Abstract
Fluorescent fusion proteins are exceedingly useful for monitoring protein localization in situ or visualizing protein behavior at the single molecule level. Unfortunately, some proteins are rendered inactive by the fusion. To circumvent this problem, we fused a hyperactive RecA protein (RecA803 protein) to monomeric red fluorescent protein (mRFP1) to produce a functional protein (RecA-RFP) that is suitable for in vivo and in vitro analysis. In vivo, the RecA-RFP partially restores UV resistance, conjugational recombination, and SOS induction to recA− cells. In vitro, the purified RecA-RFP protein forms a nucleoprotein filament whose kcat for single-stranded DNA-dependent ATPase activity is reduced ∼3-fold relative to wild-type protein, and which is largely inhibited by single-stranded DNA-binding protein. However, RecA protein is also a dATPase; dATP supports RecA-RFP nucleoprotein filament formation in the presence of single-stranded DNA-binding protein. Furthermore, as for the wild-type protein, the activities of RecA-RFP are further enhanced by shifting the pH to 6.2. As a consequence, RecA-RFP is proficient for DNA strand exchange with dATP or at lower pH. Finally, using single molecule visualization, RecA-RFP was seen to assemble into a continuous filament on duplex DNA, and to extend the DNA ∼1.7-fold. Consistent with its attenuated activities, RecA-RFP nucleates onto double-stranded DNA ∼3-fold more slowly than the wild-type protein, but still requires ∼3 monomers to form the rate-limited nucleus needed for filament assembly. Thus, RecA-RFP reveals that its attenuated biological functions correlate with a reduced frequency of nucleoprotein filament nucleation at the single molecule level.
The fusion of native proteins to various fluorescent proteins has found widespread use in biology. If the fusion protein retains proper function, then the behavior and localization of the protein can be followed in living cells (1). Complementing the single-cell analysis, it is now possible to image the behavior of a fluorescent protein at the single molecule level (2–8). However, despite the growing popularity of fusion protein studies, a detailed biochemical analysis of the fusion protein is much less common, even though such examination is crucial for molecular interpretations. Thus, an in vivo and in vitro analysis of the function of a fusion protein relative to the wild-type protein is an essential prerequisite.
Homologous recombination is an important process not only for generating genetic variation, but also for maintaining genomic integrity through the repair of DNA breaks. In Escherichia coli, recombinational repair of double-stranded DNA (dsDNA)3 breaks is mediated by the RecBCD pathway, whereas the repair of ssDNA gaps is mediated by the RecF pathway (9). Both of these recombination pathways require the functions of RecA protein.
RecA protein is essential to recombinational DNA repair (9–11). RecA-like proteins are ubiquitous and highly conserved (12, 13). The ATP-bound form of the protein binds to ssDNA and polymerizes along the DNA to form an extended nucleoprotein filament (14–16). This is the functional form of the protein that interacts with dsDNA to search for a homologous sequence. Upon finding homology, RecA protein promotes the exchange of identical DNA strands to produce the heteroduplex joint molecules. The joint molecules can be converted into Holliday junctions and resolved by the RuvABC proteins to produce recombinant DNA products (17).
The binding of RecA protein to ssDNA is competitive with the ssDNA binding (SSB) protein (18, 19). The assembly of RecA protein onto ssDNA that is complexed with SSB protein is a kinetically slow process, which is catalyzed by so-called mediator or loading proteins (20). RecBCD enzyme is one such RecA-loading protein (21, 22), but an additional set of loading proteins are the RecF, RecO, and RecR proteins that can form various subassemblies to facilitate the RecA-mediated displacement of SSB from ssDNA (23–26). In addition, a class of mutations that map to recA itself were isolated as suppressors of RecF function (srf) that produced mutant RecA proteins with an enhanced intrinsic ability to displace SSB from ssDNA (27). One such mutant is the RecA803 protein, in which valine 37 is mutated to methionine (28, 29). This mutant RecA protein displays a higher intrinsic rate of nucleoprotein filament assembly on ssDNA, which is responsible for its enhanced capacity to displace DNA-bound SSB protein.
RecA protein was successfully fused to green fluorescent protein (GFP) and was visualized in living bacteria (30). The RecA-GFP protein foci were seen to appear after UV irradiation and to be dependent on the recB and recF gene products. Although this protein is clearly functional in vivo, it was unfortunately, largely insoluble in vitro, thereby limiting large scale purification.4 Therefore, to facilitate biochemical use, an alternative fusion protein was constructed. In the present study, the monomeric red fluorescent protein (mRFP1 (31)) was fused to the carboxyl terminus of the RecA803 protein (referred to as RecA-RFP). The hyperactive RecA803 was used because it assembles on ssDNA more rapidly and competes better with SSB than the wild-type proteins and, as will be shown below, fusion to mRFP1 resulted in attenuated activity; thus, fusion to a hyperactive RecA protein permitted retention of at least partial function. The purified fluorescent protein binds to DNA but shows attenuated ATP and dATP hydrolysis activities. Although nucleoprotein filament assembly is inhibited by SSB protein under typical reaction conditions, we found that nucleoprotein filament formation and enzymatic activities are restored when dATP is substituted for ATP, or when the pH is lowered to 6.2. These characteristics are similar to those of the partially defective RecA142 mutant protein (32, 33), thereby showing that the RFP fusion converted a hypermorphic protein to a hypomorphic RecA fusion protein. Fortunately, because the behavior of this RecA-RFP protein closely fits the biochemical profile of a previously characterized mutant RecA protein, we could understand its behavior. By observing assembly on single molecules of dsDNA, we could see that nucleation of a RecA-RFP filament was ∼3-fold slower than for the wild-type protein. Importantly, these findings lend direct single molecule support to conclusions from ensemble studies where it was shown that biological function of the RecA protein correlates with its ability to displace SSB protein that, in turn, is related to the rate of RecA protein nucleation onto DNA (34).
EXPERIMENTAL PROCEDURES
Plasmids and Bacterial Strains
Plasmids derived from pBR322 encoding the fluorescent RecA proteins (pSJS1379 and pNG1) were constructed as described previously (30) and were confirmed by DNA sequencing. The construct on pNG1 is recAo1403, recA803, 4151::mrfp-1. The recA-mrfp-1 construct was made in the same manner as recA-gfp (30), with the linker being amino acid residues GSI. Both plasmids carry the recAo1403 mutation, which is a promoter mutation that increases recA expression; plasmids pSJS1379 and pNG1 carry recA-gfp (A206T) and recA803-mrfp-1, respectively. E. coli K12 strains, AB1157 (F−λ− supE44 thr-1 ara-14 leuB6 Δ(gpt-proA)62 lacY1 tsx-33 galK2 hisG4 rfbD1 mgl-51 rpsL31 kdgK51 xyl-5 mtl-1 argE3 thi-1) and BIK733, a Δ(srl-recA)306::Tn10 derivative of AB1157 (35), were used for measurement of UV sensitivity. Strain SS2081, which is JC19328 (sulB103 lacMS286 ϕ80dII lacBK1 argE3 his-4 thi-1 xyl-5 mtl-1 SmR T6R Δ(srl-recA)306::Tn10 (36)) carrying pNG1, was used for RecA-RFP purification.
Media
E. coli cells were grown in L broth (1.0% Bacto-tryptone, 0.5% yeast extract, and 0.5% sodium chloride) (40). Antibiotics were used as required at the following concentrations: ampicillin (amp) at 100 μg/ml; kanamycin (kan) at 50 μg/ml; and tetracycline (tet) at 10 μg/ml.
RecA-RFP Protein Purification
RecA-RFP protein was purified as follows. SS2081 strain was grown overnight at 37 °C in L broth containing amp, kan, and tet. The cells were harvested and stored in buffer (250 mm Tris-HCl (pH 7.5) and 25% sucrose) at 1 g/ml. Until use, the harvested cells were kept frozen at −80 °C. Cells were lysed using 1.3 mg/ml lysozyme and 0.4% Brij-35 in the presence of 0.5 mm phenylmethylsulfonyl fluoride, and then centrifuged in a Beckman Ti45 at 41,000 rpm for 45 min. To the cleared lysate, ammonium sulfate (Am2SO4) was added to 35% saturation. After centrifugation, the supernatant was recovered and the fluorescent protein was precipitated by addition of Am2SO4 to 45% saturation. After centrifugation, the pellet was dissolved in R buffer (20 mm Tris-HCl (pH 7.5), 10% glycerol, 0.1 mm DTT, and 0.1 mm EDTA), and the sample was dialyzed against R buffer overnight. The solution was loaded onto a Q-Sepharose column (GE) equilibrated with R buffer. The RecA-RFP fractions were eluted by running a linear salt gradient to 500 mm NaCl in R buffer. The RecA-RFP eluted at ∼300 mm NaCl. The pooled fractions were precipitated with 55% saturated Am2SO4, centrifuged, and then dialyzed against P buffer (20 mm potassium phosphate (pH 6.5), 10% glycerol, 0.1 mm DTT, and 0.1 mm EDTA) overnight. The dialyzed sample was loaded onto an ssDNA-cellulose column that was equilibrated with P buffer, and the fluorescent protein was eluted by washing with P buffer containing 200 mm NaCl and 1 mm ATP. The pooled protein fractions were dialyzed overnight against TEDS buffer (20 mm Tris-HCl (pH 7.5), 0.1 mm EDTA, 0.1 mm DTT, and 100 mm NaCl). The dialyzed sample was loaded onto Mono Q HR10/10 column (GE) equilibrated with TEDS buffer and was eluted with a salt gradient (100–1000 mm NaCl). The pool after the Mono Q column was concentrated by dialysis against storage buffer (20 mm Tris-HCl (pH 7.5), 1 mm DTT, 0.1 mm EDTA, and 10% glycerol). The concentration of the fluorescent RecA protein was determined using the Bradford assay (Bio-Rad), using wild-type RecA protein as the standard. The yield of RecA-RFP was ∼4.4 mg from 4 liters of culture, and its stock concentration was 21.5 μm. A Hewlett-Packard HP8453 spectrophotometer was used.
Other Proteins and Reagents
SSB protein (37) and wild-type RecA protein (38) were purified as described previously. All chemicals were reagent grade and solutions were prepared using NanoPure water.
DNA Substrates Used for Biochemical Analysis
Poly(dT) (∼220 nucleotides in length) was purchased from GE Healthcare. M13 mp7 ssDNA and dsDNA were isolated as follows. At an A600 of ∼0.2, a fresh culture (400 ml) of XL-1 blue was infected with 2.4 × 1011 M13 mp7 phage. After 3 h of shaking at 37 °C, cells were pelleted by centrifugation. The supernatant was used for preparation of the ssDNA (below). The pellet was washed with 20 ml of 50 mm Tris-HCl (pH 7.5) containing 10% sucrose, and M13 dsDNA was purified using QIAtip100 columns (Qiagen) according to the manufacturer's instructions. The M13 dsDNA was precipitated in ethanol and suspend in 100 μl of TE buffer (10 mm Tris-HCl (pH 7.5) and 1 mm EDTA).
M13 mp7 ssDNA was prepared from the supernatant described above. The filtered (0.2 μm) supernatant (50 ml) was ultracentrifuged in a Beckman 70Ti at 25,000 rpm for 60 min at 4 °C. The resulting clear pellet was suspended in 500 μl of 10 mm Tris-HCl (pH 7.5), 10 mm MgSO4, and 50 mm NaCl; then, 20 μl of 0.5 m EDTA (pH 8.0), 25 μl of 10% SDS, and 2.5 μl of Proteinase K (1 mg/ml) were added. After mixing, the solution was incubated at 65 °C for 30 min. The cooled solution was then extracted with an equal volume of phenol, followed by phenol/chloroform, and then ether. The DNA was precipitated in ethanol and suspended in 200 μl of TE buffer. Oligonucleotides and EcoRI-linearized M13 dsDNA were labeled at the 5′-end by T4 polynucleotide kinase using [γ-32P]ATP; unincorporated [γ-32P]ATP was removed using a MicroSpin S-200 HR column (GE).
UV Sensitivity Measurement
Cultures of exponentially growing cells (in L broth with amp, kan, and tet for selection of the plasmid and the host drug marker linked to the recA deletion) were diluted in L broth and spread on L agar plates. The plates were irradiated with UV light (254 nm) for various times. Colonies were scored after growth at 37 °C for 20 h in the dark.
SOS Induction
Strains were grown in L broth at 37 °C, washed in phosphate buffer, and UV irradiated for 10 s at 0.5 J/m2. The cells were then diluted into L broth and assayed for β-galactosidase activity (in duplicate) at the indicated times. The isogenic strains are derivatives of DM4000 and the different alleles were introduced by P1 transduction (39).
Conjugational Recombination
Conjugal matings were performed as described previously (39). The isogenic strains used were: wild-type RecA (JC13509; recAo+ recA+), RecA-RFP (SS2009; recAo1403 recA803,4151::rfpI), and RecA-GFP (SS1741; recAo1403 recA4136::gfp); their full genotype was described previously (30).
ATP and dATP Hydrolysis Assays
Both ATPase and dATPase activity were measured by following the procedure described previously (19, 25, 40) in a buffer containing 20 mm TrisOAc (pH 7.5), 10 mm Mg(OAc)2, 0.1 mm DTT, 0.1 mg/ml bovine serum albumin, 5% glycerol, 1 mm ATP or dATP, 1.5 mm phosphoenolpyruvate, 0.75 mm nicotine adenine dinucleotide (NADH), 10 units/ml pyruvate kinase, 10 units/ml lactate dehydrogenase, and 5% glycerol at 37 °C. Reactions were initiated by addition of 5 μm (nucleotides) of poly(dT) or M13 ssDNA. For experiments conducted at pH 6.2, 25 mm MES buffer (pH 6.2) was used instead of TrisOAc (pH 7.5). Where indicated, SSB protein (1 or 0.45 μm) was added subsequent to RecA protein to ongoing reactions. The rates of any DNA-independent ATP hydrolysis were subtracted from the reported DNA-dependent rates.
DNA Strand Exchange Assay
The procedure previously described was used (32), except for the buffer. Standard buffer (25 mm TrisOAc (pH 7.5), 10 mm Mg(OAc)2, and 0.1 mm DTT) containing 3 mm phosphoenolpyruvate, 80 units/ml pyruvate kinase, 1 mm ATP, 5 μm (nucleotides) M13 ssDNA, 0.45 μm SSB, 10 μm (nucleotides) linear M13 dsDNA (linearized using EcoRI restriction endonuclease, and 5′-end labeled with [γ-32P]ATP), and 3 μm of either wild-type or fluorescent RecA protein were used. The reaction was initiated by addition of the linear dsDNA. Samples were withdrawn at the indicated times, and analyzed by agarose gel (0.8%) electrophoresis in TAE buffer at 25 volts for 16–17 h.
Single Molecule Visualization of RecA-RFP Nucleoprotein Filaments
The experimental procedure was similar to that reported, with slight modifications (8). A three-channel flow cell was used to generate separate laminar flow channels of ∼1.5 mm width each. Bacteriophage λ DNA, ligated to a 3′-biotinylated oligonucleotide complementary to cosR, was attached to streptavidin-coated 1-μm polystyrene beads (Bangs Laboratories, Inc.). The DNA-bead complex was trapped in the first channel in 40 mm TrisOAc (pH 8.2), 15% sucrose, and 30 mm DTT. The trapped DNA-bead complex was then moved to the third channel containing the indicated concentration of RecA-RFP protein in 40 mm MES (pH 6.2), 15% sucrose, 30 mm DTT, 2 mm Mg(OAc)2, and 1 mm ATPγS. The DNA-bead complex was incubated in the third channel for 5 min to form the nucleoprotein filament; the resultant fluorescent RecA-DNA-bead complex was then moved back to the second (observation) channel that contained the same solution, but lacked the fluorescent RecA protein. The flow rate was 120 μm/s; the temperature was 37 ± 1 °C. For nucleation studies, the buffer composition in each channel was changed to permit comparison to previously published nucleation frequencies (8): the capture channel contained 20 nm YOYO-1 (Invitrogen), 20 mm TrisOAc (pH 8.2), 20% sucrose, and 30 mm DTT; the buffer in channel 2 contained 20 mm TrisOAc (pH 8.2), 20% sucrose, 30 mm DTT, 5 mm Mg(OAc)2, and 0.5 mm ATPγS; and the buffer in channel 3 contained 20 mm MES (pH 6.2), 20% sucrose, 30 mm DTT, 1 mm Mg(OAc)2, and 0.5 mm ATPγS. The nucleation reactions were performed at 30 ± 1 °C. Molecule lengths and fluorescence intensity profiles of RecA-RFP protein clusters formed on the DNA were determined using ImageJ; a cluster forming on the ssDNA end of any λ DNA molecule was excluded from the nucleation rate determinations.
RESULTS
RecA-RFP Partially Complements the Phenotypic Deficiencies of a recA Null Strain
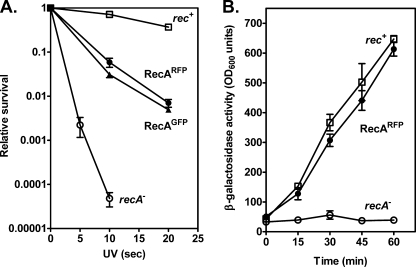
A fusion of RecA protein to green fluorescent protein (GFP) resulted in a protein that partially restored recombination and survival to cells after UV irradiation, and was used to visualize RecA focus formation at sites of DNA damage in individual living E. coli cells (30). To define the phenotype of the RecA-RFP protein, and to permit comparison to the RecA-GFP protein previously described, these functions were also examined. The recA803-mrfp1 (hereafter referred to as RecA-RFP) construct partially complements the UV-repair deficiency of a recA null strain to a level comparable with that of the wild-type recA-gfp construct (Fig. 1A).
FIGURE 1.
Fluorescent proteins fused to RecA partially suppress the UV sensitivity and SOS induction defects of a recA− strain. A, fresh log-phase cultures were diluted, plated onto L plates, and irradiated at various doses of UV light. Open squares, AB1157 (wild-type); open circles, BIK733 (ΔrecA) with the pBR322 empty vector; closed circles, BIK733 with pNG1 (recA803-mrfp1); closed triangles, BIK733 with pSJS1379 (recA-gfp). B, cells were assayed (in duplicate) for β-galactosidase activity originating from a fusion to the sulA gene at the indicated times. The isogenic strains are derivatives of DM4000 (39); open squares, DM4000 (recA+); open circles, SS2060 (Δ(recA)::kan); and closed circles, SS2065 (recA803-mrfp1). The data are the mean ± S.E. of at least three independent experiments.
The partial complementation of UV survival suggests that RecA-RFP is at least partially active for recombinational DNA repair. To test recombination function directly, conjugational recombination was examined. Table 1 shows that recombination by RecA-RFP is reduced only about 2-fold relative to wild-type, and is the same, within error, as the previously characterized RecA-GFP fusion.
TABLE 1.
Relative frequency of recombination
All strains are derivatives of JC13509 and are provided under “Experimental Procedures.” Conjugal matings are the averages of three independent experiments.
| Conjugational recombination (relative frequency) | |
|---|---|
| RecA (wild-type) | 1 |
| RecA-RFP | 0.47 ± 0.19 |
| RecA-GFP | 0.64 ± 0.21 |
A third biological function of the RecA protein is induction of the SOS-response via proteolytic cleavage of the LexA repressor. This activity was measured by using a reporter gene comprising a fusion of β-galactosidase to the LexA-regulated sulA gene (39). The results show that RecA-RFP induces the expression of this reporter gene in response to UV irradiation to almost the same extent as wild-type (Fig. 1B), indicating that RecA-RFP is proficient in LexA repressor cleavage in vivo. These collective results indicated that RecA-RFP was at least partially functional in vivo; as a consequence, the protein was purified and characterized in vitro.
Purification of RecA-RFP Protein
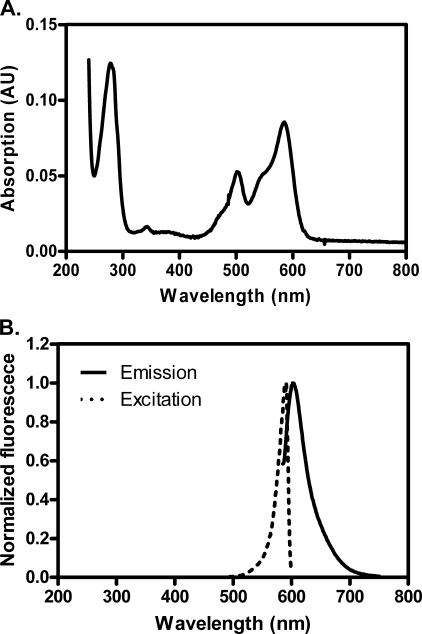
First, we attempted to purify the RecA-RFP fusion protein using a procedure that is standard for the wild-type RecA (38); however, that procedure was unsuccessful. Consequently, we modified the protocol (see “Experimental Procedures”) and succeeded in purifying the fluorescent protein. As presented in Fig. 2, the excitation and emission peaks of RecA-RFP are 582 and 608 nm, respectively. We also attempted purification of the fusion of wild-type RecA protein with green fluorescent protein, using both the same procedure and a standard RecA purification protocol (38); however, the RecA-GFP protein was less soluble, and it showed little activity in vitro.5 Therefore, we decided to focus on only the RecA-RFP protein.
FIGURE 2.
Absorption, excitation, and fluorescence emission spectra of RecA-RFP protein. A, absorption spectrum of RecA-RFP protein (4 μm) in storage buffer. B, fluorescence spectra of RecA-RFP (215 nm) in storage buffer. The excitation spectrum was obtained by measuring the fluorescence at 608 nm, and emission spectrum was as obtained by exciting at 582 nm. Each spectrum was normalized to the peak value in each scan.
RecA-RFP Protein Has ssDNA-dependent ATPase and dATPase Activities
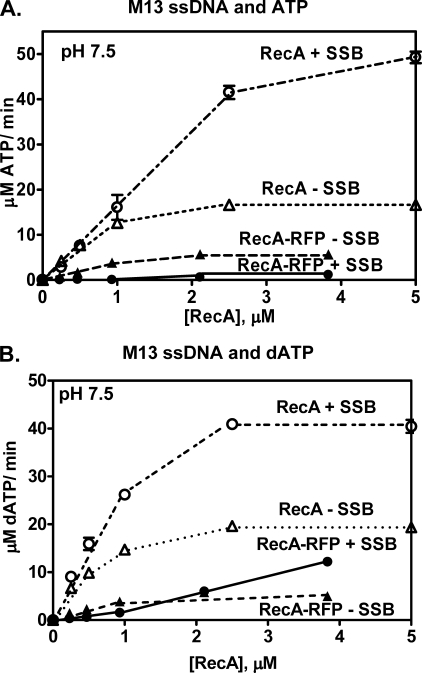
RecA protein has an ATPase activity that is stimulated by DNA. We first examined the ssDNA-dependent ATPase activity using an ssDNA, poly(dT), which is devoid of DNA secondary structure. Fig. 3A shows that the rate of ATP hydrolysis by RecA-RFP increases with increasing protein concentration, until it saturates at 1.6 μm RecA-RFP. This concentration corresponds to saturation occurring at a molar ratio of 1 RecA-RFP per ∼3.1 nucleotides, which is the canonical value for nucleoprotein filament formation by wild-type RecA protein. However, in the plateau region, the rate of ATP hydrolysis is only 13 μm ATP/min, which corresponds to a kcat value of 8.1 min−1. In comparison, the ATPase activity of the native RecA protein saturates at approximately the same protein concentration (1.5 μm), but exhibits a 3-fold greater rate of ATP hydrolysis (39 μm ATP/min), which corresponds to the expected kcat of 26 min−1.
FIGURE 3.
Purified RecA-RFP protein possesses ssDNA-dependent ATP and dATP hydrolysis activities. A, ATPase activity in the presence of poly(dT). Open triangles, wild-type RecA protein; closed triangles, RecA-RFP protein. B, dATPase activity in the presence of poly(dT). Open triangles, wild-type RecA protein; closed triangles, RecA-RFP protein. Data are mean ± S.D.
RecA protein is also a dATPase (40), and similar results were obtained when dATP hydrolysis by RecA-RFP was examined (Fig. 3B). Saturation occurs at ∼2.5 nucleotides per RecA-RFP and at a maximal rate of 16 μm dATP/min, which corresponds to a kcat of 8.0 min−1. In comparison, the activity of native RecA protein was 2.4-fold greater (38 μm dATP/min). Thus, based on the stoichiometries, all of the RecA-RFP protein seems to be active; however, the protein displays a severalfold lower intrinsic rate of ATP or dATP turnover.
RecA-RFP Protein Shows a Reduced Capacity to Compete with SSB Protein
The rate of ATP hydrolysis is known to be proportional to the steady-state level of the RecA nucleoprotein complex (19). Furthermore, kcat is also known to be unaffected by SSB protein (19). Therefore, a change in the observed rate of ssDNA-dependent ATP hydrolysis can be interpreted in terms of a proportional change in the amount of RecA-ssDNA complex formed. When ssDNA containing DNA secondary structure (e.g. M13 ssDNA) is used, ATP hydrolysis by wild-type RecA protein is stimulated by SSB protein due to disruption of the secondary structure by SSB protein. Subsequently, the SSB is displaced by RecA protein resulting in formation of more RecA-ssDNA complex, which is manifest in the increased rate of ATP hydrolysis (18, 19). Consequently, we next examined the ssDNA-dependent ATP hydrolysis activity of RecA-RFP in the presence of SSB protein (Fig. 4). When M13 ssDNA is used, wild-type RecA protein shows the well documented stimulation of activity by SSB protein (Fig. 4A) (18, 19). In contrast, the ATPase activity of RecA-RFP protein is almost completely inhibited by the SSB protein (Fig. 4A). These results show that the fluorescent fusion protein cannot displace SSB protein from ssDNA under these conditions (19).
FIGURE 4.
The M13 ssDNA-dependent ATPase activity of RecA-RFP protein is inhibited by SSB protein at pH 7.5. A, M13 ssDNA-dependent ATPase activity in the presence or absence of SSB protein (1 μm). Open triangles, wild-type RecA protein without SSB; open circles, wild-type RecA protein with SSB protein; closed triangles, RecA-RFP without SSB; closed circles, RecA-RFP with SSB protein. B, M13 ssDNA-dependent dATPase activity in the presence or absence of SSB protein (1 μm). Symbols are the same as in panel A. Data are mean ± S.D.
Because the steady-state affinity of RecA protein for ssDNA is greater in the presence of dATP than ATP, all activities of RecA proteins are enhanced when dATP is used as the nucleotide cofactor instead of, or in addition to, ATP (40, 41). In the case of RecA-RFP, its dATPase clearly does not recover to levels approaching the wild-type protein; nonetheless, activity is seen to increase (to about 12 μm/min) with increasing RecA-RFP protein concentrations even when SSB protein is present (Fig. 4B). Saturation should result in a dATP hydrolysis rate in the vicinity of 16 μm/min (based on poly(dT), Fig. 3), suggesting nucleoprotein filament formation to about 75% of maximal saturation; however, due to limitations imposed by solubility of the RecA-RFP stock solution, higher concentrations of RecA-RFP could not be tested. These findings show that RecA-RFP nucleoprotein filament formation is occurring in the presence of dATP and SSB protein.
Previously, we showed that mutant RecA proteins (RecA142 and RecA430), which displayed reduced functionality in the presence of ATP, could be partially rescued in vitro by dATP (32). We also previously reported that defect of the RecA142 protein is largely suppressed both in vitro and in vivo by decreasing the pH (33). Consequently, we next examined both ATP and dATP hydrolysis by RecA-RFP at pH 6.2 (Fig. 5). As previously reported, at this pH, the ATPase activity of wild-type RecA in the absence of SSB protein is sufficiently high that the stimulatory effect of SSB protein is modest when compared with pH 7.5 (Fig. 5A). In contrast, RecA-RFP is almost completely inhibited by SSB protein, except at the highest protein concentrations tested, when ATP is present. However, when dATP is substituted for ATP, the dATPase activity increases at all RecA-RFP concentrations (Fig. 5B). This latter finding suggests that even though dATP turnover remains reduced, dATP-RecA-RFP nucleoprotein filaments can form when SSB protein is present.
FIGURE 5.
The M13 ssDNA-dependent ATP hydrolysis of RecA-RFP protein is not inhibited by SSB protein at pH 6.2. A, M13 ssDNA-dependent ATPase activity in the presence or absence of SSB protein (1 μm). Open triangles, wild-type RecA without SSB; open circles, wild-type RecA with SSB protein; closed triangles, RecA-RFP without SSB protein; closed circles, RecA-RFP with SSB protein. B, M13 ssDNA-dependent dATPase activity in the presence or absence of SSB protein (1 μm). C, M13 ssDNA-dependent ATPase activity in the presence or absence of SSB protein (0.45 μm). D, M13 ssDNA-dependent dATPase activity in the presence or absence of SSB protein (0.45 μm). Symbols in panels B–D are the same as in panel A. Data are mean ± S.D.
To permit a more direct comparison with the DNA strand exchange experiments reported below, activity in the presence of a reduced concentration of SSB protein was also examined (Fig. 5, C and D). At this decreased SSB concentration (0.45 μm), the competitive inhibitory effect of SSB is reduced. As expected, the ATP hydrolysis activity of RecA-RFP is less inhibited (Fig. 5C), and also the dATP hydrolysis activity of RecA-RFP is greater in the presence of SSB protein (Fig. 5D). Thus, it is clear that RecA-RFP protein is forming nucleoprotein filaments at the lower pH in both the presence of ATP and dATP.
RecA-RFP Can Promote DNA Strand Exchange at pH 6.2
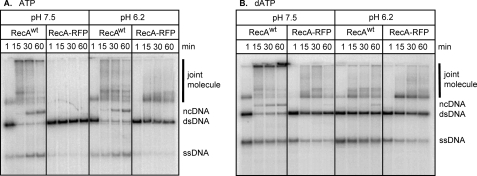
The RecA-ssDNA filament will catalyze DNA strand exchange with homologous dsDNA in vitro. To test the functionality of RecA-RFP, DNA strand exchange was examined between M13 phage circular ssDNA and linearized dsDNA. In the presence of ATP, wild-type RecA forms joint molecule intermediates and nicked circular DNA products over time (Fig. 6A, left panel); however, at pH 7.5, the RecA-RFP protein forms neither intermediates nor products (Fig. 6A).
FIGURE 6.
The RecA-RFP promotes DNA strand exchange in the presence of ATP or dATP at pH 6.2. DNA strand exchange was carried out with 5′-end labeled linear dsDNA, circular ssDNA, and either wild-type or the fluorescent RecA protein at either pH 7.5 or 6.2. A, DNA strand exchange in the presence of ATP. B, DNA strand exchange in the presence of dATP. The bands are labeled as follows: joint molecule, homologously paired joint molecule intermediate; ncDNA, nicked circular dsDNA product; dsDNA, linear dsDNA substrate; ssDNA, the displaced ssDNA product of complete DNA strand exchange.
As mentioned above, the fluorescent RecA protein shares many characteristics with the RecA142 mutant protein. Although RecA142 protein does not catalyze DNA strand exchange at pH 7.5 (32), it promotes DNA strand exchange at slightly acidic conditions (33). Consequently, DNA strand exchange was also assayed at pH 6.2 (Fig. 6A, right panels). As predicted, RecA-RFP protein produces joint molecule intermediates at the lower pH, although it is unable to produce nicked circular heteroduplex DNA product. These finding suggest that, although functional, the RecA-RFP nucleoprotein filaments are incomplete, in agreement with the ATP hydrolysis data.
DNA strand exchange was also conducted using dATP instead of ATP (Fig. 6B). Now, the fluorescent RecA produces both joint molecule intermediates and some nicked circular DNA products, even at pH 7.5 (left panels). At the lower pH, RecA-RFP forms even more joint molecule intermediates (right panel). However, the yield of DNA strand exchange product is decreased for wild-type RecA with dATP at pH 6.2 (Fig. 6B). This seemingly unexpected result is likely due to enhanced binding of wild-type RecA protein to the dsDNA substrate under these conditions (33), causing an inhibition of DNA strand exchange (42); this inhibitory phenomenon is characteristic of, and well documented for, the eukaryotic Rad51 protein that readily binds duplex DNA to reduce DNA strand exchange (43).
RecA-RFP Can Be Used to Visualize Nucleoprotein Filament Formation on Single dsDNA Molecules
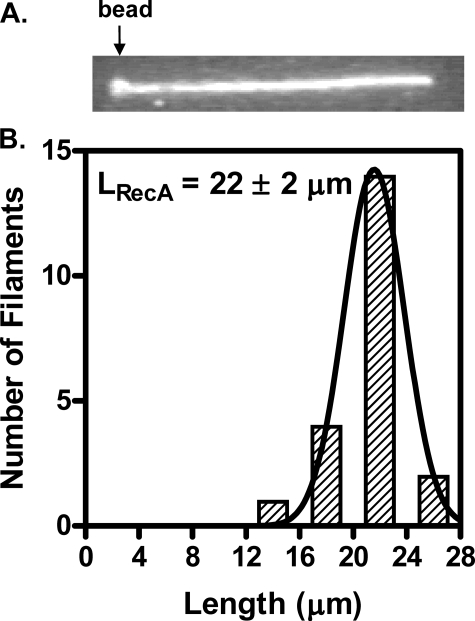
We next examined RecA-RFP nucleoprotein filament formation at the single molecule level using the protocol described (8). After incubating a DNA-bead complex in the RecA-RFP channel for 5 min in the presence of ATPγS, a fluorescent RecA nucleoprotein filament forms on the dsDNA (Fig. 7A). The length of the ATPγS-RecA-RFP nucleoprotein filament (LRecA) is 22 ± 2 μm (n = 21) (Fig. 7B). The length of the naked DNA (Lnaked) was measured to be 12.8 ± 0.6 μm in a separate single molecule assay wherein a fluorescent tag was attached to the opposite cohesive end of λ DNA. Therefore, the extension of the nucleoprotein filament (R = LRecA/Lnaked) relative to naked dsDNA is 1.7 ± 0.2. This extension is consistent with the extension (1.65 ± 0.05) measured previously for wild-type RecA protein (8), suggesting that in the presence of ATPγS, RecA-RFP can form complete and fully extended nucleoprotein filaments on dsDNA.
FIGURE 7.
RecA-RFP protein assembles on single molecules of dsDNA to form extended nucleoprotein filaments. A, an individual RecA-RFP protein filament formed on a single λ DNA molecule, imaged in the observation channel after a 5-min incubation with RecA-RFP protein (1 μm) at pH 6.2 in the presence of ATPγS (1 mm). The trapped bead is indicated by the arrow. The direction of flow is from left to right. The size of this image is 28 × 4.2 μm. B, length distribution of RecA-RFP nucleoprotein filaments (n = 21). The distribution was fit to a Gaussian function, which yielded a mean length (LRecA) = 22 ± 2 μm.
Nucleation of RecA-RFP Nucleoprotein Filaments
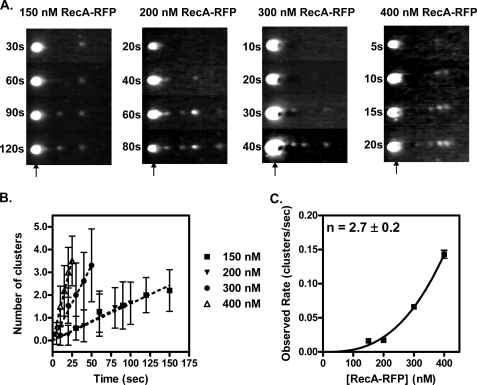
As established previously, nucleoprotein filament assembly is regulated at the nucleation step (8). To examine nucleation by RecA-RFP, the kinetics of nucleoprotein filament formation were examined. A trapped dsDNA molecule was incubated for the time indicated in the channel containing the RecA-RFP protein, and was then moved to the observation channel for analysis. Repetition of these steps permitted visualization of the time course of nucleation on the same DNA molecule (Fig. 8A). The appearance of RecA-RFP clusters was determined from an intensity profile of molecules such as those shown in Fig. 8A; the average total number of clusters on the DNA is plotted as a function of time in Fig. 8B. It is evident that the rate of nucleation increases with RecA-RFP concentration. However, compared with the wild-type protein, the rate of nucleation was only about one-third (8).5 Given that the fluorescein-RecA protein used previously displayed normal levels of enzymatic activity, and that RecA-RFP displays attenuated activity, our finding that nucleation frequencies of RecA-RFP are reduced is consistent. Previously (34), from ensemble studies, we established that the rates of RecA protein assembly onto DNA, which are nucleation-limited, parallel the biochemical and biological behavior of mutant RecA proteins, for those with either reduced or enhanced functions. Thus, our findings with RecA-RFP refine that conclusion, and are the first to reveal a correlation between a single molecule nucleation frequency and biological activity.
FIGURE 8.
Visualization of the kinetics of RecA-RFP protein nucleation on dsDNA. A, representative video frames from recordings of nucleation obtained at four different RecA-RFP protein concentrations in nucleation buffer. Flow is right to left. Each vertical strip represents the same DNA molecule repeatedly dipped into the RecA-RFP protein solution in channel 3 for the incubation times indicated. The trapped bead position is indicated by an arrow. B, clusters of RecA-RFP protein were scored and the average values were plotted as a function of time for the following concentrations of RecA-RFP protein: squares, 150 nm; inverted triangles, 200 nm; circles, 300 nm; triangles, 400 nm. C, the observed rate of cluster formation plotted as a function of RecA-RFP protein concentration. The line represents the fit to a power function, kobs = 1.2 × 10−8 [RecA]2.7, which yielded the value of 2.7 ± 0.2 for the exponent. Error bars represent the standard deviation; where the error bars are not visible, the standard deviation is smaller than the symbols.
Finally, when the observed rates of nucleation from Fig. 8B were plotted as a function of RecA-RFP protein concentration, the resultant plot (Fig. 8C) shows the expected power dependence on protein concentration (8). The exponent was 2.7 ± 0.2, and this value provides an estimate of the number of monomers required to form the nucleus of a RecA filament.
DISCUSSION
Our goal was to produce a fluorescent RecA fusion protein that could be used for visualization both in vivo and in vitro, which would permit comparisons of physiological function and biochemical behavior. Knowing that the creation of such a fusion with RecA protein often created inactive RecA protein (30),6,7 we fused monomeric red fluorescent protein to a hyperactive RecA protein, RecA803, hoping to produce a fusion protein that retained at least partial RecA function. Although RecA803 assembles on DNA more quickly and competes better with SSB than wild-type RecA protein (28, 29), we show here that the fusion with RFP results in a protein that has reduced functionality. In vivo, when overproduced, RecA-RFP displays the same phenotypic behavior as the wild-type RecA-GFP and RecA803-GFP (30). RecA-RFP confers partial restoration of UV sensitivity to a recA null strain, partial restoration of conjugal recombination, and nearly complete recovery of the SOS response. In terms of RecA focus formation upon generation of DNA damage, the RecA803-GFP protein showed a greater number of foci per cell on average than RecA-GFP (30), consistent with the enhanced assembly capacity of RecA803 protein (29). Thus, although not identical, these three fusion proteins are suitable for in vivo investigations of RecA function. The RecA-RFP protein, however, has the advantage of being more soluble and, thus, more useful for biochemical and single molecule analyses in vitro.
Biochemically, we showed that the RecA-RFP protein: 1) displays lower levels for ATP and dATP hydrolysis compared with wild-type RecA protein (kcat is reduced 2.4–3.2-fold); 2) forms nucleoprotein filaments that are sensitive to inhibition by SSB protein; 3) formed nucleoprotein filaments with a greater steady-state stability when using dATP, decreasing the pH, or lowering SSB protein concentrations; 4) is proficient in DNA strand exchange provided that dATP is present or, with ATP, that a lower pH of 6.2 is used; 5) can be used to image the assembly of individual RecA-RFP filaments on dsDNA; and finally, 6) nucleates on dsDNA with only about one-third the frequency of wild-type RecA protein.
Although the enhanced biochemical activities observed when using dATP or shifting the pH to 6.2 might seem unexpected, these are well known characteristics of RecA protein (33, 44–52). The binding of dATP to RecA protein results in a protein that has a higher affinity for DNA, assembles more quickly, and dissociates more slowly than the ATP complex (26, 40, 41, 51, 53–55). Likewise, reducing the pH to 6.2 results in similar effects (56). Both of these enhancing effects are additive; consequently, wild-type and mutant RecA proteins are most active in the presence of dATP at pH 6.2. Furthermore, we had previously established that dATP enhanced the activity of the wild-type protein, even in the presence of ATP (40, 41). Thus, all of these biochemical variables are physiologically meaningful, because both intracellular pH and nucleotide pools change with growth conditions.
The biochemical behavior of RecA-RFP protein is somewhat comparable, although not identical, to that of the mutant RecA142 protein (32, 33). The recA142 allele, in which isoleucine 225 is substituted with valine (57), is partially deficient for both recombination and SOS induction in vivo (58, 59). Although the purified RecA142 protein has ssDNA-dependent ATPase activity, it has no DNA strand exchange activity with ATP at pH 7.5 (32). Resembling the RecA-RFP protein, the ATPase activity of the RecA142 protein is inhibited by SSB, showing that nucleoprotein filament formation is disrupted by SSB protein. This failure to successfully compete with SSB protein both in vitro and in vivo is sufficient to explain all of failings of the RecA142 protein (32). Surprisingly, it was subsequently discovered that the recombination deficiency was restored at lower pH values (6.2–6.5) both in vitro and in vivo (32, 33); however, the RecA-RFP protein did not recover recombinational DNA repair function at lower pH growth conditions (data not shown). The only other major difference between RecA142 and RecA-RFP proteins is that the kcat for ATP hydrolysis by RecA142 is similar to that of the wild-type protein (32), whereas the kcat of RecA-RFP protein is reduced severalfold.
Because many characteristics of RecA-RFP protein resemble those of RecA142 protein, its defect may have a similar underlying cause. The mutation in the RecA142 protein (I225V) is located in a part of the hydrophobic core of the RecA protein that impinges upon the NH2-terminal region, which is one side of the monomer-monomer interface (33). The NH2 terminus contains a large α-helix and short β-strand involved in RecA self-assembly (60–62). The fluorescent protein used here was produced by fusing RFP to the COOH-terminal end of the hyperactive RecA803 protein, using a short amino acid linker. Although the COOH terminus does not directly contribute to the monomer-monomer interface (60, 63), nonetheless, it may occlude this interface and thereby impede the assembly process. The fact that the rate of nucleation onto DNA is reduced supports this conclusion. The RecA-RFP protein was hoped to have activity comparable with the wild-type RecA protein. Instead, the fluorescent fusion protein displays a reduced functionality. Nevertheless, the fusion protein can still be useful for in vivo experiments, provided that one bear in mind that it is a hypomorph.
We were able to visualize individual filaments of RecA-RFP forming on single DNA molecules. The length measurements showed that the RecA-RFP nucleoprotein filament is extended 1.7-fold relative to naked DNA, which is in precise agreement with our previous measurements using wild-type RecA protein that was modified with carboxyfluorescein (8), indicating that a complete nucleoprotein filament is formed. Furthermore, we measured directly the rates of filament nucleation by RecA-RFP. Compared with the wild-type protein, the nucleation frequencies are reduced. The nucleation of RecA-RFP on dsDNA is slower than the wild-type protein by a factor of ∼3. This finding is the first report of the nucleation frequency of a RecA protein with altered functional characteristics. Extensive biochemical analysis of mutant RecA proteins, and the correlation with their phenotypic behavior, led to the conclusion that the ability of a RecA protein to compete with SSB protein for DNA binding is the most predictive biochemical trait that correlates completely with its biological function (34). This ability to compete with SSB protein, in turn, was related directly to the rate at which a RecA protein assembles onto ssDNA. Using dsDNA, we previously showed that assembly of RecA protein is controlled by a rate-limiting nucleation step involving 4.5 (± 0.5) monomers. This value is close, but not identical, to the value obtained with the RecA-RFP fusion protein; although this apparent difference may reflect real differences between the two proteins, we believe that it largely reflects the difficulty of measuring the rates of nucleation at the highest concentrations of RecA, which disproportionately bias the fit to the power function. Finally, although the absolute rate of nucleation onto ssDNA and onto the SSB-ssDNA complex will unquestionably be different from the rate of nucleation onto dsDNA, if we assume that the rates will parallel one another, then our results here show that a 3-fold lower rate of nucleation is sufficient to manifest reduced function in vivo and in vitro.
Finally, even though chemical modification of the RecA protein yielded a protein that fully retained biochemical activities (8), we note that the fluorescent fusion protein described has the advantage that it can be used to analyze quantitatively both in vivo cellular function and in vitro biochemical activity for the same protein. Thus, exact correlations of in vivo and in vitro function are now possible. Given that the functions of RecA protein can be altered by physiological factors such as ATP, dATP, pH, concentration of RecA and SSB proteins, volume-excluding conditions (64), and polyvalent ions (65), comparative individual cell analysis in vivo and individual molecule analysis in vitro will be very revealing.
Acknowledgments
We thank Dr. Ichizo Kobayashi for bacterial strains and Jarukit Edward Long for technical assistance. We are grateful to members of the Kowalczykowski laboratory, Petr Cejka, Clarke Conant, Taeho Kim, Katsumi Morimatsu, Jody Plank, and Jason Wong for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant GM-62653 (to S. C. K.). This work was also supported by “Grants-in-aid for Scientific Research” from the Japan Society for the Promotion of Science (JSPS) (1770001 and 19790316) and Ministry of Education, Culture, Sports, Science and Technology (MEXT) Grants 17049008 and 19037006 (to N. H.).
- dsDNA
- double-stranded DNA
- RecA-RFP
- RecA fused to monomeric red fluorescent protein
- RecA-GFP
- RecA fused to green fluorescent protein
- ssDNA
- single-stranded DNA
- SSB
- single-stranded DNA-binding protein
- mRFP
- monomeric red fluorescent protein
- MES
- 2-(N-morpholino)ethanesulfonic acid
- DTT
- dithiothreitol
- ATPγS
- adenosine 5′-O-(thiotriphosphate).
N. Handa and S. C. Kowalczykowski, unpublished observations.
N. Handa, I. Amitani, and S. C. Kowalczykowski, unpublished observations.
S. C. Kowalczykowski, unpublished observations.
S. J. Sandler, unpublished observations.
REFERENCES
- 1.Chudakov D. M., Lukyanov S., Lukyanov K. A. (2005) Trends Biotechnol. 23, 605–613 [DOI] [PubMed] [Google Scholar]
- 2.Nie S., Chiu D. T., Zare R. N. (1994) Science 266, 1018–1021 [DOI] [PubMed] [Google Scholar]
- 3.Funatsu T., Harada Y., Tokunaga M., Saito K., Yanagida T. (1995) Nature 374, 555–559 [DOI] [PubMed] [Google Scholar]
- 4.Noji H., Yasuda R., Yoshida M., Kinosita K., Jr. (1997) Nature 386, 299–302 [DOI] [PubMed] [Google Scholar]
- 5.Ha T., Ting A. Y., Liang J., Caldwell W. B., Deniz A. A., Chemla D. S., Schultz P. G., Weiss S. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 893–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amitani I., Baskin R. J., Kowalczykowski S. C. (2006) Mol. Cell 23, 143–148 [DOI] [PubMed] [Google Scholar]
- 7.Handa N., Bianco P. R., Baskin R. J., Kowalczykowski S. C. (2005) Mol. Cell 17, 745–750 [DOI] [PubMed] [Google Scholar]
- 8.Galletto R., Amitani I., Baskin R. J., Kowalczykowski S. C. (2006) Nature 443, 875–878 [DOI] [PubMed] [Google Scholar]
- 9.Spies M., Kowalczykowski S. C. (2005) in The Bacterial Chromosome (Higgins N. P. ed) pp. 389–403, ASM Press, Washington, D.C. [Google Scholar]
- 10.Kowalczykowski S. C., Dixon D. A., Eggleston A. K., Lauder S. D., Rehrauer W. M. (1994) Microbiol. Rev. 58, 401–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuzminov A. (1999) Microbiol. Mol. Biol. Rev. 63, 751–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bianco P. R., Tracy R. B., Kowalczykowski S. C. (1998) Front. Biosci. 3, D570–D603 [DOI] [PubMed] [Google Scholar]
- 13.Lin Z., Kong H., Nei M., Ma H. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 10328–10333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kowalczykowski S. C. (1991) Annu. Rev. Biophys. Biophys. Chem. 20, 539–575 [DOI] [PubMed] [Google Scholar]
- 15.Egelman E. H., Stasiak A. (1986) J. Mol. Biol. 191, 677–697 [DOI] [PubMed] [Google Scholar]
- 16.Stasiak A., Di Capua E. (1982) Nature 299, 185–186 [DOI] [PubMed] [Google Scholar]
- 17.West S. C. (1996) J. Bacteriol. 178, 1237–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kowalczykowski S. C., Clow J., Somani R., Varghese A. (1987) J. Mol. Biol. 193, 81–95 [DOI] [PubMed] [Google Scholar]
- 19.Kowalczykowski S. C., Krupp R. A. (1987) J. Mol. Biol. 193, 97–113 [DOI] [PubMed] [Google Scholar]
- 20.Beernink H. T., Morrical S. W. (1999) Trends Biochem. Sci. 24, 385–389 [DOI] [PubMed] [Google Scholar]
- 21.Spies M., Kowalczykowski S. C. (2006) Mol. Cell 21, 573–580 [DOI] [PubMed] [Google Scholar]
- 22.Anderson D. G., Churchill J. J., Kowalczykowski S. C. (1999) J. Biol. Chem. 274, 27139–27144 [DOI] [PubMed] [Google Scholar]
- 23.Umezu K., Chi N. W., Kolodner R. D. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 3875–3879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Umezu K., Kolodner R. D. (1994) J. Biol. Chem. 269, 30005–30013 [PubMed] [Google Scholar]
- 25.Morimatsu K., Kowalczykowski S. C. (2003) Mol. Cell 11, 1337–1347 [DOI] [PubMed] [Google Scholar]
- 26.Shan Q., Bork J. M., Webb B. L., Inman R. B., Cox M. M. (1997) J. Mol. Biol. 265, 519–540 [DOI] [PubMed] [Google Scholar]
- 27.Volkert M. R., Hartke M. A. (1984) J. Bacteriol. 157, 498–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Madiraju M. V., Templin A., Clark A. J. (1988) Proc. Natl. Acad. Sci. U.S.A. 85, 6592–6596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lavery P. E., Kowalczykowski S. C. (1992) J. Biol. Chem. 267, 20648–20658 [PubMed] [Google Scholar]
- 30.Renzette N., Gumlaw N., Nordman J. T., Krieger M., Yeh S. P., Long E., Centore R., Boonsombat R., Sandler S. J. (2005) Mol. Microbiol. 57, 1074–1085 [DOI] [PubMed] [Google Scholar]
- 31.Campbell R. E., Tour O., Palmer A. E., Steinbach P. A., Baird G. S., Zacharias D. A., Tsien R. Y. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 7877–7882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kowalczykowski S. C., Burk D. L., Krupp R. A. (1989) J. Mol. Biol. 207, 719–733 [DOI] [PubMed] [Google Scholar]
- 33.Zaitsev E. N., Kowalczykowski S. C. (1999) Mol. Microbiol. 34, 1–9 [DOI] [PubMed] [Google Scholar]
- 34.Kowalczykowski S. C. (1991) Biochimie 73, 289–304 [DOI] [PubMed] [Google Scholar]
- 35.Bachmann B. J. (1987) in Escherichia coli and Salmonella typhimurium, Cellular and Molecular Biology (Neidhardt F. C., Ingraham J. L., Low K. B., Magasanik B., Schaechter M., Umbarger H. E. eds) pp. 1190–1219, American Society for Microbiology, Washington, D.C. [Google Scholar]
- 36.McCool J. D., Sandler S. J. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 8203–8210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.LeBowitz J. (1985) Biochemical Mechanism of Strand Initiation in Bacteriophage λ DNA Replication, Johns Hopkins University, Baltimore, MD [Google Scholar]
- 38.Mirshad J. K., Kowalczykowski S. C. (2003) Biochemistry 42, 5945–5954 [DOI] [PubMed] [Google Scholar]
- 39.Sandler S. J., Samra H. S., Clark A. J. (1996) Genetics 143, 5–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Menetski J. P., Kowalczykowski S. C. (1989) Biochemistry 28, 5871–5881 [DOI] [PubMed] [Google Scholar]
- 41.Menetski J. P., Kowalczykowski S. C. (1990) J. Mol. Biol. 211, 845–855 [DOI] [PubMed] [Google Scholar]
- 42.Campbell M. J., Davis R. W. (1999) J. Mol. Biol. 286, 437–445 [DOI] [PubMed] [Google Scholar]
- 43.Sung P., Robberson D. L. (1995) Cell 82, 453–461 [DOI] [PubMed] [Google Scholar]
- 44.Kowalczykowski S. C., Clow J., Krupp R. A. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 3127–3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McEntee K., Weinstock G. M., Lehman I. R. (1981) J. Biol. Chem. 256, 8835–8844 [PubMed] [Google Scholar]
- 46.Karasaki Y., Hirano H., Higashi K. (1987) Sangyo Ika Daigaku Zasshi 9, 141–1472956659 [Google Scholar]
- 47.Pugh B. F., Cox M. M. (1987) J. Biol. Chem. 262, 1337–1343 [PubMed] [Google Scholar]
- 48.Bryant F. R. (1988) J. Biol. Chem. 263, 8716–8723 [PubMed] [Google Scholar]
- 49.Brenner S. L., Zlotnick A., Stafford W. F., 3rd (1990) J. Mol. Biol. 216, 949–964 [DOI] [PubMed] [Google Scholar]
- 50.Muench K. A., Bryant F. R. (1991) J. Biol. Chem. 266, 844–850 [PubMed] [Google Scholar]
- 51.Meah Y. S., Bryant F. R. (1993) J. Biol. Chem. 268, 23991–23996 [PubMed] [Google Scholar]
- 52.Pinsince J. M., Muench K. A., Bryant F. R., Griffith J. D. (1993) J. Mol. Biol. 233, 59–66 [DOI] [PubMed] [Google Scholar]
- 53.Kowalczykowski S. C. (1986) Biochemistry 25, 5872–5881 [DOI] [PubMed] [Google Scholar]
- 54.Zaitsev E. N., Kowalczykowski S. C. (1998) Nucleic Acids Res. 26, 650–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Katz F. S., Bryant F. R. (2001) Biochemistry 40, 11082–11089 [DOI] [PubMed] [Google Scholar]
- 56.Arenson T. A., Tsodikov O. V., Cox M. M. (1999) J. Mol. Biol. 288, 391–401 [DOI] [PubMed] [Google Scholar]
- 57.Dutreix M., Bailone A., Devoret R. (1985) J. Bacteriol. 161, 1080–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Clark A. J. (1973) Annu. Rev. Genet. 7, 67–86 [DOI] [PubMed] [Google Scholar]
- 59.Horii Z., Clark A. J. (1973) J. Mol. Biol. 80, 327–344 [DOI] [PubMed] [Google Scholar]
- 60.Story R. M., Weber I. T., Steitz T. A. (1992) Nature 355, 318–325 [DOI] [PubMed] [Google Scholar]
- 61.Ogawa T., Yu X., Shinohara A., Egelman E. H. (1993) Science 259, 1896–1899 [DOI] [PubMed] [Google Scholar]
- 62.Mikawa T., Masui R., Ogawa T., Ogawa H., Kuramitsu S. (1995) J. Mol. Biol. 250, 471–483 [DOI] [PubMed] [Google Scholar]
- 63.Skiba M. C., Knight K. L. (1994) J. Biol. Chem. 269, 3823–3828 [PubMed] [Google Scholar]
- 64.Lavery P. E., Kowalczykowski S. C. (1992) J. Biol. Chem. 267, 9307–9314 [PubMed] [Google Scholar]
- 65.Lavery P. E., Kowalczykowski S. C. (1990) J. Biol. Chem. 265, 4004–4010 [PubMed] [Google Scholar]