Abstract
We have developed an algorithm (“Lever”) that systematically maps metazoan DNA regulatory motifs or motif combinations to the sets of genes that they likely regulate. Lever accomplishes this by assessing whether the motifs are enriched within cis regulatory modules (CRMs), predicted by our “PhylCRM” algorithm, in the noncoding sequences surrounding genes in a collection of gene sets. When these gene sets correspond to Gene Ontology (GO) categories, the results of Lever analysis allow the unbiased assignment of functional annotations to the regulatory motifs and also to the candidate CRMs that comprise the genomic motif occurrences. We demonstrate these methods using human myogenic differentiation as a model system, for which we statistically assessed greater than 25,000 pairings of gene sets and motifs / motif combinations. These results allowed us to assign functional annotations to candidate regulatory motifs predicted previously, and to identify gene sets that are likely to be co-regulated via shared regulatory motifs. Lever allows moving beyond the identification of putative regulatory motifs in mammalian genomes, towards understanding their biological roles. This approach is general and can be applied readily to any cell type, gene expression pattern, or organism of interest.
Of fundamental importance for understanding transcriptional regulatory networks is the functional annotation of DNA regulatory motifs (typically ~6-15 bp in length) in terms of what groups of target genes they regulate in a tissue- or temporal-specific manner in response to environmental perturbations. While effective computational methods for mapping DNA regulatory motifs exist in the yeast Saccharomyces cerevisiae, where the DNA binding sites of regulatory transcription factors (TFs) typically occur within ~600 bp upstream of genes, they cannot be applied to metazoan genomes, where genes in the same expression cluster are not necessarily co-regulated by a common mechanism, and the regulatory elements can be far from the transcription start site1.
In metazoans, regulatory motifs tend to co-occur within stretches of noncoding sequence, referred to as cis regulatory modules (CRMs), that regulate expression of the nearby gene(s). Numerous approaches have resulted in the successful identification of CRMs1-4, but such approaches do not attempt to predict ab initio the gene expression patterns or functions of the genes regulated by the CRMs. Although algorithms have been developed recently for evaluating the regulatory significance of CRM binding site composition5,6, thus far they have been unable to evaluate the vast sequence regions beyond the proximal promoter that must be considered in mammalian genomes.
Because of these complications, analyses of transcriptional regulatory elements in mammals have focused either on the prediction of CRMs starting with a collection of known co-regulatory TFs whose DNA binding specificities are available and a set of genes that the TFs may regulate2,3,7,8, or on the computational identification of `motif dictionaries'9-12. However, with the advent of high-throughput methods for assembling motif dictionaries, from either chromatin immunoprecipitations13 or protein binding microarrays14-16, the major computational problem to solve will shift from motif prediction to identifying and associating CRMs to both specific genes and biological processes17.
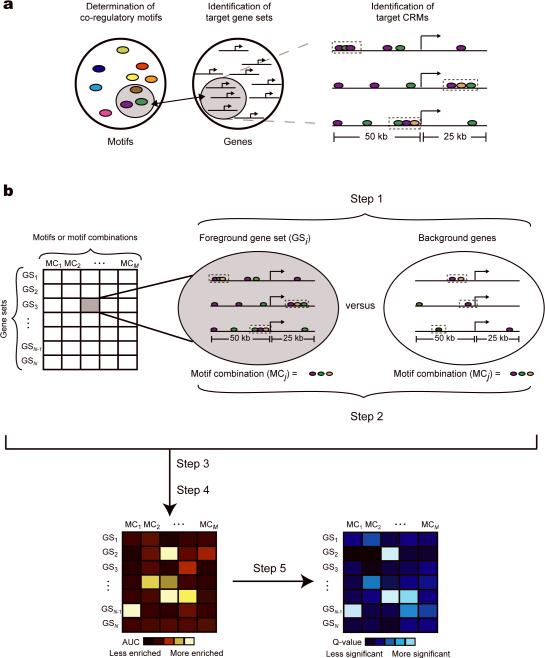
Therefore, we have developed a computational algorithm (termed “Lever”) that systematically identifies the target gene sets that are likely to be regulated by a query collection of candidate regulatory motifs. The ability to screen many gene sets with many motifs /motif combinations allows us to tackle the difficulty in a priori identification of co-regulated gene sets. Lever does not perform de novo motif discovery, but rather evaluates an input collection of motifs for enrichment within candidate CRMs in the noncoding sequences flanking various input gene sets (Figure 1a).
Figure 1. Lever schema.
(a) Lever simultaneously identifies: 1) motifs or motif combinations, 2) their sets of co-regulated genes, and 3) cis regulatory modules containing the enriched motifs or motif combinations. (b) Schematic depiction of the Lever scoring scheme. Step 1: For each gene set and motif combination pairing (“GM-pair”), search for candidate CRMs. Step 2: For each GM-pair and all corresponding background genes, rank the genes according to the PhylCRM score of each gene's single best scoring candidate CRM. Step 3: Evaluate the enrichment (AUC statistics) of a given GM-pair. Step 4: Repeat for all other GM-pairs (shown as a red and yellow matrix). Step 5: The statistical significance of each AUC (indicated by a Q-value, shown as a blue matrix) is calculated by permutation approach for multiple hypothesis correction (see Methods).
In this study, we considered 75 kb of noncoding sequence flanking each gene (50 kb upstream to 25 kb downstream of transcription start site). Lever considers a collection of user-defined gene sets; in this study, we considered GO categories and clusters of co-expressed genes as our gene sets of interest. We examined differentiation of human myoblasts into myotubes, and considered 101 myogenic gene sets and 174 candidate regulatory motifs. We define a “GM-pair” to be the pairing of an individual gene set with a particular query motif or motif combination. Specifically, for each GM-pair, Lever evaluates the degree to which the noncoding sequences surrounding the transcription start sites of the genes in the gene set are enriched for candidate CRMs comprising the given motif / motif combination under consideration, as compared to a random background set of genes.
In order to predict candidate CRMs, we developed a new tool termed “PhylCRM” (pronounced “fulcrum”), which quantifies both motif conservation18 and site clustering, across multiple genomes. We experimentally validated a number of predicted novel CRMs from among the most statistically significant GM-pairs in this study. In this study, only the highest scoring candidate CRM for each gene (see Methods) was considered by Lever, as depicted in Figure 1. Each such GM-pair can be thought of as an individual element of a gene set by motif / motif combination matrix (Figure 1b). In this study, we assessed more than 25,000 GM-pairs.
Identification of significant GM-pairs from Lever analysis allows one to assign functional annotation to motifs at the level of GO categories and gene expression patterns. Although prior studies attempted to broadly annotate motifs at the level of tissue specificity9, Lever is the first algorithm to assign specific functional annotation to metazoan motifs. Specific annotation is needed in order to go beyond the identification of putative regulatory motifs, towards understanding the biological roles of the motifs. Here, we identified the likely target gene sets for many of the motifs, and found that numerous gene sets are likely to be co-regulated via shared motifs. By identifying motifs / motif combinations that are important in co-regulating target gene sets, Lever provides an entrée into targeted experimentation aimed at understanding the logic of cis regulatory elements. Lever can be applied to any cell type, gene expression pattern, or organism of interest to connect regulatory motifs to their biological functions, and to gain insight into the architecture of transcriptional regulatory networks.
Results
Identification of CRMs by PhylCRM
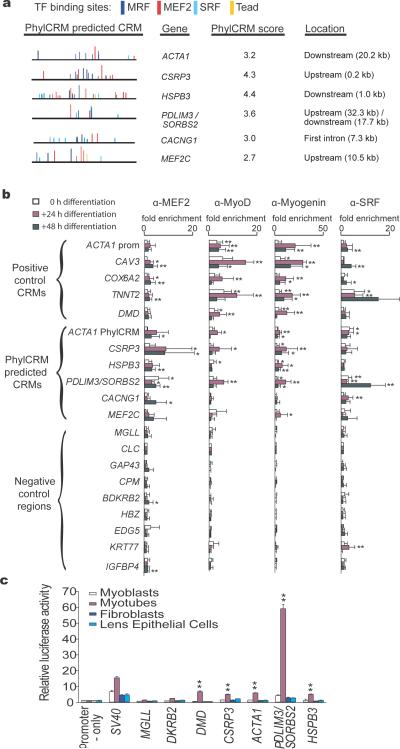
Candidate CRMs are first identified and scored with a new tool termed “PhylCRM” (pronounced “fulcrum”; Supplementary Figures 1-3 online), which scans the genomes of interest for matches to an input set of regulatory motifs. PhylCRM combines data for individual motif occurrences scored on an alignment using the previously described MONKEY scoring scheme18 into a single CRM prediction (see Methods). PhylCRM can scan very long (here, 75-kb) genomic sequences for candidate CRMs by quantifying both motif clustering and conservation across arbitrarily many genomes using an evolutionary model consistent with the phylogeny of the genomes. In the Lever analyses described in this study, we utilized the phylogenetic tree containing all 8 sequenced mammalian genomes (human, chimp, macaque, mouse, rat, dog, cow, and opossum) (Supplementary Figure 4 online)19. Significantly scoring candidate CRMs of varying lengths, ranging from 20 to 500 bp, are identified and scored to identify the maximum scoring window for each gene (“Step 1” in Figure 1b; Figure 4a). PhylCRM can also be used as a stand-alone program for CRM prediction.
Figure 4. Experimental validation of computationally predicted CRMs.
(a) Predicted human CRMs. PhylCRM scores are -log10(PhylCRM P-value) of the given sequence window and the MRF AND MEF2 motif combination, which showed greatest enrichment among the sarcomeric gene set. Window locations are relative to transcriptional start or transcriptional stop of the nearest gene(s); intronic window locations are relative to transcription start (b) Predicted CRMs are enriched for TF occupancy during myogenic differentiation. Anti-MEF2, anti-MyoD, anti-myogenin, and anti-SRF antibodies were used in biological triplicate ChIP assays. Fold-enrichment was calculated relative to mock ChIPs using anti-IgG. * P ≤ 0.05; ** P ≤ 0.005. “ACTA1 prom” is a previously described muscle CRM; “ACTA1 PhylCRM” was newly predicted. (c) Luciferase reporter assays for predicted novel CRMs indicate activity in myotubes. MGLL and BDKRB2 are negative control regions; DMD is a positive control muscle CRM; CSRP3, ACTA1, PDLIM3/SORBS2, and HSPB3 are four predicted novel CRMs. ** significant (P ≤ 0.005) increase in luciferase activity relative to the empty vector negative control.
Scoring GM-pairs by Lever
CRM scores for all genes in the genome (predicted by PhylCRM), and a collection of gene sets, are then input into Lever. In order to evaluate GM-pairs, Lever first assigns, to each gene in the “foreground” gene set of interest, which is input by the user, and to each gene in the automatically created, length-matched “background”, the PhylCRM score of the best scoring CRM. Considering all the genes in the foreground gene set and all background genes, the genes are then ranked according to the PhylCRM score of each gene's single best scoring candidate CRM (“Step 2” in Figure 1b). Then, for each entry in the GM-pair matrix, Lever calculates both the value of the corresponding area under the curve in a receiver operator characteristic (ROC) plot (“AUC score”) (“Steps 3 and 4” in Figure 1b) and its corresponding Q-value (“Step 5” in Figure 1b). The AUC score indicates the probability that a randomly chosen member of the foreground gene set will rank higher than a randomly chosen background gene, while the Q-value indicates the false discovery rate (Figure 1b; see Methods). In initial positive control analyses, we considered the four well-known myogenic TF binding site motifs for the transcriptional activators20 MEF2, Serum Response Factor (SRF), Tead, and the myogenic regulatory factors (MRFs) MyoD, Myogenin, Myf5 and Myf6, and showed that a statistically significant motif enrichment can be detected when scanning 75-kb regions of genomic sequence (Supplementary Figure 4 online).
Identification of myogenic gene sets to be examined by Lever
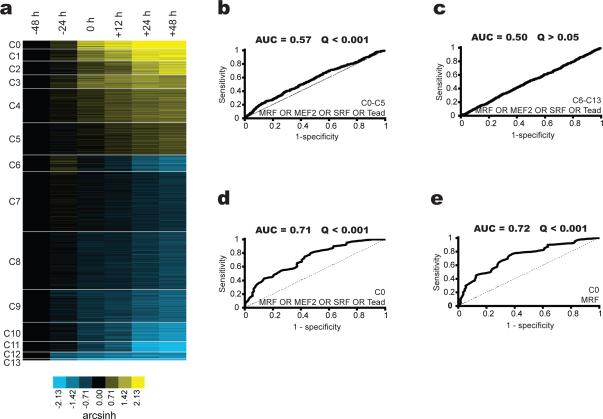
We considered two sources of gene sets: (1) clusters of co-expressed genes, and (2) GO categories. To identify appropriate gene expression clusters for examining the functions of motifs during myogenic differentiation, we first performed expression profiling over a time course of the differentiation of primary human skeletal myoblasts into myotubes at -24, -12, 0, +12, +24, and +48 hours relative to stimulation of differentiation (see Methods). We discovered 591 up-regulated and 1,070 down-regulated genes at a false discovery rate (FDR) of 5% (see Methods). Using k-means clustering, we partitioned these genes into 14 expression clusters (Figure 2a; Supplementary Table 1 online; see Methods), many of which showed enrichment for GO annotation terms consistent with myogenic differentiation (Supplementary Table 2 online). We excluded cluster C13 from Lever analyses because it contained only 12 genes. As additional gene sets to be examined by Lever, we identified the GO categories that were significantly enriched within either the up- or down-regulated genes during the time-course of myogenic differentiation, and took their intersection with either the up- or down-regulated genes, yielding a final total of 101 gene sets (see Methods). We did not utilize Gene Ontology categories alone as gene sets in this study.
Figure 2. Analysis of time course of human skeletal muscle differentiation.
(a) Expression clusters from gene expression profiling data for human adult primary skeletal muscle cells at the indicated time points with respect to stimulation of differentiation,. Arcsinh values are relative to the -48 hrs time point. Shown here are the genes that are differentially expressed at a false discovery rate of 5%. (b-e) Evaluation of enrichment using as a foreground sequence set the 75-kb regions surrounding transcription start for the (b) MRF OR MEF2 OR SRF OR Tead motifs for all genes in clusters C0 through C5, (c) MRF OR MEF2 OR SRF OR Tead motifs for all genes in clusters C6 through C13, (d) MRF OR MEF2 OR SRF OR Tead motifs for all genes in cluster C0, (e) MRF motif for all genes in cluster C0.
Evaluation of Lever using four myogenic motifs and gene expression clusters
We first applied Lever to systematically analyze each of the myogenic differentiation expression clusters considering all four of the myogenic motifs MRF, MEF2, SRF and Tead individually, and also in Boolean (“AND”, “OR”, and “NOT”) combinations (see Methods). In evaluating the degree of enrichment for motifs within gene sets, we simultaneously considered the AUC and Q-value. For example, when we examined the collection of all ~500 up-regulated genes (i.e., “C0-C5” in Figure 2a) using all four myogenic motifs, we observed only slight but significant enrichment (Figure 2b, AUC = 0.57 ± 0.01; Q ≤ 0.001). Thus, we can be highly confident that targets of these four motifs exist within the set of all up-regulated genes, yet finding specific target genes within this set would be difficult. Conversely, when we examined the set of all down-regulated genes (“C6-C13”), we observed no enrichment at all with the 4-way OR combination of these four motifs (AUC = 0.50 ± 0.01; Q > 0.05; Figure 2c). We observed strongest enrichment for these four motifs among the most up-regulated genes (cluster C0) (AUC = 0.71 ± 0.05; Q ≤ 0.001; Figure 2d). Within cluster C0, the MRF motif alone showed slightly greater enrichment (AUC = 0.72 ± 0.04; Q ≤ 0.001; Figure 2e) than all four motifs together, indicating that most of the enrichment from the 4-way “OR” combination of motifs was likely due to the MRF motif.
We generally observed greatest enrichment of these four motifs within up-regulated expression clusters (Supplementary Figure 5 online; Supplementary Table 3a online), with the notable exception of the C12 cluster of down-regulated genes which contains many genes involved in cell cycle function (Supplementary Table 2 online). The enrichment observed here is consistent with an observation from another group suggesting the existence of MRF targets involved in cell cycle progression and proliferation21. Results of additional Lever analysis controls are shown in Supplementary Results online.
Lever screen of 174 candidate regulatory motifs across 101 myogenic gene sets
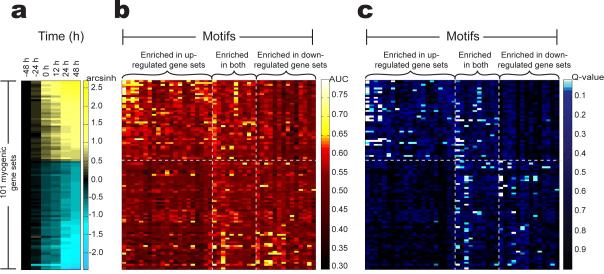
In order to identify additional motifs that might be involved in the regulation of myogenic gene sets, we performed a Lever analysis of the 101 myogenic gene sets (Figure 3a) using a dictionary of 174 candidate human regulatory motifs computationally predicted by Xie et al.9 considering 4-kb proximal promoter regions. Out of these 17,574 GM-pairs, we observed a total of 173 statistically significant (Q ≤ 0.05) GM-pairs, involving a total of 45 distinct motifs and 61 distinct gene sets (Figure 3b-c; Supplementary Table 3c online). These 45 motifs could be broadly classified into 3 categories: 1) 21 motifs enriched among only up-regulated gene sets (Figure 3b-c), 2) 10 motifs enriched among both up-regulated and down-regulated gene sets (Figure 3b-c), and 3) 14 motifs enriched among only down-regulated gene sets (Figure 3b-c).
Figure 3. Lever screen of 101 myogenic gene sets using a dictionary of 174 motifs.
(a) Median signal intensity throughout the time-course of gene expression profiling for each of 101 gene sets. (b) AUC scores for each GM-pair when considering each of the 174 motifs from Xie et al.9. (c) FDR Q-value for each GM-pair. We note that in the heat maps shown in (b) and (c), only the 45 motifs with statistically significant enrichment (Q ≤ 0.05) in at least one of the 101 myogenic gene sets are displayed. The columns of matrices (b) and (c) are sorted by decreasing overall correlation with gene expression at time +48 h. The rows of the heat maps shown in (a-c) were sorted in order of decreasing median expression arcsinh values at time point +48 h (relative to -48 h).
Several of the motifs that were part of statistically significant GM-pairs resulting from this Lever analysis correspond to the DNA binding site motifs of TFs known to function during myogenesis, including AP-1 (ref. 22), Elk-1 (ref. 23), and Pitx2 (ref. 24). We note that this dictionary of candidate regulatory motifs contained matches to the MRF, MEF2 and Tead motifs, all of which were again observed to be statistically significantly enriched in various gene sets (see Supplementary Table 3 online). For example, all of the motifs that we observed to be enriched within the sarcomeric gene set corresponded to discretized versions of either the MRF (CAGCTG, GCAnCTGnY), MEF2 (YTATTTTnR, TAAWWATAG, CTAWWWATA) or Tead (WGGAATGY) motifs.
In examining the results from this Lever analysis, we identified some interesting connections between gene sets. For example, we found that the motif GATTGGY (corresponding to the NF-Y motif) is enriched among the up-regulated lipid biosynthesis genes, the down-regulated chromatin genes, various down-regulated organelle gene sets, and a number of down-regulated gene sets involved in the cell cycle. Likewise, we found that the down-regulated plasma membrane genes appear to be co-regulated via the motif TGAnTCA (corresponding to the AP-1 motif) with a number of gene sets including response to stress, cell proliferation, and regulation of cell proliferation, and the up-regulated plasma membrane genes appear to be co-regulated via the motif CTAWWWATA (corresponding to the MEF2 motif) with a number of up-regulated gene sets involving structural properties of muscle cells, including cytoskeletal protein binding, contractile fiber, structural constituent of muscle, and actin cytoskeleton.
Interestingly, we see that certain motifs appear to regulate a large cohort of gene sets. For example, we see that the motif GATTGGY (corresponding to the NF-Y motif) co-regulates a rather large number of gene sets involved in the cell cycle. The suppression of NF-Y function has been shown previously to be important for the inhibition of several cell cycle genes and the induction of the early muscle-specific program in post-mitotic muscle cells25. Similarly, the motif TGAnTCA (annotated by Xie et al.9 as the AP-1 motif) co-regulates a number of gene sets pertaining to cell proliferation and the plasma membrane. AP-1 complexes previously have been shown to be involved in the control of duration of myoblast proliferation and fusion efficiency22.
Validation of novel CRMs that drive expression during human myogenesis
We experimentally tested 6 CRMs predicted by PhylCRM (schematized in Supplementary Figure 7 online) and consisting of the MRF AND MEF2 motif combination (Figure 4a; Supplementary Figure 6 online). We sampled CRMs from various genomic locations relative to transcriptional start, with a range of PhylCRM scores. Four of these six candidate CRMs were adjacent to genes with known or predicted sarcomeric function; two of these predicted CRMs (ACTA1 and PDLIM3/ SORBS2) are more than 17 kb away from their predicted target transcripts. Since Lever analysis identified significant enrichment (AUC = 0.82 ± 0.04; Q ≤ 0.001) for the Boolean motif combination MRF AND MEF2 in the set of sarcomeric genes (Supplementary Figure 6 online), choosing two of the six candidate CRMs to be adjacent to genes not involved in sarcomeric function also allowed us to explore whether CRMs containing this particular motif combination might function for non-sarcomeric genes.
The seven genes adjacent to these six predicted CRMs were up-regulated during differentiation (Supplementary Figure 8 online), and myogenic TFs were differentially expressed at the protein level during differentiation (Supplementary Figure 9 online). Chromatin immunoprecipitation assays followed by region-specific quantitative PCR (see Methods) showed that four of the six candidate CRMs were significantly enriched for binding by MEF2 (P ≤ 0.05), MyoD (P ≤ 0.05) and myogenin (P ≤ 0.005) (Figure 4b). Interestingly, of the six tested CRMs, the four that showed significant binding by MEF2, MyoD, and myogenin were the ones that are located next to genes involved in sarcomeric function, whereas the two that did not show significant binding by these factors are not. Although this does not tell us what sequence features distinguish the active from the inactive CRMs, it does suggest that the choice of the likely target gene sets is important in predicting CRMs that are active in a given condition (here, myogenic differentiation).
We performed luciferase assays for the four novel, candidate CRMs that were enriched for in vivo TF binding. All four of these candidate CRMs resulted in statistically significant (P ≤ 0.05) activation of luciferase expression during myogenic differentiation, but not in either fibroblasts or lens epithelial cells (Figure 4c). ShRNA knockdowns of MEF2D, myogenin, or SRF (Supplementary Figure 10 online) confirmed that these four candidate CRMs drive expression specifically in response to myogenic differentiation (Supplementary Figure 11a-c online). Results for a synthetic CRM suggest that there are further sequence requirements aside from the MRF and MEF2 motifs (Supplementary Figure 12 online). A detailed description of these experimental validations is provided in Supplementary Results online.
Functional annotation of regulatory motifs
The identification of statistically significant GM-pairs involving GO categories allowed us to assign to a regulatory motif the functional annotation of the GO categories within which it shows statistically significant enrichment. For example, we see that a discretized form of the MEF2 DNA binding site motif) is enriched among many GO categories related to muscle contraction, including contractile fiber, muscle contraction, and actin cytoskeleton, consistent with recently published ChIP-chip results21. Importantly, Lever was able to identify these regulatory associations using only sequence data and gene expression data. In addition, while that ChIP-chip study21 identified surprisingly few MEF2/MyoD/Myogenin targets to be involved in cell cycle progression, our Lever results not only agreed with these findings, but also identified additional motifs, including AP-1, that are likely to be involved in the down-regulation of the cell cycle during myogenesis. A number of known regulatory interactions were missed because of the stringency of our statistical analyses, primarily because of our need to correct for the many hypotheses tested (over 17,500 GM-pairs) in our large Lever analysis of 174 motifs across 101 gene sets (Supplementary Table 3 online).
We can also apply this annotation method to the 13 motifs, belonging to 30 statistically significant GM-pairs, for which the trans factors that may bind them have not yet been discovered (Supplementary Table 3 online). For example, we found that the putative regulatory motif TGACATY can be annotated as being involved in the regulation of plasma membrane genes. Importantly, this level of functional annotation is much more specific than just indicating the tissue-specificity of the genes upstream of which the motif is found9. We note that these annotations indicate the functions of the motifs during myogenic differentiation, and that the motifs may serve other functions in other cell types or in response to other environmental stimuli.
Discussion
We have presented a systematic method for the unbiased inference of regulatory motif function by examining a large collection of gene sets with a dictionary of known or predicted regulatory motifs. We have shown that our algorithm is effective in a mammalian setting, where distal CRMs can exist at great distances from the transcription start sites of the genes they regulate. Importantly, our approach goes beyond recent efforts at metazoan CRM identification by identifying motifs / motif combinations and their target gene sets in an automated manner. Our approach was able to identify known myogenic regulatory motifs when examining 75-kb regions rather than just proximal promoter regions, and was also able to identify several additional motifs as enriched in muscle-related gene sets.
This level of functional annotation is an important step in moving from a listing of candidate regulatory motifs, towards a functional understanding of the biological roles of such motifs. Our approach also allows for de novo reconstruction of transcriptional regulatory networks, without any prior knowledge of the functions of the examined regulatory motifs. We anticipate that this method will also be useful for the analysis of candidate regulatory motifs and gene sets from other biological systems, including other metazoans. Indeed, with motif dictionaries being derived either computationally or experimentally by high-throughput methods for identifying TFs' DNA binding sites, the next major challenges will be the identification of the CRMs that contain those motifs and the mapping of those motifs and CRMs to the biological processes that they regulate. Lever analyses could be performed using any gene sets of interest. The utility of our computational framework will greatly increase in the coming years as expanded genome-wide motif dictionaries are both predicted computationally11 and also experimentally derived13,16 using genome-scale techniques.
In this study, we were able to choose an appropriate subset of species to consider in scoring phylogenetic conservation, based on the evaluation of Lever on a positive control set of myogenic CRMs. However, the choice of the most suitable set of species to use will not always be determined as readily, particularly in the absence of a positive control set of CRMs. Future work on identifying the gene expression patterns of orthologous TFs will provide useful data for choosing the appropriate set of species to consider in evaluating phylogenetic conservation of their corresponding DNA binding site motifs. However, even with conservation of expression of the orthologous TFs, the binding site composition and locations of CRMs may still diverge rapidly26.
This method represents a major genome-wide step in moving from a motif dictionary to understanding the language of cis regulation. Although Lever analysis does not directly inform us of what sequence features within the candidate CRMs distinguish the active from the inactive CRMs, it does suggest that the choice of the likely target gene sets is important in predicting CRMs that are active in a given condition (here, myogenic differentiation). Improved computational methods and experimental testing of both native and synthetic CRMs will be important for deciphering the `grammar' of how regulatory motifs must be organized within sequence windows in order to construct CRMs that are active in a given cellular and environmental context.
Methods
Genomic sequences utilized in this study
We obtained all genomic sequences for any scans utilized in this paper from the University of California Santa Cruz (UCSC) Genome Browser Hg17 assembly. For alignments, we utilized all genomes and alignments available at the time we began our study, corresponding to the “Multiple alignments of 8 vertebrate genomes with Human”, along with pairwise alignments for macaque, cow and opossum. For annotation of gene coordinates, we used the UCSC “refGene” and “all_mrna” files. All sequences were repeat masked using the RepeatMasking provided by UCSC. We also masked out all exonic regions (exon coordinates were obtained from the refGene files).
We obtained from the supplementary data of Wasserman et al.27 a collection of 27 muscle CRMs containing matches to at least one of the MRF, MEF2, SRF, or Tead DNA binding site motifs (we note that our “Tead” motif is the same as their “Tef” motif). Genomic coordinates of positive control CRMs, negative control regions, and PhylCRM predicted CRMs are provided in Supplementary Table 4.
PhylCRM: A computational approach for finding CRMs by quantifying motif clustering and evolutionary conservation
Briefly, PhylCRM takes as input a set of pre-defined DNA motifs, a set of aligned genomic sequences within which to search for candidate CRMs comprising a particular group of motifs, and a tree indicating the phylogeny of the genomes. PhylCRM scans for the presence of TF binding site motifs using sliding windows of continuously varying sizes, since CRMs span a wide range of lengths. For each motif, it scans the aligned sequences and quantifies the degree to which each position is a phylogenetically conserved motif match, utilizing the MONKEY scoring model18 to evaluate the degree to which that position is both a conserved and a high affinity match to the TF binding site motif (depicted as colored spikes in Figure 4a). Then, for each TF binding site motif and for each window within a user-defined size range, the summation of these motif match scores is computed, and its statistical significance is evaluated using an empirically derived probability distribution of the window scores to give a motif output score. This probability distribution depends on the TF binding site motif and on the window size and is generated by inspecting all of the genomic sequences (here, 50 kb upstream and 25 kb downstream of transcription start site) with a sliding window of fixed size (see Supplementary Methods). The motif output scores from all of the motifs are combined into one output score. This output score is computed differently depending on the Boolean motif combination that is considered. This score simultaneously reflects motif over-representation and evolutionary conservation when scoring entire windows of sequence containing multiple TF binding site motifs. Because PhylCRM provides a continuous (i.e., non-binary) measure of motif enrichment within a flanking region, we sought a similarly continuous set of logical AND, OR and NOT logical operations when combining several motifs. Therefore we utilized concepts from Fuzzy logic28, where statements have a gradual assessment of being either “true” or “false”. A complete description of the PhylCRM scoring scheme is provided in Supplementary Methods and Supplementary Figures 1-3 online.
Lever
The statistical framework of Lever is based upon principles used by other groups for gene set enrichment analysis29 and utilizes permutation-based adjustment for multiple hypothesis testing. However, in contrast to gene set enrichment analysis, in the Lever framework genes are ranked by a sequence-based, rather than an expression-based, scoring function, and each combination of motifs gives rise to a distinct scoring function. For each gene set and scoring function, the ranking power of the function is statistically assessed by calculating the enrichment for highly scoring genes within the gene set. Thus, Lever simultaneously calculates and assesses the enrichment for many gene sets across many motif combinations (i.e., GM-pairs).
Noncoding foreground and background sequence regions examined by Lever
For each gene in each of these foreground gene sets, we obtained 75 kb of genomic sequence overlapping transcription start (50 kb upstream of transcriptional start and 25 kb downstream of transcriptional start). As a background set, we obtained a collection of non-overlapping, 75-kb genomic sequences for genes that were observed to be “present” in the expression microarray data but not up- or down-regulated at a FDR less than 0.1. For each foreground gene set we selected a length-matched background set17 in order to remove the possibility that any observed enrichment for high scoring candidate CRMs could be solely due to a larger search space. For each foreground gene set a background gene set is automatically built that is as large as possible (usually 10 to 40 times as large as the foreground) so that the overall distribution of lengths in the foreground and background sets is well-matched (see Supplementary Methods).
Statistical analyses in data processing
Over-representation of GO annotation terms in various gene sets was determined using FuncAssociate, a web-based program that corrects for multiple hypothesis testing30. Significant changes in luciferase reporter arrays and ChIPs were determined by Student's unpaired two-tailed t-tests.
Additional methods
Detailed descriptions of all methods can be found online in Supplementary Methods, including: construction of length-matched background sets against which foreground gene sets are evaluated in Lever; description of PhylCRM scoring scheme; evaluation of ability of PhylCRM to identify CRMs; comparison of PhylCRM to other CRM prediction methods; Lever; further discussion of interpretation of CRM enrichment results from Lever; position weight matrices utilized in this study; details of all experimental protocols, including primer sequences.
Accession numbers
MIAME-compliant microarray data in SOFT format and complete protocols have been deposited in the Gene Expression Omnibus (GEO) database under series GSE4460.
Software availability
Upon acceptance of this manuscript for publication, the PhylCRM and Lever programs will be made publicly available by download from the Bulyk lab webpage (http://the_brain.bwh.harvard.edu).
Supplementary Material
ACKNOWLEDGMENTS
The authors thank E. Margulies and the ENCODE Multiple Sequence Alignment working group for generously allowing use of their phylogenetic tree prior to its publication; S. Asthana, S. Sunyaev, G. Kryukov, M. Berger, T. Siggers and A. Aboukhalil for helpful discussions; J. Chee, E. Mathewson and T. Sierra for technical assistance; S. Elledge, A. Friedman, T. Siggers, M. Berger and F. De Masi for critical reading of the manuscript; A. Donner (Brigham & Women's Hospital) for the generous gift of human lens epithelial cells; and K. Cichowski (Brigham & Women's Hospital) for kindly providing lentiviral reagents. This work was funded in part by a PhRMA Foundation Informatics Research Starter Grant (M.L.B.), a William F. Milton Fund Award (M.L.B.), an HST Taplin Award (M.L.B.) and NIH/NHGRI R01 HG02966 (M.L.B.). J.B.W. was supported in part by NIH Training Grant T32 HL07627 and NIH Individual NRSA F32 AR051287. A.A.P. was supported in part by a National Defense Science and Engineering Graduate Fellowship from the Department of Defense, and an Athinoula Martinos Fellowship from HST. S.A.J. was supported in part by an NSF Postdoctoral Research Fellowship in Biological Informatics. We apologize to authors whose work we could not cite due to space limitations.
Footnotes
Note: Supplementary Information is available on the Nature Methods website and on our lab website, http://the_brain.bwh.harvard.edu/.
COMPETING INTERESTS STATEMENT The authors declare that they have no competing financial interests.
References
- 1.Bulyk ML. Computational prediction of transcription-factor binding site locations. Genome Biol. 2003;5:201. doi: 10.1186/gb-2003-5-1-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blanchette M, et al. Genome-wide computational prediction of transcriptional regulatory modules reveals new insights into human gene expression. Genome Res. 2006;16:656–68. doi: 10.1101/gr.4866006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hallikas O, et al. Genome-wide prediction of mammalian enhancers based on analysis of transcription-factor binding affinity. Cell. 2006;124:47–59. doi: 10.1016/j.cell.2005.10.042. [DOI] [PubMed] [Google Scholar]
- 4.Pennacchio LA, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- 5.Thompson W, Palumbo MJ, Wasserman WW, Liu JS, Lawrence CE. Decoding human regulatory circuits. Genome Res. 2004;14:1967–74. doi: 10.1101/gr.2589004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou Q, Wong WH. CisModule: de novo discovery of cis-regulatory modules by hierarchical mixture modeling. Proc. Natl. Acad. Sci. USA. 2004;101:12114–9. doi: 10.1073/pnas.0402858101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wasserman W, Fickett J. Identification of regulatory regions which confer muscle-specific gene expression. J. Mol. Biol. 1998;278:167–181. doi: 10.1006/jmbi.1998.1700. [DOI] [PubMed] [Google Scholar]
- 8.Philippakis AA, He FS, Bulyk ML. Modulefinder: a tool for computational discovery of cis regulatory modules. Pac. Symp. Biocomput. 2005:519–30. [PMC free article] [PubMed] [Google Scholar]
- 9.Xie X, et al. Systematic discovery of regulatory motifs in human promoters and 3' UTRs by comparison of several mammals. Nature. 2005;434:338–45. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elemento O, Tavazoie S. Fast and systematic genome-wide discovery of conserved regulatory elements using a non-alignment based approach. Genome Biol. 2005;6:R18. doi: 10.1186/gb-2005-6-2-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huber BR, Bulyk ML. Meta-analysis discovery of tissue-specific DNA sequence motifs from mammalian gene expression data. BMC Bioinformatics. 2006;7:229. doi: 10.1186/1471-2105-7-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ettwiller L, et al. The discovery, positioning and verification of a set of transcription-associated motifs in vertebrates. Genome Biol. 2005;6:R104. doi: 10.1186/gb-2005-6-12-r104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bulyk ML. DNA microarray technologies for measuring protein-DNA interactions. Curr. Opin. Biotechnol. 2006;17:422–30. doi: 10.1016/j.copbio.2006.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bulyk ML, Huang X, Choo Y, Church GM. Exploring the DNA-binding specificities of zinc fingers with DNA microarrays. Proc. Natl. Acad. Sci. USA. 2001;98:7158–63. doi: 10.1073/pnas.111163698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mukherjee S, et al. Rapid analysis of the DNA-binding specificities of transcription factors with DNA microarrays. Nat Genet. 2004;36:1331–9. doi: 10.1038/ng1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berger MF, et al. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat. Biotechnol. 2006;24:1429–1435. doi: 10.1038/nbt1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Philippakis AA, et al. Expression-guided in silico evaluation of candidate cis regulatory codes for Drosophila muscle founder cells. PLoS Computational Biology. 2006;2 doi: 10.1371/journal.pcbi.0020053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moses AM, Chiang DY, Pollard DA, Iyer VN, Eisen MB. MONKEY: identifying conserved transcription-factor binding sites in multiple alignments using a binding site-specific evolutionary model. Genome Biol. 2004;5:R98. doi: 10.1186/gb-2004-5-12-r98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Margulies EH, et al. Analyses of deep mammalian sequence alignments and constraint predictions for 1% of the human genome. Genome Res. 2007;17:760–74. doi: 10.1101/gr.6034307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Messenguy F, Dubois E. Role of MADS box proteins and their cofactors in combinatorial control of gene expression and cell development. Gene. 2003;316:1–21. doi: 10.1016/s0378-1119(03)00747-9. [DOI] [PubMed] [Google Scholar]
- 21.Blais A, et al. An initial blueprint for myogenic differentiation. Genes Dev. 2005;19:553–69. doi: 10.1101/gad.1281105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daury L, et al. Opposing functions of ATF2 and Fos-like transcription factors in c-Jun-mediated myogenin expression and terminal differentiation of avian myoblasts. Oncogene. 2001;20:7998–8008. doi: 10.1038/sj.onc.1204967. [DOI] [PubMed] [Google Scholar]
- 23.Wang Z, et al. Myocardin and ternary complex factors compete for SRF to control smooth muscle gene expression. Nature. 2004;428:185–9. doi: 10.1038/nature02382. [DOI] [PubMed] [Google Scholar]
- 24.Martinez-Fernandez S, et al. Pitx2c overexpression promotes cell proliferation and arrests differentiation in myoblasts. Dev. Dyn. 2006;235:2930–9. doi: 10.1002/dvdy.20924. [DOI] [PubMed] [Google Scholar]
- 25.Gurtner A, et al. Requirement for down-regulation of the CCAAT-binding activity of the NF-Y transcription factor during skeletal muscle differentiation. Mol. Biol. Cell. 2003;14:2706–15. doi: 10.1091/mbc.E02-09-0600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ludwig MZ, Bergman C, Patel NH, Kreitman M. Evidence for stabilizing selection in a eukaryotic enhancer element. Nature. 2000;403:564–7. doi: 10.1038/35000615. [DOI] [PubMed] [Google Scholar]
- 27.Wasserman W, Palumbo M, Thompson W, Fickett J, Lawrence C. Human-mouse genome comparisons to locate regulatory sites. Nat. Genet. 2000;26:225–8. doi: 10.1038/79965. [DOI] [PubMed] [Google Scholar]
- 28.Kasabov NK. Foundations of Neural Networks, Fuzzy Systems, and Knowledge Engineering. 1998;550 [Google Scholar]
- 29.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–73. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 30.Berriz GF, King OD, Bryant B, Sander C, Roth FP. Characterizing gene sets with FuncAssociate. Bioinformatics. 2003;19:2502–4. doi: 10.1093/bioinformatics/btg363. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.