Abstract
Glycoside hydrolase family 32 (GH32) harbors hydrolyzing and transglycosylating enzymes that are highly homologous in their primary structure. Eight amino acids dispersed along the sequence correlated with either hydrolase or glycosyltransferase activity. These were mutated in onion vacuolar invertase (acINV) according to the residue in festuca sucrose:sucrose 1-fructosyltransferase (saSST) and vice versa. acINV(W440Y) doubles transferase capacity. Reciprocally, saSST(C223N) and saSST(F362Y) double hydrolysis. SaSST(N425S) shows a hydrolyzing activity three to four times its transferase activity. Interestingly, modeling acINV and saSST according to the 3D structure of crystallized GH32 enzymes indicates that mutations saSST(N425S), acINV(W440Y), and the previously reported acINV(W161Y) reside very close together at the surface in the entrance of the active-site pocket. Residues in- and outside the sucrose-binding box determine hydrolase and transferase capabilities of GH32 enzymes. Modeling suggests that residues dispersed along the sequence identify a location for acceptor-substrate binding in the 3D structure of fructosyltransferases.
Electronic supplementary material
The online version of this article (doi:10.1007/s11103-008-9404-7) contains supplementary material, which is available to authorized users.
Keywords: Invertase, Fructosyltransferase, Sucrose, Transglycosylation, Site-directed mutagenesis, Molecular modeling
Introduction
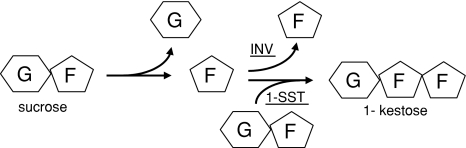
Plant vacuolar invertases and fructosyltransferases belong to glycoside hydrolase family 32 (GH32). Invertases are ubiquitous in plants whilst fructosyltransferases are restricted to several unrelated families (Hendry 1993). Protein sequences of plant fructosyltransferases generally have a very high similarity to sequences of different invertases (Sprenger et al. 1995; Vijn and Smeekens 1999). If vacuolar invertases are presented in phylogenetic trees with fructosyltransferases, fructan exo-hydrolases (FEHs) and cell-wall invertases, the vacuolar invertases cluster with fructosyltransferases whilst the FEHs cluster with cell-wall invertases (Ritsema and Smeekens 2003; Van den Ende et al. 2000; Wei and Chatterton 2001). Invertases and fructosyltransferases are also functionally related; they split off a fructosyl residue from a substrate and retain this residue in there active site while releasing the substrate, afterwards the fructosyl residue is released to water in the case of invertases and transferred to a saccharide-chain in the case of fructosyltransferases (Fig. 1).
Fig. 1.
Schematic drawing of the activities of invertase and the fructosyltransferase 1-SST. Sucrose is split into glucose and fructose, the glucose is released and fructose stays bound to the enzyme, subsequently fructose is released via hydrolysis in the case of invertase whereas fructose is coupled to another sucrose molecule in the case of 1-SST
GH32 enzymes contain three acidic amino acids (indicated in boldface) in three highly conserved motifs—NDPNG, RDP, and EC—that are the proposed active site residues (Altenbach et al. 2005; Pons et al. 2000; Reddy and Maley 1990; Reddy and Maley 1996; Ritsema et al. 2005). Recently the 3D-structures of several hydrolases of this family were resolved, starting with an invertase from the bacterium Thermotoga maritima (Alberto et al. 2004), and now also harboring a plant enzyme, a fructan-exohydrolase (FEH) from Cichorium intybus (chicory) (Verhaest et al. 2005b). Each structure of a GH32 enzyme shows a five bladed β-propeller with a deep negatively charged central pocket, representing the active site, and attached to it a β-sandwich. The three highly conserved active-site residues flank the central pocket, and can be brought in connection with the general reaction for the cleavage of glycosidic bonds according to the retaining mechanism (Alberto et al. 2004; Meng and Fütterer 2003).
Fructosyltransferases synthesize fructans (polyfructosylsucroses) from sucrose. The enzyme that converses sucrose to the shortest β(2–1) linked fructan 1-kestose is called sucrose:sucrose 1-fructosyltransferase (1-SST) (Koops and Jonker 1996a, b; Shiomi et al. 1985; Van den Ende et al. 1996b; Vijn et al. 1998). 1-SST uses two molecules of sucrose, one as fructosyl donor and the other as fructosyl acceptor for the synthesis of the trisaccharide 1-kestose (and glucose). Higher DP fructans are synthesized from 1-kestose via the action of other fructosyltransferases, the type depends on the kind of fructan synthesized by the plant (Ritsema et al. 2003; Sprenger et al. 1995; Ueno et al. 2005; Van den Ende et al. 1996a; van der Meer et al. 1998; Vijn et al. 1997).
Studying fructosyltransferases demands an appropriate expression system. The expression of these enzymes in the yeast Pichia pastoris has been shown to be ideal, since this yeast is devoid of sucrose metabolizing enzymes such as invertases. Furthermore, expressed proteins can be secreted to the medium which diminishes the presence of disturbing components (Hochstrasser et al. 1998; Lüscher et al. 2000). As a result of these advantages Pichia pastoris is currently the most used expression system to study fructosyltransferases.
The observed close relationships at both the biochemical and the molecular level between fructan synthesizing enzymes and invertases supports the idea that fructosyltransferases evolved from vacuolar invertases by relatively few mutational changes (Hendry 1993; Sprenger et al. 1995; Vijn and Smeekens 1999; Wei and Chatterton 2001). In accordance to this we recently managed to increase the fructosyltransferase capability of a vacuolar invertase by mutating two amino acids in the sucrose-binding box (Ritsema et al. 2006).
1-SST is the fructosyltransferase that is most similar to vacuolar invertase at the biochemical level, since both enzymes use sucrose as the preferred substrate; invertases for hydrolysis and 1-SSTs for transglycosylation (Fig. 1). Furthermore, 1-SSTs generally show a low but distinct hydrolase activity and vice versa invertases produce traces of 1-kestose, particularly at sucrose concentrations above 100 mM (Obenland et al. 1993; Sprenger et al. 1995; Vijn et al. 1998). Despite the close molecular and biochemical relationships of vacuolar invertases and fructosyltransferases, and the availability of crystal structures of several hydrolases, the question what structural features determine hydrolysis or transfer of the enzymes is hardly addressed. The three active-site residues can explain the cleavage of glycosidic bonds in general, but from the crystal structures it is hard to predict what components determine transferase capability. Previous biochemical and mutational studies point to a role for amino acids in the sucrose-binding box (Ritsema et al. 2004, 2006).
To address the question what amino acids determine transfructosylation or hydrolysis outside the sucrose-binding box we compared sequences of vacuolar invertases and 1-SSTs from different plants. We found eight amino acids dispersed along the sequence that correlate with either hydrolysis or transfer and did a mutational analysis to study their role in glycosyltransfer and hydrolysis. For this analysis we chose Allium cepa invertase (acINV) and Schedonorus arundinaceus (before known as Festuca arundinacea) 1-SST (saSST), two enzymes that are very efficiently expressed in the yeast Pichia pastoris and were used before in mutagenesis studies (Altenbach et al. 2004, 2005; Ritsema et al. 2006). The selected amino acids of acINV were individually mutated to the corresponding amino acid in saSST and vice versa. Analysis of the obtained mutants revealed mutations that led to an INV with an increased glycosyltransferase capacity and a 1-SST with a predominant hydrolysis capacity. Modeling the 3D structures of saSST and acINV, we show that the mutations responsible for the observed changes seem to reside close together at the entrance of the substrate-binding pocket and do not interact with sucrose; they could indicate an acceptor-substrate binding location.
Materials and methods
Comparing vacuolar invertase and 1-SST
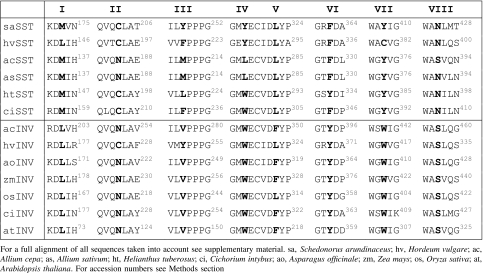
In order to identify conserved amino acid differences between invertases and 1-SST, all 1-SSTs with proven activity and many vacuolar invertases were aligned using ClustalW. We included also acid invertases from non-fructan producing plants such as Arabidopsis thaliana in order to avoid biases (Table 1 gives a representative subset of the aligned sequences). Sequences used were: Allium cepa INV (acc.nr. AJ006067), Allium cepa SST (AJ006066), Allium sativum SST (AY098442), Arabidopsis thaliana INV (S57951), Asparagus officinale INV (AF002656), Cichorium intybus INV (AJ419971), Cichorium intybus SST (U81520), Cynara scolymus SST (Y09662), Schedonorus arundinaceus (Festuca arundinacea) SST (AJ297369), Helianthus tuberosus SST (AJ009757), Hordeum vulgare INV (IAJ623275), Hordeum vulgare SST (AJ567377), Lolium perenne SST (AF492836), Oryza sativa INV (AF276703), Solanum tuberosum INV (AAA50305), Triticum aestivum INV (AJ635225), Triticum aestivum SST (AB029888), Taraxacum officinale SST (AJ250634), Vitis vinifera INV(AAB47172), Zea mays INV (U16123). A complete alignment is present in the supplemental material (Fig. S1).
Table 1.
Alignments of 1-SSTs and vacuolar invertases (INV) of different plants around the mutation sites
Cloning and mutagenesis of festuca 1-SST
Festuca 1-SST (saSST) mutants carrying single mutations at specific sites of the sequence (I, II, III, IV, V, VI, VII and VIII) were generated using the QuikChangeTM Site-Directed Mutagenesis Kit (Stratagene), based on the pK18-clone containing the tagged version of saSST (Altenbach et al. 2004). Wild type sequences and mutated sequences were cloned EcoRI/XbaI into the Pichia shuttle vector pPICZαC. All inserts in pPICZαC were sequenced after cloning and found to carry the desired mutation.
Cloning and mutagenesis of onion invertase
The onion invertase (acINV) present in pBlue (Vijn et al. 1998) was digested with SacI and XhoI and cloned into the mutagenesis vector pALTER-1 (Promega) digested with SacI and SalI (SalI and XhoI create the same overhang). Mutagenesis was performed using Altered Sites II (Promega) as described in the manual and mutations were checked by sequencing.
For cloning into the Pichia vector pPICZαC (Invitrogen) wild type invertase and mutants were PCRed with primers containing ClaI and XbaI restriction sites. After digestion of the product and the pPICZαC vector with ClaI and XbaI, they were ligated and the presence of inserts was confirmed by PCR.
Expression in Pichia pastoris and enzymatic characterization
The wild-type enzymes and all mutants were present in frame behind the yeast α-factor signal sequence of the expression vector pPICZαC, to allow entry into the secretory pathway of Pichia pastoris. Competent Pichia pastoris cells were either transformed according to the EasyComp transformation protocol (EasySelectTM Pichia Expression Kit, Invitrogen BV) or by electroporation as described in the manual and selected for Zeocine™ resistance.
Induction of protein expression in Pichia pastoris was performed as described (Altenbach et al. 2004) with minor modifications: The growth medium was supplemented by 1% (v/v) methanol at 16, 24 and 40 h of induction. After 45 h of induction the cultures were centrifuged (10 min, 3,000 rpm) and the supernatants containing the recombinant enzymes were used for all subsequent steps.
To characterize the enzymes, the supernatants were diluted 1:10 and assayed in 50 mM MES-buffer pH 5.6 with the desired sucrose concentration (1 mM to 1 M, as indicated). They were incubated for 20 min at 28°C and thereafter the reaction was stopped by heating for 2 min at 95°C. The products formed were analyzed by high performance anion exchange chromatography (HPAEC) as described before (Ritsema et al. 2006). The amounts of products were determined by comparison of peak areas with standards of known amounts.
To estimate the influence of (unknown) compounds in the medium on enzymatic activity supernatants from Pichia strains transformed with the different constructs were diluted with supernatants from wild type strains. Enzyme assays were performed with 10 μl of enzyme-containing supernatants to which 70 μl of wild type supernatants was added, instead of water. No differences were observed when water-diluted activities were compared to supernatant-diluted activities, indicating that the supernatants contain no interfering substances.
The Km of the enzymes for sucrose was calculated using a Lineweaver–Burk plot. Individual samples were measured twice and the results were averaged to obtain one independent measurement of enzyme activity at a certain sucrose concentration ranging from 1 mM till 1 M. Lineweaver–Burk plots were made per experiment and Km values determined. The error values calculated represent the standard error of the mean (SEM) resulting from the Km values calculated from three independent inductions of P. pastoris.
Molecular modeling
For acINV and saSST an initial homology model was generated with the program CPH models 2.0 (Lund et al. 2002) using as a template the crystallographic structure of a cell wall invertase from Arabidopsis thaliana (PDB code 2AC1) (Verhaest et al. 2005a). In a second step, models were modified after manual adjustments, in order to avoid side chain clashes, and using the structural information, particularly about more frequent conformations of side chains, from other homologous crystallographic structures. We used PDB codes 2ST8, fructan 1-exohydrolase IIa from Cichorium intybus; 2ADD, 1-exohydrolase IIa from Cichorium intybus; 2ADE, fructan 1-exohydrolase IIa from Cichorium intybus in complex with fructose; 2AEY, fructan 1-exohydrolase IIa from Cichorium intybus in complex with 2,5 dideoxy-2,5-imino-d-mannitol; and 2AEZ, fructan 1-exohydrolase IIa (E201Q) from Cichorium intybus in complex with 1-kestose (Verhaest et al. 2005b, 2007). Manual adjustments were performed using the program Coot (Ensley and Cowtan 2004). The final models were energetically minimized using the program CNS (Brünger et al. 1998).
Results
Selection of amino acids correlating with hydrolase or transferase activity
For the elucidation of amino acids that influence transglycosylation and hydrolysis capacity, a directed mutational approach based on sequence comparisons between acid invertases and 1-SSTs was followed. To this intend, sequences of acid invertases and 1-SSTs of monocotyledonous and dicotyledonous plants were aligned (supplementary Fig. S1). We searched for amino acids in the sequence that consistently differed between invertases in general and 1-SSTs in general. In particular, differences in amino acid polarity, hydrophobicity or hydrophily and in the type of aromatic amino acid were taken into account. Eight amino acids dispersed along the sequence could be identified which correlated either with hydrolase or glycosyl transferase activity (Sites I–VIII). Some amino acids were conserved in both monocots and dicots, others were only conserved in monocots or amongst the grass family (Table 1, supplementary material Fig. S1). Most importantly, these eight sites are located outside the highly conserved motifs NDPNG, RDP, and EC (Table 1).
Usually vacuolar invertases and fructosyltransferases consist of two subunits, a so-called large and small subunit. The large subunit has been shown to determine enzyme specificity in barley 6-SFT and festuca 1-SST (Altenbach et al. 2004, 2005). The fact that the eight amino acids correlating either with invertase or with 1-SST activity are present in the large subunit strengthens the argument that these amino acids could influence enzyme specificity.
Remarkably, half of the amino acids found to be either invertase- or 1-SST-specific turned out to be aromatic residues. Two of these four sites contain tryptophan in all aligned invertases whereas another is tyrosine in invertases (Table 1).
Functional characterization of hydrolysis and glycosyltransfer
For mutational analysis we chose as hydrolase the acid invertase of Allium cepa (acINV; accession nr. AJ006067) and as transferase the 1-SST of Schedonorus arundinaceus (saSST; accession nr. AJ297369), two enzymes that are known to be very active after expression in Pichia (Altenbach et al. 2004, 2005; Ritsema et al. 2006). The amino acids at the eight selected sites of acINV were mutated to the corresponding amino acids in saSST and vice versa (Tables 1 and 2). For both enzymes the different mutations were analyzed separately.
Table 2.
Sites I–VIII in saSST and acINV, mutations made as well as amino acid number are shown for both enzymes
| Site | saSST | activity | acINV | Activity |
|---|---|---|---|---|
| I | M173L | Wild type | L201M | Wild type |
| II | C203N | >Hydrolysis | N251C | Wild type |
| III | Y248V | None | V276Y | None |
| IV | Y317W | Wild type | W343Y | Wild type |
| V | L322F | Wild type | F348L | Wild type |
| VI | F362Y | >Hydrolysis | Y394F | None |
| VII | Y408sW | Low | W440Y | >Transferase |
| VIII | N425S | ≫hydrolysis | S457N | Wild type |
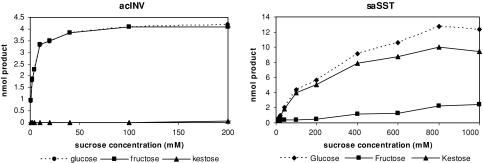
Initially, recombinant wild-type saSST and acINV were functionally characterized in order to subsequently detect possible activity shifts upon introducing the mutations. Invertases have Km-values from 2-20 mM and are generally saturated at higher sucrose concentrations. When recombinant acINV was incubated with increasing amounts of sucrose, glucose and fructose were formed. Saturation of hydrolysis activity was already seen at about 20 mM sucrose (Fig. 2). The Km was determined to be 5.8 ± 1.9 mM for sucrose, which is in agreement with Km values reported before (Ritsema et al., 2006). At sucrose concentrations ≥100 mM, a distinct but very small amount of 1-kestose was found (Fig. 2), indicating that this hydrolase is able to transfer fructose at higher sucrose concentrations, a phenomenon that was observed before (Obenland et al., 1993; Ritsema et al., 2006; Vijn et al., 1998).
Fig. 2.
Saturation curves of acINV and saSST for sucrose. Production of glucose, fructose and kestose is determined at different sucrose concentrations with the recombinant enzyme preparations incubated at 28°C for 20 min. For AcINV sucrose concentrations up to 200 mM are used, whereas saSST was tested for up to 1 M of sucrose. AcINV is saturated at relatively low sucrose concentrations, whereas saSST shows increased product formation with higher sucrose concentrations
In contrast to invertases, fructosyltransferases do not show Michaelis-Menten kinetics and are essentially non saturable for their substrate (Cairns et al., 1989). The Km has to be estimated and is generally considered to be several hundred mM (Koops and Jonker 1996a, 1996b; Lüscher et al., 2000; Van den Ende et al., 1996b). Incubation of recombinant saSST with increasing amounts of sucrose led to elevated production of glucose and 1-kestose. Also minor amounts of fructose were observed, indicative for hydrolase activity (Fig. 2). This hydrolase activity was relatively low at all offered sucrose concentrations as compared to the glycosyltransferase activity. The maximum amount of fructose produced from sucrose was around 20% leaving 80% for the transfer product 1-kestose. Substrate saturation appeared only at sucrose concentrations ≥800 mM and the Km could not be determined. The differences in the saturation characteristics for sucrose with the two enzymes, and their relatively low side activities, are the ideal basis for analyzing possible shifts in enzymatic activity upon mutational changes.
A single amino acid mutant of the transferase 1-SST renders it a hydrolase
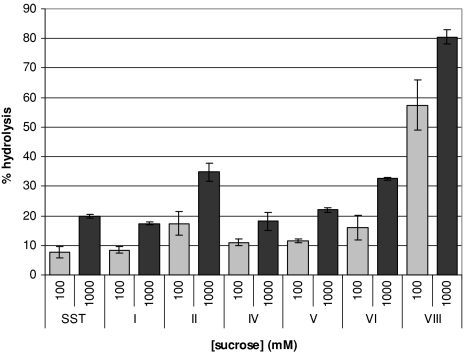
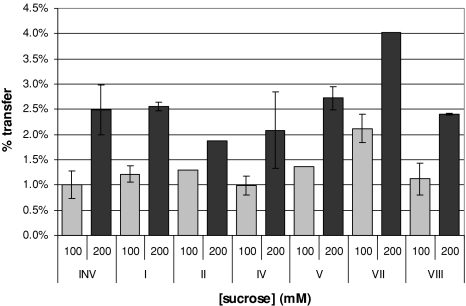
Most saSST mutants, except the one at site III, gave rise to active enzyme (Table 2). saSST mutated at site VII showed only a low residual activity at high sucrose concentrations and after prolonged incubations, for that reason it was not taken into account in our measurements (data not shown, Table 2). The active saSST mutants were incubated with sucrose for 20 min. at concentrations of 10, 20, 40, 100, 200, 400, 600, 800, and 1,000 mM. As can be seen in Fig. 3 for 100 and 1,000 mM, mutants I, IV, and V revealed no change in relative transferase (80–90%, depending on the sucrose concentration) or hydrolase (10–20%) activities as a percentage of the total of hydrolysis plus transfer activity and as compared to the recombinant wild-type saSST (Fig. 3, Table 2).
Fig. 3.
Hydrolysis as a percentage of total hydrolysis plus transfer activity by the transferase saSST and its mutants at sites I–VIII (Table 2) incubated at 28°C for 20 min with different sucrose concentrations. Hydrolysis is measured through the fructose released and increases with increasing sucrose concentrations. Measurements from three independent protein expression experiments were analyzed, the error bars represent the standard error
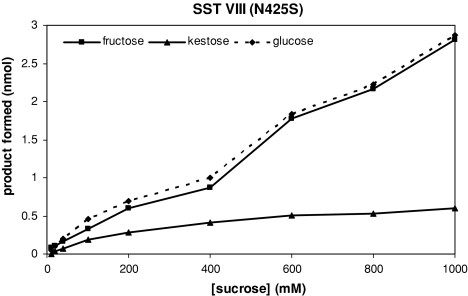
The saSST mutants at sites II (C203N) and VI (W362Y) had an almost doubled relative hydrolytic activity in comparison to wild type saSST (Fig. 3). This occurred at all sucrose concentrations offered. Of another order is the change in products made by saSST mutant VIII (N425S). In comparison to wild type saSST, this mutant had almost inverted ratios of hydrolysis versus glycosyltransfer (Figs. 3, 4). The enzyme’s relative hydrolytic activity was approximately 80% at 1 M sucrose, leaving 20% of glycosyltransfer. For wild type saSST the relative hydrolytic activity was only 20% at 1 M sucrose. Although the saSST mutant carrying the mutation at site VIII became a hydrolase, it did not become a typical invertase. Its capacity to synthesize 1-kestose is still higher then that of INV (5–6% at 1 M sucrose; data not shown). In addition, the mutant shows lack of substrate saturation, and in that respect still resembles wild type saSST (Figs. 1, 4).
Fig. 4.
Saturation curves for sucrose of saSST mutated at position VIII. Production of glucose, fructose and kestose is determined after 20 min incubation at 28°C with sucrose concentrations up to 1 M. SST mutation VIII (N425S) shows high fructose production (i.e. sucrose hydrolysis) at all sucrose concentrations tested
A single mutant of the hydrolase INV doubles its glycosyltransferase capability
Most acINV mutants were active, except those at site III and at site VI (Table 2). For saSST site III was also inactive, but site VI increased hydrolysis. The acINV mutant at site VII was quite active, this in contrast to saSST where the reciprocal mutation led to an almost inactive protein.
The active acINV mutants were incubated with sucrose for 20 min. at concentrations of 1, 2, 4, 10, 20, 40, 100, and 200 mM. acINV mutated at sites I, II, IV, V, and VIII showed the same biochemical properties as the wild-type invertase with respect to percentage of glycosyltransfer and Km (Fig. 5, Table 2, Km data not shown). The activity of mutations in acINV at sites I (L201M), IV (W343Y), and at site V (F348L) correlated with the reciprocal mutations in saSST since all behave as wild type enzymes (Fig. 5, Table 2). At site II amino acid 251 changed from asparagine to cysteine. It is surprising that this did not cause an activity shift since the reciprocal mutation in saSST did approximately double hydrolysis (Fig. 5, Table 2). The overall activity of this mutant seemed however reduced, as only small amounts of products were detected (data not shown). Even more surprising was the unchanged activity of the acINV mutant at site VIII (S457N), since the reciprocal mutation in saSST caused a dramatic increase in hydrolysis activity (Figs. 3, 5; Table 2).
Fig. 5.
Transfer as a percentage of total hydrolysis plus transfer activity by the hydrolase acINV and its mutants at sites I–VIII (Table 2) incubated at 28°C for 20 min with different sucrose concentrations. Transfer is measured through the kestose produced and increases with increasing sucrose concentrations. Measurements from three independent protein expression experiments were analyzed, the error bars represent the standard error
The invertase mutated at site VII (W440Y), showed almost doubled transglycosylation activity when compared to wild-type acINV (Fig. 5). The enzyme showed still wild type acINV characteristics with respect to substrate saturation (Table 2). The Km for sucrose was calculated to be 8.8 ± 2.4 mM, which is slightly higher then 5.8 ± 1.9 mM calculated for wild-type acINV but not noticeably increased.
Several mutations reside close together at the rim of the substrate pocket
For plant fructosyltransferases and vacuolar invertases no crystal structure is yet available. The structure of acINV and saSST were modeled using the 3D-structures of the GH32 family member cell wall invertase from Arabidopsis thaliana (PDB code 2AC1)(Verhaest et al., 2005a). For adjustments different crystal structures of Cichorium intybus FEHIIa in complex with several substrates were used (Verhaest et al., 2007; Verhaest et al., 2005b).
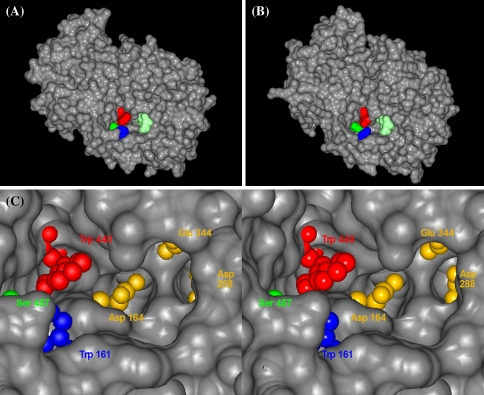
Interestingly, the amino acids that showed the largest effects on hydrolysis versus glycosyltransfer in saSST (site VIII representing Asn425) and acINV (site VII representing Trp440) are located at the entrance of the central pocket. They have similar locations and within the acINV and saSST models sites VII and VIII are residing very close together (Fig. 6a, b). These residues are also in close proximity to an earlier identified determinant of glycosyltransfer in acINV Trp161 (Fig. 6a). As can be seen from solvent accessible representations, the three residues are accessible and form a patch at the rim of the substrate pocket (Fig. 6c). It seems that those three amino acids locate to a hotspot for the determination of transfer and hydrolysis in GH32 enzymes. These amino acids do not interact with the donor sucrose substrate (Fig. 6a, b) and are not in close proximity of the catalytic residues (Fig. 6c), indicating that they do not directly influence the catalytic residues themselves. However, they are ideally located to influence the binding of the acceptor sucrose substrate in the active site once the fructose that is to be transferred is bound.
Fig. 6.
Solvent accessible surface for the homology models of (a) vacuolar invertase of onion (Allium cepa); residues Trp161 (blue), Trp440 (red) and Ser457 (green); and (b) 1-SST of festuca (Schedonorus arundinaceus); residues Tyr133 (blue), Tyr408 (red) and Asn425 (green). The active site is marked for reference with a sucrose molecule obtained by superposing the coordinates from β-fructosidase from Thermotoga maritima (PDB code 1W2T, light green). Colored residues were subtracted from each surface calculation in order to stress their spatial distribution. (c) Stereo-view of the substrate pocket of INV in yellow the three catalytic residues are indicated. The figure was prepared with CCP4 mg (Potterton et al. 2002)
Discussion
A fructosyltransferase, more specifically a 1-SST, and a vacuolar invertase were studied to discover amino acids that determine hydrolysis and glycosyltransfer in family GH32. We found a single amino acid in the fructosyltransferase saSST that when mutated renders the enzyme a hydrolase. In the hydrolase acINV we found one amino acid that doubles transferase activity when mutated, it is a tryptophan in vacuolar invertases (Trp440 in acINV) and a tyrosine or phenylalanine in fructosyltransferases (see supplemental information). Two mutations of the sucrose-binding box in acINV that were identified before also double transferase capacity (Ritsema et al., 2006). One of these amino acids was also a tryptophan in invertases (acINV Trp161) and a tyrosine or phenylalanine in fructosyltransferases. Mutating both tryptophan residues approximately doubled the transfer capability of acINV, but in contrast to the mutation in the sucrose-binding box, our novel mutant did not show an increased Km for sucrose. The reciprocal mutation of the previously identified acINV(W161Y) is in saSST(Y133W). We tested this mutation in saSST, and saw no change in activity; the mutant showed wild-type saSST activity for hydrolysis ratio’s and substrate saturation (data not shown). This phenomenon was seen before for the homologous mutation in the 6G-FFT fructosyltransferase from onion where mutation Y82W resulted also in wild-type activity (Ritsema et al., 2005).
Some mutations tested resulted in inactive enzymes. This might be due to misfolding of the proteins, especially in the case of mutation III, since no protein could be detected on Western blot for this mutation (data not shown). Inactive proteins were not used to evaluate changes in enzyme activities. Also the Y408W mutation in saSST was not taken into account. The activity was so low that no product levels could be determined after 20 min incubations. After prolonged incubations the activity of this mutant seemed wild-type.
Amino acid changes in saSST that showed doubled hydrolase activity are at site II (C203N) and at site VI (F362Y) (Tables 1 and 2). Although Cys203 is not a conserved residue in enzymes from the GH32 family it can be involved in forming a disulfide bond especially in SSTs. Therefore, this alteration can cause the loss of a disulfide bond, which could lead to a conformational change resulting in altered activity. If there is no disulfide bond present in saSST, the mutation still represents the loss of a thiol group. The saSST mutated at site VI had Phe362 changed to the in invertases omnipresent tyrosine. This mutation represents an alteration from a hydrophobic aromatic to a polar aromatic residue. Both mutations seem not to reside in proximity of the substrate-binding pocket. It is therefore probable that they influence activity via a change in the conformation of the enzyme.
To our surprise, the amino acids that influence hydrolysis and glycosyltransfer are not on similar positions in the transferase and hydrolase tested (Table 2 and saSST(Y133W) which is discussed above). When we, however, modeled the positions of the amino acids in saSST and acINV according to available 3D structures of family GH32, we discovered that several of those that influence hydrolysis versus glycosyltransfer ratios seem to reside very close together at the rim of the substrate-binding pocket. Here they indicate a hotspot for amino acids that determine transfer capability of enzymes in GH32 (Fig. 6).
The GH32 enzymes of which 3D structures are available are all hydrolases. In these 3D structures the conserved tryptophan of the sucrose-binding box that we studied before is surrounded by other aromatic amino acids (Fig. 6). One of these surrounding amino acids represents site VII, 2 others are overall conserved in 1-SSTs and invertases; Trp188/160 and Trp438/406 (amino acid number in acINV and saSST, respectively). These amino acids are also present at the entrance of the substrate-binding pocket. Interestingly, amino acids present at the variable sites VII and Trp161 (acINV) are hydrophobic tryptophan residues in invertases and hydrophilic tyrosine residues in the fructosyltransferases. Aromatic amino acids are often implied to act in sugar binding, but from our modeling it follows that the donor sucrose substrate is not bound by these amino acids (Fig. 6a, b). Combining the observation that altering the amino acids at those sites influences transglycosylation capacity of two GH32 enzymes with their location at the rim of the substrate-binding pocket leads to the proposition that these amino acids are involved in acceptor substrate binding.
The active site could be organized in a similar way as proposed for bacterial fructosyltransferase enzymes (Ozimek et al. 2006). Here, the active site binds sucrose at subsites −1 and +1, retains the fructose to-be transferred at −1, and recognizes three sugar residues of the acceptor substrate at +1, +2, +3. The glycosyltransferase SST would need only a +1 and +2 for binding of the acceptor sucrose, whereas the hydrolase INV would not even need a +2 subsite to carry out the hydrolysis reaction. Based on our results we suggest that the +2 subsite of SSTs could be identical to the hotspot present at the rim of the active site pocket that determines hydrolysis versus transglycosylation ratios and that is influenced by sites VII, VIII and the equivalent of acINV Trp161.
Our findings indicate that a distinguishing feature in the evolution of fructosyltransferases from invertases is the achievement of a location for acceptor substrate binding and that this binding-site is assembled from several amino acids that form a patch at the rim of the (donor) substrate-binding pocket.
Electronic supplementary material
Acknowledgements
We want to thank Agustin Lopez-Munguia for his advice and support. This work was supported by the Swiss National Science Foundation.
Open Access This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s11103-008-9404-7) contains supplementary material, which is available to authorized users.
References
- Alberto F, Bignon C, Sulzenbacher G, Henrissat B, Czjzek M (2004) The three-dimensional structure of invertase (β-fructosidase) from Thermotoga maritima reveals a bimodular arrangement and an evolutionary relationship between retaining and inverting glycosidases. J Biol Chem 279:18903–18910. doi:10.1074/jbc.M313911200 [DOI] [PubMed]
- Altenbach D, Nüesch E, Meyer AD, Boller T, Wiemken A (2004) The large subunit determines catalytic specificity of barley sucrose:fructan 6-fructosyltransferase and fescue sucrose:sucrose 1-fructosyltransferase. FEBS Lett 567:214–218. doi:10.1016/j.febslet.2004.04.064 [DOI] [PubMed]
- Altenbach D, Nüesch E, Ritsema T, Boller T, Wiemken A (2005) Mutational analysis of the active center of plant fructosyltransferases: Festuca 1-SST and barley 6-SFT. FEBS Lett 579:4647–4653. doi:10.1016/j.febslet.2005.07.034 [DOI] [PubMed]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW et al (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54:905–921. doi:10.1107/S0907444998003254 [DOI] [PubMed]
- Cairns AJ, Winters A, Pollock CJ (1989) Fructan biosynthesis in excised leaves of Lolium temulentum L.III. A comparison of the in vitro properties of fructosyl transferase activities with the characteristics of in vivo fructan accumulation. New Phytol 112:343–352. doi:10.1111/j.1469-8137.1989.tb00322.x [DOI]
- Ensley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi:10.1107/S0907444904019158 [DOI] [PubMed]
- Hendry GAF (1993) Evolutionary origins and natural functions of fructans—a climatological, biogeographic and mechanistic appraisal. New Phytol 123:3–14. doi:10.1111/j.1469-8137.1993.tb03753.x [DOI]
- Hochstrasser U, Lüscher M, De Virgilio C, Boller T, Wiemken A (1998) Expression of a functional barley sucrose–fructan 6-fructosyltransferase in the methylotrophic yeast Pichia pastoris. FEBS Lett 440:356–360. doi:10.1016/S0014-5793(98)01487-2 [DOI] [PubMed]
- Koops AJ, Jonker HH (1996a) Purification and characterization of the enzymes of fructan biosynthesis in tubers of Helianthus tuberosus Colombia. II. Purification of sucrose:sucrose 1-fructosyltransferase and reconstitution of fructan synthesis in vitro with purified sucrose:sucrose 1-fructosyltransferase and fructan:fructan 1-fructosyltransferase. Plant Physiol 110:1167–1175 [DOI] [PMC free article] [PubMed]
- Koops AJ, Jonker HH (1996b) Purification and characterization of the enzymes of fructan biosynthesis in tubers of Helianthus tuberosus Colombia. II. Purification of sucrose:sucrose 1-fructosyltransferase and reconstitution of fructan synthesis in vitro with purified sucrose:sucrose 1-fructosyltransferase and fructan:fructan 1-fructosyltransferase. Plant Physiol 110:1167–1175 [DOI] [PMC free article] [PubMed]
- Lund O, Nielsen M, Lundegaard C, Worning P (2002) X3M a computer program to extract 3D models. Abstract at the CASP5 conference, A102. http://www.cbs.dtu.dk/services/CPHmodels/
- Lüscher M, Hochstrasser U, Vogel G, Aeschbacher R, Galati V, Nelson CJ et al (2000) Cloning and functional analysis of sucrose:sucrose 1-fructosyltransferase from tall fescue. Plant Physiol 124:1217–1228. doi:10.1104/pp.124.3.1217 [DOI] [PMC free article] [PubMed]
- Meng G, Fütterer K (2003) Structural framework of fructosyl transfer in Bacillus subtilis levansucrase. Nat Struct Biol 10:935–941. doi:10.1038/nsb974 [DOI] [PubMed]
- Obenland DM, Simmen U, Boller T, Wiemken A (1993) Purification and characterization of three soluble invertases from barley (Hordeum vulgare L.) leaves. Plant Physiol 101:1331–1339. doi:10.1104/pp.101.4.1331 [DOI] [PMC free article] [PubMed]
- Ozimek LK, Kralj S, Van Der Maarel MJ, Dijkhuizen L (2006) The levansucrase and inulosucrase enzymes of Lactobacillus reuteri 121 catalyse processive and non-processive transglycosylation reactions. Microbiology 152:1187–1196. doi:10.1099/mic.0.28484-0 [DOI] [PubMed]
- Pons T, Hernandez L, Batista FR, Chinea G (2000) Prediction of a common beta-propeller catalytic domain for fructosyltransferases of different origin and substrate specificity. Protein Sci 9:2285–2291 [DOI] [PMC free article] [PubMed]
- Potterton E, McNicholas S, Krissinel E, Cowtan K, Noble M (2002) The CCP4 molecular-graphics project. Acta Cryst D58:1955–1957 [DOI] [PubMed]
- Reddy A, Maley F (1990) Identification of an active-site residue in yeast invertase by affinity labeling and site-directed mutagenesis. J Biol Chem 265:10817–10820 [PubMed]
- Reddy A, Maley F (1996) Studies on identifying the catalytic role of Glu-204 in the active site of yeast invertase. J Biol Chem 271:13953–13957. doi:10.1074/jbc.271.24.13953 [DOI] [PubMed]
- Ritsema T, Smeekens S (2003) Fructans: beneficial for plants and humans. Curr Opin Plant Biol 6:223–230. doi:10.1016/S1369-5266(03)00034-7 [DOI] [PubMed]
- Ritsema T, Joling J, Smeekens S (2003) Fructan patterns synthesized by onion fructan:fructan 6G-fructosyltransferase (6G-FFT) expressed in tobacco BY2 cells—is fructan:fructan 1-fructosyltransferase (1-FFT) needed in onion? New Phytol 160:61–67. doi:10.1046/j.1469-8137.2003.00858.x [DOI] [PubMed]
- Ritsema T, Verhaar A, Vijn I, Smeekens S (2004) Fructosyltransferase mutants specify a function for the β-fructosidase motif of the sucrose-binding box in specifying the fructan type synthesized. Plant Mol Biol 54:853–863. doi:10.1007/s11103-004-0276-1 [DOI] [PubMed]
- Ritsema T, Verhaar A, Vijn I, Smeekens S (2005) Using natural variation to investigate the function of individual amino acids in the sucrose-binding box of fructan:fructan 6G-fructosyltransferase (6G-FFT) in product formation. Plant Mol Biol 58:597–607. doi:10.1007/s11103-005-6504-5 [DOI] [PubMed]
- Ritsema T, Hernandez L, Verhaar A, Altenbach D, Boller T, Wiemken A et al (2006) Developing fructan-synthesizing capability in a plant invertase via mutations in the sucrose-binding box. Plant J 48:228–237. doi:10.1111/j.1365-313X.2006.02862.x [DOI] [PubMed]
- Shiomi N, Kido H, Kiriyama S (1985) Purification and properties of sucrose:sucrose 1F-β-d-fructisyltransferase in onion seeds. Phytochemistry 24:695–698. doi:10.1016/S0031-9422(00)84878-5 [DOI]
- Sprenger N, Bortlik K, Brandt A, Boller T, Wiemken A (1995) Purification, cloning, and functional expression of sucrose:fructan 6-fructosyltransferase, a key enzyme of fructan synthesis in barley. Proc Natl Acad Sci USA 92:11652–11656. doi:10.1073/pnas.92.25.11652 [DOI] [PMC free article] [PubMed]
- Ueno K, Onodera S, Kawakami A, Yoshida M, Shiomi N (2005) Molecular characterization and expression of a cDNA encoding fructan:fructan 6G-fructosyltransferase from asparagus (Asparagus officinalis). New Phytol 165:813–824. doi:10.1111/j.1469-8137.2004.01294.x [DOI] [PubMed]
- Van den Ende W, De Roover J, Van Laere A (1996a) In vitro synthesis of fructofuranosyl-only oligosaccharides from inulin and fructose by purified chicory root fructan:fructan fructosyltransferase. Physiol Plant 97:346–352. doi:10.1034/j.1399-3054.1996.970219.x [DOI]
- Van den Ende W, Van Wonterghem D, Dewil E, Verhaert P, De Loof A, Van Laere A (1996b) Purification and characterization of 1-SST, the key enzyme initiating fructan biosynthesis in young chicory roots (Cichorium intybus L.). Physiol Plant 98:455–466. doi:10.1111/j.1399-3054.1996.tb05699.x [DOI]
- Van den Ende W, Michiels A, De Roover J, Verhaert P, Van Laere A (2000) Cloning and functional analysis of chicory root fructan1-exohydrolase I (1-FEH I): a vacuolar enzyme derived from a cell-wall invertase ancestor? Mass fingerprint of the 1-FEH I enzyme. Plant J 24:447–456. doi:10.1046/j.1365-313x.2000.00890.x [DOI] [PubMed]
- van der Meer I, Koops AJ, Hakkert JC, van Tunen AJ (1998) Cloning of the fructan biosynthesis pathway of Jerusalem artichoke. Plant J 15:489–500. doi:10.1046/j.1365-313X.1998.00230.x [DOI] [PubMed]
- Verhaest M, Le Roy K, Sansen S, De Coninck B, Lammens W, De Ranter CJ et al (2005a) Crystallization and preliminary X-ray diffraction study of a cell-wall invertase from Arabidopsis thaliana. Acta Crystallogr F61:766–768 [DOI] [PMC free article] [PubMed]
- Verhaest M, Van den Ende W, Roy KL, De Ranter CJ, Van Laere A, Rabijns A (2005b) X-ray diffraction structure of a plant glycosyl hydrolase family 32 protein: fructan 1-exohydrolase IIa of Cichorium intybus. Plant J 41:400–411 [DOI] [PubMed]
- Verhaest M, Lammens W, Le Roy K, De Ranter CJ, Van Laere A, Van den Ende W (2007) Insights into the fine architecture of the active site of chicory fructan 1-exohydrolase: 1-kestose as substrate vs. sucrose as inhibitor. New Phytol 174:90–100. doi:10.1111/j.1469-8137.2007.01988.x [DOI] [PubMed]
- Vijn I, Smeekens S (1999) Fructan: more than a reserve carbohydrate? Plant Physiol 120:351–359. doi:10.1104/pp.120.2.351 [DOI] [PMC free article] [PubMed]
- Vijn I, van Dijken A, Sprenger N, van Dun K, Weisbeek P, Wiemken A et al (1997) Fructan of the inulin neoseries is synthesized in transgenic chicory plants (Cichorium intybus L.) harbouring onion (Allium cepa L.) fructan:fructan 6G-fructosyltransferase. Plant J 11:387–398. doi:10.1046/j.1365-313X.1997.11030387.x [DOI] [PubMed]
- Vijn I, van Dijken A, Lüscher M, Bos A, Smeets E, Weisbeek P et al (1998) Cloning of sucrose:sucrose 1-fructosyltransferase from onion and synthesis of structurally defined fructan molecules from sucrose. Plant Physiol 117:1507–1513. doi:10.1104/pp.117.4.1507 [DOI] [PMC free article] [PubMed]
- Wei JZ, Chatterton NJ (2001) Fructan biosynthesis and fructosyltransferase evolution: expression of the 6-SFT (sucrose:fructan 6-fructosyltransferase) gene in crested wheatgrass (Agropyron cristatum). J Plant Physiol 158:1203–1213. doi:10.1078/0176-1617-00241 [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.