Abstract
Background
Environmental conditions, such as water supply, temperature and salinity, strongly affect plant growth and development. Extremes of these conditions (abiotic stresses) adversely affect many different mechanisms associated with plant responses and adaptation to stress: photosynthetic mechanisms, e.g. stomatal control of CO2 diffusion, photosystem II repair, ribulose bisphosphate carboxylase/oxygenase (Rubisco) activity and scavenging of reactive oxygen species (ROS), are susceptible to damage, and photosynthetic efficiency can be greatly decreased. Responses and adaptations require differential gene expression, which is regulated by specific transcription factors (TFs).
Scope
The role and regulation of several TFs involved in abiotic stress response pathways are considered, with emphasis on new findings regarding expression of genes related to both stomatal and non-stomatal limitations to CO2 photosynthetic assimilation.
Conclusions
Many TFs, belonging to different families (e.g. MYB, bZIP and DREB), have been related to abiotic stress responses; however, only a few are known to regulate the expression of photosynthesis-related genes in response to stress. Several TFs belonging to the MYB family play an important role in both stomatal and non-stomatal responses by regulation of stomatal numbers and sizes, and metabolic components, respectively. To obtain more insight into this area of potentially large agronomic impact, it is essential to identify and functionally characterize new TFs that mediate the stress responses regulating the expression of genes associated with photosynthesis and related metabolism.
Key words: Transcription factors, photosynthesis, stomata, abiotic stress, abiotic stress signalling, cold, drought, salt, ABA, MYB, AP2/EREBP
INTRODUCTION
General effects of abiotic stress on plant growth and function
Extremes of temperature, salinity and water supply, i.e. conditions outside the normal range in which plants function efficiently, are probably the most important environmental conditions (‘abiotic stresses’) that limit growth and productivity. These effects are caused by altered morphology and physiology, resulting from changes to processes, such as inhibition of cell division, and effects on metabolism, including photosynthesis. Particular environmental conditions may affect specific mechanisms. For example, low temperature severely hampers reproductive development: exposure of rice plants to chilling temperature at anthesis (floral opening stage) leads to male sterility (Imin et al., 2006; Mamun et al., 2006). More extreme cold stress mainly results in disruption of membrane integrity and solute leakage, leading to severe cellular dehydration and osmotic imbalance (Thomashow, 1999). Drought and salinity are primarily manifested as increased cellular osmotic concentrations (‘osmotic stress’), resulting in disruption of homeostasis and ion distribution in cells (Zhu, 2002).
Increased production of reactive oxygen species (ROS) is a common consequence of exposure to drought, salinity and low temperature. This results from excitation of the light reactions of photosynthesis [photosystems I and II (PSI and PSII)] inducing water splitting and electron transport: when the latter exceeds the requirements of normal metabolism, oxygen is reduced. Such overexcitation of the system is characteristic of stress conditions. The light reactions occur in the thylakoid membranes which are susceptible to damage (Moller et al., 2007; Takahashi and Murata, 2008). ROS cause peroxidation and de-esterification of membrane lipids, and also lead to protein denaturation (Bowler et al., 1992) as well as other forms of photo-oxidative damage.
The physiological and biochemical changes in plants under particular stress conditions are related to altered gene expression. Onset of a stress triggers some (mostly unknown) initial sensors, which then activate cytoplasmic Ca2+ and protein signalling pathways, leading to stress-responsive gene expression and physiological changes (Bressan et al., 1998; Xiong et al., 2002). Also, accumulation of abscisic acid (ABA) plays an important role in abiotic stress signalling and transduction pathways, mediating many responses (Wasilewska et al., 2008). Regarding gene and protein expression modulated by stress, it is well documented that abiotic stresses in general, through regulation of both gene expression and protein turnover, alter the abundance of many transcripts and proteins (Seki et al., 2002; Wong et al., 2006; Yan et al., 2006; Jiang et al., 2007), indicating that transcriptional and post-transcriptional regulation play an essential role in the adaptation of cellular functions to the environmental changes. The transcript level (relative abundance) of some genes encoding proteins with antioxidant functions, such as glutathione reductase (GR) and ascorbate peroxidase (APX), is higher during recovery from water stress and may play a role protecting the cellular machinery against photo-oxidation by ROS (Ratnayaka et al., 2003).
Photosynthesis and related mechanisms affected by abiotic stress
Photosynthesis plays a central role as an energy source for plant metabolism, and its efficiency may be drastically reduced due to abiotic stresses. During evolution, plants have developed many ‘strategies’ to acclimate to adverse environments. The main goal, it is supposed, is to maintain the photosynthetic efficiency at as high a level as possible, avoiding the energy imbalance resulting from the abiotic stress and the consequent photo-oxidative damage. Abiotic stress interferes with photosynthesis at different points, e.g. CO2 diffusion, PSII efficiency, electron transport, ROS formation, ribulose-1,5-bisphosphate (RuBP) content (dependent on ATP and NADPH supply), ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) activity and photorespiration.
Drought, salinity and cold are well known to induce stomatal closure (Wilkinson et al., 2001; Zhu, 2002; Chaves et al., 2003), slowing CO2 assimilation and consequently reducing the photosynthetic rate (Fig. 1). In addition, it has been shown that reduced mesophyll conductance is an equally important cause of lower CO2 diffusion under water (Flexas et al., 2002; Warren et al., 2004) and salt (Centritto et al., 2003) stress. Moreover, plasma membrane aquaporins have been found to be closely related to mesophyll conductance (Uehlein et al., 2003; Hanba et al., 2004). Therefore, the expression and/or regulation of aquaporins can mediate the environmental effects on CO2 diffusion. The mesophyll conductance and relationship with CO2 diffusion, in terms of plant response to stress, is, however, not completely understood. Regarding the control of stomatal aperture, this process is mediated by ABA and possibly by other signals generated in response to abiotic stress. The regulation of gene expression related to guard cell movement is known to involve both ABA-dependent and ABA-independent signalling (Cominelli et al., 2005; Liang et al., 2005).
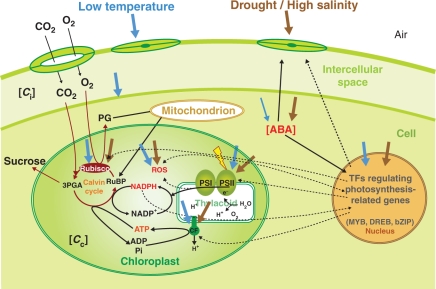
Fig. 1.
A simplified scheme of the photosynthetic-related mechanisms that can be affected by cold, drought and salinity. Blue and brown arrows correspond to cold and drought/salinity signals, respectively. Adverse environmental conditions that cause osmotic stress induce stomatal closure, which determines the rate of CO2 assimilation. Abscisic acid (ABA) is involved in most drought and high salt responses and can mediate some low temperature effects. Arrow thickness represents signal contribution. Dashed lines represent possible interactions. PSI, photosystem I; PSII, photosystem II; ROS, reactive oxygen species; 3PGA, 3-phosphoglycerate; RuBP, ribulose-1,5-biphosphate; PG, phosphoglycolate; CF, coupling factor; Ci, internal CO2; Cc, CO2 in the chloroplast.
The causes of decreased photosynthetic rate under different environmental stresses (and at different intensities) are still not established, with substantial controversy about the main physiological targets responsible for photosynthetic impairment. Stomatal closure is considered the primary response to cold, drought and salinity (Flexas and Medrano, 2002). However, low temperatures, salinity and drought alter metabolism in many ways which affect the photosynthetic rate. For example, low temperature alters ATP/ADP and ATP/reductant ratios (Savitch et al., 1997) probably as a consequence of altered sucrose metabolism (Savitch et al., 2000). However, cold stress (5°C, mild stress) in Arabidopsis was not associated with a limitation in ATP, but with an increase in the ATP/NADPH ratio due to altered reductant metabolism (Savitch et al., 2001). Drought can lead to a limitation in the supply of ATP in the chloroplast as a consequence of decreased synthetic capacity (Tezara et al., 1999) caused by loss of ATP synthase (see Lawlor and Tezara, 2009). Both temperature and drought may, for different reasons, decrease the synthesis of RuBP, which depends on ATP and NADPH concentrations and activity of enzymes of the Calvin cycle (Fig. 1), and thus reduce photosynthetic CO2 assimilation rates. As Tezara et al. (1999) showed, and as later emphasized by Flexas and Medrano (2002), stomatal closure is the earliest response to drought and the dominant limitation to photosynthesis at mild to moderate stress, but as Tezara et al. (1999) and Lawlor (2002) concluded there is a progressive reduction in the biochemical processes that becomes dominant under severe stress, leading to reduced photosynthetic CO2 assimilation.
When plants are exposed to environmental stresses and the availability of CO2 within the leaf (Ci) is restricted and/or the synthesis of ATP is impaired, the activity of the Calvin cycle is reduced, but PSII remains active. In these conditions, the concentration of the final electron acceptor NADP+ is generally very low (Fig. 1). This leads to an excess of excitation energy in the photosystems. High energy states may be dissipated by either non-photochemical quenching (e.g. xanthophyll cycle) or alternative processes, such as photorespiratory metabolism (Niyogi, 2000). If not dissipated, electrons accumulate in the electron transport chain and are transferred to oxygen (Mehler reaction): such processes generate ROS, e.g. superoxide. Because of their high reactive potential, ROS react with and damage many cellular components (e.g. proteins, DNA and lipids), constituting oxidative stress, so they must then be detoxified by scavenging mechanisms such as superoxide dismutase (SOD) and the glutathione–ascorbate cycle (Noctor and Foyer, 1998). ROS also inactivate the photochemical reaction centre of PSII, causing photoinhibition. Recently, it has been proposed that most environmental stresses inactivate PSII by inhibiting the mechanisms for repairing photodamage rather than by directly attacking it (Murata et al., 2007, and references therein). ROS generated by different stresses inhibit PSII repair by suppressing the transcription and translation of psbA genes encoding D1, one of the proteins of the PSII complex (Nishiyama et al., 2001, 2004; Allakhverdiev et al., 2002).
The stroma redox state is known to be involved in the regulation of gene expression (Pfannschmidt et al., 1999). The redox-reactive molecules (e.g. ROS, thioredoxin and reduced glutathione) generated by environmental adverse conditions act as signals to activate/inactivate the expression of chloroplast and nuclear genes. Interestingly, all genes reported as being redox regulated are related to photosynthesis (Pfannschmidt, 2003). Since the redox state can regulate kinase activity, which in turn controls the activity of plastid transcription factors (TFs; Baginsky et al., 1999), this might be the link between redox state and regulation of gene expression. Although no redox-sensitive TF has yet been identified in plants, in animals a TF has been found that can be activated by redox-dependent stimuli (Balogun et al., 2003).
In addition to the effects on CO2 diffusion, ATP synthesis and reductant status, abiotic stresses can also negatively affect the Calvin cycle by reducing the content and activity of photosynthetic carbon reduction cycle enzymes, including the key enzyme, Rubisco, thus limiting CO2 assimilation. It was shown that both low and high temperature slow down the operation of Rubisco activase, a molecular chaperone that activates Rubisco through an ATP-dependent reaction (Kingston-Smith et al., 1997). Very severe drought conditions also result in limited photosynthesis; evidence that this is related to decreased RuBP supply and not to lack of CO2 (Tezara et al., 1999; Lawlor, 2002; Lawlor and Cornic, 2002) conflicts with the view (Flexas et al., 2006) that it is due to the lower CO2 availability in the chloroplast and consequently a decline in Rubisco activity.
Given that some of the short-term and most of the long-term effects caused by abiotic stresses to photosynthesis involve regulation of gene expression, it is clear that TFs play an important role in stress acclimation, modulating the expression of stress-responsive genes. TFs are themselves regulated by abiotic stress signals, such as ABA, the redox state and the ATP/NADPH content. It is known that most plants share orthologous genes involved in abiotic stress responses, although different plants show different magnitudes of stress tolerance. These phenotypes are mostly explained by differences in stress-responsive gene expression.
TRANSCRIPTION FACTORS INVOLVED IN ABIOTIC STRESS RESPONSES
Responses to abiotic stress require the production of important metabolic proteins such as those involved in synthesis of osmoprotectants and of regulatory proteins operating in the signal transduction pathways, such as kinases or TFs. Given that most of these responses imply control of gene expression, TFs play a critical role in the abiotic stress response (Chaves and Oliveira, 2004). TFs are proteins with a DNA domain that binds to the cis-acting elements present in the promoter of a target gene. They induce (activators) or repress (repressors) the activity of the RNA polymerase, thus regulating gene expression. TFs can be grouped into families according to their DNA-binding domain (Riechmann et al., 2000). A group of genes controlled by a certain type of TF is known as a regulon. In the plant response to abiotic stresses, at least four different regulons can be identified (Fig. 2): (1) the CBF/DREB regulon; (2) the NAC (NAM, ATAF and CUC) and ZF-HD (zinc-finger homeodomain) regulon; (3) the AREB/ABF (ABA-responsive element-binding protein/ABA-binding factor) regulon; and (4) the MYC (myelocytomatosis oncogene)/MYB (myeloblastosis oncogene) regulon. The first two regulons are ABA independent, and the last two are ABA dependent. It is explained below how these regulons are controlled and how TFs may be involved in the regulation of photosynthesis as a response to abiotic stress.
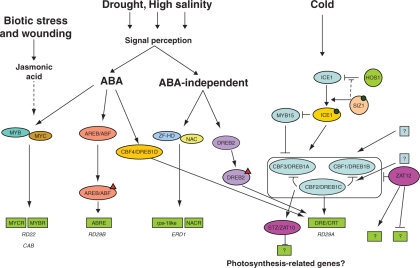
Fig. 2.
Transcriptional network of abiotic stress responses. Transcription factors are shown in ovals. Transcription factor-modifying enzymes are shown in circles. The small triangles correspond to post-translational modifications. Blue squares with question marks represent putative MYC ICE1-like transcription factors that may activate CBF1/DREB1B and CBF2/DREB1C. The green boxes represent the cis-elements present in stress-responsive genes. The green boxes with question marks represent putative cis-elements on the promoter of stress-responsive genes. The black dot corresponds to the sumoylation modification by SIZ1 of the ICE1 transcription factor. The dashed black line from SIZ1 to HOS1 represents competition for binding places on the ICE1 transcription factor. SIZ1 blocks the access of HOS1 to the ubiquitination sites on the ICE1. CBF4/DREB1D is a DRE cis-element binding factor that is ABA dependent.
The CBF/DREB regulon
This regulon is mainly involved in cold stress response and is probably the one that has attracted most attention. It is conserved throughout the plant kingdom, including in plants that do not cold-acclimate (e.g. tomato and rice) (Dubouzet et al., 2003). In 1994, Yamaguchi-Shinozaki and Shinozaki identified a novel cis-acting element that, in addition to the ABA-responsive element (ABRE), is also present in the promoter of the RESPONSIVE TO DEHYDRATION 29A (RD29A), a gene induced by drought, high salinity and cold. This new element was named C-repeat/dehydration-responsive element (CRT/DRE) and characterized as ABA independent. The core motif of this cis-acting element is CCGAC and the TFs that bind to it were named CRT-binding factors or DRE-binding proteins 1 (CBF/DREB1) (Gilmour et al., 1998; Liu et al., 1998). CBF/DREB1 gene expression is quickly and transiently induced by cold stress, and in turn CBF/DREB1 TFs activate the expression of several other genes (e.g. encoding proteins involved in production of osmoprotectants and antioxidants). Interestingly, it was shown that the low temperature induction of the CBF1–CBF3 genes is gated by the circadian clock, suggesting that the regulation of these genes has aspects in common with the regulation of Arabidopsis CAB genes (Fowler et al., 2005). Concerning the DREB2 genes, they are constitutively expressed (under normal and stress conditions), although their target genes (e.g. RD29A, RD29B, RD17 and LEA14) are only induced upon dehydration. This indicates that DREB2 factors are activated through post-translational modifications in order to regulate downstream genes (Sakuma et al., 2006).
The over-expression of CBF/DREB1 genes in Arabidopsis resulted in plants with improved survival rates when exposed to salt, drought and low temperatures (Jaglo-Ottosen et al., 1998; Kasuga et al., 1999). This improved tolerance was correlated with both altered relative abundance of transcripts encoding proteins associated with stress adaptation and increased sugar contents (Gilmour et al., 2004). When CBF/DREB1 genes from Arabidopsis were over-expressed in other plants, the result was similar to that in Arabidopsis (Hsieh et al., 2002a; Pellegrineschi et al., 2004), revealing a conserved signalling and response mechanism even between dicots and monocots. Various studies have demonstrated that improved stress tolerance by over-expression of CBF/DREB1 genes is associated with sustained photochemical efficiency and photosynthetic capacity as compared with wild-type plants (Hsieh et al., 2002b; Savitch et al., 2005; Oh et al., 2007). These plants normally show a dwarf phenotype that can be reverted through the exogenous application of gibberellins (GAs). However, the microarray expression analysis of these plants did not reveal any gene encoding GA enzymes affected by the over-expression of CBF/DREB1 (Fowler and Thomashow, 2002). Instead, most genes related to carbohydrate metabolism and photosynthesis were repressed and thus contributed to reduced growth. Caution must be exercised when evaluating abiotic stress tolerance in transgenic plants showing a dwarf phenotype, as the improvement may be mainly due to reduced size rather than metabolic changes leading to intrinsic tolerance.
A TF that acts downstream of CBF3/DREB1A (Fig. 2), STZ/ZAT10, functions as a repressor through an essential DLN/EAR-like repression motif present in its C-terminal region (Nakashima and Yamaguchi-Shinozaki, 2006). In Arabidopsis, it was observed that the over-expression of STZ represses several genes involved in photosynthesis and related metabolism. This means that STZ factor may be involved in growth retardation through repression of photosynthesis and carbohydrate metabolism genes observed in both the wild-type plants under abiotic stress and plants over-expressing CBF/DREB1 genes (e.g. CBF3/DREB1A). It would be interesting to analyse all the promoters of the genes related to photosynthesis and carbohydrate metabolism and search for cis-elements where STZ may bind (Sakamoto et al., 2004). To overcome growth retardation, CBF/DREB1 genes have been expressed in transgenic plants under the control of a stress-inducible promoter, RD29A (Kasuga et al., 2004). These plants have also shown enhanced abiotic stress tolerance without totally compromising the yield (Pino et al., 2007). However, it seems that the use of the Arabidopsis RD29A promoter is more efficient in driving the expression of CBF/DREB1 genes in dicots rather than in monocots, or at least in rice (Ito et al., 2006; T. Lourenço et al., unpubl. res.).
The control of the CBF/DREB regulon is not as simple as one might expect (Fig. 2). The Arabidopsis mutant cbf2/dreb1c has revealed that CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A gene expression (Novillo et al., 2004). However, CBF2/DREB1C shares several target genes with ZAT12 (Vogel et al., 2005), a TF that can be in a parallel regulon to the CBFs/DREBs. Plants over-expressing ZAT12 had a small but consistent increase in freezing tolerance, and the induction of the CBF/DREB genes in response to cold is reduced. This indicates that ZAT12 also plays a role in the negative regulatory circuit that leads to a decline in expression of CBF/DREB. The CBF/DREB1 regulon is controlled upstream by the INDUCER OF CBF EXPRESSION 1 (ICE1) protein (Chinnusamy et al., 2003). The ICE1 protein is a MYC-type bHLH (basic helix–loop–helix) TF that regulates the expression of CBF3/DREB1A. The ICE1 protein is present at normal growth temperatures but its activation requires cold-induced post-translational modification(s) (e.g. phosphorylation). In addition, the ICE1 protein is negatively regulated by the HIGHER EXPRESSION OF OSMOTICALLY RESPONSIVE GENES 1 (HOS1) protein. HOS1 is a RING E3 ligase that targets the ICE1 protein for ubiquitination and subsequent degradation (Dong et al., 2006). Under normal conditions, HOS1 is a cytoplasmic protein, but, upon low temperature, HOS1 is translocated to the nucleus where it targets ICE1 for degradation. Recently, it was found that ICE1 ubiquitination can be blocked by SIZ1-dependent sumoylation (Miura et al., 2007), a process that conjugates SUMO (for small ubiquitin-related modifier) to a protein substrate. SIZ1 is a SUMO E3 ligase that mediates ICE1 sumoylation (binds SUMO to a target protein). This modification activates and/or stabilizes ICE1 protein, thus facilitating its activity controlling the expression of the CBF3/DREB1A gene. The mechanism by which ICE1 protein is activated by sumoylation through SIZ1 is still not fully understood. Another TF with a regulatory function in this process is MYB15 (Agarwal et al., 2006). MYB15 is a negative regulator of the CBF/DREB1 genes, possibly through interaction with their promoter regions. This TF seems to be negatively regulated by a sumoylated ICE1 form, as a modification affecting the sumoylation site of ICE1 leads to an increased MYB15 transcript level and reduced CBF3/DREB1A expression (Fig. 2). The cold response through the CBF/DREB1 regulon is thus a strictly regulated mechanism that may have evolved to avoid unwanted negative effects in plants. In fact, uncontrolled expression of CBF/DREB1 in certain environments may lead to dwarf phenotypes and reduced yields.
The NAC and ZF-HD regulon
An ABA-independent pathway was unveiled when it was observed that EARLY RESPONSIVE TO DEHYDRATION STRESS 1 (ERD1) gene transcripts accumulated before any increase of ABA in response to dehydration and high salinity, suggesting the presence of an ABA-independent pathway (Nakashima et al., 1997). Promoter analysis of ERD1 revealed TFs belonging to the NAC family and zinc finger homeodomain (ZF-HD) as essential to the activation of the ERD1 gene (Tran et al., 2007). However, over-expression of NAC genes in Arabidopsis enhanced drought tolerance without activation of the ERD1 gene, suggesting that other interacting factors may be necessary to control the expression of ERD1 under stress conditions (Tran et al., 2004).
Recently, the STRESS-RESPONSIVE NAC1 (SNAC1) gene was isolated from an upland rice variety and over-expressed in a lowland rice (‘Nipponbare’; Hu et al., 2006). SNAC1 encodes a NAM, ATAF and CUC (NAC) TF with transactivation activity and is induced by drought, predominantly in guard cells (Hu et al., 2006). When compared with the wild type, rice plants over-expressing SNAC1 showed drought tolerance at anthesis and increased drought and salt tolerance at the vegetative stage. The plants over-expressing SNAC1 did not show the common, unwanted, dwarf phenotype of those over-expressing CBF/DREB1 (Ito et al., 2006), revealing a different stress response mechanism. The increased drought tolerance may be in part due to the reduced transpiration rate (increased stomatal closure) and to an increased ABA sensitivity. Interestingly, the photosynthesis rate was not significantly affected by the over-expression of the SNAC1 gene. It is claimed that usually rice leaves may function with more open stomata than necessary to have a normal photosynthetic rate. The strong induction of SNAC1 gene expression by drought in guard cells suggests an effect in stomatal closure (Hu et al., 2006). Two R2R3-MYB TFs (AtMYB60 and AtMYB61) are known to be directly involved in stomatal dynamics in Arabidopsis (Cominelli et al., 2005; Liang et al., 2005). In addition, the over-expression of SNAC1 upregulates a rice R2R3-MYB gene (UGS5) with a NAC recognition site in its promoter region (Hu et al., 2006). However, the relationship between SNAC1 and the TFs implicated in stomatal closure is not known. This connection needs to be investigated further to understand the regulatory mechanisms underlying stomatal movement under drought stress. SNAC1 also induced the expression of genes encoding proteins related to both osmotic adjustment (such as a sorbitol transporter and exoglucanase) and stability of cell membranes, which can be related to the stress response (Hu et al., 2006).
The AREB/ABF regulon
The over-expression of key enzymes in ABA biosynthesis (e.g. 9-cis-epoxycarotenoid dioxygenase; NCED) or mutation in ABA-degrading enzymes (e.g. cytochrome P450 CYP707A family member) resulted in transgenic plants with enhanced drought tolerance (Shinozaki and Yamaguchi-Shinozaki, 2007). The ABRE motif is a cis-acting element present in the ABA-responsive genes. The ABFs or AREBs are bZIP (basic leucine zipper) TFs that bind to the ABRE motif and activate ABA-dependent gene expression (Choi et al., 2000). Some of these TFs, such as AREB1 and AREB2, require a post-translational modification for their maximum activation (Uno et al., 2000). This post-translational modification is probably an ABA-dependent phosphorylation.
A family of protein kinases, the Snf1-related kinases family, has been implicated in the ABA signal transduction pathway. Members of this family (SnRK2) play an important role in controlling stomatal closure and are activated by drought, salinity and ABA (Mustilli et al., 2002; Yoshida et al., 2002). The over-expression of SRK2C caused hypersensivity to ABA and improved drought tolerance with reduced transpiration rate (Umezawa et al., 2004). These data suggest that SnRK2-type protein kinases activate TFs influencing osmotic stress-responsive genes. Recently, Baena-Gonzalez and co-workers (2007) have implicated other members (KIN10 and KIN11 from the SnRK1 group) of the Snf1-related protein kinase family with a pivotal role in the sensing of sugar and energy depletion due to photosynthesis inhibition in response to diverse stresses and conditions, such as hypoxia, herbicide treatment and darkness. Promoter analysis of DARK INDUCED 6 (DIN6), a KIN10-activated gene, revealed that the G-box (CACGTG) is essential to the DIN6 activation by KIN10. The authors screened for bZIP TFs that bind to the G-box cis-element in Arabidopsis and found that the co-expression of the KIN10 and the G-BOX BINDING FACTOR 5 (GBF5) had a synergistic effect on DIN6 expression. These results indicate that this family of Snf1-related protein kinases may play an important role in controlling the activation of stress-related TFs.
The MYC/MYB regulon
Expression of the drought-inducible gene RESPONSIVE TO DEHYDRATION 22 (RD22) from Arabidopsis was found to be induced by ABA (Abe et al., 2003). The promoter region of RD22 contains MYC (CANNTG) and MYB (C/TAACNA/G) cis-element recognition sites. MYC and MYB TFs only accumulate after accumulation of ABA. In Arabidopsis, it was found that for activation of RD22 gene expression, both AtMYC and AtMYB have to work co-operatively. Over-expression of these TFs resulted in enhanced sensitivity to ABA and drought tolerance. Microarray studies in transgenic plants over-expressing these TFs revealed that not only were ABA-related stress genes differentially regulated, but also jasmonic acid-related genes (Fig. 2), thus indicating a cross-talk pathway between abiotic and biotic stress responses (Abe et al., 2003).
Transcription factors with no relationship to known regulons
Although many of the TFs identified are involved in the described regulons, some TFs are involved in other response mechanisms. In recent years, two new genes, HIGHER EXPRESSION OF OSMOTICALLY RESPONSIVE GENES 9 and 10 (HOS9 and HOS10), have been associated with cold stress response (Zhu et al., 2004, 2005). HOS9 is a homeodomain TF with similarity to the Arabidopsis proteins WUSCHEL (WUS) and PRESSED FLOWER (PRS), and HOS10 is an R2R3-type MYB protein (Van Buskirk and Thomashow, 2006). Both mutants hos9 and hos10 show freezing hypersensitivity, but, interestingly, also have enhanced expression of the RD29A gene and other cold-responsive genes without changes in the CBF/DREB1 regulon. It might be expected that HOS9 and HOS10 act as negative regulators of cold stress-responsive genes, but the increased Arabidopsis sensitivity to cold rules out this hypothesis. The absence of the respective transcripts in the mutants probably resulted in expression of cold-responsive genes in order to cope with the increased cold sensitivity. The hos10 mutant has reduced NCED gene expression compared with the wild type and consequently plants do not accumulate ABA, revealing a critical role for this TF regarding different abiotic stresses. Nevertheless, additional studies have to be performed in order to clarify the function of these TFs in the abiotic stress response.
HARDY (HRD) is an AP2-EREBP-type TF isolated from Arabidopsis. The HRD gene is expressed mainly in the inflorescence tissue, most probably to protect this tissue from desiccation in a very important and sensitive stage of the plant life cycle. Rice plants over-expressing HRD showed drought tolerance and improved water use efficiency (WUE; Karaba et al., 2007). Interestingly, when grown under normal greenhouse conditions, they do not show reduced growth, seed yield or germination rate; instead they have a larger leaf canopy with more tillers. The transgenic plants also have more root biomass under drought stress; this is considered a drought adaptation to absorb the scarce water in the soil. Whether higher root biomass is associated with a faster water uptake or with a larger volume exploited is not known. These HRD lines showed a reduced transpiration rate (due to lower stomatal conductance) and a higher than wild-type net carbon assimilation rate under drought and well-irrigated conditions, corresponding to an increased WUE. No difference was observed for the maximum quantum efficiency of PSII (Fv/Fm) between wild type and transgenic plants; however, the efficiency of the PSII reaction centres (Fv′/Fm′) was higher in the HRD-over-expressing plants than in the wild-type plants. This agrees with the improved photosynthetic capacity observed in the transgenic plants. The increased number of bundle sheath cells in the transgenic plants can support the improved photosynthetic assimilation.
HRD belongs to the AP2-EREBP IIIb group, while the related CBF/DREB genes belong to the AP2-EREBP IIIc group (Nakano et al., 2006). Microarray analysis revealed that HRD over-expression induces genes repressed by drought stress, suggesting a protective influence on essential processes, such as protein biosynthesis and carbohydrate metabolism. Despite some similarities, these transgenic plants induce different clusters of genes when compared with the CBF/DREB-over-expressing plants, which may account for the differences observed (absence of stunted growth) and the unique responses to stress of these plants.
ENVIRONMENTAL CUES REGULATE PHOTOSYNTHESIS THROUGH THE ACTION OF SPECIFIC TRANSCRIPTION FACTORS
Several studies have demonstrated that plants under drought, high salt and cold stress conditions downregulate the expression of genes involved in photosynthesis and carbohydrate metabolism (Seki et al., 2002; Hannah et al., 2006; Wong et al., 2006). Enhanced freezing tolerance in Arabidopsis has been associated with the downregulation of genes related to photosynthesis and upregulation of genes related to biosynthesis of flavonoids (Hannah et al., 2006). However, when several Arabidopsis ecotypes were studied, the greatest downregulation of photosynthetic-related genes in response to freezing did not correspond to the ecotype with the highest capacity to cold-acclimate (Hannah et al., 2006). Hence, we can conclude that photosynthesis is only one of several different mechanisms that need to be modulated to improve cold acclimation.
The importance of stomatal vs. non-stomatal limitations to CO2 photosynthetic assimilation has been discussed in the Introduction. Stomatal closure plays a major role in limiting carbon assimilation under mild drought stress (Cornic, 2000, and references therein), but there is also metabolic limitation of photosynthesis (Tezara et al., 1999). Hence, it seems that both stomatal and non-stomatal changes limit the photosynthetic rate and capacity under stress. Many expression studies performed on plants subjected to different abiotic stresses have shown that a large number of genes are differentially expressed under stress, involving both stomatal and non-stomatal photosynthesis mechanisms. Thus, it is understandable that TFs modulating the expression of these genes play a critical role in plant response/adaptation to environmental cues.
Transcription factors involved in regulation of stomatal aperture by abiotic stresses
Until a decade ago, it was traditionally thought that control of stomata closure/opening did not involve transcriptional regulation (Hetherington and Quatrano, 1991). The first indication that TFs could be involved in stomata regulation appeared when it was found that ABI3 (an ABI3/VP1 B3-type TF, also involved in controlling accumulation of chlorophyll and anthocyanins) suppressed the effect of the abi1 mutation on stomatal regulation (Parcy and Giraudat, 1997). While abi1 plants do not regulate their stomata correctly (leading to increased transpiration) and are less sensitive to ABA, abi1 plants ectopically expressing ABI3 showed a wild-type phenotype. Meanwhile, it has been reported that modulation of transcription plays an important role in controlling guard cell activity. Recently two MYB-type TFs were identified as regulators of stomatal movements (Fig. 3). AtMYB60, a R2R3-MYB gene of Arabidopsis, was shown to be specifically expressed in guard cells, and its expression regulated by light conditions, ABA and water stress (Cominelli et al., 2005). However, elevated CO2 concentrations, which are known to induce stomatal closure, do not modulate AtMYB60 expression. The expression of this gene is negatively regulated under drought, concomitantly with stomatal closure. Accordingly, the atmyb60-1 null mutant shows a constitutive reduction in stomatal opening and decreased wilting under water stress conditions. Interestingly, only a limited number of genes is altered in the mutant and most of them are downregulated (Cominelli et al., 2005). Many of these genes (e.g. Aquaporin, ERD10, ERD13 and ERF) were already described as being involved in plant response to water stress (Cominelli et al., 2005).
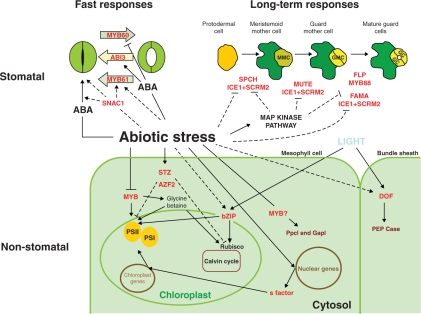
Fig. 3.
Transcription factors known to be involved in stomatal and non-stomatal limitations to CO2 photosynthetic assimilation. Line arrows represent a positive effect while lines ending with a bar indicate a negative effect. Block arrows show the direction of the stomatal movement mediated by the transcription factor. Dashed lines represent possible interactions. Additional transcription factors as well as the existence of other links cannot be ruled out. Transcription factors are in red. ABA, abscisic acid; PSI, photosystem I; PSII, photosystem II; ABI3, ABA-insensitive 3; SNAC1, stress-responsive NAC 1; SPCH, Speechless; FLP, Four Lips; ICE1, Inducer of CBF Expression 1; SCRM2, SCREAM2; MYB, myeloblastosis oncogene; Ppcl, CAM-specific isozyme of phosphoenolpyruvate carboxylase; GapI, NAD-dependent glyceraldehyde-3-phosphate dehydrogenase.
A second member of the Arabidopsis thaliana family of R2R3-MYB TFs, AtMYB61, is also specifically expressed in guard cells in a manner consistent with its involvement in the regulation of stomatal aperture (Liang et al., 2005). However, it has been shown that AtMYB60 and AtMYB61 have distinct expression patterns and functions. While AtMYB60 gene expression is induced by light, leading to an increased stomatal aperture, AtMYB61 transcription is repressed by light, although it has the same effect on stomatal movement. It was demonstrated that AtMYB61 expression is both sufficient and necessary to cause reductions in stomatal aperture. Moreover, Liang et al. (2005) showed that AtMYB61 gene expression was not altered in plants treated with ABA, salt and drought, known to induce stomatal closure. Thus, AtMYB61 seems to act via a mechanism parallel to that responsible for closing stomata in response to water deficit. However, a post-transcriptional/translational regulation of AtMYB61 cannot be ruled out. In addition, the loss-of-function mutant atmyb61 (larger stomatal aperture) was shown to remain responsive to increasing concentrations of ABA (Liang et al., 2005). Given that ABA signalling mutants, such as ost1, exhibit reduction in stomatal sensitivity to exogenously applied ABA (Mustilli et al., 2002), it seems that, in contrast to AtMYB60, the pathway through which AtMYB61 regulates stomatal behaviour is indeed different from the signalling pathway involving ABA (Fig. 3). Altogether, reported data indicate that AtMYB61 mediates stomatal aperture through an ABA-independent signalling pathway. The fact that stomatal closure can be induced by Cd2+ in the ABA-insensitive mutant abi1-1 (Perfus-Barbeoch et al., 2002) strengthens the hypothesis that stomatal movements can be regulated through an ABA-independent pathway, where AtMYB61 is integrated. It would be very interesting to investigate whether ABA-independent stomata regulation is associated with any abiotic stress response. Nevertheless, given that AtMYB61 can also be involved in the deposition of pectin (Penfield et al., 2001), which in guard cell walls has been shown to be important for stomatal movements (Jones et al., 2003), it is possible that AtMYB61 plays a second non-signalling role controlling stomatal aperture.
The control of stomata aperture may also involve the activity of SNAC1 (a TF described in a previous section; Fig. 3). Transgenic rice plants over-expressing SNAC1 are more sensitive to ABA and lose water more slowly by closing more stomata, leading to improvements in performance under drought and salinity.
Does abiotic stress control the transcription factors involved in stomatal development?
The physiological control of stomata movements by different environmental stresses is very well documented; however, little is known concerning the possible effects of abiotic stresses on stomatal development. This is an important issue that deserves thorough investigation, as the number of stomata affects both photosynthesis and WUE (Chaerle et al., 2005). It is essential that plants are able to sense environmental cues, such as CO2 concentration, light and UV-B irradiation, which have been shown to influence stomatal development and density (Lake et al., 2001; Gitz et al., 2005). Besides that, it has been shown that ABA-treated Transdescantia virginiana plants have significantly smaller stomata and more stomata per unit area of their lower epidermis (Franks and Farquhar, 2001). This suggests that ABA not only regulates short-term CO2 assimilation and water loss, but, through its effect on stomatal development, it may also permanently alter leaf photosynthesis in the direction of improving WUE. Given that ABA concentration increases upon osmotic and drought stress conditions, it is expected that prolonged abiotic stresses will have an effect on stomatal development in new leaves. It was shown that water stress and temperature may affect photosynthesis through alterations in stomatal number (Xu and Zhou, 2005). Stress responses often initiate transient physiological, biochemical and molecular alterations that lead to stable long-term adaptations reflecting complex developmental responses to adverse environmental conditions.
Stomata development requires a strict control over differentiation and cell division so that stomata morphology, distribution and behaviour respond properly to the environment, ensuring maximum plant performance. Recent work in Arabidopsis has substantially advanced our understanding of how stomata are built (Barton, 2007, and references therein). Five TFs were identified as essential for cell transitions leading to stomatal formation (Fig. 3). Together, SPEECHLESS (SPCH), MUTE and FAMA, all belonging to the bHLH family of TFs, control stomatal development at three consecutive steps: initiation, meristemoid differentiation and guard cell morphogenesis, respectively (Pillitteri et al., 2007). In addition, FOUR LIPS (FLP; MYB124) and MYB88 (both R2R3 MYB-type TFs) function in generating normal stomatal patterning (Lai et al., 2005). Recently, two paralogous proteins, SCREAM and SCREAM2 were found to interact directly with and specify the sequential actions of SPCH, MUTE and FAMA (Kanaoka et al., 2008). Surprisingly, SCREAM was found to be INDUCER OF CBF EXPRESSION 1 (ICE1), a master regulator of freezing tolerance, suggesting a potential link between the transcriptional regulation of environmental adaptation and stomatal development (Fig. 3).
Although there is no direct evidence, besides ICE1, that TFs controlling stomata development are under the control of environmental cues, it is known that stomatal development is related to environmental conditions (Chaerle et al., 2005). The question that remains to be answered is how the regulators of stomata formation themselves are regulated by different environmental conditions. Mitogen-activated protein kinase kinase (MAPKK) 4 and 5, and MAPK3 and 6 have previously been shown to play a role in the environmental stress response, with MAPK3 and 6 directly involved in osmotic stress (Nakagami et al., 2005). Wang and co-workers (2007) have recently suggested that the MKK4/MKK5–MPK3/MPK6 module is the long-sought molecular hub at which environmental signals impinge on the stomatal development pathway and influence stomatal development. Activation of this kinase cascade causes the suppression of asymmetric cell division, absence of meristemoid mother cell formation and consequent lack of stomatal differentiation (Fig. 3). The signalling events between this MAPK cascade and the downstream TFs controlling stomata development are still unknown, but their elucidation will help to clarify the regulatory mechanisms underlying environmental control of stomata formation.
Transcription factors mediating the non-stomatal responses to abiotic stress
In addition to stomatal regulation, abiotic stress conditions reduce the photosynthetic rate by controlling the expression of genes involved in non-stomatal processes associated with photosynthesis and carbohydrate metabolism. Among the genes whose expression is controlled by the environmental cues are those that encode chlorophyll a/b-binding (CAB) proteins, PSI and PSII subunits, oxygen-evolving complex and Rubisco subunits.
Most genes associated with photosynthesis are under the control of a transcriptional regulatory network evolved to control plant response to external stimuli. Transcriptional profiling studies have shown that although some are upregulated, many photosynthesis-related genes and genes for carbohydrate metabolism are downregulated under abiotic stress conditions. Those genes may be the target for TFs such as the Cys2/His2-type zinc-finger proteins STZ and AZF2. These TFs were shown to function as transcription repressors under drought, cold and high salinity stress conditions (Sakamoto et al., 2004). The expression of both STZ and AZF2 genes is induced mainly in leaves under drought stress, supporting the hypothesis that they play a role regulating photosynthesis-related genes. Transgenic plants constitutively over-expressing CBFs show higher induction of STZ, which may repress genes involved in photosynthesis and carbohydrate metabolism, thus accounting for growth retardation.
TFs acting as activators are also involved in the modulation of photosynthesis under abiotic stress. A good example is the regulation of the genes encoding CAB proteins of PSII. Two MYB-like TFs from barley, HvMCB1 and HvMCB2, were shown to bind specifically to defined regions of CAB promoters derived from barley and wheat. These TFs have characteristic features of transcriptional activators and are required for maximal CAB gene expression, but are not necessary for expression related to light and the circadian clock. Interestingly, transcription of both genes HvMCB1 and HvMCB2 is regulated by environmental factors. When barley leaves were subjected to salt, osmotic and oxidative stress, the transcripts of both MYB-encoding genes decreased significantly (Churin et al., 2003). HvMCB1 and HvMCB2 contain only one MYB repeat, while most MYB-related proteins that have been characterized belong to the R2R3-MYB protein family characterized by two MYB repeats (Jin and Martin, 1999). However, several MYB-related TFs with a single MYB repeat have also been identified in plants, including CIRCADIAN CLOCK-ASSOCIATED 1 (CCA1), a MYB-related protein from Arabidopsis that binds to a CAB promoter (Wang and Tobin, 1998). Whether CCA1 gene expression is regulated by environmental stress remains to be determined. It has been suggested that R2R3-MYB, the most abundant MYB protein family in plants, has evolved from the R1R2R3-MYB family (which is the norm in animals), to regulate plant-specific processes, including secondary metabolism, responses to plant hormones and the identity of specific cell types (Riechmann et al., 2000). MYB-type TFs with a single repeat may also be implicated in the regulation of plant-specific genes. In addition to the MYB TFs mentioned above, GLK1 and GLK2 are MYB TFs known to regulate chloroplast photosynthetic development (Fitter et al., 2002). In Brassica napus it was shown that the gene expression of both GLK1 and GLK2 is regulated by cold stress (Savitch et al., 2005). The involvement of several MYB-type TFs in the regulation of photosynthesis-related genes in response to abiotic stress indicates that this TF family is highly implicated in this process.
Other TFs, such as LONG HYPOCOTYL 5 (HY5), are known to be involved in the regulation of CAB gene expression by light, but may also play a role in the abiotic stress response. HY5 is a bZIP-type TF that binds to CAB upstream factor-1, being required for DET1 control of chlorophyll a/b-binding protein 2 (CAB2) expression (Maxwell et al., 2003). Additional studies suggested that HY5 also regulates the transcription of other photosynthesis-related genes, such as the ribulose bisphosphate carboxylase small subunit (RbcS1A) (Chattopadhyay et al., 1998; Lee et al., 2007). Given that HY5 appears to regulate the expression of several Arabidopsis genes known to respond to abiotic stress conditions [e.g. CBF1, DREB2A, RD20 and MYB59 (Lee et al. (2007)], it is inferred that HY5 may also be involved in the regulation of photosynthesis by adverse environmental conditions. Although there is no direct evidence showing that expression of photosynthesis-related genes is regulated by HY5 in response to abiotic stress, it appears that extreme temperatures (mainly cold, but also heat) induce HY5 gene expression (www.genevestigator.ethz.ch).
The regulation of photosynthesis-related genes by an abiotic stress response can also involve an indirect action of specific TFs. For instance, it is known that over-expression of OsMYB4 enhances compatible solute [e.g. glycine betaine (GB)] accumulation and increases stress tolerance of A. thaliana (Mattana et al., 2005). In addition, GB stabilizes Rubisco conformation under high salinity stress (Sakamoto and Murata, 2002). GB also protects the oxygen-evolving PSII complex against heat-induced inactivation and stimulates the repair of the PSII complex during photoinhibition. Corroborating these data, Vannini et al. (2004) have reported that Arabidopsis plants over-expressing OsMYB4 show improved PSII stability and higher tolerance to photoinhibition. Thus, one can say that despite a given TF not directly regulating the expression of a photosynthesis-related gene, it can indirectly cause an increase of the activity of the protein involved in photosynthesis. Given that OsMYB4 gene transcription is known to be regulated by abiotic stresses, namely cold, in an ABA-independent way (Vannini et al., 2004), it is possible that OsMYB4 plays a role in regulation of photosynthesis by environmental cues.
It has been proposed that the expression of photosynthetic genes in maize, a C4 plant, is at least in part controlled by two C2C2-DOF (DNA binding with One Finger)-type TFs, DOF1 and DOF2. Expression studies revealed that DOF1 might serve as an activator of transcription, whereas DOF2 may act as a tissue-specific repressor (Yanagisawa and Sheen, 1998). DOF1 can interact specifically with the maize C4 phosphoenolpyruvate carboxylase (PEPCase) gene promoter, enhancing its activity and giving PEPCase a light-regulated expression pattern matching DOF1 activity. It was then anticipated that the evolutionarily conserved DOF proteins can function as transcriptional activators or repressors of tissue-specific and light-regulated gene expression in plants (Yanagisawa and Sheen, 1998). Transcripts for the DOF1 TF involved in the activation of photosynthetic genes in maize were found in leaf tissues performing both C4 and C3 photosynthesis, with the greatest accumulation in C4 mesophyll cells, whereas the homologous DOF2 gene was expressed neither in C4 nor in C3 mesophyll cells (Hahnen et al., 2003). These data suggest that C3 and C4 photosynthetic mechanism are not controlled by the differential expression of DOF genes. A different type of TFs may also be associated with the regulation of C4 photosynthetic genes. The homeobox protein ZF-HD might be involved in the establishment of the characteristic expression pattern of the C4 PEPCase gene (Windhovel et al., 2001). Little is known about the role of the DOF-type TFs in mediating abiotic stress responses. Recently, AtDOF4;2 was shown to be involved in the regulation of phenylpropanoid metabolism in a cold-specific manner (Skirycz et al., 2007), indicating that DOF-type TFs can mediate abiotic stress responses. Whether they modulate the expression of photosynthetic-related genes in response to stress is still not known.
Crassulacean acid metabolism (CAM), an adaptation of photosynthetic carbon fixation to water-limited environments, is characterized by nocturnal CO2 fixation and diurnal CO2 re-assimilation. Normally this adaptation results in improved WUE. The primary mechanism responsible for enhanced expression of CAM-specific genes following water deficit or salt stress is transcriptional regulation. Transcriptional activation of CAM-specific genes by high salt stress is mediated through multiple interactions of cis-acting DNA sequences and trans-acting factors that increase in abundance, or in DNA-binding affinity following salt stress. The transcription of the Ppcl and Gapl genes encoding a CAM-specific isozyme of PEPCase and NAD-dependent glyceraldehyde-3-phosphate dehydrogenase, respectively, is increased by salinity stress. The promoters of both genes include regions shown to be essential for regulation of gene expression by salinity. Within these regions, several common sequence motifs resemble consensus binding sites for the MYB class of TFs (Schaeffer et al., 1995), suggesting that the regulation of these mentioned photosynthesis-related genes by high salinity may be mediated by MYB-type TFs.
Another interesting point is the transcriptional regulation of plastid-encoded photosynthesis genes by environmental stimuli. The expression of these genes is controlled through nuclear-encoded transcription regulators, as none of the genes in the higher plant chloroplast encodes TFs. Most of the chloroplast-encoded photosynthesis genes are induced at an early stage of light-induced chloroplast development and repressed in mature chloroplasts. However, the transcription of genes, such as psbA and psbD encoding the PSII reaction centre protein D1 and D2, respectively, is differentially regulated (Christopher and Mullet, 1994). In Arabidopsis, light-induced expression of psbD is controlled by AtSig5, a nuclear-encoded σ factor (Tsunoyama et al., 2004), while AtSig6, another nuclear-encoded σ factor, regulates early chloroplast development (Ishizaki et al., 2005). It would be particularly important to know how the transcription of the chloroplast-encoded genes is regulated by abiotic stresses. In the cyanobacterium Synechocystis sp. PCC 6803, salt stress inhibits transcription and translation of psbA genes and encoded proteins, thus limiting the efficiency of photosynthesis through inactivation of PSII (Allakhverdiev et al., 2002), but nothing is known concerning the TFs involved. However, it is likely that TFs regulating chloroplast-encoded genes mediate abiotic stress responses.
Although some TFs may mediate the photosynthetic responses to different abiotic stresses, little is still known. In order to understand better the effect of abiotic stress on the regulation of the photosynthesis-related genes, it would be essential to identify and characterize the function of novel TFs involved. This could be achieved using yeast-one-hybrid (Y1H) screenings and/or chromatin immunoprecipitation (ChIP) assays, followed by TF function analysis.
OVER-EXPRESSION OF TRANSCRIPTION FACTORS CONFERS ABIOTIC STRESS TOLERANCE AND PHOTOSYNTHESIS IMPROVEMENT
Tolerance of plants to abiotic stresses is well known to be a multigenic trait. For that reason, plant improvement using genes that play a role in the abiotic stress response is frequently insufficient to improve stress tolerance significantly. To overcome this, TFs that regulate several stress-responsive genes (e.g. the AP2/EREBP family) have often been used to manipulate plants in order to have a broader response. The results obtained in terms of stress tolerance are often much better than using a single gene encoding a non-regulatory protein and the observed effects on photosynthesis efficiency or photosynthetic machinery are normally positive. Table 1 shows a list of TFs transformed into various plants and conferring stress tolerance improvement associated with improved photosynthetic parameters. The interpretation of these results must, however, be cautious, as only a few parameters are evaluated and sometimes the methods are poor. In addition, abiotic stress tolerance has mostly been evaluated in laboratory conditions and little is known concerning the plant responses to adverse conditions in the field.
Table 1.
Abiotic stress-related transcription factors constitutively expressed in different plants confer stress tolerance and improve photosynthesis
| Over-expressed TF | TF family | Transgenic plant | Stress tolerance | Effect on photosynthesis under stress conditions | Reference |
|---|---|---|---|---|---|
| NtTsi1 | AP2/EREBP | Tobacco | High salt | Lower loss of chlorophyll contents | Park et al. (2001) |
| AtCBF1 | AP2/EREBP | Tomato | Chilling | Improved maximum quantum efficiency of PSII/chlorophyll accumulation | Hsieh et al. (2002b) |
| AtCBF1 | AP2/EREBP | Rice | No cold tolerance | No effect on maximum quantum efficiency of PSII | Lee et al. (2004) |
| SHN | AP2/EREBP | Arabidopsis | Drought tolerance and recovery | Reduced stomatal density (probable reduced transpiration) | Aharoni et al. (2004) |
| BNCBF5 and BNCBF17 | AP2/EREBP | Brassica napus | Freezing | Increased CO2 assimilation/increased photochemical efficiency | Savitch et al. (2005) |
| AtCBF3 | AP2/EREBP | Rice | Drought/high salt/low temperature | Improved maximum quantum efficiency of PSII | Oh et al. (2005) |
| CaPF1 | AP2/EREBP | Pine | Oxidative stress | Lower loss of chlorophyll contents | Tang et al. (2006) |
| TaERF1 | AP2/EREBP | Tobacco | High salt | Higher chlorophyll content | Xu et al. (2007) |
| JcERF | AP2/EREBP | Arabidopsis | High salt/freezing | Higher chlorophyll content | Tang et al. (2007) |
| HvCBF4 | AP2/EREBP | Rice | Drought/high salt/ low temperature | Improved maximum quantum efficiency of PSII | Oh et al. (2007) |
| NtOPBP1 | AP2/EREBP | Tobacco | High salt | Lower loss of chlorophyll contents | Guo et al. (2004) |
| AtHRD | AP2/EREBP | Rice | Drought/highsalt | Lower stomatal conductance/enhanced photosynthesis assimilation and efficiency | Karaba et al. (2007) |
| GhDREB1 | AP2/EREBP | Tobacco | Low temperature | Higher chlorophyll fluorescence/higher net photosynthetic rate | Shan et al. (2007) |
| AtABP9 | bZIP | Arabidopsis | Drought/heat shock | Improved photosynthetic capacity | Zhang et al. (2008) |
| SNAC1 | NAC | Rice | Drought/high salt | Loses water more slowly by closing more stomatal pores/no effect on photosynthesis rate | Hu et al. (2006) |
| AtNFXL1 | NF-X1 | Arabidopsis | Salt stress | Improved maximum quantum efficiency of PSII | Lisso et al. (2006) |
| AtNF-YB1 | NF-Y (HAP) | Arabidopsis | Drought | Higher water potential and photosynthesis rates than controls | Nelson et al. (2007) |
| ZmNF-YB2 | NF-Y (HAP) | Maize | Drought | Higher chlorophyll index, higher photosynthesis rate and higher stomatal conductance | Nelson et al. (2007) |
| GmSCOF-1 | C2H2 zinc finger | Tobacco | Cold | Faster recovery of chlorophyll content | Kim et al. (2001) |
| OsMYB4 | MYB | Arabidopsis | Cold/freezing | Improved PSII stability. Tolerance to photoinhibition | Vannini et al. (2004) |
CONCLUSIONS
Extremes of environmental conditions, such as drought, cold and high salinity, induce stress in plants and decrease growth and productivity. Photosynthesis and the related metabolism are among the processes most strongly affected by these abiotic stresses. Interestingly, both stomatal and non-stomatal responses to abiotic stress involve transcriptional regulation and consequently the involvement of many TFs. Some of these TFs can mediate the regulation of photosynthetic-related gene expression by the redox state. In contrast to what was previously thought, stomatal movements involve gene transcription regulation. Several TFs have been shown to regulate stomatal aperture and so mediate responses to adverse environmental conditions. Expression of genes coding for components regulating many aspects of photosynthetic metabolism is affected by stress, and TFs have a substantial role in regulating them and thus carbon metabolism. Adjustment to stress conditions may be improved by modifying the TFs in plants. However, knowledge about the TFs involved in the regulation of photosynthesis-related genes (stomatal and non-stomatal responses) by the different abiotic stresses is still limited. Investigation in this area is urgently required. Many transgenic plants over-expressing TFs show improved abiotic stress tolerance related to enhanced photosynthetic parameters. However, these results must be carefully analysed, as few photosynthetic parameters are usually evaluated and the growth /stress conditions are normally different from natural field conditions.
ACKNOWLEDGEMENTS
We thank Dr David Lawlor and Dr Manuela Chaves for critical reading and suggestions given to improve this manuscript. The work of our research group was supported by a grant from the Fundação para a Ciência e Tecnologia (POCI/BIA-BCM/56063/2004; POCI2010-FEDER). N.S. (SFRH/BPD/26636/2006) and T.L. (SFRH/BD/10615/2002) are indebted to the Fundação para a Ciência e Tecnologia for their fellowships.
LITERATURE CITED
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. The Plant Cell. 2003;15:63–78. doi: 10.1105/tpc.006130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal M, Hao Y, Kapoor A, et al. A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. Journal of Biological Chemistry. 2006;281:37636–37645. doi: 10.1074/jbc.M605895200. [DOI] [PubMed] [Google Scholar]
- Aharoni A, Dixit S, Jetter R, Thoenes E, van Arkel G, Pereira A. The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. The Plant Cell. 2004;16:2463–2480. doi: 10.1105/tpc.104.022897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allakhverdiev SI, Nishiyama Y, Miyairi S, et al. Salt stress inhibits the repair of photodamaged photosystem II by suppressing the transcription and translation of psbA genes in Synechocystis. Plant Physiology. 2002;130:1443–1453. doi: 10.1104/pp.011114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-Gonzalez E, Rolland F, Thevelein JM, Sheen J. A central integrator of transcription networks in plant stress and energy signalling. Nature. 2007;448:938–942. doi: 10.1038/nature06069. [DOI] [PubMed] [Google Scholar]
- Baginsky S, Tiller K, Pfannschmidt T, Link G. PTK, the chloroplast RNA polymerase-associated protein kinase from mustard (Sinapis alba), mediates redox control of plastid in vitro transcription. Plant Molecular Biology. 1999;39:1013–1023. doi: 10.1023/a:1006177807844. [DOI] [PubMed] [Google Scholar]
- Balogun E, Hoque M, Gong P, et al. Curcumin activates the Haem oxygenase-1 gene via regulation of Nrf2 and the antioxidant-responsive element. Biochemical Journal. 2003;371:887–895. doi: 10.1042/BJ20021619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton MK. Making holes in leaves: promoting cell state transitions in stomatal development. The Plant Cell. 2007;19:1140–1143. doi: 10.1105/tpc.107.051177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler C, Vanmontagu M, Inze D. Superoxide-dismutase and stress tolerance. Annual Review of Plant Physiology and Plant Molecular Biology. 1992;43:83–116. [Google Scholar]
- Bressan RA, Hasegawa PM, Pardo JM. Plants use calcium to resolve salt stress. Trends in Plant Science. 1998;3:411–412. [Google Scholar]
- Centritto M, Loreto F, Chartzoulakis K. The use of low [CO2] to estimate diffusional and non-diffusional limitations of photosynthetic capacity of salt-stressed olive saplings. Plant, Cell and Environment. 2003;26:585–594. [Google Scholar]
- Chaerle L, Saibo N, Van Der Straeten D. Tuning the pores: towards engineering plants for improved water use efficiency. Trends in Biotechnology. 2005;23:308–315. doi: 10.1016/j.tibtech.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay S, Ang LH, Puente P, Deng XW, Wei N. Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. The Plant Cell. 1998;10:673–683. doi: 10.1105/tpc.10.5.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves MM, Oliveira MM. Mechanisms underlying plant resilience to water deficits: prospects for water-saving agriculture. Journal of Experimental Botany. 2004;55:2365–2384. doi: 10.1093/jxb/erh269. [DOI] [PubMed] [Google Scholar]
- Chaves MM, Maroco JP, Pereira JS. Understanding plant responses to drought – from genes to the whole plant. Functional Plant Biology. 2003;30:239–264. doi: 10.1071/FP02076. [DOI] [PubMed] [Google Scholar]
- Chinnusamy V, Ohta M, Kanrar S, et al. ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes and Development. 2003;17:1043–1054. doi: 10.1101/gad.1077503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H, Hong J, Ha J, Kang J, Kim SY. ABFs, a family of ABA-responsive element binding factors. Journal of Biological Chemistry. 2000;275:1723–1730. doi: 10.1074/jbc.275.3.1723. [DOI] [PubMed] [Google Scholar]
- Christopher DA, Mullet JE. Separate photosensory pathways co-regulate blue-light/ultraviolet-A-activated psbD–psbC transcription and light-induced D2 and CP43 degradation in barley (Hordeum vulgare) chloroplasts. Plant Physiology. 1994;104:1119–1129. doi: 10.1104/pp.104.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churin Y, Adam E, Kozma-Bognar L, Nagy F, Borner T. Characterization of two Myb-like transcription factors binding to CAB promoters in wheat and barley. Plant Molecular Biology. 2003;52:447–462. doi: 10.1023/a:1023934232662. [DOI] [PubMed] [Google Scholar]
- Cominelli E, Galbiati M, Vavasseur A, Conti L, Sala T, Vuylsteke M, et al. A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Current Biology. 2005;15:1196–1200. doi: 10.1016/j.cub.2005.05.048. [DOI] [PubMed] [Google Scholar]
- Cornic G. Drought stress inhibits photosynthesis by decreasing stomatal aperture – not by affecting ATP synthesis. Trends in Plant Science. 2000;5:187–188. [Google Scholar]
- Dong CH, Agarwal M, Zhang Y, Xie Q, Zhu JK. The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proceedings of the National Academy of Sciences; USA. 2006. pp. 8281–8286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubouzet JG, Sakuma Y, Ito Y, et al. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. The Plant Journal. 2003;33:751–763. doi: 10.1046/j.1365-313x.2003.01661.x. [DOI] [PubMed] [Google Scholar]
- Fitter DW, Martin DJ, Copley MJ, Scotland RW, Langdale JA. GLK gene pairs regulate chloroplast development in diverse plant species. The Plant Journal. 2002;31:713–727. doi: 10.1046/j.1365-313x.2002.01390.x. [DOI] [PubMed] [Google Scholar]
- Flexas J, Medrano H. Drought-inhibition of photosynthesis in C3 plants: stomatal and non-stomatal limitations revisited. Annals of Botany. 2002;89:183–189. doi: 10.1093/aob/mcf027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flexas J, Bota J, Escalona JM, Sampol B, Medrano H. Effects of drought on photosynthesis in grapevines under field conditions: an evaluation of stomatal and mesophyll limitations. Functional Plant Biology. 2002;29:461–471. doi: 10.1071/PP01119. [DOI] [PubMed] [Google Scholar]
- Flexas J, Ribas-Carbo M, Bota J, et al. Decreased Rubisco activity during water stress is not induced by decreased relative water content but related to conditions of low stomatal conductance and chloroplast CO2 concentration. New Phytologist. 2006;172:73–82. doi: 10.1111/j.1469-8137.2006.01794.x. [DOI] [PubMed] [Google Scholar]
- Fowler S, Thomashow MF. Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. The Plant Cell. 2002;14:1675–1690. doi: 10.1105/tpc.003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler SG, Cook D, Thomashow MF. Low temperature induction of Arabidopsis CBF1, v2, and 3 is gated by the circadian clock. Plant Physiology. 2005;137:961–968. doi: 10.1104/pp.104.058354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks PJ, Farquhar GD. The effect of exogenous abscisic acid on stomatal development, stomatal mechanics, and leaf gas exchange in Tradescantia virginiana. Plant Physiology. 2001;125:935–942. doi: 10.1104/pp.125.2.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour SJ, Zarka DG, Stockinger EJ, Salazar MP, Houghton JM, Thomashow MF. Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. The Plant Journal. 1998;16:433–442. doi: 10.1046/j.1365-313x.1998.00310.x. [DOI] [PubMed] [Google Scholar]
- Gilmour SJ, Fowler SG, Thomashow MF. Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Molecular Biology. 2004;54:767–781. doi: 10.1023/B:PLAN.0000040902.06881.d4. [DOI] [PubMed] [Google Scholar]
- Gitz DC, Liu-Gitz L, Britz SJ, Sullivan JH. Ultraviolet-B effects on stomatal density, water-use efficiency, and stable carbon isotope discrimination in four glasshouse-grown soybean (Glyicine max) cultivars. Environmental and Experimental Botany. 2005;53:343–355. [Google Scholar]
- Guo ZJ, Chen XJ, Wu XL, Ling JQ, Xu P. Overexpression of the AP2/EREBP transcription factor OPBP1 enhances disease resistance and salt tolerance in tobacco. Plant Molecular Biology. 2004;55:607–618. doi: 10.1007/s11103-004-1521-3. [DOI] [PubMed] [Google Scholar]
- Hahnen S, Joeris T, Kreuzaler F, Peterhansel C. Quantification of photosynthetic gene expression in maize C3 and C4 tissues by real-time PCR. Photosynthesis Research. 2003;75:183–192. doi: 10.1023/A:1022856715409. [DOI] [PubMed] [Google Scholar]
- Hanba YT, Shibasaka M, Hayashi Y, et al. Overexpression of the barley aquaporin HvPIP2;1 increases internal CO2 conductance and CO2 assimilation in the leaves of transgenic rice plants. Plant Cell Physiology. 2004;45:521–529. doi: 10.1093/pcp/pch070. [DOI] [PubMed] [Google Scholar]
- Hannah MA, Wiese D, Freund S, Fiehn O, Heyer AG, Hincha DK. Natural genetic variation of freezing tolerance in Arabidopsis. Plant Physiology. 2006;142:98–112. doi: 10.1104/pp.106.081141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetherington AM, Quatrano RS. Mechanisms of action of abscisic acid at the cellular level. New Phytologist. 1991;119:9–32. doi: 10.1111/j.1469-8137.1991.tb01004.x. [DOI] [PubMed] [Google Scholar]
- Hsieh TH, Lee JT, Charng YY, Chan MT. Tomato plants ectopically expressing Arabidopsis CBF1 show enhanced resistance to water deficit stress. Plant Physiology. 2002;a 130:618–626. doi: 10.1104/pp.006783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh TH, Lee JT, Yang PT, et al. Heterology expression of the Arabidopsis C-repeat/dehydration response element binding factor 1 gene confers elevated tolerance to chilling and oxidative stresses in transgenic tomato. Plant Physiology. 2002;b 129:1086–1094. doi: 10.1104/pp.003442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Dai M, Yao J, et al. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proceedings of the National Academy of Sciences; USA. 2006. pp. 12987–12992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imin N, Kerim T, Weinman JJ, Rolfe BG. Low temperature treatment at the young microspore stage induces protein changes in rice anthers. Molecular and Cellular Proteomics. 2006;5:274–292. doi: 10.1074/mcp.M500242-MCP200. [DOI] [PubMed] [Google Scholar]
- Ishizaki Y, Tsunoyama Y, Hatano K, et al. A nuclear-encoded sigma factor, Arabidopsis SIG6, recognizes sigma-70 type chloroplast promoters and regulates early chloroplast development in cotyledons. The Plant Journal. 2005;42:133–144. doi: 10.1111/j.1365-313X.2005.02362.x. [DOI] [PubMed] [Google Scholar]
- Ito Y, Katsura K, Maruyama K, et al. Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant and Cell Physiology. 2006;47:141–153. doi: 10.1093/pcp/pci230. [DOI] [PubMed] [Google Scholar]
- Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science. 1998;280:104–106. doi: 10.1126/science.280.5360.104. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Yang B, Harris NS, Deyholos MK. Comparative proteomic analysis of NaCl stress-responsive proteins in Arabidopsis roots. Journal of Experimental Botany. 2007;58:3591–3607. doi: 10.1093/jxb/erm207. [DOI] [PubMed] [Google Scholar]
- Jin H, Martin C. Multifunctionality and diversity within the plant MYB-gene family. Plant Molecular Biology. 1999;41:577–585. doi: 10.1023/a:1006319732410. [DOI] [PubMed] [Google Scholar]
- Jones L, Milne JL, Ashford D, McQueen-Mason SJ. Cell wall arabinan is essential for guard cell function. Proceedings of the National Academy of Sciences; USA. 2003. pp. 11783–11788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaoka MM, Pillitteri LJ, Fujii H, et al. SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to Arabidopsis stomatal differentiation. The Plant Cell. 2008;20:1775–1785. doi: 10.1105/tpc.108.060848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaba A, Dixit S, Greco R, et al. Improvement of water use efficiency in rice by expression of HARDY, an Arabidopsis drought and salt tolerance gene. Proceedings of the National Academy of Sciences; USA. 2007. pp. 15270–15275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nature Biotechnology. 1999;17:287–291. doi: 10.1038/7036. [DOI] [PubMed] [Google Scholar]
- Kasuga M, Miura S, Shinozaki K, Yamaguchi-Shinozaki K. A combination of the Arabidopsis DREB1A gene and stress-inducible rd29A promoter improved drought- and low-temperature stress tolerance in tobacco by gene transfer. Plant and Cell Physiology. 2004;45:346–350. doi: 10.1093/pcp/pch037. [DOI] [PubMed] [Google Scholar]
- Kim JC, Lee SH, Cheong YH, et al. A novel cold-inducible zinc finger protein from soybean, SCOF-1, enhances cold tolerance in transgenic plants. The Plant Journal. 2001;25:247–259. doi: 10.1046/j.1365-313x.2001.00947.x. [DOI] [PubMed] [Google Scholar]
- Kingston-Smith AH, Harbinson J, Williams J, Foyer CH. Effect of chilling on carbon assimilation, enzyme activation, and photosynthetic electron transport in the absence of photoinhibition in maize leaves. Plant Physiology. 1997;114:1039–1046. doi: 10.1104/pp.114.3.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai LB, Nadeau JA, Lucas J, et al. The Arabidopsis R2R3 MYB proteins FOUR LIPS and MYB88 restrict divisions late in the stomatal cell lineage. The Plant Cell. 2005;17:2754–2767. doi: 10.1105/tpc.105.034116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake JA, Quick WP, Beerling DJ, Woodward FI. Plant development. Signals from mature to new leaves. Nature. 2001;411:154. doi: 10.1038/35075660. [DOI] [PubMed] [Google Scholar]
- Lawlor DW. Limitation to photosynthesis in water-stressed leaves: stomata vs. metabolism and the role of ATP. Annals of Botany. 2002;89:871–885. doi: 10.1093/aob/mcf110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawlor DW, Cornic G. Photosynthetic carbon assimilation and associated metabolism in relation to water deficits in higher plants. Plant, Cell and Environment. 2002;25:275–294. doi: 10.1046/j.0016-8025.2001.00814.x. [DOI] [PubMed] [Google Scholar]
- Lawlor DW, Tezara W. Causes of decreased photosynthetic rate and metabolic capacity in water-deficient leaf cells: a critical evaluation of mechanisms and integration of processes. Annals of Botany. 2009;103:561–579. doi: 10.1093/aob/mcn244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, He K, Stolc V, et al. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. The Plant Cell. 2007;19:731–749. doi: 10.1105/tpc.106.047688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, Huh KW, An K, An G, Kim SR. Ectopic expression of a cold-inducible transcription factor, CBF1/DREB1b, in transgenic rice (Oryza sativa L.) Molecular Cells. 2004;18:107–114. [PubMed] [Google Scholar]
- Liang YK, Dubos C, Dodd IC, Holroyd GH, Hetherington AM, Campbell MM. AtMYB61, an R2R3-MYB transcription factor controlling stomatal aperture in Arabidopsis thaliana. Current Biology. 2005;15:1201–1206. doi: 10.1016/j.cub.2005.06.041. [DOI] [PubMed] [Google Scholar]
- Lisso J, Altmann T, Mussig C. The AtNFXL1 gene encodes a NF-X1 type zinc finger protein required for growth under salt stress. FEBS Letters. 2006;580:4851–4856. doi: 10.1016/j.febslet.2006.07.079. [DOI] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, et al. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. The Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamun EA, Alfred S, Cantrill LC, Overall RL, Sutton BG. Effects of chilling on male gametophyte development in rice. Cell Biology International. 2006;30:583–591. doi: 10.1016/j.cellbi.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Mattana M, Biazzi E, Consonni R, et al. Overexpression of Osmyb4 enhances compatible solute accumulation and increases stress tolerance of Arabidopsis thaliana. Physiologia Plantarum. 2005;125:212–223. [Google Scholar]
- Maxwell BB, Andersson CR, Poole DS, Kay SA, Chory J. HY5, Circadian Clock-Associated 1, and a cis-element, DET1 dark response element, mediate DET1 regulation of chlorophyll a/b-binding protein 2 expression. Plant Physiology. 2003;133:1565–1577. doi: 10.1104/pp.103.025114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Jin JB, Lee J, et al. SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. The Plant Cell. 2007;19:1403–1414. doi: 10.1105/tpc.106.048397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller IM, Jensen PE, Hansson A. Oxidative modifications to cellular components in plants. Annual Review of Plant Biology. 2007;58:459–481. doi: 10.1146/annurev.arplant.58.032806.103946. [DOI] [PubMed] [Google Scholar]
- Murata N, Takahashi S, Nishiyama Y, Allakhverdiev SI. Photoinhibition of photosystem II under environmental stress. Biochimica et Biophysica Acta. 2007;1767:414–421. doi: 10.1016/j.bbabio.2006.11.019. [DOI] [PubMed] [Google Scholar]
- Mustilli AC, Merlot S, Vavasseur A, Fenzi F, Giraudat J. Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. The Plant Cell. 2002;14:3089–3099. doi: 10.1105/tpc.007906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami H, Pitzschke A, Hirt H. Emerging MAP kinase pathways in plant stress signalling. Trends in Plant Science. 2005;10:339–346. doi: 10.1016/j.tplants.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiology. 2006;140:411–432. doi: 10.1104/pp.105.073783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Yamaguchi-Shinozaki K. Regulons involved in osmotic stress-responsive and cold stress-responsive gene expression in plants. Physiologia Plantarum. 2006;126:62–71. [Google Scholar]
- Nakashima K, Kiyosue T, Yamaguchi-Shinozaki K, Shinozaki K. A nuclear gene, erd1, encoding a chloroplast-targeted Clp protease regulatory subunit homolog is not only induced by water stress but also developmentally up-regulated during senescence in Arabidopsis thaliana. The Plant Journal. 1997;12:851–861. doi: 10.1046/j.1365-313x.1997.12040851.x. [DOI] [PubMed] [Google Scholar]
- Nelson DE, Repetti PP, Adams TR, et al. Plant nuclear factor Y(NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proceedings of the National Academy of Sciences; USA. 2007. pp. 16450–16455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama Y, Yamamoto H, Allakhverdiev SI, Inaba M, Yokota A, Murata N. Oxidative stress inhibits the repair of photodamage to the photosynthetic machinery. EMBO Journal. 2001;20:5587–5594. doi: 10.1093/emboj/20.20.5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama Y, Allakhverdiev SI, Yamamoto H, Hayashi H, Murata N. Singlet oxygen inhibits the repair of photosystem II by suppressing the translation elongation of the D1 protein in Synechocystis sp. PCC 6803. Biochemistry. 2004;43:11321–11330. doi: 10.1021/bi036178q. [DOI] [PubMed] [Google Scholar]
- Niyogi KK. Safety valves for photosynthesis. Current Opinion in Plant Biology. 2000;3:455–460. doi: 10.1016/s1369-5266(00)00113-8. [DOI] [PubMed] [Google Scholar]
- Noctor G, Foyer CH. Ascorbate and glutathione: keeping active oxygen under control. Annual Review in Plant Physiology and Plant Molecular Biology. 1998;49:249–279. doi: 10.1146/annurev.arplant.49.1.249. [DOI] [PubMed] [Google Scholar]
- Novillo F, Alonso JM, Ecker JR, Salinas J. CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in stress tolerance in Arabidopsis. Proceedings of the National Academy of Sciences; USA. 2004. pp. 3985–3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SJ, Song SI, Kim YS, et al. Arabidopsis CBF3/DREB1A and ABF3 in transgenic rice increased tolerance to abiotic stress without stunting growth. Plant Physiology. 2005;138:341–351. doi: 10.1104/pp.104.059147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SJ, Kwon CW, Choi DW, Song SI, Kim JK. Expression of barley HvCBF4 enhances tolerance to abiotic stress in transgenic rice. Plant Biotechnology Journal. 2007;5:646–656. doi: 10.1111/j.1467-7652.2007.00272.x. [DOI] [PubMed] [Google Scholar]
- Parcy F, Giraudat J. Interactions between the ABI1 and the ectopically expressed ABI3 genes in controlling abscisic acid responses in Arabidopsis vegetative tissues. The Plant Journal. 1997;11:693–702. doi: 10.1046/j.1365-313x.1997.11040693.x. [DOI] [PubMed] [Google Scholar]
- Park JM, Park CJ, Lee SB, Ham BK, Shin R, Paek KH. Overexpression of the tobacco Tsi1 gene encoding an EREBP/AP2-type transcription factor enhances resistance against pathogen attack and osmotic stress in tobacco. The Plant Cell. 2001;13:1035–1046. doi: 10.1105/tpc.13.5.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrineschi A, Reynolds M, Pacheco M, et al. Stress-induced expression in wheat of the Arabidopsis thaliana DREB1A gene delays water stress symptoms under greenhouse conditions. Genome. 2004;47:493–500. doi: 10.1139/g03-140. [DOI] [PubMed] [Google Scholar]
- Penfield S, Meissner RC, Shoue DA, Carpita NC, Bevan MW. MYB61 is required for mucilage deposition and extrusion in the Arabidopsis seed coat. The Plant Cell. 2001;13:2777–2791. doi: 10.1105/tpc.010265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perfus-Barbeoch L, Leonhardt N, Vavasseur A, Forestier C. Heavy metal toxicity: cadmium permeates through calcium channels and disturbs the plant water status. The Plant Journal. 2002;32:539–548. doi: 10.1046/j.1365-313x.2002.01442.x. [DOI] [PubMed] [Google Scholar]
- Pfannschmidt T, Nilsson A, Allen JF. Photosynthetic control of chloroplast gene expression. Nature. 1999;397:625–628. [Google Scholar]
- Pfannschmidt T. Chloroplast redox signals: how photosynthesis controls its own genes. Trends in Plant Science. 2003;8:33–41. doi: 10.1016/s1360-1385(02)00005-5. [DOI] [PubMed] [Google Scholar]
- Pillitteri LJ, Sloan DB, Bogenschutz NL, Torii KU. Termination of asymmetric cell division and differentiation of stomata. Nature. 2007;445:501–505. doi: 10.1038/nature05467. [DOI] [PubMed] [Google Scholar]
- Pino MT, Skinner JS, Park EJ, et al. Use of a stress inducible promoter to drive ectopic AtCBF expression improves potato freezing tolerance while minimizing negative effects on tuber yield. Plant Biotechnology Journal. 2007;5:591–604. doi: 10.1111/j.1467-7652.2007.00269.x. [DOI] [PubMed] [Google Scholar]
- Ratnayaka HH, Molin WT, Sterling TM. Physiological and antioxidant responses of cotton and spurred anoda under interference and mild drought. Journal of Experimental Botany. 2003;54:2293–2305. doi: 10.1093/jxb/erg251. [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, et al. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- Sakamoto A, Murata N. The role of glycine betaine in the protection of plants from stress: clues from transgenic plants. Plant, Cell and Environment. 2002;25:163–171. doi: 10.1046/j.0016-8025.2001.00790.x. [DOI] [PubMed] [Google Scholar]
- Sakamoto H, Maruyama K, Sakuma Y, et al. Arabidopsis Cys2/His2-type zinc-finger proteins function as transcription repressors under drought, cold, and high-salinity stress conditions. Plant Physiology. 2004;136:2734–2746. doi: 10.1104/pp.104.046599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma Y, Maruyama K, Osakabe Y, et al. Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. The Plant Cell. 2006;18:1292–1309. doi: 10.1105/tpc.105.035881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitch LV, Gray GR, Huner NPA. Feedback-limited photosynthesis and regulation of sucrose-starch accumulation during cold acclimation and low-temperature stress in a spring and winter wheat. Planta. 1997;201:18–26. [Google Scholar]
- Savitch LV, Harney T, Huner NPA. Sucrose metabolism in spring and winter wheat in response to high irradiance, cold stress and cold acclimation. Physiologia Plantarum. 2000;108:270–278. [Google Scholar]
- Savitch LV, Barker-Astrom J, Ivanov AG, et al. Cold acclimation of Arabidopsis thaliana results in incomplete recovery of photosynthetic capacity, associated with an increased reduction of the chloroplast stroma. Planta. 2001;214:295–303. doi: 10.1007/s004250100622. [DOI] [PubMed] [Google Scholar]
- Savitch LV, Allard G, Seki M, et al. The effect of overexpression of two Brassica CBF/DREB1-like transcription factors on photosynthetic capacity and freezing tolerance in Brassica napus. Plant and Cell Physiology. 2005;46:1525–1539. doi: 10.1093/pcp/pci165. [DOI] [PubMed] [Google Scholar]
- Schaeffer HJ, Forstheoefel NR, Cushman JC. Identification of enhancer and silencer regions involved in salt-responsive expression of Crassulacean acid metabolism (CAM) genes in the facultative halophyte Mesembryanthemum crystallinum. Plant Molecular Biology. 1995;28:205–218. doi: 10.1007/BF00020241. [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Ishida J, et al. Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. The Plant Journal. 2002;31:279–292. doi: 10.1046/j.1365-313x.2002.01359.x. [DOI] [PubMed] [Google Scholar]
- Shan DP, Huang JG, Yang YT, et al. Cotton GhDREB1 increases plant tolerance to low temperature and is negatively regulated by gibberellic acid. New Phytologist. 2007;176:70–81. doi: 10.1111/j.1469-8137.2007.02160.x. [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. Gene networks involved in drought stress response and tolerance. Journal of Experimental Botany. 2007;58:221–227. doi: 10.1093/jxb/erl164. [DOI] [PubMed] [Google Scholar]
- Skirycz A, Jozefczuk S, Stobiecki M, et al. Transcription factor AtDOF4;2 affects phenylpropanoid metabolism in Arabidopsis thaliana. New Phytologist. 2007;175:425–438. doi: 10.1111/j.1469-8137.2007.02129.x. [DOI] [PubMed] [Google Scholar]
- Takahashi S, Murata N. How do environmental stresses accelerate photoinhibition? Trends in Plant Science. 2008;13:178–182. doi: 10.1016/j.tplants.2008.01.005. [DOI] [PubMed] [Google Scholar]
- Tang M, Sun J, Liu Y, Chen F, Shen S. Isolation and functional characterization of the JcERF gene, a putative AP2/EREBP domain-containing transcription factor, in the woody oil plant Jatropha curcas. Plant Molecular Biology. 2007;63:419–428. doi: 10.1007/s11103-006-9098-7. [DOI] [PubMed] [Google Scholar]
- Tang W, Newton RJ, Lin JX, Charles TM. Expression of a transcription factor from Capsicum annuum in pine calli counteracts the inhibitory effects of salt stress on adventitious shoot formation. Molecular Genetics and Genomics. 2006;276:242–253. doi: 10.1007/s00438-006-0137-5. [DOI] [PubMed] [Google Scholar]
- Tezara W, Mitchell VJ, Driscoll SD, Lawlor DW. Water stress inhibits plant photosynthesis by decreasing coupling factor and ATP. Nature. 1999;401:914–917. [Google Scholar]
- Thomashow MF. Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annual Review of Plant Physiology and Plant Molecular Biology. 1999;50:571–599. doi: 10.1146/annurev.arplant.50.1.571. [DOI] [PubMed] [Google Scholar]
- Tran LS, Nakashima K, Sakuma Y, et al. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. The Plant Cell. 2004;16:2481–2498. doi: 10.1105/tpc.104.022699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran LS, Nakashima K, Sakuma Y, et al. Co-expression of the stress-inducible zinc finger homeodomain ZFHD1 and NAC transcription factors enhances expression of the ERD1 gene in Arabidopsis. The Plant Journal. 2007;49:46–63. doi: 10.1111/j.1365-313X.2006.02932.x. [DOI] [PubMed] [Google Scholar]
- Tsunoyama Y, Ishizaki Y, Morikawa K, et al. Blue light-induced transcription of plastid-encoded psbD gene is mediated by a nuclear-encoded transcription initiation factor, AtSig5. Proceedings of the National Academy of Sciences; USA. 2004. pp. 3304–3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehlein N, Lovisolo C, Siefritz F, Kaldenhoff R. The tobacco aquaporin NtAQP1 is a membrane CO2 pore with physiological functions. Nature. 2003;425:734–737. doi: 10.1038/nature02027. [DOI] [PubMed] [Google Scholar]
- Umezawa T, Yoshida R, Maruyama K, Yamaguchi-Shinozaki K, Shinozaki K. SRK2C, a SNF1-related protein kinase 2, improves drought tolerance by controlling stress-responsive gene expression in Arabidopsis thaliana.. Proceedings of the National Academy of Sciences; USA. 2004. pp. 17306–17311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proceedings of the National Academy of Sciences; USA. 2000. pp. 11632–11637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Buskirk HA, Thomashow MF. Arabidopsis transcription factors regulating cold acclimation. Physiologia Plantarum. 2006;126:72–80. [Google Scholar]
- Vannini C, Locatelli F, Bracale M, et al. Overexpression of the rice Osmyb4 gene increases chilling and freezing tolerance of Arabidopsis thaliana plants. The Plant Journal. 2004;37:115–127. doi: 10.1046/j.1365-313x.2003.01938.x. [DOI] [PubMed] [Google Scholar]
- Vogel JT, Zarka DG, Van Buskirk HA, Fowler SG, Thomashow MF. Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. The Plant Journal. 2005;41:195–211. doi: 10.1111/j.1365-313X.2004.02288.x. [DOI] [PubMed] [Google Scholar]
- Wang H, Ngwenyama N, Liu Y, Walker JC, Zhang S. Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. The Plant Cell. 2007;19:63–73. doi: 10.1105/tpc.106.048298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Tobin EM. Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell. 1998;93:1207–1217. doi: 10.1016/s0092-8674(00)81464-6. [DOI] [PubMed] [Google Scholar]
- Warren CR, Livingston NJ, Turpin DH. Water stress decreases the transfer conductance of Douglas-fir (Pseudotsuga menziesii) seedlings. Tree Physiology. 2004;24:971–979. doi: 10.1093/treephys/24.9.971. [DOI] [PubMed] [Google Scholar]
- Wasilewska A, Vlad F, Sirichandra C, et al. An update on abscisic acid signaling in plants and more. Molecular Plant. 2008;1:198–217. doi: 10.1093/mp/ssm022. [DOI] [PubMed] [Google Scholar]
- Wilkinson S, Clephan AL, Davies WJ. Rapid low temperature-induced stomatal closure occurs in cold-tolerant Commelina communis leaves but not in cold-sensitive tobacco leaves, via a mechanism that involves apoplastic calcium but not abscisic acid. Plant Physiology. 2001;126:1566–1578. doi: 10.1104/pp.126.4.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windhovel A, Hein I, Dabrowa R, Stockhaus J. Characterization of a novel class of plant homeodomain proteins that bind to the C4 phosphoenolpyruvate carboxylase gene of Flaveria trinervia. Plant Molecular Biology. 2001;45:201–214. doi: 10.1023/a:1006450005648. [DOI] [PubMed] [Google Scholar]
- Wong CE, Li Y, Labbe A, et al. Transcriptional profiling implicates novel interactions between abiotic stress and hormonal responses in Thellungiella, a close relative of Arabidopsis. Plant Physiology. 2006;140:1437–1450. doi: 10.1104/pp.105.070508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Schumaker KS, Zhu JK. Cell signaling during cold, drought, and salt stress. The Plant Cell. 2002;14(Suppl):S165–S183. doi: 10.1105/tpc.000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu ZS, Xia LQ, Chen M, et al. Isolation and molecular characterization of the Triticum aestivum L. ethylene-responsive factor 1 (TaERF1) that increases multiple stress tolerance. Plant Molecular Biology. 2007 doi: 10.1007/s11103-007-9237-9. [DOI] [PubMed] [Google Scholar]
- Xu ZZ, Zhou GS. Effects of water stress and high nocturnal temperature on photosynthesis and nitrogen level of a perennial grass Leymus chinensis. Plant and Soil. 2005;269:131–139. doi: 10.1007/s00425-006-0281-5. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. The Plant Cell. 1994;6:251–264. doi: 10.1105/tpc.6.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan SP, Zhang QY, Tang ZC, Su WA, Sun WN. Comparative proteomic analysis provides new insights into chilling stress responses in rice. Molecular and Cellular Proteomics. 2006;5:484–496. doi: 10.1074/mcp.M500251-MCP200. [DOI] [PubMed] [Google Scholar]
- Yanagisawa S, Sheen J. Involvement of maize DOF zinc finger proteins in tissue-specific and light-regulated gene expression. The Plant Cell. 1998;10:75–89. doi: 10.1105/tpc.10.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida R, Hobo T, Ichimura K, et al. ABA-activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis. Plant and Cell Physiology. 2002;43:1473–1483. doi: 10.1093/pcp/pcf188. [DOI] [PubMed] [Google Scholar]
- Zhang X, Wollenweber B, Jiang D, Liu F, Zhao J. Water deficits and heat shock effects on photosynthesis of a transgenic Arabidopsis thaliana constitutively expressing ABP9, a bZIP transcription factor. Journal of Experimental Botany. 2008;59:839–848. doi: 10.1093/jxb/erm364. [DOI] [PubMed] [Google Scholar]
- Zhu J, Shi H, Lee BH, et al. An Arabidopsis homeodomain transcription factor gene, HOS9, mediates cold tolerance through a CBF-independent pathway. Proceedings of the National Academy of Sciences; USA. 2004. pp. 9873–9878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Verslues PE, Zheng X, et al. HOS10 encodes an R2R3-type MYB transcription factor essential for cold acclimation in plants. Proceedings of the National Academy of Sciences; USA. 2005. pp. 9966–9971. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhu JK. Salt and drought stress signal transduction in plants. Annual Review of Plant Biology. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]