Abstract
Brucella species are highly monomorphic, with minimal genetic variation among species, hindering the development of reliable subtyping tools for epidemiologic and phylogenetic analyses. Our objective was to compare two distinct multiple-locus variable-number tandem-repeat analysis (MLVA) subtyping methods on a collection of 101 Brucella melitensis isolates from sporadic human cases of brucellosis in Egypt (n = 83), Qatar (n = 17), and Libya (n = 1). A gel-based MLVA technique, MLVA-15IGM, was compared to an automated capillary electrophoresis-based method, MLVA-15NAU, with each MLVA scheme examining a unique set of variable-number tandem repeats. Both the MLVAIGM and MLVANAU methods were highly discriminatory, resolving 99 and 101 distinct genotypes, respectively, and were able to largely separate genotypes from Egypt and Qatar. The MLVA-15NAU scheme presented higher strain-to-strain diversity in our test population than that observed with the MLVA-15IGM assay. Both schemes were able to genetically correlate some strains originating from the same hospital or region within a country. In addition to comparing the genotyping abilities of these two schemes, we also compared the usability, limitations, and advantages of the two MLVA systems and their applications in the epidemiological genotyping of human B. melitensis strains.
Brucellosis is a common zoonotic disease causing infections in economically important livestock, such as cattle, goats, sheep, and pigs (22). In humans, this highly diverse illness, also known as Malta or undulant fever, initially presents as a fever, malaise, and myalgia and may later develop into a chronic illness affecting various organs and tissues (21, 25). Brucellosis is usually transmitted to humans through consumption of contaminated and untreated milk products or by direct contact with infected animals. There are nine phenotypically recognized species in the Brucella genus: Brucella melitensis, B. abortus, B. suis, B. ovis, B. canis, and B. neotomae; two new marine species, B. ceti and B. pinnipedialis (11); and B. microti (27), isolated from the common vole. Each species has distinctive host preferences, pathogenicity, and epidemiology.
Although some developed countries have eradicated B. abortus from cattle through vaccination campaigns, B. melitensis, B. abortus, and B. suis remain the principal causes of human brucellosis worldwide and are major public health problems, primarily in developing countries (7). Brucellosis is prevalent in the Mediterranean basin, the Middle East, Africa, Asia, and areas of Latin America where people are economically dependent on ruminant livestock (23, 28). Over the past decade, the global epidemiology of human brucellosis has changed, due in part to the implementation of national and international surveillance programs, animal vaccination campaigns, and socioeconomic changes (22).
Current microbiological (4) and low-resolution molecular typing methods are useful for identifying Brucella isolates and determining species and biovar designations; however, they have limited value for epidemiological trace-back investigations (6, 15). High-resolution genetic subtyping tools that could provide important information about disease transmission patterns and the molecular epidemiology of Brucella have been difficult to develop and implement due to the genetically monomorphic nature of Brucella species (12). Since the genome sequencing of four Brucella strains, B. abortus 2308 (26), B. abortus 9-941 (14), B. suis 1330 (24), and B. melitensis 16M (9), multilocus sequence typing (30) and multiple-locus variable-number tandem-repeat analysis (MLVA) have been developed (16, 18, 29). Recent studies have shown that MLVA, targeting multiple repeat regions with higher mutation rates than other genomic markers, demonstrates higher genetic resolution when applied to Brucella species than assays targeting other, more monomorphic molecular markers, such as the outer membrane proteins (8, 10), the rpoB gene (19), and insertion sequence regions (6).
In efforts to improve epidemiological surveillance and better evaluate the utility of MLVA as a genotyping tool for Brucella strains, the present study compared the discriminatory power of two distinct MLVA methods (16, 18) in their ability to distinguish geographic origin and resolve relationships within a group of 101 human B. melitensis isolates from Egypt, Qatar, and Libya. Each MLVA typing technique examined polymorphisms at 15 different tandem repeat loci and employed two different separation techniques. This study also compares the usability of the two MLVA systems with regard to resources, technological needs, scientific expertise, and time consumption.
MATERIALS AND METHODS
The laboratory work for the gel-based MLVA (MLVA-15IGM [MLVA with 15 VNTR loci from the Institut de Génétique et Microbiologie]) method was performed at the NAMRU-3 laboratory facilities in Cairo, Egypt, while data from the automated MLVA (MLVA-15NAU [MLVA with 15 VNTR loci from Northern Arizona University]) assay were generated at the Centers for Disease Control and Prevention (CDC) in Atlanta, GA.
Strain collection and template preparation.
All 101 isolates used in the study were of human origin and were identified as B. melitensis by the AMOS-PCR assay (6). They originated from Egypt (n = 83), Qatar (n = 17), and Libya (n = 1). The Egyptian isolates were collected from an acute-febrile-illness laboratory-based surveillance program implemented in a network of infectious-disease hospitals throughout Egypt from 1999 to 2003 (1, 2), and the Libyan isolate was recovered from a patient who traveled to Libya in 2006 (20). Human B. melitensis isolates from Qatar, collected in August 2006, were donated by the Hamad Medical Corporation, Doha, Qatar. The details of all isolates used in this study are presented in Table 1. Bacterial isolates were cultured on Trypticase soy agar with 5% sheep blood (BBL Microbiology Systems, Cockeysville, MD) at 37°C for 48 to 72 h. Total genomic DNA was extracted either using a Qiagen (Valencia, CA) DNeasy blood and tissue kit by following the manufacturer's protocol for extraction of genomic DNA from gram-negative bacteria or by whole-cell lysis as described by Gee et al. (13). The resulting lysates were stored at −20°C until needed.
TABLE 1.
Brucella melitensis isolates examined in this study
| Country of origin and region | Hospital locationa | No. of isolates |
|---|---|---|
| Egypt | ||
| Upper Egypt | AST | 5 |
| SOH | 2 | |
| QEN | 4 | |
| ASW | 7 | |
| Delta region | SHB | 16 |
| ZAG | 2 | |
| BEN | 2 | |
| FAY | 4 | |
| MAL | 13 | |
| Cairo metropolitan area | ABS | 12 |
| Coastal region | ALX | 15 |
| PRS | 1 | |
| Libya | 1 | |
| Qatar | Doha | 17 |
| Total | 101 |
ABS, Abbassia; ALX, Alexandria; ASW, Aswan; AST, Assiut; BEN, Benha; FAY, Fayoum; MAL, Mahalla; PRS, Port Said; QEN, Qena; SHB, Shebin; SOH, Sohag; ZAG, Zagazig.
VNTR identification.
The methods for selecting the variable-number tandem repeats (VNTRs) used in the MLVA-15NAU and MLVA-15IGM techniques have been described previously (16, 18). Table 2 shows the characteristics of the VNTRs examined in both MLVA schemes. All primers used in this study were obtained from Sigma Genosys (The Woodlands, TX).
TABLE 2.
Characteristics of the VNTRs of the two MLVA-15 assays
| MLVA-15NAU VNTR characteristics
|
MLVA-15IGM VNTR characteristics
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| VNTR | Genomic location in B. melitensis 16Ma | Repeat size (bp) | No. of alleles | Simpson's DI | VNTR | Genomic location in B. melitensis 16Ma | Repeat size (bp) | No. of alleles | Simpson's DI |
| BruceVNTR 16 | 339714-339950 | 8 | 4 | 0.49 | Bruce 18 | 339785-339930 | 8 | 6 | 0.79 |
| BruceVNTR 3 | 1249996-1250345 | 8 | 19 | 0.93 | Bruce 07 | 1250093-1250250 | 8 | 8 | 0.77 |
| BruceVNTR 28 | 1543941-1544121 | 8 | 9 | 0.79 | Bruce 04 | 1543967-1544118 | 8 | 6 | 0.64 |
| BruceVNTR 30 | 588314-588862 | 8 | 12 | 0.89 | Bruce 09 | 588256-588681 | 8 | 9 | 0.82 |
| BruceVNTR 27 | 548529-548905 | 8 | 9 | 0.84 | Bruce 16 | 548571-548722 | 8 | 7 | 0.64 |
| BruceVNTR 1 | 1938531-1938766 | 8 | 16 | 0.91 | Bruce 21 | 328936-329083 | 8 | 2 | 0.18 |
| BruceVNTR 2 | 1940192-1940778 | 8 | 17 | 0.92 | Bruce 30 | 1505073-1505223 | 8 | 2 | 0.32 |
| BruceVNTR 7 | 275628-275961 | 8 | 1 | 0 | Bruce 06 | 1322650-1323057 | 134 | 2 | 0.20 |
| BruceVNTR 21 | 574937-575041 | 6 | 2 | 0.06 | Bruce 08 | 1134883-1135230 | 18 | 3 | 0.25 |
| BruceVNTR 29 | 1915916-1916471 | 8 | 2 | 0.02 | Bruce 11 | 211361-211617 | 63 | 1 | 0 |
| BruceVNTR 31 | 344488-344993 | 5 | 6 | 0.77 | Bruce 12 | 73619-74010 | 15 | 2 | 0.26 |
| BruceVNTR 33 | 18596-19246 | 8 | 19 | 0.93 | Bruce 42 | 424317-424855 | 125 | 4 | 0.25 |
| BruceVNTR 14 | 396453-396581 | 9 | 1 | 0 | Bruce 43 | 379369-379550 | 12 | 2 | 0.37 |
| BruceVNTR 20 | 542226-542683 | 12 | 7 | 0.27 | Bruce 45 | 233374-233524 | 18 | 2 | 0.37 |
| BruceVNTR 25 | 216642-217140 | 15 | 1 | 0 | Bruce 55 | 2066378-2066650 | 40 | 3 | 0.25 |
Determined by NCBI nucleotide primer BLAST.
VNTR analysis by the MLVA-15NAU technique.
PCR amplification of 15 VNTR loci was performed as described previously (16) with the following modifications. Forward primers were designed with either a 6-carboxyfluorescein (FAM) or a HEX (N-hexachloro-fluorescein cyanoethyl phosphoramidite) fluorescent label. Four multiplex PCRs (AI, BI, AII, and BII) were performed to amplify 15 VNTR loci in a final volume of 10 μl containing 1× PCR buffer, 2 mM MgCl2, 2 mM deoxynucleoside triphosphate mix, 0.04 U Platinum Taq DNA polymerase (Invitrogen, Carlsbad, CA), 1 μl DNA template, and the following primer combinations: for AI, Bruce VNTR 21 F-HEX-Bruce VNTR 21 R, Bruce VNTR 28 F-FAM-Bruce VNTR 28 R, and Bruce VNTR 16 F-HEX-Bruce VNTR 16 R; for BI, Bruce VNTR 03 F-FAM-Bruce VNTR 03 R, Bruce VNTR 20 F-HEX-Bruce VNTR 20 R, Bruce VNTR 31 F-FAM-Bruce VNTR 31 R, Bruce VNTR 29 F-HEX-Bruce VNTR 29 R, and Bruce VNTR 33 F-FAM-Bruce VNTR 33 R; for AII, Bruce VNTR 14 F-FAM-Bruce VNTR 14 R, Bruce VNTR 01 F-HEX-Bruce VNTR 01R, Bruce VNTR 07 F-FAM-Bruce VNTR 07R, and Bruce VNTR 27 F-HEX-Bruce VNTR 27 R; and for BII, Bruce VNTR 25 F-FAM-Bruce VNTR 25 R, Bruce VNTR 30 F-HEX-Bruce VNTR 30 R, and Bruce VNTR 02 F-FAM-Bruce VNTR 02 R. All reactions were performed on an ABI 9700 thermocycler using the primer concentrations and thermal cycling conditions reported previously (16).
Multiplex PCRs were pooled and diluted to a final dilution of 1:20 with molecular-grade water. PCR products were denatured and resolved by capillary electrophoresis on an ABI Prism 3130 automated fluorescent capillary DNA sequencer (Applied Biosystems, Foster City, CA). Fragments were sized by comparison to a ROX (carboxy-X-rhodamine)-labeled molecular ladder (MapMaker 1000; BioVentures Inc., Murfreesboro, TN) with GeneMapper, version 4.0 (Applied Biosystems, Foster City, CA), software. Appropriate VNTR designations of the fragments were assigned based on size calling through internal software binning capabilities.
VNTR analysis by the MLVA-15IGM technique.
PCR primers for the MLVA-15IGM technique have been reported previously (18). PCR amplification of the 15 VNTR loci occurred in a final volume of 25 μl containing final concentrations of 2.5 mM MgCl2, 1× PCR buffer, 0.4 mM deoxynucleoside triphosphate nucleotide mixture, 1.25 U Taq DNA polymerase (Promega Corporation, Madison, WI), 0.5 μl DNA template, and 0.5 μM forward and reverse primers. All PCRs were performed on an MJ Research PTC-200 thermocycler with the thermal cycling parameters reported previously. PCR amplicons were analyzed by gel electrophoresis as described previously (3, 18); agarose gels were then normalized; and band sizes were estimated using BioNumerics version 4.61 (Applied Maths, Belgium). Band size estimates were then converted to repeat units by following the published allele numbering system that can be found at http://mlva.u-psud.fr/.
Analysis of MLVA data.
Simpson's index of diversity was calculated (5) for each tandem-repeat locus based on a sample size of 101 B. melitensis isolates (Table 2).
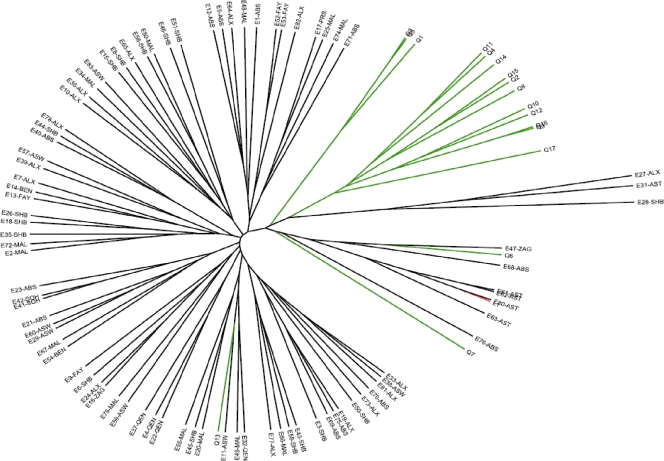
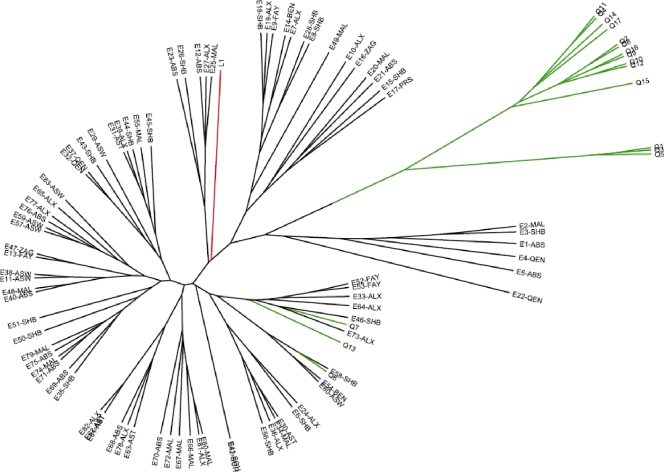
Data from both MLVA schemes were converted into character data sets within the BioNumerics software (version 5.1; Applied Maths, Saint-Martens-Latem, Belgium). Cluster analysis was performed on both data sets using the categorical coefficient and UPGMA (unweighted-pair group method using arithmetic averages) analysis. The resulting UPGMA similarity matrices were used to generate unrooted trees in FigTree, version 1.1.2 (http://tree.bio.ed.ac.uk/software/figtree) (see Fig. 1 and 2).
FIG. 1.
UPGMA analysis of 101 human B. melitensis isolates examined by the MLVA-15NAU scheme. Isolate designations indicate the country of origin (E, Egypt; Q, Qatar; L, Libya), followed by the location within the country (ABS, Abbassia; ALX, Alexandria; ASW, Aswan; AST, Assiut; BEN, Benha; FAY, Fayoum; MAL, Mahalla; PRS, Port Said; QEN, Qena; SHB, Shebin; SOH, Sohag; ZAG, Zagazig).
FIG. 2.
UPGMA analysis of 101 human B. melitensis isolates examined by the MLVA-15IGM scheme. For isolate designations, see the legend to Fig. 1.
RESULTS AND DISCUSSION
The two MLVA schemes evaluated in our study were selected based on the availability of published MLVA schemes at the time of the study design, their abilities to type human B. melitensis isolates, and the availability of the resources, technology, and technical expertise required to perform the techniques. As mentioned above, the gel-based MLVA assay was carried out in Cairo, Egypt, and data from the automated MLVA system were collected at the CDC in Atlanta, GA. Thus far, there has been no study demonstrating the typing abilities of different MLVA systems for a common collection of Brucella isolates. The objective of our study was to compare the resolving powers of two different MLVA techniques on a common collection of 101 clinical B. melitensis isolates from the Middle East and to provide an assessment of the limitations, advantages, and field applications of the two MLVA assays for genotyping B. melitensis isolates.
The MLVA techniques compared in this study were developed for different purposes, which are reflected in the unique combination of tandem-repeat markers that makes up each scheme. Originally intended for forensic purposes, the MLVA-15NAU assay contains 12 markers that have small repeat units less than or equal to 8 bp; 8 of these markers have diversity indexes (DIs) greater than 0.75 in our study. The MLVA-15IGM assay, developed for epidemiological typing, contains two panels: a panel of eight conserved markers with larger repeat units (>8 bp) (panel 1), exhibiting DIs less than 0.40 in the strains we examined, and a panel of seven more-diverse loci (panel 2) with small repeat units (<8 bp).
Overall, the VNTR markers in the MLVA-15NAU assay presented a broader number of alleles and higher DIs than the markers examined by the MLVA-15IGM scheme. Six of the 15 VNTR loci in the MLVA-15NAU scheme exhibited DIs of >0.80 for the 101 B. melitensis isolates examined, with allele numbers ranging from 9 to 20 (Table 2). The Bruce VNTR 21 and Bruce VNTR 29 markers display very low genetic DIs of 0.06 and 0.02, respectively, while the Bruce VNTR 7, Bruce VNTR 14, and Bruce VNTR 25 loci were monomorphic in the strains we examined. The invariance we observed at these loci is most likely due to the localized geographic origins of our isolates and represents a level of strain-to-strain conservation within a geographic area that can be better evaluated in a larger population of isolates from similar locations. The monomorphic nature of the Bruce VNTR 7 and Bruce VNTR 25 loci in the Middle Eastern strains could represent geographically specific alleles, since these markers have been reported to exhibit higher DIs in other B. melitensis strains, from the United States (n = 85) (16). The Bruce VNTR 14 locus is consistently monomorphic in our strains and in other B. melitensis strains examined by Huynh et al. (16) and could be examined as a species-specific marker. Bruce VNTR 02, 20, 28, 31, and 33 were initially difficult to amplify in certain isolates. However, all VNTR loci were successfully amplified in all isolates except for Bruce VNTR 33 in isolate E5 and Bruce VNTR 02 in isolate E53.
Only one of the 15 VNTR markers from the MLVA-15IGM assay had a DI of >0.80 in the 101 strains we examined. The Bruce 04, 07, 09, 16, and 18 VNTR markers exhibit the highest DIs, ranging from 0.64 to 0.82, among the 15 markers (Table 2). The remaining 10 tandem-repeat markers ranged in diversity from 0.18 to 0.37. The Bruce 11 VNTR marker is the only monomorphic marker in the Middle Eastern strains we examined. Two of the 101 B. melitensis isolates (E21 and E26) exhibited null alleles at the Bruce 09 locus; however, all other markers were successfully amplified in all isolates.
The two MLVA schemes evaluated in this study were able to correlate similar genetic relationships among the 101 B. melitensis strains we examined. The MLVA-15NAU scheme, exhibiting greater DIs, resolved 101 unique genotypes, while the MLVA-15IGM assay discriminated 99 genotypes. Dendrograms generated by UPGMA clustering analysis (Fig. 1 and 2) show similar groupings of B. melitensis strains by the MLVA methods, such as the clustering of Qatar isolates Q1, Q3, and Q5 with each other and the genetic similarity of Q6, Q7, and Q13 strains to the Egyptian genotypes. Also, the Libyan strain consistently clusters with the Egyptian genotypes by both MLVA methods. Because none of the strains we examined were epidemiologically linked, since they were sporadic cases from various regions of Egypt and Qatar, we did not expect to see a great deal of homogeneity by either MLVA method. However, by the MLVA-15IGM scheme, two strains, E41 and E42, which were isolated in the same hospital in Sohag, Egypt, 6 weeks apart from each other, had identical genotypes. Two other strains, E61 and E62, isolated 2 years apart from each other from another hospital, in Assiut, Egypt, also had identical genotypes. The same strains (E41 and E42; E61 and 62) did not have identical genotypes by the MLVANAU assay but diverged by a mutation at one or both of the hypervariable markers VNTR 01 and VNTR 33. By both MLVA schemes, it is possible to see some strains originating from the same hospital location or the same region (see Table 1 for regions) within Egypt that have very similar but not identical genotypes, which is expected for nonoutbreak strains.
The MLVA schemes evaluated in this study have different logistical and technical requirements. The gel-based MLVA method requires more time for typing from start to finish, requires less technical expertise, and involves less-expensive materials, although it does require BioNumerics software, which is quite costly and requires technical training. The use of multiple individual PCRs and agarose gels for typing one strain is cumbersome and requires considerable subjectivity in the stages of analysis of the raw data. The automated system is much faster and has a high throughput capacity; however, the instrumentation and proprietary reagents make this method more expensive. The use of an automated sequencer and the genetic analysis software require a skilled technician; however, the data output is more reliable, and once properly optimized, the method is very reproducible.
Both MLVA schemes examined in this study can be used to epidemiologically type human B. melitensis strains and can be used for high- and low-resolution typing by the inclusion and exclusion of highly variable markers. We evaluated the two MLVA schemes and compared their abilities to resolve relationships between geographically localized strains from the Middle East, where B. melitensis is the primary etiologic agent of human brucellosis. Other studies have looked at globally diverse and localized Brucella strains using the MLVAIGM assay (3, 17-19). However, it would be interesting to demonstrate the capability of the MLVANAU method for differentiating global genotypes in Brucella species. Although we currently utilize the MLVA-15NAU assay for genotyping Brucella isolates in our reference lab at the CDC and prefer the automated platform for its reproducibility, the MLVA-15IGM scheme is used widely in the European community and collaborating laboratories. The comparable geographic and in-country resolution of the two MLVA systems, targeting different VNTR regions, validates the utility of a combination analysis system, such as MLVA, for genotyping genetically monomorphic bacteria. With the increasing use of multiple MLVA systems for typing Brucella species and the increasing need to share data, it would be effective to investigate the discriminatory value of all the currently utilized VNTR markers in these combination systems by using statistical methods such as principal-component analysis in order to establish one streamlined MLVA system that serves the global need for an epidemiological genotyping tool to improve brucellosis surveillance.
Acknowledgments
This work was supported by DoD GEIS Work Unit number 847705-25GB-3906.
We are grateful to Mohammad A. Maksoud for laboratory assistance.
The opinions and assertions contained herein are the private opinions and assertions of the authors and are not to be construed as official or reflecting the views of the U.S. Navy Department.
Footnotes
Published ahead of print on 13 May 2009.
REFERENCES
- 1.Afifi, S., K. Earhart, M. A. Azab, F. G. Youssef, H. El Sakka, M. Wasfy, H. Mansour, S. El Oun, M. Rakha, and F. Mahoney. 2005. Hospital-based surveillance for acute febrile illness in Egypt: a focus on community-acquired bloodstream infections. Am. J. Trop. Med. Hyg. 73392-399. [PubMed] [Google Scholar]
- 2.Afifi, S., M. O. Wasfy, M. A. Azab, F. G. Youssef, G. Pimentel, T. W. Graham, H. Mansour, N. Elsayed, K. Earhart, R. Hajjeh, and F. Mahoney. 2007. Laboratory-based surveillance of patients with bacterial meningitis in Egypt (1998-2004). Eur. J. Clin. Microbiol. Infect. Dis. 26331-340. [DOI] [PubMed] [Google Scholar]
- 3.Al Dahouk, S., P. L. Fleche, K. Nockler, I. Jacques, M. Grayon, H. C. Scholz, H. Tomaso, G. Vergnaud, and H. Neubauer. 2007. Evaluation of Brucella MLVA typing for human brucellosis. J. Microbiol. Methods 69137-145. [DOI] [PubMed] [Google Scholar]
- 4.Alton, G. G., L. M. Jones, and D. E. Pietz. 1975. Laboratory techniques in brucellosis. Monogr. Ser. World Health Organ. 19751-163. [PubMed] [Google Scholar]
- 5.Bikandi, J., R. San Millán, A. Rementeria, and J. Garaizar. 2004. In silico analysis of complete bacterial genomes: PCR, AFLP-PCR and endonuclease restriction. Bioinformatics 20798-799. [DOI] [PubMed] [Google Scholar]
- 6.Bricker, B. J., and S. M. Halling. 1994. Differentiation of Brucella abortus bv. 1, 2, and 4, Brucella melitensis, Brucella ovis, and Brucella suis bv. 1 by PCR. J. Clin. Microbiol. 322660-2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bricker, B. J., D. R. Ewalt, and S. M. Halling. 2003. Brucella ‘HOOF-Prints’: strain typing by multi-locus analysis of variable number tandem repeats (VNTRs). BMC Microbiol. 315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cloeckaert, A., J. M. Verger, M. Grayon, and O. Grepinet. 1995. Restriction site polymorphism of the genes encoding the major 25 kDa and 36 kDa outer-membrane proteins of Brucella. Microbiology 1412111-2121. [DOI] [PubMed] [Google Scholar]
- 9.DelVecchio, V. G., V. Kapatral, R. J. Redkar, G. Patra, C. Mujer, T. Los, N. Ivanova, I. Anderson, A. Bhattacharyya, A. Lykidis, G. Reznik, L. Jablonski, N. Larsen, M. D'Souza, A. Bernal, M. Mazur, E. Goltsman, E. Selkov, P. H. Elzer, S. Hagius, D. O'Callaghan, J. J. Letesson, R. Haselkorn, N. Kyrpides, and R. Overbeek. 2002. The genome sequence of the facultative intracellular pathogen Brucella melitensis. Proc. Natl. Acad. Sci. USA 99443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ficht, T. A., H. S. Husseinen, J. Derr, and S. W. Bearden. 1996. Species-specific sequences at the omp2 locus of Brucella type strains. Int. J. Syst. Bacteriol. 46329-331. [DOI] [PubMed] [Google Scholar]
- 11.Foster, G., B. S. Osterman, J. Godfroid, I. Jacques, and A. Cloeckaert. 2007. Brucella ceti sp. nov. and Brucella pinnipedialis sp. nov. for Brucella strains with cetaceans and seals as their preferred hosts. Int. J. Syst. Evol. Microbiol. 572688-2693. [DOI] [PubMed] [Google Scholar]
- 12.Gándara, B., A. L. Merino, M. A. Rogel, and E. Martinez-Romero. 2001. Limited genetic diversity of Brucella spp. J. Clin. Microbiol. 39235-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gee, J. E., B. K. De, P. N. Levett, A. M. Whitney, R. T. Novak, and T. Popovic. 2004. Use of 16S rRNA gene sequencing for rapid confirmatory identification of Brucella isolates. J. Clin. Microbiol. 423649-3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Halling, S. M., B. D. Peterson-Burch, B. J. Bricker, R. L. Zuerner, Z. Qing, L. L. Li, V. Kapur, D. P. Alt, and S. C. Olsen. 2005. Completion of the genome sequence of Brucella abortus and comparison to the highly similar genomes of Brucella melitensis and Brucella suis. J. Bacteriol. 1872715-2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hinić, V., I. Brodard, A. Thomann, Z. Cvetnic, P. V. Makaya, J. Frey, and C. Abril. 2008. Novel identification and differentiation of Brucella melitensis, B. abortus, B. suis, B. ovis, B. canis, and B. neotomae suitable for both conventional and real-time PCR systems. J. Microbiol. Methods 75375-378. [DOI] [PubMed] [Google Scholar]
- 16.Huynh, L. Y., M. N. Van Ert, T. Hadfield, W. S. Probert, B. H. Bellaire, M. Dobson, R. J. Burgess, R. S. Weyant, T. Popovic, S. Zanecki, D. M. Wagner, and P. Keim. 2008. Multiple locus variable number tandem repeat (VNTR) analysis (MLVA) of Brucella spp. identifies species specific markers and insights into phylogenetic relationships, p. 47-54. In V. St. Georgiev, K. A. Western, and J. J. McGowan (ed.), National Institute of Allergy and Infectious Disease, NIH, vol. 1. Frontiers in research. Humana Press, Totowa, NJ. [Google Scholar]
- 17.Kattar, M. M., R. F. Jaafar, G. F. Araj, P. Le Flèche, G. M. Matar, R. Abi Rached, S. Khalife, and G. Vergnaud. 2008. Evaluation of a multilocus variable-number tandem-repeat analysis scheme for typing human Brucella isolates in a region of brucellosis endemicity. J. Clin. Microbiol. 463935-3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le Flèche, P., I. Jacques, M. Grayon, S. Al Dahouk, P. Bouchon, F. Denoeud, K. Nockler, H. Neubauer, L. A. Guilloteau, and G. Vergnaud. 2006. Evaluation and selection of tandem repeat loci for a Brucella MLVA typing assay. BMC Microbiol. 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marianelli, C., F. Ciuchini, M. Tarantino, P. Pasquali, and R. Adone. 2006. Molecular characterization of the rpoB gene in Brucella species: new potential molecular markers for genotyping. Microbes Infect. 8860-865. [DOI] [PubMed] [Google Scholar]
- 20.Mohamady, H., H. I. Shaheen, J. Klena, I. Nakhla, M. Weiner, and A. Armstrong. 2007. Case report: traveler's Brucella-specific IgA and IgM antibodies as early serodiagnosis markers of infection. Am. J. Trop. Med. Hyg. 77(Suppl. 5)259. [Google Scholar]
- 21.Pappas, G., N. Akritidis, M. Bosilkovski, and E. Tsianos. Brucellosis. 2005. N. Engl. J. Med. 3522325-2336. [DOI] [PubMed] [Google Scholar]
- 22.Pappas, G., P. Papadimitriou, N. Akritidis, L. Christou, and E. V. Tsianos. 2006. The new global map of human brucellosis. Lancet Infect. Dis. 691-99. [DOI] [PubMed] [Google Scholar]
- 23.Pappas, G., and Z. A. Memish. 2007. Brucellosis in the Middle East: a persistent medical, socioeconomic and political issue. J. Chemother. 19243-248. [DOI] [PubMed] [Google Scholar]
- 24.Paulsen, I. T., R. Seshadri, K. E. Nelson, J. A. Eisen, J. F. Heidelberg, T. D. Read, R. J. Dodson, L. Umayam, L. M. Brinkac, M. J. Beanan, S. C. Daugherty, R. T. Deboy, A. S. Durkin, J. F. Kolonay, R. Madupu, W. C. Nelson, B. Ayodeji, M. Kraul, J. Shetty, J. Malek, S. E. Van Aken, S. Riedmuller, H. Tettelin, S. R. Gill, O. White, S. L. Salzberg, D. L. Hoover, L. E. Lindler, S. M. Halling, S. M. Boyle, and C. M. Fraser. 2002. The Brucella suis genome reveals fundamental similarities between animal and plant pathogens and symbionts. Proc. Natl. Acad. Sci. USA 9913148-13153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Probert, W. S., K. N. Schrader, N. Y. Khuong, S. L. Bystrom, and M. H. Graves. 2004. Real-time multiplex PCR assay for detection of Brucella spp., B. abortus, and B. melitensis. J. Clin. Microbiol. 421290-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sánchez, D. O., R. O. Zandomeni, S. Cravero, R. E. Verdun, E. Pierrou, P. Faccio, G. Diaz, S. Lanzavecchia, F. Aguero, A. C. Frasch, S. G. Andersson, O. L. Rossetti, O. Grau, and R. A. Ugalde. 2001. Gene discovery through genomic sequencing of Brucella abortus. Infect. Immun. 69865-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scholz, H. C., Z. Hubalek, I. Sedlacek, G. Vergnaud, H. Tomaso, S. Al Dahouk, F. Melzer, P. Kampfer, H. Neubauer, A. Cloeckaert, M. Maquart, M. S. Zygmunt, A. M. Whatmore, E. Falsen, P. Bahn, C. Gollner, M. Pfeffer, B. Huber, H. J. Busse, and K. Nockler. 2008. Brucella microti sp. nov., isolated from the common vole Microtus arvalis. Int. J. Syst. Evol. Microbiol. 58375-382. [DOI] [PubMed] [Google Scholar]
- 28.Thimm, B., and W. Wundt. 1976. The epidemiological situation of brucellosis in Africa. Dev. Biol. Stand. 31201-217. [PubMed] [Google Scholar]
- 29.Whatmore, A. M., S. J. Shankster, L. L. Perrett, T. J. Murphy, S. D. Brew, R. E. Thirlwall, S. J. Cutler, and A. P. MacMillan. 2006. Identification and characterization of variable-number tandem-repeat markers for typing of Brucella spp. J. Clin. Microbiol. 441982-1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whatmore, A. M., L. L. Perrett, and A. P. MacMillan. 2007. Characterisation of the genetic diversity of Brucella by multilocus sequencing. BMC Microbiol. 734. [DOI] [PMC free article] [PubMed] [Google Scholar]