Abstract
Antibody class switching occurs in mature B cells in response to antigen stimulation and costimulatory signals. It occurs by a unique type of intrachromosomal deletional recombination within special G-rich tandem repeated DNA sequences [called switch, or S, regions located upstream of each of the heavy chain constant (CH) region genes, except Cδ]. The recombination is initiated by the B cell–specific activation-induced cytidine deaminase (AID), which deaminates cytosines in both the donor and acceptor S regions. AID activity converts several dC bases to dU bases in each S region, and the dU bases are then excised by the uracil DNA glycosylase UNG; the resulting abasic sites are nicked by apurinic/apyrimidinic endonuclease (APE). AID attacks both strands of transcriptionally active S regions, but how transcription promotes AID targeting is not entirely clear. Mismatch repair proteins are then involved in converting the resulting single-strand DNA breaks to double-strand breaks with DNA ends appropriate for end-joining recombination. Proteins required for the subsequent S-S recombination include DNA-PK, ATM, Mre11-Rad50-Nbs1, γH2AX, 53BP1, Mdc1, and XRCC4-ligase IV. These proteins are important for faithful joining of S regions, and in their absence aberrant recombination and chromosomal translocations involving S regions occur.
Keywords: AID, UNG, AP endonuclease, DNA Pol β, DNA-PK, XRCC4-DNA ligase IV, ATM, Mre11-Nbs1-Rad50, 53BP1, γH2AX, germline transcripts, R-loops, end joining
INTRODUCTION AND OVERVIEW
Antibody class, or isotype, is determined by the heavy chain constant (CH) region, which is important for determining the antibody’s effector function. The CH region is bound by cell-surface receptors, e.g., Fc receptors on many cell types and poly immunoglobulin (Ig) receptors on mucosal epithelial cells, and also by complement. Different CH regions have different affinities for these proteins, thus greatly influencing antibody function and determining whether antibody-antigen complexes will activate cells that help to eliminate pathogens, e.g., macrophages, NK cells, or mast cells. Also, the CH region determines whether an antibody can be transcytosed through epithelial membranes at mucosal surfaces, can diffuse into tissues, and will polymerize and thereby have a greater avidity. The CH region also influences the stability of the antibody (reviewed in 1). The membrane-bound forms of the various iso-types differ in their intracytoplasmic carboxy termini; the different termini result in varying abilities to associate at cell membranes with intracellular signaling proteins, although the biological roles of these differences are not yet understood (2–4).
Ig isotype switching occurs by an intrachromosomal deletional recombination event, diagrammed in Figure 1 for the mouse H chain locus. The human H chain locus is similarly organized but not identical. Class switch recombination (CSR) occurs between switch (S) regions located upstream of each of the CH regions except Cδ and results in a change from IgM and IgD expression by naive B cells to expression of one of the downstream isotypes. IgD expression occurs by alternative transcription termination/splicing of the Cμ-Cδ genes. S regions consist of tandem repeats of short G-rich sequences (20–80 bp), which differ for each isotype, with an overall length varying from ~1 kb to 12 kb, and CSR can occur anywhere within or near the S regions (5, 6). CSR occurs by an end-joining type of recombination, rather than by homologous recombination (7, 8).
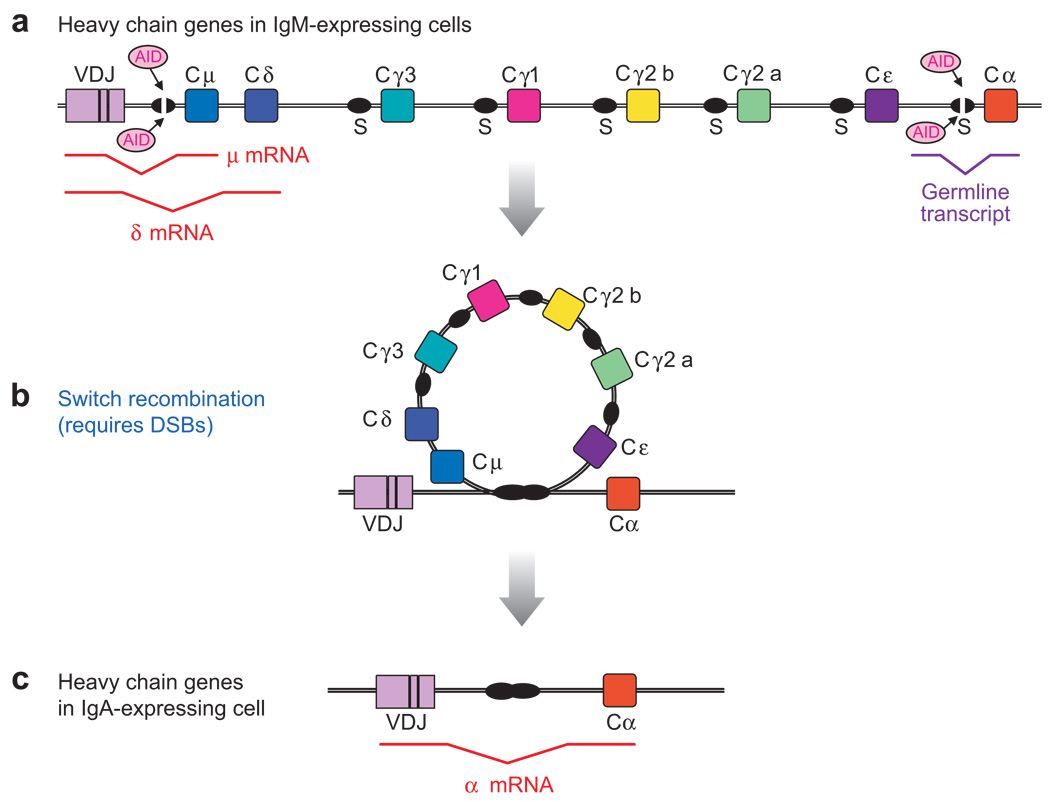
Figure 1.
Diagram of Ig class switch recombination (CSR) to IgA. (a) The mouse IgH locus in B cells expressing IgM and IgD (by alternative RNA transcription/processing). During CSR, activation-induced cytidine deaminase (AID) deaminates dC residues in the top and bottom strands of transcriptionally active S regions (Sμ and Sα in the diagram shown), initiating a process (described in the text) that results in double-strand DNA breaks (DSBs) in both S regions and CSR by intrachromosomal deletion (b). (c) The IgH locus after CSR to IgA. Splicing diagrams of the μ, δ mRNAs and the germline α transcript are indicated below the diagram of the locus. Similar germline transcripts are induced from unrearranged Cγ,Cε, and Cα genes, depending on the cytokine stimulation received by the B cell.
CSR and somatic hypermutation (SHM) are initiated by activation-induced cytidine deaminase (AID), which converts cytosines in S regions and Ig variable regions to uracils by deamination (9–14). Subsequent repair of the dU residues leads to single-strand DNA breaks (SSBs) that must be converted to double-strand breaks (DSBs) within the donor Sμ region and within an acceptor Sx region, to initiate the process of intrachromosomal DNA recombination. This review focuses mainly on the overall mechanism of CSR, which is discussed in the next section. Although there are interesting similarities and differences between CSR and SHM, we do not discuss them owing to space constraints. SHM is reviewed in another article in this volume by M.D. Scharff (15). Also, we do not extensively review all the information available about AID, as this protein is extensively discussed in the Scharff article (15) and in several other reviews (16–19).
B cells undergo antibody, or Ig, class switching in vivo after immunization or infection or upon appropriate activation in culture. Engagement of the CD40 receptor on B cells by CD154 (CD40L) or, specifically for mouse B cells, the Toll-like receptor 4 (TLR4) by lipopolysaccharide (LPS), provides crucial signaling for CSR. AID expression is induced in mouse splenic B cells activated to switch in culture, and also in vivo, with especially high levels detected in germinal center (GC) B cells, which are undergoing SHM and probably CSR (9, 20, 21). Most investigations into the roles of various genes in CSR examine their effects in mouse splenic B cells induced to switch in culture. This model allows one to use the numerous mouse gene knockout models and also ensures that the effects of the genes are B cell intrinsic and not due to effects on other cell types.
CSR requires cell proliferation, appearing to require a minimum of two complete rounds of cell division for IgG and IgA CSR and perhaps additional rounds for IgE CSR (22–25). This requirement appears to be at least partly due to the requirements for induction of AID expression (25). Transcription of AID mRNA is induced synergistically by IL-4 and CD40 signaling via induction of Stat6 and NF-κB transcription factors (26). However, these signals are very rapid. Pax5 is also essential for AID mRNA transcription, and Pax5 binds to the AID promoter in LPS+IL-4-treated splenic B cells (27). Most interestingly, binding of Pax5 to the AID promoter is not detected until two days after addition of the activators, suggesting that the kinetics of Pax5 binding might be important for explaining the requirement for cell division for AID induction. Furthermore, AID function is regulated by active export from the nucleus (28–30), which might also contribute to the delay in CSR.
Naive B cells have the potential to switch to any isotype, and cytokines secreted by T cells and other cells direct the isotype switch (reviewed in 7, 31, 32). Although there is more to be discovered, the predominant mechanism for regulating isotype specificity is by regulation of transcription through S regions, and only transcriptionally active S regions undergo CSR. The regulation of isotype specificity is further discussed in the section on “Regulation of Switching.”
MECHANISM OF CSR
Recently, the greatest progress in the field of CSR has occurred in identifying and understanding the roles of the enzymes and proteins involved in both creating DNA breaks and recombining the S regions. This, then, is the focus of our review. In this section, we start with the enzymes that create the DSBs required for CSR and then discuss the joining mechanism and its regulation. We restrict our discussion to proteins known to contribute to CSR and do not mention proteins shown not to be involved or for which the contribution is only suggestive or might have only minor effects. Pan-Hammarstrom et al. (33) comprehensively reviewed which DNA repair proteins are involved in V(D)J, CSR, and/or SHM. Other recent reviews discuss the mechanism and regulation of CSR (18, 34–37).
Creation of DSBs for CSR
A reasonably convincing model for how single-strand breaks (SSBs) are introduced into S regions and how they are converted to the DSBs required for CSR has been developed in recent years. After initiation of the process by AID, the base excision repair (BER) pathway creates the SSBs, and if the SSBs on opposite strands are sufficiently near, this results in a DSB. However, conversion of more distal SSBs to DSBs appears to require another DNA repair pathway, mismatch repair (MMR). The proteins and their function at each step in this process are discussed in this section.
Activation-induced cytidine deaminase (AID)
The finding that AID is essential for both CSR and SHM was a major break-through for these fields (9, 10). Although originally investigators postulated that AID is an RNA-editing enzyme, owing to its homology with the RNA-editing cytidine deaminase APOBEC-1 (9), a second major break-through came with the discovery that the role of AID is to initiate these processes by deamination of dC nucleotides within S regions and antibody variable regions (11–14, 38, 39). In 2000, a model was proposed by Poltoratsky et al. (40) for how deamination of dC in DNA could lead to the SSBs required for CSR, and shortly thereafter Petersen-Mahrt et al. (11) provided evidence for this model. This model is now supported by numerous studies. Using the technique of linker-ligation-mediated PCR (LM-PCR), researchers demonstrated that AID is required for generation of S region DSBs in both mouse and human B cells (21, 41, 42). Also, localization of γH2AX foci at the Ig locus during CSR is AID-dependent (43). Figure 2a presents the portion of this model that is relevant for CSR.
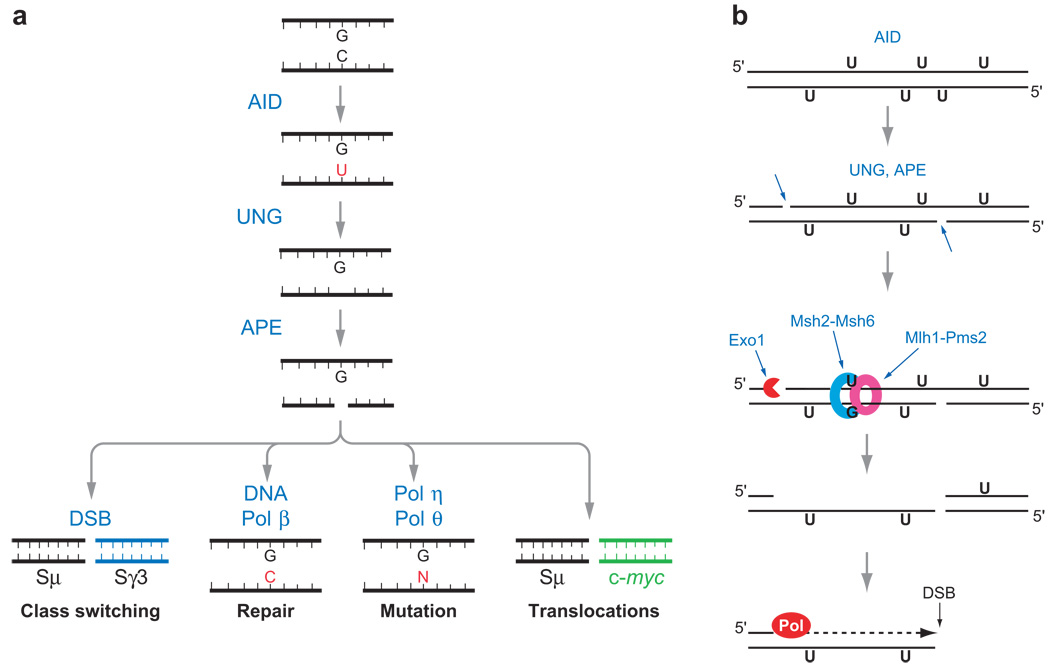
Figure 2.
Diagrams of (a) model for generation of DNA breaks, mutations, and translocations in IgS regions by AID-UNG-APE and (b) model for conversion of SSBs to DSBs by mismatch repair. (a) AID deaminates dC, resulting in dU bases, which are excised by one of the uracil DNA glycosylases, UNG. Abasic sites are cut by AP-endonuclease (APE1 and APE2) (74), creating SSBs that can spontaneously form DSBs if they are near each other on opposite DNA strands, or, if not, mismatch repair activity converts them to DSBs (see b). Alternatively, DNA Pol β can correctly repair the nick, preventing CSR, or error-prone translesion polymerases can repair the nick but introduce mutations. Finally, the DNA breaks can lead to aberrant recombinations/translocations. (b) AID is hypothesized to introduce several dU residues in S regions during one cell cycle. Some of the dU residues are excised by UNG, and some of the abasic sites are nicked by APE. The U:G mismatches that remain would be substrates for Msh2-–Msh6 (100). Msh2-–Msh6, along with Mlh1-Pms2, recruit Exo1 (and accessory proteins) to a nearby 5′ nick, from where Exo1 begins to excise toward the mismatch (90, 91), creating a DSB with a 5′ single-strand overhang, which can be filled in by DNA polymerase. Fill-in synthesis is probably performed by translesion polymerases owing to the presence of abasic sites. Alternatively, the overhang is removed by a 5′ flap endonuclease (Fen1) or by Exo1.
Purified recombinant AID converts dC to dU nucleotides in single-stranded (ss) DNA and in supercoiled transcribed plasmids (12–16, 18, 44–46). It is clear that ssDNA but not dsDNA is the AID substrate. When transcribed duplex DNA substrates are used in vitro or in Escherichia coli, the nontranscribed strand (top strand if the DNA sequence is oriented with the 5′ side to the left) is preferentially targeted. This may be because the nontranscribed strand in these substrates is single-stranded at the small bubble formed by RNA polymerase, whereas the transcribed strand is transiently hydrogen bonded to RNA (12, 14, 44, 47). However, both strands can be targeted by AID in vitro in transcribed supercoiled plasmid DNA, presumably because single-strand extrusions can occur in super-coiled plasmids (45, 46). It is clear that in B cells, dC on both transcribed and nontranscribed strands is equally mutated in proportion to its distribution in V genes and S regions, indicating that AID attacks both strands (48–50).
AID preferentially, but not exclusively, deaminates the underlined dC in WRC(W = A or T, R = purine, Y = pyrimidine), the AID hotspot target (51–53), in transcribed targets in vitro (54), in oligonucleotide substrates (55), and in vivo (50). S regions contain numerous AID hotspot targets. However, experiments using purified recombinant AID and plasmid DNA substrates suggest that AID is highly processive (14, 45, 54), and in this process AID deaminates many dC nucleotides that are not in WRC motifs. Furthermore, studies in ung−/− msh2−/− B cells, in which the initial AID-induced lesions cannot be repaired and thus can be examined, also suggest that AID acts processively across S regions in vivo (50).
The regulation of AID function is just beginning to be studied. AID is phosphorylated by protein kinase A at S38 and T27, and alanine mutations at either of these sites nearly eliminate the ability of AID to initiate CSR when retrovirally transduced into aid−/− splenic B cells (56, 57, 58). Phosphorylation by protein kinase A is required for binding of AID by replication protein A, which enables AID to deaminate transcribed duplex DNA. Replication protein A appears to increase the interaction of AID with DNA. Another very interesting but not understood discovery about AID is that the C-terminal 10 amino acids of AID are required for CSR but not for SHM (59, 60). The C terminus may be required for interaction with a protein essential for CSR but not for SHM.
UNG
Removal of the dU residues by enzymes within the BER pathway is required to introduce the DNA breaks necessary for CSR (11, 21, 38). BER consists of highly active ubiquitous DNA repair pathways for repairing oxidized and deaminated bases, which are generated more than 104 times per cell per day by oxidation, especially during inflammation, and by spontaneous hydrolysis (61). There are four mammalian uracil DNA glycosylases in the BER pathway (i.e., enzymes that excise dU bases), UNG, SMUG1, MBD4, and TDG. CSR is reduced by 95% in B cells from UNG-deficient mice and in patients that have deleterious mutations in UNG (21, 38, 62). Furthermore, S region DSBs are also greatly reduced in ung−/− mouse splenic B cells induced to undergo CSR (21). Thus, it is reasonable to conclude that UNG is the uracil DNA glycosylase that excises the dU residue created by AID activity. Two other uracil glycosylases (MBD4 and SMUG1) appear to have no role in CSR (38, 63). However, when SMUG1 is greatly overexpressed by retroviral transduction, it can support a limited amount of CSR in ung−/− cells, although in the presence of UNG, SMUG1 actually inhibits CSR (64). These data suggest that UNG may be specifically recruited to sites of AID activity and perhaps has a unique property that makes it the preferred uracil DNA glycosylase for generation of the DNA breaks required for CSR.
Apurinic/apyrimidinic endonuclease (AP endonuclease/APE)
The BER enzyme that repairs the abasic sites left by UNG activity is APE, which incises the phosphate backbone of DNA at abasic sites, producing SSBs (61). In mammals, there are three enzymes with AP endonuclease activity, APE1 and APE2, which share homology (65), and a very different, recently discovered enzyme, PALF/APLF/XIP-1 (66–68). The main AP endonuclease known to be involved in BER is APE1. APE1 endonuclease activity is essential for early embryonic development and for viability of human cell lines (69, 70). Much less is known about APE2, which is encoded on the X chromosome. APE2-deficient mice show a slight growth defect and have a twofold reduction of white blood cells in the periphery, mainly affecting T and B cells (71). Enzyme assays using abasic site-containing oligonucleotide substrates showed that purified recombinant human APE2 has weaker AP endonuclease activity than APE1 (65, 72, 73).
Examination of APE1- and/or APE2-deficient mouse splenic B cells activated to switch in culture demonstrated that both of these enzymes contribute to CSR (74). Because APE1 is an essential gene, the investigators used ape1+/− mice, which have DNA repair defects (75, 76), although the phenotype was probably less severe than if ape1−/− mice could have been studied. Splenic B cells from mice deficient for APE2 and heterozygous for APE1 switch 60%–80% as well as wild-type B cells, depending on the isotype, and have very few S region DSBs, almost as few as in aid−/− cells, which have approximately 10% of wild-type (77). These cells divide in culture as well as wild-type cells, so the reduced CSR is not due to poor proliferation (74). Although CSR is modestly reduced, the much greater reduction in Sμ DSBs suggests that Sμ DSBs are not limiting for CSR.
In wild-type cells, AID-dependent blunt and staggered DSBs in the Sμ region occur preferentially at G:C bp and at AID hotspot targets, with p values for blunt DSBs ≤ 0.002 relative to random (21, 74). This indicates that SSBs and DSBs occur at the dC nucleotides that are targeted by AID, as predicted by the DNA-deamination model (Figure 2a). In aid−/− or ung−/− or APE-deficient cells, DSBs do not specifically occur at AID hotspot targets, although B cells deficient in only one of the APEs maintain some specificity for AID targets (21, 74). These data suggest that APE1 and APE2 both serve as endonucleases to incise abasic sites introduced by AID and UNG. However, whether the third AP endonuclease PALF/APLF/XIP-1 also has a role is unknown.
DNA polymerase β (Pol β)
In the canonical BER pathway, the single-nucleotide gap generated by the action of UNG and APE is filled in by DNA polymerase β (Pol β) (78, 79). A multiprotein complex that can perform BER and that contains UNG2 (nuclear form of UNG), APE1, Pol β, replicative DNA polymerases δ and ε, XRCC1, and DNA ligase I has been isolated from both proliferating and growth-arrested HeLa cells and also from human peripheral blood lymphocytes (80, 81). Furthermore, physical interactions among BER enzymes increase repair efficiency (82–84). These findings suggest that BER will proceed to completion once it is initiated by UNG. Therefore, Pol β activity could reduce SSBs and therefore reduce CSR.
Hence, an intriguing question arises as to how S region breaks are spared from faithful repair. One appealing hypothesis is that BER components downstream of UNG and APE might be downregulated in cells undergoing CSR or specifically prevented from accessing S region lesions. As Pol β is recruited by APE1 (78), the levels of Pol β or its activity may be inhibited during CSR, or APE2 may not recruit Pol β, which could explain why APE2 is used for CSR. Alternatively, the introduction of numerous S region lesions may overwhelm the BER machinery, although BER activity is not inhibited during CSR. A recent report supports the second alternative (85). Pol β levels were increased in nuclear extracts from mouse splenic B cells induced to undergo CSR, and chromatin immunoprecipitation showed that Pol β associates specifically with the Sμ region, but not with Cμ or Cα genes, in switching B cells.
Although Pol β–deficient mice die in utero, one can obtain polβ−/− splenic B cells by transfer of polβ −/− fetal liver cells into irradiated wild-type mice. Using polβ −/− and polβ+/+ splenic B cells obtained by fetal liver transfer, investigators found that polβ−/− cells actually show modestly increased CSR (1.5–to 1.7-fold) to a subset of isotypes (IgG2a, IgG2b, and IgG3) relative to polβ+/+ controls. Furthermore, LM-PCR experiments showed that polβ−/− cells have a two- to threefold increase in Sμ and Sγ3 DSBs and a twofold increase in mutations in the germline (GL) Sμ and recombined Sμ segments. These data indicate that Pol β indeed functions to accurately repair AID-instigated SSBs but cannot repair them all, and the authors hypothesize that Pol β only inhibits switching when SSBs are limiting (85).
If B cells were to downregulate BER during CSR, this could be dangerous, given the great amount of reactive oxygen species produced during B cell activation and proliferation (86). Therefore, it is plausible that instead a mechanism is adopted that endows S regions with such numerous AID targets that the ability of BER to repair them is overwhelmed, rather than abrogating overall BER ability and thus jeopardizing the integrity of the B cell genome. Consistent with this hypothesis is the finding that, in ung−/−msh2−/− B cells, AID introduces many more lesions into the Sμ region than result in mutations in wild-type cells, most likely because they are correctly repaired in wild-type cells (50). Furthermore, recent experiments show that artificially introduced I-SceI sites in Sμ and Sγ1 regions mediate CSR to IgG1. These experiments suggest that only a single DSB in the donor and acceptor S region is sufficient for CSR (87). Introduction of numerous dU residues increases the likelihood of DSBs occurring in the donor and acceptor S regions simultaneously. In conclusion, Pol β may function normally during CSR to repair AID-initiated DNA lesions, but the numerous AID lesions overwhelm it, and thus some breaks remain unrepaired.
Role of mismatch repair in CSR: to convert SSBs to DSBs
A second repair pathway, mismatch repair (MMR), contributes to CSR but is not essential. The major role of MMR in all cells is to correct misincorporated nucleotides during DNA synthesis (88). This process involves recognition of the mismatch by a heterodimer of Msh2-–Msh6 (for nucleotide substitutions and small loops) or Msh2–Msh3 (for larger loops), followed by recruitment of the Mlh1-Pms2 heterodimer (88). The combined heterotetramer recruits replication factor C, the processivity factor proliferating cell nuclear antigen (PCNA), and Exonuclease1 (Exo1) to a nearby nick, and together they excise the single-strand segment containing the mutated nucleotide (89, 90). The excised single-strand patch can be hundreds of nucleotides long in vitro, but the length in vivo is unknown. MMR specifically repairs the newly synthesized DNA strand, probably because of its predilection to excise and resynthesize the nicked DNA strand (91). In mice that lack one of the MMR genes (Msh2, Msh6, Mlh1, Pms2, or Exo1), CSR is reduced by two- to sevenfold, depending on the gene and the Ig isotype (92–98).
The most attractive model for the role of MMR during CSR is to convert SSBs that are not near each other on the opposite DNA strands to DSBs (77, 99). If the SSBs that are introduced by AID-UNG-APE are near each other on opposite DNA strands, they can spontaneously form a DSB, but if not, the SSBs do not destabilize the duplex and are simply repaired. As S regions are large and the breaks appear to occur anywhere within S regions (5, 6, 21), it seems unlikely that the SSBs would be sufficiently proximal to form a DSB most of the time. MMR could convert these distal SSBs to the DSBs that are required for CSR. Figure 2b presents a model for how MMR could do this (99). Msh2–Msh6 can recognize and bind U:G mismatches created by AID activity (100). Mlh1-Pms2 would be recruited and Exo1 would excise from the nearest 5′ SSB created by AID-UNG-APE activity toward the mismatched dU:dG. Exo1 is hypothesized to continue past the mismatch until it reaches a SSB on the other strand, thus creating a DSB.
Several additional experimental results support this model. First, B cells in which the tandem repeats of Sμ have been deleted (SμTR−/−), which thus have very few AID hotspot targets, only show a ~twofold reduction in CSR (101). However, in these B cells, CSR is nearly ablated in the absence of Msh2 or Mlh1 (77, 102; J. Eccleston, C.E. Schrader, J. Stavnezer & E. Selsing, manuscript in preparation). Second, IgG2a, the isotype with the fewest AID hotspot targets in its S region, is the isotype most dependent upon MMR (93). Third, the great majority of S-S junctions in msh2−/− B cells occurs within the Sμ tandem repeat region, whereas in wild-type cells they can also occur upstream of Sμ, where the AID hotspot targets are infrequent (92). Fourth, the S-S junctions differ between wild-type and MMR-deficient B cells as to the lengths of microhomology between the donor (Sμ) and acceptor (down-stream) S regions. This suggests that MMR is involved in end processing from the sites of the SSBs, resulting in different lengths of single-stranded tails that can participate in homology search during S-S recombination (94, 95, 98, 103). Fifth and most importantly, LM-PCR experiments show that MMR-deficient B cells have fewer S region DSBs than do wild-type B cells (77).
End processing and mutations
After DSB formation, 5′ or 3′ single-strand overhangs remain. These tails must be either excised or filled in to create blunt, or nearly blunt, DSBs appropriate for an end-joining recombination with the other S region. The structure-specific endonuclease ERCC1-XPF excises 3′ single-strand tails at the junction with dsDNA and has a minor role in CSR (104). MMR normally recruits PCNA and replicative DNA polymerase for fill-in synthesis of 5′ overhangs (88). However, replicative Pol cannot replicate past an abasic site, resulting in the recruitment of error-prone translesion Pols. Msh2–Msh6 recruit the translesion Pol η (100), which is the most important translesion Pol for introducing mutations at A:T bps into V region genes and into both unrecombined (GL) Sμ and recombined S-S junction regions during CSR (105, 106). There are numerous mutations at both A:T and G:C bp in the regions surrounding S-S junctions (107, 108, 145). The mutations at G:C bp could be due to fill-in DNA synthesis across dU nucleotides by replicative Pols or across abasic sites, probably by the translesion DNA Pol θ (109–111). Additionally, not all mutations in S regions are likely to be due to fill-in DNA synthesis after creation of DSBs. It is likely that sometimes the excision by Exo1 does not lead to DSBs, but instead the single-strand patch is simply repaired by translesion Pols. Also, sometimes DSBs will form that do not successfully synapse with acceptor downstream S regions and that result in internal Sμ deletions rather than S-S recombination (112, 113).
Joining of Donor and Acceptor S Regions
After formation of the DSBs in the donor and acceptor S regions, the S regions are recombined using ubiquitous proteins that perform nonhomologous end-joining (NHEJ) in all cell types. These proteins are observed within a complex visible by in situ immunofluorescence in cells that have been treated with gamma radiation to induce DSBs, which is consistent with the fact that these proteins are involved in repair of DSBs after gamma radiation as well as repair of DSBs generated during CSR. Many of these proteins are also involved in V(D)J recombination during lymphocyte development.
S-S recombination occurs by an end-joining type of recombination
DNA DSBs can be induced by ionizing radiation or during repair of oxidative damage or replication. Numerous ubiquitous repair proteins exist that rapidly repair DSBs in all cells, many of which are also involved in CSR. DSBs produced during DNA replication or during G2 phase of the cell cycle are generally repaired by homologous recombination, as there is a chromosomal homolog that can be copied. However, S region DSBs in B cells induced during CSR are generated and resolved during G1 phase (77), and S regions lack sufficient homology to undergo homologous recombination.
Four proteins known to be essential for NHEJ, Ku70, Ku80, and the two-protein ligase complex XRCC4-ligase IV, are very important for CSR (113–120). Ku70–Ku80 binds to the DNA ends and serves as a tool belt for the end-joining reaction by recruiting enzymes that effect the recombination. Ku70–Ku80 improves the binding of XRCC4-ligase IV to DNA ends (121–123). Although Ku-deficient cells apoptose upon induction of CSR, Reina-San-Martin et al. (113) showed by CFSE staining that, at each cell division, CSR in the few viable ku80−/− cells is nearly ablated. These investigators also increased cell viability by expressing Bcl2 from a transgene in ku80−/− cells and again showed that CSR is nearly ablated.
XRCC4 or ligase IV deficiency is incompatible with life owing to problems during brain development, although human patients with hypomorphic mutations have been described, and mice with deficiencies have been created (120, 124, 125). Mice entirely lacking XRCC4 were produced by mating xrcc4+/− mice with p53-deficient mice, allowing survival of xrcc4−/− mice, which have ~25% of normal levels of CSR, indicating that XRCC4 is important but not essential for CSR (125). Also, humans with mutations in ligase IV have fewer peripheral blood cells that have undergone CSR than normal controls (119). Examination of the S-S junctions in the xrcc4−/− mouse B cells and in the human patients demonstrated that the S-S junctions are aberrant, as they have greatly increased lengths of junctional microhomology (119, 125). Numerous studies have shown that SS junctions in wild-type mice or normal individuals show very little microhomology, or identity, between the donor Sμ and acceptor S regions at the junction, usually 0 or 1 bp of identity (5), consistent with recombination by NHEJ. In contrast, in the xrcc4−/− mice and human ligase IV hypomorphs, many junctions have up to 10 bp or more of identity. Taken together, the data indicate that CSR occurs by NHEJ but can also occur by an alternative type of end-joining reaction that favors the use of microhomologies. XRCC4-ligase IV in the presence of Ku70–Ku80 ligates incompatible ends, consistent with the lack of micro-homology at S-S junctions in wild-type cells (126). It is unknown, however, whether in wild-type cells the alternative pathway is actually used.
A fundamental unanswered question regarding the joining process is whether the donor and acceptor S regions are in close proximity before AID attacks the donor (Sμ) or acceptor (downstream) S region. It is attractive to expect that they are preassociated in order to increase the likelihood of correct S-S recombination. This association could be directed to the correct S region by GL transcription. Another reasonable possibility is that the association occurs immediately after AID attacks Sμ. In situ studies suggest that the Ig loci form loops, apparently bringing the V and J genes in proximity prior to V(D)J recombination (127, 128).
Mre11-Rad50-Nbs1
Accumulating evidence indicates that this complex of three proteins (MRN complex) travels along the DNA duplex scanning for DNA breaks and that MRN binds DSBs within seconds of their formation (129, 130). However, owing to its great abundance, Ku is likely to bind DSBs even faster during CSR (see the section below, Ku70–Ku80-DNA-PKcs). When a DSB is encountered, the conformation of Mre11 and Rad50 changes, resulting in unwinding the DNA ends at the break. Mre11 is a globular protein associated with the DNA, whereas Rad50 forms a long coiled-coil that at its middle abruptly reverses direction, forming a loop with a zinc hook at its apex. The hooks from two Rad50 molecules associate homotypically, and this is thought to be important for holding two DNA duplexes together at the DSB (131, 132). Once bound to a DSB, the kinase ataxia telangiectasia mutated (ATM) binds the complex via Nbs1, becomes activated, and phosphorylates several substrates, among which are Nbs1, 53BP1, p53, Chk2, and H2AX, causing a further accumulation of MRN and several other repair proteins and also activating cell-cycle checkpoints (133–135) (Figure 3). Thus, MRN is upstream of a cascade of events that function to sense the DSB, resulting in repair by end joining or by homologous recombination. Null mutations in any component of MRN are lethal, and hypomorphic mutations result in aberrant chromosomes and translocations (136) and the disease syndromes Nijmegen breakage syndrome (Nbs-1 mutations) and ataxia-telangiectasia-like disorder (Mre11 mutations), characterized by increased sensitivity to ionizing radiation and other DSB-inducing agents (137, 138).
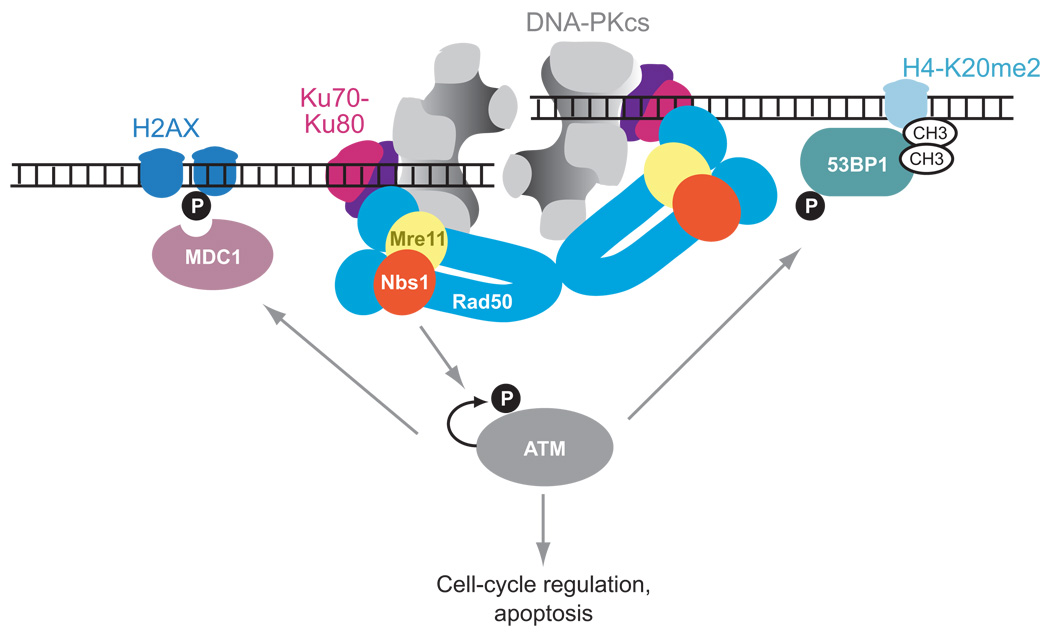
Figure 3.
Proteins that bind to DSBs that are involved in CSR. The evidence supporting this diagram is discussed in the text. DNA-PK, which consists of Ku70–Ku80 and DNA-PKcs, most likely binds DSB ends first. Mre11-Rad50-Nbs1 (MRN) probably binds next, perhaps helping to hold the DNA ends together, and MRN recruits and activates ATM (ataxia telangiectasia mutated), which phosphorylates H2AX, 53BP1, and Mdc1, resulting in accumulation of large amounts of these proteins and MRN around the DSB sites. After the DNA ends are brought sufficiently near by activities of these other proteins, DNA-PK can hold the DNA ends in the correct position for recombination. These proteins are all important for correct S-S recombination, and they inhibit aberrant recombinations and translocations. ATM also phosphorylates proteins that regulate the cell cycle and induce apoptosis, but nothing is known about how important this is during CSR.
Two groups have shown that CSR is reduced two- to threefold in cultured splenic B cells from mice in which the Nbs1 gene is inactivated by a conditional mutation (136, 139). The reduced CSR was analyzed in cells stained with CFSE, measuring switched cells at each cell division, as cell viability and proliferation are reduced in the mutant cells. Because some Nbs1 remains in the cells, these are most likely underestimates of the importance of Nbs1 for CSR. When wild-type mouse splenic B cells are treated with switch inducers in culture, the Sμ region undergoes rare translocations with the c-myc gene, which is located on a different chromo-some (140–142). This translocation is AID-dependent and, most interestingly, occurs six times more frequently in Nbs1-hypomorph B cells than in wild-type B cells (140, 143).
Patients with hypomorphic mutations in Nbs1 or Mre11 also have a lower percentage of peripheral blood lymphocytes that have undergone CSR, as assayed by detection of Sμ-Sα junctions in these cells (33, 144, 145). Further evidence for the participation of Nbs1 in CSR is the demonstration by immunocytochemistry-fluorescence in situ hybridization (immuno-FISH) that Nbs1 foci colocalize with the IgH loci, but not with Igκ loci, in wild-type, but not aid −/−, splenic B cells induced to undergo CSR (43). The finding of increased translocations with c-myc in MRN-deficient B cells suggests that the MRN complex is involved in organizing efficient and accurate S-S recombination.
Ataxia telangiectasia mutated (ATM)
ATM is a ser/thr protein kinase, a member of the phosphoinositol 3-kinase-like kinase (PIKK) family, which includes DNA-PKcs and ATR and which is involved in cell-cycle control and DNA damage responses. Upon activation by MRN, ATM accumulates at repair foci, orchestrates repair of the DSB, and initiates a cell-cycle checkpoint until repair is complete (146, 147). Recently, investigators demonstrated that V(D)J recombination is abnormal and that DSBs resulting from incomplete V(D)J recombination during lymphocyte development are maintained over several cell generations in atm−/− mice (148–150). Thus, both repair and the cell-cycle checkpoints are compromised, as atm−/− lymphocytes continue to replicate despite chromosomal breaks (142, 143).
CSR is reduced about threefold at each cell division cycle in cultured atm−/− splenic B cells relative to wild-type B cells (151, 152). Also, lower titers of switched antibodies are detected in sera after immunization. However, GL transcripts are expressed at normal levels. In B cells induced to switch in culture, the Sμ region undergoes translocations with the c-myc gene eight times more frequently in atm−/− cells than in wild-type cells (143). Thus, it seems likely that when atm−/− B cells are activated to switch, DSBs are generated as usual, are maintained longer than usual, and do not undergo normal Sμ-Sx recombination; this can result in aberrant recombinations with other chromosomes. Ataxia telangiectasia patients often have IgA and IgG deficiencies and have peripheral blood lymphocytes with fewer Sμ-Sα junctions than normal individuals, similar to patients with mutated Nbs1 or Mre11 and consistent with reduced CSR (33, 144). Thus, during CSR, ATM likely organizes the repair complex and perhaps halts the cell cycle, and it contributes to the correct positioning of DSBs together during the long-range interaction required for accurate S-S recombination. In the absence of ATM, prolonged duration of unrepaired DSBs and aberrant recombination events result in translocations.
53BP1
53BP1 was first discovered in a yeast two-hybrid screen as a protein that binds p53 (153). 53BP1 is a transcriptional coactivator for p53, binding through its tandem Tudor domains to p53 dimethylated at lysine 370 (154). However, this is not its only function. 53BP1 accumulates at DSBs within 2 min after ionizing radiation treatment (155). Its initial recruitment to DSBs does not depend on any other known protein, including Nbs1, ATM, or DNA-PK (156), although its subsequent accumulation in foci does depend on γH2AX and Mdc1 (157–159). 53BP1 functions to induce phosphorylation of ATM and ATR (160), and thus it might also increase MRN activity and MRN’s accumulation at DSBs.
Strikingly, CSR is reduced about 90% in cultured 53bp1−/− splenic B cells relative to wild-type B cells, which is not due to decreased cell proliferation nor to reduced GL transcripts. Serum IgG, IgE, and IgA isotypes are reduced even more, although IgM levels are normal (161, 162). The S-S junctions are normal.
53BP1-deficient cells do not have a dramatic increase in general chromosome instability, unlike atm−/− and h2ax−/− cells, but a much higher proportion of the chromosomal aberrancies in 53bp1−/− cells involve the IgH locus, suggesting that 53BP1 has a special role at this locus (142, 159). Another hint about the role of 53BP1 in CSR comes from the finding that there is a threefold increase in internal deletions within S regions in IgM hybridomas produced from 53bp1−/− B cells induced to undergo CSR in culture in comparison to IgM hybridomas from wild-type mice (163). Interestingly, these deletions are not increased in atm−/− or h2ax−/− B cells, despite the general increase in unrepaired DSBs in the genome. These results suggest that 53BP1 might also be important for bringing together, or synapsing, the donor Sμ and downstream S regions (159, 161). How 53BP1 performs this role, however, is completely unknown. This role would be consistent with the lack of a role for 53BP1 during V(D)J joining because the RAG complex itself possesses synaptic activity (161, 162).
Mammalian 53BP1 binds dimethyl-lysine 20 of histone H4 (H4-K20-me2), but not mono- or trimethyl K20, via its Tudor domains (164). Most interestingly, 53BP1 with single amino acid mutations within the Tudor domains that prevent H4-K20-me2-binding and RNAi-mediated knockdown of PrSet7/Set8, the histone methyltransferase that monomethylates K20 (a prerequisite for the dimethylation), decrease 53BP1 foci (164). Association of 53BP1 with irradiation-induced foci is disrupted by RNase A treatment, suggesting that there may be an RNA component involved in its binding (155). Taken together, H4-K20-me2 may be important for recruiting 53BP1 to chromatin, perhaps to the IgH locus, although this modification would probably need to be present before induction of DSBs during CSR because 53BP1 binding to DSBs is so rapid. The GL RNA transcribed from S regions may help 53BP1 recruitment. This hypothesis requires that the H4-K20-me2 mark is associated with actively transcribed regions on the IgH locus during G1 phase, which would differ from its distribution on bulk chromatin.
γH2AX (phosphorylated form of H2AX)
H2AX is a variant of histone H2A, representing about 15% of the cellular pool of H2A. It is randomly incorporated into nucleosomes (165). Within seconds after formation of a DSB induced by ionizing radiation or by a restriction enzyme, the extended C-terminal tail of H2AX is phosphorylated by a PIKK kinase, most frequently ATM (166, 167), and this phosphorylation spreads over a region estimated to span up to a megabase surrounding the break (147, 168). In fact, the peak accumulation of γH2AX (phosphorylated H2AX) is not directly at the DSB but instead located at 8–10 kb on either side in mammals (168). ATM also phosphorylates 53BP1, Nbs1, and Mdc1, which then all bind the phosphorylated tail of γH2AX, which serves as a docking site for these proteins. This phosphorylation results in a rapid assembly of these factors, plus Mre11, Rad50, and Brca1, into a large multiprotein complex. γH2AX is required for the accumulation of these proteins into foci near DSBs (169, 170). However, mice lacking H2AX can still repair DSBs, although with lower efficiency, and they can still induce cell-cycle checkpoints (169), probably because the initial assembly of repair proteins, including MRN, ATM, and 53BP1, does not depend on γH2AX (157).
CSR is markedly reduced in H2AX-deficient mice. In vitro CSR to IgG3 and IgG1 is reduced to ~25%–30% of wild-type, and this is not due to defective cell proliferation. The antigen-specific IgG1 response to immunization is reduced to about 30% of wild-type mice, and nonimmune serum levels of IgG1, IgG3, and IgA are all reduced to 15%–50% of wild-type (43, 113, 142, 169).
Similar to cells deficient in either ATM, 53BP1, or Mdc1 or cells having Mre11 or Nbs1 hypomorphic mutations,h2ax −/− B cells show numerous chromosome breaks and aberrant recombination events (142, 143, 169, 171). In h2ax−/− B cells induced to switch in culture, there is a greater than tenfold increase in AID-dependent chromosome breaks within the IgH locus relative to wild-type cells, resulting in separation of the V genes and 3′ end of the CH genes and translocations in metaphase chromosome spreads (142). The breaks occur on both chromatids, indicating that they occur prior to S phase, consistent with evidence that S region DSBs are observed in G1 phase (77). Also, foci containing γH2AX (and Nbs1) colocalize with the IgH locus in mouse splenic B cells induced to switch, in wild-type cells in the G1/early S phase of the cell cycle, but not in aid−/− cells, as detected by immuno-FISH (43). Taken together, these data suggest that γH2AX, like MRN, Mdc1, ATM, and 53BP1, is involved in holding AID-initiated DSBs in a structure that promotes accurate synapsis between Sμ and the downstream acceptor S region and also prevents recombinations with DSBs on other chromosomes.
Mdc1 (mediator of DNA damage checkpoint protein 1)
Mdc1 is a mediator protein that is recruited to DSBs by binding to Nbs1 and is phosphorylated by ATM (172, 173). Mdc1 subsequently recruits ATM to γH2AX and is required for accumulation of γH2AX at DSBs (167, 173). Without Mdc1, the initial recruitment by MRN of ATM and γH2AX occurs, but the complexes are unstable and do not form the large repair foci observed in wild-type cells. CSR in cultured mdc1−/− B cells is only mildly reduced, to about 50%–70% of wild-type B cells (167).
Ku70–Ku80, DNA-PKcs
The proteins discussed above are all involved in repairing DSBs both by end joining and by homologous recombination. However, Ku70 and Ku80 are only involved in end joining and appear to prevent the use of homologies during recombination. Ku70–Ku80 binds DNA ends at DSBs and telomeres and mediates synapsis of the two DNA ends, positioning the ends to allow end processing and direct end-to-end joining (174, 175). Ku70–Ku80 forms a ring with a broad base that encircles DNA and can only dissociate at an end (176). After binding, Ku slides away from the ends, allowing the catalytic subunit, the kinase DNA-PKcs, to bind to each end (177). Ku binds the nuclear matrix, and this binding might localize the DSBs and telomeres to the matrix (178). DNA-PKcs is transphosphorylated by the other DNA-PKcs bound at the other end of the DSB and also phosphorylates Ku70, Ku80, and XRCC4 (175, 179). Thus, DNA-PK appears to be acting both as an activator and scaffold during the actual ligation event. DNA-PKcs has several transautophosphorylation sites, and mutation studies demonstrate that their phosphorylation regulates the accessibility of the DNA ends to end-processing activities required for recombination (179).
All three components of the DNA-PK holocomplex are involved in CSR, as they are essential for NHEJ, although Ku70 and Ku80 are much more important than DNA-PKcs. All three proteins are essential for V(D)J recombination, which occurs by NHEJ, so mice lacking any of these proteins do not develop B cells, unless they are supplied with transgenic (Tg) recombined heavy (H) and light (L) chain genes. Ku70- and Ku80-deficient B cells reconstituted with a Tg L chain gene and a H chain gene knocked into the endogenous locus do not have switched isotypes in their serum nor do they undergo CSR when induced to switch in culture, although they have normal levels of GL transcripts (113, 117, 118). Unlike wild-type cells, internal Sμ deletions do not occur in ku80−/− cells (113). These data suggest that Ku80-deficient B cells that sustain DNA breaks owing to AID activity cannot recombine these DSBs, even by internal Sμ deletions, and therefore die. This would also explain why cells with mutations in the unrecombined Sμ segment are not observed in ku80−/− cells (113). Although the investigators did not demonstrate that ku80−/− cells express AID, this is highly likely. It is puzzling, however, that transition mutations that result from dU bases being replicated prior to UNG and APE activity, which would not lead to DNA breaks, were not observed in the GL Sμ. Perhaps Sμ regions with dU bases also have UNG-APE induced DNA breaks, resulting in death of these Ku-deficient cells.
It is surprising that Ku is essential for CSR, although XRCC4 is not, given the evidence that CSR can occur by an alternative end-joining pathway. This suggests that Ku might have another function and perhaps even function in the alternative pathway. Also, Ku appears to be more important than MRN for CSR. Ku is one of the most abundant nuclear proteins (~4 × 105 molecules per cell) and has a very high affinity for DNA ends (5 × 10−10) (174). Thus, it might bind prior to MRN. Perhaps MRN is more important for recruiting additional proteins involved in chromatin accessibility and perhaps for cell-cycle regulation, whereas Ku is bound at the ends, positioning them precisely for recombination and for end processing by nucleases and polymerases and for recruiting XRCC4-DNA ligase-IV (121, 122). It is interesting that Ku is required for V(D)J recombination, but MRN is not, suggesting that the synaptic activity of RAG cannot replace the role of Ku but can replace MRN.
By contrast, DNA-PKcs is not essential for CSR, and its importance is controversial, with different results obtained by three different groups. DNA-PKcs deletion was reported to eliminate CSR in cultured B cells to every isotype except IgG1 (180, 181). Two groups (182, 183) studied B cells from scid mice, which have a deletion in the DNAPKcs gene resulting in loss of the C-terminal 83 amino acids, in barely detectable protein levels, and in no detectable kinase activity. These two groups found a smaller reduction in CSR. It was reduced to 25%–50% of wild-type for all isotypes in the study by Cook et al. (183), but CSR occurred at 50%–100% of wild-type, depending on the isotype, in the Bosma et al. (182) study. There was no difference in proliferation between the wild-type and DNA-PKcs-deficient B cells induced to switch, but there was more cell death in the mutant cell cultures (183). It is difficult to reconcile the different results among the three groups, except that Manis et al. (180, 181) studied mice with no DNA-PKcs, whereas the Bosma (182) and Cook (183) groups studied scid mice, suggesting that the tiny amount of protein present that lacks kinase activity has some function, perhaps as a scaffolding protein. The differences in CSR between the scid mice studied by the two groups might be due to different amounts of back-crossing, as B cells from different mouse strains have different abilities to switch (184).
The role of DNA-PKcs in V(D)J recombination appears to be to stimulate the hairpin cutting activity of Artemis (185), which is consistent with its importance for joining the coding ends but not the signal ends. As CSR does not involve hairpin ends, and Artemis appears to have no role in CSR (181), DNA-PKcs must have another role in CSR. This may involve its ability to regulate end processing, which may help recombination at some S-S junctions and possibly involves its ability to phosphorylate other proteins (174).
REGULATION OF SWITCHING
Germline (GL) Transcripts
As described above, CD40 and/or TLR stimulation provides essential activation for B cells to undergo CSR. Additionally, cytokines produced by T helper cells and dendritic cells determine the isotype to which B cells will switch by inducing transcription from GL promoters located upstream, i.e., 5′ to each acceptor S region. The resulting GL transcripts do not encode proteins and are therefore also referred to as sterile RNAs. Figure 1 shows the transcription unit and splicing diagram for an example, GL α RNA. The exon located 5′ to the S region is called the I exon. Each GL transcript has a similar transcription and splicing pattern (31). There is also a similar GL Sμ RNA, initiating near the μ intron enhancer. Gene-targeting experiments in which a single I exon and/or promoter for a specific GL transcript is deleted showed that GL transcription of the acceptor S region is required for CSR to that isotype and that the GL transcription only functions in cis, i.e., not on the other chromosome (186, 187). Surprisingly, deletion of the GL γ1 exon splice donor also prevents IgG1 CSR, although there is no known role for the spliced transcript (31, 188, 189). The role of splicing is an intriguing unanswered question.
GL promoters have cytokine-responsive elements within them. GL γ1 and ∈ promoters, which are induced by IL-4, have binding sites for the IL-4-induced transcriptional activator Stat6. Several promoters have binding sites for NF-κB, which is induced in response to both CD40 and TLR signaling. GL γ2b and α promoters have binding sites for Smad and Runx, two factors induced by TGF-β, which induces CSR to IgG2b and IgA. A thorough review of GL transcriptional regulation was recently published (32).
The function of GL transcription appears to be to direct AID to a specific S region and to make the S region a suitable substrate for AID. There are at least three possible roles for GL transcription, which are not mutually exclusive. First, the substrate for AID is ssDNA, and the act of transcription creates short regions of ssDNA at the transcription bubble. In addition, RNA transcribed from S regions is G-rich and therefore can form an RNA-DNA hybrid (R-loop) with the bottom C-rich DNA strand. This hybrid formation leaves the top G-rich DNA strand single-stranded over long stretches (190, 191), thereby making the top strand a target for AID. However, as AID appears to target both top and bottom strands equally in vivo (50), this makes the importance of R-loops unclear. Furthermore, when the Xenopus Sμ region, which is A:T rich and cannot form R-loops, is substituted into the Sγ1 locus by gene targeting, CSR is reduced only about twofold relative to a Sγ1 segment of the same length (192, 193). However, as R-loops form at both the Sμ and acceptor S regions normally, the effect of both R-loops might be to increase CSR by fourfold, which should be physiologically significant. Evidence suggests that R-loops at the Sμ region begin upstream of the tandem repeats, at a particular sequence GGGGCTGGG, which is within a zone that has a high content of G (50%) (190), and, interestingly, S-S junctions often involve sequences 5′ to Sμ, although many also occur 5′ to this sequence (5, 102). Mice with a targeted deletion of the Sμ tandem repeats retain this sequence and have R-loops and extensive single-strand regions on the nontranscribed strand, consistent with their modest reduction in CSR efficiency (101, 190).
A likely explanation for why both the top and bottom strands are equally targeted during SHM of Ig V regions comes from the recent demonstration by Ronai et al. (194) that in human B cell lines undergoing SHM both the top and bottom VH region strands have single-strand patches averaging ~11 nucleotides in length. These single-strand patches were detected by treating fixed, permeabilized nuclei with sodium bisulfite, which deaminates dC bases within ssDNA. This group also showed that in these cell lines V regions are transcribed in both directions, and the length of the patches are consistent with being caused by the bubble that forms at the site of transcription. However, no one has shown that S regions are transcribed in both directions, and thus it is not known if this explains why the top and bottom S region strands are equally targeted by AID. Ronai et al. (194) found that if they first deproteinized the V region DNA they could not detect the single-strand patches, and thus far experiments to detect single-strand patches in S regions have always used deproteinized DNA. Thus, undetected single-strand patches may exist on both strands at S regions. However, the putative antisense transcripts would not be G-rich and therefore should not form R-loops.
The second likely role for GL transcription is to recruit AID. AID co-immunoprecipitates with RNA polymerase II in splenic B cells undergoing CSR (195). This hypothesis is also supported by the finding that AID-induced dU lesions are found in a region beginning ~150 bp 3′ to the GL RNA initiation site and extending over several kb downstream, with more mutations near the 5′ end of the S regions and fewer at the 3′ ends (50). Thus, investigators have proposed that AID is recruited to RNA Pol II either at the initiation phase of transcription or when it switches to the elongation phase at ~150 bp 3′ to the initiation site (18, 19, 49). AID may leave the transcription complex stochastically as it progresses through the S region.
A third possible role is that transcription can alter histone modification of the transcribed region, which might make the DNA more accessible to AID (196–198). Transcription clearly does alter chromatin accessibility, but whether the histone modifications affect AID binding has not yet been shown.
Roles of IgH Intron Enhancer and 3′ Enhancers
There are two enhancer regions in the IgH locus. The μ intron enhancer is located 3′ to JH4 and just 5′ to the Sμ region and is essential during B cell development for normal V(D)J recombination. Consistent with the fact that the Iμ promoter lies within the enhancer core, deletion of the enhancer core reduces CSR on that allele by about twofold (199). As neither GL Sμ transcription nor DJ transcription is eliminated, this might explain why CSR, albeit at a reduced frequency, still occurs. There are matrix attachment regions located 5′ and 3′ to the core enhancer, but this same study found no role for the matrix attachment regions in CSR.
A second set of enhancers is located 3′ to the Cα gene in mouse and 3′ to each of the two Cα genes in human. The mouse 3′ IgH enhancers are spread over 30 kb and contain four DNase hypersensitive regions, termed (from 5′ to 3′) hs3A, hs1,2, hs3B, and hs4. Although the entire 30-kb enhancer region has not been successfully deleted, various segments have been deleted by gene targeting using Cre-mediated deletion after insertion of loxP sites. Deletion of hs3A or hs1,2 has no effect on CSR (200); however, combined deletion of hs3B and hs4 greatly reduces CSR to all isotypes except IgG1 (201). Neither hs3B nor hs4 has been individually deleted. The hs3B-hs4 deletion eliminates GL transcripts (except for GLγ 1 RNA). Thus far, the only known role for hs3B-hs4 during CSR is to enhance GL transcription. The levels of Ig μ mRNA are also reduced in these mice. Further evidence that the role of the 3′ IgH enhancer is to stimulate GL transcription was provided by transient transfection experiments in which a DNA segment containing all four human 3′ DNase-hypersensitive sites was found to stimulate human GL γ3 RNA promoter activity (202). There is some evidence from transgene experiments that the region just 5′ to Sγ1 has enhancer activity for GL γ1 RNA, perhaps explaining the independence of IgG1 and GLγ1 transcripts from hs3B-hs4 (203, 204).
As the hs3B-hs4 enhancers are located far downstream from the GL promoters, their ability to stimulate GL transcription likely involves formation of a loop between the 3′ IgH enhancers and GL promoters (32). This might involve complex formation between transcription-activating factors bound to the enhancers and promoters, as has been demonstrated between the T cell receptor Dβ promoter and the 3′ T cell receptor β enhancer (205). This complex would most likely recruit histone acetylases and other chromatin modifiers to increase accessibility of the promoters to RNA polymerase and also position the donor and acceptor S regions near each other. Direct evidence for the existence of loops between the 3′ IgH enhancers and the DNA segments containing the promoters for specific GL transcripts in splenic B cells under conditions that induce CSR to that specific isotype has recently been obtained using the chromo-some conformation capture technique (206). Most interestingly, the loops depend on the hs3B-hs4 enhancer segment and are reduced in aid−/− cells. It will be very interesting and important to determine which sequences and proteins are involved.
Regulation of Isotype Specificity by S Regions
Although GL transcription is clearly essential for CSR, a few reports suggest that iso-type specificity is also regulated by the S region sequence. The evidence for this was obtained by using transiently transfected plasmid switch substrates (207). The acceptor S region isotype determines whether the plasmid will switch in particular B cell lines or in splenic B cells activated under conditions that induce CSR on the chromosome to specific isotypes. For example, plasmids containing an acceptor Sγ1 or Sγ3 sequence will not switch in two different B cell lines that switch on their endogenous chromosome to IgA, but not to IgG1 or IgG3. However, plasmids with acceptor Sα sequences will undergo Sμ-Sα recombination in these cell lines. Likewise, a plasmid with the Sγ1 acceptor S region will undergo Sμ-Sγ1 recombination in splenic B cells treated with LPS+IL-4, which induces IgG1 CSR on the chromosome, but not if the B cells are treated with LPS alone, which induces IgG3 but not IgG1 CSR. Likewise, the plasmid with an Sγ3 acceptor S region will undergo CSR in B cells induced with LPS, but not if the cells are treated with LPS+IL-4. Kenter et al. (208) also showed that mutations of 3 bp within a repeat unit of the Sγ1 sequence to the sequence found in Sγ3 allowed the plasmid to switch in the absence of IL-4, further supporting the hypothesis that isotype specificity is also controlled by switch sequences. The element that was mutated is part of a NF-κB binding motif, which therefore might be involved in determining the isotype specificity. It was not determined whether the mutations affected transcription across the plasmid S regions, although there are no Ig isotype–specific promoters in these plasmids, so this is an unlikely explanation.
Another example of isotype specific regulation by a S region comes from the finding that plasmid Sμ-Sα recombination in cell lines and splenic B cells can be stimulated threefold and tenfold, respectively, by the histone methyltransferase Suv39h1, which trimethylates histone H3 on lysine 9. Suv39h1 does not stimulate switch recombination in plasmids with any other acceptor S region (209). Mice deficient in Suv39h1 show a 50% reduction in chromosomal CSR to IgA, but to no other isotype. This reduction is not accompanied by a reduction in GL α transcripts, suggesting that the Sα sequence itself is responding to Suv39h1. The Suv39h1 target in these switching cells is unknown. Furthermore, the K9-trimethyl mark is repressive and is usually found on heterochromatin associated with centromeres, making the stimulatory role of Suv39h1 on IgA switching even more puzzling. Taken together, these two sets of results suggest that isotype specificity is not regulated solely by GL transcription but is also regulated by S region sequences by an unknown mechanism.
ISOTYPE SWITCHING OCCURS PRIOR TO GERMINAL CENTER (GC) FORMATION, AND PERHAPS ALSO IN GCs
The GC provides a unique environment that allows rapid proliferation of cells despite sustaining DNA damage initiated by AID activity. Bcl-6 is upregulated in GC cells and is required for GC formation (210, 211). Ranuncolo et al. (212) suggest that GC centrob-lasts are uniquely able to withstand the DNA damage caused by AID because of the repression of the ATR damage-sensing pathway by Bcl-6. Protection from cell death in GC cells had previously been thought to be due to the downregulation of p53 by Bcl-6, as demonstrated in Ramos cells (213), but Ranuncolo et al. (212) found that primary human centroblasts express p53 and that its expression is not affected by downregulation or inhibition of Bcl-6.
Although both isotype-switched cells and high levels of AID are found in GCs, class switching clearly can occur very early after antigen exposure, prior to GC formation. By adoptive transfer of B cell receptor Tg B cells and carrier-specific Tg CD4 T cells into normal recipients, Pape et al. (214) were able to visualize very early stages in the antibody response, tracking the B cells with anti-idiotype antibody. Tg+ isotype-switched B cells (IgG2a+) appeared in splenic B cell follicles as early as two days after immunization with cognate antigen and peaked on days 3 and 4, prior to formation of GCs. The B cells had divided at least three times and appeared to have migrated away from the Tg T cells. Their appearance was antigen-and T cell–dependent. By day 4, progeny of the Tg+ IgG2a+ follicular B cells could be found in the outer edges of the periarteriolar lymphoid sheath near the red pulp, in the marginal zone, and in pre-GCs. By day 10, GCs contained PNA+ IgG2a+ Tg B cells that showed evidence of many cell divisions. IgM+Tg B cells were abundant in follicles and the marginal zone at this time but were not in GCs. As only IgG2a+ Tg B cells were found in GCs, this suggests that CSR does not occur in GCs in this model, but instead occurs prior to GC formation. In another model, normal mice infected with attenuated Salmonella typhimurium also showed very rapid (day 4) T-dependent IgG2a isotype switching, occurring much before GC formation (215). Also, AID is expressed in large proliferating B cells in the extrafollicular areas of human tonsil and lymph node (216). However, Kolar et al. (217) have recently suggested that a population that expresses IgD+CD38−CD23−FSChiCD71+in human tonsil may be the initial GC cell to express AID. V region mutation frequency places these cells between naive and GC cells. This finding is consistent with previous assumptions that CSR also occurs in GCs.
CSR can also occur independently of T cell help. B cell activating factor (BAFF) can synergize with IL-4 to induce AID expression in CD40−/− B cells in culture (218). Near normal levels of gut IgA were detected in CD40−/− mice despite a lack of GCs (219). The site where this T-independent IgA switching occurred was not identified in this study, although the authors excluded the gut-associated lymphoid tissue (GALT), the lamina propria, and the peritoneal cavity. Mucosal epithelial cells lining the crypts of human tonsils can support class switching through the production of BAFF and IL-10 upon TLR stimulation. These epithelial cells also secrete thymic stromal lymphopoietin (TSLP), which stimulates production of BAFF by dendritic cells (220, 221). Furthermore, CSR is detectable in pre-B and immature B cells isolated from bone marrow, as assayed by AID expression, detection of transcripts from excised DNA circles owing to S-S recombination, and detection of postswitch (Iμ-Cx) transcripts (222, 223). AID expression, circle transcripts, and postswitch transcripts were also observed in developing B cells from nude mice, further indicating T cell independence. The expression of AID in both wild-type and nude mice depends on B cell receptor signaling through Bruton’s agammaglobulinemia tyro-sine kinase (BTK) in immature B cells and on TLR signaling in both pre-B and immature B cells (222).
REMAINING QUESTIONS
Numerous interesting questions remain in the field of CSR. For example, how does AID target the V and S regions specifically? CSR and SHM do not appear to occur simultaneously in a cell. Why not? What determines whether a cell will undergo CSR or instead undergo SHM? What proteins does AID interact with? Most interestingly, what is the role of the C terminus of AID? Why is UNG required for CSR, i.e., why do other uracil DNA glycosylases not substitute? Why is APE2 used to create SSBs in S regions, in addition to APE1? Is another AP endonuclease also involved? How is synapsis of two distal S regions achieved? What is the contribution of S region sequence to isotype specificity? What is the role of 53BP1? Does 53BP1 bind specifically to the IgH locus? If so, does it require a specific histone modification, and is this modification specific to the IgH locus? How is this regulated? Does the binding require GL transcripts? Why is Ku70–Ku80 essential for CSR but XRCC4-ligase IV is not? Is the ability of Ku to bind nuclear matrix important for CSR? Many very interesting questions are approachable with current techniques and will be addressed in the near future, thus promising much excitement for this field in the coming years.
ACKNOWLEDGMENTS
We thank Drs. Katheryn Meek for helpful information on DNA-PK and Amy Kenter for her manuscript in press. We acknowledge support by NIH to J.S. (RO1AI23283, RO1 AI63026, R21AI62738) and to C.E.S. (RO1 AI65639) and a postdoctoral fellowship from the Cancer Research Institute to J.E.J.G.
Glossary
- CSR
class switch recombination
- SHM (somatic hypermutation)
the process that introduces mostly single-nucleotide mutations into the variable regions of antibodies after antigen activation during infection or after immunization
- AID
activation-induced cytidine deaminase
- GC (germinal centers)
highly dividing B cells undergoing SHM that form in B cell follicles in spleen and lymph nodes during a T cell–dependent immune response
- BER (base excision repair)
a ubiquitous repair pathway that repairs damaged DNA bases
- UNG
one of four mammalian uracil DNA glycosylases
- APE (APE1 and APE2)
apurinic/apyrimidinic endonuclease, incises DNA at abasic sites
- MMR (mismatch repair)
a ubiquitous repair pathway that corrects DNA synthesis errors, including mismatches and deletions/insertions
- ATM (ataxia telangiectasia mutated)
a phosphoinositol 3-kinase-like kinase, which is mutated in the human syndrome Ataxia telangiectasia
- RAGs (recombination-activating genes)
initiate and are required for V(D)J recombination
- R-loops
RNA-DNA hybrids that form cotranscriptionally if the DNA is highly G-rich, which results in extensive single-strand regions on the G-rich DNA strand
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any biases that might be perceived as affecting the objectivity of this review.
Contributor Information
Janet Stavnezer, Email: Janet.Stavnezer@umassmed.edu.
Jeroen E.J. Guikema, Email: Jeroen.Guikema@umassmed.edu.
Carol E. Schrader, Email: Carol.Schrader@umassmed.edu.
LITERATURE CITED
- 1.Snapper CM, Finkelman FD. Immunoglobulin class switching. In: Paul WE, editor. Fundamental Immunology. 4th ed. Philadelphia: Lippincott-Raven; 1998. pp. 831–861. [Google Scholar]
- 2.Martin SW, Goodnow CC. Burst-enhancing role of the IgG membrane tail as a molecular determinant of memory. Nat. Immunol. 2002;3:182–188. doi: 10.1038/ni752. [DOI] [PubMed] [Google Scholar]
- 3.Waisman A, Kraus M, Seagal J, Ghosh S, Melamed D, et al. IgG1 B cell receptor signaling is inhibited by CD22 and promotes the development of B cells whose survival is less dependent on Ig α/β. J. Exp. Med. 2007;204:747–758. doi: 10.1084/jem.20062024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horikawa K, Martin SW, Pogue SL, Silver K, Peng K, et al. Enhancement and suppression of signaling by the conserved tail of IgG memory-type B cell antigen receptors. J. Exp. Med. 2007;204:759–769. doi: 10.1084/jem.20061923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunnick W, Hertz GZ, Scappino L, Gritzmacher C. DNA sequences at immunoglobulin switch region recombination sites. Nucleic Acids Res. 1993;21:365–372. doi: 10.1093/nar/21.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Min I, Rothlein L, Schrader C, Stavnezer J, Selsing E. Shifts in targeting of class switch recombination sites in mice that lack μ switch region tandem repeats or Msh2. J. Exp. Med. 2005;201:1885–1890. doi: 10.1084/jem.20042491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stavnezer J. Antibody class switching. Adv. Immunol. 1996;61:79–146. doi: 10.1016/s0065-2776(08)60866-4. [DOI] [PubMed] [Google Scholar]
- 8.Manis JP, Tian M, Alt FW. Mechanism and control of class-switch recombination. Trends Immunol. 2002;23:31–39. doi: 10.1016/s1471-4906(01)02111-1. [DOI] [PubMed] [Google Scholar]
- 9.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 10.Revy P, Muto T, Levy Y, Geissmann F, Plebani A, et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 11.Petersen-Mahrt SK, Harris RS, Neuberger MS. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 2002;418:99–104. doi: 10.1038/nature00862. [DOI] [PubMed] [Google Scholar]
- 12.Chaudhuri J, Tian M, Khuong C, Chua K, Pinaud E, Alt FW. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 2003;422:726–730. doi: 10.1038/nature01574. [DOI] [PubMed] [Google Scholar]
- 13.Dickerson SK, Market E, Besmer E, Papavasiliou FN. AID mediates hypermutation by deaminating single stranded DNA. J. Exp. Med. 2003;197:1291–1296. doi: 10.1084/jem.20030481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pham P, Bransteitter R, Petruska J, Goodman MF. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature. 2003;424:103–107. doi: 10.1038/nature01760. [DOI] [PubMed] [Google Scholar]
- 15.Peled JU, Kuang FL, Iglesias-Ussel MD, Roa S, Kalis SL, et al. The biochemistry of somatic hypermutation. Annu. Rev. Immunol. 2008;26:481–511. doi: 10.1146/annurev.immunol.26.021607.090236. [DOI] [PubMed] [Google Scholar]
- 16.Barreto VM, Ramiro AR, Nussenzweig MC. Activation-induced deaminase: controversies and open questions. Trends Immunol. 2005;26:90–96. doi: 10.1016/j.it.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 17.Ramiro A, San-Martin BR, McBride K, Jankovic M, Barreto V, et al. The role of activation-induced deaminase in antibody diversification and chromosome translocations. Adv. Immunol. 2007;94:75–107. doi: 10.1016/S0065-2776(06)94003-6. [DOI] [PubMed] [Google Scholar]
- 18.Longerich S, Basu U, Alt F, Storb U. AID in somatic hypermutation and class switch recombination. Curr. Opin. Immunol. 2006;18:164–174. doi: 10.1016/j.coi.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 19.Bransteitter R, Sneeden JL, Allen S, Pham P, Goodman MF. First AID (activation-induced cytidine deaminase) is needed to produce high affinity isotype-switched antibodies. J. Biol. Chem. 2006;281:16833–16836. doi: 10.1074/jbc.R600006200. [DOI] [PubMed] [Google Scholar]
- 20.Muramatsu M, Sankaranand VS, Anant S, Sugai M, Kinoshita K, et al. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J. Biol. Chem. 1999;274:18470–18476. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- 21.Schrader CE, Linehan EK, Mochegova SN, Woodland RT, Stavnezer J. Inducible DNA breaks in Ig S regions are dependent upon AID and UNG. J. Exp. Med. 2005;202:561–568. doi: 10.1084/jem.20050872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hodgkin PD, Lee J-H, Lyons AB. B cell differentiation and isotype switching is related to division cycle number. J. Exp. Med. 1996;184:277–281. doi: 10.1084/jem.184.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hasbold J, Lyons AB, Kehry MR, Hodgkin PD. Cell division number regulates IgG1 and IgE switching of B cells following stimulation by CD40 ligand and IL-4. Eur. J. Immunol. 1998;28:1040–1051. doi: 10.1002/(SICI)1521-4141(199803)28:03<1040::AID-IMMU1040>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 24.Deenick EK, Hasbold J, Hodgkin PD. Switching to IgG3, IgG2b, and IgA is division linked and independent, revealing a stochastic framework for describing differentiation. J. Immunol. 1999;163:4707–4714. [PubMed] [Google Scholar]
- 25.Rush JS, Liu M, Odegard VH, Unniraman S, Schatz DG. Expression of activation-induced cytidine deaminase is regulated by cell division, providing a mechanistic basis for division-linked class switch recombination. Proc. Natl. Acad. Sci. USA. 2005;102:13242–13247. doi: 10.1073/pnas.0502779102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dedeoglu F, Horwitz B, Chaudhuri J, Alt FW, Geha RS. Induction of activation-induced cytidine deaminase gene expression by IL-4 and CD40 ligation is dependent on STAT6 and NFκB. Int. Immunol. 2004;16:395–404. doi: 10.1093/intimm/dxh042. [DOI] [PubMed] [Google Scholar]
- 27.Gonda H, Sugai M, Nambu Y, Katakai T, Agata Y, et al. The balance between Pax5 and Id2 activities is the key to AID gene expression. J. Exp. Med. 2003;198:1427–1437. doi: 10.1084/jem.20030802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ito S, Nagaoka H, Shinkura R, Begum N, Muramatsu M, et al. Activation-induced cytidine deaminase shuttles between nucleus and cytoplasm like apolipoprotein B mRNA editing catalytic polypeptide 1. Proc. Natl. Acad. Sci. USA. 2004;101:1975–1980. doi: 10.1073/pnas.0307335101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McBride KM, Barreto V, Ramiro AR, Stavropoulos P, Nussenzweig MC. Somatic hypermutation is limited by CRM1-dependent nuclear export of activation-induced deaminase. J. Exp. Med. 2004;199:1235–1244. doi: 10.1084/jem.20040373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brar SS, Watson M, Diaz M. Activation-induced cytosine deaminase (AID) is actively exported out of the nucleus but retained by the induction of DNA breaks. J. Biol. Chem. 2004;279:26395–26401. doi: 10.1074/jbc.M403503200. [DOI] [PubMed] [Google Scholar]
- 31.Stavnezer J. Immunoglobulin class switching. Curr. Opin. Immunol. 1996;8:199–205. doi: 10.1016/s0952-7915(96)80058-6. [DOI] [PubMed] [Google Scholar]
- 32.Cogne M, Birshtein BK. Regulation of class switch recombination. In: Honjo T, Alt FW, Neuberger MS, editors. Molecular Biology of B Cells. London: Elsevier; 2004. pp. 289–305. [Google Scholar]
- 33.Pan-Hammarstrom Q, Zhao Y, Hammarstrom L. Class switch recombination: a comparison between mouse and human. Adv. Immunol. 2007;93:1–61. doi: 10.1016/S0065-2776(06)93001-6. [DOI] [PubMed] [Google Scholar]
- 34.Chaudhuri J, Alt FW. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nat. Rev. Immunol. 2004;4:541–552. doi: 10.1038/nri1395. [DOI] [PubMed] [Google Scholar]
- 35.Li Z, Woo CJ, Iglesias-Ussel MD, Ronai D, Scharff MD. The generation of antibody diversity through somatic hypermutation and class switch recombination. Genes Dev. 2004;18:1–11. doi: 10.1101/gad.1161904. [DOI] [PubMed] [Google Scholar]
- 36.Kenter AL. Class switch recombination: an emerging mechanism. Curr. Top. Microbiol. Immunol. 2005;290:171–199. doi: 10.1007/3-540-26363-2_8. [DOI] [PubMed] [Google Scholar]
- 37.Stavnezer J, Kinoshita K, Muramatsu M, Honjo T. Honjo T, Neuberger MS, Alt FW. Molecular Biology of B Cells. London: Elsevier; 2004. Molecular mechanisms of class switch recombination; pp. 307–326. [Google Scholar]
- 38.Rada C, Williams GT, Nilsen H, Barnes DE, Lindahl T, Neuberger MS. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr. Biol. 2002;12:1748–1755. doi: 10.1016/s0960-9822(02)01215-0. [DOI] [PubMed] [Google Scholar]
- 39.Di Noia J, Neuberger MS. Altering the pathway of immunoglobulin hypermutation by inhibiting uracil-DNA glycosylase. Nature. 2002;419:43–48. doi: 10.1038/nature00981. [DOI] [PubMed] [Google Scholar]
- 40.Poltoratsky V, Goodman MF, Scharff MD. Error-prone candidates vie for somatic mutation. J. Exp. Med. 2000;192:F27–F30. doi: 10.1084/jem.192.10.f27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Catalan N, Selz F, Imai K, Revy P, Fischer A, Durandy A. The block in immunoglobulin class switch recombination caused by activation-induced cytidine deaminase deficiency occurs prior to the generation of DNA double strand breaks in switch μ region. J. Immunol. 2003;171:2504–2509. doi: 10.4049/jimmunol.171.5.2504. [DOI] [PubMed] [Google Scholar]
- 42.Rush JS, Fugmann SD, Schatz DG. Staggered AID-dependent DNA double strand breaks are the predominant DNA lesions targeted to Sμ in Ig class switch recombination. Int. Immunol. 2004;16:549–557. doi: 10.1093/intimm/dxh057. [DOI] [PubMed] [Google Scholar]
- 43.Petersen S, Casellas R, Reina-San-Martin B, Chen HT, Difilippantonio MJ, et al. AID is required to initiate Nbs1/γ-H2AX focus formation and mutations at sites of class switching. Nature. 2001;414:660–665. doi: 10.1038/414660a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramiro AR, Stavropoulos P, Jankovic M, Nussenzweig MC. Transcription enhances AID-mediated cytidine deamination by exposing single-stranded DNA on the nontemplate strand. Nat. Immunol. 2003;4:452–456. doi: 10.1038/ni920. [DOI] [PubMed] [Google Scholar]
- 45.Shen HM, Storb U. Activation-induced cytidine deaminase (AID) can target both DNA strands when the DNA is supercoiled. Proc. Natl. Acad. Sci. USA. 2004;101:12997–13002. doi: 10.1073/pnas.0404974101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shen HM, Ratnam S, Storb U. Targeting of the activation-induced cytosine deaminase is strongly influenced by the sequence and structure of the targeted DNA. Mol. Cell. Biol. 2005;25:10815–10821. doi: 10.1128/MCB.25.24.10815-10821.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martomo SA, Fu D, Yang WW, Joshi NS, Gearhart PJ. Deoxyuridine is generated preferentially in the nontranscribed strand of DNA from cells expressing activation-induced cytidine deaminase. J. Immunol. 2005;174:7787–7791. doi: 10.4049/jimmunol.174.12.7787. [DOI] [PubMed] [Google Scholar]
- 48.Milstein C, Neuberger MS, Staden R. Both DNA strands of antibody genes are hypermutation targets. Proc. Natl. Acad. Sci. USA. 1998;95:8791–8794. doi: 10.1073/pnas.95.15.8791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Longerich S, Tanaka A, Bozek G, Nicolae D, Storb U. The very 5′ end and the constant region of Ig genes are spared from somatic mutation because AID does not access these regions. J. Exp. Med. 2005;202:1443–1454. doi: 10.1084/jem.20051604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xue K, Rada C, Neuberger MS. The in vivo pattern of AID targeting to immunoglobulin switch regions deduced from mutation spectra in msh2−/− ung−/− mice. J. Exp. Med. 2006;203:2085–2094. doi: 10.1084/jem.20061067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Betz AG, Milstein C, Gonzalez-Fernandez A, Pannell R, Larson T, Neuberger MS. Elements regulating somatic hypermutation of an immunoglobulin κ gene: critical role for the intron enhancer/matrix attachment region. Cell. 1994;77:239–248. doi: 10.1016/0092-8674(94)90316-6. [DOI] [PubMed] [Google Scholar]
- 52.Rogozin IB, Kolchanov NA. Somatic hypermutagenesis in immunoglobulin genes. II. Influence of neighbouring base sequences on mutagenesis. Biochim. Biophys. Acta. 1992;1171:11–18. doi: 10.1016/0167-4781(92)90134-l. [DOI] [PubMed] [Google Scholar]
- 53.Shapiro GS, Aviszus K, Ikle D, Wysocki LJ. Predicting regional mutability in antibody V genes based solely on di- and trinucleotide sequence composition. J. Immunol. 1999;163:259–268. [PubMed] [Google Scholar]
- 54.Bransteitter R, Pham P, Calabrese P, Goodman MF. Biochemical analysis of hypermutational targeting by wild type and mutant activation-induced cytidine deaminase. J. Biol. Chem. 2004;279:51612–51621. doi: 10.1074/jbc.M408135200. [DOI] [PubMed] [Google Scholar]
- 55.Yu K, Huang FT, Lieber MR. DNA substrate length and surrounding sequence affect the activation-induced deaminase activity at cytidine. J. Biol. Chem. 2004;279:6496–6500. doi: 10.1074/jbc.M311616200. [DOI] [PubMed] [Google Scholar]
- 56.Chaudhuri J, Khuong C, Alt FW. Replication protein A interacts with AID to promote deamination of somatic hypermutation targets. Nature. 2004;430:992–998. doi: 10.1038/nature02821. [DOI] [PubMed] [Google Scholar]
- 57.Basu U, Chaudhuri J, Alpert C, Dutt S, Ranganath S, et al. The AID antibody diversification enzyme is regulated by protein kinase A phosphorylation. Nature. 2005;438:508–511. doi: 10.1038/nature04255. [DOI] [PubMed] [Google Scholar]
- 58.McBride KM, Gazumyan A, Woo EM, Barreto VM, Robbiani DV, et al. Regulation of hypermutation by activation-induced cytidine deaminase phosphorylation. Proc. Natl. Acad. Sci. USA. 2006;103:8798–8803. doi: 10.1073/pnas.0603272103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ta VT, Nagaoka H, Catalan N, Durandy A, Fischer A, et al. AID mutant analyses indicate requirement for class-switch-specific cofactors. Nat. Immunol. 2003;4:843–848. doi: 10.1038/ni964. [DOI] [PubMed] [Google Scholar]
- 60.Barreto V, Reina-San-Martin B, Ramiro AR, McBride KM, Nussenzweig MC. C-terminal deletion of AID uncouples class switch recombination from somatic hypermutation and gene conversion. Mol. Cell. 2003;12:501–508. doi: 10.1016/s1097-2765(03)00309-5. [DOI] [PubMed] [Google Scholar]
- 61.Christmann M, Tomicic MT, Roos WP, Kaina B. Mechanisms of human DNA repair: an update. Toxicology. 2003;193:3–34. doi: 10.1016/s0300-483x(03)00287-7. [DOI] [PubMed] [Google Scholar]
- 62.Imai K, Slupphaug G, Lee WI, Revy P, Nonoyama S, et al. Human uracil-DNA glycosylase deficiency associated with profoundly impaired immunoglobulin class-switch recombination. Nat. Immunol. 2003;4:1023–1028. doi: 10.1038/ni974. [DOI] [PubMed] [Google Scholar]
- 63.Bardwell PD, Martin A, Wong E, Li Z, Edelmann W, Scharff MD. Cutting edge: the G-U mismatch glycosylase methyl-CpG binding domain 4 is dispensable for somatic hypermutation and class switch recombination. J. Immunol. 2003;170:1620–1624. doi: 10.4049/jimmunol.170.4.1620. [DOI] [PubMed] [Google Scholar]
- 64.Di Noia JM, Rada C, Neuberger MS. SMUG1 is able to excise uracil from immunoglobulin genes: insight into mutation vs repair. EMBO J. 2006;25:585–595. doi: 10.1038/sj.emboj.7600939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hadi MZ, Ginalski K, Nguyen LH, Wilson DM., 3rd Determinants in nuclease specificity of Ape1 and Ape2, human homologues of Escherichia coli exonuclease III. J. Mol. Biol. 2002;316:853–866. doi: 10.1006/jmbi.2001.5382. [DOI] [PubMed] [Google Scholar]
- 66.Kanno S, Kuzuoka H, Sasao S, Hong Z, Lan L, et al. A novel human AP endonuclease with conserved zinc-finger-like motifs involved in DNA strand break responses. EMBO J. 2007;26:2094–2103. doi: 10.1038/sj.emboj.7601663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Iles N, Rulten S, El-Khamisy SF, Caldecott KW. APLF (C2orf13) is a novel human protein involved in the cellular response to chromosomal DNA strand breaks. Mol. Cell. Biol. 2007;27:3793–3803. doi: 10.1128/MCB.02269-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bekker-Jensen S, Fugger K, Danielsen JR, Gromova I, Sehested M, et al. Human xip1 (c2orf13) is a novel regulator of cellular responses to DNA strand breaks. J. Biol. Chem. 2007;282:19638–19643. doi: 10.1074/jbc.C700060200. [DOI] [PubMed] [Google Scholar]
- 69.Xanthoudakis S, Smeyne RJ, Wallace JD, Curran T. The redox/DNA repair protein, Ref-1, is essential for early embryonic development in mice. Proc. Natl. Acad. Sci. USA. 1996;93:8919–8923. doi: 10.1073/pnas.93.17.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fung H, Demple B. A vital role for Ape1/Ref1 protein in repairing spontaneous DNA damage in human cells. Mol. Cell. 2005;17:463–470. doi: 10.1016/j.molcel.2004.12.029. [DOI] [PubMed] [Google Scholar]
- 71.Ide Y, Tsuchimoto D, Tominaga Y, Nakashima M, Watanabe T, et al. Growth retardation and dyslymphopoiesis accompanied by G2/M arrest in APEX2-null mice. Blood. 2004;104:4097–4103. doi: 10.1182/blood-2004-04-1476. [DOI] [PubMed] [Google Scholar]
- 72.Ide Y, Tsuchimoto D, Tominaga Y, Iwamoto Y, Nakabeppu Y. Characterization of the genomic structure and expression of the mouse Apex2 gene. Genomics. 2003;81:47–57. doi: 10.1016/s0888-7543(02)00009-5. [DOI] [PubMed] [Google Scholar]
- 73.Burkovics P, Szukacsov V, Unk I, Haracska L. Human Ape2 protein has a 3′–5′ exonuclease activity that acts preferentially on mismatched base pairs. Nucleic Acids Res. 2006;34:2508–2515. doi: 10.1093/nar/gkl259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guikema JEJ, Linehan EK, Tsuchimoto D, Nakabeppu Y, Strauss PR, et al. APE1 and APE2 dependent DNA breaks in immunoglobulin class switch recombination. J. Exp. Med. 2007;204:3017–3026. doi: 10.1084/jem.20071289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Meira LB, Devaraj S, Kisby GE, Burns DK, Daniel RL, et al. Heterozygosity for the mouse Apex gene results in phenotypes associated with oxidative stress. Cancer Res. 2001;61:5552–5557. [PubMed] [Google Scholar]
- 76.Raffoul JJ, Cabelof DC, Nakamura J, Meira LB, Friedberg EC, Heydari AR. Apurinic/apyrimidinic endonuclease (APE/REF-1) haploinsufficient mice display tissue-specific differences in DNA polymerase β-dependent base excision repair. J. Biol. Chem. 2004;279:18425–18433. doi: 10.1074/jbc.M313983200. [DOI] [PubMed] [Google Scholar]
- 77.Schrader CE, Guikema JEJ, Linehan EK, Selsing E, Stavnezer J. AID-dependent DNA breaks in class switch recombination occur during G1 phase and are mismatch repair-dependent. J. Immunol. 2007;179:6064–6071. doi: 10.4049/jimmunol.179.9.6064. [DOI] [PubMed] [Google Scholar]
- 78.Barnes DE, Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu. Rev. Genet. 2004;38:445–476. doi: 10.1146/annurev.genet.38.072902.092448. [DOI] [PubMed] [Google Scholar]
- 79.Beard WA, Wilson SH. Structure and mechanism of DNA polymerase β. Chem. Rev. 2006;106:361–382. doi: 10.1021/cr0404904. [DOI] [PubMed] [Google Scholar]
- 80.Akbari M, Otterlei M, Pena-Diaz J, Aas PA, Kavli B, et al. Repair of U/G and U/A in DNA by UNG2-associated repair complexes takes place predominantly by short-patch repair both in proliferating and growth-arrested cells. Nucleic Acids Res. 2004;32:5486–5498. doi: 10.1093/nar/gkh872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Parlanti E, Locatelli G, Maga G, Dogliotti E. Human base excision repair complex is physically associated to DNA replication and cell cycle regulatory proteins. Nucleic Acids Res. 2007;35:1569–1577. doi: 10.1093/nar/gkl1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wong D, Demple B. Modulation of the 5′-deoxyribose-5-phosphate lyase and DNA synthesis activities of mammalian DNA polymerase β by apurinic/apyrimidinic endonuclease 1. J. Biol. Chem. 2004;279:25268–25275. doi: 10.1074/jbc.M400804200. [DOI] [PubMed] [Google Scholar]
- 83.Parsons JL, Dianova II, Allinson SL, Dianov GL. DNA polymerase β promotes recruitment of DNA ligase IIIα-XRCC1 to sites of base excision repair. Biochemistry. 2005;44:10613–10619. doi: 10.1021/bi050085m. [DOI] [PubMed] [Google Scholar]
- 84.Liu Y, Prasad R, Beard WA, Kedar PS, Hou EW, et al. Coordination of steps in single-nucleotide base excision repair mediated by apurinic/apyrimidinic endonuclease 1 and DNA polymerase β. J. Biol. Chem. 2007;282:13532–13541. doi: 10.1074/jbc.M611295200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wu X, Stavnezer J. DNA polymerase β is able to repair breaks in switch regions and plays an inhibitory role during immunoglobulin class switch recombination. J. Exp. Med. 2007;204:1677–1689. doi: 10.1084/jem.20070756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ito K, Hirao A, Arai F, Matsuoka S, Takubo K, et al. Regulation of oxidative stress by ATM is required for self-renewal of haematopoietic stem cells. Nature. 2004;431:997–1002. doi: 10.1038/nature02989. [DOI] [PubMed] [Google Scholar]
- 87.Zarrin AA, Del Vecchio C, Tseng E, Gleason M, Zarin P, et al. Antibody class switching mediated by yeast endonuclease-generated DNA breaks. Science. 2007;315:377–381. doi: 10.1126/science.1136386. [DOI] [PubMed] [Google Scholar]
- 88.Kunkel T, Erie D. DNA mismatch repair. Annu. Rev. Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 89.Genschel J, Bazemore LR, Modrich P. Human exonuclease I is required for 5′ and 3′ mismatch repair. J. Biol. Chem. 2002;277:13302–13311. doi: 10.1074/jbc.M111854200. [DOI] [PubMed] [Google Scholar]
- 90.Genschel J, Modrich P. Mechanism of 5′-directed excision in human mismatch repair. Mol. Cell. 2003;12:1077–1086. doi: 10.1016/s1097-2765(03)00428-3. [DOI] [PubMed] [Google Scholar]
- 91.Kadyrov FA, Dzantiev L, Constantin N, Modrich P. Endonucleolytic function of MutLα in human mismatch repair. Cell. 2006;126:297–308. doi: 10.1016/j.cell.2006.05.039. [DOI] [PubMed] [Google Scholar]
- 92.Ehrenstein MR, Neuberger MS. Deficiency in Msh2 affects the efficiency and local sequence specificity of immunoglobulin class-switch recombination: parallels with somatic hypermutation. EMBO J. 1999;18:3484–3490. doi: 10.1093/emboj/18.12.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schrader CE, Edelmann W, Kucherlapati R, Stavnezer J. Reduced isotype switching in splenic B cells from mice deficient in mismatch repair enzymes. J. Exp. Med. 1999;190:323–330. doi: 10.1084/jem.190.3.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ehrenstein MR, Rada C, Jones AM, Milstein C, Neuberger MS. Switch junction sequences in PMS2-deficient mice reveal a microhomology-mediated mechanism of Ig class switch recombination. Proc. Natl. Acad. Sci. USA. 2001;98:14553–14558. doi: 10.1073/pnas.241525998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li Z, Scherer SJ, Ronai D, Iglesias-Ussel MD, Peled JU, et al. Examination of Msh6- and Msh3-deficient mice in class switching reveals overlapping and distinct roles of MutS homologues in antibody diversification. J. Exp. Med. 2004;200:47–59. doi: 10.1084/jem.20040355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Martin A, Li Z, Lin DP, Bardwell PD, Iglesias-Ussel MD, et al. Msh2 ATPase activity is essential for somatic hypermutation at A-T basepairs and for efficient class switch recombination. J. Exp. Med. 2003;198:1171–1178. doi: 10.1084/jem.20030880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Martomo SA, Yang WW, Gearhart PJ. A role for Msh6 but not Msh3 in somatic hypermutation and class switch recombination. J. Exp. Med. 2004;200:61–68. doi: 10.1084/jem.20040691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bardwell PD, Woo CJ, Wei K, Li Z, Martin A, et al. Altered somatic hypermutation and reduced class-switch recombination in exonuclease 1-mutant mice. Nat. Immunol. 2004;5:224–229. doi: 10.1038/ni1031. [DOI] [PubMed] [Google Scholar]
- 99.Stavnezer J, Schrader CE. Mismatch repair converts AID-instigated nicks to double-strand breaks for antibody class-switch recombination. Trends Genet. 2006;22:23–28. doi: 10.1016/j.tig.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 100.Wilson TM, Vaisman A, Martomo SA, Sullivan P, Lan L, et al. MSH2-–MSH6 stimulates DNA polymerase η, suggesting a role for A:T mutations in antibody genes. J. Exp. Med. 2005;201:637–645. doi: 10.1084/jem.20042066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Luby TM, Schrader CE, Stavnezer J, Selsing E. The μ switch region tandem repeats are important, but not required, for antibody class switch recombination. J. Exp. Med. 2001;193:159–168. doi: 10.1084/jem.193.2.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Min I, Schrader C, Vardo J, D’Avirro N, Luby T, et al. The Sμ tandem repeat region is critical for isotype switching in the absence of Msh2. Immunity. 2003;19:515–524. doi: 10.1016/s1074-7613(03)00262-0. [DOI] [PubMed] [Google Scholar]
- 103.Schrader CE, Vardo J, Stavnezer J. Role for mismatch repair proteins Msh2, Mlh1, and Pms2 in immunoglobulin class switching shown by sequence analysis of recombination junctions. J. Exp. Med. 2002;195:367–373. doi: 10.1084/jem.20011877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Schrader CE, Vardo J, Linehan E, Twarog MZ, Niedernhofer LJ, et al. Deletion of the nucleotide excision repair gene Ercc1 reduces Ig class switching and alters mutations near switch recombination junctions. J. Exp. Med. 2004;200:321–330. doi: 10.1084/jem.20040052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Faili A, Aoufouchi S, Weller S, Vuillier F, Stary A, et al. DNA polymerase η is involved in hypermutation occurring during immunoglobulin class switch recombination. J. Exp. Med. 2004;199:265–270. doi: 10.1084/jem.20031831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Delbos F, Aoufouchi S, Faili A, Weill JC, Reynaud CA. DNA polymerase η is the sole contributor of A/T modifications during immunoglobulin gene hypermutation in the mouse. J. Exp. Med. 2007;204:17–23. doi: 10.1084/jem.20062131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dunnick W, Wilson M, Stavnezer J. Mutations, duplications, and deletion of recombined switch regions suggest a role for DNA replication in the immunoglobulin heavy chain switch. Mol. Cell. Biol. 1989;9:1850–1856. doi: 10.1128/mcb.9.5.1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schrader CE, Bradley SP, Vardo J, Mochegova SN, Flanagan E, Stavnezer J. Mutations occur in the Ig Sμ region but rarely in Sγ regions prior to class switch recombination. EMBO J. 2003;22:5893–5903. doi: 10.1093/emboj/cdg550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Masuda K, Ouchida R, Takeuchi A, Saito T, Koseki H, et al. DNA polymerase θ contributes to the generation of C/G mutations during somatic hypermutation of Ig genes. Proc. Natl. Acad. Sci. USA. 2005;102:13986–13991. doi: 10.1073/pnas.0505636102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zan H, Shima N, Xu Z, Al-Qahtani A, Evinger AJ Iii, et al. The translesion DNA polymerase θ plays a dominant role in immunoglobulin gene somatic hypermutation. EMBO J. 2005;24:3757–3769. doi: 10.1038/sj.emboj.7600833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Masuda K, Ouchida R, Hikida M, Nakayama M, Ohara O, et al. Absence of DNA polymerase θ results in decreased somatic hypermutation frequency and altered mutation patterns in Ig genes. DNA Repair. 2006;5:1384–1391. doi: 10.1016/j.dnarep.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 112.Dudley DD, Manis JP, Zarrin AA, Kaylor L, Tian M, Alt FW. Internal IgH class switch region deletions are position-independent and enhanced by AID expression. Proc. Natl. Acad. Sci. USA. 2002;99:9984–9989. doi: 10.1073/pnas.152333499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Reina-San-Martin B, Difilippantonio S, Hanitsch L, Masilamani RF, Nussenzweig A, Nussenzweig MC. H2AX is required for recombination between immunoglobulin switch regions but not for intraswitch region recombination or somatic hypermutation. J. Exp. Med. 2003;197:1767–1778. doi: 10.1084/jem.20030569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rooney S, Chaudhuri J, Alt FW. The role of the nonhomologous end-joining pathway in lymphocyte development. Immunol. Rev. 2004;200:115–131. doi: 10.1111/j.0105-2896.2004.00165.x. [DOI] [PubMed] [Google Scholar]
- 115.Ma Y, Lu H, Schwarz K, Lieber MR. Repair of double-strand DNA breaks by the human nonhomologous DNA end joining pathway: the iterative processing model. Cell Cycle. 2005;4:1193–1200. doi: 10.4161/cc.4.9.1977. [DOI] [PubMed] [Google Scholar]
- 116.Sonoda E, Hochegger H, Saberi A, Taniguchi Y, Takeda S. Differential usage of nonhomologous end-joining and homologous recombination in double strand break repair. DNA Repair. 2006;5:1021–1029. doi: 10.1016/j.dnarep.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 117.Manis JP, Gu Y, Lansford R, Sonoda E, Ferrini R, et al. Ku70 is required for late B cell development and immunoglobulin heavy chain switching. J. Exp. Med. 1998;187:2081–2089. doi: 10.1084/jem.187.12.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Casellas R, Nussenzweig A, Wuerffel R, Pelanda R, Reichlin A, et al. Ku80 is required for immunoglobulin isotype switching. EMBO J. 1998;17:2404–2411. doi: 10.1093/emboj/17.8.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pan-Hammarstrom Q, Jones AM, Lahdesmaki A, Zhou W, Gatti RA, et al. Impact of DNA ligase IV on nonhomologous end joining pathways during class switch recombination in human cells. J. Exp. Med. 2005;201:189–194. doi: 10.1084/jem.20040772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Soulas-Sprauel P, Guyader GL, Rivera-Munoz P, Abramowski V, Olivier-Martin C, et al. Role for DNA repair factor XRCC4 in immunoglobulin class switch recombination. J. Exp. Med. 2007;204:1717–1727. doi: 10.1084/jem.20070255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chen L, Trujillo K, Sung P, Tomkinson AE. Interactions of the DNA ligase IV-XRCC4 complex with DNA ends and the DNA-dependent protein kinase. J. Biol. Chem. 2000;275:26196–26205. doi: 10.1074/jbc.M000491200. [DOI] [PubMed] [Google Scholar]
- 122.Nick McElhinny SA, Snowden CM, McCarville J, Ramsden DA. Ku recruits the XRCC4-ligase IV complex to DNA ends. Mol. Cell. Biol. 2000;20:2996–3003. doi: 10.1128/mcb.20.9.2996-3003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Costantini S, Woodbine L, Andreoli L, Jeggo PA, Vindigni A. Interaction of the Ku heterodimer with the DNA ligase IV/Xrcc4 complex and its regulation by DNA-PK. DNA Repair. 2007;6:712–722. doi: 10.1016/j.dnarep.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 124.Frank KM, Sekiguchi JM, Seidl KJ, Swat W, Rathbun GA, et al. Late embryonic lethality and impaired V(D)J recombination in mice lacking DNA ligase IV. Nature. 1998;396:173–177. doi: 10.1038/24172. [DOI] [PubMed] [Google Scholar]
- 125.Yan CT, Boboila C, Souza EK, Franco S, Hickernell TR, et al. IgH class switching and translocations use a robust nonclassical end-joining pathway. Nature. 2007;449:478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- 126.Gu J, Lu H, Tippin B, Shimazaki N, Goodman MF, Lieber MR. XRCC4:DNA ligase IV can ligate incompatible DNA ends and can ligate across gaps. EMBO J. 2007;26:1010–1023. doi: 10.1038/sj.emboj.7601559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sayegh C, Jhunjhunwala S, Riblet R, Murre C. Visualization of looping involving the immunoglobulin heavy-chain locus in developing B cells. Genes. Dev. 2005;19:322–327. doi: 10.1101/gad.1254305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Roldan E, Fuxa M, Chong W, Martinez D, Novatchkova M, et al. Locus ‘decontraction’ and centromeric recruitment contribute to allelic exclusion of the immunoglobulin heavy-chain gene. Nat. Immunol. 2005;6:31–41. doi: 10.1038/ni1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Moreno-Herrero F, de Jager M, Dekker NH, Kanaar R, Wyman C, Dekker C. Mesoscale conformational changes in the DNA-repair complex Rad50/Mre11/Nbs1 upon binding DNA. Nature. 2005;437:440–443. doi: 10.1038/nature03927. [DOI] [PubMed] [Google Scholar]
- 130.Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- 131.Hopfner KP, Karcher A, Shin DS, Craig L, Arthur LM, et al. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell. 2000;101:789–800. doi: 10.1016/s0092-8674(00)80890-9. [DOI] [PubMed] [Google Scholar]
- 132.de Jager M, Dronkert ML, Modesti M, Beerens CE, Kanaar R, van Gent DC. DNA-binding and strand-annealing activities of human Mre11: implications for its roles in DNA double-strand break repair pathways. Nucleic Acids Res. 2001;29:1317–1325. doi: 10.1093/nar/29.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Difilippantonio S, Celeste A, Fernandez-Capetillo O, Chen HT, Reina San Martin B, et al. Role of Nbs1 in the activation of the Atm kinase revealed in humanized mouse models. Nat. Cell Biol. 2005;7:675–685. doi: 10.1038/ncb1270. [DOI] [PubMed] [Google Scholar]
- 134.Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- 135.Cerosaletti K, Wright J, Concannon P. Active role for nibrin in the kinetics of atm activation. Mol. Cell. Biol. 2006;26:1691–1699. doi: 10.1128/MCB.26.5.1691-1699.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Reina-San-Martin B, Nussenzweig MC, Nussenzweig A, Difilippantonio S. Genomic instability, endoreduplication, and diminished Ig class-switch recombination in B cells lacking Nbs1. Proc. Natl. Acad. Sci. USA. 2005;102:1590–1595. doi: 10.1073/pnas.0406289102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Carney JP, Maser RS, Olivares H, Davis EM, LeBeau M, et al. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair (DBSR) to the cellular DNA damage response. Cell. 1998;93:477–486. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- 138.Varon R, Vissinga C, Platzer M, Cerosaletti KM, Chrzanowska KH, et al. Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell. 1998;93:467–476. doi: 10.1016/s0092-8674(00)81174-5. [DOI] [PubMed] [Google Scholar]
- 139.Kracker S, Bergmann Y, Demuth I, Frappart PO, Hildebrand G, et al. Nibrin functions in Ig class-switch recombination. Proc. Natl. Acad. Sci. USA. 2005;102:1584–1589. doi: 10.1073/pnas.0409191102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ramiro AR, Jankovic M, Eisenreich T, Difilippantonio S, Chen-Kiang S, et al. AID is required for c-myc/IgH chromosome translocations in vivo. Cell. 2004;118:431–438. doi: 10.1016/j.cell.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 141.Unniraman S, Zhou S, Schatz DG. Identification of an AID-independent pathway for chromosomal translocations between the Igh switch region and Myc. Nat. Immunol. 2004;5:1117–1123. doi: 10.1038/ni1127. [DOI] [PubMed] [Google Scholar]
- 142.Franco S, Gostissa M, Zha S, Lombard DB, Murphy MM, et al. H2AX prevents DNA breaks from progressing to chromosome breaks and translocations. Mol. Cell. 2006;21:201–214. doi: 10.1016/j.molcel.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 143.Ramiro AR, Jankovic M, Callen E, Difilippantonio S, Chen HT, et al. Role of genomic instability and p53 in AID-induced c-myc-Igh translocations. Nature. 2006;440:105–109. doi: 10.1038/nature04495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Pan Q, Petit-Frere C, Lahdesmaki A, Gregorek H, Chrzanowska KH, Hammarstrom L. Alternative end joining during switch recombination in patients with ataxia-telangiectasia. Eur. J. Immunol. 2002;32:1300–1308. doi: 10.1002/1521-4141(200205)32:5<1300::AID-IMMU1300>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 145.Lahdesmaki A, Taylor AM, Chrzanowska KH, Pan-Hammarstrom Q. Delineation of the role of the Mre11 complex in class switch recombination. J. Biol. Chem. 2004;279:16479–16487. doi: 10.1074/jbc.M312796200. [DOI] [PubMed] [Google Scholar]
- 146.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 147.Downs JA, Nussenzweig MC, Nussenzweig A. Chromatin dynamics and the preservation of genetic information. Nature. 2007;447:951–958. doi: 10.1038/nature05980. [DOI] [PubMed] [Google Scholar]
- 148.Bredemeyer AL, Sharma GG, Huang CY, Helmink BA, Walker LM, et al. ATM stabilizes DNA double-strand-break complexes during V(D)J recombination. Nature. 2006;442:466–470. doi: 10.1038/nature04866. [DOI] [PubMed] [Google Scholar]
- 149.Callen E, Jankovic M, Difilippantonio S, Daniel JA, Chen HT, et al. ATM prevents the persistence and propagation of chromosome breaks in lymphocytes. Cell. 2007;130:63–75. doi: 10.1016/j.cell.2007.06.016. [DOI] [PubMed] [Google Scholar]
- 150.Vacchio MS, Olaru A, Livak F, Hodes RJ. ATM deficiency impairs thymocyte maturation because of defective resolution of T cell receptor a locus coding end breaks. Proc. Natl. Acad. Sci. USA. 2007;104:6323–6328. doi: 10.1073/pnas.0611222104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lumsden JM, McCarty T, Petiniot LK, Shen R, Barlow C, et al. Immunoglobulin class switch recombination is impaired in Atm-deficient mice. J. Exp. Med. 2004;200:1111–1121. doi: 10.1084/jem.20041074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Reina-San-Martin B, Chen HT, Nussenzweig A, Nussenzweig MC. ATM is required for efficient recombination between immunoglobulin switch regions. J. Exp. Med. 2004;200:1103–1110. doi: 10.1084/jem.20041162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Iwabuchi K, Bartel PL, Li B, Marraccino R, Fields S. Two cellular proteins that bind to wild-type but not mutant p53. Proc. Natl. Acad. Sci. USA. 1994;91:6098–6102. doi: 10.1073/pnas.91.13.6098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Huang J, Sengupta R, Espejo B, Lee MG, Dorsey JA, et al. p53 is regulated by the lysine demethylase LSD1. Nature. 2007;449:105–108. doi: 10.1038/nature06092. [DOI] [PubMed] [Google Scholar]
- 155.Pryde F, Khalili S, Robertson K, Selfridge J, Ritchie AM, et al. 53BP1 exchanges slowly at the sites of DNA damage and appears to require RNA for its association with chromatin. J. Cell Sci. 2005;118:2043–2055. doi: 10.1242/jcs.02336. [DOI] [PubMed] [Google Scholar]
- 156.Schultz LB, Chehab NH, Malikzay A, Halazonetis TD. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J. Cell Biol. 2000;151:1381–1390. doi: 10.1083/jcb.151.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Celeste A, Fernandez-Capetillo O, Kruhlak MJ, Pilch DR, Staudt DW, et al. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat. Cell Biol. 2003;5:675–679. doi: 10.1038/ncb1004. [DOI] [PubMed] [Google Scholar]
- 158.Ward IM, Minn K, Jorda KG, Chen J. Accumulation of checkpoint protein 53BP1 at DNA breaks involves its binding to phosphorylated histone H2AX. J. Biol. Chem. 2003;278:19579–19582. doi: 10.1074/jbc.C300117200. [DOI] [PubMed] [Google Scholar]
- 159.Adams MM, Carpenter PB. Tying the loose ends together in DNA double strand break repair with 53BP1. Cell Div. 2006;1:19. doi: 10.1186/1747-1028-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Mochan TA, Venere M, DiTullio RA, Jr, Halazonetis TD. 53BP1, an activator of ATM in response to DNA damage. DNA Repair. 2004;3:945–952. doi: 10.1016/j.dnarep.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 161.Manis JP, Morales JC, Xia Z, Kutok JL, Alt FW, Carpenter PB. 53BP1 links DNA damage-response pathways to immunoglobulin heavy chain class-switch recombination. Nat. Immunol. 2004;5:481–487. doi: 10.1038/ni1067. [DOI] [PubMed] [Google Scholar]
- 162.Ward IM, Reina-San-Martin B, Olaru A, Minn K, Tamada K, et al. 53BP1 is required for class switch recombination. J. Cell Biol. 2004;165:459–464. doi: 10.1083/jcb.200403021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Reina-San-Martin B, Chen J, Nussenzweig A, Nussenzweig MC. Enhanced intraswitch region recombination during immunoglobulin class switch recombination in 53BP1−/− B cells. Eur. J. Immunol. 2007;37:235–239. doi: 10.1002/eji.200636789. [DOI] [PubMed] [Google Scholar]
- 164.Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 166.Burma S, Chen BP, Murphy M, Kurimasa A, Chen DJ. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J. Biol. Chem. 2001;276:42462–42467. doi: 10.1074/jbc.C100466200. [DOI] [PubMed] [Google Scholar]
- 167.Lou Z, Minter-Dykhouse K, Franco S, Gostissa M, Rivera MA, et al. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol. Cell. 2006;21:187–200. doi: 10.1016/j.molcel.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 168.Berkovich E, Monnat RJ, Jr, Kastan MB. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat. Cell Biol. 2007;9:683–690. doi: 10.1038/ncb1599. [DOI] [PubMed] [Google Scholar]
- 169.Celeste A, Petersen S, Romanienko PJ, Fernandez-Capetillo O, Chen HT, et al. Genomic instability in mice lacking histone H2AX. Science. 2002;296:922–927. doi: 10.1126/science.1069398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Fernandez-Capetillo O, Celeste A, Nussenzweig A. Focusing on foci: H2AX and the recruitment of DNA-damage response factors. Cell Cycle. 2003;2:426–427. [PubMed] [Google Scholar]
- 171.Bassing CH, Suh H, Ferguson DO, Chua KF, Manis J, et al. Histone H2AX: a dosage-dependent suppressor of oncogenic translocations and tumors. Cell. 2003;114:359–370. doi: 10.1016/s0092-8674(03)00566-x. [DOI] [PubMed] [Google Scholar]
- 172.Stewart GS, Wang B, Bignell CR, Taylor AM, Elledge SJ. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature. 2003;421:961–966. doi: 10.1038/nature01446. [DOI] [PubMed] [Google Scholar]
- 173.Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 174.Lieber MR, Ma Y, Pannicke U, Schwarz K. Mechanism and regulation of human nonhomologous DNA end-joining. Nat. Rev. Mol. Cell Biol. 2003;4:712–720. doi: 10.1038/nrm1202. [DOI] [PubMed] [Google Scholar]
- 175.Meek K, Gupta S, Ramsden DA, Lees-Miller SP. The DNA-dependent protein kinase: the director at the end. Immunol. Rev. 2004;200:132–141. doi: 10.1111/j.0105-2896.2004.00162.x. [DOI] [PubMed] [Google Scholar]
- 176.Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- 177.Spagnolo L, Rivera-Calzada A, Pearl LH, Llorca O. Three-dimensional structure of the human DNA-PKcs/Ku70/Ku80 complex assembled on DNA and its implications for DNA DSB repair. Mol. Cell. 2006;22:511–519. doi: 10.1016/j.molcel.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 178.Soutoglou E, Dorn JF, Sengupta K, Jasin M, Nussenzweig A, et al. Positional stability of single double-strand breaks in mammalian cells. Nat. Cell Biol. 2007;9:675–682. doi: 10.1038/ncb1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Meek K, Douglas P, Cui X, Ding Q, Lees-Miller SP. Trans autophosphorylation at DNA-dependent protein kinase’s two major autophosphorylation site clusters facilitates end processing but not end joining. Mol. Cell. Biol. 2007;27:3881–3890. doi: 10.1128/MCB.02366-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Manis JP, Dudley D, Kaylor L, Alt FW. IgH class switch recombination to IgG1 in DNA-PKcs-deficient B cells. Immunity. 2002;16:607–617. doi: 10.1016/s1074-7613(02)00306-0. [DOI] [PubMed] [Google Scholar]
- 181.Rooney S, Alt FW, Sekiguchi J, Manis JP. Artemis-independent functions of DNA-dependent protein kinase in Ig heavy chain class switch recombination and development. Proc. Natl. Acad. Sci. USA. 2005;102:2471–2475. doi: 10.1073/pnas.0409857102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Bosma GC, Kim J, Urich T, Fath DM, Cotticelli MG, et al. DNA-dependent protein kinase activity is not required for immunoglobulin class switching. J. Exp. Med. 2002;196:1483–1495. doi: 10.1084/jem.20001871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Cook AJ, Oganesian L, Harumal P, Basten A, Brink R, Jolly CJ. Reduced switching in SCID B cells is associated with altered somatic mutation of recombined S regions. J. Immunol. 2003;171:6556–6564. doi: 10.4049/jimmunol.171.12.6556. [DOI] [PubMed] [Google Scholar]
- 184.Kaminski DA, Stavnezer J. Antibody class switching differs among SJL, C57BL/6 and 129 mice. Int. Immunol. 2007;19:545–556. doi: 10.1093/intimm/dxm020. [DOI] [PubMed] [Google Scholar]
- 185.Ma Y, Pannicke U, Schwarz K, Lieber MR. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108:781–794. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 186.Jung S, Rajewsky K, Radbruch A. Shutdown of class switch recombination by deletion of a switch region control element. Science. 1993;259:984–987. doi: 10.1126/science.8438159. [DOI] [PubMed] [Google Scholar]
- 187.Bottaro A, Lansford R, Xu L, Zhang J, Rothman P, Alt F. I region transcription (per se) promotes basal IgE class switch recombination but additional factors regulate the efficiency of the process. EMBO J. 1994;13:665–674. doi: 10.1002/j.1460-2075.1994.tb06305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Lorenz M, Jung S, Radbruch A. Switch transcripts in immunoglobulin class switching. Science. 1995;267:1825–1828. doi: 10.1126/science.7892607. [DOI] [PubMed] [Google Scholar]
- 189.Hein K, Lorenz MG, Siebenkotten G, Petry K, Christine R, Radbruch A. Processing of switch transcripts is required for targeting of antibody class switch recombination. J. Exp. Med. 1998;188:2369–2374. doi: 10.1084/jem.188.12.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Huang FT, Yu K, Balter BB, Selsing E, Oruc Z, et al. Sequence-dependence of chromosomal R-loops at the immunoglobulin heavy chain Sμ class switch region. Mol. Cell. Biol. 2007;27:5921–5932. doi: 10.1128/MCB.00702-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Yu K, Chedin F, Hsieh CL, Wilson TE, Lieber MR. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat. Immunol. 2003;4:442–451. doi: 10.1038/ni919. [DOI] [PubMed] [Google Scholar]
- 192.Zarrin AA, Alt FW, Chaudhuri J, Stokes N, Kaushal D, et al. An evolutionarily conserved target motif for immunoglobulin class-switch recombination. Nat. Immunol. 2004;5:1275–1281. doi: 10.1038/ni1137. [DOI] [PubMed] [Google Scholar]
- 193.Zarrin AA, Tian M, Wang J, Borjeson T, Alt FW. Influence of switch region length on immunoglobulin class switch recombination. Proc. Natl. Acad. Sci. USA. 2005;102:2466–2470. doi: 10.1073/pnas.0409847102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Ronai D, Iglesias-Ussel MD, Fan M, Li Z, Martin A, Scharff MD. Detection of chromatin-associated single-stranded DNA in regions targeted for somatic hypermutation. J. Exp. Med. 2007;204:181–190. doi: 10.1084/jem.20062032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Nambu Y, Sugai M, Gonda H, Lee CG, Katakai T, et al. Transcription-coupled events associating with immunoglobulin switch region chromatin. Science. 2003;302:2137–2140. doi: 10.1126/science.1092481. [DOI] [PubMed] [Google Scholar]
- 196.Workman JL. Nucleosome displacement in transcription. Genes Dev. 2006;20:2009–2017. doi: 10.1101/gad.1435706. [DOI] [PubMed] [Google Scholar]
- 197.Woo CJ, Martin A, Scharff MD. Induction of somatic hypermutation is associated with modifications in immunoglobulin variable region chromatin. Immunity. 2003;19:479–489. doi: 10.1016/s1074-7613(03)00261-9. [DOI] [PubMed] [Google Scholar]
- 198.Wang L, Whang N, Wuerffel R, Kenter AL. AID-dependent histone acetylation is detected in immunoglobulin S regions. J. Exp. Med. 2006;203:215–226. doi: 10.1084/jem.20051774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Sakai E, Bottaro A, Alt FW. The Ig heavy chain intronic enhancer core region is necessary and sufficient to promote efficient class switch recombination. Int. Immunol. 1999;11:1709–1713. doi: 10.1093/intimm/11.10.1709. [DOI] [PubMed] [Google Scholar]
- 200.Manis JP, van der Stoep N, Tian M, Ferrini R, Davidson L, et al. Class switching in B cells lacking 3′ immunoglobulin heavy chain enhancers. J. Exp. Med. 1998;188:1421–1431. doi: 10.1084/jem.188.8.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Pinaud E, Khamlichi AA, Le Morvan C, Drouet M, Nalesso V, et al. Localization of the 3′ IgH locus elements that effect long-distance regulation of class switch recombination. Immunity. 2001;15:187–199. doi: 10.1016/s1074-7613(01)00181-9. [DOI] [PubMed] [Google Scholar]
- 202.Pan Q, Petit-Frere C, Stavnezer J, Hammarstrom L. Regulation of the promoter for human immunoglobulin γ3 germ-line transcription and its interaction with the 3′ α enhancer. Eur. J. Immunol. 2000;30:1019–1029. doi: 10.1002/(SICI)1521-4141(200004)30:4<1019::AID-IMMU1019>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 203.Cunningham K, Ackerly H, Alt F, Dunnick W. Potential regulatory elements for germline transcription in or near murine Sγ1. Int. Immunol. 1998;10:527–536. doi: 10.1093/intimm/10.4.527. [DOI] [PubMed] [Google Scholar]
- 204.Adams K, Ackerly H, Cunningham K, Dunnick W. A DNase I hypersensitive site near the murine γ1 switch region contributes to insertion site independence of trans-genes and modulates the amount of transcripts induced by CD40 ligation. Int. Immunol. 2000;12:1705–1713. doi: 10.1093/intimm/12.12.1705. [DOI] [PubMed] [Google Scholar]
- 205.Oestreich KJ, Cobb RM, Pierce S, Chen J, Ferrier P, Oltz EM. Regulation of TCRβ gene assembly by a promoter/enhancer holocomplex. Immunity. 2006;24:381–391. doi: 10.1016/j.immuni.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 206.Wuerffel R, Wang L, Grigera F, Manis J, Selsing E, et al. S/S synapsis during class switch recombination is promoted by distantly located transcriptional elements and activation-induced deaminase. Immunity. 2007;27:711–722. doi: 10.1016/j.immuni.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Shanmugam A, Shi M-J, Yauch L, Stavnezer J, Kenter AL. Evidence for class specific factors in immunoglobulin isotype switching. J. Exp. Med. 2000;191:1365–1380. doi: 10.1084/jem.191.8.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Kenter AL, Wuerffel R, Dominguez C, Shanmugam A, Zhang H. Mapping of a functional recombination motif that defines isotype specificity for μ->γ3 switch recombination implicates NF-κB p50 as the isotype-specific switching factor. J. Exp. Med. 2004;199:617–627. doi: 10.1084/jem.20031935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Bradley SP, Kaminski DA, Peters AH, Jenuwein T, Stavnezer J. The histone methyltransferase Suv39h1 increases class switch recombination specifically to IgA. J. Immunol. 2006;177:1179–1188. doi: 10.4049/jimmunol.177.2.1179. [DOI] [PubMed] [Google Scholar]
- 210.Dent AL, Shaffer AL, Yu X, Allman D, Staudt LM. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science. 1997;276:589–592. doi: 10.1126/science.276.5312.589. [DOI] [PubMed] [Google Scholar]
- 211.Ye BH, Cattoretti G, Shen Q, Zhang J, Hawe N, et al. The BCL-6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nat. Genet. 1997;16:161–170. doi: 10.1038/ng0697-161. [DOI] [PubMed] [Google Scholar]
- 212.Ranuncolo SM, Polo JM, Dierov J, Singer M, Kuo T, et al. Bcl-6 mediates the germinal center B cell phenotype and lymphomagenesis through transcriptional repression of the DNA-damage sensor ATR. Nat. Immunol. 2007;8:705–714. doi: 10.1038/ni1478. [DOI] [PubMed] [Google Scholar]
- 213.Phan RT, Dalla-Favera R. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature. 2004;432:635–639. doi: 10.1038/nature03147. [DOI] [PubMed] [Google Scholar]
- 214.Pape KA, Kouskoff V, Nemazee D, Tang HL, Cyster JG, et al. Visualization of the genesis and fate of isotype-switched B cells during a primary immune response. J. Exp. Med. 2003;197:1677–1687. doi: 10.1084/jem.20012065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Cunningham AF, Gaspal F, Serre K, Mohr E, Henderson IR, et al. Salmonella induces a switched antibody response without germinal centers that impedes the extra-cellular spread of infection. J. Immunol. 2007;178:6200–6207. doi: 10.4049/jimmunol.178.10.6200. [DOI] [PubMed] [Google Scholar]
- 216.Cattoretti G, Buttner M, Shaknovich R, Kremmer E, Alobeid B, Niedobitek G. Nuclear and cytoplasmic AID in extrafollicular and germinal center B cells. Blood. 2006;107:3967–3975. doi: 10.1182/blood-2005-10-4170. [DOI] [PubMed] [Google Scholar]
- 217.Kolar GR, Mehta D, Pelayo R, Capra JD. A novel human B cell subpopulation representing the initial germinal center population to express AID. Blood. 2007;109:2545–2552. doi: 10.1182/blood-2006-07-037150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Castigli E, Wilson SA, Scott S, Dedeoglu F, Xu S, et al. TACI and BAFF-R mediate isotype switching in B cells. J. Exp. Med. 2005;201:35–39. doi: 10.1084/jem.20032000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Bergqvist P, Gardby E, Stensson A, Bemark M, Lycke NY. Gut IgA class switch recombination in the absence of CD40 does not occur in the lamina propria and is independent of germinal centers. J. Immunol. 2006;177:7772–7783. doi: 10.4049/jimmunol.177.11.7772. [DOI] [PubMed] [Google Scholar]
- 220.Xu W, He B, Chiu A, Chadburn A, Shan M, et al. Epithelial cells trigger frontline immunoglobulin class switching through a pathway regulated by the inhibitor SLPI. Nat. Immunol. 2007;8:294–303. doi: 10.1038/ni1434. [DOI] [PubMed] [Google Scholar]
- 221.He B, Xu W, Santini PA, Polydorides AD, Chiu A, et al. Intestinal bacteria trigger T cell-independent immunoglobulin A(2) class switching by inducing epithelial-cell secretion of the cytokine APRIL. Immunity. 2007;26:812–826. doi: 10.1016/j.immuni.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 222.Han JH, Akira S, Calame K, Beutler B, Selsing E, Imanishi-Kari T. Class switch recombination and somatic hypermutation in early mouse B cells are mediated by B cell and Toll-like receptors. Immunity. 2007;27:64–75. doi: 10.1016/j.immuni.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Ueda Y, Liao D, Yang K, Patel A, Kelsoe G. T-independent activation-induced cytidine deaminase expression, class-switch recombination, and antibody production by immature/transitional 1 B cells. J. Immunol. 2007;178:3593–3601. doi: 10.4049/jimmunol.178.6.3593. [DOI] [PMC free article] [PubMed] [Google Scholar]