1. Introduction
Francisella tularensis is a Gram-negative facultative intracellular bacterium that can infect a variety of species and is the cause of the disease known as tularemia or rabbit fever (Sjostedt, 2007). Infection can result by exposure to the bacterium by contact with the skin, by ingestion, or by inhalation of aerosolized organisms (Ellis et al., 2002; Sjostedt, 2007). The precise course and kinetics of the disease varies with the Francisella strain and route of inoculation (Chen et al., 2003; Conlan et al., 2003). However, all routes of exposure can ultimately result in sepsis and widespread dissemination of the bacteria in the host (Conlan et al., 2003; Elkins et al., 2003). Inoculation with microorganisms in aerosolized form for some strains of F. tularensis has a remarkably low infectious dose (10 organisms or less) with a significant fatality rate if left untreated (Conlan et al., 2003; Twine et al., 2006). Tularemia can be treated with antibiotics if detected early (Hepburn and Simpson, 2008; Kman and Nelson, 2008). However, the possibility that the organism could be made antibiotic resistant, either through classic microbiologic means or by using recombinant DNA technology, is a considerable concern if the modified organism were then intentionally spread. The extreme virulence of certain strains, the ability to aerosolize the organisms and the ability of the organism to persist in the environment make it a potent potential bioweapon (Altman, 2002; Ellis et al., 2002). Indeed, both the former Soviet Union, as well as the United States, reportedly had a bioweapons program employing F. tularensis (Dennis et al., 2001; Fong and Alibek, 2005). Unfortunately, in comparison to other pathogenic microorganisms, the host response to F. tularensis is not yet well understood.
The immune response to Francisella tularensis appears complex. As might be anticipated for an intracellular organism, classical cellular immune responses appear to be critical. Studies using lymphocyte-deficient (CD4−, β2m−, TCR-γ−, TCR-β−, scid, nude) or lymphocyte-depleted (by using specific antibodies) mice have illustrated an important role for both CD4 and CD8 T cells (Conlan et al., 1994; Elkins et al., 1993; Elkins et al., 1996; Rhinehart-Jones et al., 1994; Yee et al., 1996). Interestingly, there is also an unusual Thy1+ αβ TCR+ CD4− CD8− NK1.1− T cell subset that has been shown to contribute to protection against F. tularensis challenge (Cowley and Elkins, 2003; Cowley et al., 2005). Recent studies have also suggested that IgA antibodies as well as CD4+ T cells may also play a role in the context of intranasal immunization with an inactivated strain of F. tularensis, in conjunction with IL-12 as an adjuvant (Baron et al., 2007). Thus, while the immune response against F. tularensis is clearly multifactorial, it seems that a cellular response, including CD4+ and CD8+ T cells, plays a critical role in protection.
There is no FDA approved vaccine for F. tularensis. During the 1940s an attenuated subsp. holarctica Live Vaccine Strain (LVS) was developed (Eigelsbach and Downs, 1961) as a vaccine candidate. This strain has proven invaluable for examining aspects of the F. tularensis-host interaction (Elkins et al., 2007). While LVS has greatly aided our understanding of F. tularensis biology and microbial host interactions, there are significant side effects to the use of LVS as a vaccine and the protection it affords is incomplete (Griffin et al., 2007; Saslaw et al., 1961a; Saslaw et al., 1961b). The drawbacks of the current LVS vaccine, and the possibility that F. tularensis might be used in a bioterror weapon, have added impetus to the identification of antigens recognized by the immune system. Currently, the nature of the protective antigens, indeed the molecular definition of any antigens in the cellular immune response, is limited. There have been only a few reports of immunostimulatory molecules for T cells in mice or humans (Golovliov et al., 1995; Lee et al., 2006; McMurry et al., 2007; Sjostedt et al., 1992; Sjostedt et al., 1991). Perhaps the best-characterized response is to the lipoprotein Tul4. Tul4 can be a target of the cellular and humoral immune response in both mice and humans (Golovliov et al., 1995; Sjostedt et al., 1992; Sjostedt et al., 1991). Mice are a natural host for F. tularensis infection and exhibit many of the same aspects of the infection in humans (Fortier et al., 1991). Interestingly, mice immunized with Salmonella typhimurium expressing Tul4 appeared to give partial protection as assessed by a decreased bacterial burden in spleen and liver (Sjostedt et al., 1992). It would be extremely valuable to define epitopes at the molecular level in mice so that the immune response could be quantitatively and qualitatively assessed. This would be a great aid in understanding the host immune response in the context of infection as well as helping to develop and assess vaccine vectors and immunization strategies. In the current study, we have defined in Tul4 a potential immunodominant epitope in B6 mice using a novel strategy and shown that it is an important epitope in the context of a F. tularensis infection in both the acute and memory immune response.
2. Materials and Methods
2.1. Mice, cell lines, and bacteria
C57BL/6J (H2b) (B6) mice were purchased from the Jackson Laboratory (Bar Harbor, Maine). MHC class II knockout mouse B6.129-H2Abtm1Gru was generously provided by Dr. Andrea Sant (University of Rochester). The T cell fusion partner BWZ.36/CD8+, which can be used to make MHC class II and class I restricted hybridomas, was kindly provided by Dr. Nilabh Shastri and maintained as previously described (Sanderson and Shastri, 1994). The F. tularensis live vaccine strain (LVS) was used in these studies (Cowley and Elkins, 2003). Bacterial viability was quantified by serial dilution on chocolate agar.
2.2. Fusion protein constructs and peptides
Recombinant proteins were generated using the bacterial expression vector pQE40, which contains a 6x histidine coding region (6x His) followed by murine dihydrofolate reductase (DHFR). The DHFR-Tul4 construct was constructed by cloning full length Tul4 from F. tularensis Live Vaccine Strain (LVS) genomic DNA using polymerase chain reaction (PCR) and the specific primers described below. All primers were constructed with 15–18 base pairs complimentary to the sequence of interest and included an enzyme restriction site and a GC clamp. The 3′ reverse primers also included a stop codon upstream of the restriction site. The Tul4 insert was cloned into the pQE40 vector using BglII and PstI restriction sites designed into the primers. DHFR-OVA encoded in the pQE40 vector was created in a similar fashion. Tul4 fusion protein deletion constructs were created by using the same 5′ primer and substituting with nine individual 3′ primers that recognize sequences starting at 384 base pairs (bp), 369 bp, 345 bp, 324 bp, 300 bp, 282 bp, 183 bp, and 89 bp. The Tul4 transplant epitope construct (DHFR-Tul4 86-110) was created by cloning out a 25 amino acid (aa) region from Tul4 using a specific primer set. This region was cloned into pQE40 using BglII and PstI sites engineered into the primers yielding a ‘transplanted’ region of Tul4 fused directly behind DHFR. Tul4 transplant epitope deletion constructs were created by using a constant 5′ primer recognizing a region found before the DHFR sequence and substituting individual 3′ primers producing the segments of Tul4 86-99 (14 aa), 86-98 (13 aa), 86-97 (12 aa), and 86-95 (10 aa) fused to DHFR. Site directed mutagenesis of a cysteine to serine at residue 97 by an additional 3′ primer produced the DHFR-Tul4 86-(C97S)-99 construct. All synthetic Tul4 peptides used in this paper Tul4 86-99 (RLQWQAPEGSKCHD), Tul4 86-(C97S)-99 (RLQWQAPEGSKSHD), Tul4 86-98 (RLQWQAPEGSKCH), Tul4 86-97 (RLQWQAPEGSKC), and Tul4 86-95 (RLQWQAPEGS) were synthesized by SynBioSci (Livermore, CA). The NP 118-126 peptide derived from LCMV (RPQASGVYM) and OVA 323-339 peptide (ISQAVHAAHAEINEAGR) served as irrelevant peptide negative controls and were synthesized by Macro-Molecular Resources (University of Colorado, Ft. Collins, CO).
2.3. Protein Production
Production of DHFR-OVA and all DHFR-Tul4 fusion proteins were performed in Escherichia coli strain M15 as described by manufacturers specifications (Qiagen, Valencia, CA). Recombinant proteins were isolated by lysing the bacterial pellets with 8M urea (pH7.5) and purified over Ni-NTA column (Qiagen). The DHFR-Tul4 was dialyzed with PBS to remove urea before injection. To couple tosylactivated M280 magnetic Dynabeads (Invitrogen, Carlsbad, CA), 1×107 beads with 8μg protein were mixed overnight at 37°C in 0.1M borate buffer, washed three times with PBS using magnetic separation, and diluted to working volume in PBS (2×105 beads/μl). Tul4 deletion construct production and characterization was confirmed by SDS-PAGE techniques and Coomassie Blue staining as described previously (Turner et al., 2001).
2.4. Generation of T cell hybridomas
T cell hybrids were produced in two ways. To ensure that the epitopes recognized by the hybrids were generated in a natural infection, mice were inoculated intradermally at the base of the tail with 1 × 105 viable F. tularensis LVS and allowed to clear the infection. At least 3 weeks later, mice were sacrificed and spleen cells restimulated in vitro using F. tularensis LVS infected spleen cells. Restimulation was performed for 5 days and the cells harvested (FACS analysis confirmed cells were predominantly T cells) and fused with BWZ.36/CD8+ cells. Colonies were selected in HAT and tested for activity using either infected spleen cells as APC, or spleen cells and soluble F. tularensis LVS extracts as previously described (Woolard et al., 2008). Activity was measured using the CPRG substrate in 96 well plates as previously described (Sanderson and Shastri, 1994). Positive wells were cloned by limiting dilution and retested. For the hybridomas generated with recombinant protein immunization, mice were injected in the footpad with approximately 12.5μg DHFR-Tul4 protein emulsified in complete Freund’s adjuvant and were boosted once with DHFR-Tul4 protein emulsified in incomplete Freund’s adjuvant before harvesting draining popliteal lymph node. Lymph node cells from the immunized mice were stimulated in vitro and after 3 rounds of in vitro restimulation with DHFR-Tul4 conjugated to tosylactivated M280 beads, lymphocytes were isolated by Lympholyte-M (Cedarlane Laboratories Lmtd, Ontario, Canada) and fused to the BWZ.36/CD8+ fusion partner containing lacZ driven by the Interleukin-2 promotor element as described (Sanderson and Shastri, 1994; Turner et al., 2001). Activity was measured using the beta-galactosidase substrate in 96 well plates as previously described (Turner et al., 2001).
2.5. Antigen presentation assay
Antigen presentation assay was adapted from the T cell antigen discovery (T-CAD) assay described by Turner et al. (Turner et al., 2001). Presentation assays were performed by incubating Tul4 specific hybridomas: 2×105 ID6 hybrids with 1×105 APCs or 4×105 FT13 1E4B4 hybrids with 4×105 APCs in 200μl total volume per well in a 96 well tissue culture plate. APCs were splenocytes isolated from naïve wild type or MHC class II knockout C57BL/6J mice as described in the figure legends. Recombinant proteins DHFR-OVA, DHFR-Tul4, or the individual Tul4 deletion constructs isolated from bacterial lysates were conjugated to tosylactivated beads as described above and used at 2×106 beads per well. Assays were also performed in the presence of synthesized peptides Tul4 86-99, Tul4 86-(C97S)-99, Tul4 86-98, Tul4 86-97, Tul4 86-95 at 10μg (SynBioSci, Livermore, CA), 2μg NP peptide, and 3.8μg F. tularensis extract. F. tularensis extract was produced by killing F. tularensis LVS in 70% ethanol (Woolard et al., 2007). Plate bound anti-CD3 (clone 500A2) (BD Pharmingen, San Jose, CA) was also included in each assay (not shown in figures) as a positive control for hybrid viability and ability to function. After 18–24 hour incubation at 37°C, cultures were developed using the beta-galactosidase assay. Briefly, the beta-galactosidase assay utilizes hybridomas containing the β-galactosidase gene (lacZ) under the control of the Interleukin-2 promotor elements (NFAT elements). Because these hybridoma have the reporter gene beta-galactosidase under the control of IL-2 promoter elements, and IL-2 indiction is an early event in T cell activation, activation of the hybridoma is accompanied by production of the enzyme beta-galactosidase that can be easily monitored using a colorimetric substrate (Sanderson and Shastri, 1994; Turner et al., 2001). When the β-galactosidase substrate X-gal (5-bromo-4-chloro-3-indolyl-β-D galactosidase) (Invitrogen, Carlsbad, CA) is administered a colorimetric substrate is produced turning activated hybridomas blue and the number of activated hybrids per well can then be enumerated by visual counts. Wells with low numbers of activated (< 1000 blue cells) the entire well was counted. Wells with high numbers (>1000 blue cells) three representative fields of view were counted, averaged, and multiplied by the number of fields per well. In some experiments, levels of beta-galactosidase expression was assayed using the soluble substrate chorophenol red-β D-galactopyranoside sodium salt (CPRG), allowing for identification of the overall reactivity of a hybridoma. This assay results in a colorimetric substrate from activated hybridomas and the absorbance can be measured using a spectrophotometer at OD570nm (Sanderson and Shastri, 1994).
2.6. Analysis of in vivo immunogenicity
Analysis of in vivo immunogenicity was performed essentially as previously described (Woolard et al., 2008). In brief, B6 mice were inoculated with 105 CFU F. tularensis LVS intradermally at the base of the tail. Spleens were harvested on day 7 to examine the acute response or, to characterize the memory response, mice were allowed to clear the infection and spleens were harvested approximately 2 months post inoculation. Splenocytes from both infected mice and uninfected controls were stimulated in vitro for 18–24 hours with 100μg/ml F. tularensis LVS extract, Tul4 86-99 peptide, or an irrelevant class II OVA 323-339 peptide. Brefeldin A was added for the last four hours of incubation. Cells were then washed and surface stained with anti-CD19 (B-D Pharmingen), PE-Cy7 (Caltag), anti-CD11c PE-Texas Red (Invitrogen), anti-CD11b APC750 (eBiosciences), anti-TCRβ Alexa 488 (Biolegend), anti-CD3 APC (eBiosciences), anti-CD4 PacBlue (Invitrogen), anti-CD8 PerCP (Biolegend) and anti-IFN-γ (B-D Biosciences) fixed and permeablized using eBioscience reagents and protocols (eBioscience, San Diego, CA), then stained with PE conjugated antibody to IFNγ and analyzed on a Cyan flow cytometer (Dako-Cytomation, Colrado Springs). The gating scheme used has been included in Fig. 6A. TCRβ+CD3+CD4+ splenocytes were analyzed for intracellular IFNγ with a minimum of 250,000 total cellular events accumulated. All FACS results were analyzed using Summit software (Dako-Cytomation).
Figure 6. Tul4 86-99 epitope is recognized in the context of natural infection.
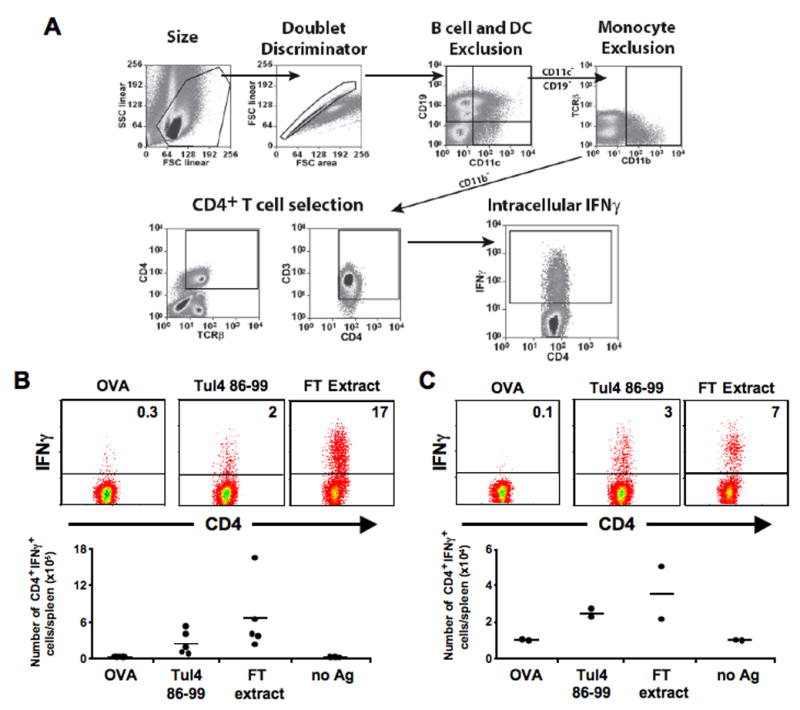
A) Gating scheme used to identify viable CD4+ interferon γ+ (IFNγ) cells and exclude B cells, DCs, and monocyte populations. B & C) Ex vivo splenocytes were activated with ethanol killed F. tularensis extracts (FT Ag), Tul4 86-99 peptide, OVA, or media alone for 24 hours. Brefeldin A was added for the last 4 hours of activation before labeling with fluorescent antibodies and FACS analysis. Representative FACS dot plots of CD4+ cells also gated for IFNγ positivity from either 7d acutely infected mice (B - top panels) or 2 months post-infection. Percentage of IFNγ+ CD4+ cells are indicated in each dot plot (C - top panels). Summary of ex vivo activation experiments, the number of CD4+/IFNγ+ cells per spleen was quantified for 7d acute infection (B - bottom panels, n = 5) and memory recall (C - bottom panels, n = 2).
3. Results
3.1. Generation and characterization of Tul4 reactive hybridomas
In the current study, we have used two different approaches to immunization in conjunction with the T-CAD technique to identify an epitope that arises against the F. tularensis lipoprotein Tul4. To identify epitopes present in both natural infection and conventional immunization with recombinant fusion protein, hybridomas were created by fusing lymphocytes derived from these two immunization methods to a variant of the BWZ.36 (lacZ+) T cell fusion partner. Hybridomas generated from these fusions that were activated by plate bound anti-CD3, reactive with DHFR-Tul4 coupled to tosylactivated beads and F. tularensis extract, and did not react with DHFR-OVA coupled to tosylactivated beads or irrelevant NP peptide were deemed as having the appropriate reactivity pattern. Based upon a combination of absolute strength of reactivity, the signal to noise ratio, and relative stability, two hybridomas were selected for detailed analysis, one from immunization with recombinant Tul4 fusion protein (ID6) and one from infection with live F. tularensis LVS (FT13 1E4B4). These hybridomas were activated by anti-CD3, F. tularensis extract, and DHFR-Tul4 indicating that these hybridomas are specific for Tul4 and the absence of reactivity with DHFR-OVA suggests that the hybridomas do not recognize the DHFR portion of the fusion protein (data not shown). Interestingly, the FT13 1E4B4 hybridoma generated from live F. tularensis infection was not only activated by F. tularensis extract but also DHFR-Tul4 which was recombinantly expressed in E. coli and was coupled to beads. These data suggest that we have generated and identified hybridomas from live F. tularensis infection and recombinant protein immunization that specifically recognize a recombinant DHFR-Tul4 in an in vitro antigen presentation assay.
3.2. Generation of Tul4 deletion constructs and identification of an epitope region within Tul4
A series of DHFR-Tul4 deletion constructs were created in the pQE40 bacterial expression vector to identify the epitope containing regions within Tul4 recognized by the hybridomas. These constructs contain the 6x His tag followed by DHFR and the C terminal deletions of Tul4 illustrated in Fig. 1A. E. coli bacterial lysates induced to express the deletion constructs were then purified over a Ni-NTA column to isolate fusion proteins via the 6x His tag and purified fusion proteins were then analyzed for appropriate molecular weight as well as quality and quantity of protein using SDS-PAGE (Fig. 1B). The predicted molecular weight of DHFR-Tul4 is 45kDa (28kDa DHFR plus 17kDa Tul4) (Fig. 1Ba) with the C terminal deletion constructs having correspondingly lower molecular weights (Fig. 1B b-i). Note that the presence of a 28kDa band in lanes a-i is likely due to DHFR protein, which may have arisen from proteolytic cleavage at the junction of DHFR-Tul4 peptides. These results show that the constructs encode fusion proteins with the appropriate molecular size, which can be successfully expressed and subsequently used for mapping studies.
Figure 1. Generation of DHFR-Tul4 deletion constructs for epitope identification.

(A) Schematic of full length Tul4 and the eight carboxy-terminal deletion constructs used in the pQE40 bacterial expression vector. The black box indicates 6 × histidine tag, the gray box indicates murine DHFR protein, and open box denotes portions of Tul4 protein fused in frame with DHFR. (B) SDS-PAGE analysis of Tul4 and deletion constructs. Constructs were purified over Ni-NTA column, separated on SDS-PAGE gel, and stained with Coomassie Blue. Gel lane headings (a-i) correspond to constructs (a-i) in panel (A). The molecular mass of full length Tul4 fused with DHFR is approximately 45kDa.
Using the hybridomas and Tul4 deletion constructs, antigen presentation experiments were conducted to identify the T cell epitopes within Tul4. Deletion constructs were individually coupled to tosylactivated beads which serves two purposes in our assay: allowing for purification of recombinant protein from a urea buffer used in the protein isolation process; as well as increasing antigen uptake and processing and presentation by APCs to the hybridomas (Storozynsky et al., 1999). Presentation assays were done as described previously in the methods section and activated hybridomas were identified by beta-galactosidase assay and quantified by visually counting blue cells (activated cells). As seen in Fig. 2, both hybridomas are activated when the first 100 amino acids of Tul4 are present but lose reactivity when 94 amino acids of Tul4 were presented by APCs. A loss of reactivity identified by a reduction in the number of activated (blue) cells suggests that the epitope recognized by these hybridomas is no longer present within the DHFR-Tul4 fusion protein. Interestingly, both hybridomas, one from conventional immunization and one from live bacterial infection, lose reactivity at the same location on the Tul4 protein suggesting that the two immunization approaches generate hybridomas that recognize the same epitope.
Figure 2. Hybridoma reactivity against DHFR-Tul4 deletion proteins.

Hybridomas were incubated with C57BL/6 splenocytes and F. tularensis extract, NP peptide, and tosylactivated beads adsorbed with the various DHFR-Tul4 deletion proteins or DHFR-OVA. After overnight incubation, a beta-galactosidase assay was done to identify activated cells. Representative data for reactivity patterns are shown (All treatments done 3 times or more).
3.3. Fine epitope mapping using expression constructs
The previous experiments using the Tul4 deletion constructs indicated that the hybridomas recognize an epitope that is lost when only 94 residues of Tul4 are present. We reasoned that an epitope could lie ~10 amino acids upstream or downstream of the 94th residue. To unequivocally identify that an epitope exists within this region of Tul4, a fusion protein was generated from full length Tul4 by PCR using primers specific to the region mentioned. This resulted in transplantation of amino acids 86-110 of Tul4 directly behind DHFR (Fig. 3A). This construct, called Tul4 transplant epitope protein, DHFR-Tul4 86-110, containing only 25 amino acids of Tul4 was capable of activating both hybridomas, illustrating that an epitope does indeed exist within this 25 amino acid sequence (Fig. 3B&C). Additionally, splenocytes from MHC class II deficient mice were not capable of activating either hybridoma indicating that the epitope recognized is class II restricted (Fig. 3C).
Figure 3. Hybridomas recognize a similar minimal epitope in a DHFR-Tul4 fusion protein.
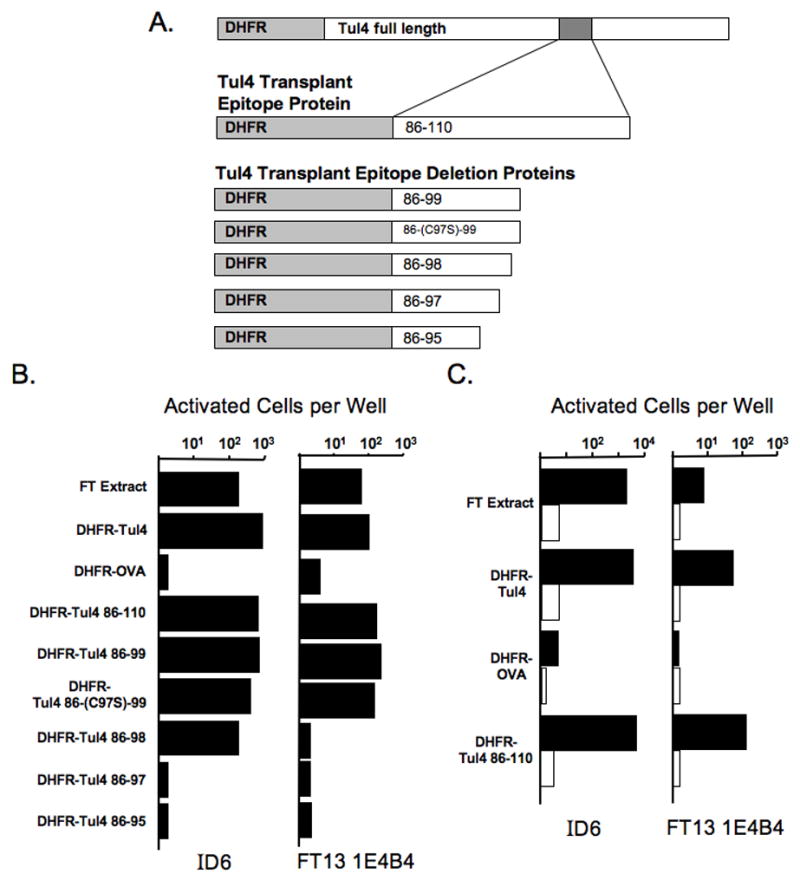
(A) Schematic of the approximate location of transplant epitope in the context of full length Tul4 and the Tul4 transplant epitope construct (25 amino acids of Tul4) along with its deletion constructs (14, 13, 12, and 10 amino acids of Tul4). (B) Hybridomas were incubated with C57BL/6 splenocytes, DHFR-Tul4 proteins, and controls. Tosylactivated beads were conjugated with DHFR-OVA, DHFR-Tul4, or the individual Tul4 transplant epitope proteins (DHFR-Tul4 86-110, -99, -(C97S)-99, -98, -97, -95). After overnight incubation, a beta-galactosidase assay was performed to identify activated cells. Representative data for reactivity patterns are shown (All treatments done 3 times or more). (C) Hybridomas were incubated with wild type (filled bars) or MHC class II knockout (open bars) splenocytes, FT extract, DHFR-Tul4, DHFR-OVA, and DHFR-Tul4 86-110 as done previously. (Similar results were obtained with 3 additional independent Tul4 reactive hybridomas.)
To further refine the minimal epitopes recognized by each hybridoma, deletion constructs of the DHFR-Tul4 86-110 epitope transplant were created. Additional residues were removed from DHFR-Tul4 86-110 as described in the Methods section to generate the Tul4 transplant epitope deletion proteins DHFR-Tul4 86-99, 86-98, 86-97, and 86-95 which were individually coupled to tosylactivated beads for use in reactivity assays (Fig. 3A&B). Hybridoma reactivity against DHFR-Tul4 86-99 is similar to that elicited by the DHFR-Tul4 86-110 even though the protein containing residues 86-99 is 10 amino acids shorter. In both hybridomas a noticeable loss of reactivity is seen with DHFR-Tul4 86-98 and 86-97 proteins. These data indicate that a similar minimal epitope for each of the hybridomas is found within amino acids 86-99 with significant residues at 97 and 98. Because the peptide contained a cysteine, which might be modified either in vitro or in vivo we also changed the cysteine at residue 97 by site directed mutagenesis to a serine, generating DHFR-Tul4 86-(C97S)-99 (Thompson et al., 2004). As seen in Fig. 3B the DHFR-Tul4 86-(C97S)-99 protein had similar stimulatory potential when compared to its wildtype counterpart in these hybrids. Together, these data indicate that an epitope of Tul4 is located within residues 86-99.
3.4. Fine epitope mapping using peptides
Synthetic peptides were used to determine the epitope of Tul4 recognized by the hybridomas. A synthetic peptide of residues 86-99 was tested and was found to be capable of activating both hybridomas showing definitively that an epitope of Tul4 is found at residues 86-99 (Fig. 4). We then sought to refine our epitope region using truncated peptides of Tul4. As seen in Fig. 4 both hybridomas are activated by peptides Tul4 86-98 and Tul4 86-97 and lose reactivity with Tul4 86-95, suggesting that the hybridomas recognize a similar minimal epitope found between residues 86 to 97. Overall, the peptide mapping experiment results nevertheless are very similar to those seen with the protein expression constructs. The peptide data does appear to differ slightly from DHFR fusion protein data regarding the reactivity of Tul4 86-98 and Tul4 98-97, which might reflect differences in processing of the fusion protein not required with the peptides. As described above, the sequence of peptide comprised by Tul4 86-99 contains a cysteine at residue 97. As before, to investigate whether or not the cysteine affects reactivity, Tul4 86-99 with a point mutation at Cys97 to Ser97 was also synthesized. The altered residue did not decrease and may slightly increase the stimulatory capability of the Tul4 86-(C97S)-99 peptide (Fig. 4). These data corroborate the fusion protein data (Fig. 3). Using synthetic peptides we have obtained similar findings to those from the fusion protein screen and have conclusively identified an epitope within residues 86 to 99 and as minimal as residues 86 to 97 of Tul4 that arise in both natural infection and classical protein immunization.
Figure 4. Fine epitope mapping with peptides indicates that hybridomas recognize a similar minimal epitope within Tul4.

The hybridomas were incubated with C57BL/6 splenocytes, Tul4 peptides, and controls. After overnight incubation, a beta-galactosidase assay was done to identify activated cells. Representative data for reactivity patterns are shown (All treatments done 3 times or more).
We also tested six hybridomas that were shown to react with Tul4 from F. tularensis LVS to determine if they react with Tul4 in extracts from the virulent F. tularensis Schu S4 strain. As can be seen in Figure 5, all of the hybridomas reacted with the Schu S4 extract indicating not only that the epitope is shared and processed similarly, but also that it is produced in sufficient amounts to stimulate these cells. While the cross-reactivity of the F. tularensis strains is illustrated by recognition of both SchuS4 and LVS extracts by all of the hybridomas, their levels of reactivity are variable, which may reflect different fine specificity amongst the hybridomas.
Figure 5. Recognition of highly virulent SchuS4 extracts by LVS hybridomas.
A small panel of hybridomas generated by infection with LVS or immunization with recombinant Tul4 from LVS were screened against LVS and SchuS4 extracts. After overnight incubation, hybridoma reactivity was measured by the CPRG assay. Hybridoma reactivity against SchuS4 extracts is shown as a percent of the response against LVS extracts after background subtraction. Representative data for reactivity patterns are shown.
3.5. In vivo validation of Tul4 as a natural epitope
To determine if Tul4 was recognized during acute infections and was incorporated into the memory pool, we examined the F. tularensis specific IFN-γ responses of mice either 7 days (acute infection) or 2 months (memory response) after intradermal inoculation with 105 F. tularensis LVS. Splenocytes from control and infected mice were isolated and stimulated in vitro for 18–24 hours with F. tularensis extracts, Tul4 86-99 peptide, and irrelevant OVA protein or media alone as negative controls. Cells were then stained using an array of cellular markers followed by analysis via multicolor flow cytometry, which allowed for identification of activated IFNγ positive CD4+ T cells and exclusion of B cell, dendritic cell, and monocyte populations (Fig. 6A). When CD4+ splenocytes from mice with acute infection (7 days post-infection) were analyzed, we saw that a large proportion were reactive with F. tularensis extract (FT Ag) (6.6×105 IFNγ+ CD4+ per spleen) and a significant portion was also reactive with Tul4 86-99 peptide (2.5×105 IFNγ+ CD4+ per spleen) with minimal background IFNγ secretion by cells activated by OVA (Fig. 6B). These data suggest that the Tul4 86-99 epitope occurs naturally during acute infection and is one of the major epitopes in an anti-F. tularensis response.
It was also of interest to examine whether or not Tul4 reactive T cells are found in the memory T cell compartment. To examine the memory response, mice were infected as previously described and were allowed to clear infection, approximately 2 months later splenocytes were isolated, activated, and subsequently analyzed by flow cytometry (Fig. 6C). A significant proportion of CD4+ cells within the memory response could be activated by F. tularensis extracts (3.6×104 IFNγ+ CD4+ per spleen) and Tul4 86-99 peptide (2.5×104 IFNγ+ CD4+ per spleen) above that of background IFNγ levels (1.0×104 IFNγ+ CD4+ per spleen) (Fig. 6C). These data further suggest that the Tul4 86-99 epitope generates CD4 T cells that arise in acute infection, persist through the contraction phase of an immune response, and survive into the memory repertoire.
4. Discussion
The current report describes a dominant class II restricted T cell epitope of a Francisella antigen in the C57BL/6 mouse model of infection. The epitope of Tul4 encoded by amino acids 86-99 is to our knowledge, the first validated murine epitope described for Tul4. While there are a number of strategies for identifying T cell epitopes (Doolan et al., 2003; Sette and Peters, 2007), here we have used the T-CAD assay (Turner et al., 2001), a cell-based functional assay which makes no a priori assumptions regarding the biochemical properties of the epitope, to define an epitope of Francisella. Interestingly, previous mouse studies using a recombinant Salmonella typhimurium vector expressing Tul4, implicated the Tul4 protein as an important antigen in the response against F. tularensis (Sjostedt et al., 1992) and our work complements and extends these studies. In that study, three B cell epitopes were described, and a proliferative response of splenocytes to Tul4 was also shown, however, T cell epitopes were not defined (Sjostedt et al., 1992). Studies from tularemic humans revealed that the Tul4 protein was capable of stimulating a proliferative and cytokine response from PBMCs and three T cell stimulating polypeptides of approximately 20-24 amino acids of Tul4 were identified (Sjostedt et al., 1990). Together these data implicate Tul4 as a significant antigen in both the murine and human responses against Francisella and suggests that Tul4 might serve as a relevant model antigen to investigate host immune responses against Francisella.
One of the most surprising findings of the current study was that the analyses of T cell responses generated from live bacterial infection and conventional immunization using purified protein revealed the same epitope. One might have predicted that antigens from these immunizations might be processed and presented very differently. During Francisella infection, the bacteria reside within APC such as macrophages and dendritic cells (Oyston et al., 2004), and thus the antigen would be from an endogenous source, whereas in the conventional immunization, the source of antigen is exogenous. Interestingly, recent data has highlighted a pathway termed autophagy, in which endogenous antigen can be effectively processed and presented in the context of class II MHC molecules (Crotzer and Blum, 2005; Strawbridge and Blum, 2007). Autophagic pathways have been hypothesized to play an important role in the response to pathogens (Crotzer and Blum, 2005; Deretic, 2006). Relevant in this regard, Francisella can be found in autophagic vesicles, providing evidence for a mechanism by which Francisella antigens could be shuttled between processing compartments (Checroun et al., 2006) and a recent study has shown these autophagic vesicles to be MHC-II positive (Hrstka et al., 2007). However, other sources of antigen might also be available due to the release of dead and dying bacteria, which might occur during the natural course of infection which then could be presented in the traditional class II antigen presentation pathway (Sant et al., 2007; Vyas et al., 2008). Whatever pathways are operational, it will be important to determine if other antigens behave the same as Tul4 and whether the epitopes are the same in the context of an infection and in a conventional protein immunization. If so, since prior immunization can dramatically affect the immunodominance hierarchy of a T cell response (Cole et al., 1997; Sant et al., 2007; Yewdell and Bennink, 1999) immunization with individual proteins may allow one to focus the immune response to epitopes which can be recognized in the context of a natural infection, but which are normally subdominant or weakly recognized. Examples of attractive targets for this strategy include proteins essential for intracellular growth or virulence, such as the proteins encoded by the pathogenicity island genes (Nano and Schmerk, 2007).
The results from the in vivo experiments have validated the Tul4 86-99 epitope and show that it is recognized by T cells in an infection model. Interestingly, we found multiple hybridomas that recognized the Tul4 86-99 peptide, which is consistent with it being a significant epitope in the response to F. tularensis. However, because the T cells were re-stimulated in culture prior to fusion, and only a few fusions were analyzed, hybridoma reactivity cannot be used to rigorously establish the frequency of responding T cells and dominance in the immune response. However, analyses of T cells from infected animals directly illustrates that Tul4 86-99 is a major epitope recognized. For example, in the acute response, a significant proportion of CD4+ splenocytes were activated by Tul4 86-99 peptide. In one mouse, as much as 20% of total acute response to F. tularensis extract can be attributed to T cells that recognize Tul4 86-99. Moreover, a significant proportion of T cells recognize Tul4 86-99 epitope two months after infection, suggesting that Tul4 reactive T cells enter the long-term memory pool, a critical property of a potential vaccine candidate. Both the ID6 and FT13 1E4B4 hybridomas were capable of being activated by both LVS and SchuS4 extracts revealing that there is significant cross-recognition of the Tul4 86-99 epitope, as one might predict given the homology of Tul4 between the strains (Titball and Petrosino, 2007). Importantly, these experiments also show Tul4 is expressed at sufficient levels by both strains, since closely related, or even identical, proteins can be regulated very differently between different bacterial strains, even altering their virulence (Beyhan et al., 2006).
Immunization with Tul4, either using recombinant S. typhimurium expressing Tul4 or Tul4 formulated with immunostimulating complexes (ISCOMs), can provide partial protection in mice (Golovliov et al., 1995). Intriguingly, Tul4 has recently been shown to function in the innate response to F. tularensis via signaling through TLR2 (Thakran et al., 2008), whether this is related to its ability to function in protection is as yet unknown. While reports suggest that Tul4 may be relevant in a protective response (Thakran et al., 2008), it is likely that a polyvalent vaccine will be required to be a fully efficacious and a number of approaches with this goal are underway (Baron et al., 2007; Duckett et al., 2005; Eyles et al., 2008; Golovliov et al., 1995; Isherwood et al., 2005; Sjostedt et al., 1992). The identification of the defined Tul4 86-99 epitope, and the recent identification of epitopes of Francisella in humans (McMurry et al., 2007) will allow the quantitative assessment of T cell responses using ELI-spot techniques, intracellular cytokine staining, or analyses using MHC tetramers, and thus will be invaluable in examining the immune response to Francisella in the context of infection or in vaccine studies.
Acknowledgments
We would like to thank Dr. Andrea Sant for the MHC class II knockout splenocytes and Dr. Nilabh Shastri for the BWZ.36 cell line. This project has been funded in part by Federal funds from the National Institutes of Health contract NO1-AI-40086, NIH/NIAID Southeast Regional Center of Excellence for Emerging Infections and Biodefense (SERCEB) (grant no. U54 AI 057157); M.V. was supported by NIAID training grant T32-AI-7362-17 and M.D.W. was supported by NIAID AI-007062.
Abbreviations
- DHFR
dihydrofolate reductase
- 6x His
6x histidine
- APC
antigen presenting cell
- bp
base pairs
- aa
amino acids
- LVS
Live Vaccine Strain
- PCR
polymerase chain reaction
- ISCOM
immunostimulating complexes
- B6
C57Bl/6J
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altman GB. Tularemia. A pathogen in nature and a biological weapon. Aaohn J. 2002;50:373–7. [PubMed] [Google Scholar]
- Baron SD, Singh R, Metzger DW. Inactivated Francisella tularensis live vaccine strain protects against respiratory tularemia by intranasal vaccination in an immunoglobulin A-dependent fashion. Infect Immun. 2007;75:2152–62. doi: 10.1128/IAI.01606-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyhan S, Tischler AD, Camilli A, Yildiz FH. Differences in gene expression between the classical and El Tor biotypes of Vibrio cholerae O1. Infect Immun. 2006;74:3633–42. doi: 10.1128/IAI.01750-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Checroun C, Wehrly TD, Fischer ER, Hayes SF, Celli J. Autophagy-mediated reentry of Francisella tularensis into the endocytic compartment after cytoplasmic replication. Proc Natl Acad Sci U S A. 2006;103:14578–83. doi: 10.1073/pnas.0601838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Shen H, Webb A, KuoLee R, Conlan JW. Tularemia in BALB/c and C57BL/6 mice vaccinated with Francisella tularensis LVS and challenged intradermally, or by aerosol with virulent isolates of the pathogen: protection varies depending on pathogen virulence, route of exposure, and host genetic background. Vaccine. 2003;21:3690–700. doi: 10.1016/s0264-410x(03)00386-4. [DOI] [PubMed] [Google Scholar]
- Cole GA, Hogg TL, Coppola MA, Woodland DL. Efficient priming of CD8+ memory T cells specific for a subdominant epitope following Sendai virus infection. J Immunol. 1997;158:4301–9. [PubMed] [Google Scholar]
- Conlan JW, Chen W, Shen H, Webb A, KuoLee R. Experimental tularemia in mice challenged by aerosol or intradermally with virulent strains of Francisella tularensis: bacteriologic and histopathologic studies. Microb Pathog. 2003;34:239–48. doi: 10.1016/s0882-4010(03)00046-9. [DOI] [PubMed] [Google Scholar]
- Conlan JW, Sjostedt A, North RJ. CD4+ and CD8+ T-cell-dependent and -independent host defense mechanisms can operate to control and resolve primary and secondary Francisella tularensis LVS infection in mice. Infect Immun. 1994;62:5603–7. doi: 10.1128/iai.62.12.5603-5607.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley SC, Elkins KL. Multiple T cell subsets control Francisella tularensis LVS intracellular growth without stimulation through macrophage interferon gamma receptors. J Exp Med. 2003;198:379–89. doi: 10.1084/jem.20030687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley SC, Hamilton E, Frelinger JA, Su J, Forman J, Elkins KL. CD4-CD8- T cells control intracellular bacterial infections both in vitro and in vivo. J Exp Med. 2005;202:309–19. doi: 10.1084/jem.20050569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotzer VL, Blum JS. Autophagy and intracellular surveillance: Modulating MHC class II antigen presentation with stress. Proc Natl Acad Sci U S A. 2005;102:7779–80. doi: 10.1073/pnas.0503088102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis DT, Inglesby TV, Henderson DA, Bartlett JG, Ascher MS, Eitzen E, Fine AD, Friedlander AM, Hauer J, Layton M, Lillibridge SR, McDade JE, Osterholm MT, O’Toole T, Parker G, Perl TM, Russell PK, Tonat K. Tularemia as a biological weapon: medical and public health management. Jama. 2001;285:2763–73. doi: 10.1001/jama.285.21.2763. [DOI] [PubMed] [Google Scholar]
- Deretic V. Autophagy as an immune defense mechanism. Curr Opin Immunol. 2006;18:375–82. doi: 10.1016/j.coi.2006.05.019. [DOI] [PubMed] [Google Scholar]
- Doolan DL, Southwood S, Freilich DA, Sidney J, Graber NL, Shatney L, Bebris L, Florens L, Dobano C, Witney AA, Appella E, Hoffman SL, Yates JR, 3rd, Carucci DJ, Sette A. Identification of Plasmodium falciparum antigens by antigenic analysis of genomic and proteomic data. Proc Natl Acad Sci U S A. 2003;100:9952–7. doi: 10.1073/pnas.1633254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckett NS, Olmos S, Durrant DM, Metzger DW. Intranasal interleukin-12 treatment for protection against respiratory infection with the Francisella tularensis live vaccine strain. Infect Immun. 2005;73:2306–11. doi: 10.1128/IAI.73.4.2306-2311.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eigelsbach HT, Downs CM. Prophylactic effectiveness of live and killed tularemia vaccines. I. Production of vaccine and evaluation in the white mouse and guinea pig. J Immunol. 1961;87:415–25. [PubMed] [Google Scholar]
- Elkins KL, Cowley SC, Bosio CM. Innate and adaptive immune responses to an intracellular bacterium, Francisella tularensis live vaccine strain. Microbes Infect. 2003;5:135–42. doi: 10.1016/s1286-4579(02)00084-9. [DOI] [PubMed] [Google Scholar]
- Elkins KL, Cowley SC, Bosio CM. Innate and adaptive immunity to Francisella. Ann N Y Acad Sci. 2007;1105:284–324. doi: 10.1196/annals.1409.014. [DOI] [PubMed] [Google Scholar]
- Elkins KL, Rhinehart-Jones T, Nacy CA, Winegar RK, Fortier AH. T-cell-independent resistance to infection and generation of immunity to Francisella tularensis. Infect Immun. 1993;61:823–9. doi: 10.1128/iai.61.3.823-829.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkins KL, Rhinehart-Jones TR, Culkin SJ, Yee D, Winegar RK. Minimal requirements for murine resistance to infection with Francisella tularensis LVS. Infect Immun. 1996;64:3288–93. doi: 10.1128/iai.64.8.3288-3293.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis J, Oyston PC, Green M, Titball RW. Tularemia. Clin Microbiol Rev. 2002;15:631–46. doi: 10.1128/CMR.15.4.631-646.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyles JE, Hartley MG, Laws TR, Oyston PC, Griffin KF, Titball RW. Protection afforded against aerosol challenge by systemic immunisation with inactivated Francisella tularensis live vaccine strain (LVS) Microb Pathog. 2008;44:164–8. doi: 10.1016/j.micpath.2007.08.009. [DOI] [PubMed] [Google Scholar]
- Fong IW, Alibek K. Bioterrorism and infectious agents: a new dilemma for the 21st century. Springer; New York: 2005. [Google Scholar]
- Fortier AH, Slayter MV, Ziemba R, Meltzer MS, Nacy CA. Live vaccine strain of Francisella tularensis: infection and immunity in mice. Infect Immun. 1991;59:2922–8. doi: 10.1128/iai.59.9.2922-2928.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovliov I, Ericsson M, Akerblom L, Sandstrom G, Tarnvik A, Sjostedt A. Adjuvanticity of ISCOMs incorporating a T cell-reactive lipoprotein of the facultative intracellular pathogen Francisella tularensis. Vaccine. 1995;13:261–7. doi: 10.1016/0264-410x(95)93311-v. [DOI] [PubMed] [Google Scholar]
- Griffin KF, Oyston PC, Titball RW. Francisella tularensis vaccines. FEMS Immunol Med Microbiol. 2007;49:315–23. doi: 10.1111/j.1574-695X.2007.00219.x. [DOI] [PubMed] [Google Scholar]
- Hepburn MJ, Simpson AJ. Tularemia: current diagnosis and treatment options. Expert Rev Anti Infect Ther. 2008;6:231–40. doi: 10.1586/14787210.6.2.231. [DOI] [PubMed] [Google Scholar]
- Hrstka R, Krocova Z, Cerny J, Vojtesek B, Macela A, Stulik J. Francisella tularensis strain LVS resides in MHC II-positive autophagic vacuoles in macrophages. Folia Microbiol (Praha) 2007;52:631–6. doi: 10.1007/BF02932193. [DOI] [PubMed] [Google Scholar]
- Isherwood KE, Titball RW, Davies DH, Felgner PL, Morrow WJ. Vaccination strategies for Francisella tularensis. Adv Drug Deliv Rev. 2005;57:1403–14. doi: 10.1016/j.addr.2005.01.030. [DOI] [PubMed] [Google Scholar]
- Kman NE, Nelson RN. Infectious agents of bioterrorism: a review for emergency physicians. Emerg Med Clin North Am. 2008;26:517–47. x–xi. doi: 10.1016/j.emc.2008.01.006. [DOI] [PubMed] [Google Scholar]
- Lee BY, Horwitz MA, Clemens DL. Identification, recombinant expression, immunolocalization in macrophages, and T-cell responsiveness of the major extracellular proteins of Francisella tularensis. Infect Immun. 2006;74:4002–13. doi: 10.1128/IAI.00257-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurry JA, Gregory SH, Moise L, Rivera D, Buus S, De Groot AS. Diversity of Francisella tularensis Schu4 antigens recognized by T lymphocytes after natural infections in humans: identification of candidate epitopes for inclusion in a rationally designed tularemia vaccine. Vaccine. 2007;25:3179–91. doi: 10.1016/j.vaccine.2007.01.039. [DOI] [PubMed] [Google Scholar]
- Nano FE, Schmerk C. The Francisella pathogenicity island. Ann N Y Acad Sci. 2007;1105:122–37. doi: 10.1196/annals.1409.000. [DOI] [PubMed] [Google Scholar]
- Oyston PC, Sjostedt A, Titball RW. Tularaemia: bioterrorism defence renews interest in Francisella tularensis. Nat Rev Microbiol. 2004;2:967–78. doi: 10.1038/nrmicro1045. [DOI] [PubMed] [Google Scholar]
- Rhinehart-Jones TR, Fortier AH, Elkins KL. Transfer of immunity against lethal murine Francisella infection by specific antibody depends on host gamma interferon and T cells. Infect Immun. 1994;62:3129–37. doi: 10.1128/iai.62.8.3129-3137.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson S, Shastri N. LacZ inducible, antigen/MHC-specific T cell hybrids. Int Immunol. 1994;6:369–76. doi: 10.1093/intimm/6.3.369. [DOI] [PubMed] [Google Scholar]
- Sant AJ, Chaves FA, Krafcik FR, Lazarski CA, Menges P, Richards K, Weaver JM. Immunodominance in CD4 T-cell responses: implications for immune responses to influenza virus and for vaccine design. Expert Rev Vaccines. 2007;6:357–68. doi: 10.1586/14760584.6.3.357. [DOI] [PubMed] [Google Scholar]
- Saslaw S, Eigelsbach HT, Prior JA, Wilson HE, Carhart S. Tularemia vaccine study. II. Respiratory challenge. Arch Intern Med. 1961a;107:702–14. doi: 10.1001/archinte.1961.03620050068007. [DOI] [PubMed] [Google Scholar]
- Saslaw S, Eigelsbach HT, Wilson HE, Prior JA, Carhart S. Tularemia vaccine study. I. Intracutaneous challenge. Arch Intern Med. 1961b;107:689–701. doi: 10.1001/archinte.1961.03620050055006. [DOI] [PubMed] [Google Scholar]
- Sette A, Peters B. Immune epitope mapping in the post-genomic era: lessons for vaccine development. Curr Opin Immunol. 2007;19:106–10. doi: 10.1016/j.coi.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Sjostedt A. Tularemia: history, epidemiology, pathogen physiology, and clinical manifestations. Ann N Y Acad Sci. 2007;1105:1–29. doi: 10.1196/annals.1409.009. [DOI] [PubMed] [Google Scholar]
- Sjostedt A, Sandstrom G, Tarnvik A. Humoral and cell-mediated immunity in mice to a 17-kilodalton lipoprotein of Francisella tularensis expressed by Salmonella typhimurium. Infect Immun. 1992;60:2855–62. doi: 10.1128/iai.60.7.2855-2862.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjostedt A, Sandstrom G, Tarnvik A, Jaurin B. Nucleotide sequence and T cell epitopes of a membrane protein of Francisella tularensis. J Immunol. 1990;145:311–7. [PubMed] [Google Scholar]
- Sjostedt A, Tarnvik A, Sandstrom G. The T-cell-stimulating 17-kilodalton protein of Francisella tularensis LVS is a lipoprotein. Infect Immun. 1991;59:3163–8. doi: 10.1128/iai.59.9.3163-3168.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storozynsky E, Woodward JG, Frelinger JG, Lord EM. Interleukin-3 and granulocyte-macrophage colony-stimulating factor enhance the generation and function of dendritic cells. Immunology. 1999;97:138–49. doi: 10.1046/j.1365-2567.1999.00741.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strawbridge AB, Blum JS. Autophagy in MHC class II antigen processing. Curr Opin Immunol. 2007;19:87–92. doi: 10.1016/j.coi.2006.11.009. [DOI] [PubMed] [Google Scholar]
- Thakran S, Li H, Lavine CL, Miller MA, Bina JE, Bina XR, Re F. Identification of Francisella tularensis lipoproteins that stimulate the toll-like receptor (TLR) 2/TLR1 heterodimer. J Biol Chem. 2008;283:3751–60. doi: 10.1074/jbc.M706854200. [DOI] [PubMed] [Google Scholar]
- Thompson LW, Hogan KT, Caldwell JA, Pierce RA, Hendrickson RC, Deacon DH, Settlage RE, Brinckerhoff LH, Engelhard VH, Shabanowitz J, Hunt DF, Slingluff CL., Jr Preventing the spontaneous modification of an HLA-A2-restricted peptide at an N-terminal glutamine or an internal cysteine residue enhances peptide antigenicity. J Immunother. 2004;27:177–83. doi: 10.1097/00002371-200405000-00001. [DOI] [PubMed] [Google Scholar]
- Titball RW, Petrosino JF. Francisella tularensis genomics and proteomics. Ann N Y Acad Sci. 2007;1105:98–121. doi: 10.1196/annals.1409.015. [DOI] [PubMed] [Google Scholar]
- Turner MJ, Abdul-Alim CS, Willis RA, Fisher TL, Lord EM, Frelinger JG. T-cell antigen discovery (T-CAD) assay: a novel technique for identifying T cell epitopes. J Immunol Methods. 2001;256:107–19. doi: 10.1016/s0022-1759(01)00436-7. [DOI] [PubMed] [Google Scholar]
- Twine SM, Shen H, Kelly JF, Chen W, Sjostedt A, Conlan JW. Virulence comparison in mice of distinct isolates of type A Francisella tularensis. Microb Pathog. 2006;40:133–8. doi: 10.1016/j.micpath.2005.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas JM, Van der Veen AG, Ploegh HL. The known unknowns of antigen processing and presentation. Nat Rev Immunol. 2008;8:607–18. doi: 10.1038/nri2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolard MD, Hensley LL, Kawula TH, Frelinger JA. Respiratory Francisella tularensis live vaccine strain infection induces Th17 cells and prostaglandin E2, which inhibits generation of gamma interferon-positive T cells. Infect Immun. 2008;76:2651–9. doi: 10.1128/IAI.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolard MD, Wilson JE, Hensley LL, Jania LA, Kawula TH, Drake JR, Frelinger JA. Francisella tularensis-infected macrophages release prostaglandin E2 that blocks T cell proliferation and promotes a Th2-like response. J Immunol. 2007;178:2065–74. doi: 10.4049/jimmunol.178.4.2065. [DOI] [PubMed] [Google Scholar]
- Yee D, Rhinehart-Jones TR, Elkins KL. Loss of either CD4+ or CD8+ T cells does not affect the magnitude of protective immunity to an intracellular pathogen, Francisella tularensis strain LVS. J Immunol. 1996;157:5042–8. [PubMed] [Google Scholar]
- Yewdell JW, Bennink JR. Immunodominance in major histocompatibility complex class I-restricted T lymphocyte responses. Annu Rev Immunol. 1999;17:51–88. doi: 10.1146/annurev.immunol.17.1.51. [DOI] [PubMed] [Google Scholar]


