer cell growth and malignant behavior (35). To evaluate transforming potential of HAS1-FL and HAS1-Vs in vitro and in vivo, non-transformed anchorage-dependent murine fibroblast NIH3T3 cells were used to select stable transfectants, as described under “Experimental Procedures.” Pools of selected colonies were maintained in selection medium for several generations, with periodic monitoring for chromosomal insertion and HAS1 family mRNA expression by genomic and reverse transcription-PCR, respectively (data not shown). Anchorage-independent growth of cell lines was monitored in soft agar for 22 days. As shown in Fig. 8A, anchorage-independent growth was observed in only HAS1-Vc-transfected cells, with similar results in four independent experiments, using both early and late passage transfectants. NIH3T3 transfectants harboring HAS1-FL, HAS-Va, HAS1-Vb, or pCDNA3 showed no or very weak colony formation. We next tested whether late passage cell lines tested in the soft agar in vitro transformation assay were also able to form subcutaneous tumors in NOD-SCID mice. Consistent with the above in vitro assay results, all of the animals (n = 5) inoculated with HAS1-Vc transfectants developed subcutaneous tumors within 21 days, as shown in Fig. 8B. Harvested tumors (D30) from mice inoculated with HAS1-Vc transfectants contained the HAS1-Vc cDNA insert in their genome, as identified by genomic PCR (data not shown).
FIGURE 8.
HAS1-Vc is transforming in vitro and tumorigenic in vivo. A, assay of anchorage-independent growth of HAS1-FL- and HAS1-Vs-transfected NIH3T3 cells in soft agar. Only HAS1-Vc-transfected NIH3T3 cells show colony formation. B, subcutaneous tumor volume (mm3) on the right flanks of female NOD-SCID mice (n = 5 in each group) measured to assess tumor progression caused by inoculation of stable NIH3T3 transfectants, as indicated. All tumor masses were genotyped to verify that they contained the expected insert.
DISCUSSION
In previous studies (5, 6), we identified three novel splice variants of the human HAS1 gene in multiple myeloma and Waldenstrom's macroglobulinemia, produced as a result of exon skipping and/or a combination of unique intronic splicing events (5). Here we analyzed molecular and cellular behaviors of transfectants constitutively expressing HAS1-Vs and/or HAS1-FL. The N-terminally tagged HAS1-FL and HAS1-Vs constructs were enzymatically active and synthesized HA molecules outside and inside the plasma membrane, respectively. In transfected cells, HAS1-FL and mHAS3 have a diffuse localization in the cell, but HAS1-Vs had a more discrete localization in deeper cytoplasmic spaces. The enzymatic activities of murine HAS2 and HAS3 were coupled to their plasma membrane residence, where a latent pool of intracellular inactive HAS continually replenishes the plasma membrane supply of active enzyme (65). Similar behaviors for HAS1 or human HASs have yet to be established. The combination of enzyme capture assay and localization data reported here suggests that HAS1-Vs are enzymatically active although they are not translocated to the plasma membrane and contain only two of seven transmembrane domains as compared with HAS1-FL.
In patient cells, HAS1-FL and HAS1-Vs appear always to be coexpressed in varying combinations (5, 6). Although HAS1-FL mRNA is always the dominant species, our work suggests that HAS1-FL protein is usually less abundant than are HAS1-Vs (Figs. 2, A and B, and 6A). This may reflect the high turnover of HAS1-FL relative to HAS1-Vs, as reported here. We examined the localizations, associations, and turnover rates in transfected cells coexpressing HAS1-FL or mHAS3 with HAS1-Vs, either singly or in combination. Provocatively, in all cases, the HAS1-Vs relocated HAS1-FL and mHAS3 to discrete accumulations in the cytoplasm, in patterns characteristic of each HAS1-V. Furthermore, coexpression with HAS1-Vs, which have slow turnover rates, prolonged the stability of intracellularly delocalized HAS1-FL. The ability of HAS1-Vs to protect HAS1-FL protein from degradation and to inhibit its plasma membrane translocation may disrupt intracellular HA dynamics and extracellular HA production and contribute to cellular dysfunction, including cellular transformation. The localization of HAS1-Vs to intracellular membranes, a fraction of these possibly anchored to the cytoskeleton, would serve to keep their enzymatically active site out of the organelle lumen, thus increasing the diffusion of synthesized HA within the cytoplasm. Moreover, the loss of five C-terminal TMDs may topologically locate the large active sites of HAS1-Vs farther away from lumen. Because they contain only two N-terminal TMDs, HAS1-Vs may be unable to pass de novo HA molecules through membrane into lumen. On the other hand, cytoplasmic HAS1-FL delocalized by HAS1-Vs and acquiring a prolonged half-life may again contribute to changes in cellular HA dynamics if, unlike mHAS3, relocalized HAS1-FL remains active. Mouse-HAS3 is active only at plasma membrane (65). This in turn may increase the concentration of HA in the cytoplasm, where it is predicted to have an impact on mitotic spindle assembly by neutralizing HA-binding proteins, which are otherwise also known to associate with the cell division apparatus (e.g. RHAMM) (66, 75–79). This may be a significant contributor to oncogenesis and disease progression, consistent with the association between human cancers and aberrant HAS1 splicing (5, 6).
Although functional activities of many membrane- and cytoskeleton-bound proteins are regulated by formation and disassembly of multimeric units, this has not been previously reported for mammalian HASs. Here, we have shown that HAS family members are associated with the cytoskeleton as multimers of HAS1-FL and/or HAS1-Vs, formed by intermolecular disulfide bonds. Previous reports for GFP fusion constructs of HAS2 and HAS3 (68) (see the GeneCards and UniProtKB sites on the World Wide Web) demonstrated a non-homogenous, patchy localization of HASs, as confirmed here, but the biochemical properties of these enzymatically active multiprotein assemblies were not analyzed. In transfected MCF7 cells, GFP-HAS3 associated with lipid rafts, mainly in membrane protrusions (67), and previous reports of enzymatic activity of murine HASs (43, 44) using crude membrane extracts of transfected cells were essentially contaminated with insoluble cytoskeleton precipitated through a sucrose cushion. The work reported here suggests that human HAS1-FL and HAS1-Vs were found to form a covalently linked “protein island” assembly, where all of the HAS1-FL and a part of intracellular HAS1-Vs are anchored to the cytoskeletal network. Splice variant isoform-dependent regulation is influenced by either differential relocation, change in activity by association, shift in turnover kinetics, differential post-translational modifications, or a combination of these. To our knowledge, this report provides the first example of splice variant isoform-dependent differential behavior among glycosaminoglycan-synthesizing enzymes.
There are very few instances where aberrant splicing, specifically intronic splicing, appears to lead to enhanced oncogenic potential rather than inactivation of tumor suppressor function (73). The work reported here suggests that HAS1-Vc may be able to transform independently of other known oncogenes. In contrast, HAS1-FL, HAS1-Va, and HAS1Vb did not initiate anchorage-independent growth and significant tumorigenesis as single agent transfectants, suggesting that they may require heteromeric interactions within the HAS1 family and/or with other oncogenic agents for their observed clinical impact. Our observation of heteromultimeric associations indicates that the HAS1 family members are unlikely to act individually in patients and that the mechanisms whereby HAS1 and HAS1-Vs influence the malignant process will be complex and likely to involve intermolecular interactions within and outside the HAS1 family. In turn, intracellular HA is predicted to regulate the activities of proteins such as RHAMM (75), which is known to confer a poor clinical outcome (76), is important in mitotic spindle assembly (77), and binds to centrosomes via a region that overlaps its HA binding domain (78). The work reported here characterizes the intermolecular interactions within the HAS1 family, confirms the oncogenic properties of HAS1-Vc, and provides the tools for further analysis of mechanistic interactions that promote and/or maintain malignancy.
Supplementary Material
This work was supported by grants from the Alberta Cancer Research Institute and the Canadian Institutes of Health Research.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
- HA
- hyaluronan
- HAS
- HA synthase
- HAS1-FL
- hyaluronan synthase 1-full-length
- HAS1-Va
- hyaluronan synthase 1-Va
- HAS1-Vb
- hyaluronan synthase 1-Vb
- HAS1-Vc
- hyaluronan synthase 1-Vc
- HAS1-Vs
- hyaluronan synthase 1-variants (all)
- HE
- hemagglutinin epitope
- GFP
- green fluorescent protein
- bHABP
- biotinylated HA-binding protein
- PBS
- phosphate-buffered saline
- HAase
- hyaluronidase
- BSA
- bovine serum albumin
- WB
- Western blot
- RIPA
- radioimmune precipitation assay
- DSS
- disuccinimidyl suberate
- TMD
- transmembrane domain
- ELISA-LA
- enzyme-linked immunosorbent assay-like analysis.
REFERENCES
- 1.Ben-Dov C., Hartmann B., Lundgren J., Valcárcel J. (2008) J. Biol. Chem. 283, 1229–1233 [DOI] [PubMed] [Google Scholar]
- 2.Venables J. P. (2006) BioEssays 28, 378–386 [DOI] [PubMed] [Google Scholar]
- 3.Venables J. P. (2004) Cancer Res. 64, 7647–7654 [DOI] [PubMed] [Google Scholar]
- 4.Kalnina Z., Zayakin P., Silina K., Linē A. (2005) Genes Chromosomes Cancer 42, 342–357 [DOI] [PubMed] [Google Scholar]
- 5.Adamia S., Reiman T., Crainie M., Mant M. J., Belch A. R., Pilarski L. M. (2005) Blood 105, 4836–4844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adamia S., Crainie M., Kriangkum J., Mant M. J., Belch A. R., Pilarski L. M. (2003) Semin. Oncol. 30, 165–168 [DOI] [PubMed] [Google Scholar]
- 7.Adamia S., Reichert A. A., Kuppusamy H., Kriangkum J., Ghosh A., Hodges J. J., Pilarski P. M., Treon S. P., Mant M. J., Reiman T., Belch A. R., Pilarski L. M. (2008) Blood 112, 5111–5121 [DOI] [PubMed] [Google Scholar]
- 8.Moon S. D., Park J. H., Kim E. M., Kim J. H., Han J. H., Yoo S. J., Yoon K. H., Kang M. I., Lee K. W., Son H. Y., Kang S. K., Oh S. J., Kim K. M., Yoon S. J., Park J. G., Kim I. J., Kang H. C., Hong S. W., Kim K. R., Cha B. Y. (2005) J. Clin. Endocrinol. Metab. 90, 878–883 [DOI] [PubMed] [Google Scholar]
- 9.Colgin L. M., Hackmann A. F., Monnat R. J., Jr. (1999) Hum. Mutat. 14, 92. [DOI] [PubMed] [Google Scholar]
- 10.Agrawal S., Pilarski R., Eng C. (2005) Hum. Mol. Genet. 14, 2459–2468 [DOI] [PubMed] [Google Scholar]
- 11.Auclair J., Busine M. P., Navarro C., Ruano E., Montmain G., Desseigne F., Saurin J. C., Lasset C., Bonadona V., Giraud S., Puisieux A., Wang Q. (2006) Hum. Mutat. 27, 145–154 [DOI] [PubMed] [Google Scholar]
- 12.Sharp A., Pichert G., Lucassen A., Eccles D. (2004) Hum. Mutat. 24, 272. [DOI] [PubMed] [Google Scholar]
- 13.Lastella P., Surdo N. C., Resta N., Guanti G., Stella A. (2006) BMC Genomics 7, 243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pagenstecher C., Wehner M., Friedl W., Rahner N., Aretz S., Friedrichs N., Sengteller M., Henn W., Buettner R., Propping P., Mangold E. (2006) Hum. Genet. 119, 9–22 [DOI] [PubMed] [Google Scholar]
- 15.Durocher F., Labrie Y., Soucy P., Sinilnikova O., Labuda D., Bessette P., Chiquette J., Laframboise R., Lépine J., Lespérance B., Ouellette G., Pichette R., Plante M., Tavtigian S. V., Simard J. (2006) BMC Cancer 6, 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Staalesen V., Falck J., Geisler S., Bartkova J., Børresen-Dale A. L., Lukas J., Lillehaug J. R., Bartek J., Lønning P. E. (2004) Oncogene 23, 8535–8544 [DOI] [PubMed] [Google Scholar]
- 17.Ludes-Meyers J. H., Bednarek A. K., Popescu N. C., Bedford M., Aldaz C. M. (2003) Cytogenet. Genome Res. 100, 101–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takahashi T., D'Amico D., Chiba I., Buchhagen D. L., Minna J. D. (1990) J. Clin. Invest. 86, 363–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nichols K. E., Houseknecht M. D., Godmilow L., Bunin G., Shields C., Meadows A., Ganguly A. (2005) Hum. Mutat. 25, 566–574 [DOI] [PubMed] [Google Scholar]
- 20.Pros E., Larriba S., López E., Ravella A., Gili M. L., Kruyer H., Valls J., Serra E., Lázaro C. (2006) Hum. Mutat. 27, 1104–1114 [DOI] [PubMed] [Google Scholar]
- 21.Serra E., Ars E., Ravella A., Sánchez A., Puig S., Rosenbaum T., Estivill X., Lázaro C. (2001) Hum. Genet. 108, 416–429 [DOI] [PubMed] [Google Scholar]
- 22.Upadhyaya M., Han S., Consoli C., Majounie E., Horan M., Thomas N. S., Potts C., Griffiths S., Ruggieri M., von Deimling A., Cooper D. N. (2004) Hum. Mutat. 23, 134–146 [DOI] [PubMed] [Google Scholar]
- 23.Schaffner C., Stilgenbauer S., Rappold G. A., Döhner H., Lichter P. (1999) Blood 94, 748–753 [PubMed] [Google Scholar]
- 24.Lee J. W., Kim M. R., Soung Y. H., Nam S. W., Kim S. H., Lee J. Y., Yoo N. J., Lee S. H. (2006) APMIS 114, 646–650 [DOI] [PubMed] [Google Scholar]
- 25.Sjöblom T., Jones S., Wood L. D., Parsons D. W., Lin J., Barber T. D., Mandelker D., Leary R. J., Ptak J., Silliman N., Szabo S., Buckhaults P., Farrell C., Meeh P., Markowitz S. D., Willis J., Dawson D., Willson J. K., Gazdar A. F., Hartigan J., Wu L., Liu C., Parmigiani G., Park B. H., Bachman K. E., Papadopoulos N., Vogelstein B., Kinzler K. W., Velculescu V. E. (2006) Science 314, 268–274 [DOI] [PubMed] [Google Scholar]
- 26.Knudsen K. E. (2006) Cell Div. 1, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knudsen K. E., Diehl J. A., Haiman C. A., Knudsen E. S. (2006) Oncogene 25, 1620–1628 [DOI] [PubMed] [Google Scholar]
- 28.Burd C. J., Petre C. E., Morey L. M., Wang Y., Revelo M. P., Haiman C. A., Lu S., Fenoglio-Preiser C. M., Li J., Knudsen E. S., Wong J., Knudsen K. E. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 2190–2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Solomon D. A., Wang Y., Fox S. R., Lambeck T. C., Giesting S., Lan Z., Senderowicz A. M., Conti C. J., Knudsen E. S. (2003) J. Biol. Chem. 278, 30339–30347 [DOI] [PubMed] [Google Scholar]
- 30.Lu F., Gladden A. B., Diehl J. A. (2003) Cancer Res. 63, 7056–7061 [PubMed] [Google Scholar]
- 31.Adamia S., Treon S. P., Mant M. J., Larratt L. M., Reiman T., Belch A. R., Pilarski L. M. (2004) Blood 104, 1363(Abstr. 1363) [Google Scholar]
- 32.Adamia S., Van Ness B., Ramos C., Reiman T., Belch A. R., Pilarski L. M. (2007) Hematologica 92, Suppl. 2, 104.(Abstr. PO127) [Google Scholar]
- 33.Buratti E., Baralle M., Baralle F. E. (2006) Nucleic Acids Res. 34, 3494–3510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Toole B. P. (2004) Nat. Rev. Cancer 4, 528–539 [DOI] [PubMed] [Google Scholar]
- 35.Tammi M. I., Day A. J., Turley E. A. (2002) J. Biol. Chem. 277, 4581–4584 [DOI] [PubMed] [Google Scholar]
- 36.Toole B. P. (2002) Glycobiology 12, 37R–42R [DOI] [PubMed] [Google Scholar]
- 37.Weigel P. H., Hascall V. C., Tammi M. J. (1997) J. Biol. Chem. 272, 13997–14000 [DOI] [PubMed] [Google Scholar]
- 38.Heldermon C. D., Tlapak-Simmons V. L., Baggenstoss B. A., Weigel P. H. (2001) Glycobiology 11, 1017–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tlapak-Simmons V. L., Baggenstoss B. A., Clyne T., Weigel P. H. (1999) J. Biol. Chem. 274, 4239–4245 [DOI] [PubMed] [Google Scholar]
- 40.Spicer A. P., Seldin M. F., Olsen A. S., Brown N., Wells D. E., Doggett N. A., Itano N., Kimata K., Inazawa J., McDonald J. A. (1997) Genomics 41, 493–497 [DOI] [PubMed] [Google Scholar]
- 41.Spicer A. P., McDonald J. A. (1998) J. Biol. Chem. 273, 1923–1932 [DOI] [PubMed] [Google Scholar]
- 42.Watanabe K., Yamaguchi Y. (1996) J. Biol. Chem. 271, 22945–22948 [DOI] [PubMed] [Google Scholar]
- 43.Itano N., Sawai T., Yoshida M., Lenas P., Yamada Y., Imagawa M., Shinomura T., Hamaguchi M., Yoshida Y., Ohnuki Y., Miyauchi S., Spicer A. P., McDonald J. A., Kimata K. (1999) J. Biol. Chem. 274, 25085–25092 [DOI] [PubMed] [Google Scholar]
- 44.Itano N., Sawai T., Atsumi F., Miyaishi O., Taniguchi S., Kannagi R., Hamaguchi M., Kimata K. (2004) J. Biol. Chem. 279, 18679–18687 [DOI] [PubMed] [Google Scholar]
- 45.Ichikawa T., Itano N., Sawai T., Kimata K., Koganehira Y., Saida T., Taniguchi S. (1999) J. Invest. Dermatol. 113, 935–939 [DOI] [PubMed] [Google Scholar]
- 46.Auvinen P., Tammi R., Parkkinen J., Tammi M., Agren U., Johansson R., Hirvikoski P., Eskelinen M., Kosma V. M. (2000) Am. J. Pathol. 156, 529–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Auvinen P. K., Parkkinen J. J., Johansson R. T., Agren U. M., Tammi R. H., Eskelinen M. J., Kosma V. M. (1997) Int. J. Cancer 74, 477–481 [DOI] [PubMed] [Google Scholar]
- 48.Anttila M. A., Tammi R. H., Tammi M. I., Syrjänen K. J., Saarikoski S. V., Kosma V. M. (2000) Cancer Res. 60, 150–155 [PubMed] [Google Scholar]
- 49.Setälä L. P., Tammi M. I., Tammi R. H., Eskelinen M. J., Lipponen P. K., Agren U. M., Parkkinen J., Alhava E. M., Kosma V. M. (1999) Br. J. Cancer 79, 1133–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu N., Gao F., Han Z., Xu X., Underhill C. B., Zhang L. (2001) Cancer Res. 61, 5207–5214 [PubMed] [Google Scholar]
- 51.Simpson M. A., Wilson C. M., Furcht L. T., Spicer A. P., Oegema T. R., Jr., McCarthy J. B. (2002) J. Biol. Chem. 277, 10050–10057 [DOI] [PubMed] [Google Scholar]
- 52.Simpson M. A., Wilson C. M., McCarthy J. B. (2002) Am. J. Pathol. 161, 849–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simpson M. A., Reiland J., Burger S. R., Furcht L. T., Spicer A. P., Oegema T. R., Jr., McCarthy J. B. (2001) J. Biol. Chem. 276, 17949–17957 [DOI] [PubMed] [Google Scholar]
- 54.Bullard K. M., Kim H. R., Wheeler M. A., Wilson C. M., Neudauer C. L., Simpson M. A., McCarthy J. B. (2003) Int. J. Cancer 107, 739–746 [DOI] [PubMed] [Google Scholar]
- 55.Itano N., Sawai T., Miyaishi O., Kimata K. (1999) Cancer Res. 59, 2499–2504 [PubMed] [Google Scholar]
- 56.Kosaki R., Watanabe K., Yamaguchi Y. (1999) Cancer Res. 59, 1141–1145 [PubMed] [Google Scholar]
- 57.Yamada Y., Itano N., Narimatsu H., Kudo T., Morozumi K., Hirohashi S., Ochiai A., Ueda M., Kimata K. (2004) Clin. Exp. Metastasis 21, 57–63 [DOI] [PubMed] [Google Scholar]
- 58.Yabushita H., Noguchi M., Kishida T., Fusano K., Noguchi Y., Itano N., Kimata K., Noguchi M. (2004) Oncol. Rep. 12, 739–743 [PubMed] [Google Scholar]
- 59.Yabushita H., Kishida T., Fusano K., Kanyama K., Zhuo L., Itano N., Kimata K., Noguchi M. (2005) Oncol. Rep. 13, 1101–1105 [PubMed] [Google Scholar]
- 60.Kanomata N., Yokose T., Kamijo T., Yonou H., Hasebe T., Itano N., Kimata K., Ochiai A. (2005) Virchows Arch. 446, 246–250 [DOI] [PubMed] [Google Scholar]
- 61.Golshani R., Hautmann S. H., Estrella V., Cohen B. L., Kyle C. C., Manoharan M., Jorda M., Soloway M. S., Lokeshwar V. B. (2007) Int. J. Cancer 120, 1712–1720 [DOI] [PubMed] [Google Scholar]
- 62.Nascimento M., Zhang W. W., Ghosh A., Houston D. R., Berghuis A. M., Olivier M., Matlashewski G. (2006) J. Biol. Chem. 281, 36257–36268 [DOI] [PubMed] [Google Scholar]
- 63.Kyossev Z., Weigel P. H. (2007) Anal. Biochem. 371, 62–70 [DOI] [PubMed] [Google Scholar]
- 64.Gordon L. B., Harten I. A., Calabro A., Sugumaran G., Csoka A. B., Brown W. T., Hascall V., Toole B. P. (2003) Hum. Genet. 113, 178–187 [DOI] [PubMed] [Google Scholar]
- 65.Rilla K., Siiskonen H., Spicer A. P., Hyttinen J. M., Tammi M. I., Tammi R. H. (2005) J. Biol. Chem. 280, 31890–31897 [DOI] [PubMed] [Google Scholar]
- 66.Adamia S., Maxwell C. A., Pilarski L. M. (2005) Curr. Drug Targets Cardiovasc. Haematol. Disorders 5, 3–14 [DOI] [PubMed] [Google Scholar]
- 67.Kultti A., Rilla K., Tiihonen R., Spicer A. P., Tammi R. H., Tammi M. I. (2006) J. Biol. Chem. 281, 15821–15828 [DOI] [PubMed] [Google Scholar]
- 68.Laurent U. B., Dahl L. B., Reed R. K. (1991) Exp. Physiol. 76, 695–703 [DOI] [PubMed] [Google Scholar]
- 69.Mian N. (1986) Biochem. J. 237, 333–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ng C. K., Handley C. J., Mason R. M., Robinson H. C. (1989) Biochem. J. 263, 761–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bansal M. K., Mason R. M. (1986) Biochem. J. 236, 515–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stewart D., Ghosh A., Matlashewski G. (2005) J. Virol. 79, 8773–8783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ghosh A., Stewart D., Matlashewski G. (2004) Mol. Cell Biol. 24, 7987–7997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mohamadzadeh M., DeGrendele H., Arizpe H., Estess P., Siegelman M. (1998) J. Clin. Invest. 101, 97–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pilarski L. M., Pruski E., Wizniak J., Paine D., Seeberger K., Mant M. J., Brown C. B., Belch A. R. (1999) Blood 93, 2918–2927 [PubMed] [Google Scholar]
- 76.Maxwell C. A., Rasmussen E., Zhan F., Keats J. J., Adamia S., Strachan E., Crainie M., Walker R., Belch A. R., Pilarski L. M., Barlogie B., Shaughnessy J., Jr., Reiman T. (2004) Blood 104, 1151–1158 [DOI] [PubMed] [Google Scholar]
- 77.Maxwell C. A., Keats J. J., Belch A. R., Pilarski L. M., Reiman T. (2005) Cancer Res. 65, 850–860 [PubMed] [Google Scholar]
- 78.Maxwell C. A., Keats J. J., Crainie M., Sun X., Yen T., Shibuya E., Hendzel M., Chan G., Pilarski L. M. (2003) Mol. Biol. Cell 14, 2262–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pilarski L. M., Adamia S., Maxwell C. A., Pilarski P. M., Reiman T., Belch A. R. (2005) Hyaluronan: Structure, Metabolism, Biological Activities, Therapeutic Applications, pp. 329–338, Matrix Biology Institute, Edgewater, NJ [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.