Abstract
Folding and stability are parameters that control protein behavior. The possibility of conferring additional stability on proteins has implications for their use in vivo and for their structural analysis in the laboratory. Cyclic polypeptides ranging in size from 14 to 78 amino acids occur naturally and often show enhanced resistance toward denaturation and proteolysis when compared with their linear counterparts. Native chemical ligation and intein-based methods allow production of circular derivatives of larger proteins, resulting in improved stability and refolding properties. Here we show that circular proteins can be made reversibly with excellent efficiency by means of a sortase-catalyzed cyclization reaction, requiring only minimal modification of the protein to be circularized.
Sortases are bacterial enzymes that predominantly catalyze the attachment of surface proteins to the bacterial cell wall (1, 2). Other sortases polymerize pilin subunits for the construction of the covalently stabilized and covalently anchored pilus of the Gram-positive bacterium (3–5). The reaction catalyzed by sortase involves the recognition of short 5-residue sequence motifs, which are cleaved by the enzyme with the concomitant formation of an acyl enzyme intermediate between the active site cysteine of sortase and the carboxylate at the newly generated C terminus of the substrate (1, 6–8). In many bacteria, this covalent intermediate can be resolved by nucleophilic attack from the pentaglycine side chain in a peptidoglycan precursor, resulting in the formation of an amide bond between the pentaglycine side chain and the carboxylate at the cleavage site in the substrate (9, 10). In pilus construction, alternative nucleophiles such as lysine residues or diaminopimelic acid participate in the transpeptidation reaction (3, 4).
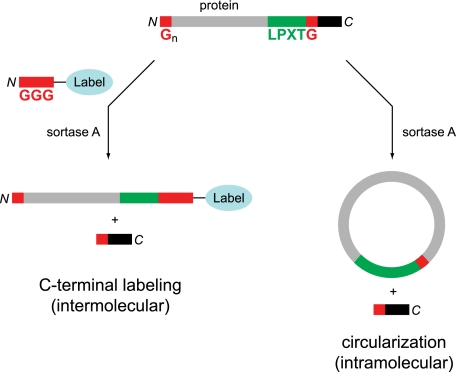
When appended near the C terminus of proteins that are not natural sortase substrates, the recognition sequence of Staphylococcus aureus sortase A (LPXTG) can be used to effectuate a sortase-catalyzed transpeptidation reaction using a diverse array of artificial glycine-based nucleophiles (Fig. 1). The result is efficient installation of a diverse set of moieties, including lipids (11), carbohydrates (12), peptide nucleic acids (13), biotin (14), fluorophores (14, 15), polymers (16), solid supports (16–18), or peptides (15, 19) at the C terminus of the protein substrate. During the course of our studies to further expand sortase-based protein engineering, we were struck by the frequency and relative ease with which intramolecular transpeptidation reactions were occurring. Specifically, proteins equipped with not only the LPXTG motif but also N-terminal glycine residues yielded covalently closed circular polypeptides (Fig. 1). Similar reactivity using sortase has been described in two previous cases; however, rigorous characterization of the circular polypeptides was absent (16, 20). The circular proteins in these reports were observed as minor components of more complex reaction mixtures, and the cyclization reaction itself was not optimized.
FIGURE 1.
Protein substrates equipped with a sortase A recognition sequence (LPXTG) can participate in intermolecular transpeptidation with synthetic oligoglycine nucleophiles (left) or intramolecular transpeptidation if an N-terminal glycine residue is present (right).
Here we describe our efforts toward applying sortase-catalyzed transpeptidation to the synthesis of circular and oligomeric proteins. This method has general applicability, as illustrated by successful intramolecular reactions with three structurally unrelated proteins. In addition to circularization of individual protein units, the multiprotein complex AAA-ATPase p97/VCP/CDC48, with six identical subunits containing the LPXTG motif and an N-terminal glycine, was found to preferentially react in daisy chain fashion to yield linear protein fusions. The reaction exploited here shows remarkable similarities to the mechanisms proposed for circularization of cyclotides, small circular proteins that have been isolated from plants (21–23).
EXPERIMENTAL PROCEDURES
Synthesis of Triglycine Tetramethylrhodamine Peptide
The structure of GGG-TMR5 and a detailed synthetic protocol are provided in the supplemental material and supplemental Fig. S7.
Cloning and Protein Expression
Full amino acid sequences for all proteins used in this study are given in supplemental Fig. S8.
Recombinant sortase A (residues 26–206) containing an N-terminal hexahistidine tag was produced in Escherichia coli as described previously (8). Purified sortase A was stored in 10% glycerol, 50 mm Tris, pH 8.0, 150 NaCl at −80 °C until further use.
G-Cre-LPETG-His6 was cloned into the pTriEx-1.1 Neo expression vector (Novagen) using standard molecular biology techniques. The construct contains two point mutations (M117V and E340Q) and a flexible spacer (GGGGSGGGGS) inserted before the LPETG sortase recognition site. G-Cre-LPETG-His6 was expressed and purified using procedures similar to those reported previously for HTNCre (24). G-Cre-LPETG-His6 was first transformed into Tuner (DE3) pLacI cells (Novagen), and a starter culture was grown in sterile LB media supplemented with 1% (w/v) glucose, chloramphenicol (34 μg/ml), and ampicillin (100 μg/ml). This culture was used to inoculate a large scale culture of sterile LB containing chloramphenicol (34 μg/ml) and ampicillin (100 μg/ml). G-Cre-LPETG-His6 was expressed after a 3-h induction with isopropyl 1-thio-β-d-galactopyranoside (0.5 mm) at 37 °C. Cells were resuspended in 10 mm Tris, 100 mm phosphate, 300 mm NaCl, and 20 mm imidazole, pH 8.0. The suspension was adjusted to 50 μg/ml DNase I, 460 μg/ml lysozyme, and 1 mm MgCl2 and incubated at 4 °C for 1.5 h. The suspension was then sonicated and centrifuged. The clarified lysate was then treated with Ni-NTA-agarose (Qiagen) for 1 h at 4 °C. The resin was washed with 12 column volumes of 10 mm Tris, 100 mm phosphate, 300 mm NaCl, and 20 mm imidazole, pH 8.0, followed by 4 column volumes of 10 mm Tris, 100 mm phosphate, 300 mm NaCl, and 30 mm imidazole, pH 8.0. The protein was then eluted with 10 mm Tris, 100 mm phosphate, 300 mm NaCl, and 300 mm imidazole, pH 8.0. The purified protein was then dialyzed first against 20 mm Tris, pH 7.5, 500 mm NaCl followed by 50% glycerol, 20 mm Tris, pH 7.5, 500 mm NaCl. G-Cre-LPETG-His6 was then passed through a 0.22-μm filter to remove minor precipitation and stored at 4 °C.
G5-eGFP-LPETG-His6 was prepared from a previously reported eGFP construct lacking the five N-terminal glycine residues using a QuickChange® II site-directed mutagenesis kit (Stratagene) and produced in E. coli using reported procedures (11). Purified G5-eGFP-LPETG-His6 was buffer exchanged into 20 mm Tris, pH 8.0, 150 mm NaCl and stored at 4 °C. UCHL3 with the sortase recognition sequence (LPETG) substituted for amino acids 159–163 was cloned and produced in E. coli as described previously (25).
Human p97 (806 amino acids) was PCR-amplified and cloned via the NdeI and HindIII restriction sites into a pET28a+ expression vector (Novagen) to yield the G-His6-p97 construct. G-His6-p97-LPSTG-XX was generated by introducing two point mutations (G782L and Q785T) and a stop codon at position 791 using QuickChange® mutagenesis (Stratagene). Recombinant p97 was expressed at 30 °C in E. coli after induction for 3 h with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside. Cells were resuspended in buffer A (50 mm Tris, pH 8.0, 300 mm NaCl, 5% glycerol, 20 mm imidazole, and 7.1 mm β-mercaptoethanol), adjusted to 15 μg/ml lysozyme and 10 μg/ml DNase I, and lysed by two passes through a French pressure cell at 1200 p.s.i. After centrifugation for 30 min at 40,000 × g, the supernatant was bound to nickel-Sepharose resin (GE Healthcare). After washing the resin with 20 column volumes of buffer A, p97 was eluted with buffer A containing 250 mm imidazole. Hexameric rings of p97 were further purified on a Superdex 200 HR 16/60 column (GE Healthcare) using 25 mm Tris, pH 8.0, 150 mm KCl, 2.5 mm MgCl2, 5% glycerol as the mobile phase. The purified protein was snap-frozen and stored at −80 °C.
Circularization and Intermolecular Transpeptidation
Transpeptidation reactions were performed by combining the necessary proteins/reagents at the specified concentrations in the presence of sortase reaction buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 10 mm CaCl2) and incubating at 37 °C for the times indicated. Diglycine and triglycine (GGG) peptides were purchased from Sigma. Reactions were halted by the addition of reducing Laemmli sample buffer and analyzed by SDS-PAGE. Gels were visualized by staining with Coomassie Blue. Fluorescence was visualized on a Typhoon 9200 Imager (GE Healthcare). Crude reactions were also diluted into either 0.1% formic acid or water for ESI-MS analysis. ESI-MS was performed on a Micromass LCT mass spectrometer (Micromass® MS Technologies) and a Paradigm MG4 HPLC system equipped with an HTC PAL autosampler (Michrom BioResources) and a Waters symmetry 5-μm C8 column (2.1 × 50 mm, MeCN:H2O (0.1% formic acid) gradient mobile phase, 150 μl/min).
Purification and Refolding of eGFP
G5-eGFP-LPETG-His6 (50 μm) was circularized by treatment with sortase A (50 μm) in sortase reaction buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 10 mm CaCl2) for 24 h at 37 °C. The reaction was run on a 750-μl scale. The entire reaction was then diluted into 10 ml of 20 mm Tris, 500 mm NaCl, and 20 mm imidazole, pH 8.0. This solution was then applied to a column consisting of 2 ml of Ni-NTA-agarose (Qiagen) pre-equilibrated with 20 mm Tris, 500 mm NaCl, and 20 mm imidazole, pH 8.0. The flow-through was then concentrated and buffer exchanged in 20 mm Tris, 150 mm NaCl, pH 8.0, using a NAPTM 5 SephadexTM column (GE Healthcare). The concentrations of circular eGFP and linear G5-eGFP-LPETG-His6 were estimated by UV-visible spectroscopy using the absorbance of eGFP at 488 nm (extinction coefficient 55,900 m−1 cm−1) (26). Circular and linear eGFP (40 μl of 18 μm solutions) was placed in 1.5-ml microcentrifuge tubes and denatured by heating to 90 °C for 5 min. Samples were then incubated at room temperature in the dark for the times indicated. Fluorescent images were acquired using a UV gel documentation system (UVP Laboratory Products).
Reaction of Cyclic UCHL3 with Activity-based Ubiquitin Probe
UCHL3 (30 μm) was incubated with sortase A (150 μm) in sortase reaction buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 10 mm CaCl2) in the presence or absence of 90 mm GGG peptide (Sigma) on a 25-μl scale at 37 °C for 3 h. Ten microliters was withdrawn and diluted with 10 μl of labeling buffer (100 mm Tris, pH 7.5, 150 mm NaCl). Hemagglutinin epitope-tagged ubiquitin vinyl methyl ester (4 μg) was added as well as 1 mm dithiothreitol and incubated at room temperature for 1 h. Reactions were then separated on an SDS-polyacrylamide gel and visualized by Coomassie staining or α-HA immunoblot (supplemental Fig. S4). Hemagglutinin epitope-tagged ubiquitin vinyl methyl ester was prepared following published protocols (27).
MS/MS Sequencing of Proteolytic Fragments from Circular Proteins
Prior to MS/MS analysis, circular eGFP and Cre were separated from sortase A by RP-HPLC using an Agilent 1100 Series HPLC system equipped with a Waters Delta Pak 5 μm, 100 Å C18 column (3.9 × 150 mm, MeCN:H2O gradient mobile phase containing 0.1% trifluoroacetic acid, 1 ml/min). Fractions containing the circular proteins were pooled and subjected to trypsin digestion. Crude transpeptidation reactions containing circular UCHL3 were separated by SDS-PAGE followed by Coomassie staining. The band corresponding to circular UCHL3 was excised and digested with Glu-C. Crude transpeptidation reactions containing dimeric p97 were separated by SDS-PAGE followed by Coomassie staining. The transpeptidation reaction used for this purpose was incubated for only 2 h and therefore contains less oligomerization than that seen after an overnight incubation (see supplemental Fig. S6). The band corresponding to dimeric p97 was excised and digested with chymotrypsin. For all protein substrates, the peptides generated from proteolytic digestion were extracted and concentrated for analysis by RP-HPLC and tandem mass spectrometry. RP-HPLC was carried out on a Waters NanoAcquity HPLC system with a flow rate of 250 nl/min and mobile phases of 0.1% formic acid in water and 0.1% formic acid in acetonitrile. The gradient used was isocratic 1% acetonitrile for 1 min followed by 2% acetonitrile per min to 40% acetonitrile. The analytical column was 0.075 μm × 10 cm with the tip pulled to 0.005 μm and self-packed with 3 μm Jupiter C18 (Phenomenex). The column was interfaced to a Thermo LTQ linear ion trap mass spectrometer in a nanospray configuration, and data were collected in full scan mode followed by MS/MS analysis in a data-dependent manner. The mass spectral data were data base searched using SEQUEST.
Construction of Molecular Models
Molecular models were generated from published crystal structures (PDB codes 1kbu, 1gfl, 1xd3, and 3cf1) (28–31). N- and C-terminal residues were added using Coot 0.5 (32). Protein termini were repositioned using the Auto Sculpting function in MacPyMOL (DeLano Scientific LLC). Residues visible in the published crystal structures were not moved during positioning of the extended N and C termini. All protein images in this study were generated using MacPyMOL.
RESULTS
Cre Recombinase
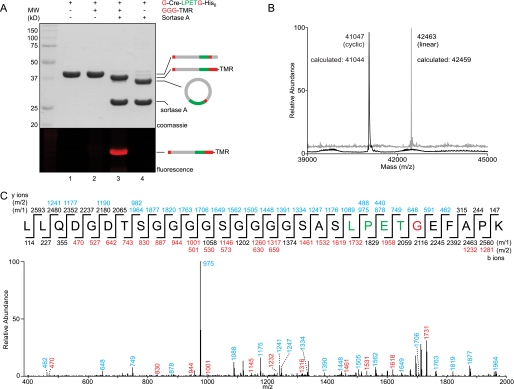
We first noticed the presence of a circular protein product when installing a C-terminal modification onto a nonfunctional mutant of Cre recombinase containing a single N-terminal glycine residue and the requisite LPETG sequence near the C terminus. The LPETG motif was separated from the native protein by a flexible amino acid linker (GGGGSGGGGS). Whereas installation of the label at the Cre C terminus proceeded efficiently when a triglycine nucleophile containing tetramethylrhodamine (GGG-TMR) was included, we observed a product that migrated more rapidly on SDS-PAGE when nucleophile was omitted from the reaction mixture (Fig. 2A). Hydrolysis of the sortase acyl enzyme is known to proceed slowly in the absence of glycine nucleophiles (19, 33, 34). However, when reaction mixtures were analyzed by ESI-MS, we consistently observed a protein species that differed from the mass expected for hydrolysis by approximately −18 Da (Fig. 2B). This mass was consistent with intramolecular nucleophilic attack, suggesting that the single N-terminal glycine residue was serving as the nucleophile in this transformation. Ultimately, MS/MS on tryptic digests of this species showed unequivocally that it consisted of a covalently closed circular product of Cre, with the N-terminal glycine fused exactly at the LPETG cleavage site in the expected position (Fig. 2C).
FIGURE 2.
Cyclization of Cre recombinase. A, G-Cre-LPETG-His6 (50 μm) was incubated with sortase A (50 μm) in the presence or absence of fluorescent GGG-TMR (10 mm) in sortase reaction buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 10 mm CaCl2) for 21.5 h at 37 °C. SDS-PAGE revealed the expected C-terminal transpeptidation product when GGG-TMR was included, whereas omission of the triglycine nucleophile resulted in clean conversion to a unique protein species with a lower apparent molecular weight. Schematic representations to the right of the gel indicate the topology of the protein species produced by transpeptidation. B, ESI-MS of linear G-Cre-LPETG-His6 and circular Cre formed by intramolecular transpeptidation. C, MS/MS spectrum of a tryptic fragment of circular Cre showing the ligation of the N-terminal residues (GEFAPK) to the C-terminal LPET motif. Expected masses for y and b ions are listed above and below the peptide sequence. Ions that were positively identified in the MS/MS spectrum are highlighted in blue or red. Only the most prominent daughter ions have been labeled in the MS/MS spectrum.
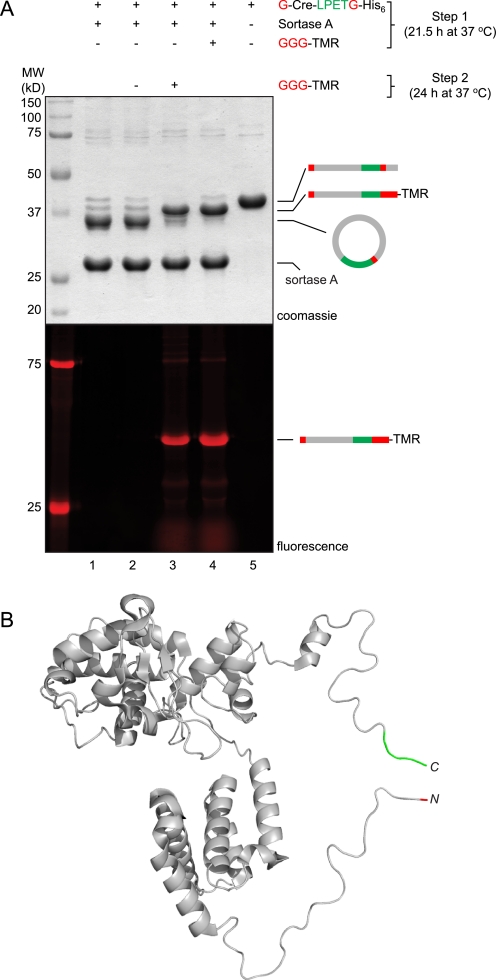
Recognizing that the LPETG motif is maintained in the cyclized Cre product, we suspected that sortase should be capable of cleaving the circular protein at this site, thus producing an equilibrium between circular and linear forms of Cre. To demonstrate this point, Cre was first incubated with sortase in the presence or absence of triglycine nucleophile (Fig. 3A). A portion of the cyclized reaction mixture (Fig. 3A, lane 1) was then treated with a large molar excess of triglycine nucleophile or left alone for a further 24 h (Fig. 3A, lanes 2 and 3). Remarkably, upon treatment with exogenous nucleophile, the pre-cyclized material yielded a reaction mixture that was nearly identical to the result obtained when nucleophile was included from the very beginning of the experiment (Fig. 3A, compare lanes 3 and 4). This result provided further evidence that cyclized Cre indeed contains the expected LPETG motif at the site of covalent closure. In addition, it suggested that hydrolysis of the acyl enzyme intermediate does not effectively compete during cyclization, because the hydrolyzed material should be unable to participate in the transpeptidation reaction.
FIGURE 3.
Cyclization of Cre is reversible. A, G-Cre-LPETG-His6 (50 μm) was circularized by treatment with sortase A (50 μm) in sortase reaction buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 10 mm CaCl2) for 21.5 h at 37 °C (lane 1). This reaction mixture was then treated with 10 mm GGG-TMR (lane 3) or simply incubated for an additional 24 h at 37 °C (lane 2). All reactions were analyzed by SDS-PAGE with visualization by Coomassie staining or in-gel fluorescence. For comparison, a C-terminal labeling reaction performed using 10 mm GGG-TMR without prior cyclization of the Cre substrate (lane 4) and a sample of linear G-Cre-LPETG-His6 incubated without sortase A or nucleophile (lane 5) are included. B, molecular model of Cre recombinase monomer (generated from PDB code 1kbu) (29) showing the proximity relationship between the N and C termini. The N-terminal glycine residue is highlighted in red, and the C-terminal LPET residues are shown in green.
The circularization reaction observed for Cre proceeded with remarkable efficiency. Conversion was estimated to be >90% by SDS-PAGE. By taking an existing crystal structure (29) of the Cre protein and modeling in those residues not visible in the structure, it was clear that the N and C termini were located in sufficiently close proximity to permit closure without significant perturbation of the native structure (Fig. 3B). We assume that these regions possess considerable flexibility because they are not resolved in the crystal structure.
eGFP
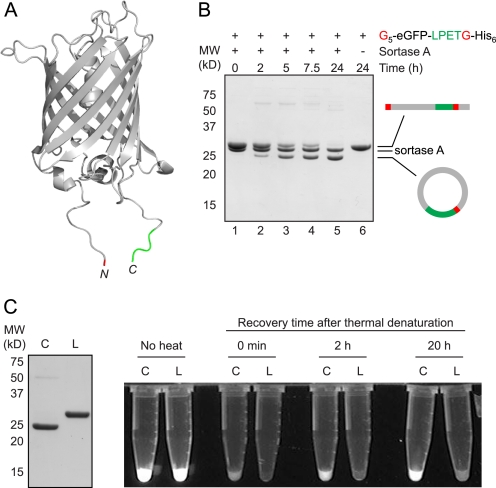
Having verified the cyclization of Cre recombinase, we sought to explore the generality of this technique. To this end we generated a derivative of eGFP containing the LPETG sequence and five N-terminal glycine residues. This construct was of particular interest because inspection of the x-ray crystal structure (31) revealed that the N and C termini were positioned on the same end of the β-barrel, suggesting that this substrate should be ideal for cyclization (Fig. 4A). Furthermore, in one of the earliest reports on the use of sortase for protein engineering, a similar eGFP substrate was described and reported to cyclize in the presence of sortase (16). In this instance, cyclization only proceeded in modest yield, and the putative cyclized product was produced as a mixture with higher molecular weight species assigned as oligomers of eGFP formed by intermolecular transpeptidation. Thus, to explore potential complications caused by intermolecular reactions, we studied the reaction of our eGFP construct in the presence of sortase.
FIGURE 4.
Circularization of eGFP. A, molecular model of eGFP (generated from PDB code 1gfl) (31) showing the proximity relationship between the N and C termini. The N-terminal glycine residue is highlighted in red, and the C-terminal LPET residues are shown in green. B, G5-eGFP-LPETG-His6 (50 μm) was circularized by treatment with sortase A (50 μm) in sortase reaction buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 10 mm CaCl2) for 24 h at 37 °C. Schematic representations to the right of the gel indicate the topology of the protein species produced by transpeptidation. C, circular eGFP (C) recovers fluorescence more rapidly than linear G5-eGFP-LPETG-His6 (L) following thermal denaturation for 5 min at 90 °C. SDS-PAGE analysis confirmed the purity and concentration of the circular eGFP (C) and linear G5-eGFP-LPETG-His6 (L) samples used for refolding.
In our hands, we observed clean conversion to a lower molecular weight species (>90% estimated conversion) with little to no evidence for oligomerization (Fig. 4B). A higher molecular weight polypeptide was observed at early time points and may represent a covalent eGFP dimer that is generated transiently over the course of the reaction. Higher molecular species, however, were only observed in trace quantities in the final reaction mixture. As in the case of Cre, evidence for circularization was provided by mass spectral characterization of the intact circular protein and MS/MS sequencing of tryptic peptides (supplemental Fig. S1). As an additional control to demonstrate that the N-terminal glycine residue was the only nucleophile participating in intramolecular transpeptidation, we analyzed the behavior of an eGFP derivative that lacked an N-terminal glycine. In this case, ESI-MS revealed products consistent with hydrolysis of the acyl enzyme intermediate, rather than intramolecular nucleophilic attack (supplemental Fig. S1).
Circularization has been shown to confer unique properties onto proteins when compared with the linear form (35–37). In the case of GFP circularized using intein-based methods, these properties include a reduced rate of unfolding when exposed to denaturants, as well as an enhanced rate of refolding following denaturation (35). We observed a similar phenomenon for eGFP circularized using sortase (Fig. 4C). Circular eGFP was first separated from residual sortase A using Ni-NTA resin. This material retained fluorescence suggesting that covalent ligation of the N and C termini had minimal impact on the structure of this substrate. Circular and linear eGFP were then subjected to simple thermal denaturation, followed by recovery at room temperature. As shown in Fig. 4C, circular eGFP regained fluorescence more rapidly than linear eGFP.
UCHL3
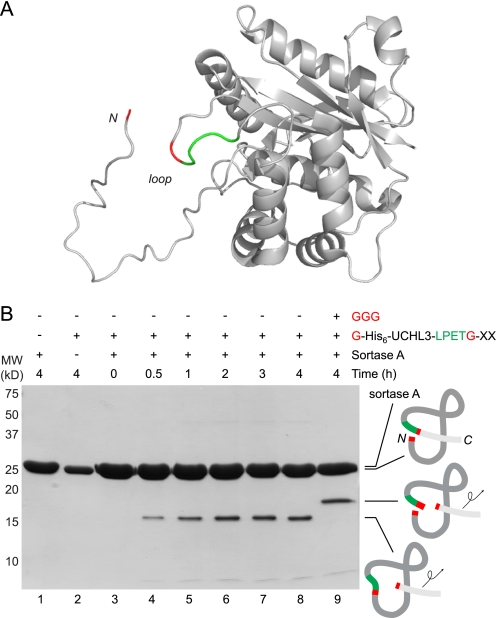
Even an internally positioned LPXTG motif was sufficient to effectuate a circularization reaction. We installed a sortase recognition site in the crossover loop of the ubiquitin C-terminal hydrolase UCHL3, and we demonstrated that the continuity of the polypeptide backbone can be disrupted with concomitant installation of a covalent modification that reports on the accuracy of cleavage and transpeptidation (25). This reaction proceeds without complete loss of activity of UCHL3, indicating that even the cleaved form of UCHL3 retains its structural integrity to a significant degree (25). This UCHL3 construct was prepared with an N-terminal glycine residue, and examination of the crystal structure of UCHL3 (30) clearly showed the close apposition of the N terminus and the crossover loop, suggesting that cyclization to yield a circular fragment containing the N-terminal portion of UCHL3 should be readily observable (Fig. 5A).
FIGURE 5.
UCHL3 with an internally positioned LPETG motif is circularized by sortase A. A, molecular model of human UCHL3 (generated from PDB code 1xd3) (30) showing the active site crossover loop bearing an LPETG substitution (LPET residues shown in green and Gly residue shown in red). The N-terminal glycine residue that serves as the nucleophile for intramolecular transpeptidation is highlighted in red. B, UCHL3 (30 μm) bearing an LPETG substitution in the active site crossover loop was incubated with sortase A (150 μm) in the absence or presence of GGG nucleophile (90 mm) in sortase reaction buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 10 mm CaCl2) for the indicated times at 37 °C and analyzed by SDS-PAGE, followed by Coomassie staining. Schematic representations to the right of the gel indicate the topology of the protein species produced by transpeptidation.
As expected, in the absence of added nucleophile, the N-terminal glycine serves as a highly efficient nucleophile to yield a circular fragment that contains the N-terminal portion of UCHL3 (Fig. 5B). The identity of the circular polypeptide was confirmed by MS/MS of the peptide containing the expected fusion of the N-terminal glycine residue with the new C terminus released from the crossover loop (see supplemental Fig. S2). Cyclization was efficiently blocked if a high concentration of triglycine (GGG) was included in the reaction, generating instead the N-terminal fragment of UCHL3 transacylated onto the triglycine nucleophile (Fig. 5B, lane 9, and supplemental Fig. S3). Cyclization could also be reversed by adding an excess of triglycine to reaction mixtures preincubated with sortase to allow cyclization. This reopening reaction was observed by both SDS-PAGE and ESI-MS (supplemental Fig. S3).
To test the functional properties of cyclic UCHL3, we incubated reaction mixtures with an activity-based probe consisting of ubiquitin equipped with an electrophilic vinyl methyl ester moiety at the C terminus (supplemental Fig. S4). Probes of this nature are able to specifically alkylate active site cysteine residues in ubiquitin-specific hydrolases such as UCHL3 (25, 27, 38). Following circularization, the active site cysteine (Cys-95) of UCHL3 is located in the circular N-terminal fragment, and indeed we observed covalent labeling of this fragment with a corresponding shift in apparent molecular weight consistent with the attachment of ubiquitin. This result suggests that despite cleavage of the polypeptide backbone, the circular N-terminal fragment of UCHL3 and the C-terminal portion released during transpeptidation remain associated and preserve the affinity of UCHL3 for ubiquitin. This result is consistent with previous observations from our laboratory demonstrating that covalent closure of the UCHL3 crossover loop is dispensable for enzyme activity (25).
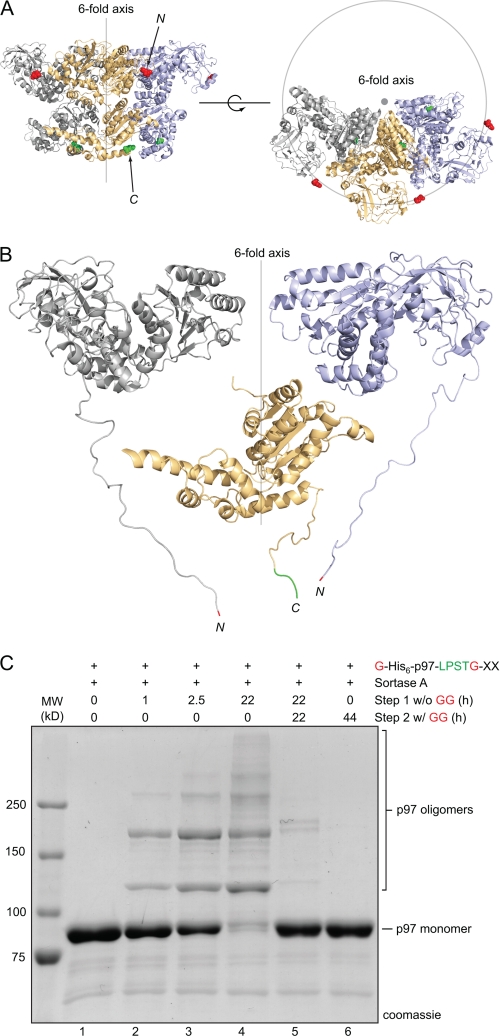
p97
The above examples concern single chain proteins whose termini are sufficiently close to allow covalent closure by means of the sortase-mediated transacylation reaction. Similar proximity relationships between protein termini should also be present on separate polypeptides that assemble into defined oligomeric structures. As an example, we examined p97, a hexameric AAA-ATPase. We generated a derivative of p97 (G-His6-p97-LPSTG-XX) containing an LPSTG motif near the C terminus, and a hexahistidine tag capped by two serine residues and a single glycine at the N terminus. The structure of a p97 trimer in the presence of ADP has been solved at 3.5 Å resolution (28), with several residues from the N and C termini not visible (Fig. 6A). When all the residues present in our modified version of p97 were modeled onto the published trimer of p97, it was evident that the N and C termini of adjacent p97 units were sufficiently close to permit covalent cross-linking (Fig. 6B). G-His6-p97-LPSTG-XX was expressed in E. coli and yielded the hexameric p97 ring, as assessed by gel filtration. As expected, this derivative of p97 was an excellent substrate for transpeptidation at its C terminus, allowing efficient installation of a label when incubated in the presence of sortase and GGG-TMR (supplemental Fig. S5). In contrast, a variant of p97 lacking the LPSTG sequence showed no labeling (supplemental Fig. S5). When G-His6-p97-LPSTG-XX was treated with sortase A in the absence of added nucleophile, we observed formation of an SDS-resistant ladder of polypeptides, as would be expected for intermolecular cross-linking of p97 monomers (Fig. 6C). We were confident that these species arise from head-to-tail ligation of p97 because introduction of excess diglycine peptide after oligomerization caused collapse of the higher molecular weight structures back to monomeric p97 (Fig. 6C, lane 5). This suggested that the higher order aggregates are held together by newly formed LPSTG units formed from the C-terminal LPST residues of one p97 monomer and the N-terminal glycine residue of a neighboring monomer. The banding pattern observed for reopening was also nearly identical to that seen when diglycine was included from the very beginning of the experiment, a scenario where installation of diglycine at the C terminus of each p97 subunit is presumed to be the major reaction pathway (Fig. 6C, lane 6). We have also been able to identify peptides consistent with intermolecular cross-linking of p97 subunits by MS/MS (supplemental Fig. S6).
FIGURE 6.
Positions of the N and C termini in the p97 hexamer are suitable for intermolecular cross-linking through sortase-catalyzed transpeptidation. A, molecular model of p97 trimer (generated from PDB code 3cf1) (28) showing the relative position of p97 monomers in the hexameric ring. The visible N and C termini from the published p97 trimer structure are indicated in red and green, respectively. B, molecular model of G-His6-p97-LPSTG-XX showing the proximity relationship between N and C termini in adjacent p97 monomers. N- and C-terminal residues not visible in the published crystal structure have been modeled onto the existing structure. N-terminal glycine residues are shown in red, and the C-terminal LPST residues are shown in green. For clarity, the C-terminal domains of the outer monomers are hidden, as is the N-terminal domain of the central monomer. C, G-His6-p97-LPSTG-XX (1.5 mg/ml) was incubated with sortase A (30 μm) in sortase reaction buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 10 mm CaCl2) for the times indicated at 37 °C. After 22 h, diglycine (GG) was added (100 mm final concentration), resulting in disappearance of the covalent oligomers (lane 5). For comparison, a control reaction containing 100 mm diglycine peptide from the outset of the experiment is shown in lane 6.
DISCUSSION
Cyclic proteins are an interesting class of polypeptides that often display unique properties because of covalent closure of the amide backbone (39, 40). Although some cyclic protein derivatives occur naturally, methods for generating cyclic proteins in the laboratory provide a means for accessing cyclic versions of proteins that only occur in linear form. Intramolecular sortase-catalyzed transpeptidation provides a straightforward method for accessing these types of cyclic proteins. The transpeptidation reaction described here bears a remarkable resemblance to the proposed biosynthesis of the largest class of naturally occurring cyclic proteins, the cyclotides (21–23). In both cases, linear protein precursors are cleaved by cysteine proteases to generate an acyl-enzyme intermediate that is subsequently resolved by nucleophilic attack from the N terminus of the linear proteins to generate the cyclic product.
In this work we have explored transpeptidation reactions using four structurally diverse protein substrates. Cyclization has been confirmed for three proteins, including an example (UCHL3) utilizing an LPXTG sequence positioned in a flexible internal loop rather than near the C terminus of the protein. Cyclization and oligomerization via sortase-mediated transpeptidation have been previously suggested to occur for an eGFP construct modified in a manner similar to that used here (16), and for a by-product from a protein purification system where the substrate circularized appears to be sortase A itself (20). In both cases, the identity of the circular products was not rigorously confirmed. Our data identify the circular or oligomeric products unambiguously by MS/MS for all substrates studied. We also find that our eGFP derivative strongly favors cyclization over oligomerization, showing little evidence for the formation of higher order structures that might be expected by the head-to-tail ligation of termini from separate eGFP monomers. Subtle differences in the structure of the eGFP constructs cannot be overlooked as a potential cause for the observed results. For example, our eGFP is extended at the N terminus by only five glycine residues, whereas the construct studied by Parthasarathy et al. (16) contains an additional 17 residues, including 3 N-terminal glycines. Future work will be required to thoroughly characterize the effect of distance relationships between protein termini on favoring intra- versus intermolecular transpeptidation.
With respect to protein cyclization, sortase-mediated circularization is efficient despite the potential for competing reaction pathways. In the absence of added oligoglycine nucleophile, these include hydrolysis of the acyl enzyme intermediate, reattachment of the C-terminal protein fragment that is lost upon initial cleavage of the protein substrate by sortase, or, as mentioned above, oligomerization of protein monomers in head-to-tail fashion. Even when oligoglycine nucleophile is added with the intent of blocking the cyclization pathway, millimolar concentrations are necessary to efficiently compete with cyclization. One factor that certainly must contribute to this observed preference for cyclization is the distance between protein termini. Inspection of the data base of PDB shows that nearly one-third of proteins with known structures have their termini in rather close apposition (within 20 Å) (40). The LPXTG sequence itself spans roughly 15 Å in an extended conformation, suggesting that circularization via sortase-catalyzed transpeptidation might be amenable to a significant fraction of proteins using the LPXTG sequence alone to bridge the gap between N and C termini. Larger distances could simply be covered by inserting flexible amino acid spacers at either termini. We also consider it likely that the circularized version of a protein will show more restricted mobility in the segment that corresponds to the newly established LPXTG connection between its termini. This fact alone may render the circular product a comparatively worse substrate for sortase and therefore assist in driving the transpeptidation reaction toward cyclization. As evidence for this point, we have observed previously that sortase fails to cleave LPXTG motifs placed in structured loops of class I major histocompatibility complex molecules (14).
Sortase-catalyzed transpeptidation provides an attractive alternative to existing methods for peptide and protein circularization. Chemical synthesis can provide access to circular polypeptides of modest size, with circularization of linear precursors having been achieved using native chemical ligation (41–43), subtiligase (44), or standard amide bond-forming reactions common to solid-phase peptide synthesis (43, 45). For larger proteins beyond the technical capabilities of solid-phase synthesis, cyclization is most often accomplished using native chemical ligation, typically in conjunction with split-intein expression constructs (35–37, 46–48). When compared with the split-intein approach, the modest modification necessary to render proteins amenable to cyclization or oligomerization is certainly an attractive feature of the sortase-catalyzed process. Proteins must simply possess a sortase recognition sequence (LPXTG) either near the C terminus or in a flexible loop and an N-terminal glycine residue to act as the nucleophile. These modifications are not anticipated to have a significant impact on protein expression or function. In contrast, protein circularization by split-intein methods requires more extensive modifications of the expression construct, a necessity that may reduce protein expression or affect protein solubility. It should be noted, however, that the number of extra amino acid residues at the site of N- to-C-terminal ligation following excision of the large intein domains can be less than the five residues (LPXTG) that remain after circularization using sortase A.
The sortase-catalyzed approach also provides additional levels of control over the ensuing transpeptidation reaction. This may be particularly useful for oligomeric species, such as the p97 example described here. Specifically, our modified p97 protein (G-His6-p97-LPSTG-XX) is produced in a form that is by itself unreactive. This allows protein expression and the subsequent assembly and purification of the hexamer to be completed first, without complications caused by premature covalent oligomerization. Cross-linking is then induced by the addition of sortase after the individual subunits have been correctly positioned in the hexameric ring. The extent of transpeptidation can be further controlled by inclusion of synthetic oligoglycine nucleophiles, either during the transpeptidation reaction or after transpeptidation is complete. The latter scenario even allows cyclization to be completely reversed. Incubating circular protein products with sortase in the presence of an oligoglycine nucleophile restores linearity to the protein product, because in the course of the initial cyclization reaction, the LPXTG motif is restored. An equilibrium between closed and open forms is thus established and can be driven toward the linear state by adding a large excess of the oligoglycine nucleophile.
The implications of protein cyclization or oligomerization for protein engineering are numerous. In the case of protein oligomerization, the ability to link protein subunits held in a defined geometry might be exploited to explore subtle changes in intersubunit interactions upon substrate engagement or recruitment of binding partners. A more detailed examination of the reaction kinetics would be required to determine, for example, whether all subunits in the hexameric ring of p97 are equally good substrates or whether subunits that lie along the 3-fold axis preferentially cross-link to yield dimers. Although in the crystal structure (28) all of the individual subunits appear identical, it remains to be determined whether this equivalency applies in solution as well. For cyclic proteins, there is compelling evidence that demonstrates improved stability of circularized proteins when compared with their linear counterparts (35–37, 40, 49). This is true for cyclic versions GFP (35), β-lactamase (36), and dihydrofolate reductase (37) generated using intein-based methods. The extension of protein cyclization to proteins of therapeutic value to improve the in vivo half-life has already been suggested (16, 39) and remains an exciting avenue for further research. Covalent closure of a protein through sortase-mediated circularization may also facilitate structural analysis of proteins whose flexible termini may interfere with crystallization.
Supplementary Material
Acknowledgments
We thank Carla Guimaraes and Mathias Pawlak for help in the preparation of the G5-eGFP-LPETG-His6 and G-Cre-LPETG-His6 substrates, respectively.
This work was supported, in whole or in part, by National Institutes of Health Grant R21-EB008875.

The on-line version of this article (available at http://www.jbc.org) contains supplemental text and reference and Figs. S1–S8.
- GGG-TMR
- triglycine tetramethylrhodamine peptide
- eGFP
- enhanced green fluorescent protein
- UCHL3
- ubiquitin C-terminal hydrolase L3
- GGG
- triglycine peptide
- ESI-MS
- electrospray ionization mass spectrometry
- RP-HPLC
- reversed-phase high performance liquid chromatography
- MS/MS
- tandem mass spectrometry
- PDB
- Protein Data Bank
- Ni-NTA
- nickel-nitrilotriacetic acid.
REFERENCES
- 1.Marraffini L. A., Dedent A. C., Schneewind O. ( 2006) Microbiol. Mol. Biol. Rev. 70, 192– 221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paterson G. K., Mitchell T. J. ( 2004) Trends Microbiol. 12, 89– 95 [DOI] [PubMed] [Google Scholar]
- 3.Budzik J. M., Marraffini L. A., Souda P., Whitelegge J. P., Faull K. F., Schneewind O. ( 2008) Proc. Natl. Acad. Sci. U. S. A. 105, 10215– 10220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Budzik J. M., Oh S. Y., Schneewind O. ( 2008) J. Biol. Chem. 283, 36676– 36686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fälker S., Nelson A. L., Morfeldt E., Jonas K., Hultenby K., Ries J., Melefors O., Normark S., Henriques-Normark B. ( 2008) Mol. Microbiol. 70, 595– 607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kruger R. G., Otvos B., Frankel B. A., Bentley M., Dostal P., McCafferty D. G. ( 2004) Biochemistry 43, 1541– 1551 [DOI] [PubMed] [Google Scholar]
- 7.Pallen M. J., Lam A. C., Antonio M., Dunbar K. ( 2001) Trends Microbiol. 9, 97– 102 [DOI] [PubMed] [Google Scholar]
- 8.Ton-That H., Liu G., Mazmanian S. K., Faull K. F., Schneewind O. ( 1999) Proc. Natl. Acad. Sci. U. S. A. 96, 12424– 12429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schneewind O., Fowler A., Faull K. F. ( 1995) Science 268, 103– 106 [DOI] [PubMed] [Google Scholar]
- 10.Ton-That H., Schneewind O. ( 1999) J. Biol. Chem. 274, 24316– 24320 [DOI] [PubMed] [Google Scholar]
- 11.Antos J. M., Miller G. M., Grotenbreg G. M., Ploegh H. L. ( 2008) J. Am. Chem. Soc. 130, 16338– 16343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Samantaray S., Marathe U., Dasgupta S., Nandicoori V. K., Roy R. P. ( 2008) J. Am. Chem. Soc. 130, 2132– 2133 [DOI] [PubMed] [Google Scholar]
- 13.Pritz S., Wolf Y., Kraetke O., Klose J., Bienert M., Beyermann M. ( 2007) J. Org. Chem. 72, 3909– 3912 [DOI] [PubMed] [Google Scholar]
- 14.Popp M. W., Antos J. M., Grotenbreg G. M., Spooner E., Ploegh H. L. ( 2007) Nat. Chem. Biol. 3, 707– 708 [DOI] [PubMed] [Google Scholar]
- 15.Tanaka T., Yamamoto T., Tsukiji S., Nagamune T. ( 2008) Chembiochem. 9, 802– 807 [DOI] [PubMed] [Google Scholar]
- 16.Parthasarathy R., Subramanian S., Boder E. T. ( 2007) Bioconjugate Chem. 18, 469– 476 [DOI] [PubMed] [Google Scholar]
- 17.Chan L., Cross H. F., She J. K., Cavalli G., Martins H. F., Neylon C. ( 2007) PLoS ONE 2, e1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clow F., Fraser J. D., Proft T. ( 2008) Biotechnol. Lett. 30, 1603– 1607 [DOI] [PubMed] [Google Scholar]
- 19.Mao H., Hart S. A., Schink A., Pollok B. A. ( 2004) J. Am. Chem. Soc. 126, 2670– 2671 [DOI] [PubMed] [Google Scholar]
- 20.Mao H. ( 2004) Protein Expr. Purif. 37, 253– 263 [DOI] [PubMed] [Google Scholar]
- 21.Gillon A. D., Saska I., Jennings C. V., Guarino R. F., Craik D. J., Anderson M. A. ( 2008) Plant J. 53, 505– 515 [DOI] [PubMed] [Google Scholar]
- 22.Saska I., Craik D. J. ( 2008) Trends Biochem. Sci. 33, 363– 368 [DOI] [PubMed] [Google Scholar]
- 23.Saska I., Gillon A. D., Hatsugai N., Dietzgen R. G., Hara-Nishimura I., Anderson M. A., Craik D. J. ( 2007) J. Biol. Chem. 282, 29721– 29728 [DOI] [PubMed] [Google Scholar]
- 24.Peitz M., Jäger R., Patsch C., Jäger A., Egert A., Schorle H., Edenhofer F. ( 2007) Genesis 45, 508– 517 [DOI] [PubMed] [Google Scholar]
- 25.Popp M. W., Artavanis-Tsakonas K., Ploegh H. L. ( 2009) J. Biol. Chem. 284, 3593– 3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsien R. Y. ( 1998) Annu. Rev. Biochem. 67, 509– 544 [DOI] [PubMed] [Google Scholar]
- 27.Borodovsky A., Ovaa H., Kolli N., Gan-Erdene T., Wilkinson K. D., Ploegh H. L., Kessler B. M. ( 2002) Chem. Biol. 9, 1149– 1159 [DOI] [PubMed] [Google Scholar]
- 28.Davies J. M., Brunger A. T., Weis W. I. ( 2008) Structure 16, 715– 726 [DOI] [PubMed] [Google Scholar]
- 29.Martin S. S., Pulido E., Chu V. C., Lechner T. S., Baldwin E. P. ( 2002) J. Mol. Biol. 319, 107– 127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Misaghi S., Galardy P. J., Meester W. J., Ovaa H., Ploegh H. L., Gaudet R. ( 2005) J. Biol. Chem. 280, 1512– 1520 [DOI] [PubMed] [Google Scholar]
- 31.Yang F., Moss L. G., Phillips G. N., Jr.( 1996) Nat. Biotechnol. 14, 1246– 1251 [DOI] [PubMed] [Google Scholar]
- 32.Emsley P., Cowtan K. ( 2004) Acta Crystallogr. Sect. D Biol. Crystallogr. 60, 2126– 2132 [DOI] [PubMed] [Google Scholar]
- 33.Huang X., Aulabaugh A., Ding W., Kapoor B., Alksne L., Tabei K., Ellestad G. ( 2003) Biochemistry 42, 11307– 11315 [DOI] [PubMed] [Google Scholar]
- 34.Ton-That H., Mazmanian S. K., Faull K. F., Schneewind O. ( 2000) J. Biol. Chem. 275, 9876– 9881 [DOI] [PubMed] [Google Scholar]
- 35.Iwai H., Lingel A., Pluckthun A. ( 2001) J. Biol. Chem. 276, 16548– 16554 [DOI] [PubMed] [Google Scholar]
- 36.Iwai H., Plückthun A. ( 1999) FEBS Lett. 459, 166– 172 [DOI] [PubMed] [Google Scholar]
- 37.Scott C. P., Abel-Santos E., Wall M., Wahnon D. C., Benkovic S. J. ( 1999) Proc. Natl. Acad. Sci. U. S. A. 96, 13638– 13643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Love K. R., Catic A., Schlieker C., Ploegh H. L. ( 2007) Nat. Chem. Biol. 3, 697– 705 [DOI] [PubMed] [Google Scholar]
- 39.Craik D. J. ( 2006) Science 311, 1563– 1564 [DOI] [PubMed] [Google Scholar]
- 40.Trabi M., Craik D. J. ( 2002) Trends Biochem. Sci. 27, 132– 138 [DOI] [PubMed] [Google Scholar]
- 41.Camarero J. A., Muir T. W. ( 1997) Chem. Commun. 1369– 1370 [Google Scholar]
- 42.Camarero J. A., Pavel J., Muir T. W. ( 1998) Angew. Chem.-Int. Ed. Engl. 37, 347– 349 [DOI] [PubMed] [Google Scholar]
- 43.Daly N. L., Love S., Alewood P. F., Craik D. J. ( 1999) Biochemistry 38, 10606– 10614 [DOI] [PubMed] [Google Scholar]
- 44.Jackson D. Y., Burnier J. P., Wells J. A. ( 1995) J. Am. Chem. Soc. 117, 819– 820 [Google Scholar]
- 45.Hartgerink J. D., Granja J. R., Milligan R. A., Ghadiri M. R. ( 1996) J. Am. Chem. Soc. 118, 43– 50 [Google Scholar]
- 46.Camarero J. A., Fushman D., Sato S., Giriat I., Cowburn D., Raleigh D. P., Muir T. W. ( 2001) J. Mol. Biol. 308, 1045– 1062 [DOI] [PubMed] [Google Scholar]
- 47.Camarero J. A., Kimura R. H., Woo Y. H., Shekhtman A., Cantor J. ( 2007) Chembiochem 8, 1363– 1366 [DOI] [PubMed] [Google Scholar]
- 48.Evans T. C., Jr., Martin D., Kolly R., Panne D., Sun L., Ghosh I., Chen L., Benner J., Liu X. Q., Xu M. Q. ( 2000) J. Biol. Chem. 275, 9091– 9094 [DOI] [PubMed] [Google Scholar]
- 49.Colgrave M. L., Craik D. J. ( 2004) Biochemistry 43, 5965– 5975 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.