Abstract
Scavenger receptors are innate immune molecules recognizing and inducing the clearance of non-host as well as modified host molecules. To recognize a wide pattern of invading microbes, many scavenger receptors bind to common pathogen-associated molecular patterns, such as lipopolysaccharides and lipoteichoic acids. Similarly, the gp340/DMBT1 protein, a member of the human scavenger receptor cysteine-rich protein family, displays a wide ligand repertoire. The peptide motif VEVLXXXXW derived from its scavenger receptor cysteine-rich domains is involved in some of these interactions, but most of the recognition mechanisms are unknown. In this study, we used mass spectrometry sequencing, gene inactivation, and recombinant proteins to identify Streptococcus pyogenes protein Spy0843 as a recognition receptor of gp340. Antibodies against Spy0843 are shown to protect against S. pyogenes infection, but no function or host receptor have been identified for the protein. Spy0843 belongs to the leucine-rich repeat (Lrr) family of eukaryotic and prokaryotic proteins. Experiments with truncated forms of the recombinant proteins confirmed that the Lrr region is needed in the binding of Spy0843 to gp340. The same motif of two other Lrr proteins, LrrG from the Gram-positive S. agalactiae and BspA from the Gram-negative Tannerella forsythia, also mediated binding to gp340. Moreover, inhibition of Spy0843 binding occurred with peptides containing the VEVLXXXXW motif, but also peptides devoid of the XXXXW motif inhibited binding of Lrr proteins. These results thus suggest that the conserved Lrr motif in bacterial proteins serves as a novel pattern recognition motif for unique core peptides of human scavenger receptor gp340.
Human gp340, also known as DMBT1 (deleted in malignant brain tumors 1), belongs to the innate immune protein family of scavenger receptor cysteine-rich (SRCR)2 proteins, all of which contain one or more evolutionarily conserved SRCR domain linked to other conserved protein domains (1, 2). Many of these proteins serve as pattern recognition receptors for innate immunity. gp340 is expressed by epithelial cells and cells of the immune system, and its expression is up-regulated after inflammatory stimuli (3, 4). It inhibits bacterial invasion to epithelial cells and the secretion of proinflammatory cytokines (5–7). Thus, it appears to be an important mediator of host immune responses to various microbes and was recently linked to Crohn disease, a human inflammatory bowel disease (8). gp340 is also found in human secretions like tears and saliva, and the salivary form has long been known as salivary agglutinin, which is an important molecule in oral biofilm formation and is suggested to have a role in dental caries development (9–12). The mechanisms of gp340 action in these different biological contexts are not known.
Common to all scavenger receptors, the ligand repertoire of gp340 is wide; it binds many different types of bacteria as well as viruses (10, 13, 14). The wide ligand recognition pattern of scavenger receptors is thought to be based on the recognition of common microbial structures, such as lipopolysaccharides and lipoteichoic acids, but in the case of gp340, specific bacterial surface proteins are reported to be involved in the interactions characterized (15–18). Because the importance of gp340/salivary agglutinin in the oral environment has been evident for a long time, most of our knowledge of gp340-microbial interactions is from studies with oral bacteria. For example, viridans streptococci, such as Streptococcus mutans and Streptococcus gordonii, interact with saliva gp340 via their surface proteins AgI/II and the sialic acid-binding Hsa or GspB adhesin. In these interactions, gp340 shows a peculiar fluid phase versus surface-adsorbed behavior, as evidenced by AgI/II polypeptides primarily mediating aggregation of bacteria by fluid phase gp340, whereas the Hsa adhesin primarily mediates adhesion of S. gordonii to surface-bound gp340 (18).
gp340 binds also to many non-oral human Gram-negative and Gram-positive pathogens, such as Helicobacter pylori and S. pyogenes (10), but these interactions are less characterized. There are few studies suggesting that both carbohydrates and the protein core of the gp340 can be involved in these interactions. For example, a VEVLXXXXW peptide derived from the SRCR domain of gp340 is shown to bind different types of bacteria (19), whereas sialic acid residues may mediate binding to, for example, influenza virus (20). However, the molecular basis of the ability of gp340 or peptides derived thereof to bind a large ligand repertoire is not understood.
The aim of the present study was to use the common human pathogen S. pyogenes as a model bacterium to identify novel bacterial proteins binding to gp340 and in this way shed light on the ligand recognition capability of gp340. We report a novel S. pyogenes-host interaction mediated by bacterial surface protein Spy0843 and gp340. gp340 binds to conserved leucine-rich repeat (Lrr) motifs in the Spy0843 and recognizes the same motif also in other bacterial proteins, from both Gram-negative and Gram-positive bacteria. Moreover, the inhibition of Lrr binding with SRCR-derived peptides differed from that previously reported, which suggests a novel mode for ligand recognition of human scavenger receptor gp340.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Culture Conditions
Streptococcus pyogenes NZ131 wild type and its rgg-deficient derivative (Δrgg) were from Dr. M. Chaussee (National Institutes of Health, Hamilton, MT) (21), and the clinical isolates were from Dr. P. Huovinen (National Public Health Institute of Finland, Turku, Finland). S. pyogenes strains were grown in Todd Hewitt broth (Difco) supplemented with 0.5% yeast extract (THY; Biokar Diagnostics) and erythromycin (3 μg/ml) or kanamycin (500 μg/ml) when needed. S. gordonii DL1 was grown in Jordan's broth containing (per liter) 5 g of trypticase, 5 g of yeast extract, 5 g of K2HPO4, 4 g of glucose, 0.5 ml of salt solution (0.8 g of MgSO4·7H2O, 0.04 g of FeSO4·7H2O, 0.019 g of MnCl2·4H2O in 100 ml of water), and 5 ml of Tween 80. Escherichia coli strains were grown in Luria-Bertani broth (LB) supplemented with ampicillin (100 μg/ml), kanamycin (30 μg/ml), or kanamycin (30 μg/ml) and chloramphenicol (30 μg/ml) when needed. All bacteria were stored at −70 °C in growth medium supplemented with 15% glycerol.
Saliva Collection and Purification of Human gp340
Human parotid saliva was collected from healthy volunteers with Lashley cups. gp340 protein was purified from freshly collected, pooled parotid saliva from six donors by gel filtration as described (10). Part of the purified gp340 was biotinylated with EZ-LinkTM Sulfo-NHS-LC-Biotin (Pierce) according to the manufacturer's instructions.
S. pyogenes Binding to gp340 in a Hydroxyapatite Assay
The adhesion of S. pyogenes to gp340 was measured by using gp340-coated hydroxyapatite beads as described (18). Protein-coated hydroxyapatite beads are widely applied in measuring interactions of salivary proteins with bacteria, because the proteins are thought to attach to the surface in natural conformation. The bacteria were metabolically labeled with [35S]methionine (20 μCi/ml) and suspended in sodium/potassium phosphate buffer (1 mm, pH 6.8) containing 50 mm KCl, 0.1 mm MgCl2, 1 mm CaCl2, and 0.5% BSA to give an A550 of 0.35. The bacteria were allowed to adhere to gp340-coated hydroxyapatite beads or to BSA-coated beads in a control assay for 60 min. In assays comparing binding of S. pyogenes NZ131 and the Δ0843 mutant, the beads were coated with fresh human parotid saliva, a natural source of gp340. To remove the unbound bacteria, the beads were washed three times with the buffer, and the amount of adhered bacteria was measured by scintillation counting. The binding was expressed as a percentage of adhered bacteria from the total amount of added bacteria.
To test the effect of r0843 to gp340 binding of S. pyogenes, the strain NZ131 and one strain from each group of clinical isolates was selected for the inhibition assay. The r0843 or rYopM (125 μl, 100 μg/ml) or the buffer in a control assay was added on gp340-coated beads for 60 min before the addition of metabolically labeled bacteria. The binding of S. pyogenes to r0843- and rYopM-treated gp340-coated beads was calculated as a percentage from the binding to non-treated gp340-coated beads.
Preparation of Bacterial Surface Extracts and Adhesin Identification by Mass Spectrometry
S. pyogenes NZ131Δrgg was grown in 50 ml of THY with no agitation overnight at 37 °C, collected by centrifugation, washed in phosphate-buffered saline (10 mm phosphate, 140 mm NaCl, pH 7.2), and resuspended in 0.5 ml of the same buffer. The bacterial suspension was digested with trypsin at a concentration of 10 μg/ml for 30 min at 37 °C. The bacteria were pelletted, and 20 μl of the supernatant was analyzed by 7.5% SDS-PAGE and stained with silver. An identical gel was run in parallel, and proteins were transferred to nitrocellulose membrane. The nonspecific binding sites were blocked with 3% BSA in TTSB buffer (TSB buffer with 0.05% Tween 20). Biotinylated gp340 (10 μg/ml) in TSB, 1 mm CaCl2, 1% BSA buffer was added and allowed to bind to bacteria for 60 min. After three washes with TTSB, 1 mm CaCl2, strepavidin-HRP conjugate (0.5 μg/ml; Amersham Biosciences) was added for 30 min. The membrane was washed three times with TTSB, 1 mm CaCl2, and the binding was detected with chemiluminescence (ECL Western blot detection kit; Amersham Biosciences).
For identification, the gp340-binding protein was cut out from the silver-stained gels, and digested in gel (22–24). The protein was reduced and alkylated before digestion with trypsin overnight at 37 °C. The resulting peptides were extracted by adding 30–40 μl of 5% formic acid to the digestion mixture, incubation at 37 °C for 30 min, and direct desalting with μ-tips (25) containing Oligo R3-material (PerSeptive Biosystems). Peptide mass fingerprinting was performed with a Voyager DE PRO MALDI-TOF instrument in the positive ion reflector mode using α-cyano-4-hydroxycinnamic acid as the matrix. The mass spectra were internally calibrated with autoproteolytic trypsin fragments, 842.50 and 2211.10 Da. Data base searches were performed with the program MS-Fit (available on the World Wide Web), and the search criteria were 0.05 Da mass accuracy and a minimum of five matching peptides.
The trypsin-extracted surface proteins were also probed with non-biotinylated gp340 (1 μg/ml) in TSB, 1 mm CaCl2, 1% BSA or human parotid saliva (1:1) diluted in TSB, 1 mm CaCl2, 1% BSA for 60 min. After three washes, monoclonal antibody (mAb) 213-6 (1:10,000; Antibodyshop) specifically recognizing gp340 was added. After 60 min of binding, the membranes were washed as above, and rabbit anti-mouse-HRP-antibody (1:5000; Dako) was added for 60 min. After three washes, the binding was detected with chemiluminescence. For inhibition of identified Spy0843-gp340 interaction with soluble Spy0843, the blotted bacterial surface-extracted Spy0843 was treated as above except that 100 μg/ml recombinant Spy0843 protein (see below) was added in saliva before it was applied on the membrane.
Insertional Inactivation of spy0843
An internal fragment of spy0843 was amplified from S. pyogenes NZ131 genomic DNA by PCR using the primers Spy0843–5′-KO and Spy0843–3′-KO (Table 1). The resulting 1338-bp PCR product was digested with EcoRI and PstI and cloned into the EcoRI-PstI-digested pSF151 to generate pSpy0843-KO. The pSpy0843-KO was transformed into S. pyogenes NZ131 and S. pyogenes NZ131Δrgg as described (26), and KanR-colonies were selected. Insertion of the pSpy0843-KO into the genomic spy0843 was verified by PCR using the primers Spy0843-PROM (annealing upstream of the spy0843), and pSF151-MCS (annealing to the vector DNA).
TABLE 1.
Primers used in this study
| Primer | Sequence 5′–3′ (restriction enzyme and/or vector-compatible sites underlined) |
|---|---|
| Spy0843-5′-KO | TTTTTTCTGCAGTGACCCTCACTACAGTATCGG |
| Spy0843-3′-KO | TTTTTTGAATTCAATAGTAACACCATTGTGCTG |
| Spy0843-PROM | GTATCAGCCAATCAAATGTG |
| pSF151-MCS | TTAGCTCACTCATTAGGCAC |
| 0843F1 | GACGACGACAAGATGAAGAAACATCTTAAAACAGTTG |
| 0843R1 | GAGGAGAAGCCCGGTTTATATTGCAGAGTGTCGTCCTCT |
| F0 | CGACACTCTGCAATATAAACC |
| R164 | TACGAGATGATCAGTTTGAG |
| F644 | GCTTAGATGATAATGATGG |
| ThrR | AGAACCGCGTGGCACCAGACC |
| R580 | TAAGGTCTTAACCTTACTTCC |
| LrrGF | GACGACGACAAGATGGTATATGGATTAGAAAGAGAGG |
| LrrGR | GAGGAGAAGCCCGGTTTATTTTCTTGCTCGTTTTCC |
| Lrr_agaF | GGGGGAGATCTATCACACTTGGTTTTACCA |
| Lrr_agaR | GGGGGAGCTCTGATTCCTTTTGAAAGCCTC |
| YerF2 | GGGGGAGATCTTAAATCTAAGACTGAATATTA |
| YerR | GGGGGAGCTCTCCATCCGAAGATCTTCCA |
Binding of Spy0843-deficient Mutants to gp340
The S. pyogenes NZ131, NZ131Δrgg, and the Spy0843-deficient mutants were grown 16 h in THY at 37 °C. Part of the bacteria was collected for binding assays and part was used to inoculate fresh broth. The bacteria were cultured at 37 °C and samples withdrawn as indicated. The bacteria were washed with TSB, 1 mm CaCl2 buffer and suspended in the same buffer to give an A600 of 0.60. The purified gp340 was immobilized on a Biacore CM3 sensor chip (Biacore) with amine coupling according to manufacturer's instructions to yield a surface of 465 resonance units (RU). Bacterial binding to gp340 was tested in TSB, 1 mm CaCl2 at a flow rate of 20 μl/min with BiacoreX (Biacore). The activated-deactivated surface without any immobilized protein was used as a control. After each run, the chip was regenerated with 100 mm EDTA (50 μl) and 10 mm NaOH (10 μl).
Cloning and Expression of Recombinant Proteins
The primers used are shown in Table 1. They were designed based on the published genomic sequences. The genomic DNA was purified from S. pyogenes NZ 131, S. agalactiae CCUG 4209, and Yersinia enterocolitica 8081 (O:8), as described (27). To produce recombinant Spy0843 (r0843) spy0843 gene was PCR-amplified with the primers 0843F1 and 0843R1 using Vent-polymerase (New England Biolabs). The PCR product was cloned into pET-30 LIC expression vector (Novagen) according to the manufacturer's instructions to yield plasmid pET-0843. The pET-0843 was transformed into E. coli NovaBlue Singles-competent cells (Novagen). Transformants were selected on LB-kanamycin agar and confirmed to be positive in colony PCR with primers 0843F1 and 0843RI. Plasmid DNA from one clone was purified with plasmid miniprep kit (Qiagen) and confirmed to have an insert of correct size by digestion with NdeI and EcoRI before it was sequenced and the correct sequence was confirmed. The plasmid was then transformed into the expression strain E. coli BL21(DE3)pLysS (Novagen).
Truncated forms of r0843 were generated by PCR-amplifying the pET-0843 with primers F0 and R164 or R580 to produce pET-Tr-1 and pET-Tr-3, respectively. The pET-Tr-2 was obtained from pET-0843 with primers F644 and ThrR. The PCR products were kinased, ligated, and transformed by electroporation into E. coli JM109 cells according to standard methods. For expression, the plasmids were further transformed to E. coli BL21(DE3)pLysS. To obtain the Lrr proteins from S. agalactiae (rLrrG) and Y. enterocolitica (rYopM), the respective DNA fragments were PCR-amplified using Vent-polymerase with primers LrrGF and LrrGR to clone the full-length lrrG or Lrr agaR and Lrr_agaF to obtain only the part containing the leucine-rich repeats of LrrG (rLrr_aga), and primers YerF2 and YerR to obtain yopM. The PCR products and purified pET-0843 were digested with SacI and BglII, gel-purified, and ligated to yield plasmids pET-LrrG, pET-Lrr_aga, and pET-Yer. For expression, the plasmids were transformed to E. coli BL21(DE3)pLysS. Origin of the expression plasmid for rBspA from T. forsythia is as described (28). For this study, the sequence coding amino acids 17–1080 was ligated to the pQE80 vector (Qiagen) and transformed into E. coli JM109 for expression.
The transformed cells were grown in LB-kanamycin- chloramphenicol (E. coli BL21(DE3)pLysS) or LB-ampicillin medium (E. coli JM109) at 37 °C. To induce the production of the recombinant protein, 0.4 or 1.0 mm isopropyl 1-thio-β-d-galactopyranoside for induction of BL21 or JM109, respectively, was added to the log phase cells, and the cells were grown for an additional 3 h either at room temperature (r0843, rLrrG, rLrr_aga, Tr-1, Tr-2, and Tr-3) or at 37 °C (rBspA and rYopM). The cells were collected, and the His-tagged proteins were purified by affinity purification using Ni2+-nitrilotriacetic acid-agarose (Qiagen). The purified proteins were stored at +4 °C.
Binding of r0843 to gp340 and Other Glycoproteins
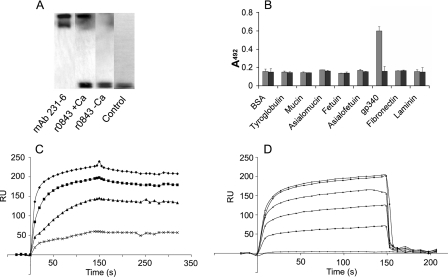
An aliquot of fresh parotid saliva, containing gp340, was applied on SDS-polyacrylamide gels in non-reducing conditions and blotted onto PVDF membrane. After blotting, the nonspecific binding sites on the membrane were blocked with 3% BSA. The purified r0843 (10 μg/ml) was added on the membrane in either TSB or TSB, 1 mm CaCl2 buffer and allowed to bind for 60 min. After the membrane was washed three times with TTSB supplemented with 1 mm CaCl2, the r0843 binding was detected with peroxidase-conjugated S-protein (Novagen) diluted 1:2000 in TTSB, 1% BSA, 1 mm CaCl2 and chemiluminescence (ECL detection kit; Amersham Biosciences). mAb 213-6, specifically recognizing the gp340, together with HRP-conjugated rabbit anti-mouse immunoglobulin, was used as a positive control.
The binding of r0843 to other glycoproteins was measured in an ELISA. Human plasma fibronectin (Chemicon), human laminin, bovine submaxillary mucin, asialomucin, fetuin, asialofetuin, bovine thyroglobulin, or BSA (all from Sigma) were coated onto 96-well microtiter plates (Nunc) at a concentration of 5 μg/ml in sodium carbonate buffer (50 mm; pH 9.6) and incubated overnight at 4 °C. The plates were washed with TTSB and blocked with 3% BSA for 60 min, after which r0843 was added at a concentration of 10 μg/ml in TTSB, 1% BSA, 1 mm CaCl2. After incubation of 60 min, the wells were washed three times with TTSB, 1 mm CaCl2, and peroxidase-conjugated S-protein, diluted 1:2000 in TTSB, 1% BSA, 1 mm CaCl2 was added into the wells. After a 60-min incubation, the wells were washed three times with TTSB, 1 mm CaCl2, and the amount of bound protein was detected colorimetrically with o-phenylenediamine. The assay was also performed in an identical way with buffers without CaCl2.
The gp340 binding was also confirmed in BiacoreX. Various amounts of the r0843 protein were allowed to bind to a CM3-chip coated with gp340 at flow rate of 20 μl/min as described above for whole bacteria. The calcium requirement of the gp340-r0843 interaction was confirmed in Biacore binding assays by dissolving the r0843 (40 μg/ml) in TSB buffer with different amounts of CaCl2 and using TSB buffer without CaCl2 as running buffer.
Inhibition of r0843 Binding
Inhibition of r0843 binding to gp340 by monosaccharides was tested in an ELISA. Equal volumes of the inhibitor solution (final concentration 50 mm) and r0843 (final concentration 10 μg/ml) were mixed. The mixture was incubated for 5 min and transferred into microtiter plate wells precoated with gp340 (10 μg/ml in 50 mm sodium carbonate), and the binding was measured as described above. To test the effect of sialidase treatment and NaIO4 oxidation on r0843 binding, immobilized gp340 was treated either with 0.1 units/ml sialidase from Vibrio cholerae (Roche Applied Science) in sodium acetate buffer (10 mm, pH 5.5) for 90 min at room temperature or oxidized with 10 mm NaIO4 in sodium acetate buffer (100 mm, pH 4.5) for 60 min at room temperature, followed by neutralization and reduction (50 mm NaBH4, 30 min). The control wells were treated with buffers only.
Six immuno grade peptides (Table 3) were purchased from Thermo Labs (Thermo Hybaid GmbH, Ulm, Germany) and dissolved in distilled water. Of the six peptides, only one (VNGGDRCAGRVEVLY) was not soluble in water or mild acetic acid or ammonia. The peptide was insoluble also to other tested solvents (DMSO, isopropyl alcohol, and acetonitrile) even after resynthesis and therefore could not be used in the experiments. The soluble peptides were added at the concentrations indicated with biotinylated gp340 (1 μg/ml) to r0843 (10 μg/ml)-coated microtiter plate wells, and binding of gp340 was detected in an ELISA with HRP-conjugated strepavidin and o-phenylenediamine.
TABLE 3.
Effect of synthetic peptides (100 μm) derived from the SRCR domain of gp340 on the binding of r0843 to gp340
The amino acids on SRCRP2 that are previously reported to be important in bacterial binding are underlined.
* Binding of r0843 to gp340 in the absence of any peptide was set as 100%. Data is expressed as mean ± SD from at least three experiments.
# ns, not soluble in solvents tested (water, DMSO, isopropyl alcohol, acetonitrile).
gp340 Binding of Truncated r0843 and Other Lrr Proteins
For binding analysis, the proteins were overexpressed as His-tagged recombinant proteins in E. coli, applied on SDS-polyacrylamide gel, and transferred to PVDF membrane. The nonspecific binding sites on the membrane were blocked with 3% BSA. For the truncated forms of r0843, biotinylated human gp340 (1 μg/ml in TTSB, 1% BSA, 1 mm CaCl2) or monoclonal anti-polyhistidine antibody (1:5000; Sigma) was added. After 60 min of binding, the membranes were washed three times with TTSB, 1 mm CaCl2. The membranes with gp340 or the polyhistidine antibody were stained with strepavidin-HRP (0.1 μg/ml) or rabbit anti-mouse HRP antibody (1:5000), respectively. After three washes, the binding was detected with ECL. Not all of the recombinant Lrr proteins from the other bacteria stained with the anti-polyhistidine antibody; therefore, Hisprobe-HRP (0.8 μg/ml; Pierce) was used. gp340 binding of the Lrr proteins on the PVDF membrane was tested by incubating the membranes for 60 min with fresh parotid saliva (diluted 1:2 in TTSB, 1% BSA, 1 mm CaCl2) and stained with mAb 213-6 antibody and rabbit anti-mouse-HRP antibody as above.
SRCR1A Inhibition of gp340 Binding to Lrr Proteins
The recombinant proteins were blotted to the membrane, and nonspecific binding sites were blocked as above. The SRCR1A (200 μm) peptide was added on the membrane and incubated for 60 min, after which an equal volume of fresh parotid saliva was added. After a 60-min incubation, the membrane was washed, and the amount of bound gp340 was measured with mAb 213-6 and ECL as above. Binding intensity was compared with the intensity of a membrane treated otherwise identically except that SRCR1A peptide was replaced with buffer.
RESULTS
Common Binding of Clinical S. pyogenes Isolates to gp340
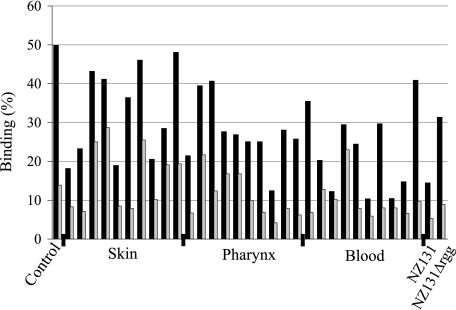
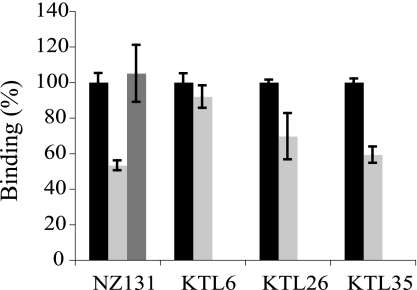
The gp340 binding among clinical isolates of S. pyogenes from skin, pharyngeal, and bacteremic infections, 10 in each group, was studied by bacterial binding to surface-adsorbed gp340 (Fig. 1). gp340 binding occurred, although to varying degrees, for all strains. Some strains showed high binding also to BSA-coated beads. Human albumin is recognized by M proteins and protein H of S. pyogenes (29), and it is possible that cross-reactions to BSA may occur. Still, all strains bound more efficiently to gp340-coated than to BSA-coated beads.
FIGURE 1.
Binding of S. pyogenes NZ131, NZ131Δrgg, and clinical isolates of different origin to human gp340. 35S-Labeled bacteria were allowed to bind to hydroxyapatite surface coated with gp340 (black bars) or BSA (gray bars) as a control. The binding is indicated as a percentage of bound bacteria from the total amount of added bacteria. Control, S. gordonii DL1.
The S. pyogenes regulatory element rgg deletion mutant strain is reported to have increased glycoprotein binding activity (30). We therefore tested the gp340 binding of S. pyogenes strain NZ131 and its rgg-deficient mutant NZ131Δrgg. Although both strains bound to gp340, the binding of the rgg-deficient mutant was stronger (Fig. 1), and it was chosen for adhesin identification studies.
Identification of Spy0843 as a gp340-binding Protein
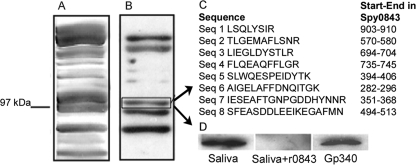
Distinct protein bands were identified in Western blots of trypsin-extracted S. pyogenes NZ131Δrgg surface proteins probed with biotinylated gp340 (Fig. 2). Mass spectrometry peptide data were obtained from one of these bands, and a data base search against the published genome of S. pyogenes serotype M1 strain SF370 identified the protein as Spy0843. The same protein band could be identified also with non-biotinylated purified gp340 as well as native gp340 in human saliva (Fig. 2D). Sequence analyses revealed a protein composed of a 23-amino acid signal peptide, the 143-amino acid N-terminal part, a middle part of ∼500 amino acids with 15 Lrr motifs, and a 360-amino acid C terminus with an LPRTG cell wall-anchoring motif. The recombinant Spy0843 protein produced in E. coli was able to inhibit the gp340 binding to the bacterial surface-extracted Spy0843 (Fig. 2D). The identity and true gp340 binding potential of the other detected protein bands remain to be solved.
FIGURE 2.
Identification of the gp340-binding protein Spy0843. A, trypsin-treated cell surface extract of S. pyogenes NZ131Δrgg was applied on SDS-polyacrylamide gel. B, extract was transferred to nitrocellulose membrane and probed with biotinylated gp340. C, peptide data from one of the strongly stained bands identified it as Spy0843. D, binding of trypsin-extracted cell surface Spy0843 with native gp340 in human saliva in the absence or presence of soluble recombinant Spy0843 protein (r0843) and with non-biotinylated gp340. All bindings were detected with monoclonal anti-gp340 antibody.
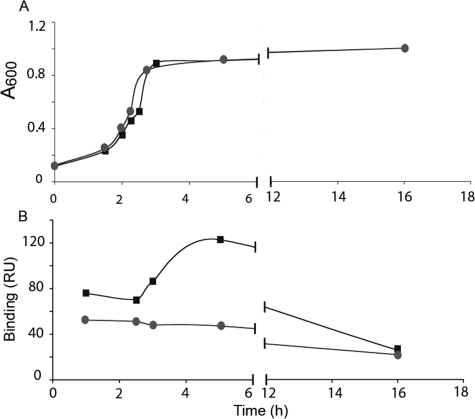
Inactivation of spy0843 did not affect the growth rate of the S. pyogenes NZ131 strain in THY medium at 37 °C (Fig. 3A). In a surface plasmon resonance assay, gp340 binding of Spy0843-deficient mutant was markedly affected when assayed in different growth phases (Fig. 3B). The binding of the wild type strain to gp340 increased during the growth and was highest at the early stationary phase. Late stationary phase bacteria showed the lowest binding. The mutant strain lacking Spy0843 also displayed low stationary phase binding, but the binding remained low during all growth phases. Similar results were obtained also in hydroxyapatite binding assay with stationary phase and logarithmic bacteria (not shown). Although it cannot be concluded that increased binding resulted from increased expression of Spy0843 on bacterial surface, there are previous data showing increased transcription of Spy0843 during the logarithmic phase of growth (31). The S. pyogenes NZ131Δ rgg mutant strain gave a high background binding to control surface, and therefore the experiments were run only with the S. pyogenes strains NZ131 and NZ131Δspy0843.
FIGURE 3.
Growth and gp340 binding of S. pyogenes wild type (NZ 131) and Δ0843. A, the growth of wild type (♦) and Δ0843 mutant (●) bacteria in THY medium at 37 °C was followed by recording A600. B, binding of bacteria to gp340 was measured by surface plasmon resonance. RU, resonance units.
Binding of Recombinant r0843 and Its Leucine-rich Repeat Region to gp340
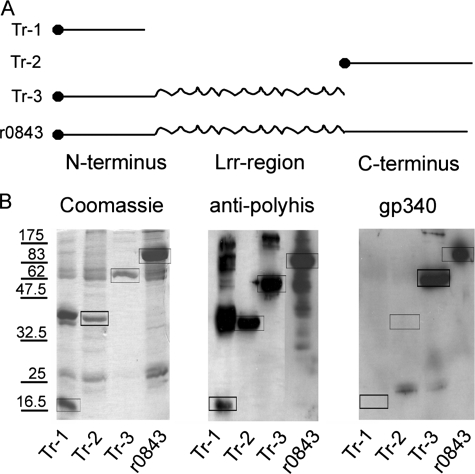
A recombinant Spy0843, r0843, and its truncated forms were expressed in E. coli to yield His/S-tagged proteins. The full-size protein (117 kDa) and truncates containing N-terminal (Tr-1; 17 kDa), C-terminal (Tr-2; 38 kDa), and N-terminal plus middle Lrr portion of the protein (Tr-3; 63 kDa) showed the expected molecular masses upon gel electrophoresis (Fig. 4). These proteins also stained with anti-polyhistidine antibody, which indicated that they were products of the transformed plasmids (Fig. 4B). In the Tr-1 sample, there was a strong staining of higher molecular weight proteins, the masses of which correspond to multimers of Tr-1. Binding of gp340 was observed only to full-size r0843 and to Tr-3, containing the N-terminal part including the Lrr-region. No binding could be seen with the truncated proteins containing only the N- or C-terminal fragments, indicating that the Lrr region of Spy084 is needed for gp340 binding.
FIGURE 4.
Expression and gp340 binding of r0843 and its truncated forms. A, r0843 and three different truncated forms were designed and expressed as His-tagged recombinant proteins in E. coli. B, the affinity-purified cell extracts were applied to SDS-polyacrylamide gels. The gels were stained with Coomassie Blue for proteins, or proteins were blotted on PVDF membrane probed with anti-polyhistidine antibody or biotinylated gp340. The boxes indicate proteins with the correct molecular mass.
Inhibition of S. pyogenes Binding to gp340 by Lrr Proteins
The effect of r0843 on the adhesion of S. pyogenes NZ131 and one clinical isolate from each type of infection (skin, pharyngeal, and bacteremic) was tested. Treatment of gp340-coated surfaces with r0843 inhibited the binding of S. pyogenes NZ131 to gp340, whereas incubation with the non-gp340 binding Lrr protein rYopM (see below) had no effect (Fig. 5). The efficacy of r0843 to inhibit the binding was significant but not complete with the tested concentration (up to 100 μg/ml). This is in accordance with the mass spectrometry analysis (Fig. 2), in which several proteins were found to bind to gp340. Complete inhibition with r0843 may not be achieved when additional adhesins are involved. The effect of r0843 on gp340 binding of clinical group A streptococci isolates varied; although the binding of KTL6 strain from skin infection was not affected, the binding of strains KTL26 and KTL35 from pharyngitis and bacteremia, respectively, were inhibited almost to the same level as S. pyogenes NZ131. All clinical isolates were spy0843-positive in PCR analysis (not shown), but the expression of the protein in these bacteria may vary. Thus, it appears that although Spy0843 mediates binding of many strains of S. pyogenes, it is not the only adhesin involved in gp340-S. pyogenes interactions.
FIGURE 5.
Inhibition of binding of different S. pyogenes strains to gp340-coated hydroxyapatite beads by leucine-rich repeat proteins r0843 (light gray) or rYopM (dark gray). The binding of bacteria to gp340 in the absence of leucine-rich repeat proteins is marked as 100% (black bars).
Glycoprotein Binding Properties of r0843
We next investigated the specificity of r0843 binding to gp340. In a Western blot assay with human parotid saliva, r0843 bound specifically to the same high molecular weight protein as the gp340-specific mAb 213-6 and to no other salivary (glyco)protein receptors (Fig. 6A). The binding specificity was also confirmed in an ELISA, where no binding to other tested glycoproteins was detected (Fig. 6B). The binding was found to be calcium-dependent, since no binding was observed in the absence of CaCl2. In surface plasmon resonance assays, the binding of r0843 to gp340 was dose-dependent, and at least 0.5 mm CaCl2 was required for optimal binding (Fig. 6, C and D).
FIGURE 6.
Binding characteristics of r0843. A, fresh human parotid saliva was subjected to SDS-PAGE, transferred to membrane, and probed with r0843 or the gp340-specific monoclonal antibody mAb 213-6 in the presence or absence of 1 mm CaCl2. Binding was detected with S-protein-HRP or with HRP-conjugated anti-mouse antibody. Control, staining with S-protein-HRP only. B, the binding of r0843 to gp340 and other glycoproteins in an ELISA in the presence (gray bars) and absence (black bars) of 1 mm calcium. C, different amounts (1, 4, 20, and 40 μg/ml from bottom to top) of r0843 were allowed to bind to the gp340-coated surface in a surface plasmon resonance assay. D, binding of r0843 (40 μg/ml) at different CaCl2 concentrations (0, 0.01, 0.02, 0.05, 0.5, and 1.0 mm from bottom to top) to the gp340-coated surface. RU, resonance units.
Inhibition of gp340 Binding
Both the carbohydrate portions of gp340 and its peptide core have been reported to mediate microbial binding (18, 32). In order to characterize the structures involved in Spy0843 binding, we first tested the effects of various monosaccharides and treatments affecting the glycopart of gp340 on r0843 binding. No clear evidence of the involvement of carbohydrates was observed in these experiments (Table 2).
TABLE 2.
Effects of monosaccharides (50 mm) and sialidase or NaIO4 treatments of gp340 on r0843 binding to gp340
The binding was determined by ELISA.
| Monosaccharide/treatment | Bindinga |
|---|---|
| % | |
| Glucose | 105 ± 17 |
| Galactose | 103 ± 19 |
| Fucose | 90 ± 3 |
| Mannose | 110 ± 19 |
| GalNAc | 103 ± 11 |
| Sialidase treatment | 99 ± 4 |
| NaIO4 oxidation | 89 ± 2 |
a Binding of r0843 in the absence of any treatment was set as 100%. Data are expressed as mean ± S.D. from at least three experiments.
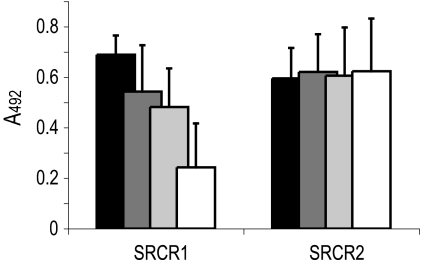
The role of the peptide core of gp340 in r0843 binding was next studied. A 16-mer peptide, SRCRP2, and especially the amino acids VEVLXXXXW in it, derived from the consensus sequence of the SRCR domains of gp340 has been previously reported to inhibit many microbial interactions of gp340 (19). SRCRP2 was found to inhibit the binding of r0843 in an ELISA (Table 3). To further investigate the essential peptide segments for r0843 binding, we designed two overlapping 15-mer peptides ranging 7 amino acids toward the amino and carboxyl terminus of the SRCR domain from the SRCRP2. Only one of these peptides, SRCR1A, inhibited the binding of r0843 to gp340. The SRCR1A peptide contains the VEVL sequence but is devoid of the XXXXW sequence. Consequently, the inhibitory motif of the peptide is different from the previously reported consensus sequence.
A protein of the group A of the SRCR family, MARCO, has been suggested to utilize the RXR motif of its SRCR domain in microbial binding (33). To evaluate a possible involvement of the bacterial RXR recognition motif in Spy0843 binding, we first demonstrated that replacement of glutamine in RXQXR with arginine to an RXRXR sequence (SRCR1), present in three SRCR domains of gp340, retained inhibitory activity (Table 3 and Fig. 7). In addition, two simultaneous Ala substitutions (AXAXR) abolished activity in SRCR1C, but a single Ala substitution (RXAXR) generated an insoluble peptide, SRCR1B, hampering further conclusions on the role of the RXR motif in Spy0843 binding.
FIGURE 7.
Inhibition of gp340 binding to r0843 in an ELISA by synthetic peptides derived from the SRCR domain of gp340 (Table 3). Black, 0 μm; dark gray, 10 μm; gray, 20 μm; white, 100 μm peptide concentration.
Characterization, Expression, and Binding of other Lrr Proteins
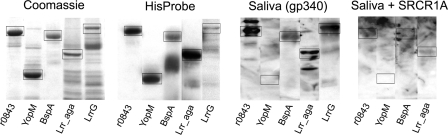
Three additional Lrr proteins, LrrG from S. agalactiae, BspA from T. forsythia, and YopM from Y. enterocolitica, were evaluated for binding to gp340 (Fig. 8). The proteins were expressed in E. coli as a full-size form. In addition, a truncated form of LrrG containing only the Lrr region of the protein was created (rLrr_aga). All partially purified cell extracts contained a protein with the expected molecular mass. These proteins also stained with HisProbe-HRP, although the staining of LrrG was weak. When probed with fresh human parotid saliva, a natural source of gp340, LrrG from S. agalactiae, BspA from T. forsythia, and r0843 from S. pyogenes bound to gp340, whereas YopM from Y. enterocolitica showed no binding. gp340 bound also to the truncated form of LrrG having only the Lrr region of the protein, further supporting the importance of Lrrs in gp340 binding. All binding Lrr proteins appeared to recognize the same epitope on gp340, since the peptide SRCR1A that efficiently inhibited binding of r0843 to gp340 (Table 3) also reduced the binding of gp340 to the other Lrr proteins (Fig. 8).
FIGURE 8.
Expression and gp340 binding of Lrr proteins. Recombinant proteins were expressed as His-tagged forms in E. coli and affinity-purified. The proteins were applied on SDS-polyacrylamide gels and stained with Coomassie Blue or transferred to PVDF membranes and stained with HisProbe or probed with human parotid saliva, a natural source of gp340, and mAb 213-6. For inhibition, the membrane was incubated with 200 μm SRCR1A peptide before adding parotid saliva. The boxes indicate proteins with the expected molecular mass of the respective protein.
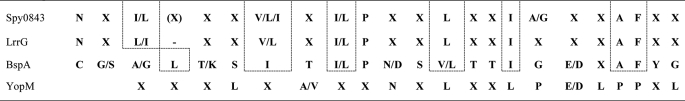
The LrrG protein of S. agalactiae has 16 complete Lrr repeats (34) and has 75% identity to Spy0843. Especially strong is the similarity in the Lrr region. The BspA protein of T. forsythia (28) has 21 Lrr repeats, but they show much lower similarity to those of Spy0843 (27% identity, 55% similarity). The arrangement of (iso)leucines in the repeats is, however, conserved in these proteins (Table 4), and they thus belong to same subgroup of Lrr proteins (35). The nature of the additional amino acids in the repeats varies in Spy0843 and LrrG, whereas they are more conserved in the repeats of BspA. The YopM protein of Y. enterocolitica is a representative of a different type of Lrr repeats, which are shorter and have an arrangement of leucines different from that of Spy0843 (Table 4).
TABLE 4.
Typical sequences of leucine rich repeats in TpLrr type Spy0843 from S. pyogenes, LrrG from S. agalactiae, BspA from T. forsythia and bacterial type YopM from Y. enterocolitica
Amino acids are indicated by one-letter code and are included in the sequence if present in half or more of the repeats.
DISCUSSION
gp340 appears to have diverse functions, being involved in cancer and tissue differentiation as well as in host innate defense and microbial diseases, such as Crohn disease and dental caries (4, 8, 12, 36). In innate immunity, gp340 functions as a scavenger receptor and is suggested to have a role in maintaining mucosal homeostasis by inhibiting bacterial invasion and down-regulating inflammation (5). Although some of the microbial interactions of gp340 are characterized in detail, the basis of its wide ligand recognition capacity has remained unclear. In the present study, we report that gp340 can recognize Lrr proteins both from the Gram-positive S. pyogenes and S. agalactiae and the Gram-negative T. forsythia. The binding appeared to be mediated by the Lrr motifs of the proteins. Recognition of such a common and conserved pattern on bacterial surface proteins may explain the wide ligand recognition ability of human scavenger receptor gp340.
Many pattern recognition receptors recognize molecular patterns present on bacterial surfaces. Lipopolysaccharides and lipoteichoic acids are such structures, but also surface proteins display repeated molecular patterns that can be widely dispersed among bacteria. Lrr motifs are currently found in more than 6000 proteins of eukaryotic and prokaryotic organisms (37). According to the length and consensus sequence of the repeats, seven subgroups of Lrr proteins have been identified (35). Two of these groups, “TpLrr” and “bacterial,” consist mainly of prokaryotic proteins, but bacterial proteins are found also in additional groups. For example, the InlA of Listeria monocytogenes and the Slr protein from S. pyogenes contain repeats similar to the yeast protein “SDS22” group (38, 39). Our results show that at least the TpLrr type repeats were recognized by gp340, whereas the one bacterial type protein tested was not. The recognition of additional Lrr-type repeats remains to be solved. Due to the conserved structure of the domains, binding activity to even one type of Lrr motifs enables binding to several types of ligands (e.g. Gram-negative and -positive bacteria), as shown in the present study.
Another interesting finding of the present study was the identification of gp340 as the host receptor for the Spy0843 surface protein of S. pyogenes. The Spy0843 protein is expressed during acute pharyngitis (40), and it is antigenic in humans (31). No host ligands for the protein have been characterized before. Although the biological function of Spy0843 is not known, antibodies raised against the protein are protective in a mouse infection model (31). The other two TpLrr type proteins, LrrG from S. agalactiae and BspA from T. forsythia, are surface proteins that mediate adhesion to cell surface receptors, such as fibronectin (28, 34). In our in vitro binding assay, Spy0843 did not bind to fibronectin, which suggests differences in host ligand recognition of these proteins. Although the functions of these proteins may differ, Lrr repeats appear to be conserved. Thereby, they are likely to be critical for bacterial survival and relatively invariant despite negative selective pressure by the immune system. Thus, they appear to be ideal ligands for a pattern recognition receptor. Interestingly, BspA interacts also with other pattern recognition receptors, such as the Toll-like receptor TLR2 (41), and recent findings suggest that Lrr domains are needed for this interaction (42). Thus, it may be that Lrr domains can serve as recognition motifs for other pattern recognition receptors as well.
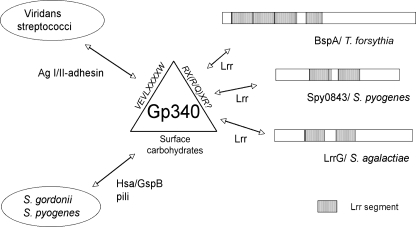
The gp340 scavenger protein contains multiple SRCR domains interspersed with O-glycosylated Ser- and Thr-rich SID (SRCR-interpersed domain) segments typical of mucins (3). Previous studies have identified bacterial adhesins binding to both carbohydrates as well as the peptide core of gp340 (summarized in Fig. 9). The sialic acid residues on gp340 mediate binding of some bacteria and viruses (18, 43, 44), and specific sialic acid adhesins, Hsa and GspB, have been identified in S. gordonii (45). Also, the AgI/II adhesins of viridans (oral) streptococci mediate binding to gp340 (17, 18, 46, 47). The peptide core of SRCR domains of gp340, and especially the amino acid sequence VEVLXXXXW, has been identified to be important in these interactions (19). Our peptide inhibition experiments showed that the peptides RCRGRVEVL and RCQGRVEVL, devoid of the XXXXW motif, were good inhibitors for Spy0843 and other Lrr protein binding, which suggested unique interaction of these proteins with the SRCR domains. Interestingly, the peptides contain an RX(R/Q) motif, which is hypothesized to be mediating the bacterial binding of MARCO, a group A SRCR protein, in its microbial binding (33). TLR2 also contains the motifs RXQ and QXQ, but their involvement in ligand binding is not known.
FIGURE 9.
Hypothetical model for bacteria-gp340 interactions. Viridans streptococci utilize the AgI/II adhesin to bind the VEVLXXXXW-epitope on gp340. In addition to AgI/II family polypeptides, some streptococci express carbohydrate-binding adhesins, like the sialic acid-binding adhesin Hsa or GspB of S. gordonii and pili proteins in S. pyogenes. The SRCR epitope, devoid of XXXXW and possibly utilizing the RX(R/Q)XR motif, is involved in recognition of conserved leucine-rich repeats in different bacterial surface proteins.
Data presented in this report suggest, however, that the Spy0843 is not the only protein mediating S. pyogenes-gp340 interaction. In fact, S. pyogenes pili proteins were recently reported to bind to gp340 (48). This interaction is inhibited by specific lectins but not by peptides containing the motif VEVLXXXXW, suggesting that group A streptococci pili might recognize complex carbohydrates on the gp340 surface. Thus, S. pyogenes appears to have multiple surface proteins that are differentially interacting with human gp340, as shown also in the restricted capacity of r0843 to inhibit gp340 binding of some group A streptococci isolates. This resembles the situation with S. gordonii, where at least three different surface molecules, recognizing both carbohydrate and protein epitopes on gp340, are involved in gp340 interactions (18, 47) (Fig. 9). In S. gordonii, these adhesins are differentially involved in interactions with surface-immobilized and liquid phase gp340. This can be true also for S. pyogenes, in which inactivation of the multiple gene regulator mga increased the binding of bacteria to liquid phase gp340 but decreased binding to surface-immobilized gp340 (18). It could be hypothesized that at different stages of growth, bacteria express surface molecules that interact differently with host immune molecules, such as gp340. In this way, bacteria could promote interactions with these molecules in stages where adhesion is required and escape interactions when needed. Expression pattern of differentially interacting surface molecules could also target the bacteria to different host sites. In that context, it is interesting to note that the expression of both Spy0843 and LrrG is increased in vivo during early phases of host-bacteria interaction (40, 49, 50).
In conclusion, the current study reveals a novel type of ligand recognition of human gp340, in which the scavenger receptor recognizes a common and conserved protein pattern, the Lrr motif, present in Gram-positive and Gram-negative bacteria. Such recognition provides a molecular basis for the binding of gp340 to many different types of ligands and helps us thus to understand how this molecule exhibits its wide ligand binding capacity. In addition, it appears that SRCR domains may expose a variety of binding motifs for recognition of different ligands, which further expands the binding capacity of the molecule. More generally, it can be hypothesized that the wide array of Lrr proteins among diverse bacteria and the conserved nature of both Lrr and the SRCR protein family could mean a novel mode of host pattern recognition based on bacterial Lrr protein families. Lrr-based recognition phenomena involving SRCR proteins, such as gp340, may thus play important roles in a wide array of diseases and host innate immune defense.
Acknowledgments
We thank Jukka Karhu for skillful technical assistance and Elise Pinta for help with Y. enterocolitica.
This work was supported by Grant 114100 from the Academy of Finland.
- SRCR
- scavenger receptor cysteine-rich
- Lrr
- leucine-rich repeat
- THY
- Todd Hewitt yeast extract broth
- BSA
- bovine serum albumin
- HRP
- horseradish peroxidase
- PVDF
- polyvinylidene difluoride
- mAb
- monoclonal antibody
- ELISA
- enzyme-linked immunosorbent assay.
REFERENCES
- 1.Sarrias M. R., Grønlund J., Padilla O., Madsen J., Holmskov U., Lozano F. (2004) Crit. Rev. Immunol. 24, 1–37 [DOI] [PubMed] [Google Scholar]
- 2.Holmskov U., Lawson P., Teisner B., Tornoe I., Willis A. C., Morgan C., Koch C., Reid K. B. (1997) J. Biol. Chem. 272, 13743–13749 [DOI] [PubMed] [Google Scholar]
- 3.Holmskov U., Mollenhauer J., Madsen J., Vitved L., Gronlund J., Tornoe I., Kliem A., Reid K. B., Poustka A., Skjodt K. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 10794–10799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mollenhauer J., Herbertz S., Holmskov U., Tolnay M., Krebs I., Merlo A., Schrøder H. D., Maier D., Breitling F., Wiemann S., Gröne H. J., Poustka A. (2000) Cancer Res. 60, 1704–1710 [PubMed] [Google Scholar]
- 5.Rosenstiel P., Sina C., End C., Renner M., Lyer S., Till A., Hellmig S., Nikolaus S., Fölsch U. R., Helmke B., Autschbach F., Schirmacher P., Kioschis P., Hafner M., Poustka A., Mollenhauer J., Schreiber S. (2007) J. Immunol. 178, 8203–8211 [DOI] [PubMed] [Google Scholar]
- 6.Mueller A., O'Rourke J., Grimm J., Guillemin K., Dixon M. F., Lee A., Falkow S. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 1292–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kang W., Nielsen O., Fenger C., Madsen J., Hansen S., Tornoe I., Eggleton P., Reid K. B., Holmskov U. (2002) Clin. Exp. Immunol. 130, 449–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Renner M., Bergmann G., Krebs I., End C., Lyer S., Hilberg F., Helmke B., Gassler N., Autschbach F., Bikker F., Strobel-Freidekind O., Gronert-Sum S., Benner A., Blaich S., Wittig R., Hudler M., Ligtenberg A. J., Madsen J., Holmskov U., Annese V., Latiano A., Schirmacher P., Amerongen A. V., D'Amato M., Kioschis P., Hafner M., Poustka A., Mollenhauer J. (2007) Gastroenterology 133, 1499–1509 [DOI] [PubMed] [Google Scholar]
- 9.Schulz B. L., Oxley D., Packer N. H., Karlsson N. G. (2002) Biochem. J. 366, 511–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prakobphol A., Xu F., Hoang V. M., Larsson T., Bergstrom J., Johansson I., Frängsmyr L., Holmskov U., Leffler H., Nilsson C., Borén T., Wright J. R., Strömberg N., Fisher S. J. (2000) J. Biol. Chem. 275, 39860–39866 [DOI] [PubMed] [Google Scholar]
- 11.Ligtenberg T. J., Bikker F. J., Groenink J., Tornoe I., Leth-Larsen R., Veerman E. C., Nieuw Amerongen A. V., Holmskov U. (2001) Biochem. J. 359, 243–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jonasson A., Eriksson C., Jenkinson H. F., Källestål C., Johansson I., Strömberg N. (2007) BMC Infect. Dis. 7, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hartshorn K. L., White M. R., Mogues T., Ligtenberg T., Crouch E., Holmskov U. (2003) Am. J. Physiol. Lung Cell Mol. Physiol. 285, L1066–L1076 [DOI] [PubMed] [Google Scholar]
- 14.Wu Z., Van Ryk D., Davis C., Abrams W. R., Chaiken I., Magnani J., Malamud D. (2003) AIDS Res. Hum. Retroviruses 19, 201–209 [DOI] [PubMed] [Google Scholar]
- 15.Crowley P. J., Brady L. J., Piacentini D. A., Bleiweis A. S. (1993) Infect. Immun. 61, 1547–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oho T., Yu H., Yamashita Y., Koga T. (1998) Infect. Immun. 66, 115–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holmes A. R., Gilbert C., Wells J. M., Jenkinson H. F. (1998) Infect. Immun. 66, 4633–4639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loimaranta V., Jakubovics N. S., Hytönen J., Finne J., Jenkinson H. F., Strömberg N. (2005) Infect. Immun. 73, 2245–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bikker F. J., Ligtenberg A. J., End C., Renner M., Blaich S., Lyer S., Wittig R., van't Hof W., Veerman E. C., Nazmi K., de Blieck-Hogervorst J. M., Kioschis P., Nieuw Amerongen A. V., Poustka A., Mollenhauer J. (2004) J. Biol. Chem. 279, 47699–47703 [DOI] [PubMed] [Google Scholar]
- 20.White M. R., Crouch E., van Eijk M., Hartshorn M., Pemberton L., Tornoe I., Holmskov U., Hartshorn K. L. (2005) Am. J. Physiol. Lung Cell Mol. Physiol. 288, L831–L840 [DOI] [PubMed] [Google Scholar]
- 21.Chaussee M. S., Ajdic D., Ferretti J. J. (1999) Infect. Immun. 67, 1715–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosenfeld J., Capdevielle J., Guillemot J. C., Ferrara P. (1992) Anal. Biochem. 203, 173–179 [DOI] [PubMed] [Google Scholar]
- 23.Shevchenko A., Jensen O. N., Podtelejnikov A. V., Sagliocco F., Wilm M., Vorm O., Mortensen P., Shevchenko A., Boucherie H., Mann M. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 14440–14445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nyman T. A., Matikainen S., Sareneva T., Julkunen I., Kalkkinen N. (2000) Eur. J. Biochem. 267, 4011–4019 [DOI] [PubMed] [Google Scholar]
- 25.Kussmann M., Lässing U., Stürmer C. A., Przybylski M., Roepstorff P. (1997) J. Mass Spectrom. 32, 483–493 [DOI] [PubMed] [Google Scholar]
- 26.Pulliainen A. T., Haataja S., Kähkönen S., Finne J. (2003) J. Biol. Chem. 278, 7996–8005 [DOI] [PubMed] [Google Scholar]
- 27.Segers R. P., Kenter T., de Haan L. A., Jacobs A. A. (1998) FEMS Microbiol. Lett. 167, 255–261 [DOI] [PubMed] [Google Scholar]
- 28.Sharma A., Sojar H. T., Glurich I., Honma K., Kuramitsu H. K., Genco R. J. (1998) Infect. Immun. 66, 5703–5710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Akesson P., Schmidt K. H., Cooney J., Björck L. (1994) Biochem. J. 300, 877–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hytönen J., Haataja S., Finne J. (2003) Infect. Immun. 71, 784–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reid S. D., Green N. M., Sylva G. L., Voyich J. M., Stenseth E. T., DeLeo F. R., Palzkill T., Low D. E., Hill H. R., Musser J. M. (2002) J. Bacteriol. 184, 6316–6324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bikker F. J., Ligtenberg A. J., Nazmi K., Veerman E. C., van't Hof W., Bolscher J. G., Poustka A., Nieuw Amerongen A. V., Mollenhauer J. (2002) J. Biol. Chem. 277, 32109–32115 [DOI] [PubMed] [Google Scholar]
- 33.Brännström A., Sankala M., Tryggvason K., Pikkarainen T. (2002) Biochem. Biophys. Res. Commun. 290, 1462–1469 [DOI] [PubMed] [Google Scholar]
- 34.Seepersaud R., Hanniffy S. B., Mayne P., Sizer P., Le Page R., Wells J. M. (2005) Infect. Immun. 73, 1671–1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kobe B., Kajava A. V. (2001) Curr. Opin. Struct. Biol. 11, 725–732 [DOI] [PubMed] [Google Scholar]
- 36.Kang W., Reid K. B. (2003) FEBS Lett. 540, 21–25 [DOI] [PubMed] [Google Scholar]
- 37.Matsushima N., Tanaka T., Enkhbayar P., Mikami T., Taga M., Yamada K., Kuroki Y. (2007) BMC Genomics 8, 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gaillard J. L., Berche P., Frehel C., Gouin E., Cossart P. (1991) Cell 65, 1127–1141 [DOI] [PubMed] [Google Scholar]
- 39.Reid S. D., Montgomery A. G., Voyich J. M., DeLeo F. R., Lei B., Ireland R. M., Green N. M., Liu M., Lukomski S., Musser J. M. (2003) Infect. Immun. 71, 7043–7052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Virtaneva K., Graham M. R., Porcella S. F., Hoe N. P., Su H., Graviss E. A., Gardner T. J., Allison J. E., Lemon W. J., Bailey J. R., Parnell M. J., Musser J. M. (2003) Infect. Immun. 71, 2199–2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hajishengallis G., Martin M., Sojar H. T., Sharma A., Schifferle R. E., DeNardin E., Russell M. W., Genco R. J. (2002) Clin. Diagn. Lab. Immunol. 9, 403–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Onishi S., Honma K., Liang S., Stathopoulou P., Kinane D., Hajishengallis G., Sharma A. (2008) Infect. Immun. 76, 198–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.White M. R., Crouch E., Vesona J., Tacken P. J., Batenburg J. J., Leth-Larsen R., Holmskov U., Hartshorn K. L. (2005) Am. J. Physiol. Lung Cell Mol. Physiol. 289, L606–L616 [DOI] [PubMed] [Google Scholar]
- 44.Hartshorn K. L., Ligtenberg A., White M. R., Van Eijk M., Hartshorn M., Pemberton L., Holmskov U., Crouch E. (2006) Biochem. J. 393, 545–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takamatsu D., Bensing B. A., Prakobphol A., Fisher S. J., Sullam P. M. (2006) Infect. Immun. 74, 1933–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jenkinson H. F., Demuth D. R. (1997) Mol. Microbiol. 23, 183–190 [DOI] [PubMed] [Google Scholar]
- 47.Jakubovics N. S., Kerrigan S. W., Nobbs A. H., Strömberg N., van Dolleweerd C. J., Cox D. M., Kelly C. G., Jenkinson H. F. (2005) Infect. Immun. 73, 6629–6638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Edwards A. M., Manetti A. G., Falugi F., Zingaretti C., Capo S., Buccato S., Bensi G., Telford J. L., Margarit I., Grandi G. (2008) Mol. Microbiol. 68, 1378–1394 [DOI] [PubMed] [Google Scholar]
- 49.Graham M. R., Virtaneva K., Porcella S. F., Barry W. T., Gowen B. B., Johnson C. R., Wright F. A., Musser J. M. (2005) Am. J. Pathol. 166, 455–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mereghetti L., Sitkiewicz I., Green N. M., Musser J. M. (2008) PLoS ONE 3, e3143. [DOI] [PMC free article] [PubMed] [Google Scholar]