Abstract
Polyubiquitylation targets multiple proteins for degradation by the proteasome. Typically, the first ubiquitin is linked to lysine residues in the substrate for degradation via an isopeptide bond, although rarely ubiquitin linkage to the N-terminal residue has also been observed. We have recently shown that Neurogenin (NGN), a basic helix-loop-helix transcription factor that plays a central role in regulating neuronal differentiation, is degraded by ubiquitin-mediated proteolysis. We have taken a biochemical and mutagenesis approach to investigate sites of ubiquitylation of NGN, initially using extracts of eggs from the frog Xenopus laevis as a source of ubiquitylation and degradation components. NGN can be targeted for destruction by ubiquitylation via lysines or the N terminus. However, we see that a modified NGN, where canonical lysine ubiquitylation and N-terminally linked ubiquitylation are prevented, is nevertheless ubiquitylated and degraded by the proteasome. We show that polyubiquitin chains covalently attach to non-canonical cysteine residues in NGN, and these non-canonical linkages alone are capable of targeting NGN protein for destruction. Importantly, canonical and non-canonical ubiquitylation occurs simultaneously in the native protein and may differ in importance for driving degradation in interphase and mitosis. We conclude that native NGN is ubiquitylated on multiple canonical and non-canonical sites by cellular ubiquitin ligases, and all types of linkage can contribute to protein turnover.
The selective degradation of proteins by ubiquitin-mediated proteolysis is a crucial regulator of diverse cellular processes in eukaryotes. Ub4 is classically conjugated to the ϵ-amino group of a substrate lysine by an isopeptide bond (1). This serves as an anchor point to build a polyubiquitin chain that results in subsequent targeting to the proteasome for destruction. In a small number of proteins, Ub has been shown to be conjugated to the N-terminal α-amino group of a protein via a peptide linkage used as an anchor site for assembly of a lysine-linked polyubiquitin chain (2).
The conjugation of ubiquitin to substrate proceeds when a nucleophilic substrate amino group attacks the labile E2-ubiquitin thioester bond. This mechanism does not exclude the possibility that ubiquitylation can occur on other substrate nucleophilic groups on amino acid side chains, such as the thiol groups present on cysteines. The resultant thioester-anchored ubiquitin chains would, however, be predicted to be less stable than the isopeptide/peptide bonds formed between ubiquitin and lysine/N-terminal amino groups (3, 4). Based on this, cysteine residues have generally been considered to be unlikely targets for ubiquitin modification (4).
However, there is recent evidence that ubiquitylation can occur on substrate cysteine residues, albeit in a very limited number of cases, largely regulating intracellular protein movements and usually driven by viral E3 ligases (3, 5–7). Only one substrate, the N-terminal fragment of the Bcl2-related protein Bid, has been described thus far to undergo both non-canonical ubiquitylation and proteasomal degradation by cellular E3 ligases (8), but mutational analyses failed to demonstrate a link between removing the non-canonical ubiquitylated residues and increased stability. In addition, the E2 UBC7 undergoes auto-polyubiquitylation via cysteine and proteasomal degradation (7).
We have been studying the degradation of the proneural transcription factor Neurogenin (NGN), which plays a pivotal role in the neural versus glial decision during embryonic central nervous system formation (9). To this end, we have exploited the highly versatile Xenopus egg extract system. Centrifugal extracts of Xenopus egg cytoplasm contain a large stockpile of enzymes required for early stages of embryogenesis including multiple components of the ubiquitin-proteasome system (UPS) and have been used extensively to explore the biochemistry of ubiquitylation (10). Using Xenopus eggs and embryos and mammalian cultured cells, we have demonstrated that NGN is a very short-lived protein whose rapid turnover is controlled by the UPS (11).
In this study, we demonstrate that NGN is one of the small number of proteins that can be targeted for destruction by ubiquitylation on both the N terminus and the internal lysines, and both contribute to its rapid degradation. Strikingly, we also found that a mutant NGN protein, where Ub conjugation to either lysines or the N terminus had been prevented, was still degraded by the UPS. Under these circumstances, ubiquitylation of NGN can occur on non-canonical cysteine sites. We show that polyubiquitylation on both canonical and non-canonical residues also simultaneously occurs in the wild-type native protein, and the relative contribution of these Ub linkages to speed of protein degradation may vary according to cell cycle stage. Moreover, in the absence of other potential sites for ubiquitylation, Ubs can also be added to serines and/or threonines, demonstrating the remarkable flexibility of the ubiquitylation machinery.
EXPERIMENTAL PROCEDURES
Plasmids and Constructs
NGN (also known as X-NGNR1a) and NeuroD constructs were described previously (11). NGNKO, NGNCO, and NGNKOCO mutants were generated using the QuikChange® multisite-directed mutagenesis kit (Stratagene). AC1, AC2, AC3 NGN, and NGN mutant constructs were generated by PCR and cloned into the pCS2 vector. Ub and UbKO were amplified by PCR without the last two glycine codons to prevent cleavage of the fusion by cytoplasmic deubiquitinating enzymes (12) and fused in-frame to NGNKO by a polar tetrapeptide linker (13).
In Vitro Translation
TnT® SP6 quick coupled transcription/translation system (Promega), in the presence of [35S] methionine (Amersham Biosciences), was carried out according to the manufacturer's instructions.
Mitotic Extracts
Activated interphase egg extracts were prepared as described previously (11) and were then supplemented with 5 μg of recombinant non-degradable cyclin B delta 90 per 100 μl of extract. Extracts were incubated at room temperature for 20 min to enable extracts to enter mitosis. The effectiveness of this treatment in driving extracts into mitosis was determined initially by examining the morphology of sperm nuclei added to the extract.
Degradation Assays
Activated interphase and mitotic egg extracts were prepared as described previously (11). Extracts were supplemented with ubiquitin (from bovine erythrocytes, Sigma) at a final concentration of 1.25 mg/ml. 35S-IVT protein (protein generated by in vitro translation in the presence of [35S]methionine) was added to the extract (8 μl of 35S-IVT protein per 36 μl of extract) and incubated at 21 °C. Aliquots were taken at various time points and mixed with 2× Laemmli sample buffer containing 100 mm β-mercaptoethanol. Samples were denatured for 3–5 min at 95 °C and separated by SDS-PAGE (15% Tris-glycine). The half-life of the protein was calculated by plotting the concentration of protein against time and finding the first order rate constant for degradation.
Where appropriate, proteasome inhibitors were added to extracts at the concentrations indicated, 5 min prior to the addition of 35S-IVT protein. Mg132 and epoxomicin were obtained commercially (BIOMOL) and used at the concentrations indicated (200 and 100 μm, respectively, or adding DMSO (1% (v/v)) in place of proteasome inhibitors in controls. Where appropriate, extracts supplemented with a final concentration of 10 mg/ml methylated ubiquitin (BIOMOL) were used in degradation assays where indicated with ubiquitin from bovine erythrocytes at a final concentration of 10 mg/ml as a control.
Ubiquitylation Assays
Ubiquitylation assays were performed in accordance with the protocol described by Salic et al. (10), with minor alterations. 5 μl of 35S-IVT protein (or IVT reaction with pCS2+ vector as control) was added as indicated to 23 μl of activated interphase or mitotic egg extracts, supplemented with 200 μm MG132 and either 2.5 mg/ml His-ubiquitin (N-terminally hexahistidine tagged ubiquitin, Sigma) or 1.25 mg/ml ubiquitin. Extract reactions were incubated at 20 °C for 1 h. Following incubation, the reactions were diluted 10-fold in His buffer (100 mm Tris/HCl (pH 7.4), 1% (v/v) Nonidet P-40, 8 m urea, 20 mm imidazole, 600 mm NaCl and 10% (v/v) ethanol) and supplemented with 15 μl (∼5% (v/v)) of Ni2+-nitrilotriacetate-agarose beads (Qiagen). Reactions were incubated for 90 min at room temperature, rotating at 12 rpm, and then washed five times with His buffer prior to elution with 2× Laemmli sample buffer supplemented with 100 mm β-mercaptoethanol. Samples were heat-treated for 5 min at 95 °C prior to separation on 15% Tris-glycine SDS-PAGE gels.
Reducing Treatments
Reducing ubiquitylation assays were performed as described above except that at the elution step, samples were eluted with either reducing 2× Laemmli sample buffer (2× Laemmli sample buffer (pH 10.5) + 10% (v/v) β-mercaptoethanol) or non-reducing 2× Laemmli sample buffer (2× Laemmli sample buffer (pH 6.8) + 10% (v/v) water), see Fig. 4, B and C). Sample buffers also included 100 mm imidazole in the case of Fig. 6A and were rocked at room temperature for 20 min after sample buffer addition but before SDS-PAGE.
FIGURE 4.
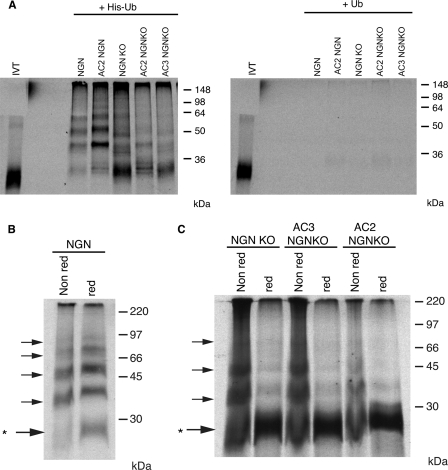
N-terminally blocked NGN lacking lysines is still ubiquitylated in egg extracts. A, ubiquitylation assays were performed using NGN, Ac2NGN, NGNKO, Ac2NGNKO, and Ac3NGNKO in the presence of His-ubiquitin or ubiquitin. B, ubiquitylation assays were performed with NGN. Reactions were eluted either under non-reducing (Non red) or under reducing/pH 10 conditions (red). C, ubiquitylation assays were performed using NGNKO, Ac2NGNKO, and Ac3NGNKO. Reactions were eluted either under non-reducing or under reducing/pH 10 conditions. Note that NGN and NGN mutant samples were from the same experiment with identical conditions and exposures. Non-modified NGN is marked by *.
FIGURE 6.
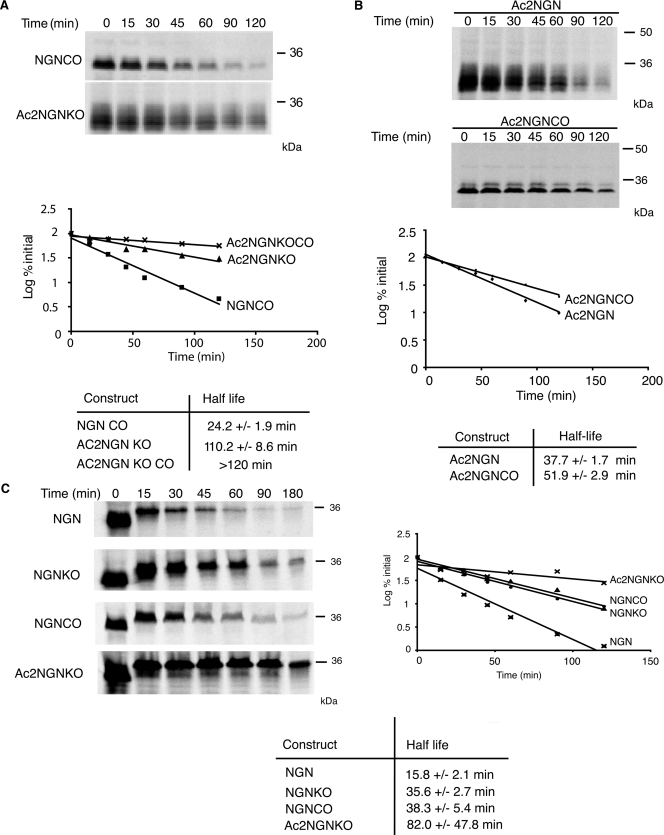
NGN lacking canonical ubiquitylation sites is still ubiquitylated and degraded in a proteasomal-dependent manner. Degradation assays in interphase egg extract of [35S]methionine-radiolabeled constructs of NGN were performed using NGNCO, Ac2NGNKO, and Ac2NGNKOCO (A) or Ac2NGN and Ac2NGNCO (B). Assays were analyzed by autoradiography and quantitative phosphorimaging analysis. C, degradation assays in mitotic egg extract were performed using NGN, NGNKO, NGNCO, and Ac2NGNKO and analyzed by autoradiography and quantitative phosphorimaging analysis.
Blocking N-terminal Acetylation
The blocking of N-terminal acetylation in reticulocyte lysates was based on procedures described previously (14, 15). Briefly, 120 μl of quick coupled in vitro transcription/translation system (Promega) was supplemented with 1 mm oxaloacetic acid (Sigma) and 50 units/ml of citrate synthase (from porcine heart, Sigma), incubated at 25 °C for 7 min, and then supplemented with desulfo CoA lithium salt (Sigma) to give a final concentration of 1 mm after the addition of plasmid DNA and [35S]methionine. Reactions were incubated for 2 h at 30 °C. Oxaloacetate and desulfo CoA were removed using Zeba microspin columns (Pierce). A control reaction was included to which citrate synthase only was added.
ATP Depletion
8 μl of in vitro translated 35S-methione-labeled protein was combined with 40 μl of high speed interphase-activated egg extract, cleared of ribosomes by spinning at 107,400 × g for 40 min, supplemented with 200 μg/ml cycloheximide, 10 μg/ml leupeptin, pepstatin, and chymostatin, 1.5 mg/ml ubiquitin, and 2 μl of Energy Regeneration mix (20 μl of 100 mm EGTA; 20 μl of 1 m MgCl2; 150 μl of 1 m phosphocreatine (Sigma); 200 μl of 100 mm adenosine 5′-triphosphate (GE Healthcare)) or 0.8 μl of 1600 units/ml hexokinase with 0.8 μl of 1 m glucose. Samples were used as in a normal degradation assay, described above.
RESULTS
NGN Can Be Ubiquitylated on Both Lysines and the N Terminus
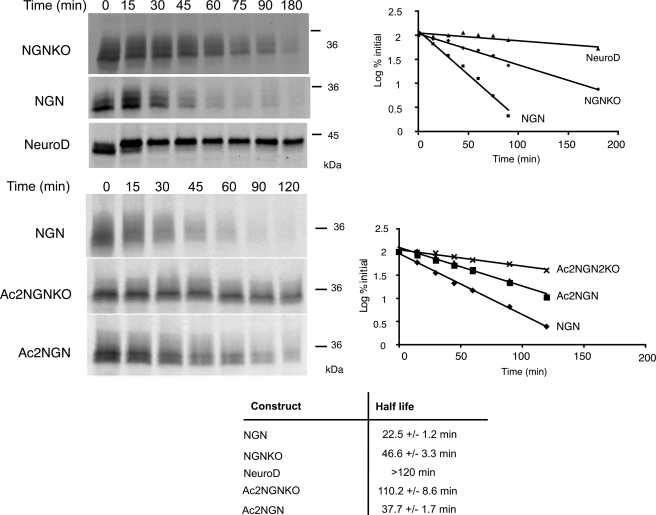
We have previously demonstrated that NGN is turned over extremely rapidly in Xenopus eggs and embryos via the UPS, whereas a related proneural protein NeuroD is stable (11). In an attempt to generate a stabilized form of NGN, its 8 lysine residues were mutated to arginines to produce a lysine-less protein (NGNKO). Arginine cannot be ubiquitylated but preserves the basic character of the lysine residues they replace (16). Both NGNKO and wild-type protein (NGN) were in vitro translated (IVT) in rabbit reticulocyte lysates in the presence of [35S]methionine and added to Xenopus egg extracts, and the half-life was determined after SDS-PAGE analysis at increasing time points in a standard degradation assay. NGNKO had approximately double the half-life of the wild-type protein (46.6 ± 3.3 min versus 22.5 ± 1.2 min, Fig. 1) but was still degraded. We investigated whether NGN could join the small number of proteins thus far identified as being ubiquitylated on the N terminus.
FIGURE 1.
Neurogenin is degraded by both a lysine-dependent pathway and an N-terminal-dependent pathway. Degradation assays in interphase egg extract were performed using [35S]methionine-radiolabeled NGN, NGNKO, Ac2NGNKO, Ac2NGN, and NeuroD, as indicated, and analyzed by autoradiography (left panels) and quantitative phosphorimaging analysis (right graphs). Half-lives for degradation were calculated using first order kinetics (bottom table).
Bulky N-terminal tags, such as six consecutive Myc tags, are thought to impede the accessibility of the α-amino group to the ubiquitylation machinery and so prevent N-terminal ubiquitylation (17). However, adding bulky tags (Myc tags or green fluorescent protein) to both the N termini and the C termini of NGN resulted in its stabilization (supplemental Fig. 1A and data not shown). Although this observation may have important implications for the use of tags on short-lived transcription factors, it led us to seek other ways to prevent N-terminal ubiquitylation.
Blocking the N Terminus by Promoting N-terminal Acetylation
A free, unmodified α-amino group of the substrate protein is required for N-terminal ubiquitylation. N-terminal cotranslational acetylation of the α-amino group is a common protein modification in eukaryotes, occurring in up to 80% of cytosolic proteins (18). The presence of an acetyl group at the N terminus prevents the attachment of ubiquitin (19), and this modification is thought to be irreversible (18). Primary sequence requirements for N-terminal acetylation in eukaryotes have been identified (18) and can be used to predict whether a protein with a given N-terminal sequence will be cotranslationally acetylated (20). We used an online prediction tool (TermiNator, available from the CNRS) to determine the likelihood that NGN is cotranslationally acetylated. This analysis revealed that NGN is highly likely to be expressed with a free, unmodified N terminus (supplemental Fig. 1B). We then devised two alterations to the N terminus of NGN (MVLLK to MSESK and to MAESK, forming Ac1NGN and Ac2NGN, respectively) to promote its cotranslational acetylation (18). Additionally, a mutant was made in which the penultimate residue of NGN was replaced with proline (MVLLK to MPLLK, forming Ac3NGN), which completely blocks its N-terminal acetylation and therefore represents an appropriate control for our experiments (18).
Using our degradation assay, we compared the stability of Ac1NGN, Ac2NGN, and Ac3NGN with the wild-type protein. As we would predict whether N-terminal ubiquitylation contributes to NGN destruction, Ac1NGN and Ac2NGN had enhanced stability when compared with wild-type NGN (33.7 ± 1.7 min and 37.7 ± 1.7 min versus 22.5 ± 1.2 min). Ac3NGN had a half-life similar to the wild-type NGN protein (26.1 ± 0.6 min versus 22.5 ± 1.2 min).
To see whether ubiquitylation on lysines and the N terminus target for destruction additively or redundantly, we constructed NGNKO mutants where the N terminus was additionally modified to block ubiquitylation to produce Ac1NGNKO and Ac2NGNKO. The half-life of Ac1NGNKO and Ac2NGNKO was substantially increased (103.4 ± 3.9 min and 110.2 ± 8.6 min, respectively) when compared with NGN (22.5 ± 1.2 min), NGNKO (46.6 ± 3.3 min), and Ac2NGN (37.7 ± 1.7 min) (Fig. 1). Thus, ubiquitylation on internal lysines and the N terminus both contribute to the rapid degradation of NGN protein.
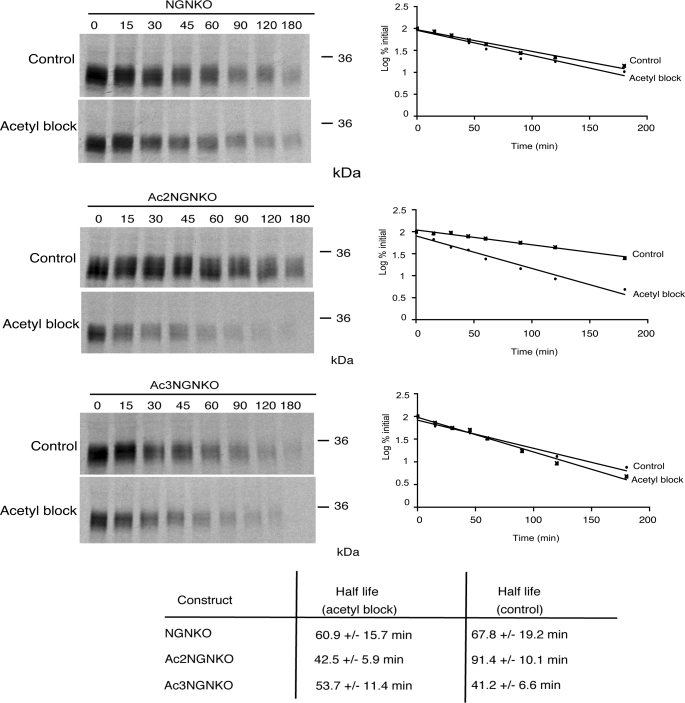
To confirm that these sequence alterations block degradation by promoting cotranslational acetylation, we examined the degradation of NGN, Ac3NGNKO, and Ac2NGNKO where cotranslational acetylation had been enzymatically inhibited. Methods for inhibiting translational acetylation in reticulocyte lysates have been previously described (14, 15). These involve enzymatic depletion of free acetyl CoA in reticulocyte lysates using citrate synthase and oxaloacetate followed by in vitro translation in the presence of a dead end analogue of acetyl CoA to competitively inhibit acetyl CoA-dependent N-terminal acetyltransferases. When Ac2NGNKO is in vitro translated under these conditions (Fig. 2), a marked destabilization in egg extract is observed (half-life of 42.5 ± 5.9 min after blocking acetylation when compared with 91.4 ± 10.1 min in the control). In contrast, NGNKO and Ac3NGNKO were found to be refractory to this treatment (Fig. 2), consistent with the non-acetylated status of the N termini of these proteins under normal in vitro translation conditions.
FIGURE 2.
Stability of acetylation mutants of NGN is affected by acetylation-blocking. Degradation assays in interphase egg extract were performed on NGN constructs, as indicated, in vitro translated either in the absence (Control) or in the presence of citrate synthase, oxaloacetate, and desulfo CoA (Acetyl block). Assays were analyzed by autoradiography (left panels) and quantitative phosphorimaging analysis (right graphs).
NGN Is Stabilized by Truncation at the N Terminus
It has been shown that several of the substrates of N-terminal ubiquitylation can be stabilized by the deletion of 5–30 amino acids from the N terminus. We took this approach with NGN as a further line of evidence for N-terminal ubiquitylation. We observed that an N-terminally truncated form of NGNKO where the first 20 amino acids had been removed (NGNKO Δ1–20) was also degraded more slowly than NGNKO (63.4 ± 4.4 min versus 45.3 ± 4 min, (supplemental Fig. 2A), again indicative of N-terminal ubiquitylation.
To mimic N-terminal ubiquitylation, we generated non-cleavable fusions of Ub and UbKO to NGNKO and examined the degradation kinetics. Consistent with N-terminal ubiquitylation contributing to NGN turnover, UbNGNKO was degraded faster than wild-type NGN (half-life of 10.5 ± 1 min versus 22.5 ± 1.2 min, respectively). In contrast, when we blocked N-terminal polyubiquitylation using an N-terminal fusion of ubiquitin where lysines had been mutated to arginines, this UbKONGNKO form of NGN was considerably stabilized when compared with both wild-type NGN and NGNKO (half-lives of 110.8 ± 10.7 min versus 22.5 ± 1.2 min and 46.6 ± 3.3 min, respectively, and see supplemental Fig. 2B). This further indicates that N-terminal ubiquitylation contributes to NGN instability. However, we were intrigued to see that, like Ac2NGNKO, slow degradation of UbKONGNKO was still observed.
In summary, the evidence presented above shows that NGN is ubiquitylated on the N terminus and on internal lysines, and both contribute to the instability of the protein. However, it is clear that, even when these two canonical modes of ubiquitylation are prevented, NGN is still subject to slow proteolysis. We have explored this further.
NGN Is Ubiquitylated on Cysteines
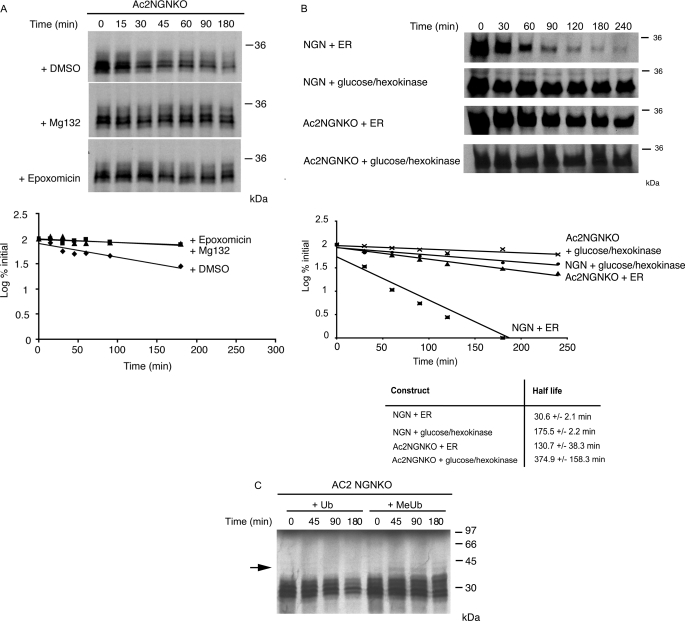
Despite the apparent absence of canonical ubiquitylation sites, Ac2NGNKO is still degraded in Xenopus egg extracts. Mg132 addition to the extract resulted in stabilization of Ac2NGNKO, as did the addition of a different proteasomal inhibitor epoxomicin (Fig. 3A; at 180 min on average, 32% of the Ac2NGNKO protein remained after the addition of DMSO when compared with 74 and 84% after the addition of MG132 and epoxomicin, respectively), indicating degradation by the proteasome. Furthermore, depletion of ATP by the addition of glucose and hexokinase, which would inactivate ATP-dependent proteasomal-mediated degradation, also resulted in stabilization of both wild-type NGN and Ac2NGNKO (Fig. 3B).
FIGURE 3.
N-terminally blocked NGN lacking lysines is still degraded in a ubiquitin-dependent manner in egg extracts. A, degradation assays in interphase egg extract of [35S]methionine-radiolabeled Ac2NGNKO were performed in the presence of the proteasome inhibitor Mg132 (200 μm), epoxomicin (100 μm) or DMSO alone and analyzed by autoradiography and quantitative phosphorimaging analysis. B, degradation assays in interphase egg extract of [35S]methionine-radiolabeled NGN or Ac2NGNKO were performed in the presence (+ER) or absence (+glucose/hexokinase) of ATP, where ER represents Energy Regeneration mix. C, degradation assays of Ac2NGNKO was performed in the presence of ubiquitin or methylated ubiquitin (MeUb).
To determine whether degradation of Ac2NGNKO requires polyubiquitylation, we added methyl-Ub, a form of Ub that cannot support chain formation (21), to the extract. As expected, methylated Ub competes with an endogenous pool of free Ub in the extract resulting in stabilization of Ac2NGNKO (Fig. 3C); at 180 min, an average of 34% of the Ac2NGNKO protein still remained after the addition of Ub when compared with 76% after the addition of methylated Ub. To confirm that ubiquitylation was occurring even in the absence of canonical ubiquitylation sites, we performed an assay to detect ubiquitylated forms of NGN directly.
IVT [35S]methionine-labeled NGN and mutants thereof were incubated in egg extract in the presence of Mg132 to prevent protein degradation by the 26 S proteasome and allow ubiquitylation to be observed. Proteins covalently bound to His-Ub (with untagged Ub as a control) were then pulled down using Ni2+-nitrilotriacetate-agarose, in the presence of 8 m urea. Proteins covalently attached to His-Ub were separated by SDS-PAGE and subjected to autoradiography (Fig. 4A). After His-Ub pull down, as expected, multiple slower migrating forms of NGN and Ac2NGN were visible as a ladder specifically in the presence of His-Ub, consistent with multiply ubiquitylated forms of the protein (Fig. 4A), presumably linked to lysines. A similar ladder was seen using NGNKO, where ubiquitylation on the N terminus is possible. However, Ac2NGNKO (Fig. 4A), which does not have canonical Ub sites, was also precipitated using His-Ub, although the ladder at higher molecular weights tended to be somewhat reduced. Significantly, in addition to this faint ladder of polyubiquitylated forms, a band was observed running below 36 kDa, the size at which unmodified NGN protein migrates on SDS-PAGE. Moreover, this band appears more prominent in NGNKO mutants, although it is clearly detectable even in the wild-type NGN sample. We reasoned that unconjugated NGN protein would be released from Ub chains if attachment were via linkages labile to the conditions used in SDS-PAGE, which involve sample preparation at high temperature and include reducing agents, conditions that would break thioester bonds.
To test whether NGN can be conjugated to Ub via such labile linkages, we performed a standard ubiquitylation assay but separated bound proteins by SDS-PAGE either without reducing agents (non-reducing (Fig. 4B (Non red)) or with reducing agents at high pH (reducing (red) conditions), based upon conditions that have previously been used to disrupt labile Ub thioester linkages to cysteines (3) (Fig. 4B). NGN showed a ladder of polyubiquitylated species in both reducing and non-reducing conditions (Fig. 4B), indicating that most Ubs are indeed conjugated to lysine, an isopeptide linkage insensitive to these conditions. However, in reducing conditions, we saw a prominent band at the size corresponding to unmodified NGN that is barely present with non-reducing sample preparation (Fig. 4B, marked with *), indicating that a proportion of ubiquitylated wild-type NGN is conjugated through labile linkages.
In contrast to wild-type NGN, forms of NGN lacking lysines and precipitated by His-Ub (NGNKO, Ac2NGNKO, Ac3NGNKO) appeared strikingly different when separated by SDS-PAGE in non-reducing and reducing conditions (Fig. 4C). In non-reducing conditions, a ladder of polyubiquitylated species is present, but this is substantially diminished under reducing conditions, and this coincides with the appearance of a strong band of unconjugated NGN. This is consistent with substantial ubiquitylation of NGNKO via linkages that are labile to reducing conditions, which would include thioester linkages between Ub and cysteine.
To investigate this further, we generated an NGN mutant where all 7 cysteines had been mutated to alanine (NGNCO). Importantly, this mutant version of NGN, NGNCO, as well as the other mutant forms of NGN assayed above, are all able to bind to DNA and to activate an NGN reporter construct after expression in Xenopus embryos (Fig. 5). Functionality indicates largely intact structures of the mutant forms. We did note that NGNCO mutants do run as a tighter band than NGN on SDS-PAGE (Fig. 6). To determine whether the presence of cysteine residues contributes to NGN instability, we tested the half-life of NGNCO in interphase, which was similar to that of wild-type NGN (24.2 ± 1.9 min versus 22.5 ± 1.2 min). We constructed forms of NGN with other potential ubiquitylation sites mutated (see above) in a background where cysteines were also mutated (Ac2NGNCO, NGNKOCO, and Ac2NGNKOCO). Half-life analyses (Fig. 6, A and B) show that mutation of cysteines results in slower degradation of all these modified forms of NGN. In particular, in interphase, when the N-terminal ubiquitylation pathway is blocked, ubiquitylation on lysines cannot fully compensate for the loss of cysteines; Ac2NGNCO is degraded more slowly than Ac2NGN, although lysines are intact in both (Fig. 6B). However, although ubiquitylation on canonical residues and on cysteines can both target for degradation in interphase, redundancy between ubiquitylation pathways exists under these conditions as cysteines are not absolutely required for rapid degradation (Fig. 6A).
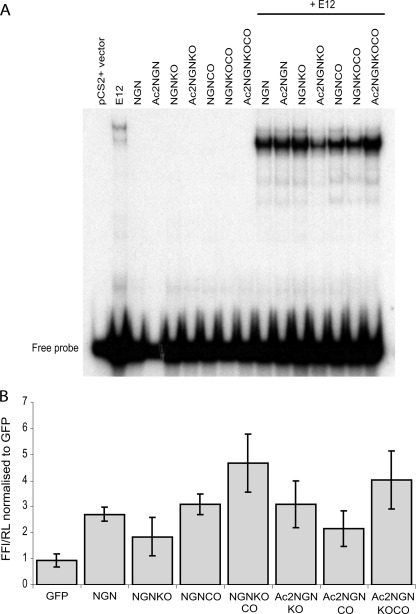
FIGURE 5.
Mutant NGN protein shows wild-type DNA binding. A, NGN constructs, as indicated, were translated in vitro and incubated with 32P-labeled E-box DNA probe in the presence or absence of the heterodimeric binding partner, E12. In the absence of E12, neither wild-type nor mutant NGN proteins are able to bind the probe. In the presence of E12, all NGN derivatives show wild-type binding to the probe. The experiment has been performed in triplicate; slight variations in binding efficiency seen here were not consistently observed. B, mutant NGN constructs activate a reporter gene at wild-type levels in vivo. Xenopus laevis embryos were injected with 50 pg of NGN mRNA as indicated, together with 200 pg of Beta2 reporter Firefly luciferase construct (FFL), as a direct downstream target of NGN, and 6 pg of constitutively expressed Renilla luciferase (RL), as an internal control. There is no significant difference between activation of the reporter gene by wild-type NGN or by mutant NGN constructs. Values obtained are from independent duplicate experiments (each sample in triplicate) and show mean ± S.E. GFP, green fluorescent protein.
NGN protein has a short half-life in interphase extracts, but the control of degradation of proteins can often vary between interphase and mitosis (22). Hence, we investigated the half-life of NGN in egg extracts that had been rendered mitotic by the addition of a non-degradable form of cyclin B (23) (Fig. 6C). Strikingly, in mitosis, NGN is degraded even more rapidly than in interphase, with a half-life of 15.8 ± 2.1 min. We then compared the contribution of lysines and cysteines with the degradation of NGN in mitosis. NGNKO had an increased half-life of 35.6 ± 2.7 min in mitotic extracts. Importantly and in contrast to interphase, NGNCO was also significantly stabilized when compared with wild-type NGN, with a half-life, 38.3 ± 5.4 min, almost three times that of the wild-type protein. These results indicate that ubiquitylation on cysteine and lysine both contribute to the very rapid rate of degradation seen in mitosis. N-terminal ubiquitylation can also contribute to degradation in mitosis; Ac2NGNKO had a half-life of 82.0 ± 47.8 min when compared with 35.6 ± 2.7 min for NGNKO.
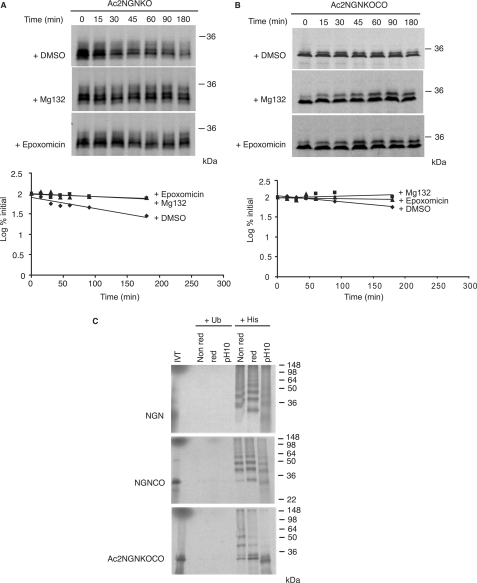
We then determined whether NGN lacking both canonical and cysteine ubiquitylation sites was fully stable, investigating the degradation of Ac2NGNKOCO, where the N terminus was blocked by acetylation and both lysines and cysteines were mutated to arginines and alanines, respectively. When compared with NGNKO, we observed that Ac2NGNKOCO was dramatically stabilized, with a half-life of greater than 180 min, and appeared more stable than Ac2NGNKO, yet we still observed very slow degradation (Fig. 7). Moreover, the stability of Ac2NGNKOCO was further enhanced by the presence of Mg132 (Fig. 7B) and epoxomicin (Fig. 7B). Under these condition, ubiquitins may still form ester linkages to serines and/or threonines, which have been predicted to be broken by high pH alone (4, 24). To investigate these potential linkages further, we performed ubiquitylation assays using wild-type NGN, NGNCO, and Ac2NGNKOCO either in the absence or in the presence of reducing agents or at high pH (Fig. 7C).
FIGURE 7.
NGN lacking canonical ubiquitylation sites and non-canonical cysteines is still ubiquitylated and degraded by the proteasome. Degradation assays in interphase egg extract of [35S]methionine-radiolabeled constructs of NGN were performed using Ac2NGNKO (A) and Ac2NGNKOCO (B) in the presence or absence of proteasome inhibitors Mg132 and epoxomicin. Assays were analyzed by autoradiography and quantitative phosphorimaging analysis. C, ubiquitylation assays were performed in interphase extract using NGN, NGNCO, and Ac2NGNKOCO in the presence of His-Ub or Ub, and samples prepared for SDS-PAGE under non-reducing conditions (Non red), reducing conditions (red), or high pH (pH10) conditions (with samples heated to 95 °C for 5 min prior to loading in all cases).
NGN Can Be Ubiquitylated on Serines and/or Threonines
To determine whether Ub was linked to Ac2NGNKOCO via ester bonds, we performed ubiquitylation assays and separated samples by SDS-PAGE either in the presence of reducing agents (with sample heating) or at high pH (Fig. 7C). In the absence of reducing agents, His-Ub precipitation of polyubiquitylated NGN, as expected, showed a multitude of higher molecular weight forms, corresponding to polyubiquitylated NGN protein. Unconjugated NGN was visible particularly after the addition of dithiothreitol, indicating that a population of wild-type NGN is linked to Ub via thioester linkages, as described above. A polyubiquitylated ladder of NGNCO was also visible, where presumably most Ubs are linked to lysines via isopeptide bonds. Interestingly, when ubiquitylated NGNCO was run after sample treatment at high temperature and pH, conditions that would break ester bonds, free NGNCO is released, indicating that in the absence of cysteines, ubiquitylation can occur on serines and/or threonines instead. We did note that when NGNCO samples were run in the presence of reducing agents, we still observed higher forms of NGNCO, although some release of free NGNCO did occur. This is most likely due to breaking of highly labile ester linkages on sample heating as cysteine residues are not available for thioester bond formation.
In contrast to NGN and NGNCO, His-Ub pull down of Ac2NGNKOCO showed reduced very high molecular mass forms of the proteins, but instead, we saw a prominent short ladder of NGN conjugates between 30 and 60 kDa. When samples were run in the presence of reducing agents, we still observed higher forms of wild-type NGN, although again some release of free Ac2NGNKOCO did occur, most likely due to breaking of labile ester linkages on sample heating. However, raising the sample pH as well as heating, which results in efficient alkaline hydrolysis of ester bonds, led to substantial collapse of slower migrating forms of NGN, releasing almost entirely unconjugated Ac2NGNKOCO. Thus, in the absence of a free N terminus, lysines, and cysteines, Ubs can still conjugate via ester linkages to serines and/or threonines (Fig. 7C), and these linkages alone can result in very slow turnover of the protein (Fig. 7). We would note that high pH treatment alone of ubiquitylated wild-type NGN did not release substantial free NGN protein, indicating that Ser/Thr ubiquitylation of NGN does not usually occur to a great extent.
DISCUSSION
For the first time, we demonstrate here that ubiquitylation of an intact and active cellular protein occurs at three distinct types of site: Ub conjugation to the N terminus, Ub conjugation to lysines, and Ub conjugation via non-canonical linkages to cysteines. In addition, in the absence of other available sites, ubiquitylation can be redirected to serines and/or threonine residues. Ubiquitylation on cysteine residues has been theoretically postulated for many years, yet it has only been very recently shown to occur in a cellular setting, almost exclusively to regulate intracellular movement. We show here that it can occur and act in parallel with canonical ubiquitylation to target a protein for destruction. Moreover, non-canonical ubiquitylation via cysteines may play a greater role in NGN turnover in mitosis, where degradation rate is faster than in interphase, indicating that different Ub acceptors may change in prominence under different cellular conditions. Although different modes of ubiquitylation may individually contribute to small differences in protein stability, nevertheless, this may lead to a significant difference in the steady state level of the protein (25).
Several studies (3, 5, 6, 8) have reported very unusual non-canonical ubiquitylation including on cysteines, but none have demonstrated this modification directly by mass spectroscopy. It has been proposed that one explanation is the small amount of ubiquitylated molecules available for purification (3). In addition, thioester and ester bonds are very labile and may not withstand sample preparation. We have also encountered technical problems purifying sufficient NGN monomeric protein for mass spectroscopy analysis. Hence, for these reasons, we have instead used biochemical methods to demonstrate non-canonical ubiquitylation.
Non-canonical ubiquitylation does occur on the wild-type NGN protein, but we did observe that these non-canonical linkages assumed more experimental prominence in the absence of lysines; we would note that very few proteins have had ubiquitylation analyzed after all lysine residues have been mutated. Cysteine ubiquitylation may be more important under these circumstances or where native lysines are not available for ubiquitylation due to other modifications such as acetylation. However, it is quite possible that non-canonical ubiquitylation may occur more widely than has previously been supposed, but these modifications could have been overlooked by current experimental approaches, particularly as assay and analysis conditions very commonly including sample preparation involving high temperature and reducing agents, conditions that will break these non-canonical linkages.
If simultaneous canonical and non-canonical ubiquitylation occurs on other proteins and site prominence varies at different cell cycle phases, this would substantially increase the complexity of the UPS system and require reanalysis of, for instance, some proteins thought to be targeted to the proteasome in the absence of ubiquitylation (for a review, see Ref.26). Possible candidates for non-canonical ubiquitylation include other basic helix-loop-helix proteins such as MyoD, which shows different modes of canonical ubiquitylation to NGN (27, 28). In addition, naturally lysineless proteins such as p14ARF (29, 30) may also undergo ubiquitylation on non-canonical sites such as cysteine to target for proteasomal degradation.
Non-canonical ubiquitylation of NGN does lead to Ub chains of sufficient length and with the correct linkage to target for destruction. The ability to ubiquitylate on multiple canonical and non-canonical sites demonstrates remarkable flexibility of the ubiquitylation apparatus. Ubiquitin ligases for NGN have yet to be identified and, at present, it is not clear whether the same enzymes that catalyze ubiquitylation of lysines can also target the N terminus and/or cysteine residues and indeed also serines and threonines in the absence of other available ubiquitylation sites. There clearly is much flexibility in the machinery that ubiquitylates NGN; we have seen that no single lysine in NGN is indispensable for canonical ubiquitylation (supplemental Fig. 3). Thus, how specificity of these ubiquitylation reactions is controlled remains to be established.
Another important point to note is that, in our system, ubiquitylation on canonical and non-canonical sites can be brought about entirely by cellular ubiquitin ligases found in the Xenopus egg extract. We have preliminary data that ubiquitylation on non-canonical residues may also occur in mammalian cells (data not shown), indicating that non-canonical ubiquitylation is a more widespread phenomenon. In summary, our analyses demonstrate that both canonical and non-canonical ubiquitylation brought about by the cellular ubiquitylation machinery occur simultaneously, and both can drive proteasomal degradation. We postulate that such regulation of a single protein by multiple canonical and non-canonical ubiquitylation events may be found to be more widespread on closer study of other proteins and systems and that the importance of these two modes of ubiquitylation may differ for the same protein under differing cellular conditions.
Supplementary Material
Acknowledgments
We thank Helen Wise for preliminary findings on activity of NGN mutants, Catherine Wilson for technical support, and Francois Guillemot, Laura Itzhaki, Shin-ichi Ohnuma, Ashok Venkitaraman, and Ron Laskey for helpful discussions. Ubiquitin constructs were a kind gift of C. Holt.
This work was supported by training awards from the MRC (to J. V. and G. M.), MRC Grants GO500101 (to C. F.-H.) and G0700758 (R. K.), and Biotechnology and Biological Sciences Research Council Grant BB/C 004 108/1.

The on-line version of this article (available at http://www.jbc.org) contains threesupplemental figures.
- Ub
- ubiquitin
- UPS
- ubiquitin-proteasome system
- E2
- ubiquitin carrier protein
- E3
- ubiquitin-protein isopeptide ligase
- NGN
- Neurogenin
- IVT
- in vitro translated
- DMSO
- dimethyl sulfoxide
- NeuroD
- neurogenic differentiation protein.
REFERENCES
- 1.Glickman M. H., Ciechanover A. ( 2002) Physiol. Rev. 82, 373– 428 [DOI] [PubMed] [Google Scholar]
- 2.Ciechanover A., Ben-Saadon R. ( 2004) Trends Cell Biol. 14, 103– 106 [DOI] [PubMed] [Google Scholar]
- 3.Cadwell K., Coscoy L. ( 2005) Science 309, 127– 130 [DOI] [PubMed] [Google Scholar]
- 4.Breitschopf K., Bengal E., Ziv T., Admon A., Ciechanover A. ( 1998) EMBO J. 17, 5964– 5973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carvalho A. F., Pinto M. P., Grou C. P., Alencastre I. S., Fransen M., Sá-Miranda C., Azevedo J. E. ( 2007) J. Biol. Chem. 282, 31267– 31272 [DOI] [PubMed] [Google Scholar]
- 6.Williams C., van den Berg M., Sprenger R. R., Distel B. ( 2007) J. Biol. Chem. 282, 22534– 22543 [DOI] [PubMed] [Google Scholar]
- 7.Ravid T., Hochstrasser M. ( 2007) Nat. Cell Biol. 9, 422– 427 [DOI] [PubMed] [Google Scholar]
- 8.Tait S. W., de Vries E., Maas C., Keller A. M., D'Santos C. S., Borst J. ( 2007) J. Cell Biol. 179, 1453– 1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun Y., Nadal-Vicens M., Misono S., Lin M. Z., Zubiaga A., Hua X., Fan G., Greenberg M. E. ( 2001) Cell 104, 365– 376 [DOI] [PubMed] [Google Scholar]
- 10.Salic A., Lee E., Mayer L., Kirschner M. W. ( 2000) Mol. Cell 5, 523– 532 [DOI] [PubMed] [Google Scholar]
- 11.Vosper J. M., Fiore-Heriche C. S., Horan I., Wilson K., Wise H., Philpott A. ( 2007) Biochem. J. 407, 277– 284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butt T. R., Khan M. I., Marsh J., Ecker D. J., Crooke S. T. ( 1988) J. Biol. Chem. 263, 16364– 16371 [PubMed] [Google Scholar]
- 13.Raiborg C., Bache K. G., Gillooly D. J., Madshus I. H., Stang E., Stenmark H. ( 2002) Nat. Cell Biol. 4, 394– 398 [DOI] [PubMed] [Google Scholar]
- 14.Rubenstein P., Smith P., Deuchler J., Redman K. ( 1981) J. Biol. Chem. 256, 8149– 8155 [PubMed] [Google Scholar]
- 15.Palmiter R. D. ( 1977) J. Biol. Chem. 252, 8781– 8783 [PubMed] [Google Scholar]
- 16.Chau V., Tobias J. W., Bachmair A., Marriott D., Ecker D. J., Gonda D. K., Varshavsky A. ( 1989) Science 243, 1576– 1583 [DOI] [PubMed] [Google Scholar]
- 17.Ciechanover A. ( 2005) in Ubiquitin-Proteasome Protocols ( Patterson C., Cyr D. M. eds) pp. 255– 270, Humana Press, New York [Google Scholar]
- 18.Polevoda B., Sherman F. ( 2003) J. Mol. Biol. 325, 595– 622 [DOI] [PubMed] [Google Scholar]
- 19.Kuo M. L., den Besten W., Bertwistle D., Roussel M. F., Sherr C. J. ( 2004) Genes Dev. 18, 1862– 1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meinnel T., Peynot P., Giglione C. ( 2005) Biochimie 87, 701– 712 [DOI] [PubMed] [Google Scholar]
- 21.Hershko A., Ganoth D., Pehrson J., Palazzo R. E., Cohen L. H. ( 1991) J. Biol. Chem. 266, 16376– 16379 [PubMed] [Google Scholar]
- 22.Lindon C., Albagli O., Domeyne P., Montarras D., Pinset C. ( 2000) Mol. Cell. Biol. 20, 8923– 8932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murray A. W., Kirschner M. W. ( 1989) Nature 339, 275– 280 [DOI] [PubMed] [Google Scholar]
- 24.Passmore L. A., Barford D. ( 2004) Biochem. J. 379, 513– 525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lingbeck J. M., Trausch-Azar J. S., Ciechanover A., Schwartz A. L. ( 2003) J. Biol. Chem. 278, 1817– 1823 [DOI] [PubMed] [Google Scholar]
- 26.Hoyt M. A., Coffino P. ( 2004) Cell Mol Life Sci. 61, 1596– 1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sadeh R., Breitschopf K., Bercovich B., Zoabi M., Kravtsova-Ivantsiv Y., Kornitzer D., Schwartz A., Ciechanover A. ( 2008) Proc. Natl. Acad. Sci. U. S. A. 105, 15690– 15695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tintignac L. A., Lagirand J., Batonnet S., Sirri V., Leibovitch M. P., Leibovitch S. A. ( 2005) J. Biol. Chem. 280, 2847– 2856 [DOI] [PubMed] [Google Scholar]
- 29.Rodway H., Llanos S., Rowe J., Peters G. ( 2004) Oncogene 23, 6186– 6192 [DOI] [PubMed] [Google Scholar]
- 30.Pollice A., Vivo M., La Mantia G. ( 2008) FEBS Lett. 582, 3257– 3262 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.