Abstract
Polycystic ovary syndrome (PCOS) is characterized by excessive theca cell androgen secretion, dependent upon LH, which acts through the intermediacy of 3′,5′-cyclic adenosine monophosphate (cAMP). cAMP signaling pathways are controlled through regulation of its synthesis by adenylyl cyclases, and cAMP degradation by phosphodiesterases (PDEs). PDE8A, a high-affinity cAMP-specific PDE is expressed in the ovary and testis. Leydig cells from mice with a targeted mutation in the Pde8a gene are sensitized to the action of LH in terms of testosterone production. These observations led us to evaluate the human PDE8A gene as a PCOS candidate gene, and the hypothesis that reduced PDE8A activity or expression would contribute to excessive ovarian androgen production. We identified a rare variant (R136Q; NM_002605.2 c.407G > A) and studied another known single nucleotide polymorphism (SNP) (rs62019510, N401S) in the PDE8A coding sequence causing non-synonymous amino acid substitutions, and a new SNP in the promoter region (NT_010274.16:g.490155G > A). Although PDE8A kinetics were consistent with reduced activity in theca cell lysates, study of the expressed variants did not confirm reduced activity in cell-free assays. Sub-cellular localization of the enzyme was also not different among the coding sequence variants. The PDE8A promoter SNP and a previously described promoter SNP did not affect promoter activity in in vitro assays. The more common coding sequence SNP (N401S), and the promoter SNPs were not associated with PCOS in our transmission/disequilibrium test-based analysis, nor where they associated with total testosterone or dehydroepiandrosterone sulfate levels. These findings exclude a significant role for PDE8A as a PCOS candidate gene, and as a Las major determinant of androgen levels in women.
Keywords: PDE8A, polycystic ovary syndrome, androgens, theca, SNP
Introduction
Polycystic ovary syndrome (PCOS) is a common endocrine and metabolic disorder that is characterized by increased circulating androgen levels, anovulatory infertility and frequently, insulin resistance and hyperinsulinemia (Franks, 1995; Legro, 2001; Ehrmann, 2005). A hallmark of PCOS is excessive theca cell androgen secretion, which is directly linked to the symptoms and pathophysiology of PCOS. Adrenal androgen excess (e.g. dehydroepiandrosterone sulfate—DHEAS) is also frequently found in women with PCOS.
Theca cell androgen synthesis is dependent upon LH, which acts through the intermediacy of the second messenger, 3′,5′-cyclic adenosine monophosphate (cAMP). cAMP signaling pathways are controlled through regulation of the synthesis of cAMP by adenylyl cyclases, and cAMP degradation by phosphodiesterases (PDEs) (Schwartz, 2001; Beavo and Brunton, 2002; Vasta et al., 2006; Kamenetsky et al., 2006). Theca cells isolated from ovaries of women with PCOS secrete increased amounts of androgens and progestins compared with normal theca cells in response to stimulation with forskolin, a diterprene activator of adenylate cyclases, suggesting that they have a heightened response to cAMP (Wood et al., 2003, Wickenheisser et al., 2006). Family based studies strongly suggest that the hyperandrogenemia associated with PCOS is an inherited trait, raising the possibility of a genetic predisposition to excessive ovarian androgen production in PCOS, which could be related to genetic differences in cAMP signaling (Legro et al., 1998). Moreover, steroid levels in blood in humans, including androgens, are heritable traits (Hong et al., 2001; Ukkola et al., 2002).
The PDE component of the cAMP signaling pathway, represented by a large superfamily that includes 21 mammalian genes that encode more than 100 isozymes, ensures the proper intensity and spatiotemporal distribution of cAMP signaling (Soderling and Beavo, 2000; Conti, 2002; Nikolaev et al., 2005; Zaccolo et al., 2006). PDEs including PDE3A, PDE4B and PDE4D are present in distinct compartments in the rodent ovary (Tsafriri et al., 1996).The mammalian PDE8 family includes PDE8A, a high-affinity cAMP-specific PDE, that is expressed in ovary, testis and other tissues including liver, kidney and heart (Soderling et al., 1998a, b; Mehats et al., 2002; Ahlström et al., 2005; Bender and Beavo, 2006; Sasseville et al., 2009). Leydig cells from mice with a targeted mutation in the Pde8a gene are sensitized to the action of LH in terms of testosterone production. These observations led us to evaluate the human PDE8A gene as a PCOS candidate gene, based on the hypothesis that reduced PDE8A activity or expression would contribute to excessive ovarian androgen production. Here we report new polymorphisms in the PDE8A coding sequence and promoter. The more common of these variants were not associated with PCOS or with testosterone and DHEAS levels.
Materials and Methods
Definition of PCOS
In this study, as in our previous work, the diagnosis was made by history of oligomenorrhea or amenorrhea (six or less menses per year) and biochemical evidence of hyperandrogenemia (Legro et al., 1998; Urbanek et al., 2005). Probands and their sisters were considered affected if they had six or fewer menses per year and either elevated total testosterone ( > 58 ng/dl; 2 nmol/l) or elevated non-sex hormone binding globulin-bound testosterone ( > 15 ng/dl; 0.5 nmol/l). Evidence for a PCO ovarian morphology was not required. Potential phenocopies (non-classical 21-hydroxylase deficiency, hyperprolactinemia, androgen-secreting tumors) were ruled out by appropriate tests. Sisters who were unaffected or considered to have an ‘unknown’ phenotype were not included in this analysis.
Theca cell cultures
The isolation and culture methods for normal and PCOS theca cells utilized in these studies have been previously described (Nelson et al., 1999; Wickenheisser et al., 2000; Wood et al., 2003).
Reverse transcription-PCR and PCR
Reverse transcription was carried out with MMLV-reverse transcriptase using RETROscript (Ambion, Austin, TX, USA) in a two step procedure. Briefly, total RNA (1.5 µg) from five normal theca cell preparations and five PCOS theca cell preparations, each from different subjects, was reverse transcribed in the presence of random decamers. The resulting cDNA was then used to carry out PCR amplification.
A PDE8A full coding sequence was amplified by PCR with a forward primer sequence of 5′-GGATCCCATGGGCTGTGCCCCGAGCATC-3′ containing a BamHI sequence (underlined) and a reverse primer 5′-GCGGCCGCTCTGGGTGGTGTCTCCCACT-3′ containing a NotI sequence, yielding a 2522 bp product. PCR was performed in a 50 µl reaction volume with the Expand long template PCR system (Roche Applied Science, Mannheim, German) in a Mastercycler ep thermal cycler (Eppendorf). After initial denaturation at 95°C for 3 min, PCR was performed for 35 cycles of denaturation at 95°C for 30 s annealing at 65°C for 30 s and extension at 72°C for 2 min and 45 s followed by a final 10 min elongation at 72°C. The PDE8A cDNAs were subjected to DNA sequence analysis. The PCR products were also cloned into the TOPO TA cloning vector (PCR2.1-TOPO, Invitrogen, Carlsbad, CA, USA), and four to five separate clones derived from each subject’s PCR amplification were sequenced.
To confirm the G596A (R136Q) variant, we carried out PCR using a forward primer sequence of 5′-GTGTTTACCAAAGAAGATAACCAATG-3′ and a reverse primer 5′-CATTTATAGGCACTTCTCCTAACTCC-3′ yielding a 547 bp product. After initial denaturation at 95°C for 3 min, PCR was performed for 35 cycles of denaturation at 95°C for 30 s, annealing at 51°C for 30 s and extension at 72°C for 1 min followed by a final 10 min elongation at 72°C. The PCR products were used for restriction fragment length polymorphism (RFLP) analysis with the restriction endonuclease TaqI (New England Biolabs) for genotyping.
Quantitative real-time PCR
RNA isolated from the theca cells were reverse transcribed to cDNA from women who had the coding sequence variants (MC03 and MC09) and from normal women (MC02 and MC31). cDNA diluted 1:10 and aliquots were subjected to quantitative real-time PCR. iTaq™ SYBR® Green Supermix with ROX reagent (Bio-Rad, Hercules, CA, USA) was used to detect amplicons of human PDE8A cDNA (forward primer 5′-GCCTGTTTCCTGGACAAAC-3′ and reverse primer 5′-GCATTCGGACA ACT CTT CTC-3′). The relative abundance of each cDNA were normalized with 18S rRNA gene (RNA primers forward: 5-GGCCCTGTAATTGGAATGAGTC-3′ and reverse 5′-CCAAGATCCAACTACGAGCTT-3′). Before amplification, samples were denatured at 95°C for 2.5 min. The amplification consisted of 40 cycles of denaturation at 95°C 15 s, annealing at the 57°C 30 s, and extension at 68°C 30 s. All reactions were run in triplicate.
Construction of expression plasmids
PCR products were cloned into the pCR2.1 TOPO vector (Invitrogen). The inserts were digested with NotI and BamHI and were subcloned into the pTarget vector.
The QuickChange II XL Site-directed Mutagenesis kit (Stratagene, La Jolla, CA, USA) was used to create cDNAs representing the various PDE8A variant sequences.
Construction of plasmids expressing PDE8A fused to enhanced green fluorescent protein
Polymerase chain reactions were carried out with the following primers containing restriction enzyme sites (underlined): 5′- CTCGAGCGCCAGCATGGGCTGTGCCCCGAGCATCCACATTTC (XhoI) and reverse primer 5′-GGATCCCTTCAGGAGGTGGTCGGAGGTTCCGCAGCTTC-3′ (BamHI). The plasmids containing the various PDE8A variant sequences were used as templates. After initial denaturation at 95°C for 3 min, PCR was performed for 18 cycles of denaturation at 95°C for 30 s, annealing at 68°C for 30 s and extension at 72°C for 2 min 45 s followed by a final 10 min elongation at 72°C. The PCR products were cloned into pCR2.1 TOPO vector; after sequencing, the inserts were subcloned into the mammalian expression pEGFP-N2 vector.
PDE8A promoter sequence variation
We examined genomic DNA from 20 Caucasian women; 10 with a diagnosis of PCOS and 10 normal women. Their genomic DNA was amplified by PCR to obtain a DNA fragment of the PDE8A promoter. Primers to amplify the PDE8A promoter including 5′-upsteam 1297 and 66 bp from the transcription start site, yielding a 1363 bp product were designed using reference sequences from the National Center for Biotechnology Information (GenBank accession NM_002605). The forward primer was 5′-CCACCAAGAAGTTAAGTGACTGGCCC-3′ and reverse primer was 5′-GCGGATCTCGCGTCAGGAAACG-3′.
PCR was performed in a 50 µl reaction volume with a GC-rich PCR system (Roche). Amplification conditions were: denaturation at 95°C for 3 min; followed by 10 cycles comprised of 30 s each at 95, 65 and 70 s at 72°C, then additional 25 cycles comprised of 30 s each at 95, 65 and 75 s at 72°C; followed by a final elongation step at 72°C for 7 min. Amplification products were run on 1% agarose and purified using QIAquick Gel extractraction kit (Qiagen, Valencia, CA, USA). The amplified promoter fragments were ligated into a TA cloning vector (Invitrogen). The PCR products were then subjected to sequence analysis.
Promoter reporter plasmid construction
The PDE8A promoter fragments were amplified by PCR with a forward primer: 5′-GGTACCCCACCAAGAAGTTAAGTGACTGGCCC-3′ (KpnI) and a reverse primer 5′-CTCGAGGCGGATCTCGCGTCAGGAAACG-3′ (XhoI). The amplified promoter fragments were ligated into a TA cloning vector and then subcloned into the luciferase reporter PGL3 basic vector (Promega, Madison, WI, USA) after KpnI and XhoI digestion. The DNA sequences of the PCR template and clones were confirmed.
Cell culture and transfection
The day before transfection, COS-1 cells and Leydig tumor cells (MA-10) were seeded into 12-well culture plates. COS-1 cells were cultured in DMEM medium (Invitrogen) supplemented with 10% fetal bovine serum and antibiotics [100 units/ml penicillin G, 100 units/ml streptomycin sulfate (Gibco/BRL, Gaithersburg, MD, USA)]. MA-10 cells kindly provided by Dr Mario Ascoli, University of Fowa, were maintained in Weymouth MB 752/1 medium modified to contain 1.1 g/l of NaHCO3, 20 mM HEPES, 50 µg/ml of gentamicin, and 15% horse serum (Invitrogen). All cells were maintained at 37°C in a water-saturated atmosphere under 5% CO2 in air.
Cells were transfected using FuGENE 6 transfection reagent (Roche) with 0.5 µg of plasmids DNA. Empty pGL3 plasmid was transfected as a control. The medium was changed 24 h after transfection. The cells were incubated for an additional 48 h before they were harvested. A Renilla luciferase plasmid was co-transfected to control per transfection efficiency.
Western blot analysis
After 48 h, whole cell lysates were collected from transfected COS-1 cells with complete lysis-M buffer (Roche). To detect PDE8A protein in these samples, 25 µg of total protein were separated by SDS-PAGE, transferred to Immobilon P polyvinylidene difluoride membrane (Millipore), and probed with a 1:500 dilution of PDE8A 121AP (C-terminal Ab IgG, 98-102 kDa) antibody (FabGennix). After extensive washing, membranes were incubated with a secondary anti-rabbit horseradish peroxidase-linked whole antibody Ab at 1:2000 (GE healthcare). PDE8A protein was detected using the SuperSignal West Femto Sensitivity reagent (Pierce, Rockford, IL, USA).
PDE8A assays
Cells were harvested with complete lysis-M buffer and centrifuged for 10 min at 13 000 × g in a microcentrifuge. A total of 10 µl aliquots of the supernatant (normalized to the 100 ng/μl protein) were assayed for PDE activity in the presence of cAMP as substrate (0.1–10 µM) and 10 nM [3H]cAMP (Thompson and Appleman, 1971). 3-Isobutyl-methylxanthine (IBMX, 100 µM) was added to inhibit PDEs except PDE8. Kinetic constants were calculated from means of replicate assay points.
Luciferase assays
Luciferase activity were assayed with Dual-Luciferase reporter system (Promega Corp., Madison, WI, USA), and luminescence was determined using a Lumat LB9507 luminometer (Berthold Systems, Pittsburgh, PA, USA). Promoter activities were expressed as the ratio between Photinus luciferase and Renilla luciferase activities. Each experiment was carried out in triplicate and each experiment was repeated a minimum of three times.
Cell culture and transfection for confocal microscopy
COS-1 cells were cultured in two-well chamber slides. The cells were transfected with pEGFP-N2 plasmids using FuGENE 6. Forty-eight hours after transfection, some cells were lysed with complete lysis-M buffer (Roche), and western blotting was performed to verify protein expression. A 1:500 dilution of PDE8A 121AP antibody (FabGennix, Frisco, TX, USA) directed against the PDE8A C-terminus and 1:1000 dilutions of anti-green fluorescent protein (anti-GFP, Roche) were be used in our studies. Living and fixed transfected cells were visualized with a Leica TCS-SP2 AOBS confocal laser scanning microscope.
Genetic studies
Genotypes of the PDE8A single nucleotide polymorphisms (SNPs) were determined using Assay By Design technology and an ABI Prism 3730 Sequence Detection System (Applied Biosystems).
Family material
SNPs were genotyped in 454 families with PCOS: 44 multiplex families (parents and two or more affected daughters) and 410 simplex families (one affected daughter and parents). There were a total of 454 affected probands plus 49 affected sisters. The self-identified ethnicities of probands in the 454 families were: 90% white, 3% Hispanic, 2% black and 5% other or unknown. This study was approved by the institutional review boards of the University of Pennsylvania, Pennsylvania State University College of Medicine, Brigham and Women's Hospital and Northwestern University. Written informed consent was obtained from all adult subjects and from a parent or guardian for minor subjects.
Genotyping and analysis
The five SNPs identified in this study plus four additional SNPs in PDE8A were genotyped using Applied Biosystems Taqman SNP Genotyping Assays. Four of these assays, 596G > A (R136Q), 352C > G (L55V), 1391A > G (N401S) and -576G > A used Taqman Custom Genotyping Assays (see Table I for primer and probe sequences). Allelic PCR products were separated using the Applied Biosystems 7900HT Sequence Detection System with SDS 2.2 software. Genotypes were auto-called by SDS 2.2 software with quality value set at 0.95. Two individuals from the Centre d'Etude du Polymorphisme Humain (CEPH, Dausset et al., 1990) collection were typed on each of 16 96-well plates; no discrepancies were found for any of the seven SNPs.
Table I.
Primer and probe sequences for Applied Biosystems custom Taqman assays
| SNP ID | Forward primer | Reverse primer | VIC probe | FAM probe |
|---|---|---|---|---|
| 352C > G (L55V), rs11540803 | CCCGCAGGGCCAGAAG | CCGGCGCCCCTTACCT | AGTCGGAGCTTCGCGAC | AGTCGGAGGTTCGCGAC |
| 596G > A (R136Q) | TTCCTGGACAAACATCATGACATTATCA | CTTGGGCTTACCTGCACAGT | CAGAAATCCTCGACAGCT | CAGAAATCCTCAACAGCT |
| 1391A > G (N401S), rs62019510 | TGAAACTCAAGTATTTTCTTTCCATAATGTAGGT | CAGGCATGGGACTACTTTCCT | TCAATATTATCAATGCTGCCC | AATATTATCAGTGCTGCCC |
| -576G > A | CCCGGCCCGGATCAC | CGCACCCAGGAGTTGCT | CGGAGCCAGAGCC | CGGAGCCGGAGCC |
Error-checking of genotypes was performed with Merlin software (Abecasis et al., 2002) and families with one or more Mendelian discrepancies for a marker were excluded in analysis of that marker. Linkage disequilibrium between SNPs and PCOS was tested with the transmission/disequilibrium test (TDT, Spielman et al., 1993). The principle of the TDT is the following: under the null hypothesis of no association and no linkage between diseases and candidate gene, the two alleles in the heterozygote for a candidate gene will be transmitted to affected offspring with equal frequency. If there are unaffected offspring they are ignored, but multiple affected (sibs) can be included, provided the test is interpreted as a test of linkage.
The quantitative TDT (QTDT version 2.4.6; Abecasis et al., 2000) was used to test for association of the SNPs in PDE8A with PCOS-related quantitative traits. Unbound testosterone, total testosterone and DHEAS were tested in families using the quantitative TDT.
Statistical analysis
GraphPad Prism 5 software (GraphPad, San Diego, CA, USA) was used for statistical analyses. Comparisons among multiple groups were made using Tukey’s multiple comparison test with P < 0.05 being considered statistically significant.
Results
Polymorphism in the PDE8A coding sequence
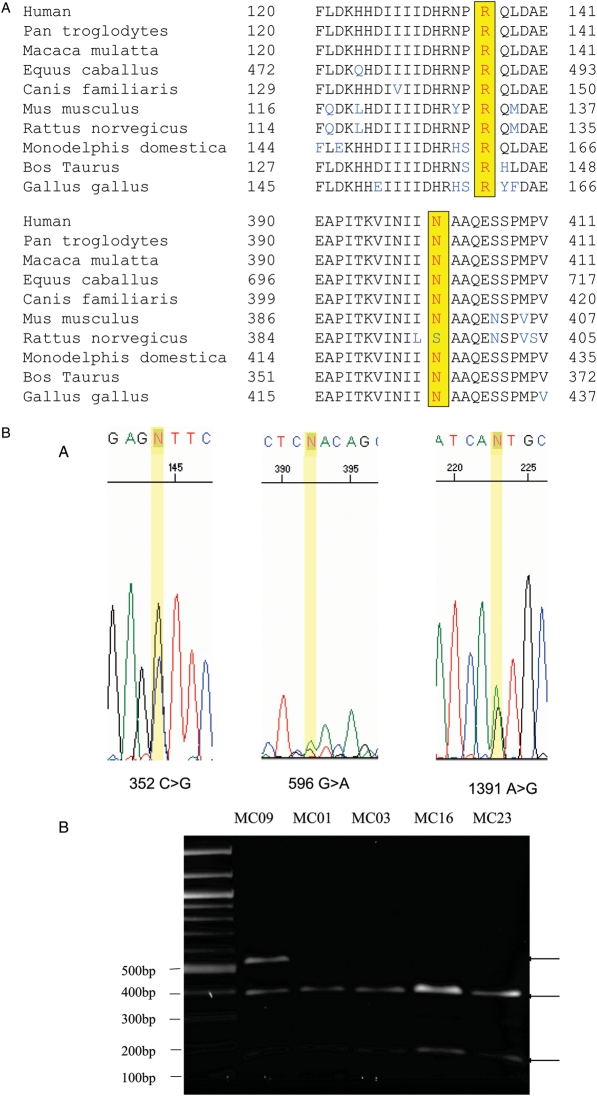
In an analysis of PDE8A coding sequences from five normal theca cell preparations and five PCOS theca cell preparations from different subjects, we identified three SNPs that resulted in non-synonymous amino acid substitutions 596G > A (R136Q, NM_002605.2 c.407G > A), 352C > G (L55V) and 1391A > G (N401S) (Table II). These were discovered in two different PCOS theca cell preparations by sequence analysis of PCR products and confirmed by RFLP in the case of the G596A SNP (Fig. 1B). The 55 V substitution, which results from a previously identified SNP (rs11540803), was present in the theca cells containing both the 136Q and 401S (rs62019510) substitutions, but it was not found on the same chromosome based on DNA sequence analysis of cloned PCR products.
Table II.
PDE8A coding sequence variation
| Samples | Variation | |
|---|---|---|
| Normal | MC02 | – |
| MC11 | – | |
| MC31 | – | |
| MC40 | – | |
| MC50 | – | |
| PCOS | MC09 | G596A(R136Q), C352G (L55V) (rs11540803) |
| MC03 | C352G(L55V), A1391G(N401S) (rs62019510) | |
| MC01 | ||
| MC16 | ||
| MC23 |
Three SNPs resulted in non-synonymous amino acid substitutions 596G > A (R136Q), 352C > G (L55V) and 1391A > G (N401S). These were discovered in two different PCOS theca cell preparations. The 55V substitution was present in the theca cells containing both the 136Q and 401S substitutions.
Figure 1.
(A) Conservation of R 136 and NHO1 in PDE8A.
The Arg at position 136, or its equivalent position in other species, is conserved in all known PDE8A sequences including the chimpanzee, macaque, cat, cow, mouse, rat chicken and opossum. The Asp at position 401 is conserved in all known PDE8A sequences except in the rat, where it is a Ser. (B-A) Confirmation of PDE8A variants. A PDE8A full coding sequence was amplified by PCR with a forward primer sequence of 5′-GGATCCCATGGGCTGTGCCCCGAGCATC-3′ containing a BamHI sequence (underlined) and a reverse primer 5′-GCGGCCGCTCTGGGTGGTGTCTCCCACT-3′ containing a NotI sequence, yielding a 2522 bp product. PDE8A variants in coding sequences were determined by ABI Prism 3730 Sequence Detection System. Three SNPs, 596G > A, 352C > G and 1391A > G were identified. (B-B) Confirmation of PDE8A G596A variants by RFLP. PCR was carried out using a forward primer sequence of 5′-GTGTTTACCAAAGAAGATAACCAATG-3′ and a reverse primer 5′-CATTTATAGGCACTTCTCCTAACTCC-3′ yielding a 547 bp product. The G596A variant was identified using the ABI Prism 3730 Sequence Detection System. The G596A variant was confirmed by RFLP. The 547 bp PCR products was digested with the restriction endonuclease TaqI, which yielded two fragment with size of 153 and 394 bp for the 596G allele, and one for the 596A allele.
The Arg at position 136, or its equivalent position in other species, is conserved in all known PDE8A sequences including the chimpanzee, macaque, cat, cow, mouse, rat chicken and opossum (Fig. 1A top). The Asp at position 401 is conserved except in the rat, where it is a Ser (Fig. 1A bottom). The Leu at position 55 is not conserved, and is a Pro in the macaque, cow and rat. Confirmation of PDE8A sequences were performed by PCR and RFLP (Fig. 1B) An examination of genomic DNA from 69 other subjects, 66 females with the diagnosis of PCOS and three CEPH subjects revealed the presence of Arg at position 136 in all samples. Combined with the theca cell analysis, 156 chromosomes were interrogated, and the estimated allele frequency of the 596A allele was 0.0064. Thus, the 596A allele is a rare variant, or possibly a new mutation. The 1391G causing the N401S substitution allele was found in four PCOS women and one CEPH DNA sample and combined with the one PCOS theca sample gives an allele frequency of 0.038 in the 156 chromosomes examined.
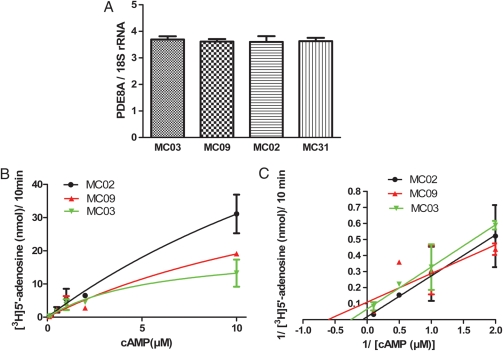
We measured the steady-state levels of PDE8A mRNA in the theca cell preparations that we used to identify the coding sequence variants by quantitative real-time PCR and found that they were similar, suggesting that the sequence variation does not have a major effect on PDE8A gene transcription, RNA processing or mRNA turnover (Fig. 2A). We then assayed PDE8A activity in lysates of the cells in which the two coding sequence variants were identified and compared activity to that in a theca cell preparation from a normal woman. All other PDE activity was inhibited by the addition of IBMX, which does not inhibit PDE8A. We found that the PDE8A activity (Vmax) was reduced in the 136Q (MC09) and 401S (MCO3) variant-containing cells compared with the normal theca cells (MC02) without these variants (Fig. 2B).
Figure 2.
(A) PDE8A mRNA levels in theca cells.
PDE8A mRNA levels were similar in the theca cells from women who had the coding sequence variants (MC03 and MC09) compared with theca cells from normal women (MC02 and MC31) by quantitative real-time PCR. cDNA diluted 1:10 and aliquots were subjected to quantitative real-time PCR. SYBR Green reagent was used to detect amplicons of human PDE8A cDNA. The relative abundance of each cDNA was normalized with 18S rRNA. All reactions were run in triplicate. Values presented are measn ± SD. (B) PDE8A activity in theca cells. The PDE assay was performed by the radiolabeled nucleotide method as previously described (Thompson and Appleman, 1971). Km and relative Vmax values of the human PDE8A activities were derived from Michaelis–Menten (left) and Lineweaver–Burk plots (right) using cAMP as substrate (0.1–10 µM). The best fit of Km value of PDE8A for MC02, MC03 and MC09 were 26.4, 19.3 and 5.7 µM. Best-fit of Vmax value of PDE8A for MC02, MC03 and MC09 were 113.0, 55.9 and 20.8 nmol/10 min.
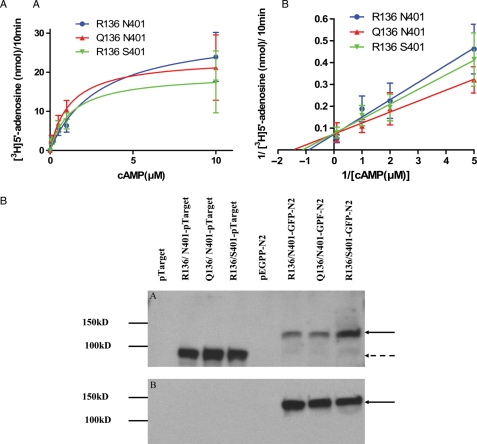
To further investigate the apparent functional differences, we prepared expression plasmids representing each of the R136Q and N401S alleles and tested the expressed proteins for PDE activity in COS-1 cells, which do not express this gene (Fig. 3A). In cells expressing equivalent amounts of PDE8A protein, as assessed by western blot analysis, we found that the different enzyme proteins had essentially similar activity (Fig. 3B). This finding was contrary to the expectations based on the analysis of activity in cell lysates and the notion that the two variants (136Q, 401S) would contribute to PCOS as a result of diminished activity. Collectively, these observations demonstrate that the two coding sequence variants display kinetic properties that would reduce cAMP catabolism in the context of human theca cells, which could be a reflection of the presence of another coding sequence variant (55 V) on the other chromosome. However, when studied in isolation in a different cell context, these kinetic differences are not evident so we must conclude that the 136Q and 401S alleles do not unto themselves impair enzyme function in a cell-free system. We did not examine the L55V alleles in this study because our genetic analysis, described below, indicated that this variant is not associated with PCOS.
Figure 3.
(A) PDE8A activity of coding variants.
Expression plasmids representing each allele were constructed with pTarget vector and transfected in COS-1 cells. The cell lysates were used to perform the PDE assay by the radiolabeled nucleotide method using cAMP as substrate (0.1–10 µM). Km and relative Vmax values of PDE8A activities were derived from Michaelis–Menten (A) and Lineweaver–Burk plots (B). The best fit of Km value of PDE8A for the major allele R136 N401, Q136 N401 (136Q variant) and R136 S401 (401S variant) were 3.0, 1.2 and 1.3 µM. The best-fit of Vmax value for R136 N401, Q136 N401 and R136 S401 were 31, 24 and 20 nmol/10 min. (B) Western blot analysis were detected with 25 μg of total protein in each sample probed with a 1:500 dilution of PDE8A 121AP antibody (A) upper panel and 1:1000 dilution of anti-GFP (B) lower panel. A single band at 98 kDa was observed with pTarget vector and 140 kDa with the GPF-N2 vector.
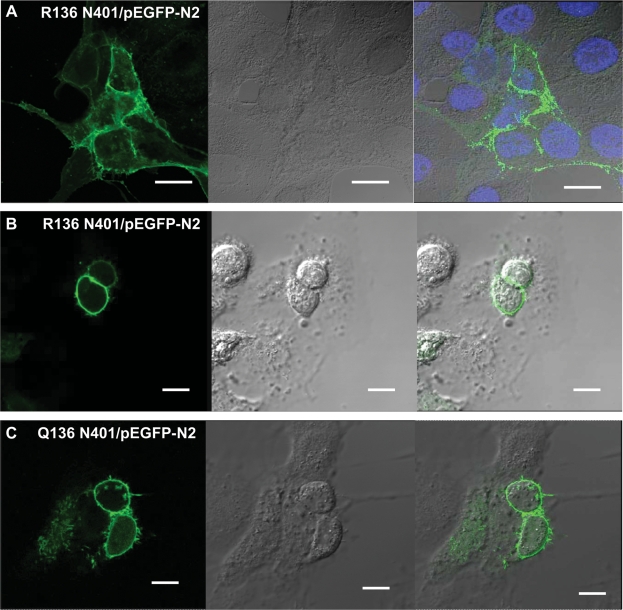
To determine if the minor alleles influenced cellular compartmentalization, we studied protein localization in COS-1 cells using tagged enzyme molecules. When expression plasmids for the different PDE8A variants coupled to GFP at the protein N-terminus were transfected into COS-1 cells, the GFP-tagged proteins were localized to the cell periphery at the plasma membrane. There appeared to be no differences in distribution among the variants (Fig. 4). This localization pattern, which was previously unknown, positions PDE8A to control cAMP levels generated in response to tropic stimulation of cell surface receptors coupled to adenylate cyclase.
Figure 4.
Localization of PDE8A variants in COS-1 cells.
The expression plasmids for the different PDE8A variants coupled to GFP at the protein N-terminus were transfected into COS-1 cells. Forty-eight hours after transfection, Living transfected cells and fixed transfected cells were visualized with a Leica TCS-SP2 AOBS confocal laser scanning microscope. Living COS-1 cell transfected images with R136 N401/pEGFP-N2 plasmids were captured by confocal microscopy and stained with DAPI (A) (×2.5 magnification, bar = 20 µm). Forty-eight hours after transfection, COS-1 cells transfected with R136 N401/pEGFP-N2 plasmids (B) and Q136 N401/pEGFP-N2 (C) were fixed with methanol and captured by confocal microscopy. (×2.5 magnification, bar = 16 µm). The GFP-tagged proteins were localized to the cell periphery at the plasma membrane. There appeared to be no differences in distribution among the variants.
Polymorphism in the PDE8A promoter sequence
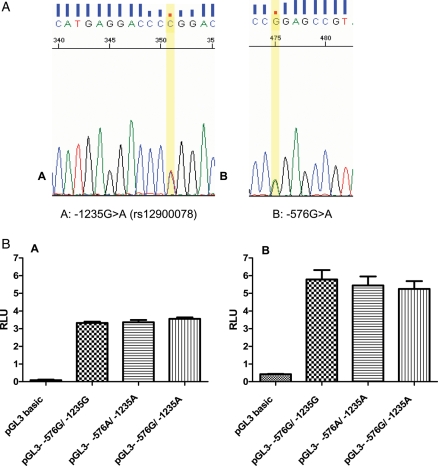
An analysis of the PDE8A promoter sequence of genomic DNA from PCOS and normal women revealed two variants (-1235G > A, -576G > A, NT_010274.16:g.490155G > A) (Table III and Fig. 5A), which occurred in four PCOS and two unaffected subjects. The -1235 SNP has been previously identified (rs12900078). These variants did not have a significant impact on function of the PDE8A promoter when tested in COS-1 and MA-10 cell hosts (Fig. 5B). The promoter activities in human theca cells with the -576G/ -1235G, -576G/-1235A and -576A/-1235G alleles were 0.23 ± 0.03, 0.28 ± 0.05 and 0.30 ± 0.05 (means ± SE, N = 3) based on luciferase assays normalized with β-galactosidase. The -576G/-1235A and -576A/-1235G alleles were 118 ± 16% and 129 ± 25%, respectively, of the promoter -576G/-1235G allele.
Table III.
PDE8A promoter sequences variation
| Samples | Variation | |
|---|---|---|
| Normal | 1 | |
| 2 | ||
| 3 | ||
| 4 | ||
| 5 | ||
| 6 | ||
| 7 | -576G > A, -1235G > A (rs12900078) | |
| 8 | -576G > A, -1235G > A (rs12900078) | |
| 9 | ||
| 10 | ||
| PCOS | 1 | -576G > A, -1235G > A (rs12900078) |
| 2 | ||
| 3 | -576G > A, -1235G > A (rs12900078) | |
| 4 | ||
| 5 | ||
| 6 | ||
| 7 | -576G > A, -1235G > A (rs12900078) | |
| 8 | ||
| 9 | ||
| 10 | -576G > A, -1235G > A (rs12900078) |
Two variants (-576G > A, -1235G > A) occurred in four PCOS and two unaffected subjects. The -1235G > A SNP has been previously identified (rs12900078).
Figure 5.
Two SNPs were identified in the PDE8A promoter sequence of genomic DNA from four PCOS and two normal women.
(A-A) The -1235G > A SNP is shown in the reverse strand, which has been previously identified (rs12900078). (A-B) A new SNP -576G > A (-576 from genomic reference sequence, NT_010274.16:g.490155G > A) was identified (Forward strand). (B) Functional activity of PDE promoters in COS-1 and MA-10 cells. COS-1 cells and MA-10 cell were transfected with reporter plasmids containing PDE8A promoter fragments representing the -1235G(g.489496G) and -576G(g.490155G) alleles. The relative Photinus luciferase activities (n = 3 separate experiments performed with triplicate wells for each cell host) standardized to Renilla luciferase. Luciferase activities were compared using the Tukey–Kramer test. These variants did not have a significant impact on function of the PDE8A promoter when tested in COS-1 cell (B-A) and MA-10 cell (B-B) hosts.
Lack of association of common variants with PCOS
We used the TDT to test for linkage and association between PCOS and each SNP described above as risk alleles for PCOS (Table IV). The coding sequence variant 596A > G (R136Q) was not polymorphic in our sample families and therefore was not evaluated. The two promoter SNPs -1235G > A and -576G > A were in near total linkage disequilibrium in a subset of the probands; therefore only -1235G > A (rs12900078) was genotyped in all the families. We also typed four additional SNPs in PDE8A: rs1109065 located ∼10 kb upstream of the gene, rs930716 in intron 1, and two SNPs near a region of splice site variation, rs8032301 and rs2304418. None of these PDE8A SNPs was significantly associated with PCOS. These genetic studies rule out a significant contribution of the 401S coding sequence variant to PCOS as well as the promoter SNPs.
Table IV.
TDT results in 454 families having at least one offspring with PCOS
| SNP location | SNP ID | MAFa | Over-transmitted allele | Tb | not T | Total: T + not T | Fraction T | c2 | P-value |
|---|---|---|---|---|---|---|---|---|---|
| 352C > G (L55V) | rs11540803c | G: 0.10 | G | 94 | 91 | 185 | 0.508 | 0.049 | 0.82 |
| 596G > A (R136Q) | NAc | Monomorphic | |||||||
| 1391A > G (N401S) | rs62019510 c | G: 0.03 | G | 30 | 26 | 56 | 0.536 | 0.286 | 0.59 |
| Promoter -576G > A | NAc | Near-complete LD with rs12900078 | |||||||
| Promoter -1235G > A | rs12900078 | A: 0.13 | G | 106 | 94 | 200 | 0.530 | 0.720 | 0.4 |
| ∼10 kb upstream | rs1109065 | C: 0.34 | T | 213 | 194 | 407 | 0.523 | 0.887 | 0.35 |
| Intron 1 | rs930716 | A: 0.41 | G | 229 | 210 | 439 | 0.522 | 0.822 | 0.36 |
| Near sites of splice variation | rs8032301 | C: 0.41 | T | 253 | 218 | 471 | 0.537 | 2.601 | 0.11 |
| rs2304418 | C: 0.26 | T | 192 | 183 | 375 | 0.512 | 0.216 | 0.64 | |
aMinor allele frequency (MAF) calculated from SNP genotypes in parents.
bT: number of transmissions in TDT analysis.
cSee Table I for Taqman assay primer and probe sequences.
Lack of association of PDE8A variants with PCOS or with the level of testosterone and DHEAS
We used the quantitative TDT (Abecasis et al., 2000) to test whether there was an association between any of the PDE8A SNPs and the level of testosterone (total and unbound) and DHEAS in the 454 PCOS families. None of the SNPs were significantly associated with hormone levels, with all P-values being ≥0.13.
Discussion
Although PDE8A appeared to be a viable candidate gene for PCOS, and new variants in the PDE8A coding sequence and promoter were identified, these variants do not appear to be functionally significant and a genetic analysis using the TDT did not reveal any evidence for significant association/linkage with PCOS. The fact that the 136Q variant is extremely rare also makes it highly unlikely that it is a significant risk allele as well, although we cannot exclude it from having a pathophysiological role. It should be noted that our findings do not exclude a potential role for variation in the activities of other PDEs, including members of the PDE4 family which are expressed in the rodent ovary (Tsafriri et al., 1996) in the pathophysiology of PCOS.
Although there is evidence for heritability of androgen levels in man (Hong et al., 2001), PDE8A variants were not associated with either testosterone or DHEAS levels in our study. No evidence for linkage on chromosome 15q was detected by performing a genome-wide linkage scan to identify loci affecting steroid concentrations (Ukkola et al., 2002), which is consistent with a lack of strong influence of the PDE8A gene on androgen levels. We, therefore, conclude that variation in the PDE8A gene is not a major contributor to the PCOS phenotype or serum androgen levels in women. This study has, however, identified new PDE8A variants and documented that PDE8A is expressed in human theca cells and that it is localized at a strategic position in the cell, the plasma membrane, to control cAMP levels. Despite the lack of evidence for genetic variation in PDE8A as a major risk factor for PCOS, pharmacologic interventions that increase either expression or activity of PDE8A might be of value in reducing hyperandrogenemia of ovarian origin.
Funding
This research was supported by National Institutes of Health grant 2U54 HD034449.
References
- Abecasis GR, Cardon LR, Cookson WO. A general test of association for quantitative traits in nuclear families. Am J Hum Genet. 2000;66:279–292. doi: 10.1086/302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abecasis GR, Cheryn SS, Cookson WO, Cardon LR. Merlin–rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- Ahlström M, Pekkinen M, Huttunen M, Lamberg-Allardt C. Cyclic nucleotide phosphodiesterases (PDEs) in human osteoblastic cells; the effect of PDE inhibition on cAMP accumulation. Cell Mol Biol Lett. 2005;10:305–319. [PubMed] [Google Scholar]
- Beavo JA, Brunton LL. Cyclic nucleotide research—still expanding after half a century. Nat Rev Mol Cell Bio. 2002;3:710–718. doi: 10.1038/nrm911. [DOI] [PubMed] [Google Scholar]
- Bender AT, Beavo JA. Cyclic nucleotide phosphodiesterases: molecular regulation to clinical use. Pharmacol Rev. 2006;58:488–520. doi: 10.1124/pr.58.3.5. [DOI] [PubMed] [Google Scholar]
- Conti M. Specificity of the cyclic adenosine 3′,5′-monophosphate signal in granulosa cell function. Biol Reprod. 2002;67:1653–1661. doi: 10.1095/biolreprod.102.004952. [DOI] [PubMed] [Google Scholar]
- Dausset J, Cann H, Cohen D, Lathrop M, Lalouel JM, White R. Centre d'etude du polymorphisme humain (CEPH): collaborative genetic mapping of the human genome. Genomics. 1990;6:575–577. doi: 10.1016/0888-7543(90)90491-c. [DOI] [PubMed] [Google Scholar]
- Ehrmann DA. Polycystic ovary syndrome. N Engl J Med. 2005;352:1223–1236. doi: 10.1056/NEJMra041536. [DOI] [PubMed] [Google Scholar]
- Franks S. Polycystic ovary syndrome. N Engl J Med. 1995;333:853–861. doi: 10.1056/NEJM199509283331307. [Erratum (1995) 333:1435] [DOI] [PubMed] [Google Scholar]
- Hong Y, Gagnon J, Rice T, Perusse L, Leon AS, Skinner JS, Wilmore JH, Bouchard C, Rao DC. Familial resemblance for free androgens and androgen glucuronides in sedentary black and white individuals; the HERITAGE Family Study. Health, risk factors, exercise training and genetics. J Endocrinol. 2001;170:485–492. doi: 10.1677/joe.0.1700485. [DOI] [PubMed] [Google Scholar]
- Kamenetsky M, Middelhaufe S, Bank EM, Levin LR, Buck J, Steegborn C. Molecular details of cAMP generation in mammalian cells: a tale of two systems. J Mol Biol. 2006;362:623–639. doi: 10.1016/j.jmb.2006.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legro RS. Polycystic ovary syndrome: the new millenium. Mol Cell Endocrinol. 2001;184:87–93. doi: 10.1016/s0303-7207(01)00640-2. [DOI] [PubMed] [Google Scholar]
- Legro RS, Droscoll D, Strauss JF, III, Fox J, Dunaif A. Evidence for a genetic basis for hyperandrogenemia in polycystic ovary syndrome. Proc Natl Acad Sci USA. 1998;95:14956–14960. doi: 10.1073/pnas.95.25.14956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehats C, Andersen CB, Filopanti M, Jin SL, Conti M. Cyclic nucleotide phosphodiesterases and their role in endocrine cell signaling. Trends Endocrinol Metab. 2002;13:29–35. doi: 10.1016/s1043-2760(01)00523-9. [DOI] [PubMed] [Google Scholar]
- Nelson VL, Legro RS, Strauss JF, 3rd, McAllister JM. Augmented androgen production is a stable steroidogenic phenotype of propagated theca cells from polycystic ovaries. Mol Endocrinol. 1999;13:946–957. doi: 10.1210/mend.13.6.0311. [DOI] [PubMed] [Google Scholar]
- Nikolaev VO, Gambaryan S, Engelhardt S, Walter U, Lohse MJ. Real-time monitoring of the PDE2 activity of live cells: hormone-stimulated cAMP hydrolysis is faster than hormone-stimulated cAMP synthesis. J Biol Chem. 2005;280:1716–1719. doi: 10.1074/jbc.C400505200. [DOI] [PubMed] [Google Scholar]
- Sasseville M, Albuz FK, Côté N, Guillemette C, Gilchrist RB, Richard FJ. Characterization of novel phosphodiesterases in the bovine ovarian follicle. Biol Reprod. 2009 doi: 10.1095/biolreprod.108.074450. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz JH. The many dimensions of cAMP signaling. Proc Natl Acad Sci USA. 2001;98:13482–13484. doi: 10.1073/pnas.251533998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderling SH, Beavo JA. Regulation of cAMP and cGMP signaling: new phosphodiesterases and new functions. Curr Opin Cell Biol. 2000;12:174–179. doi: 10.1016/s0955-0674(99)00073-3. [DOI] [PubMed] [Google Scholar]
- Soderling SH, Bayuga SJ, Beavo JA. Cloning and characterization of a cAMP-specific cyclic nucleotide phosphodiesterase. Proc Natl Acad Sci USA. 1998;a 95:8991–8996. doi: 10.1073/pnas.95.15.8991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderling SH, Bayuga SJ, Beavo JA. Cloning and characterization of a cAMP-specific cyclic nucleotide phosphodiesterase. Proc Natl Acad Sci USA. 1998;b 95:8991–8996. doi: 10.1073/pnas.95.15.8991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielman RS, McGinnis RE, Ewens WJ. Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus IDDM. Am J Hum Genet. 1993;52:506–516. [PMC free article] [PubMed] [Google Scholar]
- Thompson WJ, Appleman MM. Multiple cyclic nucleotide phosphodiesterase activities from rat brain. Biochemistry. 1971;10:311–316. [PubMed] [Google Scholar]
- Tsafriri A, Chun SY, Zhang R, Hsueh AJ, Conti M. Oocyte Maturation involves compartmentalization and opposing changes of cAMP levels in follicular somatic and germ cells: studies using selective phosphodiesterase inhibitors. Dev Biol. 1996;178:393–402. doi: 10.1006/dbio.1996.0226. [DOI] [PubMed] [Google Scholar]
- Ukkola O, Rankonen T, Gafnon J, Leon AS, Skinner JS, Wilmore JH, Rao DC, Bouchard C. A genome-wide linkage scan for steroids and SHBG levels in black and white families: the HERITAGE family study. J Clin Endocrinol Metab. 2002;876:3708–3720. doi: 10.1210/jcem.87.8.8725. [DOI] [PubMed] [Google Scholar]
- Urbanek M, Woodroffe A, Ewens KG, Diamanti-Kandarakis E, Legro RS, Strauss JF, III, Dunaif A, Spielman RS. Candidate gene region for polycystic ovary syndrome on chromosome 19p13.2. J Clin Endocrinol Metab. 2005;90:6623–6629. doi: 10.1210/jc.2005-0622. [DOI] [PubMed] [Google Scholar]
- Vasta V, Shimizu-Albergine M, Beavo JA. Modulation of Leydig cell function by cyclic nucleotide phosphodiesterase 8A. Proc Natl Acad Sci USA. 2006;103:19925–19930. doi: 10.1073/pnas.0609483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickenheisser JK, Quinn PG, Nelson VL, Legro RS, Strauss JF, 3rd, McAllister JM. Differential activity of the cytochrome P450 17alpha-hydroxylase and steroidogenic acute regulatory protein gene promoters in normal and polycystic ovary syndrome theca cells. J Clin Endocrinol Metab. 2000;85:2304–2311. doi: 10.1210/jcem.85.6.6631. [DOI] [PubMed] [Google Scholar]
- Wickenheisser JK, Nelson-DeGrave VL, McAllister JM. Human ovarian theca cells in culture. Trends Endocrinol Metab. 2006;17:65–71. doi: 10.1016/j.tem.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Wood JR, Nelson VL, Ho C, Jansen E, Wang CY, Urbanek M, McAllister JM, Mosselman S, Strauss JF., 3rd The molecular phenotype of polycystic ovary syndrome (PCOS) theca cells and new candidate PCOS genes defined by microarray analysis. J Biol Chem. 2003;278:26380–26390. doi: 10.1074/jbc.M300688200. [DOI] [PubMed] [Google Scholar]
- Zaccolo M, Di Benedetto G, Lissandron V, Mancuso L, Terrin A, Zamparo I. Restricted diffusion of a freely diffusible second messenger: mechanisms underlying compartmentalized cAMP signalling. Biochem Soc Trans. 2006;34:495–497. doi: 10.1042/BST0340495. [DOI] [PubMed] [Google Scholar]