Summary
Dscam encodes a family of cell surface proteins required for establishing neural circuits in Drosophila. Alternative splicing of Drosophila Dscam can generate 19,008 distinct extracellular domains containing different combinations of three variable immunoglobulin domains. To test the binding properties of many Dscam isoforms, we developed a high-throughput ELISA-based binding assay. We provide evidence that 95% (>18,000) of Dscam isoforms exhibit striking isoform-specific homophilic binding. We demonstrate that each of the three variable domains binds to the same variable domain in an opposing isoform and identify the structural elements that mediate this self-binding of each domain. These studies demonstrate that self-binding domains can assemble in different combinations to generate an enormous family of homophilic binding proteins. We propose that this vast repertoire of Dscam recognition molecules is sufficient to provide each neuron with a unique identity and homotypic binding specificity, thereby allowing neuronal processes to distinguish between self and non-self.
Introduction
Neurons can distinguish between self and non-self in the peripheral nervous systems (PNS) of both vertebrates and invertebrates (Kidd and Condron, 2007; Zinn, 2007). Self-recognition occurs between sister neurites (i.e. axonal and dendritic branches extending from the same cell) and results in self-avoidance through contact-dependent repulsion (Baker and Macagno, 2007; Hughes et al., 2007; Matthews et al., 2007; Soba et al., 2007). Importantly, while sister neurites are repelled, non-sister neurites (i.e. from different cells) do not recognize one another as self and are not repelled from each other. In this way, self-avoidance ensures that sister branches segregate from one another to achieve uniform coverage of receptive fields while allowing neurites of different neurons to overlap.
Self-avoidance was first described for axonal processes in the leech (Kramer et al., 1985; Kramer and Kuwada, 1983; Kramer and Stent, 1985) and has subsequently been described for highly branched axonal processes in the Zebrafish (Sagasti et al., 2005) and for dendritic branches of neurons in Drosophila (Grueber et al., 2002; Hughes et al., 2007; Matthews et al., 2007; Soba et al., 2007). In the early 1980s Kramer and Kuwada proposed that self-avoidance is more generally required for patterning axonal and dendritic processes in the central nervous system (CNS) (Kramer and Kuwada, 1983). Given the vast number of neurons in the CNS with overlapping dendritic and axonal processes, it seems likely that many cell surface molecules would be necessary to allow processes to distinguish between self and non-self.
Previous studies led us to propose that the Ig superfamily proteins encoded by the Drosophila Down Syndrome Cell Adhesion Molecule (Dscam) gene are cell surface molecules that mediate self avoidance in the developing CNS (Neves et al., 2004; Wojtowicz et al., 2004; Zhan et al., 2004; Zipursky et al., 2006) (Wang et al., 2004; Wang et al., 2002). Dscam encodes 38,016 cell surface proteins with both variable and constant Ig domains (Figure 1A) (Schmucker et al., 2000). These isoforms are generated through alternative splicing. Each isoform contains a large ectodomain with 10 Ig domains and 6 fibronectin type III repeats. Of these, 3 Ig domains, Ig2, Ig3 and Ig7, contain variable sequences. Each variable domain is encoded by a block of alternatively utilized exons containing 12, 48 and 33 exons for Ig2, Ig3, and Ig7, respectively. As splicing within each block is independent of the other two, the Dscam locus encodes 19,008 different ectodomains (i.e. 12×48×33) linked to one of two alternative transmembrane domains. Previously we demonstrated that 11 Dscam isoforms exhibit homophilic binding (Wojtowicz et al., 2004). By contrast, little if any heterophilic binding was seen between 12 pairs examined. These preferential homophilic interactions occur in trans between molecules expressed on opposing cell surfaces (Matthews et al., 2007).
Figure 1. An ELISA-Based Assay for Dscam Binding Specificity.
(A) The Dscam gene contains four blocks of alternative exons coding for the first halves of Ig2 (red) and Ig3 (blue), all of Ig7 (green), and the transmembrane region (yellow). All isoforms have the same domain structure. Horseshoes represent Ig domains and black rectangles represent fibronectin type III domains. (B) Two models for preferential homophilic binding are shown. Previous studies(Wojtowicz et al., 2004), data presented here (see Figure 3) and the crystal structure by Meijers et al (Meijers et al., 2007) support the modular model for homophilic binding. In the lower part of the panel, we schematically represent how we envision modular binding occurs. (C) Schematic of ELISA-based binding assay to examine binding between ectodomains. Binding between Dscam ectodomain fused to AP (“receptor”) and Dscam ectodomain fused to Fc (“ligand”) is tested. Dscam-AP receptor is captured onto the plate by an anti-AP antibody. The binding of Dscam-Fc ligand to the receptor is detected by an anti-Fc antibody conjugated to horseradish peroxidase (HRP). HRP activity is measured using a colorimetric assay as a direct readout of binding between ligand and receptor (see Experimental Procedures).
Recent studies have demonstrated that Dscam is required for self-avoidance in the Drosophila PNS (Hughes et al., 2007; Matthews et al., 2007; Soba et al., 2007). Here the dendrites of four classes of dendritic arborization sensory neurons elaborate overlapping receptive fields. Whether sister dendrites and the dendrites of neighboring neurons grow across (i.e. overlap with) one another can be readily assessed in this system due to the two dimensional pattern of these processes within the body wall. While sister dendrites do not overlap, the dendrites of different neurons overlap extensively. In Dscam mutant neurons, sister dendrites lose self-avoidance and remain associated with each other. Gain-of-function studies support a model for Dscam-mediated homophilic repulsion. Ectopic expression of the same Dscam isoform in two neurons, which normally share overlapping receptive fields, causes their dendrites to recognize one another as self (Hughes et al., 2007; Matthews et al., 2007; Soba et al., 2007). This leads to avoidance of non-sister dendrites and the formation of mutually exclusive receptive fields. By contrast, deletion of the Dscam cytoplasmic domain results in adhesion of dendrites rather than repulsion (Matthews et al., 2007). Based on these studies, we proposed that Dscam-mediated repulsion proceeds in two steps. First, homophilic binding occurs between identical Dscam isoforms expressed on sister dendrites. And second, cytoplasmic domain dependent signaling promotes receptor downregulation and repulsion. Gain- and loss-of-function phenotypes are consistent with Dscam-mediated self-avoidance in both dendrites and axons in the developing CNS (Hattori et al., 2007; Hummel et al., 2003; Wang et al., 2004; Wang et al., 2002; Zhan et al., 2004; Zhu et al., 2006).
For Dscam to provide self recognition each neuron must express different isoforms. Studies by Chess and colleagues suggest that this is achieved in a stochastic fashion (Neves et al., 2004). Each neuron is proposed to express a random set of some 10-50 Dscam isoforms. Due to the large repertoire of Dscam isoforms, it is unlikely that processes from different neurons, which encounter each other within the developing brain, will express an appreciable number of the same isoforms. In this way, each neuron has a unique Dscam identity, which endows its processes with the ability to recognize self.
To ensure the fidelity of self-recognition, it is essential that the vast majority of Dscam isoforms exhibit isoform-specific homophilic binding. This degree of recognition specificity would be unprecedented outside the vertebrate immune system. Based on the homophilic binding properties of 11 isoforms, or 0.06% of the isoforms encoded by the locus, we speculated that isoform-specific homophilic binding is a feature shared by all 19,008 isoforms (Wojtowicz et al., 2004). We proposed that this specificity is achieved in a modular fashion: Ig2 in one molecule binds to an identical Ig2 in an opposing molecule, Ig3 binds to an identical Ig3 and Ig7 binds to an identical Ig7 (Figure 1B). Thus, isoforms sharing the same combination of variable domains will selectively bind to each other.
In this paper we present evidence that >18,000 of the 19,008 Dscam ectodomain isoforms exhibit homophilic binding specificity. We describe a high-throughput ELISA-based binding assay which allowed us to screen interactions between thousands of Dscam isoform pairs. We provide evidence that homophilic binding is achieved in a modular fashion with each variable domain exhibiting highly specific self-binding. By swapping specificity determinants between domains, we show that each of the three variable domains achieves specificity through variations in sequence within a discrete structural element. Together these data reveal a novel strategy for generating a vast array of cell recognition molecules with different specificities through the mixing and matching of variable modules.
Results
Development of a High-Throughput Dscam Binding Assay
In order to systematically test the binding specificities of Dscam isoforms, we developed an ELISA-based binding assay that provides an efficient method to assess interactions between thousands of isoform pairs (Figure 1C). The advantages of this assay, when compared with other binding assays, are outlined in Figure S1. The ELISA-based assay allows the binding properties of proteins to be assessed without purification and is quantitative over a 70-fold range (data not shown). Dscam ectodomains were tested for binding directly from the cell culture medium into which they are secreted following small-scale transient transfection. Ectodomains were generated in two different C-terminally fused versions to 1) alkaline phosphatase (Dscam-AP) and 2) human IgG1 Fc (Dscam-Fc). These proteins were quantified (see Experimental Procedures), the levels of Dscam-AP and Dscam-Fc were normalized and binding between them was tested in an ELISA plate format. Clustering of both Dscam-AP and Dscam-Fc was essential to detect binding (Figure S2), thereby suggesting that avidity compensates for low affinity interactions between monomers. Experiments were conducted in a grid format wherein homophilic interactions are tested along the grid diagonal while heterophilic interactions are tested off-diagonal. This assay has also been used to evaluate the binding properties of other cell surface proteins (M.Y. Pecot and S.L.Z., unpublished observations).
Dscam Variable Domains Support Homophilic Binding
Our previous studies (Wojtowicz et al., 2004), the data presented in this paper (see below) and the structure of Ig1-4 (containing Ig2.1 and Ig3.34 variable domains) determined by Meijers et al. (Meijers et al., 2007), strongly argue that binding specificity is determined in a modular fashion wherein binding at each pair of identical variable Ig domains occurs independent of the identity of the other variable Ig domains (Figure 1B). Thus, whether or not the majority of Dscam isoforms exhibit preferential homophilic binding specificity depends upon whether each of Dscam's 93 different variable domains (i.e. 12 different Ig2's, 48 different Ig3's and 33 different Ig7's) preferentially binds to an identical domain in an opposing molecule.
To assess the binding specificity of each variable domain, isoforms comprising the N-terminal 9 Ig domains and 1 fibronectin type III repeat (a region of the ectodomain which contains all three variable Ig domains and has previously been shown to be sufficient for homophilic binding (Wojtowicz et al., 2004)) were made as both Fc and AP fusions in three sets: 1) All 12 variable Ig2 domains with a constant Ig3 and Ig7; 2) 47 of the 48 Ig3 domains with a constant Ig2 and Ig7; and 3) All 33 Ig7 domains with a constant Ig2 and Ig3 (Figure 2A). Variable domain Ig3.11 was not tested as cDNAs encoding it were not identified. All pair-wise combinations within each set were tested for binding (i.e. 12×12 for Ig2, 47×47 for Ig3 and 33×33 for Ig7). As shown in Figure 2B-D, all 12 Ig2 variable domains, all 47 Ig3 variable domains, and 32 of 33 variable Ig7 domains support preferential homophilic binding. Ig7.33 was the only domain that did not support homophilic binding.
Figure 2. Testing All Dscam Variable Ig Domain Binding Specificities.
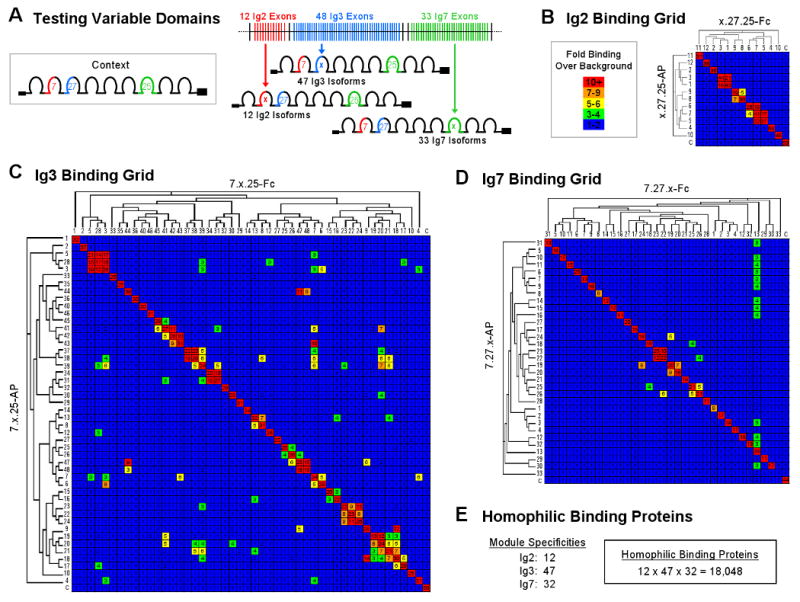
(A) Three sets of Dscam isoforms were generated as both AP and Fc fusions for the ELISA-based binding assay. Ig3.11, believed to be a pseudo exon, was not tested. (B-D) Binding properties of all Ig2, Ig3 and Ig7 variable domains. Each set of isoforms was tested for binding to all other members of the set. Variable Ig domains are arranged on the grid axes according to their sequence relatedness as shown in the dendrograms. Binding is indicated as fold over background by a color scale and the number in each block. The unrelated control isoform 1.30.30 (denoted “C”) was used to provide a value for background binding. The levels of all AP and Fc proteins were normalized (see Experimental Procedures). The average results of duplicate experiments are shown. (E) Summary of variable domain binding experiments showing the number of Ig2, Ig3 and Ig7 variable domains that exhibit preferential self-binding. From the self-binding properties of these domains, the number of preferential homophilic binding proteins encoded by the Dscam gene is estimated.
Some variable Ig domains supported heterophilic binding, which in all cases, was weaker than their homophilic binding. Heterophilic binding occurred largely between variable domains encoded by closely-related alternative exons as illustrated by the dendrograms in Figure 2. In some cases (i.e. 24/137) reciprocal binding pairs gave different results. The reason for this is unknown. Each heterophilic pair was tested in a reciprocal fashion (i.e. A-AP to B-Fc and B-AP to A-Fc). There were 8 examples of heterophilic binding out of the 132 Ig2 pairs tested (6%), 110 out of the 2,162 Ig3 pairs tested (5%), and 19 out of the 1,058 Ig7 pairs tested (2%).
Variable Ig Domain Binding is Modular
The Ig2, Ig3 and Ig7 grid binding experiments support the modular model for homophilic binding (i.e. three separate interfaces), however, they do not rule out the possibility that all three variable domains contribute jointly to the generation of a single interface (see Figure 1B). If the modular model is correct, then the binding profile of each variable domain (i.e. the domains with which it exhibits heterophilic interactions and the domains with which it does not) will be independent of the identity at the other two variable domains (Figure 3A). This will not be the case if all three variable domains combine to form a single interface.
Figure 3. Modular Variable Ig Domain Interactions Give Rise to Homophilic Binding Specificity.
(A) According to the modular model, the binding properties of each variable domain are independent of the identity of the other two variable domains. Therefore, domains should exhibit the same binding properties (i.e. the same pattern of homophilic and heterophilic binding) regardless of the identity of the other two variable domains (i.e. context). Different Ig7 variants are represented by different shapes and different shades of green. Different Ig2 and Ig3 variants are denoted by different shades of gray and different shapes. (B) The binding properties of a subset of Ig2, Ig3 and Ig7 variable domains were tested in two different contexts. The data in the left hand column (i.e. context #1) were taken from the grids in Figure 2. Binding is indicated as fold over background by a color scale and the number in each block. In context #1 experiments, the context #2 isoform was used as a negative control (denoted “C”) for binding and vice versa. The average results of duplicate experiments are shown.
We selected a subset of variable domains from the Ig2, Ig3 and Ig7 binding grids that support heterophilic binding to some variants but not to others and tested them for binding in a different context. For example, Ig2 domain variants were placed in the context of a different Ig3 and Ig7 from those used in the initial grid experiments in Figure 2B. We observed that the unique binding profiles of individual variable domains are exhibited independent of the context of the other two variable domains (as indicated by the identical grid patterns in Figure 3B). These findings strongly argue that binding specificity arises from a modular molecular strategy of Ig2 to Ig2, Ig3 to Ig3, and Ig7 to Ig7 binding.
In summary, the vast majority of variable domains (i.e. 91/93) exhibit preferential self-binding. This binding occurs independent of the identity at the other variable domains for all domains tested (i.e. 15/93). Modular, self-binding at each variable domain provides a molecular strategy for achieving homophilic specificity. These findings argue that the vast majority of Dscam isoforms (i.e. 12×47×32=18,048) exhibit preferential homophilic binding.
The Molecular Logic of Variable Ig Domain Self-Binding
To achieve highly-specific self-binding at each of the three variable domains, one molecular strategy for self recognition may have evolved which was then utilized by all three variable domains or, alternatively, each of the three variable domains may have evolved a unique strategy. We, therefore, sought to assess how self-binding is achieved at each of the three variable domains. We reasoned that 1) if closely-related domains bind weakly or not at all to one other, then one or more of the residues that differ between them must be critical for determining binding specificity and 2) specificity residues would be surface exposed and localized within a discrete region or element within the domain.
To identify candidate specificity-determining residues we used primary amino acid sequence analysis, the binding properties of closely related Ig domains, molecular modeling (for Ig7) and the crystal structure of Ig1-Ig4 (containing Ig2.1 and Ig3.34) solved by Meijers et al. (Meijers et al., 2007) and communicated to us prior to publication. If amino acid differences within the specificity-determining elements of the variable domains confer unique binding properties to isoforms, then “swapping” these differences between variable domains should swap their binding specificity (Figure 4A). The specificity swapping approach enabled us to identify sequences that are not only necessary but sufficient to confer unique binding properties to each of the three variable Ig domains. Representative examples of specificity swapping are shown for all three variable domains in Figures 4 and 5. Many additional examples are shown in supplementary Figures S4-S6.
Figure 4. Characterization of the Ig2 and Ig3 Specificity Elements.

(A) Experimental design to assess variable Ig domain specificity determinants. (B) Spacefill models of wild type variable Ig2 domains and A′ β-strand swapped Ig2 domains are shown. Different Ig2 variants are represented by different shades of red. The sequence alignment shows the A′ β-strand sequences (residues 105-114) for these Ig2 domains. The binding properties of isoforms containing wild type and strand-swapped Ig2 domains were tested. Binding is indicated as fold over background by a color scale and the number in each block. The unrelated control isoform 1.30.30 (denoted “C”) was used to provide a value for background binding. The average results of duplicate experiments are shown. Many additional strand swaps (>10) show the same results (see Figure S3). (C) Spacefill models of wild type variable Ig3 domains and A-A′ segment swapped Ig3 domains are shown. Different Ig3 variants are represented by different shades of blue. The sequence alignment shows the A-A′ segment sequences (residues 214-224) for these Ig3 domains. Other segment-swapped Ig3 domains also show swapped binding specificity (see Figure S4). Specificity strands predicted according to Ig2.1 and Ig3.34 interfaces in the Ig1-4 structure (Meijers et al., 2007).
Figure 5. The Specificity Element in Ig7 Comprises Multiple β-Strands.
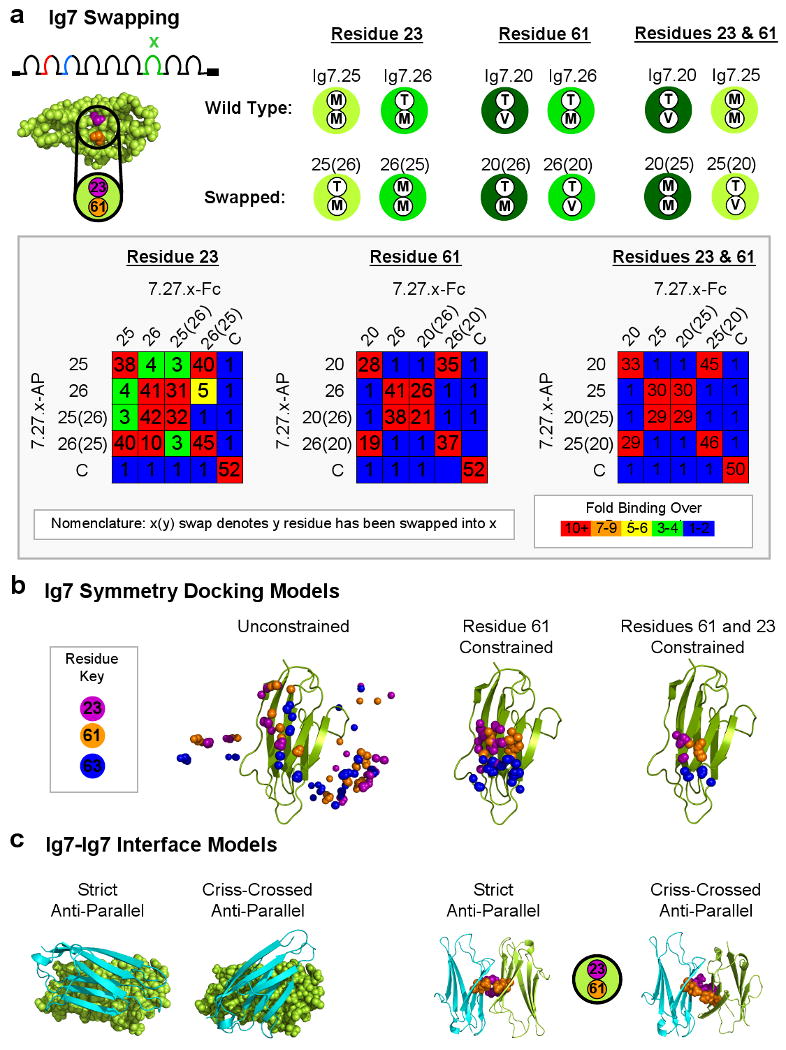
(A) Spacefill model of Ig7.25 generated using homology fold recognition servers (i.e. PHRYE, FUGUE, and ESyPred3D) showing two candidate specificity-determining residues (purple and orange) located on adjacent β-strands. A key shows the identity of these two residues in each wild type and residue-swapped Ig7 domain. Different Ig7 variants are represented by different shades of green. Residue 23 was swapped between Ig7.25 and Ig7.26 (note that these Ig7 variants engage in low levels of heterophilic binding with each other). Residue 61 was swapped between Ig7.20 and Ig7.26. Residues 23 and 61 were swapped between Ig7.20 and Ig7.25. The binding properties of isoforms containing wild-type and residue-swapped Ig7 domains were tested. Binding is indicated as fold over background by a color scale and the number in each block. The unrelated control isoform 1.30.30 (denoted “C”) was used to provide a value for background binding. The average results of duplicate experiments are shown. (B) Distribution of binding modes resulting after symmetrical protein-protein docking of homology models of Ig7.25. Ten different models generated using Rosetta (Das et al., 2007) are used as a starting point for symmetrical docking (Andre et al., 2007). The lowest energy models from each docking experiment are collected and the position of Cα atoms of residue 23 (purple) and 61 (orange) (in the center of the proposed binding region) in one monomer are shown. Residue 63 (blue) is included as a reference point. Left panel, Unconstrained docking models. Middle panel, Docking models constrained using a distance constraint of 8 Å between residue 61 in both monomers. Right panel, Docking models additionally filtered to include models in which residue 23 is also within 8.0 Å of itself in the other monomer. (C) Two docking models for Ig7.25 are shown. One Ig7.25 monomer is shown in cartoon (cyan) and the other is shown in spacefill (green). Two images on the right show the corresponding specificity-determining residues 23 and 61 at the center of the interface of the Ig7.25 monomers in both docking models.
Ig2 and Ig3 Specificity-Determining Residues Reside on Single Strands
Ig1-4 fragments of Dscam form dimers in the crystal structure as determined by Meijers et al. (Meijers et al., 2007). The binding interface is defined by an anti-parallel pair-wise matching of Ig2 to Ig2 and Ig3 to Ig3. The Ig2-Ig2 binding interface occurs between the same β-strand, called the A′ β-strand, in each monomer (Figure 4B). The Ig3-Ig3 interface occurs along a segment between two β-strands, called the A-A′ segment, which loops away from each domain much like a teapot handle (Figure 4C). We reasoned that if swapping Ig2 A′ β-strands and Ig3 A-A′ segments swaps binding specificity in diverse Ig2 and Ig3 variants then, in all likelihood, the variation within these discrete structural elements determines the specificity of Ig2 and Ig3 domains.
To assess whether residues along the A′ β-strand are sufficient to confer the binding specificity of Ig2 variants, we first swapped the surface exposed strand residues which form direct contacts between interacting Ig2 monomers in the crystal structure. Surprisingly, isoforms containing surface swapped Ig2 domains did not have swapped binding specificity. Instead, they exhibited both homophilic and robust, promiscuous heterophilic binding (Figure S3). We reasoned that the inward facing residues, along with the surface exposed residues, on the A′ β-strand contribute to specificity. Therefore, we swapped the entire A′ β-strand between pairs of variable Ig2 domains and the entire A-A′ segment between pairs of variable Ig3 domains. Strand swaps were performed for all 12 Ig2 variable domains and segment swaps were performed for 7 highly diverse Ig3 domains sharing as low as 22% amino acid sequence identity.
All entire strand and segment swapped isoforms bound to isoforms with which they shared strand or segment identity regardless of the identity of the remainder of the variable domain (i.e. domain backbone) (Figures 4B, C, S4, S5). They bound to themselves (i.e. homophilically). They did not bind to the wild type isoform from which they were generated (i.e. isoform with the same domain backbone but different strand or segment). Importantly, swapped isoforms now bound to the wild type isoform with which they shared strand or segment identity in spite of a differing domain backbone. This argues that the A′ β-strand and the A-A′ segment are the specificity elements for most, if not all, Ig2 and Ig3 domains, respectively. Thus, these short contiguous polypeptide stretches are both necessary and sufficient for homophilic binding specificity within the Ig2 and Ig3 domains.
Ig7 Specificity-Determining Residues Reside on Multiple Strands
As a crystal structure of Ig7 is not available, we used a combined biochemical and modeling approach to define the specificity determinants in Ig7. Candidate Ig7 specificity-determining residues were identified using three closely-related Ig7 domains which exhibited little or no heterophilic binding to each other. There are 7-9 amino acid differences between each pair. They all differ at residue 23, 61, or both (Figure 5A). Molecular modeling using homology fold recognition servers (i.e. PHRYE, FUGUE, and ESyPred3D) placed residues 23 and 61 adjacent to each other on neighboring β-strands on one face of the domain. To test whether they are specificity determining residues, we swapped them between domains and assessed the binding properties of new isoforms resulting from the swaps. Swapped isoforms exhibited the following binding properties: 1) Each bound to itself (i.e. homophilic binding); 2) Each lost the ability to bind to the wild-type isoform from which it was derived (i.e. sharing the same domain backbone but a different specificity-determining residue(s)) and; 3) Each acquired the ability to bind to the other wild type isoform which contained identical residues at the swapped positions. Thus, swapping these residues swapped the binding specificity of Ig7 domains. Interestingly, for Ig7 variants that differed at both residues 23 and 61, swapping only one residue did not swap binding specificity but instead generated Ig7 variants with new preferential homophilic binding specificity (Figure S6A). These data demonstrate that residues 23 and 61 are Ig7 specificity-determining residues for these Ig7 variants and argue that they are localized at the Ig7-Ig7 interface.
Molecular Modeling of the Ig7-Ig7 Interface
To gain insight into how self-binding of Ig7 variants may occur, we used the Rosetta (Das et al., 2007) program to build homology models of two Ig7 variants (Ig7.25 and Ig7.20) and then generated symmetric homodimeric complexes through protein-protein docking (Andre et al., 2007). Models were filtered based on distance constraints forcing contacts between specificity-determining residues 23 and/or 61 across the interface and the lowest energy structures were selected (Figure 5B and S7). The conformations adopted by these low energy structures range from a strict anti-parallel orientation, which is most favored, to a criss-crossed anti-parallel orientation (Figure 5C). Both binding modes place amino acids 23 and 61 adjacent to each other on neighboring β-strands in the center of the Ig7-Ig7 interface. The docked complexes provide a basis for interpreting the experimental data, and can be evaluated and further refined with input of additional experimental constraints.
The docked Ig7 complexes suggest a speculative model (Figure S8) for the physical origins of self-binding for the Ig7 variants differing at residues 23 and/or 61. Both of these switches involve a change of a Met residue to a β-branched amino acid, either Thr or Val. Inspection of the docked complexes shows that interdigitated Met-Met pairs and face-on-face β-branched Val-Val or Thr-Thr residue pairs can pack quite well, while heterotypic Met-Val or Met-Thr pairs are considerably less optimal as the β-branching prevents interdigitation of the long linear Met residue.
We sought to assess whether the docked Ig7.25 and Ig7.20 complexes can be generalized to other Ig7 domains. Not all Ig7 pairs differ at residues 23 and 61 suggesting that additional residues at the Ig7-Ig7 interface contribute to binding specificity. By swapping interface residues that differ between other pairs of Ig7 variants, additional residues on three neighboring β-strands at the docking interface were shown to affect binding. For two pairs of Ig7 variants we observed a complete swap in binding specificity indicating that some or all of the swapped residues are at the interface. For two other pairs of Ig7 variants binding was affected, but specificity was not swapped. In these cases we observed partial swaps in specificity or reductions in homophilic binding suggesting that some or all of these residues are also at the interface (Figure S6B and data not shown). As six of the Ig7 domains used in these swapping experiments are not closely related to the Ig7 domains used for modeling, it is likely that the modeled interface represents the interface for many, if not all, variable Ig7 domains.
Discussion
In this paper we provide evidence that the vast majority of Dscam isoforms exhibit preferential homophilic binding and thus provide an enormous repertoire of cell recognition proteins. We demonstrate that preferential homophilic binding specificity arises through a molecular strategy that involves modular interactions between distinct structural elements within each of the three variable Ig domains. It is likely that the in vitro binding properties we have described here translate into cell recognition events in vivo as two populations of cells in culture expressing isoforms differing by only a single amino acid at the Ig7 interface readily segregate into isoform-specific aggregates (Matthews et al., 2007).
How is the remarkable binding specificity of Dscam isoforms achieved?
The proliferation of different homophilic specificities demonstrated in this paper is, to our knowledge, unprecedented. While there are other families of homophilic recognition proteins, they are several orders of magnitude smaller than Dscam (e.g. ∼20 different proteins for the classical cadherins in vertebrates) (Uemura, 1998). Structural and biochemical studies have provided insights into the mechanisms underlying cadherin specificity (Boggon et al., 2002; Patel et al., 2006). Here specificity is determined by sequences within a single self-binding domain. This self-binding domain mediates homophilic binding between opposing molecules. The domains orient in a parallel fashion and a strand is swapped between them to stabilize the homophilic interaction. Our studies reported here and the crystal structure reported by Miejers et al (Meijers et al., 2007) argue that the structural basis for Dscam isoform-specific homophilic binding is fundamentally different. In Dscam, binding specificity is determined by three self-binding domains, each oriented in an anti-parallel fashion.
Deciphering the molecular mechanisms that underlie this multitude of anti-parallel self-binding domains is a fascinating challenge for structural biophysics. In the crystal structure of Ig2.1 and Ig3.34 (Meijers et al., 2007), the anti-parallel interfaces are formed between opposing A′ β-strands and A-A′ segments, respectively. Interactions between these anti-parallel sequences form a 2-fold symmetric network of hydrogen bonds and salt bridges. When compared to one another, the 12 different Ig2 A′ β-strands and the 47 different Ig3 A-A′ segments have multiple amino acid differences. Our findings strongly suggest that they all mediate self-binding. We propose that self-binding for all Ig2 and Ig3 variable domains is specified by unique 2-fold symmetric networks of interactions between amino acids within these short polypeptide sequences.
Modeling and mutagenesis studies suggest that Ig7 domains also associate in an anti-parallel fashion. In contrast to Ig2 and Ig3, Ig7 specificity is determined by a unique combination of residues on multiple β-strands comprising one face of the Ig domain. If the Ig7 interface is oriented in a strictly anti-parallel fashion, then like Ig2 and Ig3, binding will be determined by a 2-fold symmetric network of interactions. Whether the Ig7 interface is oriented in a strictly anti-parallel or in a criss-cross anti-parallel orientation, it is surprising that swapping a single amino acid at the center of the interface swaps binding specificity. In the case of heterophilic interactions in other proteins, alterations in binding specificity involve compensatory changes such as swapping a big-small residue pair for a small-big residue pair. This cannot be the case for changes in binding specificity at the center of symmetric homodimeric interfaces as the same residue change occurs in both monomers. One way specificity could be achieved is illustrated in Figure S8. Here, the complete specificity swaps brought about by changes at the center of the interface (i.e. Met to Val and Met to Thr; see Figure 5 and S6), could result from favorable interactions between opposing long/thin Met residues which can interdigitate and between opposing short/fat β-branched Val and Thr residues which can pack face-on-face. By contrast, unfavorable packing between opposed Met and β-branched Val or Thr residues disfavor heterodimer formation.
How can a single residue change in only one of the three variable Ig domains prevent binding between isoforms? Differences in binding specificity between isoforms differing at only a single residue suggest that binding energy differences favoring self-binding need not be large. As detection of homophilic binding specificity requires oligomerization, avidity effects arising from the multivalency of Dscam interactions magnify these small differences in binding energy. That oligomerization of weakly interacting molecules can lead to strong binding has previously been described for other cell surface molecules (Shapiro et al., 1995; Shapiro et al., 1996). We propose that the vast repertoire of Dscam homophilic binding specificities arises through a combination of multiple low energy interactions at each of the three variable domains and avidity.
The evolution of Dscam homophilic binding specificities
As thousands of neurites from many different neurons intermingle in densely packed regions of the brain, the vast majority of Dscam isoforms must exhibit homophilic binding specificity for Dscam to mediate the phenomenon of self-avoidance. The binding studies presented here show that 1) Dscam homophilic binding is modular, 2) 91 of 93 variable domains exhibit preferential self-binding and 3) only a small fraction (i.e. 2-6%) of variable domains show any non-self binding. Together these data argue that >18,000 Dscam isoforms exhibit preferential homophilic binding. This molecular self-recognition provides a robust mechanism for neurite self-recognition and avoidance in the developing brain. In this way, Dscam molecules function as identification tags for individual neurons. Whether Dscam isoforms also function as tags to mediate interactions between neurons, as envisioned by Sperry, remains to be critically addressed (Sperry, 1963).
How did so many self-binding variable domains arise? Each block of alternative exons encoding the Dscam variable domains presumably evolved by exon duplication followed by sequence divergence (Graveley et al., 2004). Isoforms containing new variable domains may have lost the ability to bind to isoforms containing the variable domain from which they diverged, exhibited promiscuous binding, or acquired a new homophilic binding specificity. Indeed, in our swapping experiments we generated variable domains that exhibited each of these properties. These data strongly argue that alternative exons diverged until self-binding variable domains arose. We propose that the essential role for efficient self-avoidance in the CNS provided the selective pressure to maintain them.
Experimental Procedures
Plasmid Construction
Generation of Dscam-Fc and Dscam-AP constructs
Dscam 7.27.25 EC10 containing the N-terminal 9 Ig domains followed by 1 fibronectin type III repeat was subcloned into the pIB-V5/His vector (Invitrogen) in frame with 1) the Fc region of human IgG1 or 2) human placental alkaline phosphatase (AP) as follows: 1) The vast majority of the pIB-V5/His MCS was destroyed by double restriction digestion with HindIII and EcoRV, Klenow treatment and religation to generate pIBΔMCS; 2) The Fc region of human IgG1 was PCR amplified from an EST using an upstream primer containing 5′ XhoI and SpeI sites and a downstream primer containing a 5′ XbaI site; 3) AP was PCR amplified from APtag-2 (Flanagan and Leder, 1990) using primers containing the same sites; 4) Fc and AP PCR fragments were digested with XhoI and XbaI and subcloned into XhoI and XbaI sites in the MCS of pIBΔMCS to generate pIBΔMCS-Fc and pIBΔMCS -AP, respectively; and 5) Dscam 7.27.25 EC10 (including signal peptide) was PCR amplified using an upstream primer containing a 5′ NotI site and a downstream primer containing a 5′ SpeI site and subcloned in-frame and upstream of Fc and AP to generate pIBΔMCS 7.27.25 EC10-Fc and pIBΔMCS 7.27.25 EC10-AP, respectively.
To generate a universal pIBΔMCS 7.27.25 EC10-Fc vector that would allow variable Dscam exons to be easily swapped, unique SacI and HpaI restriction sites were introduced by silent point mutations in exons 5 and 7, respectively, using site-directed mutagenesis. Exon 3 contains a unique AatII site, thereby allowing the swapping of variable exon 4 by AatII-SacI double digest. SacI-HpaI sites in exons 5 and 7 were used to swap variable exon 6. The two XhoI sites in pIBΔMCS 7.27.25 EC10-Fc reside in exons 8 and 10 flanking variable exon 9 and were used to swap exon 9, followed by PCR amplification to check for orientation. Nearly all of the variable exons were present in Dscam exon3-10 cDNA library clones (Schmucker et al., 2000; Zhan et al., 2004) (D. Hattori and S.L.Z. unpublished) and those not present were amplified from cDNA in two PCR steps using primers that annealed within the variable exon. 92 out of the 93 Ig2 (exon 4), Ig3 (exon 6) and Ig7 (exon 9) encoding variable exons were obtained. Each variable exon was PCR amplified using primers containing the unique restriction sites described above and purified to generate a library of variable exons for subcloning into pIBΔMCS 7.27.25 EC10-Fc to generate the Ig2, Ig3, and Ig7 series containing all variable exons.
To generate Dscam-AP constructs, the EC10 region from the Dscam-Fc clones for each isoform was subsequently subcloned into the pIBΔMCS-AP vector by NotI-SpeI double digestion. All isoforms used in this study were prepared by swapping exons into the universal pIBΔMCS 7.27.25 EC10-Fc vector. All mutagenesis was performed using the QuickChange Site-Directed Mutagesis kit (Stratagene) according to the manufacturer's protocol. Primer sequences available upon request.
Protein Expression, Detection and Quantification
Dscam EC10-Fc and Dscam EC10-AP proteins were expressed by transient transfection of Drosophila S2 cells as follows: 5×106 cells/well were seeded in 6-well cell culture plates in 1.6 ml Schneider's Drosophila medium (Invitrogen) containing 10% Ultra-Low IgG FCS (Invitrogen) and pen-strep (Invitrogen) at 25° C and allowed to grow overnight. Cells were transfected with 0.8 μg pIBΔMCS Dscam EC10-Fc or pIBΔMCS Dscam EC10-AP using Effectene reagents (6.4 μl Enhancer and 40 μl Effectene; Qiagen) and incubated for 6-7 days at 25° C during which time secreted Dscam EC10-Fc and Dscam EC10–AP proteins accumulated in the culture medium. Culture medium was harvested, spun 10 min at 1,000 rpm in a tabletop centrifuge to pellet cells, filtered (0.2 μm) and stored at 4° C. Proteins (including AP activity) remained stable in culture medium at 4° C for >2 years without appreciable loss of binding activity.
Dscam EC10-Fc and Dscam EC10-AP proteins were analyzed by SDS-PAGE and immunoblotting for Fc or AP to test for expression and size. Quantitative immunoblotting using a Typhoon scanner (GE Healthcare) and ImageQuant software was performed with known concentrations of purified Dscam EC10-Fc (purified as in (Wojtowicz et al., 2004)) to determine the concentrations of Dscam EC10-Fc in culture media. The levels of Dscam EC10-AP in culture media were quantified by AP enzymatic activity following addition of substrate (Pierce) relative to the activity of purified calf intestinal phosphatase (CIP) (Pierce). Absorbance at 605 nm was measured using the SpectraMax 340 PC microplate spectrophotometer (Molecular Devices). Following quantification, protein levels were normalized to 8 ng/ml (Dscam EC10-Fc) and 4 U/μl (Dscam EC10-AP; where a unit (U) is equivalent to the activity of 10 pg purified CIP) by addition of mock transfected culture media.
ELISA-Based Binding Assay
Binding between Dscam EC10-Fc and Dscam EC10-AP was tested in an ELISA-based format. Each well of Nunc Immunosorp 96-well plates was incubated overnight at 4° C with 50 μl 3 μg/ml of a mouse antibody to AP (IgGAb-1 clone 8B6.18) (NeoMarkers) in 1× PBS, pH 7.4. Wells were washed 3 times for 1-3 min at room temperature with 400 μl 1× PBS, pH 7.4 + 0.05% Tween-20 (PBST). Wells were incubated for 1-2 hr at room temperature with 400 μl 1% casein in 1× PBS, pH 7.4. The 1% casein block was removed. This was followed by the addition of 20 μl Dscam EC10-Fc (8 ng/ul) and 20 μl Dscam EC10-AP (4 u/μl) culture medium containing monoclonal mouse anti-human IgG1-HRP (2 μg/ml; Serotec). Plates were covered and incubated 4 hr at room temperature protected from light. Wells were washed 3 times for 1-3 min at room temperature with PBST. 1-Step Ultra TMB-ELISA HRP substrate (100 μl; Pierce) equilibrated to room temperature was added and plates were incubated 1 hr at room temperature. Absorbance at 450 nm was measured using the SpectraMax 340 PC microplate spectrophotometer (Molecular Devices).
All assays were done under the same conditions. Conditions were established using previously characterized isoforms (Wojtowicz et al., 2004) such that homophilic binding was 20-60 fold higher than background. Binding between isoforms differing at all three variable Ig domains was equivalent to background levels obtained in the absence of ligand. Therefore, in all binding assays an unrelated control isoform was included that differed by at least two, and usually three, variable Ig domains from those tested. The binding level between the control isoform and the tested isoforms provided an average background binding value. The concentrations of Dscam EC10-Fc, Dscam EC10-AP and monoclonal mouse anti-human IgG1-HRP used in the assay were optimized so that binding between two previously characterized isoforms that are identical at variable domains Ig2 and Ig3 and differ only by 7 residues in variable domain Ig7 (i.e. 7.27.25 and 7.27.26) was readily observed (i.e. ∼5-fold over background).
Immunoprecipitation
Pull-downs were preformed in 200 μl RIPA buffer (150 mM NaCl, 10% glycerol, 0.5% deoxycholate, 50 mM Tris, Ph 8.0, 0.4 mM EDTA, pH 8.0, 1% NP-40, 0.1% SDS) and contained all or some of the following: 20 μl ImmunoPure Immobilized Protein G (Pierce), 150 ng/μl purified 1.30.30 Dscam EC16-Fc containing the entire extracellular domain of Dscam fused to Fc (purified as in (Wojtowicz et al., 2004)), 500 ng/μl purified 1.30.30 or 7.27.25 Dscam EC8-His containing the first eight immunoglobulin domains of the extracellular domain (a region sufficient for homophilic bidning) fused to a 6×His tag (Caltech Protein Expression Facility), and 10 ng/μl α-His conjugated with HRP (Penta-His HRP; Qiagen; Lot # 12183222). Pull-downs were incubated overnight at 4° C on a Nutator and washed three times for 5 min with 200 μl RIPA buffer at 4° C on a Nutator. Wash buffer was removed, 25 μl 2× SDS loading buffer (containing β-mercaptoethanol) was added, samples were boiled 5 min and 10 μl was analyzed on 8% SDS-PAGE gels. Semi-dry transfer was preformed for 35 min at 15V onto Immobilon-P membrane (Miilipore). Membranes were blocked in 1× TBST containing 5% milk (α-Fc immunoblot) or 10 mM Tris, pH 7.7, 150 mM NaCl, 1% casein, 0.1% Tween-20 (α-His IgG immunoblot) for 1 hr at room temperature. Immunoblotting was preformed in blocking buffer with 1:5000 goat α-human IgG-HRP (Bio-Rad) or α-His IgG-HRP (Penta-His HRP; Qiagen; Lot # 12183222). Membranes were washed 6 times 10 min in blocking buffer, rinsed with 1× TBST, incubated 5 min with SuperSignal West Pico Chemiluminescent Substrate (Pierce), exposed and developed.
Molecular Modeling
Homology modeling
The amino-acid sequence to be modeled was threaded onto the best template backbone, and regions containing insertions or deletions relative to the template were built using an improved version of the Rosetta loop modeling protocol (Das et al., 2007) which incorporates CCD closure followed by gradient based energy minimization. Side chains were modeled using a combinatorial search through an extended version of the Dunbrack rotamer library supplemented with side-chain conformations from the template using Monte Carlo sampling. Full-chain, all-atom refinement was then carried out with the Rosetta all-atom energies. Several thousand structure models were generated using the above protocol, and the structures with the lowest Rosetta all-atom energies were used as the predicted models.
Protein-Protein Docking
For Ig7.20 and Ig7.25 one member from each of the 10 lowest scoring clusters from homology modeling was selected for symmetrical protein-protein docking in order to produce models of homophilic interactions. The method carries out simultaneous optimization of side-chain rigid-body degrees of freedom while restricting the search space to symmetrical conformations of backbone and side-chains (Andre et al., 2007). Around 105 models were generated per starting. In a second simulation, models were given an energy bonus score if a distance of less than 8 Å between residues 61 across the interface was observed and only models meeting the distance criteria were collected. These models were then filtered to select those models that additionally had less than 8 Å between residues 23 across the interface and to find models that place both residue pairs together.
Supplementary Material
Acknowledgments
We especially wish to thank Jia-huai Wang and Dietmar Schmucker for communication of data prior to publication and for helpful discussions. We thank Owen Witte, David Eisenberg, Mike Sawaya and members of the Zipursky lab for comments on the manuscript and helpful discussion. We acknowledge Owen Witte for use of the SpectraMax 340 PC microplate spectrophotometer. We acknowledge Daisuke Hattori for providing purified Dscam EC10-Fc protein and Dscam cDNA clones and Kartik Pappu for help with binding data presentation. We thank Dr. Jost Vielmetter of the Caltech Protein Expression Facility for production of Dscam-His protein. The authors would like to note that all biochemistry experiments were performed in the laboratory of S.L.Z. and all Rosetta modeling and protein-protein docking was performed in the laboratory of D.B. All mutageneses and binding studies with mutant proteins were performed by W.W. This work was supported by an NIH predoctoral training grant (USPHS National Research Service Award GM07185, W.M.M.), The Knut and Alice Wallenberg Foundation (I.A.), The American Leukemia and Lymphoma Society Fellowship (B.Q.). S.L.Z. and D.B. are investigators of the Howard Hughes Medical Institute.
Footnotes
Supplemental Data: Supplemental data include eight figures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andre I, Bradley P, Wang C, Baker D. Prediction of the Structure of Symmetrical Protein Assemblies. PNAS. 2007 doi: 10.1073/pnas.0702626104. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker MW, Macagno ER. In vivo imaging of growth cone and filopodial dynamics: evidence for contact-mediated retraction of filopodia leading to the tiling of sibling processes. J Comp Neurol. 2007;500:850–862. doi: 10.1002/cne.21228. [DOI] [PubMed] [Google Scholar]
- Boggon TJ, Murray J, Chappuis-Flament S, Wong E, Gumbiner BM, Shapiro L. C-cadherin ectodomain structure and implications for cell adhesion mechanisms. Science. 2002;296:1308–1313. doi: 10.1126/science.1071559. [DOI] [PubMed] [Google Scholar]
- Das R, Qian B, Raman S, Vernon R, Thompson J, Bradley P, Khare S, Tyka MD, Bhat D, Chivian DC, et al. Structure prediction for CASP7 targets using extensive all-atom refinement with Rosetta@home. Proteins: Structure, Function and Bioinformatics. 2007 doi: 10.1002/prot.21636. in press. [DOI] [PubMed] [Google Scholar]
- Flanagan JG, Leder P. The kit ligand: a cell surface molecule altered in steel mutant fibroblasts. Cell. 1990;63:185–194. doi: 10.1016/0092-8674(90)90299-t. [DOI] [PubMed] [Google Scholar]
- Graveley BR, Kaur A, Gunning D, Zipursky SL, Rowen L, Clemens JC. The organization and evolution of the dipteran and hymenopteran Down syndrome cell adhesion molecule (Dscam) genes. RNA. 2004;10:1499–1506. doi: 10.1261/rna.7105504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grueber WB, Jan LY, Jan YN. Tiling of the Drosophila epidermis by multidendritic sensory neurons. Development. 2002;129:2867–2878. doi: 10.1242/dev.129.12.2867. [DOI] [PubMed] [Google Scholar]
- Hattori D, Demir E, Kim HW, Viragh E, Zipursky SL, Dickson BJ. Dscam Diversity is Essential for Neuronal Wiring and Self-Recognition. Nature. 2007 doi: 10.1038/nature06099. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes ME, Bortnick R, Tsubouchi A, Baumer P, Kondo M, Uemura T, Schmucker D. Homophilic Dscam interactions control complex dendrite morphogenesis. Neuron. 2007;54:417–427. doi: 10.1016/j.neuron.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel T, Vasconcelos ML, Clemens JC, Fishilevich Y, Vosshall LB, Zipursky SL. Axonal targeting of olfactory receptor neurons in Drosophila is controlled by dscam. Neuron. 2003;37:221–231. doi: 10.1016/s0896-6273(02)01183-2. [DOI] [PubMed] [Google Scholar]
- Kidd T, Condron B. Avoiding the SCAMs. Neuron. 2007;54:350–352. doi: 10.1016/j.neuron.2007.04.018. [DOI] [PubMed] [Google Scholar]
- Kramer AP, Goldman JR, Stent GS. Developmental arborization of sensory neurons in the leech Haementeria ghilianii. I. Origin of natural variations in the branching pattern. J Neurosci. 1985;5:759–767. doi: 10.1523/JNEUROSCI.05-03-00759.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer AP, Kuwada JY. Formation of the receptive fields of leech mechanosensory neurons during embryonic development. J Neurosci. 1983;3:2474–2486. doi: 10.1523/JNEUROSCI.03-12-02474.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer AP, Stent GS. Developmental arborization of sensory neurons in the leech Haementeria ghilianii. II. Experimentally induced variations in the branching pattern. J Neurosci. 1985;5:768–775. doi: 10.1523/JNEUROSCI.05-03-00768.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews BJ, Kim ME, Flanagan JJ, Hattori D, Clemens JC, Zipursky SL, Grueber WB. Dendrite sef-avoidance is controlled by Dscam. Cell. 2007;129:593–604. doi: 10.1016/j.cell.2007.04.013. [DOI] [PubMed] [Google Scholar]
- Meijers R, Puettmann-Holgado R, Skiniotis G, Liu JH, Walz T, Wang JH, Schmucker D. Structural Basis of Dscam Isoform Specificity. Nature. 2007 doi: 10.1038/nature06147. submitted. [DOI] [PubMed] [Google Scholar]
- Neves G, Zucker J, Daly M, Chess A. Stochastic yet biased expression of multiple Dscam splice variants by individual cells. Nat Genet. 2004;36:240–246. doi: 10.1038/ng1299. [DOI] [PubMed] [Google Scholar]
- Patel SD, Ciatto C, Chen CP, Bahna F, Rajebhosale M, Arkus N, Schieren I, Jessell TM, Honig B, Price SR, Shapiro L. Type II cadherin ectodomain structures: implications for classical cadherin specificity. Cell. 2006;124:1255–1268. doi: 10.1016/j.cell.2005.12.046. [DOI] [PubMed] [Google Scholar]
- Sagasti A, Guido MR, Raible DW, Schier AF. Repulsive interactions shape the morphologies and functional arrangement of zebrafish peripheral sensory arbors. Curr Biol. 2005;15:804–814. doi: 10.1016/j.cub.2005.03.048. [DOI] [PubMed] [Google Scholar]
- Schmucker D, Clemens JC, Shu H, Worby CA, Xiao J, Muda M, Dixon JE, Zipursky SL. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell. 2000;101:671–684. doi: 10.1016/s0092-8674(00)80878-8. [DOI] [PubMed] [Google Scholar]
- Shapiro L, Fannon AM, Kwong PD, Thompson A, Lehmann MS, Grubel G, Legrand JF, Als-Nielsen J, Colman DR, Hendrickson WA. Structural basis of adhesion by cadherins. Nature. 1995;374:327–337. doi: 10.1038/374327a0. [DOI] [PubMed] [Google Scholar]
- Shapiro L, Doyle JP, Hensley P, Colman DR, Hendrickson WA. Crystal structure of the extracellular domain from P0, the major structural protein of pheripheral nerve myelin. Neuron. 1996;17:435–449. doi: 10.1016/s0896-6273(00)80176-2. [DOI] [PubMed] [Google Scholar]
- Soba P, Zhu S, Emoto K, Younger S, Yang SJ, Yu HH, Lee T, Jan LY, Jan YN. Drosophila sensory neurons require Dscam for dendritic self-avoidance and proper dendritic field organization. Neuron. 2007;54:403–416. doi: 10.1016/j.neuron.2007.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperry RW. Chemoaffinity in the orderly growth of nerve fiber patterns and connections. Proc Natl Acad Sci USA. 1963;50:703–710. doi: 10.1073/pnas.50.4.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T. The cadherin superfamily at the synapse: more members, more missions. Cell. 1998;93:1095–1098. doi: 10.1016/s0092-8674(00)81452-x. [DOI] [PubMed] [Google Scholar]
- Wang J, Ma X, Yang JS, Zheng X, Zugates CT, Lee CH, Lee T. Transmembrane/juxtamembrane domain-dependent Dscam distribution and function during mushroom body neuronal morphogenesis. Neuron. 2004;43:663–672. doi: 10.1016/j.neuron.2004.06.033. [DOI] [PubMed] [Google Scholar]
- Wang J, Zugates CT, Liang IH, Lee CH, Lee T. Drosophila Dscam is required for divergent segregation of sister branches and suppresses ectopic bifurcation of axons. Neuron. 2002;33:559–571. doi: 10.1016/s0896-6273(02)00570-6. [DOI] [PubMed] [Google Scholar]
- Wojtowicz WM, Flanagan JJ, Millard SS, Zipursky SL, Clemens JC. Alternative splicing of Drosophila Dscam generates axon guidance receptors that exhibit isoform-specific homophilic binding. Cell. 2004;118:619–633. doi: 10.1016/j.cell.2004.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan XL, Clemens JC, Neves G, Hattori D, Flanagan JJ, Hummel T, Vasconcelos ML, Chess A, Zipursky SL. Analysis of Dscam diversity in regulating axon guidance in Drosophila mushroom bodies. Neuron. 2004;43:673–686. doi: 10.1016/j.neuron.2004.07.020. [DOI] [PubMed] [Google Scholar]
- Zhu H, Hummel T, Clemens JC, Berdnik D, Zipursky SL, Luo L. Dendritic patterning by Dscam and synaptic partner matching in the Drosophila antennal lobe. Nat Neurosci. 2006;9:349–355. doi: 10.1038/nn1652. [DOI] [PubMed] [Google Scholar]
- Zinn K. Dscam and neuronal uniqueness. Cell. 2007;129:455–456. doi: 10.1016/j.cell.2007.04.024. [DOI] [PubMed] [Google Scholar]
- Zipursky SL, Wojtowicz WM, Hattori D. Got diversity? Wiring the fly brain with Dscam. Trends Biochem Sci. 2006;31:581–588. doi: 10.1016/j.tibs.2006.08.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.




