Abstract
Nasopharyngeal carcinoma (NPC) is one of the most common cancers among Chinese living in South China, Singapore, and Taiwan. At present, its etiological factors are not well defined. To identify which genetic alterations might be involved in NPC pathogenesis, we identified genes that were differentially expressed in NPC cell lines and normal nasomucosal cells using subtractive hybridization and microarray analysis. Most NPC cell lines and biopsy specimens were found to have higher expression levels of the gene encoding nucleolar and coiled-body phosphoprotein 1 (NOLC1) as compared with normal cells. Severe combined immunodeficiency mice bearing NPC xenografts derived from NOLC1-short hairpin-RNA-transfected animals were found to have 82% lower levels of tumor growth than control mice as well as marked tumor cell apoptosis. Measuring the expression levels of genes related to cell growth, apoptosis, and angiogenesis, we found that the MDM2 gene was down-regulated in the transfectants. Both co-transfection and chromatin immunoprecipitation experiments showed that tumor protein 53-regulated expression of the MDM2 gene requires co-activation of NOLC1. These findings suggest that NOLC1 plays a role in the regulation of tumorigenesis of NPC and demonstrate that both NOLC1 and tumor protein 53 work together synergistically to activate the MDM2 promoter in NPC cells.
Nasopharyngeal carcinoma (NPC) is a malignant tumor with specific racial and geographic distribution patterns. Although it is common in southern China, Taiwan, Singapore, and southeastern Asia, it is rare in Western countries and in neighboring Asian countries, such as Japan.1 The incidence of NPC in southern China, especially in Guangdong, has been reported to be 25 to 50 per 100,000 individuals.2 Emigrants from endemic countries to nonendemic areas, such as the United States, maintain this high risk, whereas second- and third-generation offspring have slightly lower risk.2 The etiology of NPC is multifactorial, but to date, not well defined. However, it has been suggested that environmental factors such as the long-term consumption of salted fish in Hong Kong3,4 and Malaysian Chinese5 and the long-term exposure to sulfuric acid vapor in Taiwan,6,7 can induce the formation of NPC. Genetic factors may also play some role in its development,7 though until now no gene has been associated with the carcinogenesis of NPC.8 The Epstein-Barr virus (EBV) has, however, been closely associated with its progression.2,9,10,11,12,13,14,15,16,17,18
Tumor markers for NPC are urgently needed, but the molecular mechanisms of NPC tumorigenesis remain obscure.2,9,18 Suppression subtractive hybridization (SSH) has been proven powerful in isolation of differential expressed genes, especially in isolation of rare transcripts.19,20,21 Combination of SSH and microarray provides an advantage in the global investigation of changes in gene expression in the biological system.22,23 In this study, we performed these two methods to investigate the differentially expressed genes between NPC and normal nasomucosal (NNM) cells and found high expressions of the gene encoding nucleolar and coiled-body phosphoprotein 1, NOLC1, previously also called hNOPP140 gene, in most NPC cell lines, but low expressions in NNM cells.
Human NOLC1 has a high degree (72% to 73%) of sequence homology with the well-characterized rat homologue, the nucleolar phosphoprotein NOPP140.24 This protein contains a nuclear localization signal binding sequence and is thought to shuttle between the nucleolus and the cytoplasm.24 A previous study found NOLC1 to have transcription factor-like activity.25 By binding to the transcription factor C/EBPβ (also known as AGP/EBP or NF-IL6), NOPP140 can indirectly activate the transcription of the α-1 acid glycoprotein gene.25 Overexpression of the partial or whole NOLC1 cDNA resulted in mislocalization of nucleolar proteins, improper formation of the nucleolus, and inhibition of rRNA gene transcription. These observations suggest that hNopp140 is crucial for normal cell growth. We were not compelled to study NOLC1 because of these reasons, but because it was overexpressed in NPC cells and may be associated the tumorigenesis of NPC.
The MDM2 gene is a cellular proto-oncogene, that is often amplified in ∼7% of all human cancers.26 Two promoters have been identified in MDM2 gene structure: a constitutive promoter and a TP53-response intronic promoter (P2).27,28 From our previous study, the expression of MDM2 gene can be indirectly enhanced in the EBV-infected NPC cells through enhancement of TP53 activation.29
Using RNA interference in vivo to examine the role of NOLC1 in the pathogenesis of NPC, we found that NOLC1 was crucial for NPC cell growth and that reduction of its expression in transfected xenografts resulted in retardation of tumor growth and apparent apoptosis and necrosis. We subsequently examined several genes related to this function and found that the depletion of NOLC1 resulted in a reduction of the MDM2 expression. Moreover, we found that NOLC1 and TP53 synergistically co-regulated MDM2 expression in NPC cells.
Materials and Methods
Cell Lines, Tissues, and Surgical Specimens
Fourteen NPC cell lines were grown in Dulbecco’s modified Eagle’s medium supplemented with 5% fetal calf serum (Gibco BRL, Gaithersburg, MD). They included NPC-TW01, 02, 03, 04, 05, 06, 07, 08, 09, and 10, all established in our laboratory30,31; NPC-CGBM-1, a gift from Dr. S. K. Liao (Chang-Gung University, Taoyuan, Taiwan)32; and three other lines CNE1, CNE2, and HONE-1 cells, all originating from China.33,34 The NPC-TW06 cell line contains a heterozygous point mutation in the tumor protein 53 (TP53) gene; the TP53 protein is retained in the cytoplasm and lost the transcriptional activity.35 TW01, 02, 05, and 08 are keratinizing squamous cell carcinoma lines (World Health Organization Type I), and TW03 is an undifferentiated carcinoma (World Health Organization Type III), also called lymphoepitheliometous carcinoma. The other cell lines were all undifferentiated carcinoma types (World Health Organization Type III). The four NNM cell cultures, NNM-9, NNM-11, NNM-12, and NNM-13, were primary cultured cells from nasal polyps as described previously,17 and were grown in Dulbecco’s modified Eagle’s medium supplemented with 20% fetal calf serum. NPC paraffin blocks were obtained from the archives of the Department of Pathology, National Taiwan University Hospital, Taipei, Taiwan. The use of human specimens in this research was approved by the Institutional Review Board (IRB-926170459) of National Taiwan University Hospital.
RNA Isolation and SSH
SSH was used to isolate genes present in the NPC-TW04 or NNM cells. Total RNAs from these cells were isolated using the acid guanidinium thiocyanate-phenol-chloroform method (TRIzol; Invitrogen Life Technologies, Carlsbad, CA). The mRNA from NPC-TW04 cells was used as the “tester” and the mRNA from NNM cells was used as the “driver” for cDNA subtraction. The construction was performed following the SSH procedure using a PCR-select cDNA subtraction kit (Clontech, Palo Alto, CA). Briefly, equal amounts of mRNA from the tester and driver populations were converted to double-stranded cDNA by reverse transcription followed by digestion with RsaI separately. The digested tester cDNA was subdivided into two populations, each ligated with a different adaptor. Ligation efficiency was evaluated using PCR using primers specific to chicken glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA and the adaptor sequences. Following ligation, two hybridization steps was performed. For the first hybridization, an excess of driver was added to each tester, which was denatured, and allowed to anneal. The target sequences in the testers became enriched for differentially expressed genes in NPC-TW04 cells (NPC-TW04-NNM cDNA). The same procedure was repeated using mRNA from NNM cells as tester and mRNA from NPC-TW04 cells as a reference for cDNA subtraction. This produced other target sequences that also became enriched for differentially expressed in NNM cells (NNM-NPC-TW04 cDNA). In the second hybridization step, the subtracted target cDNAs were specifically amplified by nested PCR using adaptor-specific primer pairs and labeled with the addition of digoxigenin- or biotin-labeled nucleotides (Roche Molecular Biochemicals, Indianapolis, Ind) to obtain the digoxigenin-NPC-TW04-NNM cDNA and biotin-NNM-NPC-TW04 cDNA, respectively.
cDNA Microarray Analysis and Screening of the cDNA Library with Subtracted Probes
A nylon microarray membrane containing 9600 cloned expressed sequence tag DNAs was constructed using cDNA clones from the National Taiwan University Hospital Microarray Core Facility for Genomic Medicine, Taiwan. The DNA microarray analysis was based on the colorimetry detection method on human eye recognition. The probes derived from the subtraction experiments were added to the membrane, the substrate was added, and the two colors developed. The image of each DNA dot was digitized by scanning on a high-resolution flat bed scanner (Umax Magic Scan at 3000 dpi). The digitized images were separated into cyan and magenta colors. The most significantly different hybridizing cDNAs were selected for further analysis. Clones containing the chosen expressed sequence tag cDNA were used to screen an NPC cDNA library prepared from the NPC-TW01 cell line in 1998 by Dr. P. Ouyang of the Department of Anatomy, Chang-Gung University, Taiwan, following a standard procedure.36 The full-length nucleotide sequences were compared with sequences in the database using nucleotide BLAST from the National Center for Biotechnology Information BLAST website (http://www.ncbi.nlm.nih.gov/BLAST/).
Touchdown and Quantitative Reverse-Transcription PCR analysis
RNA was obtained from 14 NPC, three NNM, NS (nonspecific)-short hairpin (sh)RNA-NPC-TW03, shNOLC1-1-NPC-TW03, and shNOLC1-2-NPC-TW03 cell lines, and four xenograft tumors, as described above in the RNA isolation method. Reverse transcription was performed using the SuperScript First-Strand Synthesis Kit (Invitrogen Life Technologies, Carlsbad, CA). Touchdown PCR was performed for each of 29 genes (Table 1) following procedures described in Don et al,37 with denaturation performed at 95°C for 30 seconds, anneling of 10 cycles at 65–56° C and 20 cycles at 55°C for 30 sec per cycle, and extension at 72°C for 1 minute. The PCR products were analyzed on a 1.2% agarose gel. The endogenous reference gene was ACTB (encoding β-actin). We performed quantitative reverse transcription (QRT-PCR) analysis using the comparative threshold cycle method using an ABI PRISM 7700 Sequence Detector System and SYBR Green PCR Master Mix Kit (Perkin Elmer, Applied Biosystems, Wellesley, MA) , according to the manufacturers’ instructions and as described in our recent paper.17 The endogenous reference gene used was GAPDH.
Table 1.
The Primers Used for RT-PCR and QRT-PCR
|
A. For RT-PCR
|
|
|
|
|---|---|---|---|
| UniGene number | Gene symbol | Official full name | Primers |
| Hs.467020 | BCL-2 | B-cell CLL/lymphoma 2 | 5′-ACTTGTGGCCCAGATAGGCACCCAG-3′ |
| 5′-CGACTTCGCCGAGATGTCCAGCCAG-3′ | |||
| Hs.631546 | BAX | BCL2-associated X protein | 5′-GCTCTGAGCAGATCATGAAGACAG-3′ |
| 5′-CACAAAGATGGTCACGGTCTGC-3′ | |||
| Hs.2490 | CASP1 | caspase 1, apoptosis-related cysteine peptidase (interleukin 1, beta, convertase) | 5′-TTTGATTGACTCCGTTATTC-3′ |
| 5′-TCTCTGCCGACTTTTGTTTC-3′ | |||
| Hs.23960 | CCNB1 | cyclin B1 | 5′-GAAGCTACTGGAAACATG-3′ |
| 5′-CTACAGCTCGTTGTATGA-3′ | |||
| Hs.153752 | CDC25B | cell division cycle 25B | 5′-CACGCCCGTGCAGAATAAGC-3′ |
| 5′-ATGACTCTCTTGTCCAGGCTACAGG-3′ | |||
| Hs.370771 | CDKN1A | cyclin-dependent kinase inhibitor 1A (p21, Cip1) | 5′-ATGTCAGAACCGGCTGGGGATG-3′ |
| 5′-GCAGGCCAAGGCCCCGCAC-3′ | |||
| Hs.644056 | CSNK2A1 | casein kinase 2, alpha 1 | 5′-ATGACCACCAGTCACGGCTTAC-3′ |
| 5′-GGTTCAGACACGGTGCTTCTG-3′ | |||
| Hs.488293 | EGFR | epidermal growth factor receptor | 5′-CATAGACGACACCTTCCTCC-3′ |
| 5′-GGGTCTAAGAGCTAATGCGG-3′ | |||
| Hs.244139 | FAS | Fas (TNF receptor superfamily, member 6) | 5′-TAGCTCCTATATTTTCGGCTT-3′ |
| 5′-CTCACCAGCAACACCAAGTGC-3′ | |||
| Hs.396530 | HGF | hepatocyte growth factor | 5′-ACTGGCTCTTTTAGGCACTGACTC-3′ |
| 5′-TGTTCCCTTGTAGCTGCGTCCTTT-3′ | |||
| Hs.132966 | MET | Met proto-oncogene (hepatocyte growth factor receptor) | 5′-ACTCCCCCTGAAAACCAAAGCC-3′ |
| 5′-GGCTTACACTTCGGGCACTTAC-3′ | |||
| Hs.567303 | MDM2 | Mdm2, transformed 3T3 cell double minute 2, p53 binding protein (mouse) | 5′-GCAGGGGAGAGTGATACAGAT-3′ |
| 5′-GATGGCTGAGAATAGTCTTCA-3′ | |||
| Hs.513617 | MMP2 | Matrix metallopeptidase 2 | 5′-GGGGCCTCTCCTGACATT-3′ |
| 5′-CATTCCCTGCAAAGAACACA-3′ | |||
| Hs.297413 | MMP9 | Matrix metallopeptidase 9 | 5′-TGGGCTACGTGACCTATGAC-3′ |
| 5′-CAAAGGTGAGAAGAGAGGGC-3′ | |||
| Hs.202453 | MYC | v-myc myelocytomatosis viral oncogene homolog | 5′-GTGGCACCTCTTGAGGACCA-3′ |
| 5′-TGGTGCTCCATGAGGAGACA-3′ | |||
| Hs.463456 | NME1 | Non-metastatic cells 1, protein (NM23A) | 5′-TGCTGCGAACCACGTGGGT-3′ |
| 5′-ATGTGGTCTGCCCTCCTGT-3′ | |||
| Hs.645227 | TGFB1 | Transforming growth factor, beta 1 | 5′-CTCCGAGAAGCGGTACCTGAAC-3′ |
| 5′-CACTTGCAGTGTGTTATCCCT-3′ | |||
| Hs.522632 | TIMP-1 | TIMP metallopeptidase inhibitor 1 | 5′-CTGGAAAACTGCAGGATGGA-3′ |
| 5′-CGCTGAGCTAAGGTCAGGCT-3′ | |||
| Hs.633514 | TIMP-2 | TIMP metallopeptidase inhibitor 2 | 5′-CTCATTGCAGGAAAGGCCGA-3′ |
| 5′-TGGGTGGTGCTCAGGGTGTC-3′ | |||
| Hs.591665 | TIMP-4 | TIMP metallopeptidase inhibitor 4 | 5′-CCAGAGGTCAGGTGGTAA-3′ |
| 5′-ACAGCCAGAAGCAGTATC-3′ | |||
| Hs.241570 | TNF | Tumor necrosis factor (TNF superfamily, member 2) | 5′-CTTCTGCCTGCTGCACTTTGGA-3′ |
| 5′-TCCCAAAGTAGACCTGCCCAGA-3′ | |||
| Hs.81791 | TNFRSF11B | Tumor necrosis factor receptor superfamily, member 11b (osteoprotegerin) | 5′-AGGAAATGCAACACACGACAAC-3′ |
| 5′-AGGAACAGCAAACCTGAAGAATG-3′ | |||
| Hs.156346 | TOP2A | topoisomerase (DNA) II alpha 170kDa | 5′-TGGTCAGAAGAGCATATGAT-3′ |
| 5′-CTCACAATCTGATCAGCTAC-3′ | |||
| Hs.408312 | TP53 | Tumor protein p53 | 5′-CTATGTCGAAAAGTGTTTCTGTCATC-3′ |
| 5′-CAGCCAAGTCTGTGACTTGCACGTAC-3′ | |||
| Hs.111779 | SPARC | Secreted protein, acidic, cysteine-rich (osteonectin) | 5′-ACTGAAGCTTCCCAGCACCATG-3′ |
| 5′-GAGAGGATCCGGTACTGTGG-3′ | |||
| Hs.585572 | SOX5 | SRY (sex determining region Y)-box 5 | 5′-CAACCTTGGTGCTGCTGTATCT-3′ |
| 5′-GTCTTGGGTTTAGCTGATAGGTTCA-3′ | |||
| Hs.73793 | VEGF | Vascular endothelial growth factor A | 5′-CGATCGTTCTGTATCAGTCTTTCC-3′ |
| 5′-GAAGTGGTGAAGTTCATGGATGTC-3′ | |||
| Hs.523238 | NOLC1 | Nucleolar and coiled-body phosphoprotein 1 | 5′-AGAAAAGAAAAAGGCGGCAG-3′ |
| 5′-TCCTCATCAAGACCCTCACC-3′ | |||
| Hs.520640 | ACTB | Actin, beta | 5′-CACTCTTCCAGCCTTCCTTC-3′ |
| 5′-GCCATGCCAATCTCATCTTG-3′ | |||
| Hs.544577 | GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | 5′-CGGGAAGCTTGTGATCCATGG-3′ |
| 5′-GGCAGTGATGGCATGGACTG-3′ | |||
| (table continues) | |||
Table 1A.
Continued
|
B. For QRT-PCR
|
|
|---|---|
| Gene | Primers |
| ARF | 5′-GAAGCCAAGGAAGAGGAATGAG-3′ |
| 5′-CAAATATGTTCCCCCCTTCAGA-3′ | |
| BAX | 5′-ATGTTTTCTGACGGCAACTTCA-3′ |
| 5′-CAGTTCCGGCACCTTGGT-3′ | |
| CASP1 | 5′-GCAGGACAACCCAGCTATGC-3′ |
| 5′-TCTGCCGACTTTTGTTTCCAT-3′ | |
| MMP9 | 5′-AGTTTGCCGGATACAAACTGGTA-3′ |
| 5′-GAAACACTCCAACAAAAAACAAAGGT-3′ | |
| NOLC1 | 5′-GACTGCATCTTCTCGTTTTTTACAGTATA-3′ |
| 5′-GATCAGTGATTCTCAACCATGTAGGA-3′ | |
| MDM2 | 5′-CCTTAGCTGACTATTGGAAATGCA-3′ |
| 5′-CAGGAAGCCAATTCTCACGAA-3′ | |
| TNF | 5′-CCTGCCCCAATCCCTTTATT-3′ |
| 5′-CCCTAAGCCCCCAATTCTCTT-3′ | |
| TP53 | 5′-GGGTTAGTTTACAATCAGCCACATT-3′ |
| 5′-GGGCCTTGAAGTTAGAGAAAATTCA-3′ | |
| VEGF | 5′-TCTACCTCCACCATGCCAAGT-3′ |
| 5′-CTGCGCTGATAGACATCCATGA-3′ | |
| β-actin | 5′-ACGTGGACATCCGCAAAGAC-3′ |
| 5′-CTCAGGAGGAGCAATGATCTTGAT-3′ | |
| GAPDH | 5′-TGGTATCGTGGAAGGACTCA-3′ |
| 5′-AGTGGGTGTCGCTGTTGAAG-3′ | |
| TP53 binding site of MDM2 promoter | 5′-TAGTCTGGGCGGGATTG-3′ |
| 5′-TGCAGTTTCGGAACGTG-3′ | |
| Exson 2 of MDM2 promoter | 5′-TGGCGATTGGAGGGTAGA-3′ |
| 5′-ACCTGGATCAGCAGAGAA-3′ | |
| GAPDH promoter | 5′-TCCAAGCGTGTAAGGGT-3′ |
| 5′-GAAGGGACTGAGATTGGC-3′ | |
| Beta-Hemoglobulin promoter | 5′-ATCTGAGCCAAGTAGAAGACCTTTTC-3′ |
| 5′-TCTGCCTGGACTAATCTGCAAG-3′ |
Immunohistochemical Staining
Both NPC cell lines and NPC biopsy specimens were subjected to routine immunohistochemical staining using a monoclonal antibody directed against NOLC1,38 according to a previously described method.13 Immunoreactivity, defined as the number of positive tumor cells over total tumor cells, was scored independently by two researchers. The number of NOLC1-positive and negative NPC cells was counted under light microscope at a magnification of ×400, with only the cells displaying brown nucleoli on the section considered NOLC1-positive. For each slide, 7 to 10 microscopic fields were randomly chosen. Positive scores were the categorized into weak staining (only one nucleolus was stained), moderate staining (more than one nucleolus was stained), and strong staining (both nucleus and nucleolus of the tumor cell staining). The average percentage of NOLC1-positive NPC cells was then calculated for each group.
Western Blot Analysis
Lysates from the cultured cells were subjected to routine Western blotting as described previously.39 The antibodies used were monoclonal anti-mouse antibodies against NOLC1,38 TP53, MDM2, and α-tubulin, and polyclonal rabbit antibodies against MMP9 and CASP1. Antibodies of TP53, MDM2, MMP9, α-tubulin, and CASP1 were purchased from Lab Vision Co. (Fremont, CA). The results shown are representative of two independent experiments.
Establishment of Stable shRNA Transfectants
The shRNA constructs described in Table 2 were purchased from Open Biosystems (Huntsville, AL). When the NPC cultured cells had reached 70% to 80% confluence, the shRNA constructs were transfected into the NPC cells using the Arrest-In Transfection Reagent for RNAi (Open Biosystems). After incubation for 48 hours, the cells were selected with puromycin to establish of two stable lines: shNOLC1-1-NPC-TW03 and shNOLC1-2-NPC-TW03 line. To avoid the individual clonal variation of gene expression, we used mixed clones of all of the candidate cells that were successfully selected after adding the antibiotics. Then we checked the RNA and protein levels to confirm that these selected clones contained shRNA.
Table 2.
The Construct Sequences of shRNA
| shRNA symbol | Sequence |
|---|---|
| NOLC1-1 shRNA | 5′-TGCTGTTGACAGTGAGCGCGACATCTAAGTCTGCAGTTAATAGTGAAGCCACAGATGTATTAACTGCAGACTTAGATGTCTTGCCTACTGCCTCGGA-3′ |
| NOLC1-2 shRNA | 5′-TGCTGTTGACAGTGAGCGAACAGTTAAAGCTCAGACTAAATAGTGAAGCCACAGATGTATTTAGTCTGAGCTTTAACTGTCTGCCTACTGCCTCGGA-3′ |
| NS-shRNA | 5′-TGCTGTTGACAGTGAGCGAACCACTAAGCTTCTGTCTTAATAGTGAAGCCACAGATGTATTAAGACAGAAGCTTAGTGGTCTGCCT5′-ACTGCCTCGGA-3′ |
Tumor Growth in Severe Combined Immunodeficient Mice
To establish an animal model for the functional analysis of NOLC1, 24 six-week-old NOD/severe combined immunodeficient female mice were obtained from the National Taiwan University Hospital Experimental Animal Center. The animals were divided into four groups. We injected 1 × 107 NPC-TW03 cells, transfected with NOLC1-1 shRNA, NOLC1-2 shRNA, or the control vector, or the same number of untransfected cells, subcutaneously into the right flanks of six mice in each group separately. One week after cell transplantation, two dimensions of the tumor size were measured with the calipers. The tumor volume was estimated using the equation: length × width2 × 0.52.29 The tumor sizes were measured once per week. Values are presented as the mean values ± SEM. After 11 weeks, all animals were examined by routine autopsy and all xenografts were excised, fixed in 4% paraformaldehyde, and embedded in paraffin blocks, or stored at −80°C for QRT-PCR analysis and other experimental use. The use of animals was approved by the Institutional Animal Care Use Committee.
Transient Cotransfection for Reporter Induction
NPC-TW03, shNOLC1-1-NPC-TW03, NS-shRNA-NPC-TW03, and NPC-TW06 cells were seeded into six-well plates, grown to a density of 80% confluence, and transfected separately with a transfection mixture that included fluorescent Arrest-In Transfection Reagent. The transfection mixture, containing the appropriate reporter and effector plasmids (including pGL2-MDM2-Luc, pGL3- SV40-TP53, pGL3-SV40-TP53 null, and pGL3-CMV-βgal) and a lipophilic reagent were mixed in serum-free medium and applied to the cells according to the manufacturer’s instructions. After three hours, complete medium was added. After 48 hours, the cells were harvested for analysis. Luciferase enzyme assays and colorimetric β-galactosidase assays were performed according to the manufacturer’s instructions (Promega, Madison, WI). Luciferase activity was normalized to β-galactosidase activity to assess the transfection efficiency. Each transfection experiment was repeated three times.
Chromatin Immunoprecipitation Assay
The chromatin immunoprecipitation (ChIP) assay was performed according to the manufacturer’s instructions (Upstate, Charlottesville, VA). Briefly, 1 × 105 NPC-TW04 cells were fixed with 1% formaldehyde (Sigma, St. Louis, MO) in culture medium, and then total DNA was extracted and sonicated to an average size of 250 bp. ChIP assay was performed by adding NOLC1 or TP53 monoclonal antibodies, agarose-conjugated protein A beads, and then incubated for 2 hours. Immunoprecipitates were then washed, processed, and eluted for QRT-PCR analysis. The primers are shown in Table 1.
Effect of EBV Infection on NOLC1 Expression in NPC Cell Lines
The procedures used to isolate EBV particles and to identify viral DNA in the isolated viral solution were as described previously.13 EBV infection using IgA receptor (secretary component protein)-mediated endocytosis in the NPC-TW01, NPC-TW03, NPC-TW04, NPC-TW06, and CG-BM-1 cell lines was performed as previously reported.13,29,40 After infection, the cells were cultured for 10 days, collected, and used in the detection of the EBV by EBNA-1 immunostaining,29 and subjected to the QRT-PCR analysis of NOLC1 gene expression.
Statistical Analysis
Data were described as the means ± SE of mean of the indicated number of separate experiments. Statistical significance was determined by the paired two-tailed Student’s t-test. P values less than 0.05 were considered significant.
Results
Genes Selected by SSH and Microarray
After the mRNAs of NPC and NNM cells were subjected to each other using SSH, two populations of differentially expressed cDNA in both cell types were generated, respectively. To identify these two differentially expressed transcripts, the subtractive cDNA of digoxigenin- or biotin-labeled NPC-TW04-NNM and NNM-NPC-TW04 were monitored by screening with a cDNA microarray of expressed sequence tag clones (see supplemental Figure S1 at http://ajp.amjpathol.org). After hybridization, only red and blue spots represented that these two transcripts are only up-regulated or down-regulated genes in NPC-TW04 compared with NNM cells. To verify the expression of candidate genes in other NPC cells, candidate expressed sequence tag genes (see supplemental Table S1 at http://ajp.amjpathol.org), which were highly expressed in NPC or NNM cells, were screened by a phage λgt11 expression library of NPC genes. After sequence analysis and database searches with BLAST, three genes, NOLC1, MRPL19, and AL359844 (see supplemental Table S2 at http://ajp.amjpathol.org), were identified. Because NOLC1 was the most highly expressed gene among these three genes in NPC cells, it was chosen for further study.
NOLC1 Gene Expression is Up-Regulated in Most NPC Tumor Cells
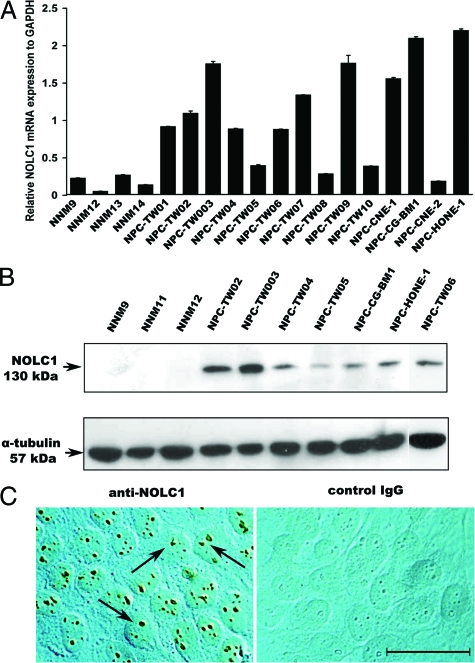
To verify the differences in expression of the selected gene, we performed QRT-PCR analysis (Figure 1A) for NOLC1 gene expression in 14 NPC and four NNM cell lines. The expression of the NOLC1 gene was elevated in most of the NPC cell lines (12/14 lines), with the highest expression of both the NOLC1 mRNA (Figure 1A) and protein (Figure 1B) in NPC-TW03 cells. NNM cells exhibited minimal or no expression (Figure 1, A and B). Immunohistochemical localization of NOLC1 protein clearly showed the reaction product in the nucleoli of all NPC cells (Figure 1C, left) but no reaction product in the nucleoli of control IgG (Figure 1C, right). These results demonstrate that NOLC1 is highly expressed in NPC cells, but in normal cells it is very weak if not nil.
Figure 1.
NOLC1 mRNA and protein expression in NPC cell lines. A: NOLC1 mRNA levels detected by QRT-PCR in 14 different NPC lines and four NNM cells. The NOLC1 gene was highly expressed in most of the NPC lines compared with the NNM cells. Experiments were repeated three times and normalized to the GAPDH control. B: Western blot analysis of NOLC1 protein expression. A 130-kDa band of NOLC1 protein is shown in the NPC cell lines. In contrast, NNM cells showed no detectable signals. α-tubulin was used as an internal control. C: Immunohistochemical localization of NOLC1 protein in NPC cells. The left panel was stained with anti-NOLC1 antibody and right panel was stained with the nonspecific negative control IgG. NOLC1 immunoreactivity was seen clearly in the nucleoli of the NPC-TW03 cells (arrows). The control IgG staining shows no reaction product. Scale bar = 10 μm.
NOLC1 Is Highly Expressed in the NPC Biopsy Specimens
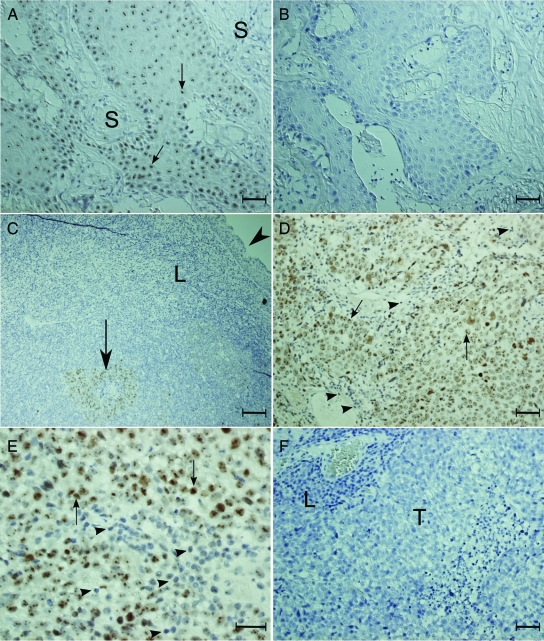
To find out if NOLC1 protein was highly expressed in NPC patients, we performed immunohistochemical localization of this protein in 30 NPC biopsies. As shown in Figure 2, the nucleoli of the tumor cells in the NPC biopsies were found to have NOLC1 immunoreactivity (Figure 2, A, C–E), while those of the control IgG showed no reaction product (Figure 2, B and F). All 30 specimens, including World Health Organization type I (keratinizing squamous cell carcinoma) and World Health Organization type III (undifferentiated carcinoma), expressed the NOLC1 proteins. Looking at all of the staining data, we found that regardless of pathological type, all expressed NOLC1 proteins, average staining of the positive-NOLC1 NPC cells being 93%. There was weak staining in 14%, moderate staining in 33%, and strong staining in 46% of the positive-NOLC1 NPC specimens. However, no reaction product was found in the stromal cells (Figure 2A, S), lymphocytes (Figure 2C, L), or normal mucosal epithelia (Figure 2C, arrowhead). These results clearly demonstrate that NOLC1 is highly expressed in NPC cells both in vitro and in vivo.
Figure 2.
Representative pictures of NOLC1 protein staining in different NPC biopsy specimens. A–B: Type I NPC biopsy specimens. C–F: Type III NPC biopsy specimens. The paraffin sections were stained with either monoclonal anti-NOLC1 (A, C–E) or the control IgG (B, F). The NOLC1 immunoreactivity was seen clearly in the nucleoli of the tumor cell (arrows) in the tumor nests, but not in the stromal cells or in the normal squamous metaplastic epithelial cells (C, arrowhead). No reaction product was seen in the control sections. S: stromal cells; T: tumor nest; L: lymphocyte. Scale bars = 25 μm (A, B, D–F); 5 μm (C).
Inhibition of NOLC1 Expression Decreases Proliferation and Increases Apoptosis in NPC Cells
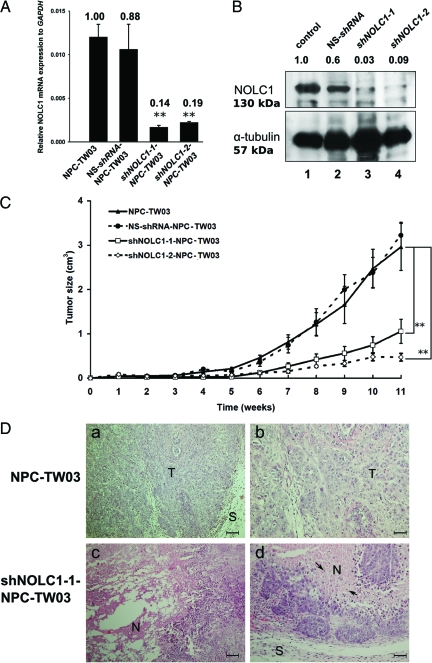
To investigate the function of NOLC1 protein in NPC cells, shRNA vectors directed against NOLC1 (shNOLC1-1 and shNOLC1-2) were transfected into NPC-TW03 cells separately to establish two stable cell lines. NPC-TW03 cells were chosen for their higher NOLC1 expression (Figure 1B). Two days after transfection, we found reduced expression of NOLC1 mRNA and protein in NOLC1- shRNA-transfected cells. The expression of NOLC1 mRNA was found to be decreased to approximately 86% and 81% in shNOLC1-1 and shNOLC1-2 transfectants, compared with control (Figure 3A), and expression of protein was found to be decrease to 97% and 91% (Figure 3B) in shNOLC1-1 and shNOLC1-2 transfectants, compared with untransfected controls, respectively.
Figure 3.
Verification of two shNOLC1-transfected NPC stable lines. A: After the transfection of two NOLC1 shRNAs plasmids (shNOLC1-1 and shNOLC1-2) separately into the NPC-TW03 cell lines, the total RNAs of each line were extracted and examined by QRT-PCR analysis. The data were obtained from triplicate experiments and normalized to GAPDH expression levels. Both shNOLC1-1 and shNOLC1-2 had a clear suppressive effect on NOLC1 mRNA expression in the transfected NPC lines. Samples of mixed clone cells were measured, each point representing the mean value ± SEM (n = 3). **P < 0.01. B: Western blot analysis of NOLC1 protein expression in NOLC1-shRNA-transfected NPC-TW03 cells. Lane 1, NPC-TW03 cells without transfection (control); lanes 2–4, stable lines transfected with NS-shRNA, shNOLC1-1, or shNOLC1-2, respectively. One 130-kDa band of NOLC1 protein was visible in lanes 1 and 2. The staining intensity of the same band was very weak in lane 3 and almost invisible in lane 4. α-tubulin a 57-kDa band was used as the internal control in each lane. C: The effect of NOLC1 shRNA transfection on NPC tumor growth in vivo. severe combined immunodeficient mice bearing xenografts of NPC-TW03 cells, including NS-shRNA-, NOLC1-shRNA (shNOLC1-1 and −2)-transfected and untransfected control NPC-TW03 cells, were observed for 11 weeks, and then the tumor masses from the four groups were removed and analyzed. Each point represents the mean value ± SEM. (n = 6) **P < 0.01. D: Histological features of tumor sections from mice bearing NPC and shNOLC1-NPC xenografts. H&E staining from a paraffin section of an NPC-TW03 xenograft showed an undifferentiated carcinoma (a,b), whereas the shNOLC1-NPC xenograft sections revealed marked tumor cell necrosis (c) and apoptotic changes (d). T: tumor cells; N: necrosis; S: stroma; arrows: apoptotic cells. Scale bars = 50 μm (a and c); 100 μm (b and d).
To determine the role of NOLC1 in NPC cell proliferation in vivo, we established an animal model using severe combined immunodeficient mice bearing NPC xenografts derived from either the shNOLC1-1- or the shNOLC1-2-transfected NPC-TW03 cell line. The tumor growth exhibited a clear reduction of tumor size when compared with that of mice bearing xenografts from untransfected NPC-TW03 cells. By 11 weeks, the tumor size of the shNOLC1-1-transfected xenografts was suppressed to 33% (67% inhibition) of the tumor sizes of the control group. In mice bearing the shNOLC1-2-NPC-TW03 xenograft, tumor growth had decreased to approximately 18% (82% inhibition) of that of the control group. The xenograft from the NS-shRNA transfectants did not exhibit any change in tumor size compared with the control group (Figure 3C). Histopathological sections of the shNOLC1-1-NPC-TW03 (Figure 3D, c and d) and shNOLC1-2-NPC-TW03 (data not shown) xenograft tumors revealed massive tumor cell necrosis and apoptosis in the tumor region when compared with untransfected xenografts (Figure 3D, a and b). Taken together, the data show that depletion of NOLC1 results in a reduction of tumor growth and the induction of necrosis/apoptosis in NPC xenografts.
NOLC1 Regulates MDM2 Gene Expression
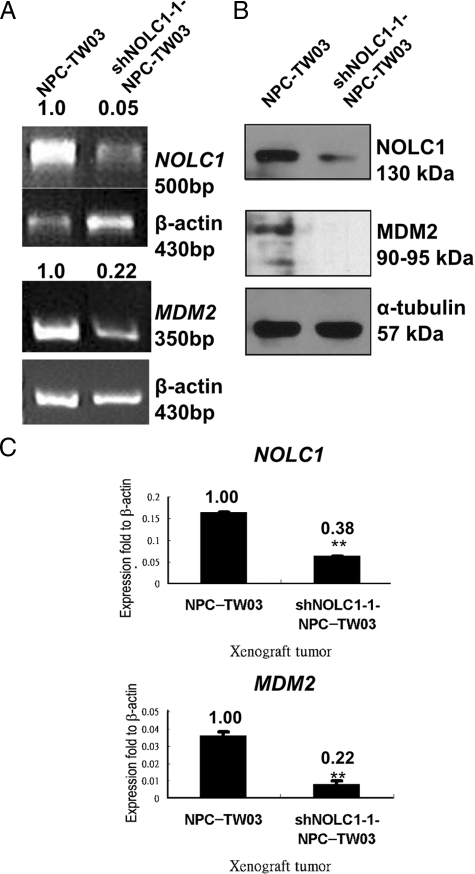
To clarify the molecular mechanisms underlying the effect of NOLC1 expression on tumor behavior, we searched the previous studies of some genes involved in cell proliferation, angiogenesis, or apoptosis,41,42 and then examined the expression of 26 genes (Table 1) by reverse transcription-PCR in shNOLC1-1-NPC-TW03 cells. The results showed that the expression of the MDM2 oncogene (Figure 4A, bottom) was significantly suppressed (0.22-fold, 78% inhibition). To confirm the alteration of the mRNA expression pattern, we also determined the expression of protein by Western blotting. We found a reduced expression of MDM2 protein in shNOLC1-1-NPC-TW03 cells (Figure 4B). To further confirm this phenomenon in vivo, we checked the gene expressions in NPC-TW03 and shNOLC1-1-NPC-TW03 xenograft tissues. Expressions of the NOLC1 and MDM2 genes (Figure 4C) were also significantly suppressed (67% and 78% inhibition, respectively). Taken together, our in vitro and in vivo findings suggest that depletion of NOLC1 results in reduced expression of MDM2. Other NOLC1-regulated downstream genes, including MMP9, VEGF, TNF-α, BAX, and CASP1, are shown in the supplemental Figure S2 at http://ajp.amjpathol.org.
Figure 4.
Depletion of NOLC1 suppressed MDM2 expression in NPC-TW03 cells. A: Knockdown of NOLC1 expression suppressed the MDM2 mRNA expression in NPC-TW03 cells. Total RNAs derived from NPC-TW03 and shNOLC1-1-NPC-TW03 cells were analyzed by reverse transcription-PCR. Both NOLC1 and MDM2 mRNA expression were suppressed. The β-actin cDNA was used as an internal control. B: Western blot analysis of NOLC1 and MDM2 protein expression in NPC-TW03 and shNOLC1–1-NPC-TW03 cells. α-tubulin was used as the internal control in each lane. C: QRT-PCR analysis of the expressions of the NOLC1 (top) and MDM2 (bottom) genes in NPC-TW03 and shNOLC1-1-NPC-TW03 xenograft tumors. The mRNA of NOLC1 and MDM2 expression in the shNOLC1-NPC xenograft was also markedly suppressed. The data were obtained from triplicate experiments and normalized to the β-actin cDNA as the control. **P < 0.01.
NOLC1 Acts Synergistically with TP53 to Up-Regulate MDM2 Expression
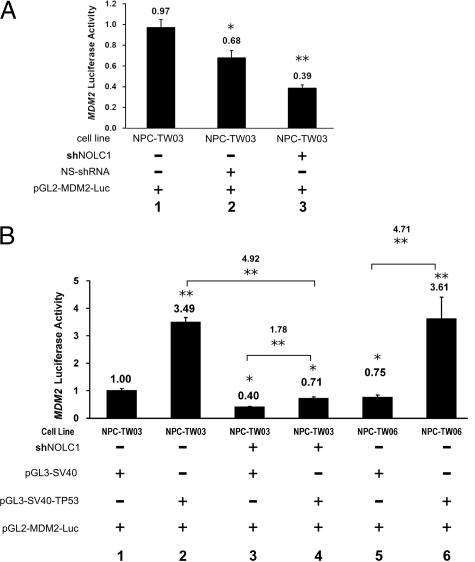
Since TP53 is a key activator of MDM2 gene expression,43 and TP53 can up-regulate MDM2 expression during NPC pathogenesis,44,45 we used the luciferase reporter gene assay to analyze NOLC1’s regulation of MDM2 gene expression. The ability of NOLC1 to activate the transcription of MDM2 was measured by the transient transfection of a reporter plasmid containing the luciferase gene under the MDM2 p2 promoter (pGL2-MDM2-Luc) into NPC-TW03, shNOLC1-NPC-TW03, or NS-shRNA-NPC-TW03 cells, and cotransfection with a β-galactosidase expression construct for normalized transfection efficiencies (Figure 5A). When NOLC1 expression was inhibited, the MDM2 promoter in the shNOLC1-NPC-TW03 cells showed reduced activity, resulting in a 0.39-fold decrease in the luciferase activity of the untransfected NPC-TW03 cells (Figure 5A, lanes 1 and 3). However, the NS-shRNA-transfected cells also displayed a reduction in luciferase activity to 0.68-fold that of the NPC-TW03 cells (Figure 5A, lane 2). These data support the hypothesis that normal MDM2 promoter activity in NPC cells requires NOLC1 regulation, even in the presence of wild-type TP53 (wtTP53). Therefore, MDM2 transcription is not entirely dependent on the function of the TP53 transcription factor.
Figure 5.
The relationship between TP53 and NOLC1 regulation of MDM2 gene expression in NPC cells. A: Depletion of NOLC1 gene expression resulted in down-regulation of MDM2-expression in NPC-TW03 cells. The MDM2 promoter reporter (pGL2-MDM2-Luc) was cotransfected with the internal control pGL3-CMV-βgal into NPC-TW03 (lane 1), NS-shRNA-NPC-TW03 (lane 2) or shNOLC1-NPC-TW03 (lane 3) cells separately for 48 hours, and MDM2 P2 promoter expression was measured by luciferase assay. B: The regulation of MDM2 gene expression by TP53 depended on NOLC1 co-activation. The MDM2 P2 promoter reporter (pGL2-MDM2-Luc) was cotransfected with either pGL3-SV40-TP53 or pGL3-SV40 (control) and an internal control pGL3-CMV-βgal into NPC-TW03 (lane 1,2), shNOLC1-NPC-TW03 (lane 3,4) or NPC-TW06 (lane 5,6) cells for 48 hours, and the MDM2 P2 promoter expression was measured by luciferase assay. The luciferase activity was measured in each cell lysate and reported in arbitrary units. The same experiment was repeated three times. All data are reported as average values ± SEM; n = 3 for each condition. *P < 0.05. **P < 0.01.
To examine the effects of increased TP53 expression on MDM2 expression, we cotransfected the plasmid constructs pGL2-MDM2-Luc and pGL3-SV40-TP53 into NPC-TW03, shNOLC1-NPC-TW03, and NPC-TW06 cells (Figure 5B). In NPC-TW06 cells, the TP53 gene contains a heterozygous point mutation causing the faulty TP53 protein to sequester any normal TP53 outside the nucleus, causing a lack of TP53 as a transcription factor.35 In NPC-TW03 cells, which have wtTP53, exogenous TP53 brought about a 3.49-fold increase in MDM2 promoter (Figure 5B, lanes 1 and 2). In the shNOLC1-NPC-TW03 cells, exogenous TP53 could only barely activate the MDM2 promoter to regulate the luciferase activity (Figure 5B, lanes 3 and 4). There was 4.92-fold higher than that of the shNOLC1-NPC-TW03 cells while transfected with pGL3-SV40-TP53 (Figure 5B, lanes 2 and 4), indicating that the exogenous TP53 had a less effect on MDM2 expression in shNOLC1 transfected cells. Thus, TP53 upregulation of the MDM2 promoter activity in NPC-TW03 cells may require the presence of NOLC1 for full up-regulation (Figure 5B, lanes 2 and 4).
To test whether NOLC1 affected TP53’s regulation of MDM2 expression, we also performed shNOLC1 transfection experiments in a TW06 cell line, which did not have the TP53 transactivation activity.35 We found that the cells with neither TP53 nor NOLC1 proteins could not survive (data not shown). However, in the NPC-TW06 cell line, which has normal NOLC1 but abnormal TP53 (Figure 5B, lane 5), we could still detect MDM2 promoter activity, 0.75 times that found in NPC-TW03 cells (Figure 5B, lane 1). However, when the NPC-TW06 cells were cotransfected with pGL3-SV40-TP53 and pGL2-MDM2-Luc, luciferase activity was up-regulated 4.71-fold times (Figure 5B, lanes 5 and 6). These results indicate the TP53 protein and NOLC1 protein work together synergistically upregulating MDM2 promoter activity. However, in the cells little or no NOLC1 protein, the TP53 protein only mildly up-regulated MDM2 promoter activity (Figure 5B, lanes 3 and 4). Taken together, these data indicate that increased levels of both wtTP53 and NOLC1 are necessary for the marked upregulation of MDM2 promoter activity in NPC-TW03 and NPC-TW06 cells.
NOLC1 Interacts with the MDM2 Promoter at the TP53 Binding Site
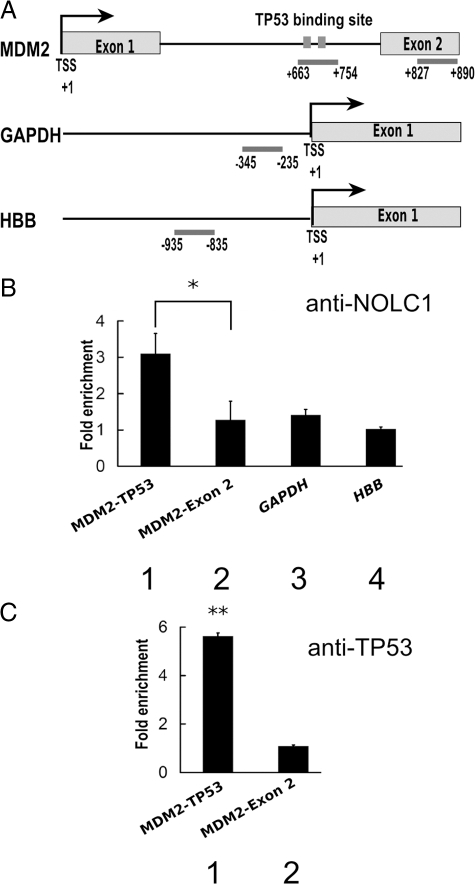
Results of the luciferase activity assay revealed that inhibition of NOLC1 markedly down-regulated the expression of MDM2 (Figure 5B, lanes 3 and 4). Therefore, we further analyzed the co-existence of the NOLC1 and the TP53 binding region of the MDM2 promoter46 using a ChIP assay in NPC cells with the NOLC1 antibody (Figure 6A and B, lane 1). The results showed that NOCL1 occupancy was specifically increased by 3 times at the TP53 binding region of the MDM2 intron 1 promoter than the control downstream Exon 2 of the MDM2 DNA sequence, which was measured by quantitative PCR (Figure 6B, lane 2). We also examined NOCL1 occupancies at the GAPDH and β-hemoglobulin (HBB) promoter as controls. Under normal physiological conditions, GAPDH was highly expressed; in contrast, HBB is strictly repressed in NPC cells (data not shown). We found no difference in NOCL1 occupancies between the two control regions and the no-antibody control (Figure 6B, lane 3 and 4). These data suggest that NOLC1 specifically targets the TP53 binding region of the MDM2 promoter in vivo. To further address whether TP53 and NOCL1 cooperatively regulate MDM2 transcription, we measured TP53 occupancies at the MDM2 promoter intron 1.46 TP53 was greatly enriched at MDM2 intron 1 but not at exon 2 (Figure 6C, lanes 1 and 2), which was also found for NOCL1 binding at these two positions (Figure 6B, lanes 1 and 2). These data suggest that TP53, a DNA binding transcription factor, may recruit or cooperate with NOCL1 to bind MDM2 promoter intron 1 and regulate MDM2 transcription synergistically.
Figure 6.
Identification of the interaction region of NOLC1 in the MDM2 P2 promoter. A: Regions amplified by QRT-PCR are indicated by bars and nucleotide number relative to the transcription start site (TSS) of the internal promoter (the TP53 binding site in intron 1) of MDM2, and Exon 2 of the MDM2, GAPDH, and HBB promoter correspondingly. B: ChIP assay of the promoters by the NOLC1 antibody. QRT-PCR was used to evaluate the precipitation of the TP53 binding site and Exon 2 of the MDM2 promoter, GAPDH and the HBB promoter region from chromatin prepared from NPC-TW04 cell line chromatin using an anti-NOLC1 monoclonal antibody. Relative enrichment was calculated by comparison with the threshold cycle value for 10% of input genomic DNA and data are expressed as the fold enrichment of DNA associated with immunoprecipitated NOLC1 relative to the no-antibody control chromatin. The error bars plotted represent the mean values ± SE of triplicate measurements. C: ChIP assay by the anti-TP53 antibody as described in B. HBB: hemoglobin β. *P < 0.05, **P < 0.01.
Discussion
Using SSH, we have identified two gene groups differentially expressed in NPC cells with high or low levels of expression. Of these genes, NOLC1 was one of the most highly expressed genes in NPC cells, though it is comparatively very weakly expressed in normal cells (Figure 1A). NOLC1 protein was abundant in both NPC cell lines and tumor cells from NPC biopsy specimens (Figures 1, B and C, and 2). To determine the functional role of NOLC1 in NPC pathogenesis, we used interference RNA to knock down NOLC1 gene expression and establish stable shNOLC1 expression in NPC cell lines. To avoid the possibility of an inconsistent result induced by a single transfection clone, we used mixed shNOLC1 transfectant clones to test the cell growth rate in vivo. The shNOLC1 suppressed the growth of shNOLC1-NPC xenografts (Figure 3C) by inducing severe necrosis and apoptosis of the tumor cells (Figure 3D). This finding supports the hypothesis that NOLC1 plays a role as an oncogene in NPC tumorigenesis. Although an increasing number of reports have shown that NOLC1 is a multiple functional protein,47 no report has indicated that it has a function associated with tumorigenesis. Results from the present experiment demonstrate that NOLC1 plays a role in enhancing NPC tumorigenesis.
We found that when NOLC1 expression was down-regulated in the shNOLC1-NPC-TW03 stable cell lines (Figure 4), the expression patterns of MDM2 (Figure 4) and the tumor-invasion-related MMP9 gene (see supplemental Figure S2 at http://ajp.amjpathol.org) were also suppressed while the expression of the apoptosis-related genes, such as TNF-α, BAX, and CASP1, were up-regulated (see supplemental Figure S3 at http://ajp.amjpathol.org). These findings suggest that NOLC1 may enhance NPC tumorigenesis by regulating of gene expression in NPC cells. The regulation of the TNF-α gene is complex at both the transcriptional and translational levels. Although TNF-α can activate caspase enzymes through its receptor, it can also mediate the activation of the transcription factor activity of NF-κB to protect the cells for survival.48,49 However, there was no change in the expression of the general apoptosis inhibitors, such as BCL2 mRNA (see supplemental Figure S4 at http://ajp.amjpathol.org), as well as other invasion related genes, such as MMP2, TIMP1, TIMP2, TIMP4, or NME1genes (see supplemental Figure S4 at http://ajp.amjpathol.org).
Several tumor growth related genes such as MDM2, VEGF, and BAX have been reported to be regulated by TP53, which itself does not undergo any changes at the mRNA level.50 We found TP53 expression to be mildly suppressed by shNOLC1 treatment (Figures 4 and see supplemental Figure S2 at http://ajp.amjpathol.org). Furthermore, our study of MDM2 promoter activation suggested that the up-regulation of MDM2 expression by exogenous TP53 in NPC cells relied on the presence of NOLC1, with the two proteins acting synergistically (Figure 5B). NOLC1 protein cannot directly bind to DNA but it can promote gene expression in a manner similar to a transcription factor.25 We used ChIP analysis to study the relationship between NOLC1 protein and the MDM2 promoter region, and found that NOLC1 may bind to certain factors that are able to cooperatively react with TP53 and bind to the MDM2 intron promoter region and regulate the expression of MDM2 (Figure 6).
EBV infection can enhance MDM2 expression and NPC tumor growth.29,44 Therefore, we also investigated the relationship between EBV infection and NOLC1 expression in NPC cells. We found mild upregulation of NOLC1 in four EBV-infected NPC cell lines (see supplemental Figure S5 at http://ajp.amjpathol.org), suggesting that NOLC1 expression may be regulated in the response of the EBV infected NPC cells. However, we did not observe any alteration in NOLC1 expression in the NPC-TW06 cell line (see supplemental Figure S5; lane 5 at http://ajp.amjpathol.org). It may result in the formation of a complex of wtTP53 and mutant TP53 (mtTP53) protein, which stays in the cytoplasm and loses TP53 transactivation activity.35 Without TP53 protein in the NPC-TW06 nucleus, the NOLC1 gene cannot be up-regulated by EBV infection, which suggests that TP53 plays some role in the regulation of NOLC1 gene expression after EBV infection in NPC cells. Previous studies have reported the development of NPC to be unlike other cancers. TP53 is moderately up expressed and less mutated in different NPC specimen21,23,44,51 and wtTP53 can act with BCL-2 synergistically to increase tumor cell growth.22,54 However, the details of the mechanism between wtTP53 and EBV infection in NPC remain unclear. As mentioned above, EBV infection can induce TP53 expression and also up-regulate MDM2.44 In this report, we demonstrated that the NOLC1 protein can bind to the MDM2 promoter of the TP53 binding region (Figure 6) and work synergistically with TP53 to regulate MDM2 expression (Figure 5). Therefore, the NOLC1 gene may play a role in regulation of NPC progression.
In our animal experiment, the xenograft from shNOLC1- NPC cells displayed marked shrinkage of tumor mass (about one sixth of the control xenografts, Figure 3C) with marked tumor necrosis and apoptosis (Figure 3D). This result suggests that it might be possible that NPC can be treated by inhibiting NOLC1 gene expression in tumor cells.
ARF has been reported to be able to induce TP53 transcriptional activity by binding and inactivating MDM2,52,53,54 and in NPC there is often decreased expression of ARF protein23,54,55 We found no difference in ARF expression between the shNOLC1 transfected and non-transfected NPC-TW03 xenografts (data not shown). Therefore, we suggest that NOLC1 is a nucleolar protein that does not affect ARF-mediated MDM2 regulation and has no effect on TP53 activity. Therefore, the regulation of MDM2 expression by NOLC1 and TP53 is not affected by ARF expression.
In summary, although the NOLC1 protein has been studied for 15 years, it has not been previously reported to play a role in regulation of cancer progression , as our work has shown. A better understanding of NOLC1 regulation and its control of gene transcription as well as its co-activation with TP53 may provide greater insight into cancer cell behavior and may lead to the development of new therapeutic strategies.
Supplementary Material
Acknowledgments
We thank Ms. Mary Wyatt and Dr. Shau-Feng Chang of Biomedical Engineering Research Laboratories, Industrial Technology Research Institute, Taiwan, for help with the manuscript writing. Dr. Konan Peck of the Institute of Biomedical Sciences, Academia Sinica, Taiwan, provided nylon microarray membranes, and Dr. Pin Ouyang of the Department of Anatomy, Chang-Gung University, Taiwan, provided the NPC cDNA library.
Footnotes
Address reprint requests to Dr. H.C. Wu, Institute of Cellular and Organismic Biology, Academia Sinica, #128 Academia Rd., Section 2, Nankang, Taipei, 11529, Taiwan. E-mail: hcw0928@gate.sinica.edu.tw, or Dr. C.T. Lin, Department of Pathology, National Taiwan University Hospital, #7 Chung-Shan S. Rd., Taipei 10002, Taiwan. E-mail: ctl@ntu.edu.tw.
Supported in part by research grants from National Science Council (NSC-96-2323-B002-015), National Health Research Institute (NHRI-96-9416BI) and a clinical research grant from National Taiwan University Hospital (NTHU-96A20), Taipei, Taiwan to C.-T.L., and NSC-96-2323-B-001-002 to H.C.W.
Supplemental material for this article can be found on http://ajp.amjpathol.org.
References
- WHO International Agency for Research on Cancer Lyon, France: IARC,; Epstein-Barr virus and Kaposi’s sarcoma herpes virus/human herpes virus 8. IARC Monographs on the Evaluation of Carcinogenic Risk to Humans. 1997:pp 1–2. [Google Scholar]
- Lo KW, To KF, Huang DP. Focus on nasopharyngeal carcinoma. Cancer Cell. 2004;5:423–428. doi: 10.1016/s1535-6108(04)00119-9. [DOI] [PubMed] [Google Scholar]
- Yu MC, Huang TB, Henderson BE. Diet and nasopharyngeal carcinoma: a case-control study in Guangzhou China. Int J Cancer. 1989;43:1077–1082. doi: 10.1002/ijc.2910430621. [DOI] [PubMed] [Google Scholar]
- Geser A, Charnay N, Day NE, de-The G, Ho HC. Environmental factors in the etiology of nasopharyngeal carcinoma: report on a case-control study in Hong Kong. IARC Sci Publ. 1978:213–229. [PubMed] [Google Scholar]
- Armstrong RW, Armstrong MJ, Yu MC, Henderson BE. Salted fish and inhalants as risk factors for nasopharyngeal carcinoma in Malaysian Chinese. Cancer Res. 1983;43:2967–2970. [PubMed] [Google Scholar]
- Ho CK, Lo WC, Huang PH, Wu MT, Christiani DC, Lin CT. Suspected nasopharyngeal carcinoma in three workers with long-term exposure to sulphuric acid vapour. Occup Environ Med. 1999;56:426–428. doi: 10.1136/oem.56.6.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YH, Du CL, Lin CT, Chan CC, Chen CJ, Wang JD. Increased morbidity from nasopharyngeal carcinoma and chronic pharyngitis or sinusitis among workers at a newspaper printing company. Occup Environ Med. 2002;59:18–22. doi: 10.1136/oem.59.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia WH, Collins A, Zeng YX, Feng BJ, Yu XJ, Huang LX, Feng QS, Huang P, Yao MH, Shugart YY. Complex segregation analysis of nasopharyngeal carcinoma in Guangdong China: evidence for a multifactorial mode of inheritance (complex segregation analysis of NPC in China). Eur J Hum Genet. 2005;13:248–252. doi: 10.1038/sj.ejhg.5201305. [DOI] [PubMed] [Google Scholar]
- Young LS, Rickinson AB. Epstein-Barr virus: 40 years on. Nat Rev Cancer. 2004;4:757–768. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- Raab-Traub N, Flynn K. The structure of the termini of the Epstein-Barr virus as a marker of clonal cellular proliferation. Cell. 1986;47:883–889. doi: 10.1016/0092-8674(86)90803-2. [DOI] [PubMed] [Google Scholar]
- Rickinson A, Kieff E. Epstein-Barr virus. Field BN, Knipe DM, Howley PM, editors. Philadelphia: Lippincott-Raven,; Fields Virology. 1996:pp 2397–2446. [Google Scholar]
- Niedobitek G, Hansmann ML, Herbst H, Young LS, Dienemann D, Hartmann CA, Finn T, Pitteroff S, Welt A, Anagnostopoulos I, Friedrich R, Lobeck H, Sam CK, Araujo I, Rickinson AB, Stein H. Epstein-Barr virus and carcinomas: undifferentiated carcinomas but not squamous cell carcinomas of the nasopharynx are regularly associated with the virus. J Pathol. 1991;165:17–24. doi: 10.1002/path.1711650105. [DOI] [PubMed] [Google Scholar]
- Lin CT, Lin CR, Tan GK, Chen W, Dee AN, Chan WY. The mechanism of Epstein-Barr virus infection in nasopharyngeal carcinoma cells. Am J Pathol. 1997;150:1745–1756. [PMC free article] [PubMed] [Google Scholar]
- zur Hausen H, Schulte-Holthausen H, Klein G, Henle W, Henle G, Clifford P, Santesson L. EBV DNA in biopsies of Burkitt tumours and anaplastic carcinomas of the nasopharynx. Nature. 1970;228:1056–1058. doi: 10.1038/2281056a0. [DOI] [PubMed] [Google Scholar]
- Helen G, Helen W. Seroepidemiology of the virus. Epstein MA, Achong BG, editors. Berlin: Springer-Verlag,; The Epstein-Barr Virus. 1979:pp 279–320. [Google Scholar]
- Klein G. The relationship of the virus to nasopharyngeal carcinoma. Epstein MA, Achong BG, editors. Berlin: Springer-Verlag,; The Epstein-Barr Virus. 1979:pp 239–350. [Google Scholar]
- Lee YC, Hwang YC, Chen KC, Lin YS, Huang DY, Huang TW, Kao CY, Wu HC, Lin CT, Huang CY. Effect of Epstein-Barr virus infection on global gene expression in nasopharyngeal carcinoma. Funct Integr Genomics. 2007;7:79–93. doi: 10.1007/s10142-006-0035-2. [DOI] [PubMed] [Google Scholar]
- Lin CT. Epstein-Barr virus: new research in epithelial carcinoma. Umar CS, editor. New York: Nova Science Publishers,; New Development in Epstein-Barr Virus Research. 2006:pp 79–93. [Google Scholar]
- Diatchenko L, Lau YF, Campbell AP, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov ED, Siebert PD. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Natl Acad Sci USA. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgos JS. Involvement of the Epstein-Barr virus in the nasopharyngeal carcinoma pathogenesis. Med Oncol. 2005;22:113–121. doi: 10.1385/MO:22:2:113. [DOI] [PubMed] [Google Scholar]
- Chen MK, Lee HS, Chang JH, Chang CC. Expression of p53 protein and primary tumour volume in patients with nasopharyngeal carcinoma. J Otolaryngol. 2004;33:304–307. doi: 10.2310/7070.2004.00304. [DOI] [PubMed] [Google Scholar]
- Niemhom S, Kitazawa S, Murao S, Kunachak S, Maeda S. Co-expression of p53 and bcl-2 may correlate to the presence of epstein-barr virus genome and the expression of proliferating cell nuclear antigen in nasopharyngeal carcinoma. Cancer Lett. 2000;160:199–208. doi: 10.1016/s0304-3835(00)00582-6. [DOI] [PubMed] [Google Scholar]
- Fan SQ, Ma J, Zhou J, Xiong W, Xiao BY, Zhang WL, Tan C, Li XL, Shen SR, Zhou M, Zhang QH, Ou YJ, Zhuo HD, Fan S, Zhou YH, Li GY. Differential expression of Epstein-Barr virus-encoded RNA and several tumor-related genes in various types of nasopharyngeal epithelial lesions and nasopharyngeal carcinoma using tissue microarray analysis. Hum Pathol. 2006;37:593–605. doi: 10.1016/j.humpath.2006.01.010. [DOI] [PubMed] [Google Scholar]
- Meier UT, Blobel G. Nopp140 shuttles on tracks between nucleolus and cytoplasm. Cell. 1992;70:127–138. doi: 10.1016/0092-8674(92)90539-o. [DOI] [PubMed] [Google Scholar]
- Miau LH, Chang CJ, Tsai WH, Lee SC. Identification and characterization of a nucleolar phosphoprotein. Nopp140, as a transcription factor. Mol Cell Biol. 1997;17:230–239. doi: 10.1128/mcb.17.1.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliner JD, Kinzler KW, Meltzer PS, George DL, Vogelstein B. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature. 1992;358:80–83. doi: 10.1038/358080a0. [DOI] [PubMed] [Google Scholar]
- Barak Y, Gottlieb E, Juven-Gershon T, Oren M. Regulation of mdm2 expression by p53: alternative promoters produce transcripts with nonidentical translation potential. Genes Dev. 1994;8:1739–1749. doi: 10.1101/gad.8.15.1739. [DOI] [PubMed] [Google Scholar]
- Zauberman A, Flusberg D, Haupt Y, Barak Y, Oren M. A functional p53-responsive intronic promoter is contained within the human mdm2 gene. Nucleic Acids Res. 1995;23:2584–2592. doi: 10.1093/nar/23.14.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HC, Lin YJ, Lee JJ, Liu YJ, Liang ST, Peng Y, Chiu YW, Wu CW, Lin CT. Functional analysis of EBV in nasopharyngeal carcinoma cells. Lab Invest. 2003;83:797–812. doi: 10.1097/01.lab.0000074896.03561.fb. [DOI] [PubMed] [Google Scholar]
- Lin CT, Wong CI, Chan WY, Tzung KW, Ho JK, Hsu MM, Chuang SM. Establishment and characterization of two nasopharyngeal carcinoma cell lines. Lab Invest. 1990;62:713–724. [PubMed] [Google Scholar]
- Lin CT, Chan WY, Chen W, Huang HM, Wu HC, Hsu MM, Chuang SM, Wang CC. Characterization of seven newly established nasopharyngeal carcinoma cell lines. Lab Invest. 1993;68:716–727. [PubMed] [Google Scholar]
- Liao SK, Perng YP, Shen YC, Chung PJ, Chang YS, Wang CH. Chromosomal abnormalities of a new nasopharyngeal carcinoma cell line (NPC-BM1) derived from a bone marrow metastatic lesion. Cancer Genet Cytogenet. 1998;103:52–58. doi: 10.1016/s0165-4608(97)00416-0. [DOI] [PubMed] [Google Scholar]
- Chinese Academy of Medical Sciences, Chungshan Medical College Establishment of an epithelial cell line and fusiform cell line from a patient with nasopharyngeal carcinoma (in Chinese). Sci Sin. 1978;21:113–118. [PubMed] [Google Scholar]
- Gu SY, Tang WP, Zeng Y, Zhao ML, Zhao EWP, Deng WH, Li K. An epithelial cell line established from poorly differentiated nasopharyngeal carcinoma (in Chinese). Chin J Cancer. 1983;2:70–72. [Google Scholar]
- Hwang JK, Lin CT. Co-localization of endogenous and exogenous p53 proteins in nasopharyngeal carcinoma cells. J Histochem Cytochem. 1997;45:991–1003. doi: 10.1177/002215549704500709. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory,; 1989:pp 11.38–11.98. [Google Scholar]
- Don RH, Cox PT, Wainwright BJ, Baker K, Mattick JS. ‘Touchdown’ PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai CY, Chen HK, Sheu HL, Yeh NH. Cell-cycle-dependent alterations of a highly phosphorylated nucleolar protein p130 are associated with nucleologenesis. J Cell Sci. 1995;108:1911–1920. doi: 10.1242/jcs.108.5.1911. [DOI] [PubMed] [Google Scholar]
- Wu HC, Lin CT. Association of heterotrimeric GTP binding regulatory protein (Go) with mitosis. Lab Invest. 1994;71:175–181. [PubMed] [Google Scholar]
- Sixbey JW, Yao QY. Immunoglobulin A-induced shift of Epstein-Barr virus tissue tropism. Science. 1992;255:1578–1580. doi: 10.1126/science.1312750. [DOI] [PubMed] [Google Scholar]
- Singhal S, Vachani A, ntin-Ozerkis D, Kaiser LR, Albelda SM. Prognostic implications of cell cycle, apoptosis, and angiogenesis biomarkers in non-small cell lung cancer: a review. Clin Cancer Res. 2005;11:3974–3986. doi: 10.1158/1078-0432.CCR-04-2661. [DOI] [PubMed] [Google Scholar]
- Huang DY, Lin YT, Jan PS, Hwang YC, Liang ST, Peng Y, Huang CY, Wu HC, Lin CT. Transcription factor SOX-5 enhances nasopharyngeal carcinoma progression by down-regulating SPARC gene expression. J Pathol. 2008;214:445–455. doi: 10.1002/path.2299. [DOI] [PubMed] [Google Scholar]
- Barak Y, Juven T, Haffner R, Oren M. mdm2 expression is induced by wild type p53 activity. EMBO J. 1993;12:461–468. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HC, Lu TY, Lee JJ, Hwang JK, Lin YJ, Wang CK, Lin CT. MDM2 expression in EBV-infected nasopharyngeal carcinoma cells. Lab Invest. 2004;84:1547–1556. doi: 10.1038/labinvest.3700183. [DOI] [PubMed] [Google Scholar]
- Chou J, Lin YC, Kim J, You L, Xu Z, He B, Jablons DM. Nasopharyngeal carcinoma–review of the molecular mechanisms of tumorigenesis. Head Neck. 2008;30:946–963. doi: 10.1002/hed.20833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, Shahab A, Yong HC, Fu Y, Weng Z, Liu J, Zhao XD, Chew JL, Lee YL, Kuznetsov VA, Sung WK, Miller LD, Lim B, Liu ET, Yu Q, Ng HH, Ruan Y. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- Lo SJ, Lee CC, Lai HJ. The nucleolus: reviewing oldies to have new understandings. Cell Res. 2006;16:530–538. doi: 10.1038/sj.cr.7310070. [DOI] [PubMed] [Google Scholar]
- Bouwmeester T, Bauch A, Ruffner H, Angrand PO, Bergamini G, Croughton K, Cruciat C, Eberhard D, Gagneur J, Ghidelli S, Hopf C, Huhse B, Mangano R, Michon AM, Schirle M, Schlegl J, Schwab M, Stein MA, Bauer A, Casari G, Drewes G, Gavin AC, Jackson DB, Joberty G, Neubauer G, Rick J, Kuster B, Superti-Furga G. A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat Cell Biol. 2004;6:97–105. doi: 10.1038/ncb1086. [DOI] [PubMed] [Google Scholar]
- Kelliher MA, Grimm S, Ishida Y, Kuo F, Stanger BZ, Leder P. The death domain kinase RIP mediates the TNF-induced NF-kappaB signal. Immunity. 1998;8:297–303. doi: 10.1016/s1074-7613(00)80535-x. [DOI] [PubMed] [Google Scholar]
- Lacroix M, Toillon RA, Leclercq G. p53 and breast cancer, an update. Endocr Relat Cancer. 2006;13:293–325. doi: 10.1677/erc.1.01172. [DOI] [PubMed] [Google Scholar]
- Sheu LF, Chen A, Lee HS, Hsu HY, Yu DS. Cooperative interactions among p53, bcl-2 and Epstein-Barr virus latent membrane protein 1 in nasopharyngeal carcinoma cells. Pathol Int. 2004;54:475–485. doi: 10.1111/j.1440-1827.2004.01654.x. [DOI] [PubMed] [Google Scholar]
- Lowe SW, Sherr CJ. Tumor suppression by Ink4a-Arf: progress and puzzles. Curr Opin Genet Dev. 2003;13:77–83. doi: 10.1016/s0959-437x(02)00013-8. [DOI] [PubMed] [Google Scholar]
- Tao W, Levine AJ. P19(ARF) stabilizes p53 by blocking nucleo-cytoplasmic shuttling of Mdm2. Proc Natl Acad Sci USA. 1999;96:6937–6941. doi: 10.1073/pnas.96.12.6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber JD, Taylor LJ, Roussel MF, Sherr CJ, Bar-Sagi D. Nucleolar Arf sequesters Mdm2 and activates p53. Nat Cell Biol. 1999;1:20–26. doi: 10.1038/8991. [DOI] [PubMed] [Google Scholar]
- Makitie AA, MacMillan C, Ho J, Shi W, Lee A, O'Sullivan B, Payne D, Pintilie M, Cummings B, Waldron J, Warde P, Irish J, Brown D, Gilbert R, Gullane P, Liu FF, Kamel-Reid S. Loss of p16 expression has prognostic significance in human nasopharyngeal carcinoma. Clin Cancer Res. 2003;9:2177–2184. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.