Abstract
Alpha-synuclein has been implicated in Parkinson’s disease, yet the mechanism by which alpha-synuclein causes cell injury is not understood. Using a transgenic mouse model, we evaluated the effect of alpha-synuclein overexpression on gene expression in the substantia nigra. Nigral mRNA from wildtype and alpha-synuclein transgenic mice was analyzed using Affymetrix gene arrays. At three months, before pathological changes are apparent, we observed modest alterations in gene expression. However, nearly 200 genes were altered in expression at nine months, when degenerative changes are more apparent. Functional genomic analysis revealed that the genes altered at nine months were predominantly involved in gene transcription. As in human Parkinson’s disease, gene expression changes in the transgenic model were also modulated by gender. These data demonstrate that alterations of gene expression are widespread in this animal model, and suggest that transcriptional dysregulation may be a disease mechanism that can be targeted therapeutically.
Keywords: Parkinson’s disease, alpha-synuclein, microarray, substantia nigra, laser capture microdissection, mouse, transcription
Introduction
Parkinson’s disease (PD) is a debilitating neurological disorder associated with dopaminergic cell loss in the substantia nigra (SN). Familial PD cases have been linked to several genes, including α-synuclein (α-syn), a 140-amino-acid protein that may function in neurotransmitter release (Abeliovich et al., 2000; Murphy et al., 2000). Families with mutated α-syn exhibit autosomal dominant PD (Athanassiadou et al., 1999; Kruger et al., 1998; Polymeropoulos et al., 1997; Zarranz et al., 2004), and gene multiplication leading to increased wildtype α-syn levels also causes disease (Singleton et al., 2003). Certain α-syn promoter polymorphisms are PD risk factors, suggesting that even modest increases in expression can predispose to disease (Farrer et al., 2001; Kruger et al., 1999; Pals et al., 2004; Tan et al., 2000; Wang et al., 2006). In patients without genetic mutations, α-syn is found in Lewy bodies and degenerating neurites (Irizarry et al., 1998; Spillantini et al., 1997).
Alpha-syn overexpression has been used to generate cellular and animal models of PD. Mutant α-syn or overexpression of wildtype α-syn induces cell death in dopaminergic cell lines and in primary dopaminergic cultures (Oluwatosin-Chigbu et al., 2003; Xu et al., 2002; Zhou et al., 2000; Zhou et al., 2002). Transgenic mice expressing mutant or wildtype α-syn show motor deficits, alterations in dopamine levels, and α-syn-positive inclusions (Hashimoto et al., 2003; Maries et al., 2003). Alpha-syn is also involved in the action of neurotoxins, such as MPTP and rotenone, used to model PD (Dauer et al., 2002; Betarbet et al., 2000).
The mechanism by which α-syn injures dopaminergic neurons remains unclear. A potential mechanism not closely examined is transcriptional dysregulation, in which interference with normal gene expression patterns leads to cellular dysfunction. Transcriptional dysregulation has been implicated in Huntington’s and Alzheimer’s diseases (Cha, 2000; Robakis, 2003). Using microarray methods, we have observed that PD causes gene expression alterations in human dopaminergic neurons (Cantuti-Castelvetri et al., 2007). Alpha-syn can also inhibit histone acetylation, a potent mechanism for altering gene expression (Kontopoulos et al., 2006).
Here we examined gene expression patterns in SN cells isolated from transgenic mice in which human wildtype α-syn is overexpressed under the platelet-derived growth factor β promoter. These mice develop neuronal α-syn inclusions in the SN and other brain regions (Masliah et al., 2000). At twelve months, they show motor impairments and decreased striatal dopaminergic terminals and tyrosine hydroxylase activity (Masliah et al., 2000). We used laser capture microdissection to isolate nigral cell RNA from transgenic and control mice, and microarray analysis to evaluate for alterations in gene expression induced by α-syn overexpression at two time points: 1) three months, when pathological changes are few, and 2) nine months, when α-syn inclusions are more widespread. Our data reveal the early effects of α-syn overexpression are modest. At the later time point, changes in gene expression are more prominent in these transgenic mice, and many of the altered genes have functions related to transcription.
Methods
Animals
α-synuclein mice were originally generated by Masliah et al. (2000). A breeding colony from the Masliah D line was set up at Charles River Laboratories to generate transgenic and wildtype littermates. The use of mice was supervised by the Massachusetts General Hospital Animal Resources Program in accordance with the PHS policy on Humane Care and Use of Laboratory Animals. Prior to sacrifice, animals were euthanized by CO2 inhalation. Six gender-matched wildtype and six transgenic littermates were sacrificed at three months of age, and another six control and six transgenic mice were sacrificed at nine months of age. Brains were immediately frozen in isopentane and stored at -80°C. Brains were sectioned at eight μm by cryostat, mounted on uncoated glass slides, and stored at -80°C.
Laser capture microdissection (LCM)
All of the following procedures were done under strict RNase-free conditions. Eight μm sections through the midbrain were first thawed and fixed with acetone for 40 seconds. To identify nigral neurons, these sections were immunostained using a primary mouse monoclonal antibody against tyrosine hydroxylase (TH; 5 minutes at 1:1500; Sigma) and a cy3-conjugated goat anti-mouse secondary antibody (5 minutes at 1:200; Jackson ImmunoResearch Labs, West Grove, PA) and then dehydrated (50, 70, 95, 100% ethanol, 5 seconds at each concentration, followed by xylene for several minutes). TH-stained substantia nigra pars compacta (SNpc) were laser captured from each section with an Arcturus PixCell II instrument (Mountain View, CA). Because the SNpc in mice is very densely packed with cells, we were unable to accurately count the number of captured TH-positive neurons and likely also captured some other cell types. To minimize differences in the amount of captured RNA, we attempted to capture cells from about eight nigral sections of comparable size for each animal. Global normalization of microarray data and normalization of quantitative PCR data with “housekeeping” genes were performed to correct for differences in the number of captured cells among animals.
Amplification and gene expression microarray
RNA from laser-captured nigral cells was extracted using the Picopure™ RNA isolation kit (Arcturus) according to manufacturer’s specifications. Purified RNA was then amplified two rounds using the RiboAmp™ RNA amplification kit (Arcturus). This kit is based on a T7-RNA polymerase method that can linearly amplify total RNA quantities as small as 1 ng. After amplification, mRNA quality was assessed by an Agilent Bioanalyzer (Agilent, Palo Alto, CA). Selected samples had similar product sizes averaging approximately 500-1000 bp. Those samples with smaller average product sizes were discarded. Appropriate amplified RNA (aRNA) samples were sent to the Harvard Partners Center for Genetics and Genomics (HPCGG) facility to generate biotinylated aRNA that was then hybridized to Mouse Expression 430 2.0 arrays (Affymetrix, Santa Clara, CA). Each biotinylated aRNA sample from an individual animal was hybridized to one array, for a total of 24 arrays.
Microarray validation
Validation PCRs were performed on a separate set of nigral neurons laser captured from wildtype and transgenic mice using the Arcturus Veritas system. RNA from these cells was extracted using the Picopure™ RNA isolation kit. The extracted RNA was reverse transcribed into first-strand cDNA using the SuperScript™ II reverse transcriptase kit (Invitrogen, Carlsbad, CA). cDNA was then precipitated using sodium acetate and ethanol with 1μg of glycogen as a carrier.
For each age group, two genes whose levels were altered in the microarray analysis were selected to validate microarray data. The two genes selected for validation were altered at both time points in the microarray analysis and were expressed at high intensities. 20mer, synthetic PCR primers were designed using Primer3 (http://frodo.wi.mit.edu). Primers against cyclin D (NM_007631) were 5’ tgaacccaaggaggaatcag 3’(forward) and 5’ gaagcccaaattcaccaaac 3’ (reverse; product size 256 bp). Primers against TCDD-inducible poly (ADP-ribose) polymerase (NM_178892) were 5’ ctggaaccctgagatccttg 3’ (forward) and 5’ gacacgatgggttgatttcc 3’ (reverse; product size 153 bp). For real-time quantitative PCR (QPCR), first strand cDNA created from extracted RNA was incubated with appropriate forward and reverse primers and SYBR® Green PCR Master Mix (Applied Biosystems, Foster City, CA) in a 96-well plate. QPCR was performed using an iQ5Cycler (BioRad, Hercules, CA) set to the following protocol: 1 cycle of denaturation at 95°C for 10 minutes; 50 cycles of denaturation at 95°C for 30 seconds, annealing at 57°C for 30 seconds, and polymerization at 72°C for 45 seconds; and finally, 80 0.5°C increases in temperature (starting at 55°C) to collect melting curve data. For each primer set, we used a standard curve with known concentrations of cDNA to calculate primer efficiency and to quantitate PCR products. The quantity of each PCR sample was calculated using the ΔΔCt method (Fink et al., 1998; Livak and Schmittgen, 2001). We used both pyruvate decarboxylase (forward primer 5’ gctggagaaggacctgattg 3’; reverse primer 5’ ccagctcaacctcaaactcc 3’; product size 191 bp) and hypoxanthine phosphoribosyltransferase (forward primer 5’ tgttgttggatatgcccttg 3’; reverse primer 5’ tgcgctcatcttaggctttg 3’; product size 107 bp) levels to normalize PCR results, as both genes were unchanged between transgenic and wildtype mice.
Microarray analysis
Microarray analysis was performed as previously described (Cantuti-Castelvetri et al., 2007). Chips were developed, scanned, and normalized using global scaling. All quality control parameters calculated by Affymetrix GCOS Software were monitored. The images of the chips were analyzed to find spotted or damaged array regions, and the graphical analysis of all chips through this step provided evidence that the data was of good quality (Quackenbush, 2002). All data were normalized using the GeneChip Robust MultiChips Analysis (GCRMA) algorithm (Cope et al., 2004) performed with ArrayAssistLite (Stratagene, La Jolla, CA). Normalized data was then statistically analyzed with multiclass analysis followed by two-tailed unpaired t-tests using the Significance Analysis of Microarrays (SAM) algorithm with SAM 2.0 plug-in for Excel (Efron and Tibshirani, 2002; Larsson et al., 2005; Tusher et al., 2001).
Any probes associated with transcripts that were no longer listed in the EntrezGene or Unigene databases were excluded, as were probes which were present for less than half the animals in a given experimental group (transgenic, wildtype, female transgenic, female nontransgenic, etc). Probes with intensities lower than background in all samples were also filtered out (log2 intensity <3). Average expression for each probe/gene was calculated for each of the following groups of animals: 1) all three-month-old transgenic animals, 2) all three-month-old wildtype animals, 3) all nine-month-old transgenic animals, 4) all nine-month-old wildtype animals, 5) three-month-old female transgenic mice, 6) three-month-old female wildtype mice, 7) three-month-old male transgenic mice, 8) three-month-old male wildtype mice, 9) nine-month-old female transgenic mice, 10) nine-month-old female wildtype mice, 11) nine-month-old male transgenic mice, and 12) nine-month-old male wildtype mice. Average expression for a given probe was calculated for all transgenic mice and for all wildtype mice, and the ratio of average transgenic mouse expression to average wildtype mouse expression was calculated for each probe. This ratio was expressed as a log2 ratio in our computer analyses programs. Probes were considered to be different between two groups if the log2 ratio between the two groups was greater than 0.5 (equivalent to 1.4-fold difference) or less than -0.5 (equivalent to 0.7-fold difference) and if the p value was less than 0.05 for the unpaired t-test. To reduce the possibility of a type I statistical error and to control for the variance within each probe set considered in the analysis, we also estimated the false discovery rates (represented by q values) using SAM and limited the final lists of differentially expressed genes to those probes with q values ≤ 20%.
Clustering
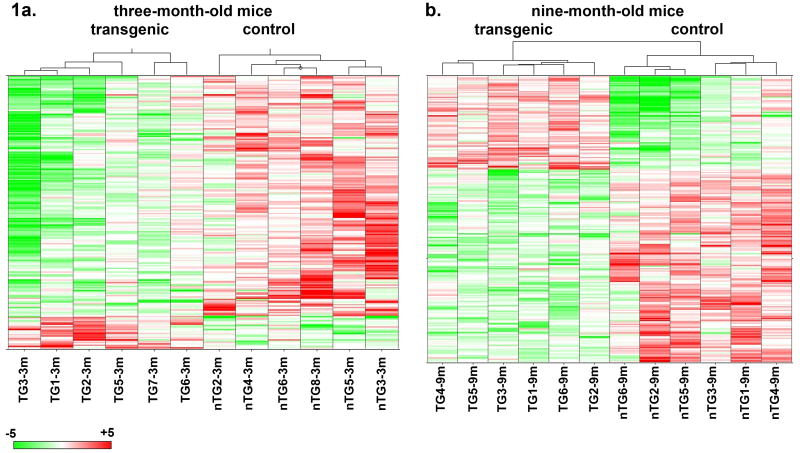
Hierarchical clustering on median normalized samples using cosine correlation with complete linkage was performed for each selected probe sets lists on all three-month samples and on all nine-month samples using SpotFire DecisionSite for Functional Genomics 8.0 (Fig. 1; Eisen et al., 1998).
Figure 1.
Hierarchical clustering on median normalized samples using cosine correlation with complete linkage was performed on all samples from a) three-month-old mice and from b) nine-month-old mice. Green lines reflect decreased expression of probes, and red lines reflect increased expression of probes. This method appropriately sorts wildtype from transgenic mice at both ages.
Functional profiling
To analyze the biological roles of the differentially expressed genes, we used the Database for Annotation, Visualization and Integrated Discovery (DAVID) 2007 (Dennis et al., 2003; Hosack et al., 2003). DAVID 2007 is a freely-available, NIH-sponsored data-mining tool that uses several databases containing gene-specific functional data, such as Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), the Protein Information Resources (PIR), and others. DAVID calculates the probability that particular functional categories are overrepresented in a given data set using a one-tailed Fisher exact probability for overrepresentation (or EASE score) using a Gaussian hypergeometric probability distribution. This distribution describes sampling without replacement from a finite population made of two types of elements: genes that belong to a particular functional category and genes that do not belong to that particular category.
For the functional analysis, we used only those differentially expressed probe sets that had a q value ≤ 20%. First, the DAVID functional annotation tool was used to generate those categories defined by GO database annotations that were overrepresented. All GO categories that had a p value ≤ 0.05 and which contained at least 3% of all regulated genes were considered significant, regardless of how broad or specific the categories were. The functional annotation clustering tool in DAVID was then used as a second level of functional analysis. This tool evaluated each annotation term by the genes that it was comprised of from a given data set and then clustered that term with similar annotations from other ontology databases based on the amount of genes that were co-associated. Only those functional clusters that gave a median p value ≤ 0.05 were considered significant.
Results
Isolation and analysis of mRNA from cells collected by laser capture microdissection
We used laser capture microdissection to isolate nigral cell RNA from both α-syn transgenic and wildtype mice. After amplification, RNA quality was assessed by an Agilent Bioanalyzer. Samples selected for microarray hybridization had similar product sizes averaging 500-1000 base pairs. Those samples with smaller-than-expected average product sizes were discarded, and more nigral cell RNA was captured and amplified. For each age group, we isolated and amplified RNA from gender-matched groups of six transgenic and six wildtype mice. The resultant aRNA samples were labeled and hybridized to Affymetrix Mouse Expression microarrays. Each aRNA sample from an individual animal was hybridized to one array, for a total of 24 arrays. Background intensity levels for all arrays were similar with a mean 58.9 ± 6.8 (SEM). Average signal for present probes across all arrays was 388.6 ± 22.2 (SEM). As a group, the 24 arrays had a mean 42.0 ± 0.8% (SEM) of probes classified as “present.”
α-syn overexpression leads to early alterations in gene expression
We initially examined nigral neurons from mice at three months of age, a point at which there is no apparent histological abnormality in the SN. Data normalized by the GCRMA algorithm (Cope et al., 2004) was statistically analyzed with multiclass analysis followed by two-tailed unpaired t-tests using the SAM algorithm (Efron and Tibshirani, 2002; Larsson et al., 2005; Tusher et al., 2001). We considered probes to be different between wildtype and transgenic mice if the log2 ratio between the groups was greater than 0.5 (equivalent to 1.4-fold difference compared to wildtype) or less than -0.5 (equivalent to 0.7-fold difference compared to wildtype) and if the p value was less than 0.05 for the unpaired t-test. 237 genes were differentially expressed between transgenic and wildtype littermates (Supp. Table 1). 180 of these genes have been identified in the Entrez Gene and Unigene databases with names instead of expressed sequence or RIKEN numbers. Hierarchical clustering performed using the data from these differentially expressed genes appropriately classified all of the samples from three-month-old mice, although it was clear that there was some variation within the groups in the strength of the phenotype defined by this set of genes (Fig. 1a).
Among this list of genes, we validated two genes by quantitative PCR. The two genes chosen were cyclin D and TCDD-inducible poly (ADP-ribose) polymerase (Tiparp), which were both expressed at high intensities at three months. We collected SNpc by laser capture microdissection from six transgenic and six wildtype mice. Expression levels were determined in cDNA samples produced from the isolated neurons without any intervening amplification steps. The expression levels of these two genes as determined by QPCR were similar to the microarray results (Table 1).
Table 1.
Quantitative PCR validation.
| Age | Gene | TG ± SEM* | WT ± SEM* | QPCR
TG/WT |
Array
TG/WT |
|---|---|---|---|---|---|
| 3 month | Cyclin D2 | 0.179±0.016 | 0.222±0.031 | 0.806 | 0.556 |
| Tiparp | 0.255±0.011 | 0.363±0.027 | 0.702 | 0.509 | |
| 9 month | Cyclin D2 | 0.180±0.017 | 0.363±0.072 | 0.496 | 0.444 |
| Tiparp | 0.471±0.088 | 0.844±0.105 | 0.558 | 0.493 | |
Comparison of microarray results with QPCR results for two genes, cyclin D2 and TCDD-inducible poly(ADP-ribose) polymerase (Tiparp), which were altered at both three and nine months.
QPCR results shown in table were normalized to pyruvate decarboxylase.
To reduce the possibility of a type I statistical error and to control for the variance within each probe set considered in the analysis, we also estimated the false discovery rate for each gene (represented by the q value) altered at three months and eliminated those genes that had a q value > 20%. By this more conservative statistical approach, we reduced the initial list of 237 genes to 29 (Table 2). Six of these genes were downregulated in the transgenic mice, while 23 were upregulated. 22 of the 29 genes were associated with gene names other than expressed sequence or RIKEN numbers. None of the altered genes in this list include molecules previously linked to PD. All upregulated genes from the initial 237 gene list were found in this more conservative list, while only six of the initial 214 downregulated genes had q values ≤ 20%. Because of the small number of genes modulated at this early time point, further classification by ontological analysis was not informative.
Table 2.
Probes whose expressions are altered by at least a log2 ratio of ±0.5 in three-month-old transgenic α-syn mice. P value for each probe is ≤0.05 and q-value is ≤20%.
| Probe Set ID | Genbank ID | Gene Name | log2 (TG/WT) | q-value |
|---|---|---|---|---|
| 1451127_at | BC024822 | EXPRESSED SEQUENCE AW146242 | -1.79 | 0 |
| 1460037_at | BF303035 | RIKEN CDNA 2610510H03 GENE | -0.91 | 0 |
| 1434735_at | BB744589 | HEPATIC LEUKEMIA FACTOR | -0.75 | 0 |
| 1448101_s_at | NM_009054 | TRIPARTITE MOTIF PROTEIN 27 | -0.74 | 0 |
| 1429097_at | BB090042 | RIKEN CDNA C030044C12 GENE | -0.67 | 0 |
| 1451240_a_at | BC024663 | RIKEN CDNA 1110008E19 GENE | -0.52 | 0 |
| 1426421_s_at | AK005802 | RIKEN CDNA 1700009P03 GENE | 0.51 | 0 |
| 1441807_s_at | AI851474 | RIKEN CDNA A830093I24 GENE | 0.53 | 0 |
| 1420296_at | T25545 | CHLORIDE CHANNEL 5 | 0.53 | 0 |
| 1452975_at | AK005060 | ALANINE-GLYOXYLATE AMINOTRANSFERASE 2-LIKE 1 | 0.56 | 0 |
| 1444064_at | AV376100 | SAM DOMAIN AND HD DOMAIN, 1 | 0.56 | 0 |
| 1450688_at | NM_009059 | RAL GUANINE NUCLEOTIDE DISSOCIATION STIMULATOR-LIKE 2 | 0.56 | 0 |
| 1416055_at | NM_009669 | AMYLASE 2, PANCREATIC | 0.57 | 0 |
| 1457443_at | BB311369 | G PATCH DOMAIN CONTAINING 2 | 0.57 | 0 |
| 1443430_at | BM234069 | CASP2 AND RIPK1 DOMAIN CONTAINING ADAPTOR WITH DEATH DOMAIN | 0.58 | 0 |
| 1420115_at | C80158 | EXOSOME COMPONENT 8 | 0.59 | 0 |
| 1441266_at | BM238307 | STRIATIN, CALMODULIN BINDING PROTEIN 3 | 0.60 | 0 |
| 1439205_at | BM122872 | NUCLEAR FACTOR OF ACTIVATED T-CELLS, CYTOPLASMIC, CALCINEURIN-DEPENDENT 2 | 0.63 | 0 |
| 1435453_at | AV343709 | RIKEN CDNA A930011O12 GENE | 0.70 | 0 |
| 1418539_a_at | U35368 | PROTEIN TYROSINE PHOSPHATASE, RECEPTOR TYPE, E | 0.78 | 0 |
| 1452831_s_at | AV305746 | PHOSPHORIBOSYL PYROPHOSPHATE AMIDOTRANSFERASE | 0.79 | 0 |
| 1418264_at | NM_021790 | SOXLZ/SOX6 LEUCINE ZIPPER BINDING PROTEIN IN TESTIS | 0.82 | 0 |
| 1445718_at | BM237480 | WD REPEAT AND FYVE DOMAIN CONTAINING 3 | 0.88 | 0 |
| 1434585_at | BB667130 | TUBBY LIKE PROTEIN 4 | 0.95 | 0 |
| 1427822_a_at | U20265 | COATOMER PROTEIN COMPLEX, SUBUNIT GAMMA 2, ANTISENSE 2 | 0.98 | 0 |
| 1457634_at | BG093687 | RIKEN CDNA 4930488E11 GENE | 0.99 | 0 |
| 1426113_x_at | U07662 | T-CELL RECEPTOR ALPHA CHAIN | 0.99 | 0 |
| 1457455_at | BB430212 | SUPPRESSOR OF HAIRY WING HOMOLOG 4 (DROSOPHILA) | 1.05 | 0 |
| 1440764_at | AI552435 | V-RAF MURINE SARCOMA 3611 VIRAL ONCOGENE HOMOLOG | 1.05 | 0 |
Late effects of α-syn overexpression on gene expression
We performed similar laser capture microdissection and array experiments with nine-month-old mice, which have widespread α-syn inclusions in the brain, to evaluate gene expression changes at a later point in disease pathogenesis. We found that the magnitude of the transcriptional dysregulation, as measured by the number of altered genes, was much larger at this late stage of the disease model, and the identities of the genes modified were different than those observed to be altered at the earlier time point.
As with the three month data, we performed an initial analysis using criteria of p value ≤ 0.05 and a log2 ratio of at least ±0.5. Using these filters, we found that 278 genes were altered between nine-month-old α-syn and wildtype mice (Supp. Table 2). 211 of these genes have been identified in the Entrez Gene and Unigene databases with names other than expressed sequence or RIKEN numbers. Hierarchical clustering using this gene set revealed that the data readily classified the transgenic and control animals (Fig. 1b). Indeed, the distinctions between the groups were much more striking than at three months, suggesting that at a later stage in the model the effect of α-syn overexpression is more uniform across the population of animals studied. Among the nine-month list of genes, we validated cyclin D and Tiparp by QPCR. The expression levels of these two genes as determined by QPCR were similar to the microarray results (Table 1).
As with the three-month microarray data, we also estimated the false discovery rate for each gene altered at nine months and eliminated those genes that had a q value > 20%. This conservative statistical approach reduced the initial list of 278 genes to 179 (Table 3). In contrast to the three-month time point, the predominant effect at nine months was a reduction of gene expression, with more genes downregulated than upregulated in the transgenic mice. The mRNA levels of 165 genes were decreased, while the mRNA levels of 14 genes were elevated in the nine-month-old α-syn mice. Some of the genes that were altered in the nine-month-old group included an E3 ubiquitin protein ligase (Cbl), a proteosomal subunit (Psmd14), and other molecules linked to PD, including the neurotrophin receptor TrkB (von Bohlen und Halbach et al., 2005), and 14-3-3θ(Berg et al., 2003; Kawamoto et al., 2002). Interestingly, among the downregulated genes was ceruloplasmin, which is decreased in Wilson’s disease (Scheinberg and Gitlin, 1952), a genetic disorder with Parkinsonian symptoms.
Table 3.
Probes whose expressions are altered by at least a log2 ratio of ±0.5 in nine-month-old transgenic α-syn mice. P value for each probe is ≤0.05 and q-value is ≤20%.
| Probe Set ID | Genbank ID | Gene Name | log2 (TG/WT) | q-value |
|---|---|---|---|---|
| 1436964_at | BB314814 | DNA SEGMENT, CHR 7, ERATO DOI 715, EXPRESSED | -2.78 | 18.07 |
| 1442568_at | BM213120 | RIKEN CDNA 3110039L19 GENE | -1.98 | 18.07 |
| 1443281_at | BB297094 | RIKEN CDNA C030017F07 GENE | -1.61 | 8.66 |
| 1442742_at | BE994454 | RIKEN CDNA 1700121J11 GENE | -1.55 | 17.17 |
| 1440773_at | BB209878 | F-BOX ONLY PROTEIN 8 | -1.37 | 8.51 |
| 1440163_at | BB392503 | RIKEN CDNA 6030490B17 GENE | -1.33 | 8.51 |
| 1430660_at | AV291471 | RIKEN CDNA 2610100B20 GENE | -1.31 | 11.70 |
| 1431248_at | BB637274 | RIKEN CDNA 5031426D15 GENE | -1.26 | 8.66 |
| 1426413_at | BM116592 | NEUROGENIC DIFFERENTIATION 1 | -1.22 | 17.17 |
| 1455512_at | AI852151 | GENE MODEL 879, (NCBI) | -1.20 | 8.66 |
| 1455956_x_at | AV310588 | CYCLIN D2 | -1.17 | 10.02 |
| 1429769_at | BI107300 | PROTEIN GERANYLGERANYLTRANSFERASE TYPE I, BETA SUBUNIT | -1.12 | 8.66 |
| 1451301_at | BB633110 | TROPOMODULIN 2 | -1.06 | 8.51 |
| 1426721_s_at | BB707122 | TCDD-INDUCIBLE POLY(ADP-RIBOSE) POLYMERASE | -1.02 | 0.00 |
| 1440304_at | BB085100 | EXPRESSED SEQUENCE BB214985 | -0.99 | 8.51 |
| 1455393_at | BB009037 | CERULOPLASMIN | -0.95 | 17.17 |
| 1422795_at | AV273804 | CULLIN 3 | -0.95 | 12.54 |
| 1434216_a_at | BG070689 | NUDIX (NUCLEOSIDE DIPHOSPHATE LINKED MOIETY X)-TYPE MOTIF 19 | -0.95 | 18.07 |
| 1432944_at | AK013658 | RIKEN CDNA 2900046L07 GENE | -0.94 | 0.00 |
| 1450061_at | BM120053 | ECTODERMAL-NEURAL CORTEX 1 | -0.93 | 8.66 |
| 1440534_at | BB177862 | RIKEN CDNA 6330403A02 GENE | -0.93 | 15.94 |
| 1457198_at | AV291009 | NEUROPILIN 1 | -0.92 | 17.17 |
| 1447314_at | AI480643 | TRANSFORMING GROWTH FACTOR, BETA RECEPTOR III | -0.91 | 10.02 |
| 1429703_at | AV154947 | RIKEN CDNA 2900072G11 GENE | -0.90 | 0.00 |
| 1442214_at | BG173293 | NUCLEAR FACTOR I/B | -0.90 | 8.51 |
| 1433184_at | AK020162 | RIKEN CDNA 6720477C19 GENE | -0.89 | 15.02 |
| 1424525_at | BC024515 | GASTRIN RELEASING PEPTIDE | -0.89 | 13.49 |
| 1454286_at | AV007471 | RIKEN CDNA 1110004M10 GENE | -0.88 | 12.54 |
| 1458263_at | BE979587 | CUG TRIPLET REPEAT, RNA BINDING PROTEIN 2 | -0.87 | 0.00 |
| 1453402_at | AV338754 | RIKEN CDNA 6430500C12 GENE | -0.86 | 10.02 |
| 1459144_at | BB518323 | HYPOTHETICAL PROTEIN LOC213817 | -0.85 | 13.49 |
| 1430808_at | AK005461 | TBC1 DOMAIN FAMILY, MEMBER 5 | -0.85 | 10.02 |
| 1420965_a_at | BM120053 | ECTODERMAL-NEURAL CORTEX 1 | -0.84 | 10.66 |
| 1445391_at | AW554652 | DIAPHANOUS HOMOLOG 1 (DROSOPHILA) | -0.83 | 19.25 |
| 1455914_at | AW554430 | EXPRESSED SEQUENCE AI987944 | -0.82 | 19.25 |
| 1435129_at | AW495875 | PROTEIN TYROSINE PHOSPHATASE 4A2 | -0.81 | 10.02 |
| 1444557_at | BG060557 | SMT3 SUPPRESSOR OF MIF TWO 3 HOMOLOG 1 (YEAST) | -0.81 | 8.66 |
| 1439940_at | AV328280 | RIKEN CDNA 2900019G14 GENE | -0.80 | 19.25 |
| 1443526_at | BM204661 | PHD FINGER PROTEIN 21A | -0.80 | 0.00 |
| 1439123_at | BB367687 | PHD FINGER PROTEIN 21A | -0.79 | 15.02 |
| 1445708_x_at | AV158652 | RIKEN CDNA 3110021A11 GENE | -0.79 | 19.25 |
| 1453068_at | BM226301 | PR DOMAIN CONTAINING 2, WITH ZNF DOMAIN | -0.78 | 19.25 |
| 1446953_at | BG073799 | TRANSCRIPTION FACTOR 4 | -0.77 | 8.66 |
| 1445037_at | AV339661 | RIKEN CDNA 6430510B20 GENE | -0.77 | 10.02 |
| 1437793_at | BB711615 | TANKYRASE, TRF1-INTERACTING ANKYRIN-RELATED ADP-RIBOSE POLYMERASE 2 | -0.76 | 8.51 |
| 1446374_at | BB460605 | RHO GUANINE NUCLEOTIDE EXCHANGE FACTOR 10 | -0.76 | 12.54 |
| 1442500_at | C77406 | EXPRESSED SEQUENCE C77406 | -0.76 | 10.02 |
| 1460186_at | AV061337 | RIKEN CDNA 1810073M23 GENE | -0.76 | 19.25 |
| 1441538_at | BB267954 | RIKEN CDNA B930011P16 GENE | -0.75 | 17.17 |
| 1442409_at | BB166984 | DNA SEGMENT, CHR 9, WAYNE STATE UNIVERSITY 90, EXPRESSED | -0.74 | 19.25 |
| 1429051_s_at | BE825056 | SRY-BOX CONTAINING GENE 11 | -0.74 | 13.49 |
| 1444472_at | BB154631 | SNF1-LIKE KINASE 2 | -0.74 | 13.49 |
| 1440728_at | BB283560 | POTASSIUM LARGE CONDUCTANCE CALCIUM-ACTIVATED CHANNEL, SUBFAMILY M, ALPHA MEMBER 1 | -0.74 | 18.07 |
| 1437180_at | BE824567 | RIKEN CDNA 6530403A03 GENE | -0.73 | 0.00 |
| 1439169_at | BB471557 | EUKARYOTIC TRANSLATION INITIATION FACTOR 4E NUCLEAR IMPORT FACTOR 1 | -0.73 | 13.49 |
| 1449068_at | X98096 | ZINC FINGER PROTEIN 148 | -0.72 | 8.66 |
| 1434835_at | BM230523 | CDNA SEQUENCE BC037674 | -0.72 | 17.17 |
| 1432352_at | AK014201 | RIKEN CDNA 5730405I09 GENE | -0.71 | 17.17 |
| 1446159_at | BB044011 | P21 (CDKN1A)-ACTIVATED KINASE 7 | -0.71 | 8.51 |
| 1454510_at | AK013612 | RIKEN CDNA 2900034C19 GENE | -0.71 | 8.51 |
| 1444785_at | AI503808 | SLOAN-KETTERING VIRAL ONCOGENE HOMOLOG | -0.71 | 12.54 |
| 1442659_at | BB244656 | PROTOCADHERIN 9 | -0.70 | 13.49 |
| 1423433_at | BG069917 | TROVE DOMAIN FAMILY, MEMBER 2 | -0.69 | 8.51 |
| 1448282_at | BC006750 | PLEIOTROPIC REGULATOR 1, PRL1 HOMOLOG (ARABIDOPSIS) | -0.69 | 11.70 |
| 1460123_at | AW541072 | G PROTEIN-COUPLED RECEPTOR 1 | -0.69 | 17.17 |
| 1453864_at | AK014097 | RIKEN CDNA 3110030G19 GENE | -0.69 | 19.25 |
| 1445847_at | BE948333 | NEUROTROPHIC TYROSINE KINASE, RECEPTOR, TYPE 2 | -0.69 | 18.07 |
| 1459219_at | BM120546 | RAP GUANINE NUCLEOTIDE EXCHANGE FACTOR 2 | -0.68 | 10.02 |
| 1458469_at | BB205662 | CASITAS B-LINEAGE LYMPHOMA B | -0.68 | 12.54 |
| 1459238_at | AV319481 | PHOSPHOLAMBAN | -0.68 | 8.51 |
| 1441072_at | BB353142 | CUG TRIPLET REPEAT, RNA BINDING PROTEIN 2 | -0.67 | 10.02 |
| 1416892_s_at | BC021353 | RIKEN CDNA 3110001A13 GENE | -0.67 | 8.51 |
| 1459734_at | BB737833 | PROTEASOME (PROSOME, MACROPAIN) 26S SUBUNIT, NON-ATPASE, 14 | -0.67 | 17.17 |
| 1430675_at | BM932606 | RIKEN CDNA 2900055J20 GENE | -0.67 | 18.07 |
| 1434378_a_at | BG868949 | MAX DIMERIZATION PROTEIN 4 | -0.67 | 8.51 |
| 1436248_at | AW120749 | RAS PROTEIN ACTIVATOR LIKE 2 | -0.67 | 8.51 |
| 1441487_at | BB197646 | TRIPARTITE MOTIF PROTEIN 2 | -0.66 | 18.07 |
| 1442679_at | BM241898 | MITOGEN ACTIVATED PROTEIN KINASE KINASE 4 | -0.66 | 17.17 |
| 1451506_at | BB280300 | MYOCYTE ENHANCER FACTOR 2C | -0.66 | 15.02 |
| 1428936_at | BI080417 | ATPASE, CA++ TRANSPORTING, PLASMA MEMBRANE 1 | -0.66 | 10.02 |
| 1438282_at | AV344830 | SYNAPTOTAGMIN I | -0.66 | 10.66 |
| 1434178_at | AV297525 | MYELOID/LYMPHOID OR MIXED-LINEAGE LEUKEMIA 3 | -0.66 | 13.58 |
| 1454424_at | AK011729 | RIKEN CDNA 2610040L17 GENE | -0.66 | 8.66 |
| 1455799_at | BB751387 | RAR-RELATED ORPHAN RECEPTOR BETA | -0.66 | 8.51 |
| 1443508_at | BB131767 | EXPRESSED SEQUENCE BB075781 | -0.65 | 17.17 |
| 1420579_s_at | NM_021050 | CYSTIC FIBROSIS TRANSMEMBRANE CONDUCTANCE REGULATOR HOMOLOG | -0.65 | 12.54 |
| 1458376_at | BE648536 | RIKEN CDNA B930025B16 GENE | -0.65 | 18.07 |
| 1458690_at | BB079254 | MEMBRANE-ASSOCIATED RING FINGER (C3HC4) 7 | -0.65 | 15.02 |
| 1439209_at | BB540782 | TRANSCRIPTION FACTOR 12 | -0.65 | 12.54 |
| 1416129_at | NM_133753 | ERBB RECEPTOR FEEDBACK INHIBITOR 1 | -0.65 | 8.66 |
| 1443260_at | BB055155 | MYELOID ECOTROPIC VIRAL INTEGRATION SITE 1 | -0.64 | 11.70 |
| 1437064_at | AV232123 | ANDROGEN RECEPTOR | -0.64 | 17.17 |
| 1418955_at | NM_009567 | ZINC FINGER PROTEIN 93 | -0.64 | 8.51 |
| 1454743_at | BQ177153 | NUCLEOPORIN 205 | -0.64 | 10.66 |
| 1435505_at | BB698273 | DYSTROPHIA MYOTONICA-CONTAINING WD REPEAT MOTIF | -0.64 | 15.02 |
| 1445340_at | BB156822 | PAM, HIGHWIRE, RPM 1 | -0.64 | 0.00 |
| 1431482_at | AV153257 | RIKEN CDNA 2900012M01 GENE | -0.63 | 18.07 |
| 1426546_at | BQ179435 | TESTIS-SPECIFIC KINASE 2 | -0.62 | 15.94 |
| 1444409_at | BQ176035 | RABPHILIN 3A-LIKE (WITHOUT C2 DOMAINS) | -0.62 | 10.66 |
| 1420136_a_at | AI427540 | OPIOID GROWTH FACTOR RECEPTOR-LIKE 1 | -0.62 | 19.25 |
| 1441231_at | BB233964 | GENE MODEL 1167, (NCBI) | -0.62 | 15.94 |
| 1456896_at | BB053380 | RIKEN CDNA 6720462K09 GENE | -0.62 | 18.07 |
| 1454922_at | BM246326 | EXPRESSED SEQUENCE AI553587 | -0.62 | 13.49 |
| 1456413_at | BB235927 | PHOSPHODIESTERASE 4D INTERACTING PROTEIN | -0.61 | 8.66 |
| 1441654_at | BB345762 | GUANINE NUCLEOTIDE BINDING PROTEIN, ALPHA 10 | -0.61 | 19.25 |
| 1428616_at | BG075140 | ZINC FINGER PROTEIN 131 | -0.61 | 17.17 |
| 1459947_at | BB007036 | THIOREDOXIN DOMAIN CONTAINING 5 | -0.61 | 15.02 |
| 1444516_at | AI503166 | PROTEIN TYROSINE PHOSPHATASE, RECEPTOR-TYPE, F INTERACTING PROTEIN, BINDING PROTEIN 2 | -0.61 | 13.58 |
| 1438245_at | BI664122 | NUCLEAR FACTOR I/B | -0.61 | 8.66 |
| 1435663_at | AI646838 | ESTROGEN RECEPTOR 1 (ALPHA) | -0.60 | 18.07 |
| 1430195_at | BF662697 | RIKEN CDNA 2810043O03 GENE | -0.60 | 15.94 |
| 1437426_at | BM240080 | RIKEN CDNA 1110067P07 GENE | -0.60 | 8.66 |
| 1437984_x_at | BB461609 | HLA-B-ASSOCIATED TRANSCRIPT 1A | -0.59 | 17.17 |
| 1437339_s_at | BB241731 | PROPROTEIN CONVERTASE SUBTILISIN/KEXIN TYPE 5 | -0.59 | 18.07 |
| 1439106_at | AV320128 | ZINC FINGER PROTEIN 462 | -0.59 | 10.66 |
| 1457019_s_at | BB241474 | RIKEN CDNA 3110030G19 GENE | -0.59 | 13.49 |
| 1417644_at | BC021484 | SARCOSPAN | -0.59 | 18.07 |
| 1437784_at | AW550878 | CBFA2T1 IDENTIFIED GENE HOMOLOG (HUMAN) | -0.59 | 17.17 |
| 1435474_at | AV117817 | TAF5 RNA POLYMERASE II, TATA BOX BINDING PROTEIN (TBP)-ASSOCIATED FACTOR | -0.59 | 18.07 |
| 1445774_at | BB479183 | POTASSIUM LARGE CONDUCTANCE CALCIUM-ACTIVATED CHANNEL, SUBFAMILY M, ALPHA MEMBER 1 | -0.59 | 8.51 |
| 1440020_at | BB212045 | EXPRESSED SEQUENCE AI195322 | -0.59 | 10.66 |
| 1438352_at | BM217851 | MICROTUBULE-ASSOCIATED PROTEIN, RP/EB FAMILY, MEMBER 2 | -0.58 | 18.07 |
| 1430058_at | AK016826 | STEM-LOOP BINDING PROTEIN | -0.58 | 15.94 |
| 1430291_at | AK004325 | RIKEN CDNA 1110060D06 GENE | -0.58 | 19.25 |
| 1452092_at | AK019474 | RIKEN CDNA 4631426J05 GENE | -0.58 | 12.54 |
| 1446670_at | BM241206 | CUG TRIPLET REPEAT, RNA BINDING PROTEIN 2 | -0.57 | 13.58 |
| 1444186_at | BB519728 | RIKEN CDNA 2900006B13 GENE | -0.57 | 15.02 |
| 1421948_a_at | BB042564 | RIKEN CDNA 2610507L03 GENE | -0.57 | 18.07 |
| 1441100_at | BB496952 | MBT DOMAIN CONTAINING 1 | -0.57 | 15.94 |
| 1446272_at | BB200545 | RIKEN CDNA 6430598J10 GENE | -0.57 | 11.70 |
| 1443161_at | BM240648 | TRICHORHINOPHALANGEAL SYNDROME I (HUMAN) | -0.57 | 8.66 |
| 1446750_at | BB524087 | IMPRINTED AND ANCIENT | -0.57 | 11.70 |
| 1439174_at | AI644595 | UNC-5 HOMOLOG C (C. ELEGANS) | -0.57 | 10.02 |
| 1431228_s_at | AA120741 | RIKEN CDNA 4930526I15 GENE | -0.56 | 15.02 |
| 1417818_at | BC014727 | WW DOMAIN CONTAINING TRANSCRIPTION REGULATOR 1 | -0.56 | 18.07 |
| 1444456_at | BB460570 | RIKEN CDNA 9030425P06 GENE | -0.56 | 13.49 |
| 1435433_at | BE688580 | EXPRESSED SEQUENCE R75364 | -0.56 | 18.07 |
| 1457716_at | BM235074 | ZINC FINGER, A20 DOMAIN CONTAINING 1 | -0.55 | 18.07 |
| 1453416_at | BE199211 | GROWTH ARREST-SPECIFIC 2 LIKE 3 | -0.55 | 15.02 |
| 1443337_at | BB531021 | RIKEN CDNA B130020M22 GENE | -0.55 | 19.25 |
| 1442587_at | BB451946 | RIKEN CDNA C030017F07 GENE | -0.55 | 19.25 |
| 1441545_at | BM243297 | RIKEN CDNA 9230115F04 GENE | -0.55 | 12.54 |
| 1437676_at | AV279478 | SPERM ASSOCIATED ANTIGEN 9 | -0.54 | 8.51 |
| 1431402_at | BB626088 | KIN OF IRRE LIKE 3 (DROSOPHILA) | -0.54 | 19.25 |
| 1447204_at | BB337014 | MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE KINASE 5 | -0.54 | 10.02 |
| 1418168_at | BB223737 | ZINC FINGER, CCHC DOMAIN CONTAINING 14 | -0.53 | 8.51 |
| 1440770_at | AI505544 | B-CELL LEUKEMIA/LYMPHOMA 2 | -0.53 | 19.25 |
| 1457781_at | BG063584 | KCNQ1 OVERLAPPING TRANSCRIPT 1 | -0.53 | 10.02 |
| 1434829_at | BB487560 | CASITAS B-LINEAGE LYMPHOMA | -0.53 | 8.51 |
| 1442924_at | BB430910 | CALSYNTENIN 2 | -0.53 | 15.02 |
| 1422486_a_at | AK004804 | MAD HOMOLOG 4 (DROSOPHILA) | -0.53 | 17.17 |
| 1454182_at | AK019946 | RIKEN CDNA 5430417C01 GENE | -0.53 | 11.70 |
| 1438535_at | BB523030 | NEURONAL DIFFERENTIATION RELATED PROTEIN | -0.52 | 19.25 |
| 1429652_at | AK009976 | RIKEN CDNA 1190002C06 GENE | -0.52 | 8.51 |
| 1439626_at | BB209799 | TROPOMODULIN 3 | -0.52 | 15.02 |
| 1439192_at | BB524078 | NEURO-ONCOLOGICAL VENTRAL ANTIGEN 2 | -0.52 | 19.25 |
| 1415925_a_at | NM_053074 | NUCLEOPORIN 62 | -0.52 | 13.58 |
| 1454852_at | BB795189 | TRANS-ACTING TRANSCRIPTION FACTOR 1 | -0.52 | 15.94 |
| 1433543_at | BI690018 | ANILLIN, ACTIN BINDING PROTEIN (SCRAPS HOMOLOG, DROSOPHILA) | -0.52 | 13.49 |
| 1442320_at | BB364132 | HYPOTHETICAL LOC553096 | -0.52 | 19.25 |
| 1453371_at | AK012860 | RIKEN CDNA 4930535B03 GENE | -0.51 | 18.07 |
| 1455225_at | AV237615 | SYNAPTIC NUCLEAR ENVELOPE 1 | -0.51 | 18.07 |
| 1433641_at | AV014063 | MAD HOMOLOG 5 (DROSOPHILA) | -0.51 | 17.17 |
| 1440311_at | BB259710 | SORBIN AND SH3 DOMAIN CONTAINING 1 | -0.51 | 19.25 |
| 1436795_at | BM247060 | RIKEN CDNA 9630058J23 GENE | -0.51 | 10.66 |
| 1433803_at | BQ032637 | JANUS KINASE 1 | -0.51 | 18.07 |
| 1458053_at | BB051811 | ABL-INTERACTOR 2 | -0.51 | 18.07 |
| 1429918_at | AK018317 | RIKEN CDNA 6530403F17 GENE | -0.51 | 11.70 |
| 1443020_at | BB027719 | RIKEN CDNA F830020C16 GENE | -0.51 | 8.66 |
| 1455854_a_at | BB053082 | SLINGSHOT HOMOLOG 1 (DROSOPHILA) | -0.50 | 19.25 |
| 1446965_at | BG067578 | RHO GUANINE NUCLEOTIDE EXCHANGE FACTOR 12 | -0.50 | 18.07 |
| 1441635_at | BM202709 | NUCLEAR RECEPTOR SUBFAMILY 6, GROUP A, MEMBER 1 | -0.50 | 18.07 |
| 1418000_a_at | NM_008410 | INTEGRAL MEMBRANE PROTEIN 2B | -0.50 | 8.51 |
| 1428368_at | BM213829 | RHO GTPASE ACTIVATING PROTEIN 21 | 0.51 | 15.94 |
| 1418506_a_at | NM_011563 | PEROXIREDOXIN 2 | 0.55 | 17.17 |
| 1416028_a_at | NM_008258 | HEMATOLOGICAL AND NEUROLOGICAL EXPRESSED SEQUENCE 1 | 0.58 | 15.02 |
| 1426448_at | BM199789 | PRAJA1, RING-H2 MOTIF CONTAINING | 0.58 | 15.02 |
| 1420828_s_at | NM_011739 | TYROSINE 3-MONOOXYGENASE/TRYPTOPHAN 5-MONOOXYGENASE ACTIVATION PROTEIN, THETA POLYPEPTIDE | 0.65 | 15.94 |
| 1435258_at | AW551717 | DNA SEGMENT, CHR 2, ERATO DOI 217, EXPRESSED | 0.66 | 15.02 |
| 1421873_s_at | NM_009000 | RAB24, MEMBER RAS ONCOGENE FAMILY | 0.67 | 15.02 |
| 1450243_a_at | NM_030598 | DOWN SYNDROME CRITICAL REGION GENE 1-LIKE 1 | 0.72 | 15.02 |
| 1433799_at | AV366849 | RETINOL DEHYDROGENASE 13 (ALL-TRANS AND 9-CIS) | 0.79 | 10.66 |
| 1415773_at | BF118393 | NUCLEOLIN | 0.85 | 10.66 |
| 1455868_a_at | BB387906 | TUBULIN, GAMMA COMPLEX ASSOCIATED PROTEIN 2 | 0.86 | 15.02 |
| 1428370_at | AK018264 | RIKEN CDNA 1500011B03 GENE | 1.02 | 7.49 |
| 1448732_at | M14222 | CATHEPSIN B | 1.25 | 15.02 |
| AFFX-GapdhMur/M32599_5_at | AFFX-GapdhMur/M3259 9_5 | SIMILAR TO GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE (GAPDH) | 1.57 | 15.02 |
We next compared the identity of the genes altered in the three-month-old and nine-month-old groups. Surprisingly, we found no genes that overlapped between the three-month and nine-month groups when analyzing those lists limited by q value ≤ 20%. When looking at the less conservative gene lists (in which genes were not eliminated by q value criterion), nine genes showed alterations in expression level at both time points. These genes included calmodulin 1, cyclin D2, disheveled associated activator of morphogenesis 1, fibronectin type III domain containing 4 (Fndc4), hematological and neurological expressed sequence 1 (Hn1), nucleoporin 62, PCTAIRE-motif protein kinase 2 (RIKEN 6430598J10), Tiparp, and expressed sequence AI55357. Seven of these genes showed concordant downregulation at both three and nine months, while Fndc4 and Hn1 showed alterations in opposing directions at the two time points.
Functional genomic analysis of the data from the nine-month-old mice was performed to evaluate whether the nine-month data set was enriched in any particular functional categories. We used the DAVID 2007 functional annotation tool (http://david.abcc.ncifcrf.gov/home.jsp; Dennis et al., 2003; Hosack et al., 2003), which classifies each gene into one or more functional categories using annotations from over 40 ontological databases, including the GO database, and then determines which functional categories are represented in the data set more frequently than expected by chance alone. We used the more conservative nine-month list of 179 genes with upregulated and downregulated genes combined to perform this analysis. We considered significantly overrepresented those functional categories in which the number of genes altered exceeded the number expected with a p value ≤ 0.05. We limited the functional annotation chart generated by DAVID to those categories defined by GO database annotations (http://www.geneontology.org/). Table 4 reveals 39 GO ontological categories that contain at least 3% of the regulated genes. We did not exclude broad categories from this ontology table and instead included all parent and children categories that revealed a p value ≤ 0.05 and contained at least 3% of all regulated genes. Categories that were enriched in our data set included GTPase regulator activity, cytoskeletal protein binding, ubiquitin cycle, protein amino acid phosphorylation, transcription, and signal transduction.
Table 4.
Functional profiling of genes altered in nine-month-old transgenic α-syn mice.
| GO annotation term | % of regulated genes | p value |
|---|---|---|
| protein binding | 28.94% | 6.81E-06 |
| regulation of biological process | 25.11% | 3.05E-05 |
| regulation of cellular process | 21.70% | 8.93E-04 |
| regulation of metabolism | 17.02% | 0.0013 |
| regulation of physiological process | 21.28% | 0.0013 |
| transcriptional activator activity | 3.40% | 0.0014 |
| biopolymer modification | 12.77% | 0.0018 |
| regulation of cellular physiological process | 20.43% | 0.0018 |
| transcription regulator activity | 10.21% | 0.0021 |
| cytoskeletal protein binding | 4.68% | 0.0022 |
| DNA binding | 13.62% | 0.0026 |
| intracellular signaling cascade | 8.94% | 0.0031 |
| biopolymer metabolism | 17.45% | 0.0034 |
| transcription factor activity | 8.09% | 0.0035 |
| regulation of transcription | 14.89% | 0.0039 |
| protein modification | 11.91% | 0.0044 |
| zinc ion binding | 12.34% | 0.0044 |
| binding | 48.09% | 0.0045 |
| regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolism | 14.89% | 0.0047 |
| transcription | 14.89% | 0.0064 |
| regulation of transcription, DNA-dependent | 14.04% | 0.0064 |
| regulation of cellular metabolism | 15.32% | 0.0076 |
| transcription, DNA-dependent | 14.04% | 0.0082 |
| cellular physiological process | 48.94% | 0.0099 |
| cellular process | 54.47% | 0.012 |
| enzyme linked receptor protein signaling pathway | 3.40% | 0.015 |
| nucleic acid binding | 17.87% | 0.017 |
| ubiquitin cycle | 4.68% | 0.021 |
| GTPase regulator activity | 3.40% | 0.021 |
| transition metal ion binding | 13.19% | 0.025 |
| primary metabolism | 34.47% | 0.026 |
| protein amino acid phosphorylation | 5.53% | 0.027 |
| nucleobase, nucleoside, nucleotide and nucleic acid metabolism | 18.30% | 0.029 |
| macromolecule metabolism | 22.55% | 0.035 |
| negative regulation of biological process | 6.38% | 0.039 |
| signal transduction | 14.47% | 0.040 |
| enzyme regulator activity | 5.11% | 0.040 |
| negative regulation of cellular process | 5.96% | 0.043 |
| cell communication | 15.32% | 0.043 |
To determine functional categories associated with α-syn overexpression, we used the DAVID 2007 functional annotation tool (http://david.abcc.ncifcrf.gov/home.jsp), which classifies each probe into one or more functional categories using annotations from over 40 ontological databases and then determines which functional categories are represented in the data set more frequently than expected by chance alone. We limited the functional annotation chart generated by DAVID by classification of genes to functional categories defined by Gene Ontology Database annotations. Only probes with a q value ≤20% were used for the ontology analysis. We considered significant those categories with a p value ≤0.05 and contained at least 3% of the regulated genes. All molecular function and biological process annotation terms that fulfilled these criteria were included in the table regardless of how broad or narrow the annotation was. Therefore, we included broad categories, such as “binding,” as well as more specific and informative categories, such as “zinc ion binding.” The website QuickGO (http://www.ebi.ac.uk/ego) describes the GO annotation categories with their meanings and hierarchical relationships.
We next performed a higher order functional analysis using the functional annotation clustering tool in DAVID, which evaluates each annotation term by the genes that it is comprised of from a given data set and then clusters that term with similar annotations from other ontology databases based on the amount of genes that are co-associated. By this clustering method, we found six functional clustering groups that had a median p value ≤ 0.05 (Table 5). Five of these six functional clusters were associated with DNA transcription and related categories, and one was associated with signaling. Among these co-clustering functional categories, 14.89% of genes altered at nine months were involved in transcription, 13.62% bind DNA, 12.34% bind zinc ions, 4.26% comprise transcription factor complexes, and 8.09% have transcription factor activity. Interestingly, 2.13% show steroid hormone receptor activity. Based on this functional annotation clustering, transcription appeared to be the predominant biological function that was altered in the nine-month group.
Table 5.
Functional annotation clustering in nine-month-old transgenic α-syn mice.
| Functional Group 1 | Median: 0.0034982 Geometric mean: 0.0022242 | ||
|---|---|---|---|
| Database | Term | % | p value |
| GOTERM_BP_ALL | regulation of biological process | 25.11% | 3.05E-05 |
| GOTERM_CC_ALL | nucleus | 24.26% | 5.12E-05 |
| SP_PIR_KEYWORDS | transcription regulation | 10.64% | 1.03E-04 |
| SP_PIR_KEYWORDS | transcription | 10.21% | 2.97E-04 |
| SP_PIR_KEYWORDS | nuclear protein | 17.45% | 6.26E-04 |
| GOTERM_BP_ALL | regulation of cellular process | 21.70% | 8.93E-04 |
| GOTERM_BP_ALL | regulation of metabolism | 17.02% | 0.0013 |
| GOTERM_BP_ALL | regulation of physiological process | 21.28% | 0.0013 |
| GOTERM_BP_ALL | regulation of cellular physiological process | 20.43% | 0.0018 |
| GOTERM_MF_ALL | transcription regulator activity | 10.21% | 0.0021 |
| GOTERM_MF_ALL | DNA binding | 13.62% | 0.0026 |
| GOTERM_MF_ALL | transcription factor activity | 8.09% | 0.0035 |
| GOTERM_BP_ALL | regulation of transcription | 14.89% | 0.0039 |
| GOTERM_BP_ALL | regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolism | 14.89% | 0.0047 |
| GOTERM_BP_ALL | transcription | 14.89% | 0.0064 |
| GOTERM_BP_ALL | regulation of transcription, DNA-dependent | 14.04% | 0.0064 |
| SP_PIR_KEYWORDS | dna-binding | 8.51% | 0.0067 |
| GOTERM_BP_ALL | regulation of cellular metabolism | 15.32% | 0.0076 |
| GOTERM_BP_ALL | transcription, DNA-dependent | 14.04% | 0.0082 |
| GOTERM_MF_ALL | nucleic acid binding | 17.87% | 0.0166 |
| GOTERM_CC_ALL | intracellular membrane-bound organelle | 28.51% | 0.0203 |
| GOTERM_CC_ALL | membrane-bound organelle | 28.51% | 0.0213 |
| GOTERM_BP_ALL | nucleobase, nucleoside, nucleotide and nucleic acid metabolism | 18.30% | 0.0287 |
| Functional Group 2 | Median: 0.00977724 Geometric mean: 0.00506997 | ||
| Database | Term | % | p value |
| SP_PIR_KEYWORDS | transcription regulation | 10.64% | 1.03E-04 |
| GOTERM_MF_ALL | transcription regulator activity | 10.21% | 0.0021 |
| GOTERM_CC_ALL | protein complex | 13.19% | 0.0043 |
| GOTERM_CC_ALL | nucleoplasm | 5.53% | 0.0070 |
| GOTERM_CC_ALL | nuclear lumen | 5.96% | 0.0125 |
| GOTERM_CC_ALL | organelle lumen | 6.38% | 0.0149 |
| GOTERM_CC_ALL | membrane-enclosed lumen | 6.38% | 0.0149 |
| GOTERM_CC_ALL | transcription factor complex | 4.26% | 0.0239 |
| Functional Group 3 | Median: 0.01106735 Geometric mean: 0.00732782 | ||
| Database | Term | % | p value |
| GOTERM_CC_ALL | nucleus | 24.26% | 5.12E-05 |
| GOTERM_CC_ALL | intracellular | 38.72% | 5.43E-04 |
| SP_PIR_KEYWORDS | nuclear protein | 17.45% | 6.26E-04 |
| GOTERM_CC_ALL | intracellular organelle | 33.19% | 0.0030 |
| GOTERM_CC_ALL | organelle | 33.19% | 0.0031 |
| GOTERM_MF_ALL | binding | 48.09% | 0.0045 |
| GOTERM_CC_ALL | cell | 49.36% | 0.0081 |
| GOTERM_BP_ALL | cellular physiological process | 48.94% | 0.0099 |
| GOTERM_BP_ALL | cellular process | 54.47% | 0.0123 |
| GOTERM_CC_ALL | intracellular membrane-bound organelle | 28.51% | 0.0203 |
| GOTERM_CC_ALL | membrane-bound organelle | 28.51% | 0.0213 |
| GOTERM_BP_ALL | primary metabolism | 34.47% | 0.0264 |
| GOTERM_BP_ALL | nucleobase, nucleoside, nucleotide and nucleic acid metabolism | 18.30% | 0.0287 |
| GOTERM_BP_ALL | cellular metabolism | 34.89% | 0.0533 |
| GOTERM_BP_ALL | physiological process | 51.06% | 0.0724 |
| GOTERM_BP_ALL | metabolism | 36.60% | 0.0770 |
| Functional Group 4 | Median: 0.0397 Geometric mean: 0.0175 | ||
| Database | Term | % | p value |
| GOTERM_BP_ALL | intracellular signaling cascade | 8.94% | 0.0031 |
| GOTERM_BP_ALL | signal transduction | 14.47% | 0.0397 |
| GOTERM_BP_ALL | cell communication | 15.32% | 0.0435 |
| Functional Group 5 | Median: 0.023 Geometric mean: 0.0285 | ||
| Database | Term | % | p value |
| SP_PIR_KEYWORDS | Zinc-finger | 8.94% | 0.0038 |
| GOTERM_MF_ALL | Zinc ion binding | 12.34% | 0.0044 |
| SP_PIR_KEYWORDS | metal binding | 11.91% | 0.0115 |
| SP_PIR_KEYWORDS | Zinc | 9.36% | 0.0209 |
| GOTERM_MF_ALL | transition metal ion binding | 13.19% | 0.0245 |
| GOTERM_MF_ALL | cation binding | 15.74% | 0.1200 |
| GOTERM_MF_ALL | metal ion binding | 16.17% | 0.1915 |
| GOTERM_MF_ALL | ion binding | 16.17% | 0.1915 |
| Functional Group 6 | Median: 0.027 Geometric mean: 0.033 | ||
| Database | Term | % | p value |
| GOTERM_MF_ALL | steroid hormone receptor activity | 2.13% | 0.0033 |
| GOTERM_MF_ALL | ligand-dependent nuclear receptor activity | 2.13% | 0.0035 |
| SMART_NAME | SM00399:ZnF_C4 | 2.13% | 0.0053 |
| SMART_NAME | SM00430:HOLI | 2.13% | 0.0063 |
| INTERPRO_NAME | IPR001628:Nuclear hormone receptor, DNA-binding | 1.70% | 0.0093 |
| INTERPRO_NAME | IPR000536:Nuclear hormone receptor, ligand-binding | 1.70% | 0.0105 |
| UP_SEQ_FEATURE | DNA-binding region:Nuclear receptor | 1.70% | 0.0107 |
| UP_SEQ_FEATURE | zinc finger region:NR C4-type | 1.70% | 0.0107 |
| GOTERM_BP_ALL | steroid hormone receptor signaling pathway | 1.28% | 0.0270 |
| GOTERM_BP_ALL | intracellular receptor-mediated signaling pathway | 1.28% | 0.0364 |
| INTERPRO_NAME | IPR001723:Steroid hormone receptor | 1.28% | 0.0727 |
| GOTERM_MF_ALL | steroid binding | 1.28% | 0.0922 |
| GOTERM_MF_ALL | sequence-specific DNA binding | 3.40% | 0.1219 |
| SP_PIR_KEYWORDS | zinc finger | 1.28% | 0.2049 |
| SP_PIR_KEYWORDS | DNA binding | 2.13% | 0.2837 |
| GOTERM_MF_ALL | lipid binding | 1.70% | 0.4329 |
| SP_PIR_KEYWORDS | receptor | 4.68% | 0.7521 |
Gender modifies the effect of α-syn on gene expression
Because gender has recently been found to play an important role in transcriptional changes in human PD (Cantuti-Castelvetri et al., 2007), we evaluated whether gender modified the gene expression changes in the α-syn transgenic mice. The primary analysis described above was conducted in a group of animals where the number of mice of each gender was balanced. To analyze the influence of gender on gene expression patterns between wildtype and transgenic mice, we compared the average gene expression of female α-syn mice to female wildtype littermates and did a similar comparison of male transgenic to male wildtype mice. The group sizes in the gender analysis were necessarily smaller than in the primary analysis, and therefore, may lack statistical power to detect gene expression alterations. For this analysis, we limited the gender-specific data sets by the following criteria: 1) p value ≤0.05, and 2) log2 ratio of at least ±0.5. At both time points, females showed a greater dysregulation in gene expression than males did. 226 genes were altered in three-month-old transgenic females compared to wildtype females, and 332 genes were altered in nine-month-old transgenic females by at least a log2 ratio of ±0.5. The majority of the genes were downregulated in the transgenic female mice compared to gender-matched controls. Expression of 191 genes was decreased, while 35 genes showed increased expression at three months. At nine months, 193 genes showed decreased expression, and 85 had elevated levels in the transgenic females.
In contrast, fewer genes showed alterations in expression in the transgenic males compared to male controls. In three-month-old transgenic males, alterations in gene expression were apparent in 173 genes, while in nine-month-old transgenic males, 188 genes were altered by at least a log2 ratio of ±0.5. Unlike the females, transgenic males had more genes that increased in expression. At three months, 73 genes had decreased expression levels, and 100 had increased levels in transgenic males. At nine months, a decrease in expression was found in 77 genes, and an increase was seen in 111 genes in transgenic males.
A comparison of the identities of the genes regulated in the two genders revealed that the difference between the males and females was not simply the result of a difference in magnitude of regulation. Few of the same genes were altered in both genders at either age. Six genes were altered in both male and female transgenic mice at three months, and 20 were altered in both genders at nine months. Because our group sizes in the gender analysis were small, interpretation of our gender-based findings is limited. While we cannot conclude which genes are necessarily crucial to gender-related differences in α-syn toxicity, this gender analysis does point to the general conclusion that gender impacts the gene expression changes observed in α-syn mice.
Discussion
In this study, we examined the effect of α-syn overexpression in mice on gene expression in the SNpc. Our data showed that α-syn overexpression caused alterations in a small number of genes early in the pathological process, before any cellular or behavioral changes were apparent in the transgenic mice. Alterations in gene expression were more pronounced at a later stage of disease when pathological changes were apparent, and these late changes affected different genes than those modified early. These late gene expression changes were focused around transcription-related processes.Our analysis also suggested that gender modified the alterations of gene expression in this model.
We found striking differences between early changes in gene expression and those observed after pathological changes had clearly developed in the α-syn model. Prior to the onset of pathological changes, gene expression changes in the α-syn transgenic mice were modest. By conservative statistical criteria, we found only 29 genes to be altered. In contrast, 179 genes were altered in older transgenic mice. The larger gene list from the nine-month-old mice analysis reflects in part a reduction in variability among the animals, which presumably is a result of a more stable state of α-syn-related pathology at this later stage. When limiting our data sets by false discovery rates, we found no genes altered at both early and late time periods. Only nine genes were found altered in transgenic mice at both time periods when we evaluated our less stringent gene lists not limited by false discovery rate. Two of these nine genes are implicated with cell cycle regulation – calmodulin 1 (Rasmussen and Means, 1989) and cyclin D2 (Jena et al., 2002). The other seven genes do not seem to share any particular biological processes, with their functions still largely unknown.
The most striking result of our study is that many of the genes altered later in the disease model were directly related to transcriptional regulation, nuclear function, and DNA binding. These data support the view that transcriptional dysregulation is an important consequence of α-syn overexpression and, therefore, may be critical to the pathogenesis of synucleinopathies. Alpha-syn has been identified within cell nuclei and has been shown to associate with histones in vitro (Goers et al., 2003; Maroteaux et al., 1988). Studies by Kontopoulos et al. (2006) have provided direct evidence that α-syn can inhibit histone acetylation in both mammalian cell culture models and transgenic Drosophila. The effect of α-syn on histone acetylation appears critical to α-syn-induced toxicity, as treatment with histone deacetylase inhibitors rescues against α-syn-induced toxicity (Kontopoulos et al., 2006). The present study confirms the transcriptional effect of α-syn in a mouse model for PD.
It is important to recognize that microarray studies do not directly assess transcription. The parameter measured is the abundance of specific mRNAs, which can be affected not only by transcription but also by alterations in RNA half-lives. While we cannot exclude an effect of α-syn on the stability of some transcripts, the large number of alterations as well as the observed changes in mRNAs for many components of the transcriptional process suggests that transcriptional dysregulation is the predominant effect underlying the changes.
Another significant finding is the confirmation in this mouse model that gender influences gene expression changes induced by α-syn overexpression. This analysis was prompted by the recent observation that gender has a marked effect on gene expression in human dopaminergic SN neurons, influencing the patterns of gene expression in both normal brain and the response to PD (Cantuti-Castelvetri et al., 2007). Other microarray studies have also revealed gender differences in gene expression in normal human brain (Vawter et al., 2004), but there is little additional data available on the interaction of gender and disease state on neuronal gene expression in either humans or animal models. The importance of gender is emphasized by the fact that the incidence of PD is nearly twice that in men as in women (Baldereschi et al., 2000; Van Den Eeden et al., 2003). Since our study was originally designed with gender-matched animal groups, we were able to examine the effect of gender directly, although the number of animals in the subgroup analysis is necessarily smaller and lacks statistical power compared to the primary analysis. Nevertheless, we found substantial gender-based differences in the response to α-syn overexpression. We observed that α-syn caused more gene changes among female mice, and most altered genes were different between genders. At present it is not known whether mice exhibit gender-based differences in vulnerability to α-syn toxicity, yet in other animal PD models, males have exhibited increased susceptibility to MPTP (Dluzen and McDermott, 2000; Freyaldenhoven et al., 1996; Miller et al., 1998) and 6-hydroxydopamine (Murray et al., 2003; Tamas et al., 2005). Our findings suggest that issues of gender should be carefully considered in the design of experimental studies using animal models of PD.
The mouse model used here was selected because it demonstrates progressive dopaminergic dysfunction with a motor phenotype (Masliah et al., 2000), but the nature of the model imposes some limitations on our study. Because the mouse SNpc is much more densely packed with cells than in the human SNpc, we dissected the entire SNpc region for this study, and not individual neurons. Thus, the transcriptional changes may reflect alterations in glia as well as neurons. It is also important to note that this transgenic α-syn mouse does not fully recapitulate all pathological aspects of PD. This mouse does show evidence of dopaminergic dysfunction and mild motor impairment, but it does not show loss of dopaminergic nigral neurons (Masliah et al., 2000). Another pathological difference is that α-syn inclusions are much more widespread in Masliah mouse brains than are Lewy bodies found in PD brains. Therefore, many of the expression changes that we saw may be more broadly applicable to other synucleinopathies, such as dementia with Lewy Bodies. All other published α-syn transgenic mice also fail to show dopaminergic neuron loss, yet these models vary in the appearance of α-syn-positive inclusions and severity of motor disability (Hashimoto et al., 2003; Maries et al., 2003). Transgenic mice expressing the A53T α-syn mutant show more severe motor impairments (Giasson et al., 2002; Lee et al., 2002), and some of the other transgenic mice have α-syn inclusions more typical of α-syn aggregates found in PD (Kahle et al., 2000; Lee et al., 2002).
While there are accepted principles for microarray analyses, different approaches to normalization and selection may lead to different outcomes regarding which genes are defined as differentially expressed (Hoffmann et al., 2002). Significance Analysis of Microarrays (SAM) is currently the method most commonly used that corrects for the large number of comparisons inherent in microarray experiments by estimating false discovery rate (Efron and Tibshirani, 2002; Larsson et al., 2005; Tusher et al., 2001). We limited false positives by excluding those genes whose q values were > 20%. However, by using this conservative statistical method, we likely eliminated some genes that are truly altered between wildtype and transgenic animals. Indeed, by QPCR we did confirm alterations in two genes identified by less stringent criteria, Tiparp and cyclin D2, yet both of these genes had high q values in the three-month animals and were thus eliminated from the more stringent three-month list. Attempts to validate directly individual genes introduce additional issues, especially with regard to the selection of genes for normalization. Although genes such as beta-actin and GAPDH have often been used for this purpose, expression of such “housekeeping” genes can vary, especially in neurodegenerative disease (Gutala and Reddy, 2004; Radonic et al., 2004; Vandesompele et al., 2002). Indeed, our array data does show that GAPDH levels are altered in the nine-month-old α-syn transgenic mice. We chose pyruvate decarboxylase and hypoxanthine phosphoribosyltransferase, both of which were unchanged in our microarray lists, to verify that similar results were obtained regardless of the gene chosen for normalization.
Given these considerations, we have decided to publish two sets of gene lists in this paper: 1) the three-month and nine-month tables (Tables 2 & 3) restricted by q values in addition to fold-change and p value criteria; and 2) the supplementary tables (Supp. Tables 1 & 2) restricted only by fold-change and p value criteria. In addition, the full gene array dataset from this study is publicly available on the Array Express site and permits further analysis and comparison with other studies.
In conclusion, we have explored the molecular changes induced by α-syn at a transcriptional level by comparing α-syn transgenic mice to wildtype mice by microarray analysis. Significant transcriptional changes occur in the SNpc of older transgenic mice, and these changes are influenced by gender. A large proportion of the altered genes are involved in transcriptional regulation. The next step is to evaluate whether some of the differentially expressed genes are protective or, alternatively, can magnify α-syn toxicity in cellular and animal models. This study reinforces the concept that targeting transcriptional dysregulation as a mechanism may be a useful therapeutic approach for PD.
Supplementary Material
Acknowledgments
Supported by the MGH/MIT Morris Udall Center of Excellence in PD Research (NIH NS38372) and the Parkinson’s Association of Alabama.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abeliovich A, et al. Mice lacking alpha-synuclein display functional deficits in the nigrostriatal dopamine system. Neuron. 2000;25:239–252. doi: 10.1016/s0896-6273(00)80886-7. [DOI] [PubMed] [Google Scholar]
- Athanassiadou A, et al. Genetic analysis of families with Parkinson disease that carry the Ala53Thr mutation in the gene encoding alpha-synuclein. Am J Hum Genet. 1999;65:555–558. doi: 10.1086/302486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldereschi M, et al. Parkinson’s disease and parkinsonism in a longitudinal study: twofold higher incidence in men. ILSA Working Group. Italian Longitudinal Study on Aging. Neurology. 2000;55:1358–1363. doi: 10.1212/wnl.55.9.1358. [DOI] [PubMed] [Google Scholar]
- Berg D, et al. Specification of 14-3-3 proteins in Lewy bodies. Ann Neurol. 2003;54:135. doi: 10.1002/ana.10621. [DOI] [PubMed] [Google Scholar]
- Betarbet R, et al. Chronic systemic pesticide exposure reproduces features of Parkinson’s disease. Nat Neurosci. 2000;3:1301–1306. doi: 10.1038/81834. [DOI] [PubMed] [Google Scholar]
- Cantuti-Castelvetri I, et al. Effects of gender on nigral gene expression and parkinson disease. Neurobiol Dis. 2007;26:606–614. doi: 10.1016/j.nbd.2007.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha JH. Transcriptional dysregulation in Huntington’s disease. Trends Neurosci. 2000;23:387–392. doi: 10.1016/s0166-2236(00)01609-x. [DOI] [PubMed] [Google Scholar]
- Cope LM, et al. A benchmark for Affymetrix GeneChip expression measures. Bioinformatics. 2004;20:323–331. doi: 10.1093/bioinformatics/btg410. [DOI] [PubMed] [Google Scholar]
- Dauer W, et al. Resistance of alpha -synuclein null mice to the parkinsonian neurotoxin MPTP. Proc Natl Acad Sci U S A. 2002;99:14524–14529. doi: 10.1073/pnas.172514599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Jr, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Dluzen DE, et al. Gender differences in neurotoxicity of the nigrostriatal dopaminergic system: implications for Parkinson’s disease. J Gend Specif Med. 2000;3:36–42. [PubMed] [Google Scholar]
- Efron B, et al. Empirical bayes methods and false discovery rates for microarrays. Genet Epidemiol. 2002;23:70–86. doi: 10.1002/gepi.1124. [DOI] [PubMed] [Google Scholar]
- Eisen MB, et al. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrer M, et al. alpha-Synuclein gene haplotypes are associated with Parkinson’s disease. Hum Mol Genet. 2001;10:1847–1851. doi: 10.1093/hmg/10.17.1847. [DOI] [PubMed] [Google Scholar]
- Fink L, et al. Real-time quantitative RT-PCR after laser-assisted cell picking. Nat Med. 1998;4:1329–1333. doi: 10.1038/3327. [DOI] [PubMed] [Google Scholar]
- Freyaldenhoven TE, et al. The dopamine-depleting effects of 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine in CD-1 mice are gender-dependent. Brain Res. 1996;735:232–238. doi: 10.1016/0006-8993(96)00598-7. [DOI] [PubMed] [Google Scholar]
- Giasson BI, et al. Neuronal alpha-synucleinopathy with severe movement disorder in mice expressing A53T human alpha-synuclein. Neuron. 2002;34:521–533. doi: 10.1016/s0896-6273(02)00682-7. [DOI] [PubMed] [Google Scholar]
- Goers J, et al. Nuclear localization of alpha-synuclein and its interaction with histones. Biochemistry. 2003;42:8465–8471. doi: 10.1021/bi0341152. [DOI] [PubMed] [Google Scholar]
- Gutala RV, et al. The use of real-time PCR analysis in a gene expression study of Alzheimer’s disease post-mortem brains. J Neurosci Methods. 2004;132:101–107. doi: 10.1016/j.jneumeth.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Hashimoto M, et al. Transgenic models of alpha-synuclein pathology: past, present, and future. Ann N Y Acad Sci. 2003;991:171–188. [PubMed] [Google Scholar]
- Hoffmann R, et al. Profound effect of normalization on detection of differentially expressed genes in oligonucleotide microarray data analysis. Genome Biol. 2002;3:RESEARCH0033. doi: 10.1186/gb-2002-3-7-research0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosack DA, et al. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry MC, et al. Nigral and cortical Lewy bodies and dystrophic nigral neurites in Parkinson’s disease and cortical Lewy body disease contain alpha-synuclein immunoreactivity. J Neuropathol Exp Neurol. 1998;57:334–337. doi: 10.1097/00005072-199804000-00005. [DOI] [PubMed] [Google Scholar]
- Jena N, et al. Critical role for cyclin D2 in BCR/ABL-induced proliferation of hematopoietic cells. Cancer Res. 2002;62:535–541. [PubMed] [Google Scholar]
- Kahle PJ, et al. Physiology and pathophysiology of alpha-synuclein. Cell culture and transgenic animal models based on a Parkinson’s disease-associated protein. Ann N Y Acad Sci. 2000;920:33–41. doi: 10.1111/j.1749-6632.2000.tb06902.x. [DOI] [PubMed] [Google Scholar]
- Kawamoto Y, et al. 14-3-3 proteins in Lewy bodies in Parkinson disease and diffuse Lewy body disease brains. J Neuropathol Exp Neurol. 2002;61:245–253. doi: 10.1093/jnen/61.3.245. [DOI] [PubMed] [Google Scholar]
- Kontopoulos E, et al. Alpha-synuclein acts in the nucleus to inhibit histone acetylation and promote neurotoxicity. Hum Mol Genet. 2006;15:3012–3023. doi: 10.1093/hmg/ddl243. [DOI] [PubMed] [Google Scholar]
- Kruger R, et al. Ala30Pro mutation in the gene encoding alpha-synuclein in Parkinson’s disease. Nat Genet. 1998;18:106–108. doi: 10.1038/ng0298-106. [DOI] [PubMed] [Google Scholar]
- Kruger R, et al. Increased susceptibility to sporadic Parkinson’s disease by a certain combined alpha-synuclein/apolipoprotein E genotype. Ann Neurol. 1999;45:611–617. doi: 10.1002/1531-8249(199905)45:5<611::aid-ana9>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Larsson O, et al. Considerations when using the significance analysis of microarrays (SAM) algorithm. BMC Bioinformatics. 2005;6:129. doi: 10.1186/1471-2105-6-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MK, et al. Human alpha-synuclein-harboring familial Parkinson’s disease-linked Ala-53 --> Thr mutation causes neurodegenerative disease with alpha-synuclein aggregation in transgenic mice. Proc Natl Acad Sci U S A. 2002;99:8968–8973. doi: 10.1073/pnas.132197599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, et al. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Maries E, et al. The role of alpha-synuclein in Parkinson’s disease: insights from animal models. Nat Rev Neurosci. 2003;4:727–738. doi: 10.1038/nrn1199. [DOI] [PubMed] [Google Scholar]
- Maroteaux L, et al. Synuclein: a neuron-specific protein localized to the nucleus and presynaptic nerve terminal. J Neurosci. 1988;8:2804–2815. doi: 10.1523/JNEUROSCI.08-08-02804.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masliah E, et al. Dopaminergic loss and inclusion body formation in alpha-synuclein mice: implications for neurodegenerative disorders. Science. 2000;287:1265–1269. doi: 10.1126/science.287.5456.1265. [DOI] [PubMed] [Google Scholar]
- Miller DB, et al. The impact of gender and estrogen on striatal dopaminergic neurotoxicity. Ann N Y Acad Sci. 1998;844:153–165. [PubMed] [Google Scholar]
- Murphy DD, et al. Synucleins are developmentally expressed, and alpha-synuclein regulates the size of the presynaptic vesicular pool in primary hippocampal neurons. J Neurosci. 2000;20:3214–3220. doi: 10.1523/JNEUROSCI.20-09-03214.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray HE, et al. Dose- and sex-dependent effects of the neurotoxin 6-hydroxydopamine on the nigrostriatal dopaminergic pathway of adult rats: differential actions of estrogen in males and females. Neuroscience. 2003;116:213–222. doi: 10.1016/s0306-4522(02)00578-x. [DOI] [PubMed] [Google Scholar]
- Oluwatosin-Chigbu Y, et al. Parkin suppresses wild-type alpha-synuclein-induced toxicity in SHSY-5Y cells. Biochem Biophys Res Commun. 2003;309:679–684. doi: 10.1016/j.bbrc.2003.08.059. [DOI] [PubMed] [Google Scholar]
- Pals P, et al. alpha-Synuclein promoter confers susceptibility to Parkinson’s disease. Ann Neurol. 2004;56:591–595. doi: 10.1002/ana.20268. [DOI] [PubMed] [Google Scholar]
- Polymeropoulos MH, et al. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- Quackenbush J. Microarray data normalization and transformation. Nat Genet. 2002;32(Suppl):496–501. doi: 10.1038/ng1032. [DOI] [PubMed] [Google Scholar]
- Radonic A, et al. Guideline to reference gene selection for quantitative real-time PCR. Biochem Biophys Res Commun. 2004;313:856–862. doi: 10.1016/j.bbrc.2003.11.177. [DOI] [PubMed] [Google Scholar]
- Rasmussen CD, et al. Calmodulin is required for cell-cycle progression during G1 and mitosis. Embo J. 1989;8:73–82. doi: 10.1002/j.1460-2075.1989.tb03350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robakis NK. An Alzheimer’s disease hypothesis based on transcriptional dysregulation. Amyloid. 2003;10:80–85. doi: 10.3109/13506120309041729. [DOI] [PubMed] [Google Scholar]
- Scheinberg IH, et al. Deficiency of ceruloplasmin in patients with hepatolenticular degeneration (Wilson’s disease) Science. 1952;116:484–485. doi: 10.1126/science.116.3018.484. [DOI] [PubMed] [Google Scholar]
- Singleton AB, et al. alpha-Synuclein locus triplication causes Parkinson’s disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- Spillantini MG, et al. Alpha-synuclein in Lewy bodies. Nature. 1997;388:839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- Tamas A, et al. Age and gender differences in behavioral and morphological outcome after 6-hydroxydopamine-induced lesion of the substantia nigra in rats. Behav Brain Res. 2005;158:221–229. doi: 10.1016/j.bbr.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Tan EK, et al. Polymorphism of NACP-Rep1 in Parkinson’s disease: an etiologic link with essential tremor? Neurology. 2000;54:1195–1198. doi: 10.1212/wnl.54.5.1195. [DOI] [PubMed] [Google Scholar]
- Tusher VG, et al. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Den Eeden SK, et al. Incidence of Parkinson’s disease: variation by age, gender, and race/ethnicity. Am J Epidemiol. 2003;157:1015–1022. doi: 10.1093/aje/kwg068. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vawter MP, et al. Gender-specific gene expression in post-mortem human brain: localization to sex chromosomes. Neuropsychopharmacology. 2004;29:373–384. doi: 10.1038/sj.npp.1300337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Bohlen und Halbach O, et al. Haploinsufficiency for trkB and trkC receptors induces cell loss and accumulation of alpha-synuclein in the substantia nigra. Faseb J. 2005;19:1740–1742. doi: 10.1096/fj.05-3845fje. [DOI] [PubMed] [Google Scholar]
- Wang CK, et al. alpha-Synuclein promoter RsaI T-to-C polymorphism and the risk of Parkinson’s disease. J Neural Transm. 2006;113:1425–1433. doi: 10.1007/s00702-006-0435-4. [DOI] [PubMed] [Google Scholar]
- Xu J, et al. Dopamine-dependent neurotoxicity of alpha-synuclein: a mechanism for selective neurodegeneration in Parkinson disease. Nat Med. 2002;8:600–606. doi: 10.1038/nm0602-600. [DOI] [PubMed] [Google Scholar]
- Zarranz JJ, et al. The new mutation, E46K, of alpha-synuclein causes Parkinson and Lewy body dementia. Ann Neurol. 2004;55:164–173. doi: 10.1002/ana.10795. [DOI] [PubMed] [Google Scholar]
- Zhou W, et al. Overexpression of human alpha-synuclein causes dopamine neuron death in rat primary culture and immortalized mesencephalon-derived cells. Brain Res. 2000;866:33–43. doi: 10.1016/s0006-8993(00)02215-0. [DOI] [PubMed] [Google Scholar]
- Zhou W, et al. Overexpression of human alpha-synuclein causes dopamine neuron death in primary human mesencephalic culture. Brain Res. 2002;926:42–50. doi: 10.1016/s0006-8993(01)03292-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.