Abstract
Crystal structures of cleaved and uncleaved forms of the YscU cytoplasmic domain, an essential component of the type III secretion system (T3SS) in Yersinia pestis, have been solved by single-wavelength anomolous dispersion and refined with X-ray diffraction data extending up to atomic resolution (1.13 Å). These crystallographic studies provide structural insights into the conformational changes induced upon auto-cleavage of the cytoplasmic domain of YscU. The structures indicate that the cleaved fragments remain bound to each other. The conserved NPTH sequence that contains the site of the N263-P264 peptide bond cleavage is found on a β-turn which, upon cleavage, undergoes a major reorientation of the loop away from the catalytic N263, resulting in altered electrostatic surface features at the site of cleavage. Additionally, a significant conformational change was observed in the N-terminal linker regions of the cleaved and noncleaved forms of YscU which may correspond to the molecular switch that influences substrate specificity. The YscU structures determined here also are in good agreement with the auto-cleavage mechanism described for the flagellar homolog FlhB and E. coli EscU.
Keywords: type III secretion, Yersinia, molecular switch, atomic resolution, YscU
Introduction
Yersinia pestis is the causative agent of plague, one of the most deadly diseases in history and a potential instrument of bioterrorism.1,2 Y. pestis and many other gram-negative bacterial pathogens use a type III secretion system (T3SS) as a protein transport apparatus to inject a small number of effector proteins through a hollow needle that extends across the inner and outer bacterial membranes and into the cytosol of eukaryotic cells.3–5 The effector Yops (Yersinia outer proteins) enable the pathogenic bacteria to defeat the immune response of the host by interfering with the signal transduction pathways that regulate the actin cytoskeleton, phagocytosis, apoptosis, and the inflammatory response.6 The export apparatus consists in part of cytoplasmic and inner-membrane proteins that identify T3SS substrates and control the switching of substrate specificity during morphogenesis and host-cell contact.3,4
YscU, an essential component of the secretion apparatus in Yersinia, is composed of 354 amino acid residues that are organized into a four-helix transmembrane N-terminal domain and a large C-terminal cytoplasmic domain separated by a long linker region that is highly conserved among YscU orthologs.7 The cytoplasmic domain of YscU undergoes auto-cleavage of the N263-P264 peptide bond at the conserved NPTH site, resulting in an N-terminal fragment, YscUCN (residues 211–263), and a C-terminal fragment, YscUCC (residues 264–354), that remain tightly intertwined and copurify together.8,9 Site-directed mutagenesis of N263 or P264 to alanine abolishes auto-cleavage of YscU, which blocks the export of translocators (LcrV, YopB, and YopD) but not effector proteins via the T3SS.8–10 Therefore, it has been proposed that auto-cleavage of the YscU cytoplasmic domain results in a conformational change that triggers the recognition and export of translocators at the proper time during the assembly of the type III secretion apparatus.9,11–13
We report here the crystal structures of two noncleaved forms of YscU, YscUN263A and YscUN263A/P264A, and an atomic resolution structure of the wild-type cleaved form, YscUcleaved, to elucidate structural changes that occur upon cleavage. These crystallographic studies reveal the conformational changes induced upon auto-cleavage of YscU and provide structural insights into the cleavage mechanism, at very high resolution, that confirm and extend the information gleaned from structures of YscU homologs from Shigella flexneri (Spa40), enteropathogenic Escherichia coli (EscU) and Salmonella typhimurium (SpaS).14,15
Results and Discussion
Structure determination
The noncleaved structure of the cytoplasmic domain of YscU (residues 211–354) was trapped by constructing two mutants, one possessing a single N263A mutation (YscUN263A) and the other a double mutation, N263A/P264A (YscUN263A/P264A), at the NPTH site.9 The YscUN263A/P264A and YscUN263A mutants yielded tetragonal crystals that diffracted X-rays to 1.30 and 1.53 Å, respectively. Hexagonal crystals of YscUcleaved (residues 220–342) were obtained that diffracted X-rays up to a resolution of 1.13 Å using synchrotron radiation. Because there were no structural homologs available at the time of structure determination, the structures of YscUcleaved and YscUN263A/N264A were solved by single-wavelength anomalous dispersion (SAD) using crystals soaked in potassium iodide and sodium bromide, respectively. The experimental electron density maps obtained from both crystal forms were of excellent quality (figure of merit = 0.66 and 0.70 after density modification, respectively) which allowed for almost complete auto-tracing of the main chain atoms and a majority of the side chains using the program SOLVE/RESOLVE.16 The structure of YscUcleaved was refined with data extending to 1.13 Å resolution and a final working R-factor of 16.4% and R-free of 16.8%. The high resolution of the data allowed for the placement of hydrogen atoms and several alternate side chain conformations in the final refined model. Data collection and refinement statistics are presented in Table I.
Table I.
Data Collection and Refinement Statistics
| Parameter | YscUcleaved | YscUcleaved (Iodide derivative) | YscUN263A/P264A | YscUN263A/N264 A (Bromide derivative) | YscUN263A |
|---|---|---|---|---|---|
| Data collection statistics | |||||
| Wavelength (Å) | 1.0 | 1.5418 (CuKα) | 1.0 | 0.91957 (peak) | 0.97921 |
| Space group | P6522 | P6522 | P43212 | P43212 | P43212 |
| Unit cell dimensions | |||||
| a. b. c (Å) | 48.2, 48.2, 190.2 | 48.1, 48.1, 190.2 | 66.0, 66.0, 70.7 | 66.3, 66.3, 70.8 | 65.8, 65.8, 66.7 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 120 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Molecules/A.U.a | 1 | 1 | 1 | 1 | 1 |
| Resolution range (Å)b | 50–1.13 (1.16–1.13) | 50–2.14 (2.22–2.14) | 50–1.30 (1.34–1.30) | 50–1.96 (2.03–1.96) | 50–1.53 (1.58–1.53) |
| Total reflections | 665037 | 144918 | 303431 | 139218 | 303021 |
| Unique reflections | 49262 | 7876 | 37461 | 11899 | 22446 |
| Completeness (%) | 97.6 (91.1) | 99.2 (92.3) | 95.5 (66.5) | 99.7 (100) | 98.4 (94.6) |
| Redundancy | 13.5 (11.3) | 18.4 (15.7) | 8.1 (3.1) | 11.7 (11.9) | 13.5 (10.6) |
| I/(σ)I | 50.4 (6.2) | 48.5 (27.1) | 19.2 (1.6) | 20.8 (4.9) | 63.4 (3.2) |
| Rsym (%) | 4.0 (39.9) | 4.8 (9.5) | 8.1 (58.7) | 10.8 (66.4) | 5.3 (61.1) |
| Refinement statistics | |||||
| Resolution range (Å) | 41.7–1.13 | 48.3–1.30 | 46.9–1.53 | ||
| R-factor (%) | 16.4 (26.7) | 21.9 (41.7) | 18.9 (20.6) | ||
| R-free (%)c | 16.8 (29.2) | 24.8 (39.0) | 21.4 (24.7) | ||
| Protein molecules/A.U. | 1 | 1 | 1 | ||
| solvent molecules/A.U. | 0 | 0 | 4 (PEG 400) | ||
| Water molecules/A.U. | 166 | 172 | 131 | ||
| r.m.s. deviations | |||||
| Bond lengths (Å) | 0.023 | 0.017 | 0.014 | ||
| Bond angles (°) | 2.23 | 1.70 | 1.70 | ||
| Ramachandran plot | |||||
| Most favored (%) | 93.2 | 91.3 | 91.2 | ||
| Additionally allowed (%) | 4.5 | 6.7 | 6.9 | ||
| Generously allowed (%) | 2.3 | 1.9 | 1.0 | ||
| Disallowed region (%) | 0 | 0 | 1.0 | ||
| PDB code | 2jli | 2jlj | 2jlh | ||
A.U. (asymmetric unit).
Values in parenthesis represent the highest resolution shell of data.
Calculated with 5% of the reflections.
The structure of YscUN263A
The structures of the two uncleaved YscU mutants (YscUN263A/P264A and YscUN263A) are virtually identical. The following discussion will focus on the YscUN263A mutant. YscUN263A adopts an α2, β3, α, β, α2 fold in which the central core of the structure is composed of a four-stranded mixed β-sheet that is surrounded by five α-helices [Fig. 1(A)]. Although this form of YscU includes the putative linker region (residues 211–245) that connects the N-terminal membrane-bound domain with the cytoplasmic C-terminal domain,7 no electron density was observed for residues 211–230 in YscUN263A, suggesting that they are disordered. Although the core of the YscUN263A structure is highly similar to its EscUN262A homolog (r.m.s.d. = 1.4 Å over 94 Cα atoms), the N-terminal region of YscUN263A differs significantly from the structure of EscUN262A [Fig. 2(A)] in that nearly all of the linker region in EscU is disordered whereas in YscUN263A residues 235–245 form a four-turn α-helix (α1) that is oriented approximately perpendicularly to the central β-strand core.15 We examined the crystal packing environment to see if the differences in the conformation of the N-terminal region may be a crystallographic artifact. Although the N-terminal helix packs against neighboring symmetry mates, there is also available solvent space that could accomodate movement of the helix. Thus, it is unlikely that the formation of a well-ordered α-helix at the N-terminus is influenced by the crystal packing environment.
Figure 1.
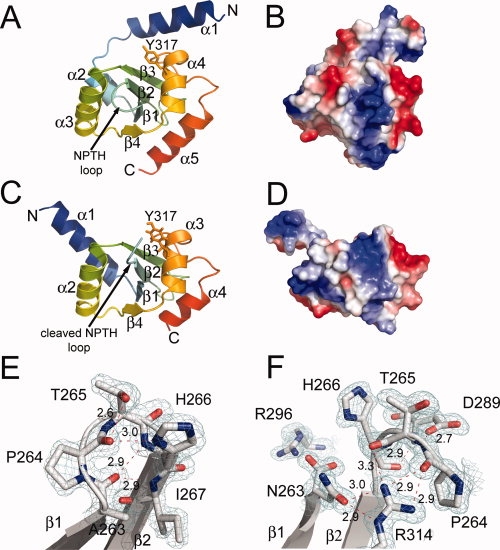
(A) Ribbon-drawing of the crystal structure of the noncleaved YscUN263A mutant and (B) the electrostatic potential mapped onto the solvent exposed surface of YscUN263A in the same orientation. (C) Ribbon drawing of the crystal structure of the cleaved form of YscU (YscUcleaved) and (D) the electrostatic potential mapped onto the surface of YscUcleaved. (E) The molecular interactions of the NPTH residues located on the β-turn between strands β1 and β2 in the YscUN263A structure and (F) after cleavage in the YscUcleaved structure with carbon (gray), nitrogen (blue), and oxygen (red) atoms shown in stick form. The 2Fo-Fc electron density maps are shown contoured at 1.5 σ level. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]:
Figure 2.
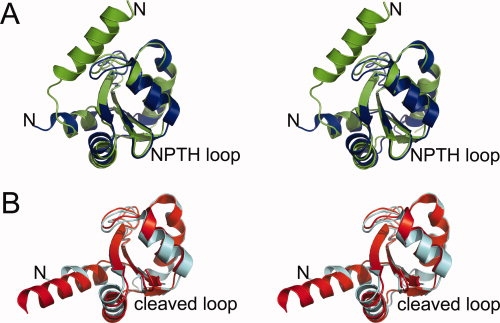
(A) Stereoview of the structural overlay of the crystal structures of YscUN263A (green; pdb code: 2jlh) and EscUN262A (blue; pdb code: 3bzp), and (B) YscUcleaved (red; pdb code: 2jli) and EscUcleaved (cyan; pdb code: 3bzo). [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]:
The conserved NPTH cleavage site is located on a solvent-exposed β-turn between strands β1 and β2 [Fig. 1(A)]. These residues are involved in an extensive hydrogen bonding network that probably serves to promote autocatalysis [Fig. 1(E)]. The H266 imidazole side chain forms a hydrogen bond with the carbonyl oxygen of P264 and there is an extensive hydrogen bonding network between the backbone atoms of A263, P264, H266, and I267. This hydrogen bonding network places the carbonyl oxygen of A263 at a distance of 3.0 Å from the carbonyl oxygen of P264, which creates an n → π* interaction that is proposed to be important for exerting electron withdrawal effects on the N263 carbonyl to promote catalysis.15,17 The NPTH backbone atoms overlay well with the homologous residues in the EscUN262A crystal structure, but the conformation of the H266 imidazole in YscUN263A differs significantly from that observed in EscUN262A. The orientation of the histidine side chain in the EscUN262A structure does not allow for the imidazole ring to participate in hydrogen bonding interactions as observed in YscUN263A. In contrast, the EscU histidine side chain protrudes out into the solvent.15 Thus, in the YscUN263A crystal structure, the H266 imidazole appears to be directly involved in the hydrogen bonding network of the NPTH β-turn in contrast to EscU, although the different conformations of the imidazole side chain in the two structures do not appear to affect catalysis.15
Ferris et al. proposed a cleavage mechanism for the flagellar paralog of YscU, FlhB, in which the protein undergoes auto-cleavage of the peptide bond between asparagine and proline at the conserved NPTH site, and Zarivach et al. have recently provided structural evidence to support such an intein-like, auto-cleavage mechanism involving asparagine cyclization by determining the crystal structure of the E. coli YscU ortholog EscU in the cleaved form along with several structures of uncleaved mutants.15,18 The auto-cleavage mechanism requires a specific conformation of the NPTH loop in which the NPTH sequence adopts a type II β-turn, giving rise to a strained conformation that is required for efficient catalysis to occur. The cleavage reaction is initiated by the lone pair of electrons associated with the side chain Nδ atom of N263 attacking the partially positively charged P264 carbonyl carbon atom to create a cyclized tetrahedral intermediate. After collapse of the tetrahedral intermediate, a proton is transferred from water to the P264 amide to create a new N-terminus at P264 and a succinimide intermediate at N263, which is then hydrolyzed back to a C-terminal asparagine residue.15 An overlay of the YscUN263A and EscUN262A crystal structures [Fig. 2(A)] indicates that both proteins share essentially the same conformation of the NPTH β-turn and many of the same intramolecular interactions. Therefore, the crystal structure of YscUN263A is in good agreement with the mechanism proposed by Zarivach et al.
The structure of wild-type YscU (YscUcleaved)
We were unable to crystallize a wild-type form of YscU with the same boundaries as the mutants (residues 211–354), but were able to obtain crystals of a shorter fragment (residues 220–342) with endpoints that correspond to the limits of visible electron density in the structure of YscUN263A. Purified YscUcleaved (residues 220–342) undergoes auto-cleavage to create a YscUCN fragment (residues 220–263) and a YscUCC fragment (residues 264–342) that remain tightly bound to each other after cleavage and copurify. The overall structure of YscUcleaved is very similar to those of the cleaved forms of EscU [Fig. 2(B)], SpaS, and Spa40,14,15 as indicated by r.m.s.d. values of 1.12 (89 Cα atoms), 1.16 (93 Cα atoms), and 0.99 (88 Cα atoms) Å, respectively. The core of the YscUcleaved structure retains essentially the same conformation after cleavage (r.m.s.d. = 0.7 Å for 78 Cα residues) [Fig. 1(C)], but there is a very large conformational change in the N-terminal linker region. In the structure of YscUcleaved, residues 220–239 are not visible in the electron density map, while residues 240–249 undergo a large shift and are incorporated into a five-turn α-helix composed of residues 240–256. The linker region, which is highly conserved among YscU orthologs, has been proposed to take on the role of a molecular switch by providing a potential point of conformational freedom for the cytoplasmic domain.15 Indeed, deletion and point mutations in the linker region of FlhB and EscU exhibited alterations in type III secretion.15,19
The structures of EscU, SpaS, Spa40, and noncleaved EscU mutants exhibit a substantial degree of variability in the conformation of their N-terminal linkers, indicative of inherent flexibility in this region of the molecules.14,15 In the structures of YscU reported here, the visible portion of the linker adopts an α-helical conformation that undergoes a radical change in its orientation with respect to the core of the molecule upon autocleavage [Figs. 1(A.C) and 2]. This conformational change could very well have a significant impact on the orientation of the cytoplasmic domain of YscU with respect to its membrane-bound N-terminal domain and may therefore correspond to the “molecular switch” that influences the chronological order in which specific classes of proteins are delivered through the type III secretion apparatus.
The cleavage of the N263-P264 peptide bond also results in a major reorientation of the P264-H266 loop in which P264 moves ∼10.7 Å away from the Cα atom of N263, as observed in the structures of Spa40, EscU, and SpaS [1(F)].14,15 This rearrangement results in a new chemical environment for P264, T265, H266, and several other residues that were buried by the β-turn between β1 and β2 in the uncleaved conformation. Additionally, the negative charge on the C-terminus of N263 created after cleavage of the peptide bond is stabilized by an interaction with the positively charged side chain of R296 located on the α3 helix. The newly formed surface is electrostatically distinct from that of the uncleaved form, and may be a critical recognition site for other T3SS proteins [Figs. 1(B,D)].14,15 Additional evidence for the importance of this region comes from the observation that the YscU Y317D mutation, located on the α2 helix directly below the NPTH loop in YscUcleaved [Fig. 1(C)], results in reduced export of the inner rod protein YscI, which is an important process for switching of substrate specificity in the Yersinia T3SS.13 The new orientation of the side chains of P264, T265, and H266 may also play an important role in the interaction of YscUcleaved with other T3SS proteins. Indeed, Zarivach et al. have previously shown that the homologous H265 residue in EscU may play a structural role in the secretion process, as their data have revealed that the H265A mutant does not inhibit the cleavage of EscU but does inhibit secretion.15 Although it has been proposed that auto-cleavage of the YscU cytoplasmic domain results in a conformational change that triggers recognition and export of translocators at the proper time during the assembly of the type III secretion apparatus,9,11–13 we were unable to detect a stable interaction between YscUcleaved and either LcrV or an LcrV/LcrG complex by gel filtration chromatography (data not shown). It is possible, therefore, that YscU plays an indirect role in the recognition of the translocators.
As expected, the structure of Y. pestis YscU is similar to those of EscU, SpaS and Spa40. However, atomic resolution data (1.13 Å) collected from crystals of YscUcleaved gave rise to an electron density map of exceptionally high quality and detail, enabling a number of hydrogen atoms to be added to the molecular model. Moreover, differences in the conformation of the linker region were observed in the structures of YscUN263A and YscUcleaved that were not seen in the structures of its orthologs but which may be of biological significance. The structures reported here should facilitate ongoing studies of the biological role of YscU in the prototypical T3SS of pathogenic Yersiniae.
Materials and Methods
Cloning, expression, and purification
Three forms of the cytosolic domain of YscU were overproduced in E. coli and purified to homogeneity: YscU220-342 (wild-type), YscU211-354 (N263A), and YscU211-354 (N263A/P264A). All YscU constructs were made as HisMBP fusion proteins with a cleavage site for tobacco etch virus (TEV) protease between the HisMBP and YscU domains by recombinational cloning, using the destination vector pDEST-HisMBP as described.20 Site-directed mutants of YscU were constructed by overlap extension PCR21 or QuickChange mutagenesis (Stratagene, La Jolla, CA) and verified by DNA sequencing. The HisMBP-YscU expression vectors were transformed into E. coli BL21(DE3) CodonPlus-RIL cells (Stratagene). Single ampicillin/chloremphenicol-resistant colonies were used to inoculate Luria broth containing 100 μg/mL ampicillin, 30 μg/mL chloramphenicol and 0.2% glucose. The cultures were grown to an OD600 of 0.4–0.6 at 37°C and then induced with 1 mM IPTG for 4 h at 30°C. The cells were harvested by centrifugation and frozen at −80°C until use.
Each YscU variant was purified in exactly the same manner and all steps were carried out at 4°C. The cell pellet was resuspended in 50 mM sodium phosphate pH 8.0, 150 mM NaCl, 25 mM imidazole, and the cells were disrupted using a APV Model G1000 homogenizer (Invensys, Roholmsvej, Denmark). The lysate was centrifuged at 15,000 rpm at 4°C using an SA-600 rotor, filtered, and then the HisMBP fusion proteins were purified by immobilized metal affinity chromatography (IMAC) as described.20 Fractions containing the fusion proteins were pooled, cleaved overnight with hexahistidine-tagged TEV protease,22 and then subjected to another round of IMAC as described.20 The flow-through fractions containing YscU were pooled and applied to an amylose column (New England Biolabs, Beverely, MA) to remove a small amount of His6-MBP that did not bind to the second Ni-NTA column. The flow-through fractions were concentrated to 5 mL, using an Amicon stirred cell with a YM10 membrane (Millipore, Billerica, MA) and applied to a 26/60 Superdex-75 preparative size exclusion column (GE Healthcare, Piscataway, NJ) equilibrated in 25 mM Tris pH 7.5, 150 mM NaCl, 2 mM Tris(2-carboxyethyl) phosphine hydrochloride (TCEP) and the peak fractions corresponding to YscU were pooled and concentrated to ∼20 mg/mL with an Amicon stirred cell (Millipore) as mentioned.
Protein crystallization
Crystallization trials were conducted using sparse-matrix screening kits from Hampton Research (Aliso Viejo, CA), Qiagen (Valencia, CA) and Axygen Biosciences (Union City, CA) at 18°C. The YscUN263A/P264A mutant was crystallized by mixing a 1:1 ratio of protein (14 mg/mL) with well solution consisting of 0.1 M Hepes pH 7.0, 1 M sodium malonate pH 7.0, and 0.75 M sodium chloride. The crystals grew within 1 week and were cryoprotected by immersion into perfluoropolyether PFO-X175/08 (Hampton Research) and flash frozen by plunging into liquid nitrogen. The YscUN263A crystals were obtained by mixing a 1:1 ratio of protein (13.6 mg/mL) and well solution consisting of 0.1 M Hepes pH 7.5 and 1 M sodium malonate, pH 7.0. The crystals were cryoprotected by soaking for 1 min in well solution supplemented with 30% (v/v) PEG 400. Crystals of YscUcleaved were obtained by mixing a 1:1 ratio of protein (19 mg/mL) and well solution consisting of 0.1 M CHES pH 9.5 and 30% (v/v) PEG 400. A single crystal with dimensions of 0.5 mm × 0.2 mm × 0.2 mm grew within 1 month and was captured with a litholoop and flash frozen in liquid nitrogen. Additional crystals of YscUcleaved were obtained in 0.1 M Tris pH 8.5 and 30% (v/v) PEG 400 for use in derivative screening by streak seeding with crystals from the prior condition. Derivatives for phasing were prepared by the quick soak halide method.23 A crystal of YscUN263A/P264A was soaked in 0.1 M Hepes pH 7.0, 1 M sodium malonate, 0.75 M sodium chloride, and 1 M sodium bromide for 30 s, dipped in perfluoropolyether PFO-X175/08 and flash frozen in liquid nitrogen. An iodide derivative of YscUcleaved was obtained by soaking a crystal in 0.1 M Tris-HCl pH 8.5, 30% (v/v) PEG 400, and 1 M potassium iodide for 30 s and directly flash freezing in liquid nitrogen.
Data collection, phasing, and structure refinement
Native data sets for YscUcleaved, YscUN263A, and YscUN263A/P264A crystals were collected at the SER-CAT 22-ID and 22-BM beamlines (Advanced Photon Source, Argonne National Laboratory). A SAD data set was collected on a single YscUN263A/P264A crystal soaked with bromide using the peak wavelength set at 0.91957 Å. X-ray diffraction data from a single YscUcleaved crystal soaked with iodide was collected at 1.5418 Å wavelength with a MAR345 image plate detector mounted on a Rigaku RU-H3R generator operated at 50 kV and 100 mA (Rigaku Corporation, The Woodlands, TX). All data sets were processed with the HKL3000 program suite24 and the processed data were submitted into the SGXPRO program platform for automatic structure solution.25 The YscUN263A/P264A structure was solved by locating eight bromide ions in the asymmetric unit with data extending to 1.96 Å using SHELXD26 and the handedness was subsequently determined by ISAS.27 The phases were further improved by refinement of heavy atoms in SOLVE.28 After density modification, 65% of the residues were automatically built by RESOLVE.16 The model was completed using COOT.29 The final model was then used to search for a molecular replacement solution using the higher resolution 1.30 Å data from the YscUN263A/P264A crystal and the 1.53 Å resolution data collected from the YscUN263A crystal with the program MOLREP from the CCP4 suite.30 The model was refined using REFMAC531 and manually corrected using COOT.
The YscUcleaved structure was solved by locating 10 iodide atoms in the asymmetric unit using the SGXPRO program platform and data to 2.14 Å resolution as described earlier. The resulting model was then used as the starting search model for molecular replacement using the 1.13 Å resolution data and was refined using REFMAC5. Anisotropic B-factor refinements were conducted and hydrogen atoms were added to the final model. During the course of refinement, the electron density maps for P264 exhibited extra electron density protruding from the amide nitrogen. Upon seeing this density, we investigated whether this might be a covalent adduct on P264 by using mass spectrometry. However, mass spectral analysis of the protein in solution and of the crystals dissolved in buffer after data collection returned molecular weights which agreed with the sequence of the protein and did not indicate any additional mass in the protein crystal (data not shown). Moreover, the original experimental electron density maps obtained after SAD phasing did not show any extra electron density on the amide nitrogen to suggest the presence of an adduct. Therefore, water molecules were placed in this density and they refined well. All refined models were validated using Molprobity32 and figures were prepared using Pymol (Delano Scientific LLC, Palo Alto, CA). Coordinates and structure factors for the YscUN263A, YscUN263A/P264A, and YscUcleaved structures were deposited in the Protein Data Bank with accession codes 2jlh, 2jlj, and 2jli, respectively.
Acknowledgments
We thank Dr. Alexander Wlodawer for technical assistance and helpful discussions. Electrospray mass spectrometry experiments were conducted on the LC/ESMS instrument maintained by the Biophysics Resource in the Structural Biophysics Laboratory, Center for Cancer Research, National Cancer Institute at Frederick. X-ray diffraction data were collected at the Southeast Regional Collaborative Access Team (SER-CAT) beamlines 22-ID and 22-BM at the Advanced Photon Source, Argonne National Laboratory. Supporting institutions may be found at-http://www.ser-cat.org/members.html.
References
- 1.Rollins SE, Rollins SM, Ryan ET. Yersinia pestis and the plague. Am J Clin Pathol. 2003;119:S78–S85. doi: 10.1309/DQM9-3R8Q-NQWB-FYU8. [DOI] [PubMed] [Google Scholar]
- 2.Ligon BL. Plague: a review of its history and potential as a biological weapon. Semin Pediatr Infect Dis. 2006;17:161–170. doi: 10.1053/j.spid.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 3.Cornelis GR. The type III secretion injectisome. Nat Rev Microbiol. 2006;4:811–825. doi: 10.1038/nrmicro1526. [DOI] [PubMed] [Google Scholar]
- 4.Galan JE, Wolf-Watz H. Protein delivery into eukaryotic cells by type III secretion machines. Nature. 2006;444:567–573. doi: 10.1038/nature05272. [DOI] [PubMed] [Google Scholar]
- 5.Blocker AJ, Deane JE, Veenendaal AK, Roversi P, Hodgkinson JL, Johnson S, Lea SM. What's the point of the type III secretion system needle? Proc Natl Acad Sci USA. 2008;105:6507–6513. doi: 10.1073/pnas.0708344105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Navarro L, Alto NM, Dixon JE. Functions of the Yersinia effector proteins in inhibiting host immune responses. Curr Opin Microbiol 2005. 2005;8:21–27. doi: 10.1016/j.mib.2004.12.014. [DOI] [PubMed] [Google Scholar]
- 7.Allaoui A, Woestyn S, Sluiters C, Cornelis GR. YscU, a Yersinia enterocolitica inner membrane protein involved in Yop secretion. J Bacteriol. 1994;176:4534–4542. doi: 10.1128/jb.176.15.4534-4542.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lavander M, Sundberg L, Edqvist PJ, Lloyd SA, Wolf-Watz H, Forsberg A. Proteolytic cleavage of the FlhB homologue YscU of Yersinia pseudotuberculosis is essential for bacterial survival but not for type III secretion. J Bacteriol. 2002;184:4500–4509. doi: 10.1128/JB.184.16.4500-4509.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sorg I, Wagner S, Amstutz M, Muller SA, Broz P, Lussi Y, Engel A, Cornelis GR. YscU recognizes translocators as export substrates of the Yersinia injectisome. Embo J. 2007;26:3015–3024. doi: 10.1038/sj.emboj.7601731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lavander M, Sundberg L, Edqvist PJ, Lloyd SA, Wolf-Watz H, Forsberg A. Characterisation of the type III secretion protein YscU in Yersinia pseudotuberculosis. YscU cleavage–dispensable for TTSS but essential for survival. Adv Exp Med Biol. 2003;529:109–112. doi: 10.1007/0-306-48416-1_20. [DOI] [PubMed] [Google Scholar]
- 11.Edqvist PJ, Olsson J, Lavander M, Sundberg L, Forsberg A, Wolf-Watz H, Lloyd SA. YscP and YscU regulate substrate specificity of the Yersinia type III secretion system. J Bacteriol. 2003;185:2259–2266. doi: 10.1128/JB.185.7.2259-2266.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riordan KE, Schneewind O. YscU cleavage and the assembly of Yersinia type III secretion machine complexes. Mol Microbiol. 2008;68:1485–1501. doi: 10.1111/j.1365-2958.2008.06247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wood SE, Jin J, Lloyd SA. YscP and YscU switch the substrate specificity of the Yersinia type III secretion system by regulating export of the inner rod protein YscI. J Bacteriol. 2008;190:4252–4262. doi: 10.1128/JB.00328-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deane JE, Graham SC, Mitchell EP, Flot D, Johnson S, Lea SM. Crystal structure of Spa40, the specificity switch for the Shigella flexneri type III secretion system. Mol Microbiol. 2008;69:267–276. doi: 10.1111/j.1365-2958.2008.06293.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zarivach R, Deng W, Vuckovic M, Felise HB, Nguyen HV, Miller SI, Finlay BB, Strynadka NC. Structural analysis of the essential self-cleaving type III secretion proteins EscU and SpaS. Nature. 2008;453:124–127. doi: 10.1038/nature06832. [DOI] [PubMed] [Google Scholar]
- 16.Terwilliger T. SOLVE and RESOLVE: automated structure solution, density modification and model building. J Synchrotron Radiat. 2004;11:49–52. doi: 10.1107/s0909049503023938. [DOI] [PubMed] [Google Scholar]
- 17.Hodges JA, Raines RT. Energetics of an n –> pi interaction that impacts protein structure. Org Lett. 2006;8:4695–4697. doi: 10.1021/ol061569t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferris HU, Furukawa Y, Minamino T, Kroetz MB, Kihara M, Namba K, Macnab RM. FlhB regulates ordered export of flagellar components via autocleavage mechanism. J Biol Chem. 2005;280:41236–41242. doi: 10.1074/jbc.M509438200. [DOI] [PubMed] [Google Scholar]
- 19.Fraser GM, Hirano T, Ferris HU, Devgan LL, Kihara M, Macnab RM. Substrate specificity of type III flagellar protein export in Salmonella is controlled by subdomain interactions in FlhB. Mol Microbiol. 2003;48:1043–1057. doi: 10.1046/j.1365-2958.2003.03487.x. [DOI] [PubMed] [Google Scholar]
- 20.Tropea JE, Cherry S, Nallamsetty S, Bignon C. Waugh DS. A generic method for the production of recombinant proteins in Escherichia coli using a dual hexahistidine-maltose-binding protein affinity tag. Methods Mol Biol. 2007;363:1–19. doi: 10.1007/978-1-59745-209-0_1. [DOI] [PubMed] [Google Scholar]
- 21.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 22.Kapust RB, Tozser J, Fox JD, Anderson DE, Cherry S, Copeland TD, Waugh DS. Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 2001;14:993–1000. doi: 10.1093/protein/14.12.993. [DOI] [PubMed] [Google Scholar]
- 23.Dauter M, Dauter Z. Phase determination using halide ions. Methods Mol Biol. 2007;364:149–158. doi: 10.1385/1-59745-266-1:149. [DOI] [PubMed] [Google Scholar]
- 24.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution–from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 25.Fu ZQ, Rose J, Wang BC. SGXPro: a parallel workflow engine enabling optimization of program performance and automation of structure determination. Acta Crystallogr D Biol Crystallogr. 2005;61:951–959. doi: 10.1107/S0907444905010541. [DOI] [PubMed] [Google Scholar]
- 26.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr D Biol Crystallogr. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 27.Wang BC. Resolution of phase ambiguity in macromolecular crystallography. Methods Enzymol. 1985;115:90–112. doi: 10.1016/0076-6879(85)15009-3. [DOI] [PubMed] [Google Scholar]
- 28.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D Biol Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 30.Vagin A, Teplyakov A. An approach to multi-copy search in molecular replacement. Acta Crystallogr D Biol Crystallogr. 2000;56:1622–1624. doi: 10.1107/s0907444900013780. [DOI] [PubMed] [Google Scholar]
- 31.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 32.Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–W619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]


