Abstract
Molecular epidemiology studies have allowed the identification of the methicillin (meticillin)-resistant (MRSA) and methicillin-susceptible (MSSA) clonal complexes (CCs) and clones of Staphylococcus aureus circulating in a Spanish hospital recently. Of 81 isolates tested, 32.1% were MRSA. Most of them carried staphylococcal cassette chromosome mec (SCCmec) IVc (88.5%) and belonged to CC5 (88.5%; multilocus sequence typing types ST125 [mainly associated with spa type t067], ST5, and ST228). A higher diversity was found among MSSA isolates (67.9%). Eighty percent shared the genetic background of major MRSA lineages (CC5 [38.2%; ST125 and ST5], CC30 [25.5%; ST30], CC45 [14.5%; ST45 and ST47], and CC8 [1.8%; ST8]), but CC12, CC15, CC51, and CC59 were also detected. Many exotoxin genes were present in each of the 81 isolates, independent of whether they were involved in sepsis (11 to 22) or other types of infections (13 to 21), and they appeared in 73 combinations. The relevant data are that (i) all isolates were positive for hemolysin and leukotoxin genes (98.8% for lukED and 25.9% for lukPV); (ii) all contained an enterotoxin gene cluster (egc with or without seu), frequently with one or more genes encoding classical enterotoxins; (iii) about half were positive for tst and 95% were positive for exfoliatin-encoding genes (eta, etb, and/or etd); and (iv) the four agr groups were detected, with agrII (55.6%) and agrIII (23.5%) being the most frequent. Taken together, results of the present study suggest a frequent acquisition and/or loss of exotoxin genes, which may be mediated by efficient intralineage transfer of mobile genetic elements and exotoxin genes therein and by eventual breakage of interlineage barriers.
Staphylococcus aureus is both a commensal bacterium and an extremely versatile pathogen that causes a wide range of diseases in humans, including superficial, deep-seated, and systemic infections, as well as a variety of toxemic syndromes, such as toxic shock syndrome (TSS), staphylococcal scalded-skin syndrome (SSSS), and staphylococcal food poisoning (36). S. aureus produces a wide range of virulence factors that mediate host colonization, invasion of damaged skin and mucosa, dissemination through the body, and evasion of host defense mechanisms (8, 12). Relevant among them are a variety of exotoxins that comprise α-, β-, γ-, and δ-hemolysins, leukotoxins (the classical LukS-PV-LukF-PV Panton-Valentine leukocidin [LukPV], LukE-LukD [LukED] and LukM-LukF′-PV [LukM]), exfoliative toxins, and pyrogenic toxin superantigens, such as the staphylococcal TSS toxin (TSST-1, first referred to as SEF) and staphylococcal enterotoxins (SEs) (14, 29, 43). Five major serological types of SEs, SEA through SEE (known as classical enterotoxins, encoded by the sea to see genes, respectively) have been initially identified. However, new types of SEs and their coding genes (seg through seu) were later reported. Several SE genes (seg, sei, sem, sen, and seo) are part of an operon termed the enterotoxin gene cluster (egc), of which a variant that contains seu instead of the two pseudogenes present in the originally described egc between sei and sen has been identified (27, 33). Both TSST-1 and SEs are potent activators of T-cell populations, leading to massive proliferation and uncontrolled release of proinflammatory cytokines (14). Expression of most virulence factors in S. aureus is under the control of the agr (accessory gene regulator) locus, which encodes a two-component signaling pathway and its activating ligand, a bacterial-density-sensing peptide termed the autoinducing peptide (37). S. aureus strains can be subdivided into four major agr groups, based on polymorphisms in the amino acid sequence of the autoinducing peptide and other components of the system (26, 28). Within a given group, each strain produces a peptide that can activate the agr response in other members of the group, whereas the autoinducing peptides produced by different groups are usually mutually inhibitory (26).
Apart from having pathogenic versatility, S. aureus can adapt rapidly to the selective pressure of antibiotics, with the emergence and spread of methicillin (meticillin)-resistant S. aureus (MRSA) isolates being a relevant example. Resistance to methicillin and other beta-lactam antibiotics is caused by the mecA gene, situated on a mobile genetic element, the staphylococcal cassette chromosome mec (SCCmec), which consists of the mec gene complex, the ccr gene complex, and the “junkyard” regions. Based on the variability of the differently combined components, several types of SCCmec and several variants of the types have been distinguished (21, 23, 24, 25, 30, 31, 38, 45, 51).
In the present work, the techniques most commonly applied in epidemiological studies of S. aureus were used to identify the prevalent and sporadic MRSA and methicillin-susceptible S. aureus (MSSA) clones that have been causing disease in a Spanish hospital (the Hospital Universitario Central de Asturias [HUCA]) over a recent time period (2005 to 2006). These methods included pulsed-field gel electrophoresis (PFGE) of SmaI-digested genomic DNA (SmaI PFGE), S. aureus protein A gene (spa) typing, multilocus sequence typing (MLST) analysis, and SCCmec typing of MRSA (4, 9, 40, 42, 50, 52). The risk for human health posed by the accumulation of virulence genes in S. aureus (34) along with the potential application of such genes for subtyping prompted the assessment of the virulence gene repertoire of the HUCA isolates, with regard to the agr group and 30 exotoxin-encoding genes.
MATERIALS AND METHODS
S. aureus isolates.
Eighty-one S. aureus isolates were analyzed in this study. Each was collected from a different patient attending the HUCA from December 2005 to December 2006. These isolates were identified in the hospital by use of standard procedures. Briefly, suspected colonies of S. aureus obtained in primary cultures were tested for agglutination with Pastorex Staph-plus (Bio-Rad Laboratories SA, Alcobendas, Madrid, Spain) and for thermonuclease (on DNase test agar; Biomedics, Madrid, Spain) and coagulase production (coagulase plasma; Becton Dickinson, San Agustín de Gualdalix, Madrid, Spain). Isolates that proved to be positive for the three tests were also evaluated for susceptibility or resistance to methicillin. S. aureus isolates were recovered from suppurative samples from spontaneous infections (5 isolates from patients with conjunctivitis, abscess in the hand, umbilical pyogenic granuloma, parotiditis, or infected ascites), surgical wounds (29 isolates associated with hand or leg amputations, hip, knee, or valvular prostheses, varicose veins, cellulitis, arthrodesis, osteomyelitis, Morton's neuroma, thoracotomy, tracheotomy, pleuritis [in two patients with esophageal cancer], or colon infection [in a patient with colon cancer]), urine (1 isolate from a patient with postcatheter urinary tract infection), tracheobronchial aspirates (11 isolates from patients with staphylococcal pneumonia, who have required intubation or assisted ventilation after cerebrovascular accident, brain injury, or polytraumatism), skin exudates (2 isolates from children with SSSS, 1 also involved in sepsis), and blood (33 isolates from patients who developed sepsis, most of them with severe underlying conditions, such as leukemia, stomach cancer, liver transplantation, cerebral vascular accident, thrombosis, aortic aneurysm, chronic renal insufficiency subjected to hemodialysis or peritoneal dialysis, osteomyelitis, pneumonia, and/or the presence of inserted catheters).
Macrorestriction-PFGE analysis.
Whole DNA from each S. aureus isolate was analyzed by SmaI PFGE using a CHEF-DRIII SYS220/240 (Bio-Rad Laboratories, S.A., Madrid, Spain) system and the consensus protocol of the European HARMONY group (42). The resulting profiles were analyzed visually, and the presence or absence of fragments larger than ca. 20 kb was recorded. Those showing one or more mismatching bands were considered different and labeled with an “S” followed by a number. Genetic similarity between profiles was determined by the unweighted-pair method with arithmetic averages and Jaccard's coefficient, using the software program MVSP version 3.1 (Multivariate Statistics Package for PCs; RockWare Inc.). NCTC 8325 was used as a reference strain for PFGE.
spa typing and MLST.
spa typing and MLST were performed as described by Shopsin et al. (50) and Enright et al. (9), respectively. PCR products were sequenced by Qiagen (Hilden, Germany). spa types were identified with the software Ridom Staphtype (Ridom GmbH), and sequence types (STs) were assigned by using the MLST website (http://www.mlst.net). Isolates were grouped into a single clonal cluster (CC) when five of the seven housekeeping genes used in the MLST scheme were identical (9).
SCCmec typing and virulence gene content.
SCCmec was typed by the multiplex PCR strategy reported by Zhang et al. (60). Primers for the exotoxin genes and the agr group have been described previously (3, 15, 16, 39, 41, 55). PCR assays were performed at least twice for each exotoxin gene and isolate, and positive and negative controls were always included. A dendrogram of similarity showing the clustering of the isolates according to virulence gene profiles was constructed by using Jaccard's coefficient of similarity (BioNumerics version 5.1; Applied Maths).
Statistical methods.
Statistical comparison between isolates giving positive and negative test results was performed with the χ2 test, using SPSS software (version 15.0; SPSS, Chicago, IL). Differences between groups were considered statistically significant if P values were <0.05. The discrimination index (DI; i.e., the probability that two unrelated isolates would be assigned to different SmaI or virulence profiles) was calculated using Simpson's index of diversity (53).
RESULTS
Genotypic typing of the isolates.
For a detailed assessment of the epidemiology of S. aureus circulating in the HUCA, the 81 isolates selected for the present study were subjected to SmaI PFGE, and representative subsets were also typed by spa typing and MLST.
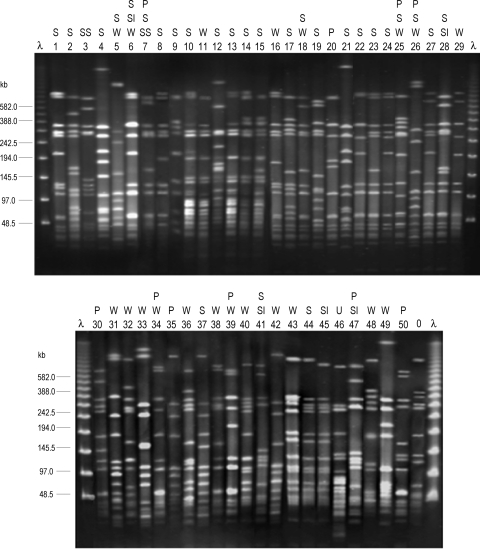
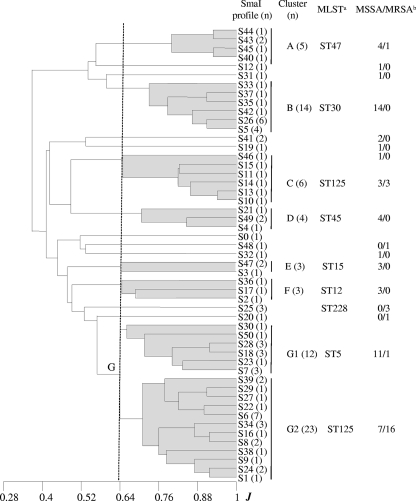
SmaI PFGE yielded a DI of 0.98 and identified 50 profiles (S1 to S50; Fig. 1), with only 15 including more than one isolate (two up to seven). On the basis of the coefficient of similarity of Jaccard, a dendrogram was constructed (Fig. 2). At a cutoff point of 0.64, two major clusters (B and G), five minor clusters (A, C, D, E, and F), and several ungrouped branches were revealed. Cluster G, which accounted for 43.2% of the isolates, could be in turn separated into subclusters G1 and G2. Thirteen isolates of cluster G were selected for spa typing (Table 1). Four of them, all belonging to G1, were assigned to t002, another two were assigned to t3734, and seven were assigned to t067, and these last nine were included in G2. MLST performed on one or two isolates of each spa type associated t002 with ST5, while the t067 and t3734 isolates were associated with ST125 (which differs from ST5 by only one base pair in the amplified region of the yqiL gene [46]). Interestingly, representative isolates of cluster C and the single S20 isolate that generated the unclustered branch closest to cluster G were also of spa type t067 and ST125, whereas the next branch included three S25 isolates that were of spa type t109 and ST228 (a two-locus variant of ST5, with 1- and 2-bp differences in the tpi and yqiL amplified regions, respectively [48]). ST5, ST125, and ST228 belong to the same CC, namely, CC5, which clearly predominated in the HUCA.
FIG. 1.
SmaI PFGE analysis of S. aureus isolates from a Spanish hospital. Lanes λ, lambda ladder PFGE marker (New England Biolabs); lane 0, SmaI profile of strain NCTC 8325 included as a quality control; lanes 1 to 50, SmaI profiles generated from clinical isolates. P, pneumonia; S, sepsis; SI, spontaneous infection; SS, scalded skin syndrome; U, urinary tract infection; W, surgical wound infection. This is a composite figure from five gels that were run under identical PFGE conditions.
FIG. 2.
Dendrogram showing the relatedness between SmaI macrorestriction fragment profiles generated from the S. aureus clinical isolates and the control strain (NCTC 8325). At a Jaccard coefficient of similarity (J) of 0.64, seven clusters (labeled A to G, the latter with subclusters G1 and G2) and several unclustered branches were detected. Footnote a, determined multilocus sequence types of representative isolates; footnote b, MSSA and MRSA. n, number of isolates.
TABLE 1.
Relationships between CC, MLST type, spa type, and PFGE clustera
| CC | MLST type(s) (n)b | spa type(s) (n)c | PFGE clusterd |
|---|---|---|---|
| CC5 | ST5 (2) | t002 (4) | G1 |
| ST125 (2) | t067 (7), t3734 (2) | G2 | |
| ST125 | t067 (4) | C | |
| ST228 | t109 (3) | - | |
| (ST125) | t067 | - | |
| CC8 | (ST8) | t024 | - |
| CC12 | ST12 | t160, t1381 | F |
| CC15 | ST15 (2) | t084 (2), t547 | E |
| CC30 | ST30 (2) | t012 (4), t021 | B |
| CC45 | ST45 | t282, t1618 (2) | D |
| ST47 | t081 (2), t383, t1494 | A | |
| CC51 | (ST121, ST427) | t159 | - |
| CC59 | ST59 | NT | - |
The numbers of isolates (n) are listed in parentheses if the number is more than one.
Determined or presumptive (according to the spa type; in parentheses) STs of representative isolates are shown.
NT, nontypeable.
-, unclustered PFGE profile.
With regard to the remaining SmaI PFGE clusters, the following observations were made. (i) Representative isolates of cluster B showed spa types t012 (four out of five tested) and t021 (the remaining one), and one isolate of each type proved to be of ST30 within CC30. (ii) Isolates belonging to cluster D were of spa types t282 and t1618 (one and two out of three tested, respectively), and one of the latter isolates was of ST45. (iii) Isolates in cluster A were of spa types t081, t383, and t1494, and the t383 isolate was of ST47. Both ST45 and ST47 (which differs from ST45 by one change in the amplified region of aroE [9]) belong to CC45, although the corresponding PFGE profiles grouped separately in the dendrogram. (iv) Two spa types and one ST were associated with clusters E (t547 and t084; both ST15) and F (t160 and t1381; ST12). Finally, the S32, S12, and S19 unclustered SmaI PFGE profiles were t024, t159, and nontypeable, respectively. The predicted ST for t024 is ST8 (CC8), t159 could be of ST121 or ST427 (CC51), and the last type was experimentally assigned to ST59 (CC59).
Incidence of MRSA and SCCmec typing.
Of the isolates under study, 55 were MSSA (67.9%) and 26 were MRSA (32.1%). The latter belonged to SmaI PFGE clusters A, C, and G or generated one of three unclustered patterns (S20, S25, and S48). However, more than half (16 out of 26) grouped in subcluster G2 (spa type t067 or t3734; ST125) within cluster G, and nearly all were members of CC5 (Fig. 2). All MRSA isolates contained SCCmec IVc, except two that carried SCCmec I and generated the unclustered S25 profile (spa type t109; ST228) and one that was nontypeable by the applied method (t067; subcluster G2).
Virulence gene profiles.
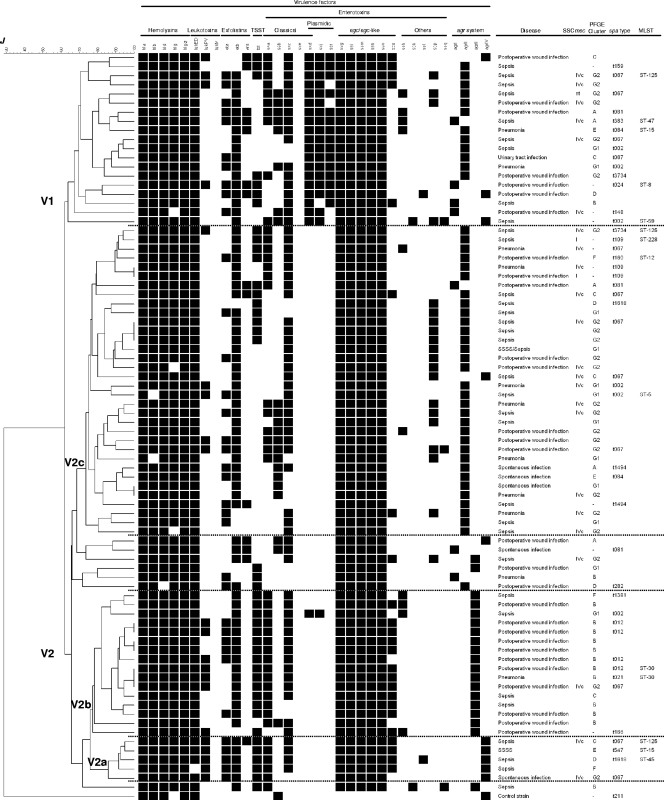
The agr group and the exotoxin gene repertoire were determined for the 81 S. aureus isolates of the HUCA and for S. aureus NCTC 8325 (included as a control). A dendrogram of similarity constructed on the basis of the presence or absence of the screened genes separated the control strain from the HUCA isolates and revealed a wide heterogeneity among the latter (Fig. 3). In fact, they were distributed into 73 virulence gene profiles, with only six being shared by more than one isolate (two or three). Relevant data were as follows. (i) The four types of agr systems were represented in the series, with agrII being the most common (55.6%), followed by agrIII (23.5%), agrIV (12.3%), and agrI (8.6%). (ii) All isolates were beta-hemolytic and positive for hla and hlg-2 (encoding α- and γ-variant hemolysins, respectively). A clearly prevalent profile (92.6%) included the five hemolysin genes tested (hla-hlb-hld-hlg-hlg-2). (iii) All isolates carried at least one leukotoxin-encoding gene, most frequently lukED (98.8%), but lukPV also occurred (25.9%). In contrast, lukM was not detected. (iv) A remarkable number of isolates (95.1%) was positive for genes encoding exfoliative toxins. etb (89.9%), found alone or in combination with eta and/or etd, was the most common. It was followed by eta and etd, carried by 43.2% and 17.3% of the isolates, respectively. Despite this, only two young children were diagnosed with SSSS, with one of the responsible isolates being positive for etb, eta, and agrIV and the other being positive for etb and agrII. (v) About half of the isolates (51.9%) carried tst, although none of the patients suffered from TSS. (vi) All isolates contained an egc or an egc-like cluster (70.4% and 29.6%, respectively), frequently together with other SE genes, including those encoding classical enterotoxins (92.6%). Of the latter, sec, sea, seb, and sed were present in 80.2%, 55.6%, 27.2%, and 24.7% of the isolates, respectively. Each of these genes appeared either alone (in a few cases) or with one or two of the other in different combinations. All HUCA isolates were negative for see. The sed, sej, and ser genes carried by plasmids were present in 24.7% of the isolates, with the three coinciding in 17.3% of them. Among other SE genes, sep was the most frequent (24.7%), but seh, sek, sel, and seq were also represented (12.3%, 2.5%, 2.5%, and 3.7%, respectively). Interestingly, typing of the isolates on the basis of the virulence gene profile yielded a DI close to 1 (0.997).
FIG. 3.
Dendrogram showing the relatedness between virulence gene profiles (for 30 exotoxin genes and the agr group) of the S. aureus isolates tested. At a Jaccard coefficient of similarity (J) of ca. 0.63, two main clusters, termed V1 and V2, and three subclusters within V2 (V2a, V2b, and V2c) are indicated. The type of infection caused by each isolate, the SmaI PFGE cluster to which it belongs, the SCCmec type (I, IVc, or not typeable [nt]) of MRSA, and the spa type as well as the MLST type of representative isolates, are shown at the right of the dendrogram.
Overall, a very high number of exotoxin genes was present in each of the HUCA isolates, independently of whether they were involved in spontaneous infections (13 up to 16), surgical wound infections (13 up to 21), pneumonia (13 up to 19), sepsis (11 up to 22), or SSSS (followed or not followed by sepsis, 14 and 18, respectively). Among the screened genes, only sep was significantly more frequent in isolates associated with sepsis (P = 0.032), and seb was more frequent in those that caused other infections (P = 0.032). Moreover, agrII and sep were more common in MRSA isolates than in MSSA isolates (P values of 0.029 and 0.048, respectively), while agrIII and tst were more common in MSSA isolates (P values of 0.021 and 0.032, respectively).
At a Jaccard coefficient of ca. 0.63, the dendrogram shown in Fig. 3 separated the HUCA isolates into two main virulence clusters (termed V1 and V2; Table 2). V1 incorporated all except one of the isolates that carried enterotoxin genes from plasmids (sed, sej, and/or ser), which were scattered among three of the four agr groups (agrII, agrI, and agrIV, with 12, 4, and 3 isolates, respectively). Most V2 isolates fell within one of three subclusters (V2a, V2b, and V2c). V2a included five agrIV isolates, all positive for lukPV, eta, etb, tst, sea, and the five hemolysin genes tested. With single exceptions, lukED, sec, and egc-like genes were also present. V2b grouped most of the agrIII isolates (16 out of 19) characterized by the occurrence of the five hemolysin genes, lukED (all isolates), etb, tst, sea (each with a single exception), and sec (two exceptions). In addition, most carried the egc-like cluster, and nearly half tested positive for lukPV. V2c included most of the agrII isolates which lacked SE genes from plasmids. The hemolysin genes, lukED, etb, sec, and egc were detected in all or most isolates of the subcluster (from ca. 80% up to 100%), while eta, tst, sea, seb, and sep were relatively common (32.4% to 44.1%). Finally, data compiled in Fig. 3 failed to reveal a correlation between virulence gene profile(s) and type of disease, with all clusters and subclusters grouping isolates involved in sepsis together with isolates that caused other types of infections. Accordingly, virulence factors apart from those screened in the present work, differential expression of the detected genes, and/or host-related factors may have been critical for the outcome of the disease (32, 35, 58).
TABLE 2.
Characteristics of the virulence clusters and subclustersa
| Virulence cluster (n) | agr type (n) | CC (n) | Cytotoxin(s) (n)b | Pyrogenic toxin superantigen gene(s) (n)b |
|---|---|---|---|---|
| V1 | agrI (4) | CC8, CC30, CC45 | lukED (4), lukPV | tst (2), sea, seb (2), sec (4) |
| eta (3), etb (4), etd (2) | sed (4), sej (3), ser (3) | |||
| seh | ||||
| egc (3), egc-like | ||||
| agrII (12) | CC5 (9), CC15, CC45 | lukED (12), lukPV (2) | tst (3), sea (7), seb (4), sec (10) | |
| eta (6), etb (11), etd (3) | sed (12), sej (12), ser (10) | |||
| seh (3), sep (3) | ||||
| egc (9), egc-like (3) | ||||
| agrIV (3) | CC5, CC45 | lukED (3), lukPV | tst (2), sea (2), seb, sec (3) | |
| eta, etb (2), etd | sed (3), sej (3), ser (2) | |||
| sek, sel, sep, seq | ||||
| egc (2), egc-like | ||||
| V2a (5) | agrIV (5) | CC5 (4), CC45 | lukED (4), lukPV (5) | tst (5), sea (5), sec (4) |
| eta (5), etb (5), etd | sel | |||
| egc, egc-like (4) | ||||
| V2b (16) | agrIII (16) | CC5 (4), CC30 (11) | lukED (16), lukPV (7) | tst (15), sea (15), seb, sec (14) |
| eta (6), etb (15) | sed, sej | |||
| seh (4) | ||||
| egc (4), egc-like (12) | ||||
| V2c (34) | agrII (32) | CC5 (28), CC15, CC45 | lukED (32), lukPV (5) | tst (10), sea (13), seb (11), sec (25) |
| eta (12), etb (28), etd (2) | seh (2), sep (14), seq | |||
| egc (30), egc-like (2) | ||||
| agrIV | CC5 | lukED, etb | sec, sep | |
| egc | ||||
| agrI | CC45 | lukED, etb, etd | tst, sea, sec | |
| egc |
The numbers of isolates (n) are listed in parentheses if the number is more than one.
All isolates carried three to five hemolysin genes; exotoxin genes detected in all or most (more than 75%) isolates of the corresponding virulence cluster or subcluster are shown in boldface.
DISCUSSION
Molecular epidemiology studies allowed the identification of the MRSA and MSSA CCs and clones that have been circulating in the HUCA during a recent time period. Overall, the frequency of MRSA isolates in this hospital (32.1%) coincided with that reported by the EPINE group, which surveys the prevalence of nosocomial infections in Spain (32.5% for the 2001-to-2003 period [2]), and was also in line with the results of a single-day surveillance study that analyzed 439 S. aureus isolates recovered from 143 Spanish hospitals during 2002 (30.5%) (5).
Most MRSA isolates from the HUCA (73%) were ST125-SCCmec IV isolates. This clone emerged in Spain during 1996, although it was first reported in 2001, and has now become predominant in Spanish hospitals (46, 47, 56). At a lower frequency, it was also found in Norway (13). As in the present study, a strong correlation between ST125 and spa type t067 was observed in both countries. Most of the remaining MRSA isolates from the HUCA were also positive for SCCmec IV but belonged to other STs (i.e., ST5, ST228, or ST47); two carried SCCmec I and were of ST228, while one was not typeable by the applied method (60). The ST5-SCCmec IV (pediatric clone) (49) and ST228-SCCmec I (southern German clone) (48) clones are major hospital-acquired MRSA clones, widely distributed worldwide (7, 46, 56). In summary, typing of MRSA isolates from the HUCA revealed that nearly all belonged to CC5 (88.5%), one of the most diversified lineages of MRSA (48), and carried SCCmec IV (also 88.5%), specifically SCCmec IVc, which in Spain accounted only for 24% of the MRSA isolates, whereas subtype IVa was the most frequent (73.3%) (47).
As previously reported by other authors (7, 10, 52), a higher diversity of CCs was found among the MSSA isolates of the HUCA, which accounted for 67.9% of the analyzed isolates. Of them, 80.0% had a genetic background common to major MRSA lineages, namely, CC5 (38.2%), CC30 (25.5%), CC45 (14.5%), and CC8 (1.8%) (7, 17, 48). As was the case for MRSA, MSSA isolates with a CC5 genetic background mostly belonged to SmaI PFGE subclusters G1 and G2 and were of either ST5 or ST125; CC30 isolates fell into SmaI PFGE cluster B and were of ST30; and ST47 and ST45, both members of CC45, were separated by SmaI PFGE into clusters A and D. Four successful MSSA lineages different from the major MRSA clones, namely, CC12, CC15 (PFGE clusters F and E), CC51, and CC59 (each with one sporadic isolate), were also detected. Isolates of these four lineages have been observed in both nosocomial and community settings in different countries (1, 18, 19, 44, 54, 57). Finally, it is worth noting that, like in other studies (4, 20), a good concordance between typing by SmaI PFGE, spa type, and MLST type was found for the HUCA isolates. In fact, most isolates with the same MLST had similar SmaI profiles and, hence, grouped within the same PFGE cluster. However, several SmaI PFGE clusters and/or branches could be associated with a certain CC and even with a certain ST, consistent with the higher discrimination of the former method.
With regard to the virulence gene repertoires, the high number of exotoxin genes carried by each of the 81 isolates analyzed in the present work (from 11 up to 22) is remarkable. All HUCA isolates harbored genes encoding four or five hemolysins, one or two leukocidins, and five up to 13 enterotoxins. All tested positive for egc with or without seu, and most were positive for one or more genes encoding classical enterotoxins, including high percentages of sec and sea. Moreover, all except four isolates contained at least one exfoliatin gene (eta, etb, and/or etd), whereas tst was present in half of them, and the leukotoxin gene lukPV was present in about one-fourth. In Spanish hospitals, the incidence of MSSA isolates carrying lukPV has increased from 0% in 2002 up to 36.4% in 2006 to 2007 (5, 47). In the present study, 66.7% of the lukPV-positive isolates were MSSA, but 33.3% were MRSA, and most belonged to the prevalent CC5. Of the 30 virulence genes screened, only lukM and see were absent from the series. Those that were detected appeared in many different combinations, with a total of 73 virulence profiles displayed by 81 isolates. A wide diversity of virulence gene combinations has also been reported for S. aureus isolates from England, Germany, and France (6, 22, 35).
Overall, the high number of exotoxin genes carried by each isolate and the rather indiscriminate distribution of these genes among different agr groups and genomic backgrounds, together with the striking diversity of exotoxin gene combinations, are consistent with a frequent acquisition and/or loss of the screened genes. It is widely accepted that mobile genetic elements carrying exotoxin genes can efficiently spread within lineages (35). In addition, they may be eventually capable of breaking interlineage barriers. An important barrier is the SauI type I restriction-modification system, which appears to be one of the major mechanisms underlying the clonal structure of S. aureus (11, 59). Nevertheless, a mobile genetic element can enter a CC from which it was originally excluded by means of a SauI restriction mutant. Once in such a mutant, the element might become modified at the specific sequences recognized by the hsdS1 and hsdS2 products characteristic of this particular lineage. Afterwards, its transfer to other members of the same lineage would be possible. In any case, the accumulation of exotoxin genes within S. aureus isolates is a matter of concern, since it may result in enhanced pathogenicity under appropriate circumstances (34). In the HUCA, the epidemiological surveillance of these potential “superbugs” could be facilitated, at least in the near future, by these findings with regard to the remarkable diversity of exotoxin gene combinations, which nearly differentiated at the strain level.
Acknowledgments
This work was supported by project FIS-06-PI052489 from the Spanish Ministry of Science and Innovation and projects BfR 45-004 and 45-005 of the Federal Institute for Risk Assessment (BfR), Germany. M. A. Argudín is the recipient of grant FPU AP-2004-3641 from the Ministry of Science and Innovation, Spain, cofunded by the European Social Fund. Part of the work was performed during a short stay of M. A. Argudín at the Department of Biological Safety of BfR, supported by the same grant.
We are grateful to Ana Fleites and Marta Landero from the Hospital Universitario Central de Asturias for supplying S. aureus isolates, to J. Hammerl and S. Hertwig (BfR) for their advice on spa typing and MLST, and to the Department of Biological Safety for their hospitality.
Footnotes
Published ahead of print on 20 May 2009.
REFERENCES
- 1.Aires de Sousa, M., T. Conceição, C. Simas, and H. de Lencastre. 2005. Comparison of genetic backgrounds of methicillin-resistant and -susceptible Staphylococcus aureus isolates from Portuguese hospitals and the community. J. Clin. Microbiol. 435150-5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asensio, A., R. Canton, J. Vaque, J. Rossello, F. Calbo, J. Garcia-Caballero, V. Dominguez, A. Hernandez, A. Trilla, and EPINE Working Group. 2006. Nosocomial and community-acquired methicillin-resistant Staphylococcus aureus infections in hospitalized patients (Spain, 1993-2003). J. Hosp. Infect. 63465-471. [DOI] [PubMed] [Google Scholar]
- 3.Becker, K., A. W. Friedrich, G. Lubritz, M. Weilert, G. Peters, and C. Von Eiff. 2003. Prevalence of genes encoding pyrogenic toxin superantigens and exfoliative toxins among strains of Staphylococcus aureus isolated from blood and nasal specimens. J. Clin. Microbiol. 411434-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cookson, B. D., D. A. Robinson, A. B. Monk, S. Murchan, A. Deplano, R. de Ryck, M. J. Struelens, C. Scheel, V. Fussing, S. Salmenlinna, J. Vuopio-Varkila, C. Cuny, W. Witte, P. T. Tassios, N. J. Legakis, W. van Leeuwen, A. van Belkum, A. Vindel, J. Garaizar, S. Haeggman, B. Olsson-Liljequist, U. Ransjo, M. Muller-Premru, W. Hryniewicz, A. Rossney, B. O'Connell, B. D. Short, J. Thomas, S. O'Hanlon, and M. C. Enright. 2007. Evaluation of molecular typing methods in characterizing a European collection of epidemic methicillin-resistant Staphylococcus aureus strains: the HARMONY collection. J. Clin. Microbiol. 451830-1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cuevas, O., E. Cercenado, E. Bouza, C. Castellares, P. Trincado, R. Cabrera, A. Vindel, and Spanish Group for the Study of Staphylococcus 2007. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in Spain: a multicentre prevalence study (2002). Clin. Microbiol. Infect. 13250-256. [DOI] [PubMed] [Google Scholar]
- 6.Dauwalder, O., G. Lina, G. Durand, M. Bes, H. Meugnier, V. Jarlier, B. Coignard, F. Vandenesch, J. Etiene, and F. Laurent. 2008. Epidemiology of invasive methicillin-resistant Staphylococcus aureus clones collected in France in 2006 and 2007. J. Clin. Microbiol. 463454-3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deurenberg, R. H., and E. E. Stobbering. 2008. The evolution of Staphylococcus aureus. Infect. Genet. Evol. 8747-763. [DOI] [PubMed] [Google Scholar]
- 8.Dinges, M. M., P. M. Orwin, and M. Schlievert. 2000. Exotoxins of Staphylococcus aureus. Clin. Microbiol. Rev. 1316-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enright, M. C., N. P. Day, C. E. Davis, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 381008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faria, N. A., J. A. Carrico, D. C. Oliveira, M. Ramirez, and H. de Lencastre. 2008. Analysis of typing methods for epidemiological surveillance of both methicillin-resistant and methicillin-susceptible Staphylococcus aureus strains. J. Clin. Microbiol. 46136-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng, Y., C.-J. Chen, L.-H. Su, S. Hu, J. Yu, and C.-H. Chiu. 2008. Evolution and pathogenesis of Staphylococcus aureus: lessons learned from genotyping and comparative genomics. FEMS Microbiol. Rev. 3223-37. [DOI] [PubMed] [Google Scholar]
- 12.Ferry, T., T. Perpoint, F. Vandenesch, and J. Etienne. 2005. Virulence determinants in Staphylococcus aureus and their involvement in clinical syndromes. Curr. Infect. Dis. Rep. 7420-428. [DOI] [PubMed] [Google Scholar]
- 13.Fossum, A. E., and G. Butcholm. 2006. Increased incidence of methicillin-resistant Staphylococcus aureus ST80, novel ST125 and SCCmecIV in the south-eastern part of Norway during a 12-year period. Clin. Microbiol. Infect. 12627-633. [DOI] [PubMed] [Google Scholar]
- 14.Fraser, J. D., and T. Proft. 2008. The bacterial superantigen and superantigen-like proteins. Inmunol. Rev. 225226-243. [DOI] [PubMed] [Google Scholar]
- 15.Fueyo, J. M., M. C. Mendoza, M. A. Alvarez, and M. C. Martin. 2005. Relationships between toxin gene content and genetic background in nasal carried isolates of Staphylococcus aureus from Asturias, Spain. FEMS Microbiol. Lett. 243447-454. [DOI] [PubMed] [Google Scholar]
- 16.Fueyo, J. M., M. C. Mendoza, M. R. Rodicio, J. Muñiz, M. A. Alvarez, and M. C. Martin. 2005. Cytotoxin and pyrogenic toxin superantigen gene profiles of Staphylococcus aureus associated with subclinical mastitis in dairy cows and relationships with macrorestriction genomic profiles. J. Clin. Microbiol. 431278-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gomes, A. R., H. Westh, and H. de Lencastre. 2006. Origins and evolution of methicillin-resistant Staphylococcus aureus clonal lineages. Antimicrob. Agents Chemother. 503237-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grundmann, H., S. Hori, M. C. Enright, C. Webster, A. Tami, E. J. Feil, and T. Pitt. 2002. Determining the genetic structure of the natural population of Staphylococcus aureus: a comparison of multilocus sequence typing with pulsed-field gel electrophoresis, randomly amplified polymorphic DNA analysis, and phage typing. J. Clin. Microbiol. 404544-4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hallin, M., O. Denis, A. Deplano, R. De Mendonça, R. De Ryck, S. Rottiers, and M. J. Struelens. 2007. Genetic relatedness between methicillin-susceptible and methicillin-resistant Staphylococcus aureus: results of a national survey. J. Antimicrob. Chemother. 59465-472. [DOI] [PubMed] [Google Scholar]
- 20.Hallin, M., A. Deplano, O. Denis, R. De Mendonça, R. De Ryck, and M. J. Strudels. 2007. Validation of pulsed-field gel electrophoresis and spa typing for long-term, nationwide epidemiological surveillance studies of Staphylococcus aureus infections. J. Clin. Microbiol. 45127-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hisata, K., K. Kuwahara-Arai, M. Yamanoto, T. Ito, Y. Nakatomi, L. Cui, T. Baba, M. Terasawa, C. Sotozono, S. Kinoshita, Y. Yamashiro, and K. Hiramatsu. 2005. Dissemination of methicillin-resistant staphylococci among healthy Japanese children. J. Clin. Microbiol. 433364-3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holtfreter, S., D. Grumann, M. Schmudde, H. T. T. Nguyen, P. Eichler, B. Strommenger, K. Kopron, J. Kotola, S. Giedrys-Kalemba, I. Steinmetz, W. White, and B. M. Bröker. 2007. Clonal distribution of superantigen genes in clinical Staphylococcus aureus isolates. J. Clin. Microbiol. 452669-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ito, T., Y. Katayama, and K. Hiramatsu. 1999. Cloning and nucleotide sequence determination of the entire mec DNA of pre-methicillin-resistant Staphylococcus aureus N315. Antimicrob. Agents Chemother. 431449-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ito, T., Y. Katayama, K. Asada, N. Mori, K. Tsutsumimoto, C. Tiensasitorn, and K. Hiramatsu. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome of MRSA strains in the world. Antimicrob. Agents Chemother. 451323-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ito, T., X. X. Ma, F. Takeuchi, K. Okuma, H. Yuzawa, and K. Hiramatsu. 2004. Novel type V staphylococcal cassette chromosome mec driven by a novel cassette chromosome recombinase, ccrC. Antimicrob. Agents Chemother. 482637-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jarraud, S., G. J. Lyon, A. M. Figueiredo, G. Lina, F. Vandenesch, J. Etienne, T. W. Muir, and R. P. Novick. 2000. Exfoliatin-producing strains define a fourth agr specificity group in Staphylococcus aureus. J. Bacteriol. 1826517-6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jarraud, S., M. A. Peyrat, A. Lim, A. Tristan, M. Bes, C. Mougel, J. Etienne, F. Vandenesch, M. Bonneville, and G. Lina. 2001. egc, a highly prevalent operon of enterotoxin gene, forms a putative nursery of superantigens in Staphylococcus aureus. J. Immunol. 166669-677. [DOI] [PubMed] [Google Scholar]
- 28.Ji, G., R. Beavis, and R. P. Novick. 1997. Bacterial interference caused by autoinducing peptide variants. Science 2762027-2030. [DOI] [PubMed] [Google Scholar]
- 29.Kaneko, J., and Y. Kamio. 2004. Bacterial two-component and hetero-heptameric pore-forming cytolytic toxins: structures, pore-forming mechanism, and organization of the genes. Biosci. Biotechnol. Biochem. 68981-1003. [DOI] [PubMed] [Google Scholar]
- 30.Katayama, Y., T. Ito, and K. Hiramatsu. 2000. A new class of genetic element, staphylococcal cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 441549-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwon, N. H., K. T. Park, J. S. Moon, W. K. Jung, S. H. Kim, J. M. Kim, S. K. Hong, H. C. Koo, Y. S. Joo, and Y. H. Park. 2005. Staphylococcal cassette chromosome mec (SCCmec) characterization and molecular analysis for methicillin-resistant Staphylococcus aureus and novel SCCmec subtype IVg isolated from bovine milk in Korea. J. Antimicrob. Chemother. 56624-632. [DOI] [PubMed] [Google Scholar]
- 32.Labandeira-Rey, M., F. Couzon, S. Boisset, E. L. Brown, M. Bes, Y. Benito, E. M. Barbu, V. Vazquez, M. Höök, J. Etienne, F. Vandenesch, and M. G. Bowden. 2007. Staphylococcus aureus Panton-Valentine leukocidin causes necrotizing pneumonia. Science 3151130-1133. [DOI] [PubMed] [Google Scholar]
- 33.Letertre, C., S. Perelle, F. Dilasser, and P. Fach. 2003. Identification of a new putative enterotoxin SEU encoded by the egc cluster of Staphylococcus aureus. J. Appl. Microbiol. 9538-43. [DOI] [PubMed] [Google Scholar]
- 34.Lindsay, J. A., and M. T. G. Holden. 2004. Staphylococcus aureus: superbug, super genome? Trends Microbiol. 12378-385. [DOI] [PubMed] [Google Scholar]
- 35.Lindsay, J. A., C. E. Moore, N. P. Day, S. J. Peacock, A. A. Witney, R. A. Stabler, S. E. Husain, P. D. Butcher, and J. Hinds. 2006. Microarrays reveal that each of the ten dominant lineages of Staphylococcus aureus has a unique combination of surface-associated and regulatory genes. J. Bacteriol. 188669-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lowy, F. D. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339520-532. [DOI] [PubMed] [Google Scholar]
- 37.Lyon, G. J., and R. P. Novick. 2004. Peptide signaling in Staphylococcus aureus and other Gram-positive bacteria. Peptides 251389-1403. [DOI] [PubMed] [Google Scholar]
- 38.Ma, X. X., T. Ito, C. Tiensasitorn, M. Jamklang, P. Chongtrakool, S. Boyle-Vavra, R. S. Daum, and K. Hiramatsu. 2002. Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob. Agents Chemother. 461147-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McClure, J. A., J. M. Conly, V. Lau, S. Elsayed, T. Louie, W. Hutchins, and K. Zhang. 2006. Novel multiplex PCR assay for detection of the staphylococcal virulence marker Panton-Valentine leukocidin genes and simultaneous discrimination of methicillin-susceptible from -resistant staphylococci. J. Clin. Microbiol. 441141-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McDougal, L. K., C. D. Steward, G. E. Killgore, J. M. Chaitram, S. K. McAllister, and F. C. Tenover. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 415113-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moore, P. C. L., and J. A. Lindsay. 2001. Genetic variation among hospital isolates of methicillin-sensitive Staphylococcus aureus: evidence of horizontal transfer of virulence genes. J. Clin. Microbiol. 392760-2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murchan, S., M. E. Kaufmann, A. Deplano, R. de Ryck, M. Struelens, C. E. Zinn, V. Fussing, S. Salmenlinna, J. Vuopio-Varkila, N. El Solh, C. Cuny, W. Witte, P. T. Tassios, N. Legakis, W. van Leeuwen, A. van Belkum, A. Vindel, I. Laconcha, J. Garaizar, S. Haeggman, B. Olsson-Liljequist, U. Ransjo, G. Coombes, and B. Cookson. 2003. Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J. Clin. Microbiol. 411574-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nishifuji, K., M. Sugai, and M. Amagai. 2008. Staphylococcal exfoliative toxins: “molecular scissors” of bacteria that attack the cutaneous defense barrier in mammals. J. Dermatol. Sci. 4921-31. [DOI] [PubMed] [Google Scholar]
- 44.Nulens, E., E. E. Stobberingh, H. van Dessel, S. Sebastian, F. H. van Tiel, P. B. Beisser, and R. H. Deurenberg. 2008. Molecular characterization of Staphylococcus aureus bloodstream isolates collected in a Dutch university hospital between 1999 and 2006. J. Clin. Microbiol. 462438-2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oliveira, D. C., A. Tomasz, and H. de Lencastre. 2001. The evolution of pandemic clones of methicillin-resistant Staphylococcus aureus: identification of two ancestral genetic backgrounds and the associated mec elements. Microb. Drug Resist. 7349-361. [DOI] [PubMed] [Google Scholar]
- 46.Pérez-Roth, E., F. Lorenzo-Díaz, N. Batista, A. Moreno, and S. Méndez-Alvarez. 2004. Tracking methicillin-resistant Staphylococcus aureus clones during a 5-year period (1998 to 2002) in a Spanish hospital. J. Clin. Microbiol. 424649-4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pérez-Vázquez, M., A. Vindel, C. Marcos, J. Oteo, O. Cuevas, P. Trincado, V. Bautista, H. Grundmann, J. Campos, et al. 2009. Spread of invasive Spanish Staphylococcus aureus spa-type t067 associated with a high prevalence of aminoglycoside-modifying enzyme ant(4′)-Ia and the efflux pump genes mrsA/mrsB. J. Antimicrob. Chem. 6321-31. [DOI] [PubMed] [Google Scholar]
- 48.Robinson, D. A., and M. C. Enright. 2003. Evolutionary models of the emergence of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 473926-3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sá-Leão, R., I. Santos Sanches, D. Dias, I. Peres, R. M. Barros, and H. de Lencastre. 1999. Detection of an archaic clone of Staphylococcus aureus with low-level resistance to methicillin in a pediatric hospital in Portugal and in international samples: relics of a formerly widely disseminated strain? J. Clin. Microbiol. 371913-1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shopsin, B., M. Gomez, S. O. Montgomery, D. H. Smith, M. Waddington, D. E. Dodge, D. A. Bost, M. Riehman, S. Naidich, and B. N. Kreiswirth. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 373556-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shore, A., A. S. Rossney, C. T. Keane, M. C. Enright, and D. C. Coleman. 2005. Seven novel variants of the staphylococcal chromosomal cassette mec in methicillin-resistant Staphylococcus aureus isolates from Ireland. Antimicrob. Agents Chemother. 492070-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Strommenger, B., C. Braulke, D. Heuck, C. Schmidt, B. Pasemann, U. Nubel, and W. Witte. 2008. spa typing of Staphylococcus aureus as a frontline tool in epidemiological typing. J. Clin. Microbiol. 46574-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Struelens, M. J., A. Bauernfeind, A. Van Belkum, D. Blanc, B. D. Cookson, L. Dijkshoorn, N. El Solh, J. Etienne, J. Garaizar, P. Gerner-Smidh, N. Legakis, H. de Lencastre, M. H. Nicolas, T. L. Pitt, U. Römling, V. Rosdahl, and W. Witte. 1996. Consensus guidelines for appropriate use and evaluation of microbial epidemiologic typing systems. Clin. Microbiol. Infect. 22-11. [DOI] [PubMed] [Google Scholar]
- 54.Takano, T., W. Higuchi, T. Otsuka, T. Baranovich, S. Enany, K. Saito, H. Isobe, S. Dohmae, K. Ozaki, M. Takano, Y. Iwao, M. Shibuya, T. Okubo, S. Yabe, D. Shi, I. Reva, L.-J. Teng, and T. Yamamoto. 2008. Novel characteristics of community-acquired methicillin-resistant Staphylococcus aureus strains belonging to multilocus sequence type 59 in Taiwan. Antimicrob. Agents Chemother. 52837-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tenover, F. C., L. K. McDougal, R. V. Goering, G. Killgore, S. J. Projan, J. B. Patel, and P. M. Dunman. 2006. Characterization of a strain of community-associated methicillin-resistant Staphylococcus aureus widely disseminated in the United States. J. Clin. Microbiol. 44108-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vindel, A., P. Trincado, E. Gómez, R. Cabrera, T. Boquete, C. Solá, S. Valdezate, and J. A. Saez-Nieto. 2006. Prevalence and evolution of methicillin resistant Staphylococcus aureus in Spanish hospitals between 1996 and 2002. J. Clin. Microbiol. 44266-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vivoni, A. M., B. A. Diep, A. C. de Gouveia Magalhães, K. R. N. Santos, L. W. Riley, G. F. Sensabaugh, and B. Moreira. 2006. Clonal composition of Staphylococcus aureus isolates at a Brazilian university hospital: identification of international circulating lineages. J. Clin. Microbiol. 441686-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Voyich, J. M., K. R. Braughton, D. E. Sturdevant, A. R. Whitney, B. Saïd-Salim, S. F. Porcella, R. D. Long, D. W. Dorward, D. J. Gardner, B. N. Kreiswirth, J. M. Musser, and F. R. DeLeo. 2005. Insights into mechanisms used by Staphylococcus aureus to avoid destruction by human neutrophils. J. Immunol. 1753907-3919. [DOI] [PubMed] [Google Scholar]
- 59.Waldron, D. E., and J. A. Lindsay. 2006. SauI: a novel lineage-specific type I restriction-modification system that blocks horizontal gene transfer into Staphylococcus aureus and between S. aureus of different lineages. J. Bacteriol. 1885578-5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang, K., J. A. McClure, E. Sameer, T. Louie, and J. M. Conly. 2005. Novel multiple PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to IV in methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 435026-5033. [DOI] [PMC free article] [PubMed] [Google Scholar]