Abstract
Dendritic cells (DCs) are professional antigen presenting cells to initiate immune response against pathogens, but mechanisms controlling the maturation of DCs are unclear. Here we report that, in the absence of recombination signal binding protein-Jκ (RBP-J, the transcription factor mediating Notch signaling), lipopolysaccharide-stimulated monocyte-derived DCs are arrested at a developmental stage with few dendrites, low major histocompatibility complex II (MHC II) expression, and reduced motility and antigen presentation ability. RBP-J null DCs had lower expression of CXCR4. Transduction with a CXCR4-expressing lentivirus rescued developmental arrest of RBP-J-deficient DCs. Activation of Notch signaling in DCs up-regulated CXCR4 expression and increased the outgrowth of dendrites and the expression of MHC II. These effects were abrogated by a CXCR4 inhibitor. Therefore, Notch signaling is essential for DCs to transit from a dendritelowMHC IIlow immature state into a dendritehighMHC IIhigh mature state, during the lipopolysaccharide-induced DC maturation, most likely through the up-regulation of CXCR4.
Dendritic cells (DCs)4 are professional antigen-presenting cells that initiate specific immune responses against pathogens such as bacteria, viruses, fungi, and parasites (1). Although several types of DCs participate in host resistance to microbial infections (2), myeloid DCs play a primary role to reject microbes invading into tissues. The precursors of myeloid DCs move to the sites of infection using signaling through chemotactic receptors. Immature DCs, which are offspring of DC precursors, recognize microbial structures (pathogen-associated molecular patterns) using pattern recognition receptors (3) such as Toll-like receptors, and capture antigens by phagocytosis, micropinocytosis, and endocytosis. Antigen uptake triggers the maturation of DCs, which is characterized by decreased endocytosis capacity, up-regulation of MHC II and co-stimulatory molecules, outgrowth of dendrites, and modulation of chemokine receptor expression patterns to facilitate the migration of antigen-loaded DCs to the T-cell zone in local lymph nodes (4). These changes finally cause DCs to prime certain types of T-cell responses against the invading microbes. However, the mechanisms controlling DC maturation have not been fully understood.
The Notch signaling pathway is an evolutionarily highly conserved pathway that mediates direct cell-cell interaction and signaling (5). Both Notch receptors and ligands are type I transmembrane proteins. When Notch receptors are triggered by ligands on neighboring cells, the intracellular domain of Notch receptors, or NIC, is released upon intra-membrane proteolysis executed by a γ-secretase-containing proteinase complex. NIC then translocates into the nuclear and associates with the DNA-binding protein recombination signal binding protein-Jκ (RBP-J) (6, 7). This protein-protein interaction transactivates downstream genes associated with cell differentiation, proliferation, and apoptosis. Although four Notch receptors have been identified in mammals, it is believed that RBP-J mediates the transcriptional activation of all four Notch receptors (8).
Notch signaling plays an important role in DC genesis. Cheng et al. (9) showed that Notch1-deficient embryonic stem cells or hematopoietic progenitor cells were unable to differentiate into DCs. These authors further showed that different Notch ligands exhibited different effects on DC differentiation: Delta-like1 promoted the generation of fully differentiated DCs, whereas Jagged1 stimulated the accumulation of DC precursors but prevented their transition into terminally differentiated DCs (10). Other studies have shown roughly consistent findings showing that Notch signaling favors DC generation. For example, Weijzen et al. (11) reported that Jagged1 was able to induce the maturation of monocyte-derived human DCs, whereas Ohishi et al. showed that Delta-like1 inhibits the differentiation of monocytes into macrophages but permits their differentiation into DCs (12, 13). Recently, using a conditional knockout mouse of RBP-J and a DC-specific Cre transgenic mouse (CD11c-Cre), Caton et al. found that the deletion of RBP-J in DCs did not preclude DC lineage commitment, but could result in the reduction of spleen DC fraction, and specifically, could lead to the loss of spleen CD8− DCs (14). In addition, Notch signaling has also been implicated in the development of plasmacytoid DCs (15, 16).
Mouse bone marrow (BM) cells can generate DCs when cultured in the presence of granulocyte macrophage-colony stimulating factor (GM-CSF) and interleukin (IL)-4, mimicking the process of differentiation of monocytes into DCs (17–20). Winzler et al. have divided this differentiation process into three stages: immature stage (stage 1), mature stage (stage 2), and apoptotic stage (stage 3) (21). In this study, using this system, we show that lipopolysaccharide (LPS)-induced maturation of monocyte-derived DCs is regulated by the Notch signaling through the up-regulation of CXCR4.
EXPERIMENTAL PROCEDURES
Mice
Mice were maintained in a specific pathogen-free environment. The RBP-J-floxed mouse was described previously (22). For the induction of RBP-J deletion, RBP-J-floxed mice and Mx-Cre mice were mated. One-month-old mice with suitable genotypes were induced with poly(I)-poly(C) as described (22). Two months after the first injection, mice were used for further analysis. All animal experiments, including maintenance, injection, BM transplantation, and so on, were approved by and performed in accordance with a guideline from the Animal Experiment Administration Committee of the Fourth Military Medical University, to comply with the international humanitarian standards.
Flow Cytometry
Mice were sacrificed humanly by cervical dislocation. Single cell suspensions were prepared from cultured cells, or collected from mouse tissues by passing minced tissues through a stainless mesh, followed by treatment with buffered 0.14 m NH4Cl. Cells (3–5 × 105) were stained with antibodies for 30 min on ice, and were analyzed using a FACSCaliburTM (BD Immunocytometry Systems, San Jose, CA). Data were analyzed using the CellQuestTM software. Dead cells were excluded by propidium iodide staining. Anti-mouse-CD11c-FITC (N418), anti-mouse-CD11c-PE (HL3), biotinylated anti-mouse-I-Ab (KH174), biotinylated anti-mouse-CD184 (CXCR4, 2B11), and streptavidin-antigen-presenting cells were from BD Pharmingen (San Diego, CA). Biotinylated anti-mouse CCR7 antibody was purchased from eBioscience (San Diego, CA).
Production of Recombinant Human Delta-like1 (hDll1)
The Delta-Serrate-Lag2 (DSL) domain (amino acids 127–225) of hDll1 was inserted in-frame into the polyclonal site of pET32a(+) (Novagen, Darmstadt, Germany), which contains 326-bp Trx coding sequence before a His tag, by routine DNA recombination technology. Production of the recombinant Trx-His-hDll1DSL (described as hDll1DSL hereafter) and the Trx-His protein (Trx, as a control) was performed following the protocol from Novagen, and the biological activity of hDll1DSL was determined essentially as described (23).
RT-PCR
DCs were sorted from BM cells using anti-CD11c magnetic beads (Miltenyi Biotec GmbH, Germany) following the recommended protocol. Sorted or cultured DCs (see below) were disrupted in the TRIzol reagent (Invitrogen), and total cellular RNA was prepared according to the manufacturer's instructions. cDNA was prepared from the total RNA using a reverse-transcription kit from TOYOBO (Osaka, Japan). Real-time PCR was performed using a kit (SYBR Premix EX Taq, TaKaRa) and the ABI PRISM 7300 Real-Time PCR system, with β-actin as a reference control. Primers used in real-time PCR were as follows: CXCR4-F, 5′-GTTGCCATGGAACCGATCA; CXCR4-R, 5′-TGCCGACTATGCCAGTCAAGA; β-actin-F, 5′-CATCCGTAAAGACCTCTATGCCAAC; and β-actin-R, 5′-ATGGAGCCACCGATCCACA.
Western Blot
Whole cell extracts were prepared by lysing cells with the RIPA buffer (50 mm Tris-HCl, pH 7.9, 150 mm NaCl, 0.5 mm EDTA, and 0.5% Nonidet P-40, 0.1 mm phenylmethylsulfonyl fluoride). Proteins were separated by 12% SDS-PAGE and were electroblotted onto polyvinylidene difluoride membrane. Membranes were probed using rabbit-anti-mouse CXCR4 (Thermo Fisher Scientific, Fremont, CA), monoclonal anti-tubulin (Tu-02, Santa Cruz Biotechnology), or monoclonal anti-β-actin (AC-74, Sigma) at appropriate dilutions, followed by incubation with horseradish peroxidase-conjugated secondary anti-rabbit or anti-mouse IgG antibody (Sigma). Blots were developed using an enhanced chemiluminescence system (Roche Applied Science).
Scanning Electron Microscope
Cells grown on coverslips were fixed in 3% glutaraldehyde overnight at 4 °C. Cells were then routinely osmicated and dehydrated. Samples were incubated in hexamethyldisilazane, mounted on stubs, splutter-coated with gold, and examined under a scanning electron microscope (SEM, S-3400N, Hitachi, Japan).
DC Culture
To culture DCs (24), mice were sacrificed humanely by cervical dislocation. BM was flushed from femurs and tibias, and single-cell suspension was obtained by passing through an 18-gauge needle and filtered with a nylon filter, followed by erythrolysis in buffered 0.14 m NH4Cl. Cells (2 × 106) were counted and cultured in 24-well plates in 1 ml of RPMI1640 medium containing 20% fetal calf serum and 2 mm l-glutamine, and supplemented with 20 ng/ml mouse GM-CSF and 10 ng/ml mouse IL-4 (Pepro Tech, Inc.) for the indicated times. LPS (1 μg/ml, Sigma) was added 12 h before the end of the culture. In some experiments, the recombinant hDll1DSL, GSI (γ-secretase inhibitor IX, Calbiochem), or a CXCR4 inhibitor AMD3100 (Sigma) was added at concentrations of 1.5 μg/ml, 75 μm, or 10 μg/ml, respectively.
Cell Migration Assays
Cultured DCs (2 × 105) were incubated with carboxyfluorescein diacetate succinimidylester (CFSE, Sigma) at 37 °C for 10 min, followed by incubating with 5 ml of ice-cold Dulbecco's modified Eagle's medium for 5 min on ice. Cells were washed with cold Dulbecco's modified Eagle's medium and were injected into the hind footpads of C57BL/6 mice. One day later, cells were collected from draining lymph nodes and then analyzed by FACS.
Chemotaxis experiments were performed in polycarbonate Transwell inserts (8-μm pore, Corning Costar Corp.). HeLa cells (2 × 105) transfected with pMikNeo-Fc-SDF1α or pMikNeo-Fc5 were planted in the lower chamber. DCs (1 × 105) were seeded on the next day in the upper compartment and were cultured at 37 °C overnight. Cells in the lower chamber were harvested and counted, and analyzed by FACS.
Contact Sensitization
Contact sensitization assay was conducted as described (25) by painting an FITC solution (10 mg in 1 ml of acetone dibutylphalate) on the ears of mice. The mandibular lymph node cells were collected 48 h later and analyzed by FACS.
Lentivirus
The coding region of the mouse CXCR4 cDNA was amplified by PCR from a mouse embryonic cDNA library using primers 5′-GCCATGGAACCGATCAGTGTGAG and 5′-TGCATAAGTGTTAGCTGGAGTG. The CXCR4 cDNA fragment was fused with an internal ribosome entry site-EGFP unit and was inserted into pLenti-EGFP (kindly provided by Dr. X.-B. Wu) to replace the EGFP gene and, thus, to generate pLenti-CXCR4. pLenti-EGFP and pLenti-CXCR4 were transfected into 293FT cells using Lipofectamine 2000TM, together with the packaging plasmids (Invitrogen), according to the manufacturer's protocols. Supernatants were collected 48 h after the transfection, and the virons were concentrated by ultracentrifugation at 70,000 × g, at 4 °C for 2 h. Pellets were resuspended in Dulbecco's modified Eagle's medium and stored at −80 °C. For infection, BM cells were co-cultured with the viron suspensions for 48 h, and then cultured in normal medium until further analysis.
Mixed Lymphocyte Reaction
BM-derived DCs (2 × 105), which were stimulated with LPS, were seeded in 24-well plates. Syngeneic T-cells (2 × 106) labeled with CFSE were mixed with irradiated allogenic BM-derived DCs, and were added to each well. Five days later, the proliferation of T-cells was detected by FACS.
In Vivo Antigen Presentation Assay
Escherichia coli (2 × 109) was lysed by ultrasound. The supernatant was collected after centrifugation, and was used as E. coli super-antigens. Mice were injected in the left hind footpad with 5 × 105 BM-derived DCs pulsed with the E. coli super-antigens. Meanwhile, BrdUrd (0.6 mg) was administered by intraperitoneal injection every 6 h. Mice were sacrificed 3 day later, and draining lymph nodes were harvested. CD3+ T-cells were stained using FITC-labeled anti-BrdUrd antibody (Sigma), and were analyzed by FACS.
Delayed-type Hypersensitivity Assay
Mice were injected at hind footpads with 5 × 105 E. coli antigen-pulsed DCs. Seven days after the initial DC injection, the mice were challenged by injection of 50 μl of E. coli super-antigen into one rear footpad, while the other rear footpad received 50 μl of phosphate-buffered saline. Twenty four hours later, footpad swelling was measured using a Vernier caliper. The magnitude of the DTH responses was determined by the footpad thickness.
Reporter Assay
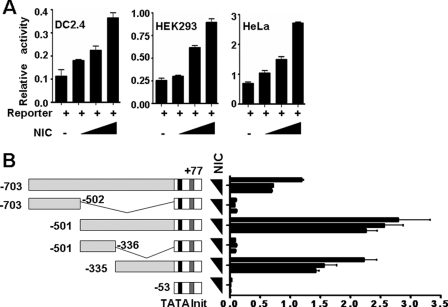
The 5′ flanking sequence (−703∼+77) of the mouse CXCR4 gene was amplified by PCR from mouse genomic DNA. The resulting fragment was inserted into pGL3-basic, to generate a reporter construct pGL-CXCR4. Different truncates of the 5′ flanking fragment, as depicted in Fig. 7B, were also generated by PCR, and fused with the basic promoter (−53∼+77) of CXCR4 to generate different truncated reporter constructs. DC2.4, HEK293, and HeLa cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and 2 mm l-glutamine. Cells (2 × 104) were transfected with 0.1 μg of reporter construct, different amounts of pEFBOS-NIC that expresses NIC, and 5 ng of Renilla luciferase vector (phRL-TK, Promega, Madison, WI) using Lipofectamine 2000TM (Invitrogen). Forty-eight hours after the transfection, luciferase activity was assessed using Luminoskan Ascent (Labsystems, Helsinki, Finland) and a Dual-Luciferase Reporter Assay Kit (Promega) according to the manufacturer's protocol. All luciferase activity was normalized to Renilla luciferase activity. NF-κB inhibitor (IκB kinase inhibitor peptide), phosphatidylinositol 3-kinase inhibitor (Ly294002), MEK inhibitor (PD98059), and p38 inhibitor (SB203580) were all from Calbiochem.
FIGURE 7.
The CXCR4 promoter was activated by Notch-RBP-J signaling. A, reporter assay. DC2.4, HEK293, and HeLa cells were transfected with pGL-CXCR4 and increasing amounts (0, 50, 100, and 150 ng) of pEFBOS-NIC (NIC). Cells were collected 48 h after the transfection, and the luciferase activity in lysates was assayed. B, different truncates of the mouse CXCR4 promoter were fused with the basic promoter and luciferase gene, and were used for reporter assay as in A. The amounts of pEFBOS-NIC added were 0, 100, and 150 ng. Bars represent means ± S.D. (n = 5).
Statistics
Statistical analysis was performed with the SPSS 12.0 program. Results were expressed as means ± S.D. Comparisons between groups were undertaken using unpaired Student's t test. p < 0.05 was considered statistically significant.
RESULTS
RBP-J Deletion Interrupts Dendrite Outgrowth and MHC II Up-Regulation during LPS-induced DC Maturation
To examine the role of Notch signaling in DC maturation, we employed homozygous RBP-J-floxed mice bearing Mx-Cre transgene (RBP-J−/−), with heterozygous mice (RBP-J+/−) as controls (22). We cultured BM cells from the poly(I)/poly(C)-induced RBP-J knockout and control mice, in the presence of GM-CSF and IL-4, and stimulated DC maturation with LPS (24). RBP-J−/− BM cells produced significantly less CD11c+ DCs (supplemental Fig. S1) after being cultured for 9 days. No differences in cell apoptosis and proliferation during the culture were found between the RBP-J knockout and control cells (data not shown).
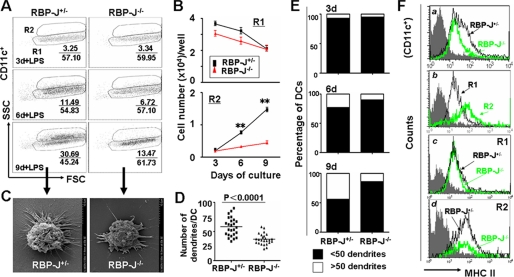
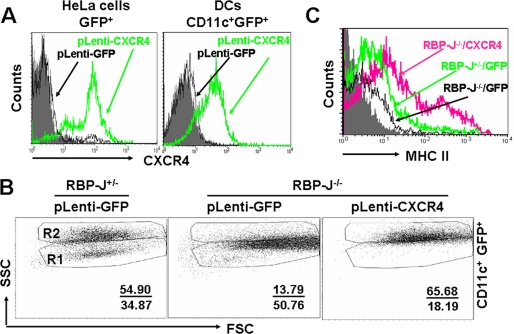
When RBP-J+/− and RBP-J−/− DCs (CD11c+) were stimulated with LPS and were plotted for forward scattering and side scattering (SSC), we found that RBP-J−/− DCs were deficient in a population of cells with high SSC (R2 in Fig. 1A). As shown in Fig. 1 (A and B), on the 3rd day of the culture, RBP-J+/− and RBP-J−/− BM cells generated similar number of DCs with low SSC (R1). On the 6th and 9th days of the culture, LPS induced an increase of SSChigh DCs derived from RBP-J+/− BM, with a concomitant decrease of SSClow DCs. In contrast, however, LPS stimulation did not induce the increase of SSChigh DCs derived from RBP-J−/− BM, although SSClow DCs decreased as in RBP-J+/− BM culture.
FIGURE 1.
RBP-J-deficient DCs showed maturation defects upon LPS stimulation. A, cells (2 × 106) were obtained from BM and cultured in 24-well plates in the presence of GM-CSF and IL-4. Aliquots of cells were stimulated with LPS for 12 h on day 3, 6, and 9 of the culture, and were analyzed by FACS. B, number of CD11c+ DCs with low SSC (R1) and high SSC (R2). The number of cells per well in R1 and R2 in A was calculated and compared between RBP-J knockout and control mice. C, typical appearance of DCs from RBP-J knockout and control mice under an SEM. D, comparison of dendrite number between DCs from RBP-J knockout and control mice. DCs were cultured as above, and the number of dendrites of each DC was counted under an SEM. The average dendrite number was compared between RBP-J knockout and control DCs (n = 25). E, RBP-J knockout and control cells were cultured as in Fig. 1A. DCs with >50 dendrites and DCs with <50 dendrites were counted under an SEM. F, LPS-stimulated DCs were analyzed by FACS using anti-MHC II. The result represents three independent experiments. Bars represent means ± S.D. (n = 5). **, p < 0.01.
SSC reflects cell irregularity, which might correlate with the outgrowth of dendrites of DCs. We then examined dendrite outgrowth of normal DCs stimulated with LPS. DCs with more than 50 dendrites and DCs with less than 50 dendrites were counted under an SEM. Indeed, as shown in supplemental Fig. S2A, LPS induced dendrite outgrowth: the number of DCs with more than 50 dendrites increased while the number of DCs with less than 50 dendrites decreased. This tendency of dendrite outgrowth correlated well with the change in SSC: LPS stimulation increased DCs with high SSC (R2) and decreased DCs with low SSC (R1) (supplemental Fig. S2B), suggesting that a transition of SSClow DCs into SSChigh DCs might reflect the increase of dendrites. We compared RBP-J−/− DCs with RBP-J+/− DCs under an SEM. As shown in Fig. 1C, a typical LPS-stimulated DC had a stellate appearance with many dendrites. However, most RBP-J−/− DCs lost their dendrites, as demonstrated by counting dendrites under an SEM (Fig. 1D). Direct counting and comparison of the number of dendrites of RBP-J+/− and RBP-J−/− DCs on different culturing days led to a similar conclusion (Fig. 1E). These data suggested that RBP-J might be essential for the maturation of DCs in terms of dendrite outgrowth, upon LPS stimulation.
In addition to dendrite outgrowth, the expression of MHC II molecules is another hallmark of DC maturation. We compared the expression of MHC II between RBP-J−/− and RBP-J+/− DCs. RBP-J−/− DCs had lower MHC II expression than RBP-J+/− DCs (Fig. 1F, panel a). A detailed analysis suggested that RBP-J+/− DCs could be divided into two populations, one with low MHC II and the other with high MHC II. In RBP-J−/− DCs, however, only the MHC II-low population was detected (Fig. 1F, panel a). To confirm that the low expression of MHC II on RBP-J−/− DCs was due to the lack of MHC IIhigh sub-population, we compared MHC II expression between DCs with low SSC and with high SSC (R1 and R2 in Fig. 1A, respectively) in the control (RBP-J+/−). The result showed that on day 9 of the culture, LPS-stimulated DCs in R2 had a significantly higher level of MHC II expression compared with DCs in R1 (Fig. 1F, panel b). When CD11c+ DCs in R1 and R2 were gated, and their MHC II expression was compared between RBP-J−/− and RBP-J+/− DCs, we failed to detect significant difference in MHC II expression between RBP-J−/− and RBP-J+/− DCs in either R1 or R2 (Fig. 1F, panels c and d). Therefore, the deletion of RBP-J might block DC maturation at a state with low dendrites and low MHC II. However, other DC maturation-related molecules, such as CD80 and CD86, did not changed significantly upon RBP-J deletion (data not shown).
RBP-J Disruption Blocks the Up-regulation of CXCR4 during LPS-induced DC Maturation
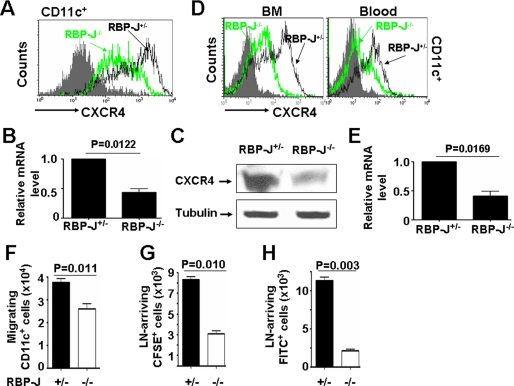
Pathogen-triggered DC maturation is accompanied by modified expression of chemokine receptors. In RBP-J-deleted DCs, whereas the expression of CCR7 was not damaged (data not shown), the expression CXCR4 (CD184) was significantly reduced upon LPS stimulation when compared with the control (Fig. 2A). At the mRNA level, quantitative RT-PCR (Fig. 2B) demonstrated that knockout of RBP-J down-regulated CXCR4 expression in DCs. This was further confirmed by Western blot analysis of cell lysates of RBP-J−/− and RBP-J+/− DCs with anti-CXCR4 antibody, which showed that RBP-J−/− DCs had lower level of CXCR4 (Fig. 2C). We further examined the expression of CXCR4 on DCs freshly isolated from RBP-J−/− mice. FACS analysis showed that, in BM and the peripheral blood of RBP-J−/− mice, CXCR4 expression was significantly reduced in CD11c+ DCs (Fig. 2D). When DCs were gated using another DC marker, CD205, we found that CD205+ cells from RBP-J−/− mice also showed reduced expression of CXCR4, as compared with the control (data not shown). We again compared the mRNA level of CXCR4 between magnetically sorted RBP-J−/− and RBP-J+/− DCs using quantitative real-time RT-PCR. Both experiments showed that CD11c+ DCs from RBP-J−/− mice had lower level of CXCR4 mRNA than the RBP-J+/− control (Fig. 2E). These results led to a conclusion that, in RBP-J knockout mice, the expression of CXCR4 was reduced in DCs.
FIGURE 2.
RBP-J knockout reduced the expression of CXCR4 in DCs. A, BM-derived DCs were stimulated with LPS and were analyzed by FACS using anti-CD11c plus anti-CD184 (CXCR4). B, total RNA was prepared from DCs, reverse-transcribed, and then analyzed by real-time PCR for relative levels of CXCR4 mRNA, with β-actin as a reference control. C, Western blot. Total cell lysates were prepared from cultured DCs and were subjected to Western blot analysis using an anti-CXCR4 antibody, with tubulin as a control. D, single cell suspension was prepared from BM and peripheral blood and was analyzed by FACS. E, real-time RT-PCR. Total RNA was prepared from magnetically isolated DCs and was analyzed as in B. F, cell migration assay. In vitro cultured DCs were seeded in the upper chamber of a Transwell culture system, with HeLa cells expressing SDF1α in the lower chamber. CD11c+ DCs migrating into the lower chamber were analyzed by FACS and cell counting. G, in vivo migration assay. DCs cultured from RBP-J knockout and control mice were labeled with CFSE and were injected into the hind foot pads of normal mice. Cells migrating into the draining lymph nodes (LN) were analyzed by FACS. H, contact sensitization assay. FITC solution was painted on mouse ears. CD11c+ FITC+ cells at the draining lymph nodes (LN) were analyzed by FACS 48 h later. The results represent three independent experiments. Bars represent means ± S.D. (n = 5).
Reduced CXCR4 expression will predict a reduced chemotactic migration capacity. We then examined the migration of LPS-stimulated RBP-J−/− and RBP-J+/− DCs in response to SDF1α, the ligand of CXCR4. In a Transwell culture system, in vitro cultured RBP-J−/− and control DCs were seeded in the upper chamber, and HeLa cells transfected with an SDF1α-expressing vector were cultured in the lower chamber. CD11c+ DCs in the lower chamber were counted 12 h after the starting of the co-culture. The result indicated that RBP-J−/− DCs had lower migrating ability in response to SDF-1α (Fig. 2F). We also studied in vivo migration of RBP-J−/− and RBP-J+/− DCs by injecting CFSE-loaded DCs into the hind footpads of normal mice, and subsequently examined DCs arriving at the draining lymph nodes by FACS. As shown in Fig. 2G, significantly less RBP-J−/− DCs arrived at the draining lymph nodes, compared with RBP-J+/− DCs. Moreover, we performed a contact-sensitization assay, which reflects the migration of mature DCs in vivo. The result showed that a similar fraction of CD11c+ DCs were loaded with FITC at the site of FITC painting (data not shown), but less FITC-loaded RBP-J−/− DCs migrated to the draining lymph nodes (Fig. 2H).
Reduced Antigen Presentation Ability of RBP-J-deficient DCs
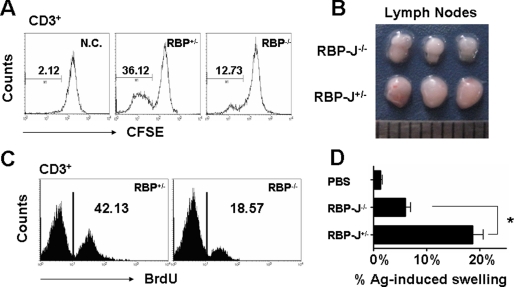
We accessed the immunological function of the RBP-J-deficient DCs using several methods. As shown in Fig. 3A, in mixed lymphocyte reaction assay, T-cells cultured with RBP-J−/− DCs, which were activated with LPS, showed remarkably less proliferation, compared with the control. To examine the in vivo antigen presentation capacity, normal mice were injected with Escherichia coli antigen-pulsed RBP-J+/− or RBP-J−/− DCs, and the proliferation of T-cells in the draining lymph nodes was examined (26, 27). The results showed that RBP-J−/− DCs stimulated significantly weaker T-cell proliferation response, as assessed by the size of draining lymph nodes (Fig. 3B) and BrdUrd incorporation by CD3+ T-cells in the draining lymph nodes (Fig. 3C). We also performed a DTH assay. The results showed that RBP-J−/− DCs invoked a weaker DTH than RBP-J+/− DCs (Fig. 3D). Therefore, RBP-J-deficient DCs had reduced antigen presentation capacity.
FIGURE 3.
RBP-J-deleted DCs showed reduced antigen presentation ability. A, mixed lymphocyte reaction. BM-derived DCs (2 × 105) were stimulated with LPS, and were co-cultured with syngeneic T-cells (2 × 106) loaded with CFSE and stimulated with irradiated allogenic BM-derived DCs. Five days later, the proliferation of T-cells was detected by FACS. N.C., negative control without stimulator. B and C, the hind footpads of normal mice were injected with DCs pulsed with E. coli super-antigens. The mice were injected with BrdUrd to label proliferating T-cells. Three days later, the draining lymph nodes were photographed (B), and T-cells in draining lymph nodes were analyzed for BrdUrd by FACS (C). The result represented three independent experiments. D, DTH assay. Mice were injected at hind footpads with E. coli antigen-pulsed DCs, and were re-challenged with E. coli super-antigen or phosphate-buffered saline 7 days later. Twenty-four hours after the re-challenge, footpad swelling was measured using a Vernier caliper. The magnitude of the DTH responses was determined by the differences in the thickness between the Ag- and phosphate-buffered saline-injected footpads. Bars represent means ± S.D. (n = 5). *, p < 0.05.
Overexpression of CXCR4 Rescues the Maturation Defects of RBP-J−/− DCs
CXCR4 was shown to influence DC differentiation (28–31). To decide whether lowered expression of CXCR4 accounted for the maturation arrest of RBP-J−/− DCs, we transduced the cultured DCs from BM of RBP-J+/− and RBP-J−/− mice with a lentivirus expressing the mouse CXCR4 and GFP, with virus expressing GFP only as a control. Virus-transduced HeLa cells and BM-derived DCs from normal mice showed the expression of GFP and the mouse CXCR4 (Fig. 4A). When CD11c+GFP+ cells were gated and compared, we found that, although the GFP expression did not influence the differentiation of either RBP-J+/− or RBP-J−/− DCs, the transduction of CXCR4-expressing vector rescued the dendrite outgrowth in RBP-J−/− DCs, as represented by DCs with high SSC (Fig. 4B). Moreover, the expression of CXCR4 also rescued MHC II expression in RBP-J−/− DCs (Fig. 4C). Therefore, RBP-J deletion-induced maturation arrest of DCs could be attributed to the down-regulation of CXCR4 expression.
FIGURE 4.
Overexpression of CXCR4 rescued the RBP-J deficiency-induced blockade of DC differentiation. A, overexpression of the mouse CXCR4 by lentivirus infection. HeLa cells or BM-derived DCs were infected with different virons, and the CXCR4 expression was assessed by FACS. B and C, RBP-J−/− and control BM cells were infected with lentivirus-expressing GFP (as control) or CXCR4 plus GFP, and were induced for DC differentiation as above. Cells were collected after 9-day culture and were analyzed by FACS. The data represent three independent experiments.
Notch Triggering Up-regulates CXCR4 Expression in DCs and Promotes DC Development
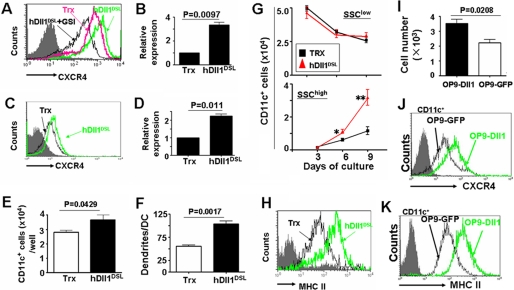
RBP-J mediates canonical Notch signaling. We generated a recombinant soluble truncated Notch ligand protein, hDll1DSL, which could trigger Notch signaling in a γ-secretase-dependent manner (Ref. 23 and data not shown). We compared the expression of CXCR4 on DCs cultured in the presence of hDll1DSL with those cultured with the control Trx. FACS analysis showed that DCs cultured with hDll1DSL had a mildly higher level of CXCR4 on their surface than DCs in the control culture (Fig. 5A). GSI, which blocks Notch signaling, abrogated hDll1DSL-induced CXCR4 up-regulation in DCs, suggesting that the hDll1DSL-induced up-regulation of CXCR4 was dependent on Notch activation. By quantitative real-time RT-PCR (Fig. 5B), we further showed that triggering of Notch receptors could up-regulate the expression of CXCR4 mRNA in DCs. Moreover, in a mouse DC line, DC2.4, hDll1DSL up-regulated the expression of CXCR4 on the cell surface and at the mRNA level, as shown by FACS analysis (Fig. 5C) and real-time RT-PCR (Fig. 5D), respectively.
FIGURE 5.
Notch stimulation up-regulated CXCR4 expression and promoted DC differentiation. A, BM cells of normal mice were cultured in the presence of GM-CSF and IL4, with recombinant hDll1DSL or Trx for 9 days. CXCR4 expression on DCs was assayed by FACS. B, total RNA was prepared from the in vitro cultured DCs in Fig. 5A, reverse-transcribed, and was analyzed by real-time PCR for CXCR4 mRNA with β-actin as a reference control. C, DC2.4 was cultured in the presence of recombinant hDll1DSL or Trx for 48 h and analyzed by FACS. D, total RNA was prepared from DC2.4 in Fig. 5C, reverse-transcribed, and analyzed by real-time PCR for CXCR4 mRNA, with β-actin as a reference control. E, BM cells of normal mice were cultured in the presence of GM-CSF and IL4, with recombinant hDll1DSL or Trx for 9 days. CD11c+ DCs were counted by cell counting and FACS analysis. F, the number of dendrites of DCs cultured in the presence of hDll1DSL or Trx. DCs were cultured in vitro as above, and were examined on day 9 under an SEM. The number of dendrites was counted and compared (n = 20). G, SSC change of CD11c+ DCs during their differentiation in the presence of hDll1DSL or Trx. The number of CD11c+ DCs with low SSC (in R1) and high SSC (in R2) at different time points was calculated and compared between cultures with hDll1DSL or Trx. H, MHC II expression in DCs cultured in the presence or absence of hDll1DSL. DCs were cultured in vitro as above, collected on day 9, and analyzed by FACS. I, OP9-Dll1 or OP9-GFP cells (2 × 105) were seeded in 24-well plates. BM cells (2 × 106) were then seeded and co-cultured in the presence of GM-CSF and IL4 for 9 days, and stimulated with LPS 12 h before the end of the culture. The number of DCs was analyzed by FACS and cell counting. J and K, CXCR4 and MHC II expression was analyzed by FACS. The result represents three independent experiments. Bars represent means ± S.D. (n = 5). *, p < 0.05; **, p < 0.01.
We then examined whether triggering of Notch signaling might promote DC maturation through increasing dendrite outgrowth and MHC II expression. When BM cells from normal mice were cultured in the presence of GM-CSF and IL-4, and stimulated with LPS, inclusion of hDll1DSL could increase the production of DCs (Fig. 5E). We also found that, in the presence of hDll1DSL, DCs grew more dendrites than DCs cultured with the control tag peptide (Fig. 5F). This was supported by monitoring dendrite outgrowth of CD11c+ DCs by SSC (Fig. 5G): a time course analysis of cell number with SSClow and SSChigh showed that, although the number of CD11c+ DCs with SSClow did not show much difference, the number of DCs with SSChigh in hDll1DSL-stimulated culture was significantly higher than DCs in the control culture after 9-day culture. These results suggested that triggering Notch receptors mainly increased DCs with more dendrites. Staining of surface MHC II expression showed that triggering of Notch receptors with hDll1DSL increased the expression of MHC II on the surface of DCs (Fig. 5H).
Finally, we co-cultured normal BM cells with OP9 cells expressing membrane-anchored hDll1 (OP9-hDll1) in the presence of GM-CSF and IL4, with OP9-GFP as a control (32). DC maturation was stimulated with LPS as above. As shown in supplemental Fig. S3 and Fig. 5I, co-culture with OP9-Dll1 generated higher percentage and number of DCs than co-culture with the control. The expression of CXCR4 (Fig. 5J) and MHC II (Fig. 5K) on DCs cultured on OP9-Dll1 were also up-regulated, compared with the control. These results further confirmed that Notch triggering promoted DC maturation.
A CXCR4 Inhibitor Abrogates Notch Activation-induced DC Maturation
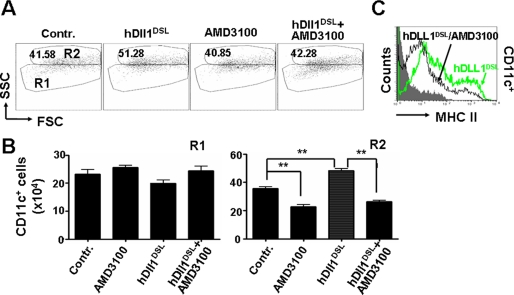
We next examined the effect of an inhibitor of CXCR4 on LPS-induced DC maturation in the presence of Notch activation. DCs were cultured from BM cells of normal mice in the presence of hDll1DSL, which promoted LPS-induced DC differentiation, as shown in Fig. 6A and described above. A CXCR4 inhibitor significantly blocked the dendrite outgrowth (Fig. 6, A and B), as well as the up-regulation of MHC II (Fig. 6C), induced by hDll1DSL. These results further confirmed that the induction of DC maturation by Notch activation could be dependent on CXCR4 activity.
FIGURE 6.
A CXCR4 inhibitor abrogated Notch activation-promoted DC maturation. DCs were cultured from normal BM cells in the presence or absence of hDll1DSL. The CXCR4 inhibitor AMD3100 was added as indicated. Cells were cultured for 9 days and analyzed by FACS. A, CD11c+ DCs were plotted for SSC and forward scattering. B, the number of CD11c+ DCs in R1 and R2 was calculated and compared. C, expression of MHC II was analyzed by FACS. Bars represent means ± S.D. (n = 5). **, p < 0.01.
Notch Signaling Activates CXCR4 Promoter
To examine whether Notch signaling could activate the CXCR4 promoter, we cloned the −703∼+77 fragment of the 5′-flanking sequence of the mouse CXCR4 gene by PCR. The fragment was inserted into the pGL3-basic vector to generate pGL-CXCR4. We co-transfected DC2.4, HEK293, or HeLa cells with pGL-CXCR4 and pEFBOS-NIC (the latter expresses NIC) and examined the transactivation of the mouse CXCR4 promoter by NIC using luciferase assay.TM As shown in Fig. 7A, in all three types of cells, co-transfection of the NIC-expressing vector induced the expression of luciferase from pGL-CXCR4 in a dose-dependent manner, suggesting that NIC could transactivate the CXCR4 promoter. To further identify the cis-element that responds to Notch signaling, we truncated the CXCR4 promoter. Reporter assay confirmed that one or more elements responding to Notch signaling were located in the −335∼−54 fragment (Fig. 7B). These results indicated that, although the exact cis-element(s) is/are unclear, the CXCR4 promoter could be transactivated directly or indirectly by Notch signaling.
DISCUSSION
Previous studies have shown that Notch signaling is not essential for lineage commitment of DCs (14). Consistently, we found that, when BM cells from RBP-J knockout mice were cultured in vitro in the presence of GM-SCF and IL-4, a substantial fraction of DCs could be generated, although at a less efficiency in terms of DC number. We further characterized phenotypic changes of these RBP-J null DCs and found that RBP-J-deficient DCs possess significantly less dendrites, lower level of surface MHC II molecules, and reduced capacity of migration and antigen presentation both in vitro and in vivo. On the other hand, cultured DCs with activated Notch signaling could increase the dendrite outgrowth and MHC II expression, indicating that Notch triggering promoted DC maturation. By monitoring the dynamic changes during DC differentiation, we found that the deficiency of RBP-J-mediated Notch signaling arrested LPS-induced DC maturation at a stage before the outgrowth of dendrites and the up-regulation of MHC II molecules, while triggering of Notch signaling promoted DC differentiation by passing through this checkpoint. Because no significant change in proliferation and apoptosis between RBP-J−/− and RBP-J+/− DCs were detected (data not shown), our results suggested that Notch signaling might be essential for the LPS-induced DC maturation.
Trafficking of DCs is closely related to their differentiation and function. Our culture system mimics the differentiation from pre-DCs, specifically from monocytes, to DCs. Both Notch ligands and receptors are cell surface-anchored proteins, and Notch signaling therefore mediates direct cell-cell communication. It would be desirable to elucidate where DC precursors receive the activation signal for Notch receptors during their differentiation. One possible cell type that provides Notch ligands for DC precursors may be stromal cells such as those within BM. Diao et al. (33) showed that DC-committed precursor populations in BM is comprised of two distinct subsets according to their B220 expression, and those DC precursors exhibited different differentiation pathways. A Notch ligand-receptor interaction might take place between BM stromal cells and DC precursor to facilitate DC maturation, as shown by Cheng et al. (10). Another potential site that DC precursors receive Notch activation might be during the trans-endothelial trafficking. Randolph et al. showed that monocytes cultured on endothelial cells differentiated into DCs within 2 days (34). Because endothelial cells express high level of Notch ligands, including Dll1, it is likely that monocytes receive Notch ligand stimulation as their trafficking through endothelial cells. Experiments are now under way to test this hypothesis.
We have tried to unveil the potential molecular mechanisms underlying the Notch signaling-mediated differentiation transition of DCs. We obtained a series of evidence showing that the chemokine receptor CXCR4 is downstream to canonical Notch signaling, either directly or indirectly, and is responsible for Notch signaling-mediated differentiation transition of DCs. Our results from RT-PCR, Western blotting, and FACS analysis using in vitro cultured DCs, freshly isolated DCs, as well as a DC line, have shown that the expression of CXCR4 is down-regulated in RBP-J-deficient DCs and up-regulated in a γ-secretase-dependent manner when DCs were stimulated with Notch ligands. By using virus-mediated overexpression, we found that the forced expression of CXCR4 rescued RBP-J deficiency-induced differentiation block of DCs. Furthermore, Notch triggering-induced DC differentiation is blocked by a specific inhibitor of CXCR4. These results established a new signaling axis from Notch receptor to chemokine receptor CXCR4, which is critical for DC differentiation. We have tried to find out the mechanism of Notch-mediated up-regulation of CXCR4. Although our reporter assays supported that Notch signaling activates the CXCR4 promoter, this transactivation is most likely to be indirect. In the −703∼+77 promoter fragment of CXCR4, which is transactivated by NIC, there is no consensus RBP-J binding site. The Notch signaling-mediated up-regulation of CXCR4 might not depend on NF-κB (35), because the blocking of NF-κB by a specific inhibitor did not abrogate up-regulation of CXCR4 in DCs by hDll1DSL. Moreover, Notch signaling might not up-regulate CXCR4 expression through p38, MEK, or phosphatidylinositol 3-kinase pathways, because inhibitors of these pathways did not abrogated Notch signaling-stimulated CXCR4 up-regulation (supplemental Fig. S4). Given the fact that Notch signaling and CXCR4 signaling have numerous overlapping functions in the regulating development of immune system, central nervous system, as well as stem cells, it is not surprising that sophisticated mechanisms might be involved in the regulation of CXCR4 by Notch signaling. Recently, Williams et al. (36) reported that in human endothelial cells and glioblastoma cells, overexpression of the human Notch ligand Dll4 results in the down-regulation of human CXCR4 expression and the repression of a human CXCR4 reporter construct. Further biochemical analysis is now under way to identify the cis- and trans-element(s) as well as regulation mechanism(s) involved in the Notch-mediated transcriptional regulation of the CXCR4 promoter.
As the strongest professional antigen-presenting cells, DCs are potentially useful in therapies of many human diseases, including viral infections and cancer. Promoting DC maturation is highly desirable in DC-involved cell therapies, because immature DCs are actually immunorepressive. Our results reported in this study have shown that Notch signaling is essential for the maturation of at least some populations of DCs, and triggering Notch receptors can promote dendrite outgrowth and MHC II expression on DCs. Both dendrite outgrowth and MHC II expression are essential for DC migration, antigen uptake, and/or antigen presentation. Therefore our findings might have important therapeutic implications in promoting DC maturation in vitro and in applying DCs in cell therapies.
Supplementary Material
Acknowledgments
We thank J. C. Zuniga-Pflucker for OP9-Dll1 and OP9-GFP and Dr. X.-B. Wu for lentivirus vectors.
This work was supported by the National Natural Science Foundation (Grants 30600544, 30830067, 30801050, and 30800454) and the Ministry of Science and Technology of China (Grants 2006AA02A111, 2009CB521706, and 2007AA02Z441).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
Y.-C. Wang, P. Zhang, and H. Han, unpublished observation.
- DCs
- dendritic cells
- RT
- reverse transcription
- MLR
- mixed lymphocyte reaction
- DTH
- delayed-type hypersensitivity
- SEM
- scanning electron microscope
- GSI
- γ-secretase inhibitor
- LPS
- lipopolysaccharide
- FACS
- fluorescence-activated cell sorting
- SSC
- side-scattering counter
- RBP-J
- recombination signal binding protein-Jκ
- BM
- bone marrow
- CFSE
- carboxyfluorescein diacetate succinimidylester
- NIC
- Notch intracellular domain
- MHC
- major histocompatibility complex
- Dll
- Delta-like
- DSL
- Delta-Serrate-Lag2
- IL
- interleukin
- GM-CSF
- granulocyte macrophage-colony stimulating factor
- NF-κB
- nuclear factor κ-light-chain-enhancer of activated B cells
- MEK
- mitogen-activated protein kinase/extracellular signal-regulated kinase
- FITC
- fluorescein isothiocyanate
- GFP
- green fluorescent protein
- EGFP
- enhanced GFP
- BrdUrd
- bromodeoxyuridine
- Trx
- thioredoxin.
REFERENCES
- 1.Moll H. ( 2003) Cell. Microbiol. 5, 493– 500 [DOI] [PubMed] [Google Scholar]
- 2.Randolph G. J., Ochando J., Partida-S'anchez S. ( 2008) Annu. Rev. Immunol. 26, 293– 316 [DOI] [PubMed] [Google Scholar]
- 3.Janeway C. A., Jr., Medzhitov R. ( 2002) Annu. Rev. Immunol. 20, 197– 216 [DOI] [PubMed] [Google Scholar]
- 4.Ardavín C., Martínez del Hoyo G., Martín P., Anjuére F., Arias C. F., Marín A. R., Ruiz S., Parrillas V., Hernández H. ( 2001) Trends Immunol. 22, 691– 700 [DOI] [PubMed] [Google Scholar]
- 5.Artavanis-Tsakonas S., Rand M. D., Lake R. J. ( 1999) Science 284, 770– 776 [DOI] [PubMed] [Google Scholar]
- 6.Jarriault S., Brou C., Logeat F., Schroeter E. H., Kopan R., Israel A. ( 1995) Nature 377, 355– 358 [DOI] [PubMed] [Google Scholar]
- 7.Tamura K., Taniguchi Y., Minoguchi S., Sakai T., Tun T., Furukawa T., Honjo T. ( 1995) Curr. Biol. 5, 1416– 1423 [DOI] [PubMed] [Google Scholar]
- 8.Honjo T. ( 1996) Genes Cells 1, 1– 9 [DOI] [PubMed] [Google Scholar]
- 9.Cheng P., Nefedova Y., Miele L., Osborne B. A., Gabrilovich D. ( 2003) Blood 102, 3980– 3988 [DOI] [PubMed] [Google Scholar]
- 10.Cheng P., Nefedova Y., Corzo C. A., Gabrilovich D. I. ( 2007) Blood 109, 507– 515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weijzen S., Velders M. P., Elmishad A. G., Bacon P. E., Panella J. R., Nickoloff B. J., Miele L., Kast W. M. ( 2002) J. Immunol. 169, 4273– 4278 [DOI] [PubMed] [Google Scholar]
- 12.Ohishi K., Varnum-Finney B., Flowers D., Anasetti C., Myerson D., Bernstein I. D. ( 2000) Blood 95, 2847– 2854 [PubMed] [Google Scholar]
- 13.Ohishi K., Varnum-Finney B., Serda R. E., Anasetti C., Bernstein I. D. ( 2001) Blood 98, 1402– 1407 [DOI] [PubMed] [Google Scholar]
- 14.Caton M. L., Smith-Raska M. R., Reizis B. ( 2007) J. Exp. Med. 204, 1653– 1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dontje W., Schotte R., Cupedo T., Nagasawa M., Scheeren F., Gimeno R., Spits H., Blom B. ( 2006) Blood 107, 2446– 2452 [DOI] [PubMed] [Google Scholar]
- 16.Olivier A., Lauret E., Gonin P., Galy A. ( 2006) Blood 107, 2694– 2701 [DOI] [PubMed] [Google Scholar]
- 17.León B., Marténez del Hoyo G., Parrillas V., Vargas H. H., Sánchez-Mateos P., Longo N., López-Bravo M., Ardavín C. ( 2004) Blood 103, 2668– 2676 [DOI] [PubMed] [Google Scholar]
- 18.León B., Lopez-Bravo M., Ardavin C. ( 2005) Semin. Immunol. 17, 313– 318 [DOI] [PubMed] [Google Scholar]
- 19.Fogg D. K., Sibon C., Miled C., Jung S., Aucouturier P., Littman D. R., Cumano A., Geissmann F. ( 2006) Science 311, 83– 87 [DOI] [PubMed] [Google Scholar]
- 20.Varol C., Landsman L., Fogg D. K., Greenshtein L., Gildor B., Margalit R., Kalchenko V., Geissmann F., Jung S. ( 2007) J. Exp. Med. 204, 171– 180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Winzler C., Rovere P., Rescigno M., Granucci F., Penna G., Adorini L., Zimmermann V. S., Davoust J., Ricciardi-Castagnoli P. ( 1997) J. Exp. Med. 185, 317– 328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han H., Tanigaki K., Yamamoto N., Kuroda K., Yoshimoto M., Nakahata T., Ikuta K., Honjo T. ( 2002) Int. Immunol. 14, 637– 645 [DOI] [PubMed] [Google Scholar]
- 23.Shi Z. X., He F., Wang L. L., Liang Y. M., Han H., Wang C. Z., Zhao Q., Geng X. D. ( 2008) Protein Expr. Purif. 59, 242– 248 [DOI] [PubMed] [Google Scholar]
- 24.Inaba K., Inaba M., Romani N., Aya H., Deguchi M., Ikehara S., Muramatsu S., Steinman R. M. ( 1992) J. Exp. Med. 176, 1693– 1702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Macatonia S. E., Edwards A. J., Knight S. C. ( 1986) Immunology 59, 509– 514 [PMC free article] [PubMed] [Google Scholar]
- 26.Bhardwaj N., Friedman S. M., Cole B. C., Nisanian A. J. ( 1992) J. Exp. Med. 175, 267– 273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ingulli E., Mondino A., Khoruts A., Jenkins M. K. ( 1997) J. Exp. Med. 185, 2133– 2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kabashima K., Sugita K., Shiraishi N., Tamamura H., Fujii N., Tokura Y. ( 2007) Biochem. Biophys. Res. Commun. 361, 1012– 1016 [DOI] [PubMed] [Google Scholar]
- 29.Kabashima K., Shiraishi N., Sugita K., Mori T., Onoue A., Kobayashi M., Sakabe J., Yoshiki R., Tamamura H., Fujii, Ninaba K., Tokura Y. ( 2007) Am. J. Pathol. 171, 1249– 1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kohara H., Omatsu Y., Sugiyama T., Noda M., Fujii N., Nagasawa T. ( 2007) Blood 110, 4153– 4160 [DOI] [PubMed] [Google Scholar]
- 31.Sallusto F., Schaerli P., Loetscher P., Schaniel C., Lenig D., Mackay C. R., Qin S., Lanzavecchia A. ( 1998) Eur. J. Immunol. 28, 2760– 2769 [DOI] [PubMed] [Google Scholar]
- 32.Schmitt T. M., Zuniga-Pflucker J. C. ( 2002) Immunity 17, 749– 756 [DOI] [PubMed] [Google Scholar]
- 33.Diao J., Winter E., Chen W., Cantin C., Cattral M. S. ( 2004) J. Immunol. 173, 1826– 1833 [DOI] [PubMed] [Google Scholar]
- 34.Randolph G. J., Beaulieu S., Lebecque S., Steinman R. M., Muller W. A. ( 1998) Science 282, 480– 483 [DOI] [PubMed] [Google Scholar]
- 35.Cheng P., Zlobin A., Volgina V., Gottipati S., Osborne B., Simel E. J., Gabrilovich D. I. ( 2001) J. Immunol. 167, 4458– 4467 [DOI] [PubMed] [Google Scholar]
- 36.Williams C. K., Segarra M., Sierra Mde L., Sainson R. C., Tosato G., Harris A. L. ( 2008) Cancer Res. 68, 1889– 1895 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.