Abstract
Osteosarcoma is the most common primary bone cancer. Mutations of the RB gene represent the most frequent molecular defect in this malignancy. A major consequence of this alteration is that the activity of the key cell cycle regulator E2F1 is unleashed from the inhibitory effects of pRb. Studies in animal models and in human cancers have shown that deregulated E2F1 overexpression possesses either “oncogenic” or “oncosuppressor” properties, depending on the cellular context. To address this issue in osteosarcomas, we examined the status of E2F1 relative to cell proliferation and apoptosis in a clinical setting of human primary osteosarcomas and in E2F1-inducible osteosarcoma cell line models that are wild-type and deficient for p53. Collectively, our data demonstrated that high E2F1 levels exerted a growth-suppressing effect that relied on the integrity of the DNA damage response network. Surprisingly, induction of p73, an established E2F1 target, was also DNA damage response-dependent. Furthermore, a global proteome analysis associated with bioinformatics revealed novel E2F1-regulated genes and potential E2F1-driven signaling networks that could provide useful targets in challenging this aggressive neoplasm by innovative therapies.
Osteosarcoma is a mesenchymal malignancy in which the cells produce bone matrix. It is the most common primary bone cancer, accounting for approximately 20% of primary bone malignancies. Osteosarcomas occur mainly during adolescence, with a peak incidence during the growth spurt, at 15 to 19 years of age. Men are more commonly affected than women with a ratio of 1.6: 1. Notably, most osteosarcomas develop at sites of highest bone growth, where cell proliferation activity is at its crest. Such areas are the metaphyseal regions of the long bones of the extremities.1
At the molecular level, one of the most frequent defects found in osteosarcomas is genomic alterations of the RB (retinoblastoma) gene. Patients with hereditary retinoblastoma have up to 103 times greater risk for developing such tumors. Among sporadic osteosarcomas, alterations of RB occur in about 70% of cases.2 The main biochemical activity of the pocket protein pRb is to control the transcription factor E2F1, a central modulator of cell cycle progression, by forming a complex with it during the G1-phase of the cell cycle. On growth stimulating signals pRb becomes hyperphosphorylated, by cyclin-dependent kinases, and releases E2F1, which then activates its target genes and thereby promotes the G1 to S-phase progression.3,4 Beyond its pivotal role in G1 to S-phase transition, E2F1 also possesses the ability to induce apoptosis in a p53-dependent or -independent manner, mainly via p73.3,5,6,7,8,9,10 The ability of a cellular factor to trigger both proliferation and apoptosis seems paradoxical. However, the rationale behind the connection of opposing functions by the same factor is that the cell consumes less time and energy to control the switch from the proliferation state to the self-destruction program in case of irreversible cellular damage, especially during DNA synthesis. A predicted biochemical consequence of deficient pRb expression is that E2F1 activity is constantly unleashed from the inhibitory effects of pRb. In such a setting the delicate balance between growth promoting and growth suppressing properties of E2F1 would be disrupted and the final outcome would possibly be dictated by the specific cellular background.3,11 The bimodal impact of E2F1 has been demonstrated in various in vitro cellular systems and in vivo animal models.3 Characteristically, loss of E2F1 in RB+/− mice increases tumor incidence within certain tissues and decreases it in others.12 A similar multifarious pattern of E2F1 response appears to apply to human cancers, as well. In certain types of carcinomas, such as those derived from the lung, breast, thyroid, and pancreas, E2F1 seems to act as an oncogene13,14,15,16 whereas, in others, such as carcinomas of the colon and prostate, an onco-suppressor role has been proposed.14 The embryological origin of the organ, as exemplified by esophageal cancer17,18 may also modulate the phenotype of E2F1-driven tumors.
In contrast to epithelial tumors, there are no data about the expression status and role of endogenous E2F1 in tumors of mesenchymal origin. This information is important in view of E2F1’s role in sensitizing cells to various chemotherapeutic agents.19 To address these questions we examined the status of E2F1 in relation to tumor kinetics [proliferation index: % of tumor cells proliferating, and apoptotic index: % of apoptotic tumor cells in a tumor, respectively], and nodal cell cycle regulators in a series of primary osteosarcoma specimens. The functional basis of the findings obtained with the clinical samples was then investigated in human osteosarcoma cell line models with inducible E2F1, harboring either wild-type or mutant p53. Finally, a global proteome and bioinformatic analysis was performed in an attempt to obtain a broader picture of the potential pathways that could follow E2F1 induction; thus helping us understand better the multifaceted nature of the E2F1-mediated response and design more rational E2F1-targeted therapeutic strategies in the future.
Materials and Methods
Tumor Specimens
Formalin-fixed, paraffin-embedded sections from 57 surgically removed osteosarcomas were analyzed, after local ethical committee approval. The samples were taken from patients that had not undergone any chemo- or radiotherapy before surgical resection. The majority of patients were diagnosed and treated in the General Hospital of Asklepeion Voula, Athens, Greece. Clinicopathological features of the patients are presented in Table 1.
Table 1.
Clinicopathological Data
| Age | Range 7–82 years | Mean 22.8 years | ||
| Sex | Male | Female | ||
| 32 (56.1%) | 25 (43.9%) | |||
| Histologic variant | Osteoblastic | Chondroblastic | Combined | |
| 42 (73.7%) | 6 (10.5%) | 9 (15.8%) | ||
| Grade | Low | Moderate | High | |
| 18 (31.6%) | 20 (35.1%) | 19 (33.3%) | ||
| Survival* | Yes | No | ||
| 13 (22.8%) | 44 (77.2%) | |||
| First symptom | Pain | Swelling | Pain and swelling | Pain, swelling, and limp |
| 22 (38.6%) | 4 (7%) | 21 (36.8%) | 10 (17.6%) | |
| Tumor location | Lower third femur | Upper third tibia | Rest femur | Other sites** |
| 22 (38.6%) | 15 (26.3%) | 11 (19.3%) | 9 (15.8%) | |
| Duration*** | ≤3 Months | 3–6 Months | >6 Months | |
| 29 (50.8%) | 22 (38.6%) | 6 (10.6%) | ||
| Necrosis | Range (%) | Mean (%) | SD (%) | |
| 20–96 | 71.2 | 25.4 | ||
| Extent | 90% | 80% | 70%–50% | ≤50% |
| 20 (35.1%) | 8 (14%) | 17 (29.9%) | 12 (21%) |
SD, standard deviation.
5 year follow-up period.
Distribution: 5 Humerus, 3 Thumb, 1 Fibula.
Time intervening between the emergence of the first symptom and the final diagnosis.
Immunohistochemistry
Antibodies
The following antibodies were used at 1:100 dilution: anti-E2F1 (KH95, Santa Cruz, Bioanalytica, Athens, Greece), anti-Ki-67 (MIB-1, DAKO, Kalifronas, Athens, Greece), anti-p53 (DO7, DAKO, Kalifronas, Athens, Greece), anti-pRB (IF8, Santa Cruz, Bioanalytica, Athens, Greece), anti-phospho-pRb (Ser795-R, Santa Cruz, Bioanalytica, Athens, Greece), anti-phospho-H2AX (Ser139, Millipore, Lab Supplies, Athens, Greece), and anti-phospho-Chk2 (Thr68, Cell Signaling, Bioline, Athens, Greece).
Method
Immunohistochemistry was performed according to the indirect streptavidin–biotin–peroxidase method, as previously described with a modification in the heat-mediated antigen retrieval method.13,14 Specifically, unmasking of the related proteins was carried out in a 10 mmol/L citrate solution with pH = 6.0 for 50 minutes at 95οC in a steamer.
Evaluation
Evaluation of the E2F1, Ki-67 (proliferation index), p53, γH2AX, and Chk2-pT68 expression was performed as previously reported,13,14,20 and included counting of an average of 500 cells at ×400 per case, for each antibody, respectively. Only nuclear staining was considered as positive. Three independent observers performed slide examination. Interobserver variability was minimal.
Controls
MCF-7 breast cancer cells were used as positive control for E2F1 expression. Sections from previously characterized tumors13,14,20 for Ki-67, p53, γH2AX, and Chk2-pT68 staining, were used as positive controls for the corresponding antibodies. Antibodies of the corresponding Ig class, but of unrelated specificity were used as negative controls.
Tdt-Mediated dUTP Nick End Labeling Assay
Method
Double-stranded DNA breaks were detected by Tdt-mediated dUTP nick end labeling assay, according to the protocol described elsewhere.14
Evaluation
Cells were considered to undergo apoptosis when nuclear staining from the fragmented DNA, without cytoplasmic background, was observed. The apoptotic index was estimated as the percentage of cancer cells with nuclear staining. Slide examination was performed by three independent observers, with minimal interobserver variability.
Controls
Sections incubated with DNase I, before treatment with TdT, were used as positive controls, while sections without TdT addition were used as negative controls.
Growth Index Estimation
The growth index of the samples was estimated by the proliferation to apoptosis index ratio, as previously described.13,14
Nucleic Acid Analysis
DNA Extraction
DNA extraction was performed according to previous protocol.13
TP53 and p73 Sequence Analysis
Genomic analysis of exons 4 to 10 of p53 and exons 2 to 14 of p73 for mutation presence, respectively, was accomplished as formerly mentioned13,14,21 on an ABI-PRISM 377 automatic sequencer (Applied Biosystems, Biosolutions, Athens, Greece).
D17S179E Allelic Imbalance Analysis
To assess allelic imbalance presence at the TP53 locus, the D17S179E polymorphic pentanucleotide marker, located within the first TP53 intron, was analyzed as previously described,14 on an ABI-PRISM 377 automatic sequencer (Applied Biosystems, Biosolutions, Athens, Greece).
MDM2 Gene Amplification
Gene amplification was assessed with differential-PCR as described elsewhere.22 Primers and annealing temperature used for MDM2 gene amplification were: 5′-TACCATGATCTACAGGAACTT-3′ and 5′-CTCAGTATGTGGTTTTAGTTC-3′ at 60°C.
Cell Lines
The U2OS E2F1-ER and Saos2 E2F1-ER cells20,23,24 were grown in Dulbeco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and incubated at 37°C and 5% CO2.
Plasmids and siRNA Transfections
The phoenix amphotropic helper-free retrovirus producer line was used to construct retroviruses containing the pRETROSUPER-short-hairpin(sh)p53, pRETROSUPER-shp14alternate reading frame(ARF) and the corresponding pRETROSUPERshLaz control.22 Infection of the U2OS E2F1-ER or Saos2 E2F1-ER cells was performed at 12 hours intervals in 60-mm dishes, as previously described.22 For ATM or p73 silencing, 3 × 105 cells were plated in 60-mm dishes and next day transfected using Lipofectamine 2000 (Invitrogen, Anti-Sel, Athens, Greece) with the appropriate Stealth Select RNAi pool or the corresponding RNAi negative control (Invitrogen, Anti-Sel, Athens, Greece), respectively, according to manufacturer’s instructions. To study the effects of p53, ATM, p73, and p14ARF on E2F1 behavior we activated the U2OS E2F1-ER or Saos2 E2F1-ER cellular systems 24 hours after siRNA interference.25 To observe effects later than 72 hours, the cells of interest were retransfected.
Immunofluorescence
Indirect immunofluorescence analysis was performed according to a previously described protocol.22 In brief, cells grown on coverslips were fixed with methanol and subsequently incubated with the primary antibody overnight at 4°C. Subsequently, cells were incubated with the Oregon Green-conjugated secondary antibody (O-6380, Invitrogen, Anti-Sel, Athens, Greece), counterstained with 4,6-diamidino-2-phenylindole (Sigma) and mounted with Fluoromount G (#0100–01, SouthernBiotech, AlterChem, Athens, Greece).
The antibodies used were: anti-E2F1 (1:100)(KH-95, Santa Cruz, Bioanalytica, Athens, Greece), anti-Cyclin B1 (1:100)(H-20, Santa Cruz, Bioanalytica, Athens, Greece), and anti-γH2AX-pS139 (1:100)(JBW301, Millipore, Lab-Supplies, Athens, Greece).
Immunoblotting Analysis
Protein Extraction
Total protein extraction from cells was performed according to protocols described previously.20,22
SDS-Polyacrylamide Gel Electrophoresis
Thirty μg of protein from total extracts from each sample were adjusted with NuPAGE LDS Sample Buffer (Invitrogen, Anti-Sel, Athens, Greece) and loaded on 4% to 12% gradient NuPAGE precast gels (Invitrogen, Anti-Sel, Athens, Greece), according to manufacturer’s instructions. Gel electrophoresis, transfer to polyvinylidene difluoride membrane (Millipore, Lab Supplies, Athens, Greece), blotting, immunodetection, and signal development were performed as previously described.20,22
Antibodies
The antibodies used were: anti-γH2AX-pS139 (1:500) (Millipore, Lab-Supplies, Athens, Greece), anti-p53-pS15 (1:500) (Cell Signaling, Bioline, Athens, Greece), anti-p53 (1:500) (DAKO, Kalifronas, Athens, Greece), anti-pChk2 T68 (1:500) (Cell Signaling, Bioline, Athens, Greece), anti-Chk2 (hybridoma supernatant),20 anti-Cyclin B1 (1:500) (SantaCruz, Bioanalytica, Athens, Greece), anti-p73 (1:300) (NeoMarkers, Bioanalytica, Athens, Greece), anti-β-actin (1:1000) (Abcam, Anti-Sel, Athens, Greece), anti-Csn8 (1:500) (SantaCruz, Bioanalytica, Athens, Greece), anti-ATM (1:500) (Abcam, Anti-Sel, Athens, Greece), anti-p14ARF (1:500) (Neomarkers, Bioanalytica, Athens, Greece), anti-BUB3 (1:500) (Abcam, Anti-Sel, Athens, Greece), anti-COP9 subunit8 (1:500) (Abcam, Anti-Sel, Athens, Greece), anti-Cdt1 (1:500) (SantaCruz, Bioanalytica, Athens, Greece), anti AuroraA (1:500) (Abcam, Anti-Sel, Athens, Greece), and anti-HNRPU (a kind gift from Dr Guialis A.).
Growth Curves
The growth of cells was monitored as mentioned elsewhere.22 Cells were seeded into 12-well plates at a density of 25 × 103 cells/ml in a total volume of 1 ml/well. One day after seeding, 4-OH Tamoxifen (Sigma Hellas, Athens, Greece) was added to the medium of half of the wells at a final concentration of 300 nmol/L. Cells were harvested at days 0, 1, 2, 3, 4, and 5 after 4-OH Tamoxifen administration. For each time point the number of cells from four wells was counted: two wells with 4-OH Tamoxifen treatment and the corresponding untreated ones. Cells were trypsinized, centrifuged, and resuspended with 1 ml of medium and counted with a Newbauer Hematocytometer. The whole procedure was repeated in triplicate and counting was performed by two independent investigators.
Fluorescence-Activated Cell Sorting Analysis
Approximately 5 × 105 cells were seeded per 100-mm dish and cultured according to the conditions mentioned above. Next day after seeding, 4-OH Tamoxifen (Sigma Hellas, Athens, Greece) was added to the medium at a final concentration of 300 nmol/L. Before and 1, 2, 3, 4, and 5 days after the beginning of the treatment, cells were trypsinized and centrifuged and the pellet was washed with 0,5 ml of PBS and fixed with 4,5 ml of ice-cold ethanol 80%. Cells were stained with propidium iodide and their DNA content was counted with the use of BD FACS Calibur (BD Hellas, Athens, Greece), as previously described.22 In addition, the percentage of apoptotic cells emerged by cell cycle analysis was confirmed with the AnnexinV: FITC apoptosis detection kit (BD Hellas, Athens, Greece).
Sa-β-Gal Staining
U2OS E2F1-ER and Saos2 E2F1-ER cells grown on coverslips with the addition of 4-OH-Tamoxifen for 7 and 10 days, as well as untreated control cells, were Sa-β-Gal stained as described elsewhere.22 Cells with cytoplasmic staining were scored as positive.
Proteomic Analysis
Materials and Reagents
Acrylamide/piperazine-di-acrylamide solution (37.5:1, w/v) was purchased from Biosolve Ltd. (Valkenswaard, the Netherlands). Immobilized pH-gradient strips, buffers, and other reagents for the polyacrylamide gel preparation were obtained from BioRad (Bioscience, Athens, Greece). 3-[(3-Cholamidopropyl)dimethylammonio]propanesulfonate was obtained from Roche Diagnostics (Mannheim, Germany), urea from AppliChem (Darmstadt, Germany), thiourea from Fluka (Buchs, Switzerland), 1,4-dithioerythritol, and EDTA from Merck (Darmstadt, Germany). Except for 3-[(3-cholamidopropyl)dimethylammonio]propanesulfonate, which was kept at 23°C, the other reagents were kept at 4°C.
Two-Dimensional Gel Electrophoresis
Protein extraction and two-dimensional gel electrophoresis was performed as previously described.26
Peptide Mass Fingerprint and Post Source Decay
Peptide analysis and protein identification were performed as described.27 Spots were automatically detected by Melanie 4.02 software (GeneBio, Geneve Bioinformatics S.A., Geneva, Switzerland) on the Coomassie blue-stained gel, excised by the Proteiner SPII (Bruker Daltonics, Bremen, Germany), destained with 30% acetonitrile in 50 mmol/L ammonium bicarbonate, and dried in a speed vacuum concentrator (MaxiDry Plus; Heto, Allered, Denmark). Each dried gel piece was rehydrated with 5 μl of 1 mmol/L ammonium bicarbonate containing 50 ng trypsin (Roche Diagnostics, Manheim, Germany) and left in the dark overnight at room temperature. Twenty μl of 50% acetonitrile, containing 0.1% trifluoroacetic acid were added to each gel piece and incubated for 15 minutes with constant shaking. The resulting peptide mixture (1 μl) was simultaneously applied with 1 μl of matrix solution, consisting of 0.8% α-cyano-4-hydroxycinnamic acid (Sigma, Athens, Greece), standard peptides des-Arg-bradykinin, (904.4681 Da; Sigma), and adrenocorticotropic hormone fragment 18–39, (2465.1989 Da; Sigma-Aldrich, Antisel, Greece) in 50% acetonitrile and 0.1% trifluoroacetic acid. Samples were analyzed for peptide mass fingerprint with matrix-assisted laser desorption-mass spectrometry in a time-of-flight mass spectrometer (Ultraflex II, Bruker Daltonics, Bremen, Germany). Laser shots (n = 400) at intensity between 40% and 60% were collected and summarized using the FlexControl v2.2 software by Bruker. Peak list was created with Flexanalysis v2.2 software (Bruker). Smoothing was applied with the Savitzky-Golay algorithm (width 0.2mz, cycle number 1). Signal to noise (S/N) threshold ratio of 2.5 was allowed. SNAP (Bruker) algorithm was used for peak picking. Matching peptides and protein searches were performed automatically, as previously described.26 Each spectrum was interpreted with Mascot Software v2.0 (Matrix Sciences Ltd., London, UK). For peptide identification, the monoisotopic masses were used and a mass tolerance of 0.0025% (25 ppm) was allowed.
All extraneous peaks, such as trypsin autodigests, matrix, and keratin peaks were not considered for the protein search. Cysteine carbamidomethylation and methionine oxidation were set as fixed and variable modifications, respectively. One miscleavage was allowed. The peptide masses were compared with the theoretical peptide masses of all available proteins from Homo sapiens using the SWISS-PROT (ftp://ftp.expasy.org/databases/uniprot/knowledgebase), IPI (ftp://ftp.ebi.ac.uk/pub/databases/IPI/current/), and MSDB (ftp://ftp.ebi.ac.uk/pub/databases/MassSpecDB) databases, which are updated bimonthly. The probability score identified by the software was used as the criterion of the identification. Samples not identified by peptide mass fingerprint (P < 0.05) were automatically selected for postsource decay mass spectroscopy (MS) analysis or matrix-assisted laser desorption time-of-flight MS. The peptide masses chosen for postsource decay-MS analysis had a signal intensity of >600 counts and were excluded from the trypsin autodigest, matrix, and keratin peaks. The resulting postsource decay spectra were also interpreted by the Mascot Software V2.0 and Mascot probability-based scores of P < 0.02 were considered significant. The identified proteins were annotated on the gel image by hand.
Bioinformatic Analysis
Two-Dimensional Image Analysis
Two-dimensional image analysis was performed using the PDQuest software from Biorad (Biorad, Bioscience, Athens, Greece), on the gels to compare for quantitative expression differences of proteins between the induced and non-induced U2OS E2F1-ER cells.
In Silico Promoter Analysis
The sequences of 1871 human promoters present in the EPD (Eukaryotic Promoter Database)28 were downloaded in Fasta format. All of the sequences were 16 kb long and aligned relative to the TSS (Transcription Start Site), which is located at position 10.000 in all sequences. The presence of the E2F1 binding motif YTTSSCGS in the promoter sequences was investigated using the GeneR package of the Bioconductor29 platform installed in the R Language (http://www.R-project.org).
In Silico Identification of Candidate E2F1–p53 and –ATM Pathways
Differentially expressed proteins from the proteomic analysis were processed using the Pathway Studio Software (Ariadne Genomics, Inc., Rockville, MD), to retrieve data from PubMed and create potential biochemical-pathway interactions between these (overexpressed and suppressed) proteins and p53 and ATM proteins, respectively. This type of analysis reveals proteins that have an indirect or direct effect on p53 and ATM (indirect effect occurs through a third protein termed: intermediate protein-gene), respectively.
Statistical Analysis
Statistical analyses, comprising Spearman correlation, were performed with the SPSS 12.0 software.
Results
The Status of p53 is a Key Determinant of the Association between E2F1 Expression and Tumor Growth in Osteosarcomas
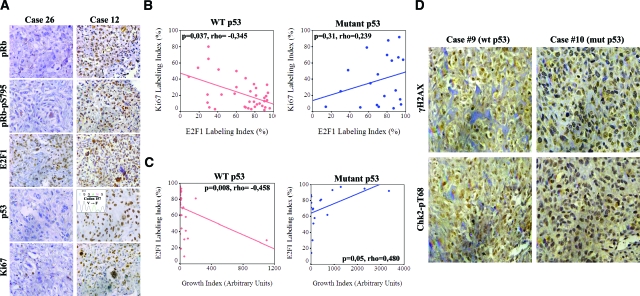
Expression of E2F1 was assessed immunohistochemically, in a series of osteosarcomas (Table 1), as the percentage of nuclear stained tumor cells. E2F1 values ranged from 9 to 97% (Table 2A, Figure 1A), with the majority of the cases (46/57) exhibiting over 50%. In the cases that did not show aberrations in pRb expression30 (Table 2A), E2F1 staining was accompanied by phosphorylation of the intact pRb protein, as assessed by the anti-phospho-Ser795-pRb antibody, indicating that E2F1 staining corresponds to the unbound and active form of the protein (Figure 1A). The initial analysis, examining overall the relationship of E2F1 with the tumor kinetic parameters of proliferation and apoptosis, as assessed by Ki-67 staining and Tdt-mediated dUTP nick end labeling assay, respectively, did not reveal an association (Table 2A). However, this general approach could mask a putative relationship of E2F1 with tumor growth in some subgroups separated according to the status of E2F1 downstream effectors. The p53 protein, a main downstream effector of E2F1, is frequently targeted in osteosarcomas, mainly by mutations and rarely by deregulation of its negative regulator, MDM231,32 (Figure 1A, and Table 2, A–C). The p73 protein, another vital downstream effector of E2F19,10 was also analyzed. However, though p53 immunohistochemical analysis significantly reflects its mutational status,33 a similar approach does not offer such information for p73. Thus, we proceeded to direct sequencing of p73 gene, but no mutations were detected (Table 2D). The above led us to assume that p53 could modulate the relationship between E2F1 overexpression and tumor kinetics. Indeed, within the group of osteosarcomas harboring wild-type p53, E2F1 expression was associated inversely with proliferation (P = 0.037, Figure 1B) and positively with apoptosis (P = 0.014, see supplemental Figure 1 at http://ajp.amjpathol.org). Furthermore, growth index—that is a cumulative indicator of the tumor’s growth potential (see Material and Methods) —was inversely correlated with E2F1 in cases with wild-type p53 and positively in those bearing mutant p53 (P = 0.008 and P = 0.05, respectively, Figure 1C). Altogether, these results suggest that deregulated E2F1 overexpression is followed mainly by a p53-dependent growth suppressing response in osteosarcomas. Of note, there were two cases that possessed p53 mutations, but scored high apoptotic indexes, implying that p53-independent apoptotic pathways also operate in this malignancy. In accordance to this notion, was that the positive correlation found between E2F1 and apoptosis in the p53 wild-type cases, was lost and not inverted in the p53 mutant cases (see supplemental Figure 1 at http://ajp.amjpathol.org).
Table 2.
Immunohistochemical Data (A), TP53 Genetic Aberrations (B), MDM2 Gene Amplifications (C), and p73 Mutations Found in the Examined Osteosarcoma Database (D).
| A)
|
|
Immunohistochemical analysis
|
|
|||
|---|---|---|---|---|---|---|
| Mean (%) | SE | n | ||||
| E2F1 | 68,43 | 3,23 | 57 | |||
| Ki67 | 27,87 | 3,3 | 57 | |||
| TUNEL | 1,68 | 0,33 | 57 | |||
| GI (PI/AI) | 339,27 | 124,72 | 57 | |||
| γH2AX | 65,72 | 4,22 | 57 | |||
| Chk2-pT68 | 59,63 | 4,53 | 57 | |||
| Negative | Positive | |||||
| p53 status | 37 (64,9%) (normal expression) | 20 (35,1%) (abnormal expression) | 57 | |||
| pRb status | 29 (52,6%) (abnormal expression) | 28 (47,4%)* (normal expression) | 57 | |||
| B)
|
|
Genetic aberrations
|
|
|||
|
|
Case
|
Exon
|
Codon
|
Base change
|
Effect
|
Allelic status*
|
| 21 | 4 | 43 | TTG/TAG | Leu → Stop | LOH | |
| 5 | 4 | 47 | CCG/CTG | Pro → Leu | No LOH | |
| 12 | 5 | 157 | GTC/TTC | Val → Phe | No LOH | |
| 31 | 5 | 175 | CGC/CAC | Arg → His | No LOH | |
| 15 | 6 | 216 | GTG/ATG | Val → Met | LOH | |
| 49 | 6 | 220 | TAT/TGT | Tyr → Cys | LOH | |
| 25 | 7 | 237 | ATG/ATA | Met → Ile | LOH | |
| 54 | 7 | 242 | TGC/TAC | Cys → Tyr | LOH | |
| 34 | 7 | 241 | 1bp deletion | Frameshift | LOH | |
| 3 | 7 | 250 | CCC/CTC | Pro → Leu | No LOH | |
| 41 | 8 | 273 | CGT/CAT | Arg → His | No LOH | |
| 11 | 8 | 281 | GAC/CAC | Asp → His | No LOH | |
| 20 | 8 | 281 | GAC/AAC | Asp → Asn | LOH | |
| 44 | 10 | 337 | CGC/TGC | Arg → Cys | LOH | |
|
|
*Analysis of the pentanucleotide marker D17S179E located in the first intron of p53
|
|
||||
| C)
|
Case
|
18
|
4.5-fold (MDM2) of amplification
|
|||
| D) | No p73 mutations were detected by sequencing analysis | |||||
SE, standard error; GI, growth index; PI, proliferation index; AI, apoptotic index.
all these cases stained positive for p-pRB.
Figure 1.
p53 status determines the association of E2F1 expression in relation to proliferation and apoptosis in osteosarcomas. A: Immunohistochemical analysis (IHC) of pRb, pRb-pS795, E2F1, p53, and Ki-67 in two representative osteosarcoma cases with different p53 status. Inset: confirmation of p53 mutation by DNA sequencing. B: Scatter plots depicting the correlation of E2F1 and Ki-67 expression according to p53 status. Note the statistically significant inverse relationship between E2F1 IHC expression and proliferation index (P = 0.037, Spearman test) in cases retaining wild-type p53. C: Scatter plots showing the relation of E2F1 expression with Growth Index in cases with intact and mutant p53 (P = 0.008 and P = 0.05 respectively, Spearman test). D: IHC analysis for γH2AX and Chk2-pT68 in representative osteosarcomas with wild-type (wt) and mutant (mut) p53, respectively.
The DNA Damage Response Pathway Is Constitutively Activated in Osteosarcomas
It has been proposed that E2F1 triggers p53-dependent apoptosis mainly via p14ARF.34,35 On the other hand, several reports suggest that ARF-independent pathways may also function in E2F1 induced p53-dependent apoptosis by using the ATM signaling pathway to activate Chk2 and p53.36,37 Moreover, according to certain studies, ARF forms a negative feedback loop with E2F1 by repressing the expression of E2F1,38 and therefore questioning ARF’s significance in E2F1-mediated apoptosis. Taking together the above and studies from our own and other laboratories, demonstrating that the DNA damage response (DDR) network may provide an intrinsic barrier against cancer progression by sensing oncogenic signals,20,23,39,40,41,42 we examined the status of central DDR markers in our clinical setting. Interestingly, intense DDR activation was evident, as demonstrated by γ-H2AX and Chk2-pT68 staining, in almost all tumor samples (Table 2A, Figure 1D). A question that arises from this finding is whether the DDR machinery participates in E2F1-related growth suppression, and whether p53 status determines such response in osteosarcomas.
The DNA Damage Checkpoint Pathway Links Induced Expression of E2F1 with Growth Suppression in p53-Wild-Type and -Deficient Osteosarcoma Cells
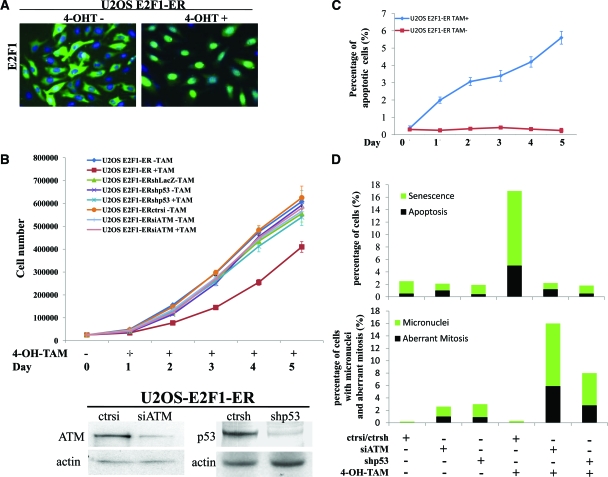
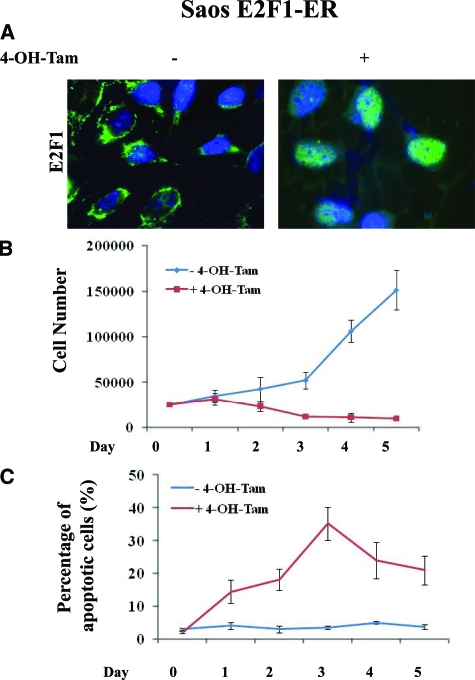
To address the significance of activated DDR in relation to E2F1 expression and examine the functional basis of the associations presented in Figure 1, we used two osteosarcoma cell lines, U2OS and Saos2, which are wild-type and deficient for p53, respectively. Both cell lines were genetically modified to express conditionally E2F1. Particularly, E2F1 was fused to the murine estrogen receptor ligand-binding domain, permitting nuclear translocation and activation of E2F1 by tamoxifen23,24 (Figure 2A). Of note, addition of tamoxifen to the control parental U2OS and Saos2 cell lines, did not evoke any of the biological effects subsequently described after E2F1-induction in the corresponding genetically modified cell lines (data not shown).
Figure 2.
E2F1 overexpression in U2OS cells raises anti-tumor barriers. A: Addition of 400 nmol/L 4-OH-Tamoxifen (4-OH-Tam) leads to nuclear translocation of E2F1. B: Growth curves of untransfected U2OS E2F1-ER cells and cells transfected with control siRNA, siATM, or shp53 before and after the addition of 4-OH-Tamoxifen. The lines are the means of three independent experiments. Immunoblots for p53 and ATM, confirming gene silencing. C: Percentage of apoptotic cells as determined by fluorescence-activated cell sorting analysis (the lines are the means of three independent experiments) in untreated and 4-OH-Tamoxifen administered U2OS E2F1-ER cells. D: Histograms depicting the activation of the anti-tumor barriers (apoptosis and senescence), the presence of micronuclei and the aberrant mitoses in the U2OS E2F1-ERshp53 and U2OS E2F1-ERsiATM cells, and their corresponding controls, before and after the addition of 4-OH-Tamoxifen.
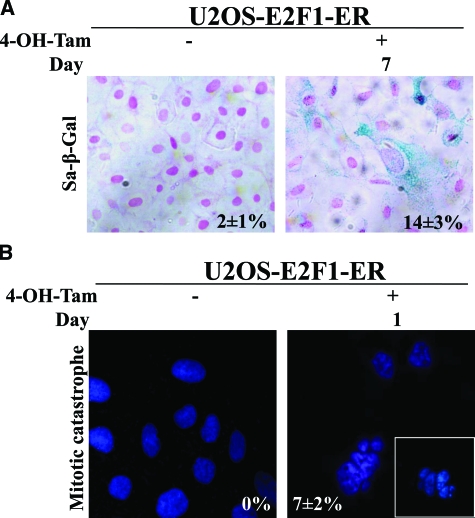
Monitoring the growth properties of the modified U2OS cells, we noticed an apparent growth retardation of the E2F1-induced cells, as compared with the non-induced ones (Figure 2B). The growth delay was accompanied, from the end of day 1 post-E2F1 induction, by a gradual increase in the percentage of apoptotic cells (Figure 2C, see supplemental Figure 2A at http://ajp.amjpathol.org), which was slightly decreased and stabilized around day 7 (data not shown). In addition, between days 5 and 10 an increase in cells acquiring a senescence phenotype, as demonstrated by SA-β-gal activity, was noticed (Figure 2D, Figure 3A). Notably, the first day after E2F1 activation, we also observed a rapid increase in cells, around 7%, dying with distinct morphological features including large size, multiple micronuclei and de-condensed chromatin (Figure 2D, 3B). According to some authors, these characteristics define cells dying from “mitotic catastrophe.”43,44 Their numbers gradually decreased during day 2 and were completely abolished after day 3.
Figure 3.
A: Sa-β-Gal staining after 7 days of 4-OH-Tamoxifen addition in U2OS E2F1-ER cells (percentages presented as an inset). B: Cells with morphological characteristics of “mitotic catastrophe” after the addition of 4-OH-Tamoxifen (percentages presented as an inset).
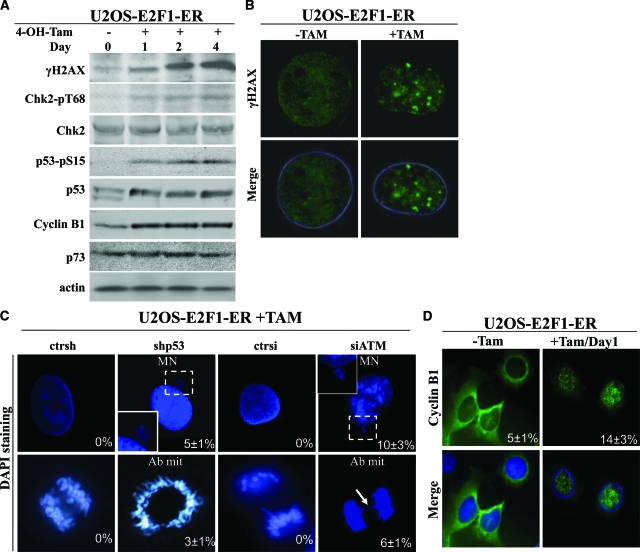
Apoptosis and senescence represent nodal aspects of the intrinsic cellular anti-tumor barrier. We hypothesized that E2F1 overexpression could induce these processes by triggering the DDR pathway20,23,39 (Figure 4A), since U2OS cells do not express the senescence-linked protein, p16INK4A or the oncogenic-sensor protein and E2F1 target, p14ARF.44,45,46,47 Furthermore, the levels of p73, which is a potent inducer of E2F1 mediated apoptosis,9,10 did not increase after E2F1 activation48 (Figure 4A), making a p73-dependent effect in this setting unlikely. Consistent with the in vivo findings (Figure 1), shRNA-mediated knock-down of p53 in the U2OS-induced E2F1 cells, resulted in restoration of the growth potential (Figure 2B) and almost complete suppression of apoptosis and senescence (Figure 2D, see supplemental Figure 2A at http://ajp.amjpathol.org). Similar results were obtained after siRNA-mediated knock-down of ATM (Figure 2, B and D, see supplemental Figure 2A at http://ajp.amjpathol.org). Moreover, down-regulation of p53 as well as ATM, by siRNA interference, resulted in the appearance of aberrant mitoses and cells characterized by micronuclei, a feature indicative of chromosomal instability22 (Figures 2D and 4, B and C). Regarding the phenomenon of “mitotic catastrophe” we considered that it was possibly mediated via the mitosis-initiating complex cyclin B1-cdc2, which was induced and translocated to nucleus in an E2F1-dependent manner (Figure 4D),48 and whose activity is known to lead to aberrant mitotic entry and subsequent death when ectopically overexpressed.49,50
Figure 4.
The effect of E2F1 overexpression in U2OS cells is mediated by DDR activation. A: Western blot analysis for γH2AX, Chk2-pT68, Chk2, p53-pS15, p53, Cyclin B1, and p73. Actin serves as loading control. B: Immunofluorescence analysis for γH2AX in U2OS E2F1-ER cells showing foci after addition of 4-OH-Tamoxifen. C: U2OS E2F1-ERshp53 and U2OS E2F1-ERsiATM cells, as well as their corresponding controls, were stained with 4,6-diamidino-2-phenylindole and analyzed for morphological indications of genomic instability (MN: micronuclei and Ab mit: aberrant mitosis). The arrow indicates a nucleoplasmic bridge. D: Immunofluorescence analysis showing Cyclin B1 nuclear translocation in the U2OS E2F1-ER cells after 4-OH-Tamoxifen induction of E2F1.
Since E2F1-triggered, p53-independent apoptotic pathways seem also to operate in osteosarcomas (see first part of the Results section) we compared the E2F1-inducible, p53-deficient Saos2 osteosarcoma cell line (Figure 5A) with the p53-proficient U2OS system, and examined whether the DDR can participate in p53-independent E2F1 mediated growth suppression. After E2F1 induction we observed a growth-retardation of the Saos2 cells compared with the non-induced control (Figure 5B). The growth delay was accompanied, from day 1 post-E2F1 induction, by a rapid increase in the percentage of cells dying from apoptosis (Figure 5C, see supplemental Figure 2B at http://ajp.amjpathol.org), and the extent of cell death was significantly higher in Saos2 compared with the E2F1-inducible U2OS model (Figure 2C, see supplemental Figure 2A at http://ajp.amjpathol.org). In contrast to E2F1-induced U2OS, however, the E2F1-induced Saos2 cells neither adopted a senescence phenotype nor presented cells with features of “mitotic catastrophe,” the phenomena seen in the U2OS cells.
Figure 5.
E2F1 overexpression in Saos2 cells raises anti-tumor barriers. A: Addition of 400 nmol/L 4-OH-Tamoxifen leads to nuclear translocation of E2F1. B: Growth curves of Saos2 E2F1-ER cells with and without the addition of 4-OH-Tamoxifen (4-OH-Tam). The lines are the means of three independent experiments. C: Percentage of apoptotic cells as determined by fluorescence-activated cell sorting analysis. The lines are the means of three independent experiments.
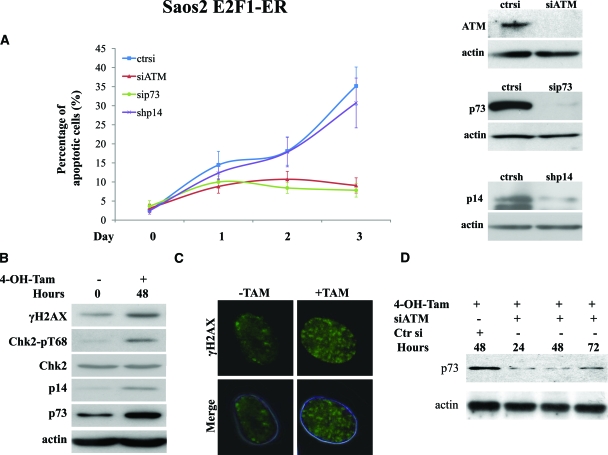
As previously demonstrated in the same cellular system, p73 is most likely responsible for the apoptotic wave produced after E2F1 expression.9,10 Indeed, apoptosis was almost abolished after p73 siRNA transfection (Figures 6A, see supplemental Figure 2B at http://ajp.amjpathol.org). In addition, E2F1 induction generated an intense DDR, as documented by the increased expression of γ-H2AX and Chk2-pT68 (Figure 6, B and C). Interestingly, knocking down ATM with siRNA reduced apoptosis and restored cell growth to levels comparable with those noticed after siRNA-mediated knock-down of p73 (Figure 6A, see supplemental Figure 2B at http://ajp.amjpathol.org). Strikingly, p73 was also down-regulated after ATM knock-down (Figure 6D), implying that the DDR pathway acts along the same axis encompassing also E2F1 and p73.
Figure 6.
The outcome of E2F1 overexpression in Saos2 cells is mediated by DDR activation. A: Apoptotic rate in the Saos2 E2F1-ERsiATM, Saos2 E2F1-ERsip73, and Saos2 E2F1-ERshp14ARF cells before and after the addition of 4-OH-Tamoxifen (4-OH-Tam), respectively. The lines are the means of three independent experiments. Immunoblots for ATM, p73, and p14ARF confirmed corresponding gene silencing. B: Western blot analysis of γH2AX, Chk2-pT68, Chk2, and p73. Actin serves as loading control. C: Immunofluorescence analysis in Saos E2F1-ER cells showing the presence of H2AX foci before and after addition of 4-OH-Tamoxifen. D: Western blot analysis for p73 expression in Saos2 E2F1-ER cells treated with 4-OH-Tamoxifen and silenced for ATM expression.
Subsequently, in light of ARF’s ability to act also in a p53-independent manner,51 we investigated the putative impact of ARF on the behavior of Saos2 cells after E2F1 activation. As previously reported,34,35 ARF expression followed E2F1 induction (Figure 6B). Suppression of ARF, by siRNA, affected neither the growth properties of the cells nor their apoptotic response (Figure 6A, see supplemental Figure 2B at http://ajp.amjpathol.org).
Proteomic Analysis of E2F1-Induced U2OS Cells Reveals Potential Novel E2F1 Targets
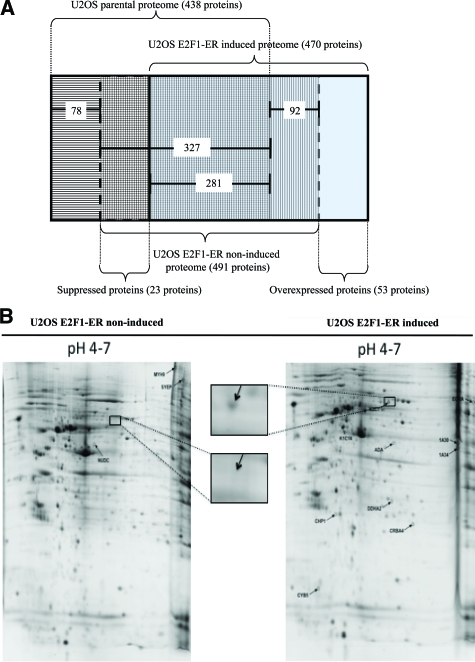
To gain a more global view of the molecular changes evoked in U2OS cells by E2F1 and to complement an analogous proteomic study in Saos2 cells52 and two E2F1-induced cDNA microarray screenings in U2OS53 and Saos2 cells,54 we performed a two-dimensional analysis of the induced and non-induced U2OS cells on immobilized pH-gradient strips with pH range 3 to 10 and 4 to 7. We chose the 48-hour postinduction time point for the experiment, because putative E2F1-independent cell cycle effects are suggested to be minimal52 (though these effects are largely avoided due to the applied algorithm described below) and the DDR pathway is fully activated (Figure 7A). The experiments were performed three times. Two approaches were used to identify qualitative and quantitative differences, respectively. According to the first one, after Coomassie Blue staining approximately 1300 protein spots were detected in each pH 3 to 10 gel (see supplemental Figure 3 at http://ajp.amjpathol.org) and 800 to 900 protein spots in each pH 4 to 7 gel (Figure 7B). All of the detected spots were excised from the gels and the proteins were identified by matrix-assisted laser desorption time-of-flight MS. The qualitative differences between the parental U2OS, previously analyzed by our group,55 and the non-induced and the induced U2OS E2F1-ER cells were detected by a subtractive algorithm (Figure 7A). According to this algorithm 53 proteins were found overexpressed and 23 suppressed, after E2F1 induction (Figure 7B, see supplemental Tables 1 and 2 at http://ajp.amjpathol.org). Representative proteins were confirmed by Western blotting (see supplemental Figure 4A at http://ajp.amjpathol.org). We should note that with this approach, overexpressed or suppressed proteins were defined as those being exclusively “present” or “absent,” respectively, in the induced U2OS E2F1-ER cells. Proteins categorized as “absent” were either expressed at very low levels or might be altered due to post-translational modifications, thereby preventing their detection at the staining process of the gels or at the matrix-assisted laser desorption-mass spectrometry time-of-flight MS identification stage. In addition, a limitation of proteomic analysis is that it reveals the presence of proteins that are relatively abundant in the cells, whereas proteins that function at low levels, such as certain transcription factors and checkpoint/regulatory proteins, are not easily identified.56 For instance, the replication licensing factor hCdt1, previously shown by our group to contain functional E2F1 responsive elements33 was induced by E2F1, as assessed by Western blot analysis in the present report (see supplemental Figure 4B at http://ajp.amjpathol.org), but was not identified by the proteomic procedure. Another example Aurora A, a mitotic checkpoint kinase,57 was found to be down-regulated in an E2F1-dependent fashion (see supplemental Figure 4C at http://ajp.amjpathol.org). According to the second method applying the two-dimensional image analysis (see Materials and Methods), we identified sixteen proteins that exhibited quantitative changes in the gels (see supplemental Tables 3, A and B at http://ajp.amjpathol.org). The expression of these proteins was defined as “increased” or “decreased,” respectively, in the induced U2OS E2F1-ER cells compared with the non-induced ones. With this procedure, 6 proteins were found increased and 10 decreased, after E2F1 induction (see supplemental Tables 3, A and B, at http://ajp.amjpathol.org).
Figure 7.
A: The subtractive algorithm used for the identification of overexpressed and suppressed proteins after E2F1 induction in U2OS E2F1-ER cells. B: Representative two-dimensional gel analysis of proteins from U2OS E2F1-ER cells either untreated (U2OS E2F1-ER non-induced) or 48 hours after 4-OH-Tamoxifen treatment (U2OS E2F1-ER induced). Proteins were extracted and separated on a non-linear immobilized pH-gradient strip pH 4 to 7, followed by a 12% SDS-polyacrylamide gel separation. The gel was stained with Coomassie blue. Identification of protein spots was performed by peptide mass fingerprint and/or postsource decay MS. Proteins found to be suppressed or overexpressed are annotated on the gels. Names and descriptions of each annotated protein are presented in supplemental Tables 3A and 3B (see http://ajp.amjpatho.org). Insets are magnified photos indicating suppression of K2C1 protein.
The spectrum of identified E2F1-regulated proteins included chaperones, metabolic enzymes, proteins associated with RNA processing, components of the protein degradation/turn-over machinery, cytoskeletal and motor/contractile related proteins, regulatory and cell signaling molecules, transport carriers and channels, as well as putative oncogenes (see supplemental Tables 1 and 2, and supplemental Figures 5 and 6 at http://ajp.amjpathol.org). Interestingly, with the exception of adenosine deaminase,58 caldesmon, hnRNPA3, cytokeratin 1, splicing factor arginine/serine-rich 1, transitional endoplasmic reticulum ATPase, prohibitin, vimentin, and hnRNAK52 (see supplemental Tables 1 and 3 at http://ajp.amjpathol.org), all of the remaining proteins appear to be potentially novel E2F1-regulated targets.
To gain a clearer picture of whether E2F1 is directly or indirectly involved in the regulation of all of the above characterized proteins we investigated their promoters for putative E2F1 responsive elements by scanning the Eukaryotic Promoter Database. The promoter sequence was available for 27 overexpressed and 13 suppressed proteins, found with the algorithmic approach (Figure 7A, see supplemental Tables 1 and 2 at http://ajp.amjpathol.org). The analysis revealed E2F1 responsive elements in genes encoding 17/27 (62.3%) of the overexpressed and 10/13 (77%) of the suppressed proteins (Table 3, see supplemental Tables 1 and 2 at http://ajp.amjpathol.org). Accordingly, E2F1 responsive elements were found in available promoter sequences of genes encoding 4/4 (100%) of the increased and 5/6 (83%) of the decreased proteins, detected with the two-dimensional image analysis (see Table 3 and supplemental Tables 3, A and B at http://ajp.amjpathol.org).
Table 3.
Deregulated Proteins after E2F1 Induction in U2OS E2F1-ER Cells that Possess E2F1-Responsive Element
| Qualitative changes (see results) | |
| Overexpressed | Suppressed |
| Carbonic anhydrase 2, Carbonate dehydratase II | Mitotic checkpoint protein BUB3 |
| Caldesmon | 10 kd heat shock protein, mitochondrial, Hsp10, CPN10 |
| Collagen alpha-1(I) chain precursor | Elongation factor 1-beta |
| Crk-like protein | Enhancer of rudimentary homolog |
| Alpha crystallin B chain | FK506-binding protein 3 |
| Cold shock domain-containing protein E1 | Heterogeneous nuclear ribonucleoprotein R, hnRNP R |
| COP9 signalosome complex subunit 8, JAB1-containing signal | Nuclear migration protein nudC |
| Cytochrome b5 | T-complex protein 1 subunit delta |
| N(G),N(G)-dimethylarginine dimethylaminohydrolase 2 | Thymidylate synthase |
| Trifunctional enzyme subunit alpha, mitochondrial precursor | UDP-glucose 6-dehydrogenase |
| Keratin, type II cytoskeletal 1 | |
| Leukotriene A-4 hydrolase | |
| Mitochondrial-processing peptidase subunit beta, mitochondrial precursor | |
| 26S proteasome non-ATPase regulatory subunit 11 | |
| Ras-related protein Rab-1A | |
| Heat shock protein 75 kd, mitochondrial precursor | |
| Spliceosome RNA helicase BAT1, DEAD box protein UAP56 | |
| Quantitative changes (see results) | |
| Increased | Decreased |
| Cathepsin B precursor | Splicing factor, arginine/serine-rich 1 |
| Eukaryotic translation initiation factor 3 subunit I | Proliferating cell nuclear antigen |
| Transitional endoplasmic reticulum ATPase, Valosin-containing protein | Vimentin |
| Prohibitin | |
| Thioredoxin-like protein 1 | |
| Chloride intracellular channel protein 1 | |
| Heterogeneous nuclear ribonucleoprotein K | |
Bioinformatic Analysis Reveals Candidate Protein Networks Connecting the Detected E2F1 Targets with Components of the DDR Machinery
To gain an insight into candidate molecular routes that could link the detected proteins and the DDR proteins, ATM and p53, shown to modulate the impact of E2F1, led us to perform a bioinformatic study. Applying the Pathway Studio Software we observed that a significant number of the identified E2F1 downstream targets are part of potential signaling cascades regulating ATM and/or p53 (see supplemental Figures 7 and 8 at http://ajp.amjpathol.org). Among the intermediate proteins were the non-receptor tyrosine kinase protein Src, the onco-proteins, myc, H- and N-ras, jun, fos, raf1, abl, rel, Akt, the growth factors, epidermal growth factor, vascular endothelial growth factor, and transforming growth factor-β 1, the epidermal growth factor receptor, the onco-suppressor proteins, RB1 and BRCA1, the anti-apoptotic protein BCL-2 along with its apoptotic counterpart BAX, the cytokines, tumor necrosis factor, interleukin (IL)-1, IL-6, IL-8, and IL-10, and members of the mitogen-activated protein kinase pathway (see supplemental Figures 7 and 8 at http://ajp.amjpathol.org). Moreover, using the same software oriented from ATM and p53 toward the identified E2F1 downstream targets we observed that only one E2F1-linked target protein, namely the succinate dehydrogenase flavoprotein mitochondrial subunit, is a putative p53 regulated target as it possesses a p53 responsive element (see supplemental Table 1B at http://ajp.amjpathol.org, data not shown).
Discussion
The bimodal behavior of E2F1, favoring either proliferation or apoptosis, is demonstrated in various carcinomas, as well as in experimental models.3 Considering that E2F1 has been projected as a potential therapeutic target,59 the determination of its action in each type of cancer is important. The absence of reports dealing with its potential role in malignancies of mesenchymal origin and the biological distinction of the later compared with epithelial ones,60 renders such a study essential.
To this end we examined the expression of E2F1 and its relationship with tumor kinetics in a clinical setting of primary human osteosarcomas. Over 80% of the cases demonstrated increased expression (more that 50% of the nuclei stained) of the pRb-unbound and active form of E2F1 (Figure 1A, Table 2A). Stratifying the cases according to the mutational status of p53 we interestingly observed that E2F1 expression was inversely related with cell proliferation, in wild-type p53 tumors (Figure 1B). In this group of patients increased expression of E2F1 was also positively correlated with apoptosis (see supplemental Figure 1 at http://ajp.amjpathol.org). Both associations were conversed in the group of patients that harbored p53 mutations (Figure 1b, see supplemental Figure 1 at http://ajp.amjpathol.org). These findings indicate that deregulated E2F1 overexpression is followed mainly by a p53-dependent growth suppressing response in osteosarcomas, though in a couple of cases p53 mutations were accompanied by high apoptotic scores, implying a p53-independent mode of E2F1 action in a small subset of patients.
The ability of E2F1 to evoke p53-dependent apoptosis has been attributed mainly to the oncogenic sensor protein and E2F1 target, p14ARF.34,35 Counter to this premise there are several reports, which suggest that ARF-independent pathways may also function in E2F1 induced p53-dependent apoptosis.36,37 According to these reports E2F1 utilizes the ATM signaling pathway to activate p53. Moreover, certain studies imply that ARF forms a negative feedback loop with E2F1 by repressing expression of E2F138; thus questioning ARF’s significance in E2F1-mediated apoptosis. Although ARF is frequently silenced by promoter methylation in osteosarcomas,61 suggesting a tumor-suppressive role in this malignancy, the aforementioned observations36,37 entail that p53 activation pathway(s), independent of ARF, may also operate in this tumor type. Recent studies from our own and other laboratories demonstrated that the DDR network also provide an intrinsic barrier against cancer progression by sensing oncogenic signals.20,23,39,40,41,42 While, this network elicits a strong anti-tumor response in early preneoplastic lesions,40 it also functions in advanced cancer stages, as depicted in various cancer cell lines,62 probably though not so efficiently as in early stages.40 As the DDR pathway is also essential for the repair of the genome, its complete loss may have lethal effects even for cancer cells, in case of severe DNA damage.
Indeed, an intense DDR was observed in almost all osteosarcoma samples, as depicted by the γ-H2AX and Chk2-pT68 staining scores (Table 2A, Figure 1D). This finding along with the high frequency of p53 mutations found in our tumor series (Figure 1A, Table 2B) supports one of the key predictions of our concept that the DDR machinery could act as a driving force selecting for p53 mutations at some time during the development of this malignancy.20,23,39,40,63 Moreover, mutations of the CKH2 gene are found more often in osteosarcomas than in other malignancies underscoring the significance of the DDR network in this cancer type.64 To examine whether a functional link between deregulated E2F1 overexpression and DDR exists, we used two E2F1-inducible osteosarcoma cell lines, U2OS and Saos2, which are wild-type and deficient for p53, respectively. Such an approach would help us understand the mechanisms underlying p53-dependent and -independent E2F1-driven effects, which appear to take place in the examined clinical material (Figure 1).
In U2OS cells, E2F1 induction was followed by growth suppression that was accompanied initially by mitotic catastrophe and then by apoptosis and senescence (Figure 2, see supplemental Figure 2A at http://ajp.amjpathol.org). Having in mind that U2OS cells do not express the senescence-linked protein, p16INK4A or the oncogenic-sensor protein and E2F1 target, p14ARF,45,46,47 we assumed that the E2F1 growth suppressive activity could be mediated by DDR. Indeed, the growth rate and the percentage of apoptotic and senescent cells, in U2OS E2F1-induced cells, reverted to control levels after silencing of ATM or p53, underscoring the significance of the DDR pathway in dictating the impact of E2F1 activity (Figure 2B). Moreover, suppression of the DDR components ATM and p53 resulted in the appearance of features indicative of chromosomal instability, such as aberrant mitoses and micronuclei (Figure 4C).22 Similar morphological phenomena were observed, by our group, after prolonged expression of the E2F1 target, hCdt1, in U2OS cells, when the DDR-induced anti-tumor barriers were bypassed.22 With regard to “mitotic catastrophe” it has been suggested that active DNA structure checkpoints and particularly Chk2 negatively regulate this type of cell death.43 Induction of cyclin B1 (Figure 4, A and D), that is probably responsible for the occurrence of this form of cell death,49,50 was more pronounced than DDR activation only during day 1 of E2F1 induction (Figure 3B). It appears that “mitotic catastrophe” took place during this narrow time window, in a small population of cells, where sudden increase of cyclin B1 was not associated by an analogous DDR. As the DNA damage response was intensified the following days a relatively rapid de-escalation of this type of mitotic cell death and a shift to apoptosis occurred.
In the Saos2 context, E2F1 induction also forced an intense DDR activation (Figure 6B, 6c), followed by growth suppression and a high apoptotic wave (Figure 6A, see supplemental Figure 2B at http://ajp.amjpathol.org). The absence of premature senescence possibly reflects the requirement for p53 for this phenomenon to occur, whereas Saos2 cells, compared with U2OS, demonstrate a constitutively activated DDR checkpoint that probably inhibits the potential “mitotic catastrophe” process.65 The apoptotic response evoked by E2F1 in the Saos2 cells was mediated by p73 (Figure 6D), in accordance with previous reports in the same cellular system.9,10 Of great interest though was that E2F1 induction did not affect either apoptosis or p73 expression levels when ATM was silenced (Figure 6, A and D, see supplemental Figure 2B at http://ajp.amjpathol.org), indicating an E2F1-ATM-p73 axis in the Saos2 cellular system. Since p73 is a direct transcriptional target of E2F1,9,10 this last result was not anticipated. Initially, after the discovery of p73, it was proposed that it had no role in response to DNA damage.66 Later it was shown that after γ-irradiation or treatment with other DNA damaging insults, p73 was post-translationally modified and activated. 65 Hence, our results show that the oncogene-induced DNA damage activates p73 as well, possibly conferring post-translational stability to the protein. The latter effect could be achieved either directly by modifying p73,67 or indirectly by modulating the p73-regulatory proteins, such as Itch and NQO1 that affect p73 degradation.65 Comparing the results from the two cellular models, a question that arises is why was p73 not induced by E2F1 in U2OS cells? A possible answer is that p73 acts as a back-up system when p53 is intact and it becomes more prominently involved when p53 is mutated. Moreover, the complex nature of p73 transcription, producing a large number of isoforms and forming tightly regulated negative feedback loops in the presence of p53, could also contribute to the inability of p73 to respond to E2F1 in the p53-proficient U2OS cells. In contrast, no alterations in E2F1-triggered apoptosis was detected after ARF silencing, possibly indicating that p53 is required for ARF’s impact on the cellular phenotype within the context of Saos2 cells (Figure 6A, see supplemental Figure 2B at http://ajp.amjpathol.org). Similarly, in mouse fibroblasts, ARF was not required for E2F1-mediated apoptosis.36
Taken together, the in vivo (Figure 1) and in vitro data (Figures 2 to 6) support the concept that E2F1 overexpression is sensed, in both p53-wild-type and p53-deficient osteosarcoma cells, as an oncogenic signal that activates the DDR pathway that eventually controls the fate of the cells. Since high E2F1 levels were found only in a subset of the clinical samples examined, and almost all cases exhibited signs of DNA damage checkpoint activation (see second part of the Results section), also some other “oncogenic” stimuli likely trigger the checkpoint within the context of osteosarcomas. Potential DDR-activating signals could derive from oncogenes such as, c-erb-B2, MET, and c-myc, reported to be involved in osteosarcoma pathogenesis.68
To obtain a large-scale picture of the proteins that took part in the E2F1 response and to complement a recent proteomic study in Saos2 cells,52 as well as cDNA microarray screenings, after E2F1 induction, in U2OS51 and Saos2,54 we performed a similar proteomic analysis in U2OS. We detected 92 deregulated proteins (with two methodological approaches) as potential E2F1 targets among which, 72% of them with available the promoter sequence, possessed E2F1 responsive elements (Table 3, see supplemental Tables 1, 2, and 3 at http://ajp.amjpathol.org). Interestingly, with the exception of adenosine deaminase,58 caldesmon, hnRNPA3, cytokeratin 1, splicing factor arginine/serine-rich 1, transitional endoplasmic reticulum ATPase, prohibitin, vimentin, and hnRNAK52 (see supplemental Tables 1 and 3 at http://ajp.amjpathol.org), all of the remaining proteins appear to be novel E2F1-regulated targets. Among the induced proteins several have been reported to play a role in tumorigenesis. For example, the Crk-like (CrkL) protein (see supplemental Table 1 at http://ajp.amjpathol.org) is an SH2 and SH3 domain-containing adaptor protein mediating Src signaling and implicated in the pathogenesis of chronic myelogenous leukemia.69 Moreover, a recent study demonstrates the involvement of CrkL in epithelial to mesenchymal transition, a process crucial for invasion and metastasis.70 The signalosome COP9 (see supplemental Table 1 at http://ajp.amjpathol.org) inactivates the oncosuppressor protein p27Kip1, by exporting it from the nucleus, and promotes ubiquitin-dependent degradation of p53.71 Interestingly, BUB3 a component of the spindle assembly checkpoint, involved in the maintenance of chromosomal stability and implicated in various malignancies including osteosarcomas,72 was found suppressed (see supplemental Table 2 and supplemental Figure 1A at http://ajp.amjpathol.org). Of note, also nucleophosmin/B23, a nucleolar associated protein that has been implicated in p14ARF and MDM2 regulation, was found decreased.73 A comparison of our present U2OS data and the previous analysis of Saos2 cells52 showed different spectra of the detected E2F1 targets. Numerous technical and cell context related reasons could account for these variations, as extensively discussed by Li et al for various high-throughput platforms.52 An interesting observation was that the proteins, caldesmon, hnRNPA3, and cytokeratin 1, were inversely expressed after E2F1 activation in the two cellular systems. This finding further denotes the effect of the cellular context on the global E2F1 response. Therefore, the information derived from different methodologies and cellular systems must be interpreted in a complementary fashion to obtain a broader and more detailed overview of the proteins/pathways/networks under investigation.
The belief that redundant signaling cascades control fundamental proteins and cellular functions led us to assume that the proteins regulated by E2F1, in the inducible U2OS cellular system, could represent elements of alternative pathways affecting constituents of the DDR checkpoint. By performing a bioinformatic analysis we identified a number of molecules that could participate in alternative pathways modulating E2F1 behavior through interaction with ATM and/or p53 (see supplemental Figures 7 and 8 at http://ajp.amjpathol.org). Whether these potential pathways take part in the activation of ATM and p53, remains to be proven. Furthermore, these putative pathways may play a role also in other malignancies than osteosarcomas, and it will be interesting to investigate such possibilities based on our present bioinformatics data.
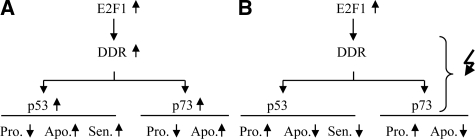
In conclusion, the current study demonstrated that the DNA damage response machinery is a major modulator of the biological impact of E2F1 activity in osteosarcomas (Figure 8, A and B), and identified novel E2F1 targets and potential signaling networks that could be useful in future basic research as well as clinical applications.
Figure 8.
Proposed model summarizing the DDR-dependent modulation of E2F1 activity in human osteosarcoma cells (A) with intact DDR response machinery, and (B) with aberrations in the DDR response machinery. DDR, DNA damage response; Pro, proliferation; Apo, apoptosis; Sen, senescence. Symbols: increase (upward pointing arrow), decrease (downward pointing arrow), aberration(s) (zigzag arrow).
Supplementary Material
Acknowledgments
We thank Dr. Euphemia Thomas-Tsagli and the Department of Pathology, General Hospital of Asklepeion Voula, Athens, Greece, for kindly providing the patients material, and Panayota Papanagnou for her valuable technical support.
Footnotes
Address reprint requests to Dr. Vassilis G. Gorgoulis, Antaiou 56 Str, Lamprini, Ano Patisia, GR-11146, Athens, Greece, E-mail: vgorg@med.uoa.gr.
Supported by the European Commission (FP7-project GENICA), NKUA - SARG grants No 70/3/8916 and 70/3/1703, the Czech Ministry of Education (MSM6198959216), the Danish Cancer Society, and the Danish National Research Foundation and the NIH, USA (grant CA118827).
M.L. and K.N. contributed to this work equally.
Supplemental material for this article can be found on http://ajp.amjpathol.org.
References
- Meyers PA, Gorlick R. Osteosarcoma. Pediatr Clin North Am. 1997;44:973–989. doi: 10.1016/s0031-3955(05)70540-x. [DOI] [PubMed] [Google Scholar]
- Wadayama B, Toguchida J, Shimizu T, Ishizaki K, Sasaki MS, Kotoura Y, Yamamuro T. Mutation spectrum of the retinoblastoma gene in osteosarcomas. Cancer Res. 1994;54:3042–3048. [PubMed] [Google Scholar]
- Tsantoulis PK, Gorgoulis VG. Involvement of E2F transcription factor family in cancer. Eur J Cancer. 2005;41:2403–2414. doi: 10.1016/j.ejca.2005.08.005. [DOI] [PubMed] [Google Scholar]
- Iaquinta PJ, Lees JA. Life and death decisions by the E2F transcription factors. Curr Opin Cell Biol. 2007;19:649–657. doi: 10.1016/j.ceb.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell LA, Ryan KM. Life and death decisions by E2F-1. Cell Death Differ. 2004;11:137–142. doi: 10.1038/sj.cdd.4401324. [DOI] [PubMed] [Google Scholar]
- Crighton D, Ryan KM. Splicing DNA-damage responses to tumor cell death. Biochim Biophys Acta. 2004;1705:3–15. doi: 10.1016/j.bbcan.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Ginsberg D. E2F1 pathways to apoptosis. FEBS Lett. 2002;529:122–125. doi: 10.1016/s0014-5793(02)03270-2. [DOI] [PubMed] [Google Scholar]
- Urist M, Tanaka T, Poyurovsky MV, Prives C. p73 induction after DNA damage is regulated by checkpoint kinases Chk1 and Chk2. Genes Dev. 2004;18:3041–54. doi: 10.1101/gad.1221004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin M, Marin MC, Phillips AC, Seelan RS, Smith DI, Liu W, Flores ER, Tsai KY, Jacks T, Vousden KH, Kaelin WG., Jr Role for the p53 homologue p73 in E2F-1-induced apoptosis. Nature. 2000;407:645–648. doi: 10.1038/35036614. [DOI] [PubMed] [Google Scholar]
- Stiewe T, Pützer BM. Role of the p53-homologue p73 in E2F1-induced apoptosis. Nat Genet. 2000;26:464–469. doi: 10.1038/82617. [DOI] [PubMed] [Google Scholar]
- La Thangue NB. The yin and yang of E2F-1: balancing life and death. Nat Cell Biol. 2003;5:587–589. doi: 10.1038/ncb0703-587. [DOI] [PubMed] [Google Scholar]
- Yamasaki L, Bronson R, Williams BO, Dyson NJ, Harlow E, Jacks T. Loss of E2F-1 reduces tumorigenesis and extends the lifespan of Rb1(+/−) mice. Nat Genet. 1998;18:360–364. doi: 10.1038/ng0498-360. [DOI] [PubMed] [Google Scholar]
- Gorgoulis VG, Zacharatos P, Mariatos G, Kotsinas A, Bouda M, Kletsas D, Asimacopoulos PJ, Agnantis N, Kittas C, Papavassiliou AG. Transcription factor E2F-1 acts as a growth-promoting factor and is associated with adverse prognosis in non-small cell lung carcinomas. J Pathol. 2002;198:142–156. doi: 10.1002/path.1121. [DOI] [PubMed] [Google Scholar]
- Zacharatos P, Kotsinas A, Evangelou K, Karakaidos P, Vassiliou LV, Rezaei N, Kyroudi A, Kittas C, Patsouris E, Papavassiliou AG, Gorgoulis VG. Distinct expression patterns of the transcription factor E2F-1 in relation to tumor growth parameters in common human carcinomas. J Pathol. 2004;203:744–753. doi: 10.1002/path.1582. [DOI] [PubMed] [Google Scholar]
- Saiz AD, Overa M, Rezk S, Florentine BA, McCourty A, Brynes RK. Immunohistochemical expression of cyclin D1. E2F-1, and Ki-67 in benign and malignant thyroid lesions. J Pathol. 2002;198:157–162. doi: 10.1002/path.1185. [DOI] [PubMed] [Google Scholar]
- Yamazaki K, Yajima T, Nagao T, Shinkawa H, Kondo F, Hanami K, Asoh A, Sugano I, Ishida Y. Expression of transcription factor E2F-1 in pancreatic ductal carcinoma: an immunohistochemical study. Pathol Res Pract. 2003;199:23–28. doi: 10.1078/0344-0338-00348. [DOI] [PubMed] [Google Scholar]
- Evangelou K, Kotsinas A, Mariolis-Sapsakos T, Giannopoulos A, Tsantoulis PK, Constantinides C, Troupis TG, Salmas M, Kyroudis A, Kittas C, Gorgoulis VG. E2F-1 overexpression correlates with decreased proliferation and better prognosis in adenocarcinomas of Barrett’s oesophagus. J Clin Pathol. 2008;61:601–605. doi: 10.1136/jcp.2007.050963. [DOI] [PubMed] [Google Scholar]
- Yamazaki K, Hasegawa M, Ohoka I, Hanami K, Asoh A, Nagao T, Sugano I, Ishida Y. Increased E2F-1 expression via tumor cell proliferation and decreased apoptosis are correlated with adverse prognosis in patients with squamous cell carcinoma of the oesophagus. J Clin Pathol. 2005;58:904–910. doi: 10.1136/jcp.2004.023127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee D, Schnieders B, Fu JZ, Adhikari D, Zhao S-C, Bertino JR. Role of E2F-1 in chemosensitivity. Cancer Res. 1998;58:4292–4296. [PubMed] [Google Scholar]
- Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, Ditullio RA, Jr, Kastrinakis NG, Levy B, Kletsas D, Yoneta A, Herlyn M, Kittas C, Halazonetis TD. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- Yoshikawa H, Nagashima M, Khan MA, McMenamin MG, Hagiwara K, Harris CC. Mutational analysis of p73 and p53 in human cancer cell lines. Oncogene. 1999;18:3415–3421. doi: 10.1038/sj.onc.1202677. [DOI] [PubMed] [Google Scholar]
- Liontos M, Koutsami M, Sideridou M, Evangelou K, Kletsas D, Levy B, Kotsinas A, Nahum O, Zoumpourlis V, Kouloukoussa M, Lygerou Z, Taraviras S, Kittas C, Bartkova J, Papavassiliou AG, Bartek J, Halazonetis TD, Gorgoulis VG. Deregulated overexpression of hCdt1 and hCdc6 promotes malignant behavior. Cancer Res. 2007;67:10899–10909. doi: 10.1158/0008-5472.CAN-07-2837. [DOI] [PubMed] [Google Scholar]
- Bartkova J, Horejsí Z, Koed K, Krämer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, Ørntoft T, Lukas J, Bartek J. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- Hershko T, Chaussepied M, Oren M, Ginsberg D. Novel link between E2F and p53: proapoptotic cofactors of p53 are transcriptionally upregulated by E2F. Cell Death Differ. 2005;12:377–383. doi: 10.1038/sj.cdd.4401575. [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- Fountoulakis MTG, Oh JE, Maris A, Lubec G. Protein profile of the HeLa cell line. J Chromatogr A. 2004;1038:247–265. doi: 10.1016/j.chroma.2004.03.032. [DOI] [PubMed] [Google Scholar]
- Berndt PHU, Langen H. Reliable automatic protein identification from matrix-assisted laser desorption/ionization mass spectrometric peptide fingerprints. Electrophoresis. 1999;20:3521–3526. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3521::AID-ELPS3521>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Schmid CD, Perier R, Praz V, Bucher P. EPD in its twentieth year: towards complete promoter coverage of selected model organisms. Nucleic Acids Res. 2006;34:D82–D85. doi: 10.1093/nar/gkj146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgoulis VG, Koutroumbi EN, Kotsinas A, Zacharatos P, Markopoulos C, Giannikos L, Kyriakou V, Voulgaris Z, Gogas I, Kittas C. Alterations of the p16-pRb pathway and the chromosome locus 9p21–22 in non-small-cell lung carcinomas: relationship with p53 and MDM2 protein expression. Am J Pathol. 1998;153:1749–1765. [Google Scholar]
- Lonardo FTU, Huvos AG, Healey J, Ladanyi M. p53 and MDM2 alterations in osteosarcomas. Cancer. 1997;79:1541–1547. [PubMed] [Google Scholar]
- Overholtzer M, Rao PH, Favis R, Lu XY, Elowitz MB, Barany F, Ladanyi M, Gorlick R, Levine AJ. The presence of p53 mutations in human osteosarcomas correlates with high levels of genomic instability. Proc Natl Acad Sci USA. 2003;100:11547–11552. doi: 10.1073/pnas.1934852100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakaidos P, Taraviras S, Vassiliou LV, Zacharatos P, Kastrinakis NG, Kougiou D, Kouloukoussa M, Nishitani H, Papavassiliou AG, Lygerou Z, Gorgoulis VG. Overexpression of the replication licensing regulators hCdt1 and hCdc6 characterizes a subset of non-small-cell lung carcinomas: synergistic effect with mutant p53 on tumor growth and chromosomal instability-evidence of E2F-1 transcriptional control over hCdt1. Am J Pathol. 2004;165:1351–1365. doi: 10.1016/S0002-9440(10)63393-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiebert SW, Packham G, Strom DK, Haffner R, Oren M, Zambetti G, Cleveland JL. E2F-1: DP-1 induces p53 and overrides survival factors to trigger apoptosis. Mol Cell Biol. 1995;15:6864–6874. doi: 10.1128/mcb.15.12.6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates S, Phillips AC, Clark PA, Stott F, Peters G, Ludwig RL, Vousden KH. p14ARF links the tumor suppressors RB and p53. Nature. 1998;395:124–125. doi: 10.1038/25867. [DOI] [PubMed] [Google Scholar]
- Rogoff HA, Pickering MT, Debatis ME, Jones S, Kowalik TF. E2F1 induces phosphorylation of p53 that is coincident with p53 accumulation and apoptosis. Mol Cell Biol. 2002;22:5308–5318. doi: 10.1128/MCB.22.15.5308-5318.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers JT, Hong S, Mayhew CN, Rogers PM, Knudsen ES, Johnson DG. E2F1 uses the ATM signaling pathway to induce p53 and Chk2 phosphorylation and apoptosis. Mol Cancer Res. 2004;2:203–214. [PubMed] [Google Scholar]
- Martelli F, Hamilton T, Silver DP, Sharpless NE, Bardeesy N, Rokas M, DePinho RA, Livingston DM, Grossman SR. p19ARF targets certain E2F species for degradation. Proc Natl Acad Sci USA. 2001;98:4455–4560. doi: 10.1073/pnas.081061398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LV, Kolettas E, Niforou K, Zoumpourlis VC, Takaoka M, Nakagawa H, Tort F, Fugger K, Johansson F, Sehested M, Andersen CL, Dyrskjot L, Ørntoft T, Lukas J, Kittas C, Helleday T, Halazonetis TD, Bartek J, Gorgoulis VG. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, Schurra C, Garre' M, Nuciforo PG, Bensimon A, Maestro R, Pelicci PG, d'Adda di Fagagna F. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- Mallette FA, Gaumont-Leclerc MF, Ferbeyre G. The DNA damage signaling pathway is a critical mediator of oncogene-induced senescence. Genes Dev. 2007;21:43–48. doi: 10.1101/gad.1487307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castedo M, Perfettini J-L, Roumier T, Andreau K, Medema R, Kroemer G. Cell death by mitotic catastrophe: a molecular definition. Oncogene. 2004;23:2825–2837. doi: 10.1038/sj.onc.1207528. [DOI] [PubMed] [Google Scholar]
- Roninson IB, Broude EV, Chang B-D. If not apoptosis, then what? Treatment-induced senescence and mitotic catastrophe in tumor cells. Drug Resistance Updates. 2001;4:303–313. doi: 10.1054/drup.2001.0213. [DOI] [PubMed] [Google Scholar]
- Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, Niki M, Koutcher JA, Scher HI, Ludwig T, Gerald W, Cordon-Cardo C, Pandolfi PP. Crucial role of p53- pendent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–730. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- Collado M, Serrano M. Cellular senescence in cancer and aging. Cell. 2007;130:223–233. doi: 10.1016/j.cell.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Russo AJ, Magro PG, Hu Z, Li WW, Peters R, Mandola J, Banerjee D, Bertino JR. E2F-1 Overexpression in U2OS cells increases cyclin B1 levels and cdc2 kinase activity and sensitizes cells to antimitotic agents. Cancer Res. 2006;66:7253–7260. doi: 10.1158/0008-5472.CAN-05-3725. [DOI] [PubMed] [Google Scholar]
- Heald R, McLoughlin M, McKeon F. Human wee1 maintains mitotic timing by protecting the nucleus from cytoplasmically activated cdc2 kinase. Cell. 1993;74:463–474. doi: 10.1016/0092-8674(93)80048-j. [DOI] [PubMed] [Google Scholar]
- Jin P, Hardy S, Morgan DO. Nuclear localization of cyclin B1 controls mitotic entry after DNA damage. J Cell Biol. 1998;141:875–885. doi: 10.1083/jcb.141.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ. Divorcing ARF and p53: an unsettled case. Nat Rev Cancer. 2006;6:663–673. doi: 10.1038/nrc1954. [DOI] [PubMed] [Google Scholar]
- Li Z, Mikkat S, Mise N, Glocker MO, Püetzer BM. Proteomic analysis of the E2F1 response in p53-negative cancer cells: new aspects in the regulation of cell survival and death. Proteomics. 2006;6:5735–5745. doi: 10.1002/pmic.200600290. [DOI] [PubMed] [Google Scholar]
- Müller H, Bracken AP, Vernell R, Moroni MC, Christians F, Grassilli E, Prosperini E, Vigo E, Oliner JD, Helin K. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev. 2001;15:267–285. doi: 10.1101/gad.864201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanelle J, Stiewe T, Theseling CC, Peter M, Pützer BM. Gene expression changes in response to E2F1 activation. Nucleic Acids Res. 2002;30:1859–1867. doi: 10.1093/nar/30.8.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niforou KM, Anagnostopoulos A, Vougas K, Kittas C, Gorgoulis VG, Tsangaris GT. The proteome profile of the human osteosarcoma U2OS cell line. Cancer Genomics Proteomics. 2008;5:63–78. [PubMed] [Google Scholar]
- Gygi SP, Corthals GL, Zhang Y, Rochon Y, Aebersold R. Evaluation of two-dimensional gel electrophoresis-based proteome analysis technology. Proc Natl Acad Sci USA. 2000;97:9390–9395. doi: 10.1073/pnas.160270797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löffler H, Lukas J, Bartek J, Krämer A. Structure meets function–centrosomes, genome maintenance and the DNA damage response. Exp Cell Res. 2006;312:2633–40. doi: 10.1016/j.yexcr.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Ma Y, Croxton R, Moorer RL, Jr, Cress WD. Identification of novel E2F1-regulated genes by microarray. Arch Biochem Biophys. 2002;399:212–224. doi: 10.1006/abbi.2002.2761. [DOI] [PubMed] [Google Scholar]
- Kaelin WG., Jr E2F1 as a target: promoter-driven suicide and small molecule modulators. Cancer Biol Ther. 2003;2:S48–S54. [PubMed] [Google Scholar]
- Berman J. Modern classification of neoplasms: reconciling differences between morphologic and molecular approaches. BMC Cancer. 2005;5:100. doi: 10.1186/1471-2407-5-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benassi MS, Molendini L, Gamberi G, Magagnoli G, Ragazzini P, Gobbi GA, Sangiorgi L, Pazzaglia L, Asp J, Brantsing C, Picci P. Involvement of INK4A gene products in the pathogenesis and development of human osteosarcoma. Cancer. 2001;92:3062–3067. doi: 10.1002/1097-0142(20011215)92:12<3062::aid-cncr10161>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- DiTullio RA, Jr, Mochan TA, Venere M, Bartkova J, Sehested M, Bartek J, Halazonetis TD. 53BP1 functions in an ATM-dependent checkpoint pathway that is constitutively activated in human cancer. Nat Cell Biol. 2002;4:998–1002. doi: 10.1038/ncb892. [DOI] [PubMed] [Google Scholar]
- Halazonetis TD. Constitutively active DNA damage checkpoint pathways as the driving force for the high frequency of p53 mutations in human cancer. DNA Repair. 2004;3:1057–1062. doi: 10.1016/j.dnarep.2004.03.036. [DOI] [PubMed] [Google Scholar]
- Miller CW, Ikezoe T, Krug U, Hofmann WK, Tavor S, Vegesna V, Tsukasaki K, Takeuchi S, Koeffler HP. Mutations of the CHK2 gene are found in some osteosarcomas, but are rare in breast, lung, and ovarian tumors. Genes Chromosomes Cancer. 2002;33:17–21. doi: 10.1002/gcc.1207. [DOI] [PubMed] [Google Scholar]
- Marabese M, Vikhanskaya F, Broggini M. p73: a chiaroscuro gene in cancer. Eur J Cancer. 2007;43:1361–1372. doi: 10.1016/j.ejca.2007.01.042. [DOI] [PubMed] [Google Scholar]
- Kaghad M, Bonnet H, Yang A, Creancier L, Biscan JC, Valent A, Minty A, Chalon P, Lelias JM, Dumont X, Ferrara P, McKeon F, Caput D. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell. 1997;90:809–819. doi: 10.1016/s0092-8674(00)80540-1. [DOI] [PubMed] [Google Scholar]
- Gonzalez S, Prives C, Cordon-Cardo C. p73alpha regulation by Chk1 in response to DNA damage. Mol Cell Biol. 2003;23:8161–71. doi: 10.1128/MCB.23.22.8161-8171.2003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Papachristou DJ, Papavassiliou AG. Osteosarcoma and chondrosarcoma: new signaling pathways as targets for novel therapeutic interventions. Int J Biochem Cell Biol. 2007;39:857–862. doi: 10.1016/j.biocel.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Nichols GL, Raines MA, Vera JC, Lacomis L, Tempst P, Golde DW. Identification of CRKL as the constitutively phosphorylated 39-kD tyrosine phosphoprotein in chronic myelogenous leukemia cells. Blood. 1994;84:2912–2918. [PubMed] [Google Scholar]
- Lamorte L, Royal I, Naujokas M, Park M. Crk adapter proteins promote an epithelial-mesenchymal-like transition and are required for HGF-mediated cell spreading and breakdown of epithelial adherens junctions. Mol Cell Biol. 2002;13:1449–1461. doi: 10.1091/mbc.01-10-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson KS, Zundel W. The emerging role of the COP9 signalosome in cancer. Mol Cancer Res. 2005;3:645–653. doi: 10.1158/1541-7786.MCR-05-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza S, David H, Gaylord GM, Miller CW. Allelic loss at 10q26 in osteosarcoma in the region of the BUB3 and FGFR2 genes. Cancer Genet Cytogenet. 2005;158:142–147. doi: 10.1016/j.cancergencyto.2004.08.035. [DOI] [PubMed] [Google Scholar]
- Lee C, Smith BA, Bandyopadhyay K, Gjerset RA. DNA damage disrupts the p14ARF-B23(nucleophosmin) interaction and triggers a transient subnuclear redistribution of p14ARF. Cancer Res. 2005;65:9834–9842. doi: 10.1158/0008-5472.CAN-05-1759. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.