Abstract
The light-activated enzyme NADPH-protochlorophyllide oxidoreductase (POR) catalyzes the trans addition of hydrogen across the C-17–C-18 double bond of protochlorophyllide (Pchlide), a key step in chlorophyll biosynthesis. Similar to other members of the short chain alcohol dehydrogenase/reductase family of enzymes, POR contains a conserved Tyr and Lys residue in the enzyme active site, which are implicated in a proposed reaction mechanism involving proton transfer from the Tyr hydoxyl group to Pchlide. We have analyzed a number of POR variant enzymes altered in these conserved residues using a combination of steady-state turnover, laser photoexcitation studies, and low temperature fluorescence spectroscopy. None of the mutations completely abolished catalytic activity. We demonstrate their importance to catalysis by defining multiple roles in the overall reaction pathway. Mutation of either residue impairs formation of the ground state ternary enzyme-substrate complex, pointing to a key role in substrate binding. By analyzing the most active variant (Y193F), we show that Tyr-193 participates in proton transfer to Pchlide and stabilizes the Pchlide excited state, enabling hydride transfer from NADPH to Pchilde. Thus, in addition to confirming the probable identity of the proton donor in Pchlide reduction, our work defines additional roles for these residues in facilitating hydride transfer through stabilization of the ground and excited states of the ternary enzyme complex.
The light-driven enzyme protochlorophyllide oxidoreductase (POR)3 (EC 1.3.1.33) catalyzes the trans addition of hydrogen across the C-17–C-18 double bond of the chlorophyll precursor protochlorophyllide (Pchlide) (see Fig. 1A) (1). This reaction is a key step in the synthesis of chlorophyll and leads to profound changes in the morphological development of photosynthetic organisms through modification and reorganization of plastid membranes (2, 3). In addition to POR, nonflowering land plants, algae, and cyanobacteria possess a light-independent Pchlide reductase, which consists of three separate subunits and allows these organisms to produce chlorophyllide in the dark (4). Together with DNA photolyase (5), POR is one of only two enzymes studied so far that exhibit a direct, natural requirement for light and because mixing strategies are no longer required to initiate the reaction, it is possible to trigger catalysis on very fast time scales and at cryogenic temperatures. Consequently, POR has proven to be an excellent model system for studying the role of protein dynamics in driving enzyme catalysis (1).
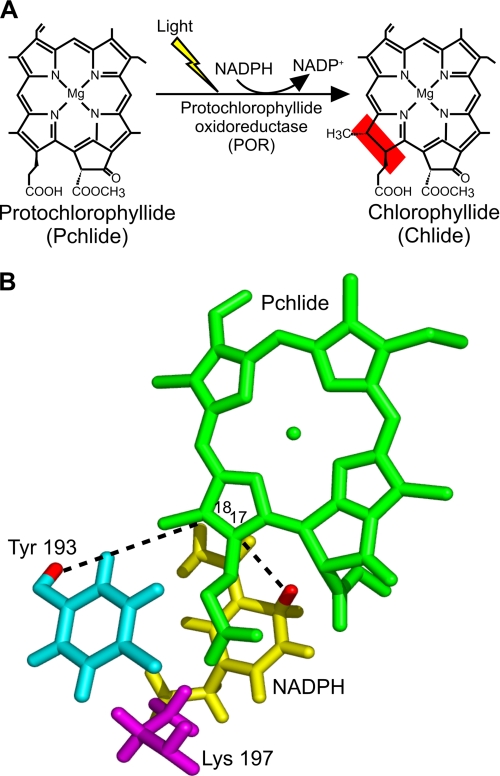
FIGURE 1.
The light-driven reduction of Pchlide. A, the trans addition of hydrogen across the C-17–C-18 double bond of Pchlide to form chlorophyllide (Chlide) in the chlorophyll biosynthesis pathway is catalyzed by the light-driven enzyme POR. B, shown is a three-dimensional model of the POR-catalyzed reaction based on the structural homology model of POR (26) and the proposed mechanism of hydride and proton transfer (8). Upon activation by light, a hydride is transferred to the C-17 position of Pchlide from the pro-S face of NADPH (shown in yellow), and the proton at the C-18 position is derived from Tyr-193 (shown in cyan). The conserved Lys-197 residue (shown in magenta) is proposed to decrease the pKa of the Tyr to facilitate the proton transfer reaction.
In the POR catalytic cycle, a ternary enzyme-NADPH-Pchlide complex is formed. Following light activation of this complex, a hydride ion is transferred from the pro-S face of NADPH to the C-17 atom of Pchlide (6, 7). The valence of the C-18 atom is satisfied by proton transfer, which is suggested to originate from an active site tyrosine residue (8). The catalytic cycle of POR has been analyzed through the trapping of intermediates at cryogenic temperatures. Following the initial light-driven reaction (9), there are a series of subsequent (slower) dark reactions (10, 11). The light-driven step involves hydride transfer from NADPH to form a charge transfer complex, which then facilitates protonation of the pigment intermediate during the first of the “dark” reactions (12). Moreover, through laser activation of catalysis, we have shown that both of these H-transfer reactions proceed by quantum mechanical tunneling coupled to motions in the enzyme-substrate complex on the submicrosecond time scale (13). The final dark steps in the reaction cycle involve a series of ordered product release and cofactor binding steps linked to conformational changes in the enzyme (10, 11, 14). Ultrafast measurements have uncovered spectral changes on the picosecond timescale that are likely to represent conformational changes prior to Pchlide reduction (15–19). Previous excitation of POR with a laser pulse leads to a more efficient conformation of the active site and an enhancement in the catalytic efficiency of the enzyme (18).
POR is a member of a large family of enzymes known as short chain dehydrogenases/reductases (SDR). These are single domain NAD(P)+- or NAD(P)H-binding oxidoreductases that exist generally as dimers or tetramers (20). A number of SDR enzymes (e.g. carbonyl reductase, alcohol dehydrogenase, and dihydrofolate reductase) have been good model systems for studying the dynamics linked to enzyme catalysis (21–23). This family of enzymes has been amenable to studies of biological H-tunneling (24–26), and in particular the unique light-activated properties of POR make it an excellent system for studying mechanisms of H-transfer and dynamics in this family of enzymes.
Structures of several SDR family members are available, and these have enabled the construction of a homology model of POR from Synechocystis (27). This model comprises a central parallel β-sheet of seven β-strands, surrounded by nine α-helices, with an additional unique 33-residue insertion between the fifth and sixth β-sheets. The NADPH cofactor binds within the N-terminal region of the enzyme, which contains a common nucleotide-binding motif with a tight βαβ fold, termed the Rossmann fold (27). Importantly, a Tyr and a Lys residue are both absolutely conserved throughout all members of the SDR family and are critical for catalysis in a number of enzymes (28–31). A common mechanism has been proposed for this group of enzymes, involving a Tyr-X-X-X-Lys motif. The Lys residue in this motif is presumed to facilitate proton donation from the Tyr hydroxyl group to substrate through favorable perturbation of the hydroxyl group pKa (8, 31). In POR, multiple turnover assays have also indicated that these Tyr and Lys residues are important for activity (8, 32, 33), leading to a proposed mechanism that involves proton transfer from the conserved Tyr residue to the C-18 position of Pchlide (8) (Fig. 1B). The close proximity of the Lys residue is thought to allow the deprotonation step to occur at physiological pH by lowering the apparent pKa of the phenolic group of the Tyr (8). However, confirmation of the exact role of these conserved residues has been compromised by the limited levels of activity observed in previous studies of the variant enzymes (8, 32, 33), and a detailed evaluation of the role of the active site Tyr and Lys residues on the chemical steps (i.e. hydride and proton transfer) has not been reported. We address this deficiency in the current work by analyzing a number of site-specific mutant forms altered at Tyr-193 and Lys-197 in a thermophilic POR from Thermosynechococcus elongatus BP-1. This was achieved using steady-state (multiple turnover) and laser photoexcitation (single turnover) methods and by trapping transient reaction intermediates by fluorescence spectroscopy performed at cryogenic temperatures.
EXPERIMENTAL PROCEDURES
Sample Preparation
All chemicals were obtained from Sigma, except NADPH (Melford Laboratories). Recombinant POR from the thermophilic cyanobacteria T. elongatus BP-1 was overexpressed in Escherichia coli and purified as described previously (11). Site-directed mutagenesis of the por gene was performed using the Phusion kit (New England Biolabs) to mutate Tyr-193 to a Phe, Ser, or Ala residue and to mutate Lys-197 to an Arg, Gln, or Ala residue. The following primers (Eurofins MWG Operon) were used to create the Tyr-193 variants: 5′-AAGTCGGGCAAGGCCTTC (for Y193F, or TCC for Y193S, and CCG for Y193A) AAAGACAGCAAGCTC-3′ (forward primer); 5′-GAGCTTGCTGTCTTTGAA (for Y193F, or GGA for Y193S, and GGC for Y193A) GGCCTTGCCCGACTT-3′ (reverse primer). The following primers were used to create the Lys-197 variants: 5′-GCCTACAAAGACAGCAGG (for K197R, or CAG for K197Q, and GCG for K197A) CTCTGCAATATGCTG-3′ (forward primer); and 5′-CAGCATATTGCAGAGCCT (for K197R, or CTG for K197Q, and CGC for K197A) GCTGTCTTTGTAGGC-3′ (reverse primer). The correct mutations were confirmed by DNA sequencing (Eurofins MWG Operon), and variant protein was produced as described previously (11). NMR measurements (details are provided in the legend to supplemental Fig. S1) were used to verify that the fold of the protein had not been compromised by the mutations (supplemental Fig. S1). The Pchlide pigment was produced and purified as described previously (11).
Multiple Turnover (Steady-state) Kinetics and Substrate Binding Measurements
Steady-state activity measurements were carried out as described previously (34) using a Cary 50 spectrophotometer (Varian). The binding of the NADPH coenzyme was monitored using fluorescence resonance energy transfer in a Cary Eclipse fluorimeter (Varian), and the binding of Pchlide was measured by following the red shift in absorbance at 642 nm, essentially as described (9, 35). The quantum efficiency for the reaction was calculated as described (9).
Laser Photoexcitation to Access Hydride and Proton Transfer
Rate constants for both the hydride and proton transfer reactions were measured using laser photoexcitation of dark assembled enzyme-NADPH-Pchlide ternary complexes. Samples were excited at 450 nm, using an optical parametric oscillator of a Q-switched Nd:YAG laser (Brilliant B, Quantel) in a 1-cm path length cuvette as described previously (13).
Low Temperature Fluorescence Studies
Low temperature fluorescence spectra of POR bound initially to NADPH and Pchilde were measured in 44% glycerol and 20% sucrose containing 50 mm Tris-HCl, pH 7.5, 0.1% Genapol, and 0.1% 2-mercaptoethanol using a Cary Eclipse fluorimeter. The excitation light was provided from a xenon arc light source at 450 nm. Excitation and emission slit widths were 3 nm. 1-ml samples were maintained in an Optistat DN liquid nitrogen cryostat at the desired temperature (Oxford Instruments, Inc.).
RESULTS
Initial Characterization of Tyr-193 and Lys-197 Enzymes
The catalytic activity of wild type POR and the active site Tyr and Lys variants were determined initially under steady-state conditions (Table 1). All of the variants showed a significant reduction in activity compared with the wild type enzyme. The Lys variants were more impaired (typically <4% wild type activity) compared with the Tyr variants.
TABLE 1.
Steady-state kinetic and substrate binding parameters for wild type and mutant forms of POR
All measurements were performed at 25 °C as described under “Experimental Procedures.”
| Enzyme | kcat | Relative activity |
Kd |
|
|---|---|---|---|---|
| NADPH | Pchlide | |||
| s−1 | % | μm | ||
| Wild type | 0.164 ± 0.002 | 100.0 ± 1.2 | 0.021 ± 0.009 | 5.6 ± 0.6 |
| Y193F | 0.032 ± 0.003 | 19.5 ± 1.8 | 0.022 ± 0.003 | 18.9 ± 1.4 |
| Y193A | 0.009 ± 0.001 | 5.5 ± 0.6 | 0.025 ± 0.005 | 254 ± 18.9 |
| Y193S | 0.004 ± 0.0002 | 2.4 ± 0.1 | 0.021 ± 0.002 | >250 |
| K197A | 0.004 ± 0.0001 | 2.7 ± 0.1 | 0.025 ± 0.001 | >250 |
| K197R | 0.004 ± 0.0001 | 2.3 ± 0.1 | 0.026 ± 0.003 | >250 |
| K197Q | 0.003 ± 0.0003 | 2.0 ± 0.1 | 0.021 ± 0.002 | >250 |
To provide a more in-depth rationale for this reduction in catalytic activity, we measured the ability of each variant form to bind the substrates NADPH and Pchlide. The dissociation constant for NADPH was determined by measuring the fluorescence resonance energy transfer signal from Trp residue(s) in the protein to the bound NADPH coenzyme (supplemental Fig. S2) (35). The apparent dissociation constant (Kd) for each of the variant enzymes was similar to that of the wild type POR (0.021 ± 0.009 μm; Table 1). The binding of Pchlide to the POR-NADPH complex was monitored by measuring the red shift in the absorbance maximum of the pigment (supplemental Fig. S3) (9, 35).4 It was only possible to determine accurate Kd values for the Y193F and Y193A enzymes (the binding constant for the remaining mutants is >250 μm; Table 1). In all cases, ternary complex formation is significantly affected (weaker binding) for all variant enzymes compared with wild type POR, indicating that both the Tyr and Lys residues are important determinants for Pchlide binding.
Mutagenesis of Tyr-193 and Lys-197 Leads to Impaired Photochemistry in the POR-NADPH-Pchlide Complex
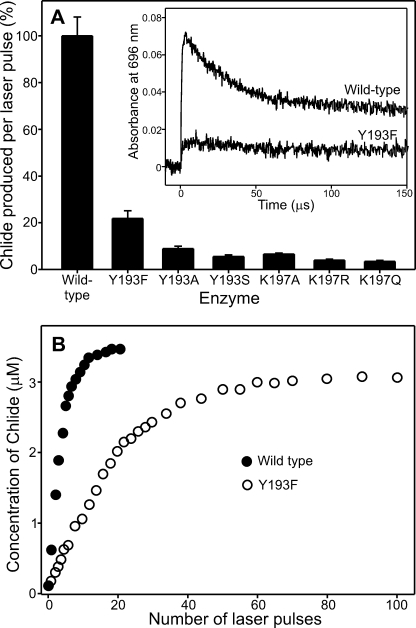
We have shown recently that the catalytic reaction of POR can be triggered by using a 6-ns laser pulse tuned to the Soret region of the Pchlide absorbance spectrum, allowing the rates of both the hydride and proton transfer steps to be determined (13). We have used similar laser photoexcitation measurements to assess the effects on hydride and proton transfer with all of the variant enzymes (Fig. 2A). To ensure that similar levels of the ternary enzyme-substrate complex were used in the experiments, it was necessary to use higher concentrations of enzyme for the mutant enzymes. The efficiency of the photochemistry is dramatically impaired for all of the variant POR enzymes compared with the wild type, as is evident from the considerable reduction in the chlorophyllide production per laser pulse (Fig. 2A). As a result, it was not possible to measure accurate rates for the sequential hydride and proton transfer steps for the variant enzymes, although visual inspection of the traces suggests that the rate of proton transfer is significantly reduced (Fig. 2A, inset). Consequently, to provide further evidence for the impaired photochemistry in the variant enzymes, we monitored the amount of chlorophyllide produced as a function of the number of laser pulses for the most active variant YI93F (Fig. 2B). By taking into account the respective Kd values for Pchlide bound to the wild type and Y193F complexes, we ensured that an identical starting concentration of ternary complex was used in these experiments. Although the amount of chlorophyllide produced in the wild type enzyme saturates after ∼20 laser pulses, it was necessary to use at least 100 laser pulses to produce a comparable level of the chlorophyllide product for the Y193F variant.
FIGURE 2.
Impaired photochemical efficiency of the mutant enzymes. A, the amount of chlorophyllide (Chlide) produced upon photoexcitation with a 6-ns laser pulse at 25 °C for the wild type and mutant enzymes. Data were collected using an equivalent concentration of ternary enzyme substrate complex for each enzyme (determined from the Kd values in Table 1), and errors were calculated from the average of 10 laser pulses. The inset shows a typical kinetic absorption trace measured at 696 nm over 150 μs for wild type and Y193F POR. B, the amount of chlorophyllide produced as a function of the number of laser pulses for the wild type and Y193F enzymes. To ensure an identical starting concentration of ternary complex, wild type POR was used at 50 μm and Y193F was used at 92.7 μm enzyme (based on the Kd for Pchlide in Table 1).
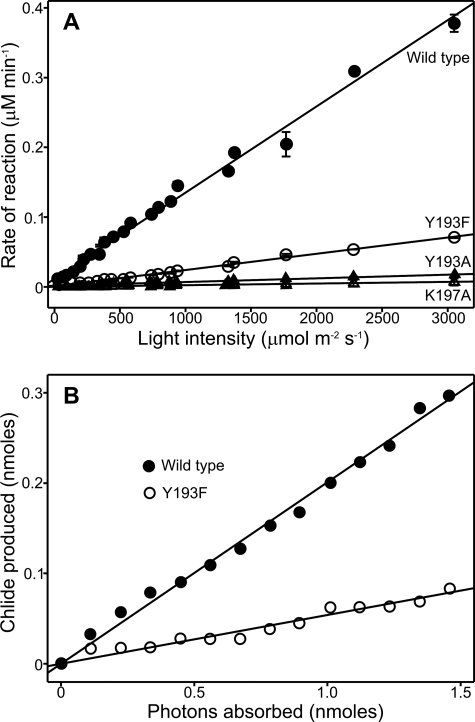
We studied the activation by light in more detail by measuring steady-state activities over a range of light intensities from 20 to 3000 μm photons s−1 m−2 for the wild type, Y193F, Y193A, and K197A enzymes (Fig. 3A). In all cases, the steady-state rate of reaction increases in a linear manner at higher light intensities. However, the wild type enzyme is more sensitive to changes in light intensity than any of the mutant enzymes, providing further evidence that the photochemistry is significantly impaired in the variant forms. Moreover, the quantum efficiency for photoreduction of Pchlide to chlorophyllide (Fig. 3B) is much lower for Y193F (5.1 ± 0.3%) compared with the wild type enzyme (19.7 ± 0.5%).
FIGURE 3.
Light intensity dependence of Pchlide reduction for Tyr and Lys mutants. A, the rate of chlorophyllide (Chlide) formation was measured under steady-state conditions using 1 μm of each enzyme in the presence of 10 μm Pchlide and 20 μm NADPH. Rates were measured at a range of light intensities from 0 to 3200 μmol/s−1 m−2. The error bars were calculated from the average of at least three traces. B, the quantum yield for the photoreduction of Pchlide was determined using 2.34 μm wild type POR (19.7 ± 0.5%) and 3.6 μm Y193F (5.1 ± 0.3%) in the presence of 19.4 μm Pchlide and 250 μm NADPH.
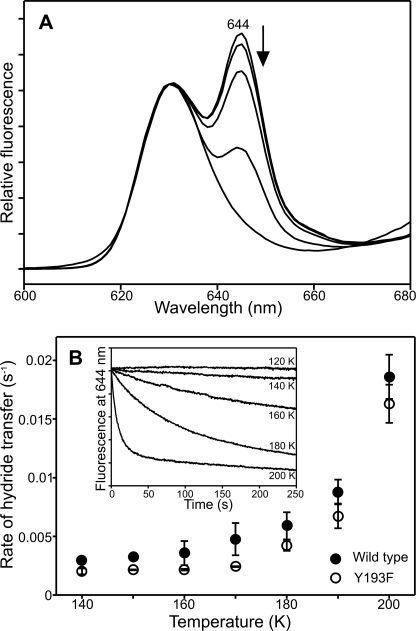
Low Temperature Fluorescence Studies of the Hydride Transfer Step
As the quantum yield of the hydride transfer step is significantly reduced in the variant enzymes, it is not possible to kinetically observe the chemical steps following a single laser pulse at room temperature. To negate this problem and to analyze the hydride and proton steps in more detail, we used low temperature fluorescence measurements to follow the formation of the catalytic intermediates under continuous illumination conditions. It has been shown previously that hydride transfer involves the formation of a nonfluorescent charge transfer species and that the rate of this step can be measured at cryogenic temperatures by monitoring the decrease in the fluorescence band for the ternary enzyme-substrate complex at 644 nm (9). We conducted similar measurements to determine the rate of hydride transfer for the Y193F variant over a range of temperatures from 120 K to 200 K, using the fluorescence excitation source to simultaneously trigger catalysis and monitor fluorescence. Our studies show that, under continuous illumination conditions, a similar level of the nonfluorescent intermediate is formed in the Y193F variant compared with the wild type enzyme (Fig. 4A). There is also no significant difference in the rate of hydride transfer for Y193F compared with wild type POR at cryogenic temperatures (Fig. 4B).
FIGURE 4.
Low temperature fluorescence measurements of the hydride transfer reaction. A, 77 K fluorescence emission spectra of 1.5 μm Pchlide and 250 μm NADPH in the presence of 57.6 μm Y193F (or 20 μm for wild type POR) was measured after illumination at increasing temperatures for 10 min. The arrow indicates the disappearance of the ternary enzyme-substrate peak at 644 nm. B, the rate of hydride transfer was measured over a range of temperatures for the wild type and Y193F enzymes. The inset shows typical kinetic traces that were obtained at each temperature.
Low Temperature Fluorescence Studies of Proton Transfer Step
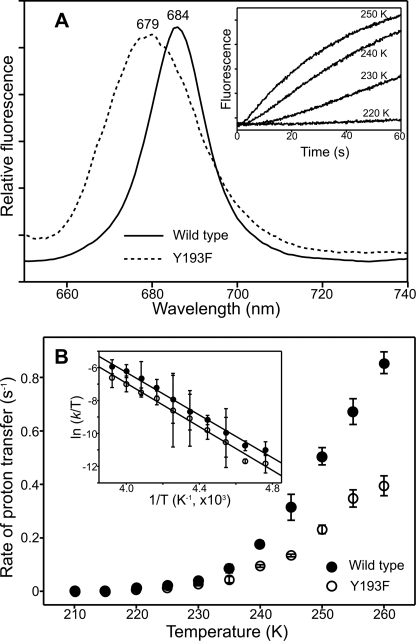
A similar approach has been used to determine the effect of the mutations on the proton transfer step. Low temperature fluorescence measurements were used to monitor the conversion of the nonfluorescent intermediate to the POR-chlorophyllide-NADP+ species that is formed during the first dark step. This intermediate has a fluorescence peak at 684 nm in the wild type enzyme but becomes slightly blue-shifted in the Y193F variant with a fluorescence maximum at 679 nm (Fig. 5A) and a relative fluorescence intensity that is much lower than that for the wild type enzyme. In addition, the rate of the proton transfer step is significantly reduced in the Y193F variant compared with wild type POR when measurements are repeated over a range of temperatures from 200 K to 260 K (Fig. 5B). The enthalpy of activation, ΔH‡, for the proton transfer step, which can be calculated from the Eyring plot, is 58.6 ± 1.4 kJ mol−1 for wild type POR and 58.2 ± 1.8 kJ mol−1 for Y193F; the entropy of activation, ΔS‡, is −14.1 ± 0.5 for wild type POR and 21.2 ± 1.2 J mol−1 K−1 for Y193F.
FIGURE 5.
Low temperature fluorescence measurements of the proton transfer reaction. A, 77 K fluorescence emission spectra of 1.5 μm Pchlide and 250 μm NADPH in the presence of 57.6 μm Y193F (or 20 μm for wild type POR) was measured after illumination at 240 K for 10 min. The inset shows typical kinetic traces that were obtained at each temperature. B, the rate of hydride transfer was measured over a range of temperatures for the wild type and Y193F enzymes. The inset shows an Eyring plot of ln(kobs/T) versus 1/T for the proton transfer step. The data shown are fitted to the Eyring equation to calculate activation enthalpies, ΔH‡, and activation entropies, ΔS‡, for the reaction. All error bars were calculated from the average of at least three traces.
DISCUSSION
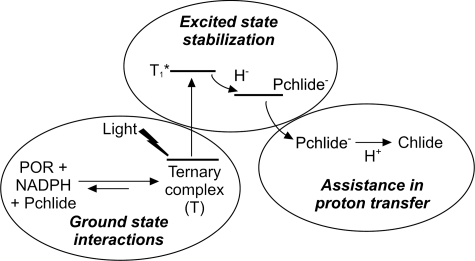
The light-driven enzyme POR catalyzes a key reaction in chlorophyll biosynthesis and is an important model system for studying the mechanism of catalysis of the large SDR superfamily of enzymes. Two conserved active site residues, a Tyr and a Lys, are known to be essential for activity and are proposed to be involved in proton transfer in the SDR family of enzymes (28–31). Although previous work has illustrated the importance of these residues for POR activity, a lack of any enzyme activity prevented a detailed demonstration of their exact role in catalysis (8, 32). We have analyzed a number of site-directed variants of the conserved Tyr-193 and Lys-197 residues in a thermophilic POR using a variety of kinetic and spectroscopic methods to probe key aspects of the catalytic cycle. Although our studies confirmed the importance of these two residues in catalysis, all of the variants retained some activity. We have shown that both residues have multiple roles in catalysis and participate in ground state and excited state processes in the overall reaction, which are summarized in Fig. 6.
FIGURE 6.
Schematic model for the role of the active site Tyr and Lys residues in the photoreduction of Pchlide by POR. Chlide, chlorophyllide.
Our data has illustrated the importance of Tyr-193 and Lys-197 in formation of the photoactive ternary enzyme-substrate complex. However, it appears that binding of the NADPH coenzyme is relatively unaffected by mutations to either residue, and it is therefore likely that the majority of the binding interactions with NADPH are provided by the conserved nucleotide binding domain, which is located near the N-terminal region of the protein (27, 32). Previous studies have suggested that both residues may interact with the ribose hydroxyl group of the NADPH molecule (32), and it may be the case that such an interaction is necessary to position the nucleotide cofactor appropriately in the active site to enable efficient operation of the photochemistry. Conversely, the binding of Pchlide substrate is significantly compromised by mutations to the conserved Tyr and Lys residues. Mutation of Lys-197 has a more dramatic effect on Pchlide binding compared with the situation following mutation of Tyr-193. This supports the notion that Lys-197 provides key interactions with the carboxyl group on the side chain attached to the C-17 position of the pigment molecule (32). It appears that both residues have a key role in forming a productive ternary complex and are responsible for maintaining effective interactions between Pchlide, NADPH, and protein within the active site.
Under conditions of identical ternary complex concentration, we have shown that excited state processes in the enzyme-bound Pchlide species are also affected by mutations to the Tyr or Lys residues. A significant decrease in the quantum yield of the reaction is observed for the mutants, although it appears that the rate of the hydride transfer reaction from the excited state is unaffected. Therefore, it is likely that these conserved residues are involved in the stabilization of the photoexcited state, thus increasing the lifetime of the excited state species and allowing the hydride transfer reaction to proceed more efficiently. The involvement of phenol groups in excited state and charge transfer processes has been reported previously in other systems (36, 37), and such a role may also be important in POR to ensure a high quantum yield for the hydride transfer reaction. It has also recently been suggested that free Pchlide can form a long lived triplet state upon photoexcitation, which is quenched by specific chromophore-protein interactions in the enzyme-substrate complex (19). Hence, it is possible that the Tyr and Lys residues are involved in quenching the triplet state, thus ensuring that this nonproductive pathway is not populated during the reaction. Alternatively, the OH group of the Tyr residue may interact with the Pchlide excited state, and removal of this group in the mutant enzymes may lead to a more rapid relaxation to the ground state, thereby reducing the lifetime of the excited state species.
It has been suggested that the conserved Tyr residue is responsible for transferring a proton to the C-18 position of the Pchlide molecule and that the Lys residue is required to lower the relative pKa of the Tyr hydroxyl group. We have shown that substitution of the Tyr residue with other residues is not sufficient to prevent proton transfer. Also, our cryogenic fluorescence spectroscopy studies have shown that the fluorescence peak of the intermediate formed after proton transfer is considerably blue-shifted compared with that formed with wild type enzyme. This likely reflects changes in the active site geometry of the ternary enzyme-product complex, as a consequence of the loss of interaction between chlorophyllide product and Tyr-193. We have also shown that the rate of the proton transfer reaction decreases significantly in the variant enzymes. Consequently, whereas Tyr-193 remains the most likely putative proton donor, the variant enzymes are still able to satisfy the valence of the C-18 atom of Pchlide by using a surrogate proton source, as is often the case when key proton donors are removed from an enzyme active site by site-directed mutagenesis (38, 39). The most likely source of the proton is solvent, although we cannot rule out other conserved residues that might be sufficiently close to the pigment molecule to enable proton transfer.
In summary, we have used a variety of spectroscopic and kinetic techniques to analyze the role of the conserved active site Tyr and Lys residues in the catalytic mechanism of the light-driven enzyme POR. We have shown that a single mutation of either residue has a dramatic effect on the reaction but is not sufficient to prevent the formation of chlorophyllide product. We find that both residues have multiple roles in catalysis, involving formation of the ground state ternary enzyme-substrate complex, stabilization of a Pchlide excited state species, and proton transfer to the reaction intermediate formed after the light reaction (hydride transfer; Fig. 6). Whether these residues have multiple roles in other members of the SDR family remains to be seen. In the absence of a crystal structure for POR, our studies are consistent with the reported homology model of the enzyme. Our studies also provide new insight into the role of these residues in facilitating both the hydride (excited state) and proton (ground state) transfer reactions through stabilization of both the excited and ground electronic states of the POR ternary complex.
Supplementary Material
This work was supported by the Biotechnology and Biological Sciences Research Council, Swindon, United Kingdom.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
4 The sequential binding mechanism dictates that Pchlide does not bind to POR in the absence of NADPH (34).
- POR
- NADPH-protochlorophyllide oxidoreductase
- Pchlide
- protochlorophyllide
- SDR
- short-chain dehydrogenase/reductase.
REFERENCES
- 1.Heyes D. J., Hunter C. N. ( 2005) Trends Biochem. Sci. 30, 642– 649 [DOI] [PubMed] [Google Scholar]
- 2.Lebedev N., Timko M. P. ( 1998) Photosynth. Res. 58, 5– 23 [DOI] [PubMed] [Google Scholar]
- 3.Masuda T., Takamiya K. ( 2004) Photosynth. Res. 81, 1– 29 [DOI] [PubMed] [Google Scholar]
- 4.Bröcker M. J., Virus S., Ganskow S., Heathcote P., Heinz D. W., Schubert W. D., Jahn D., Moser J. ( 2008) J. Biol. Chem. 283, 10559– 10567 [DOI] [PubMed] [Google Scholar]
- 5.Mees A., Klar T., Gnau P., Hennecke U., Eker A. P., Carell T., Essen L. O. ( 2004) Science 306, 1789– 1793 [DOI] [PubMed] [Google Scholar]
- 6.Valera V., Fung M., Wessler A. N., Richards W. R. ( 1987) Biochem. Biophys. Res. Commun. 148, 515– 520 [DOI] [PubMed] [Google Scholar]
- 7.Begley T. P., Young H. ( 1989) J. Am. Chem. Soc. 111, 3095– 3096 [Google Scholar]
- 8.Wilks H. M., Timko M. P. ( 1995) Proc. Natl. Acad. Sci. U.S.A. 92, 724– 728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heyes D. J., Ruban A. V., Wilks H. M., Hunter C. N. ( 2002) Proc. Natl. Acad. Sci. U.S.A. 99, 11145– 11150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heyes D. J., Ruban A. V., Hunter C. N. ( 2003) Biochemistry 42, 523– 528 [DOI] [PubMed] [Google Scholar]
- 11.Heyes D. J., Hunter C. N. ( 2004) Biochemistry 43, 8265– 8271 [DOI] [PubMed] [Google Scholar]
- 12.Heyes D. J., Heathcote P., Rigby S. E., Palacios M. A., van Grondelle R., Hunter C. N. ( 2006) J. Biol. Chem. 281, 26847– 26853 [DOI] [PubMed] [Google Scholar]
- 13.Heyes D. J., Sakuma M., de Visser S. P., Scrutton N. S. ( 2009) J. Biol. Chem. 284, 3762– 3767 [DOI] [PubMed] [Google Scholar]
- 14.Heyes D. J., Sakuma M., Scrutton N. S. ( 2007) J. Biol. Chem. 282, 32015– 32020 [DOI] [PubMed] [Google Scholar]
- 15.Heyes D. J., Hunter C. N., van Stokkum I. H., van Grondelle R., Groot M. L. ( 2003) Nat. Struct. Biol. 10, 491– 492 [DOI] [PubMed] [Google Scholar]
- 16.Dietzek B., Kiefer W., Hermann G., Popp J., Schmitt M. ( 2006) J. Phys. Chem. B 110, 4399– 4406 [DOI] [PubMed] [Google Scholar]
- 17.Zhao G. J., Han K. L. ( 2008) Biophys. J. 94, 38– 46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sytina O. A., Heyes D. J., Hunter C. N., Alexandre M. T., van, Stokkum I. H., van, Grondelle R., Groot M. L. ( 2008) Nature 456, 1001– 1004 [DOI] [PubMed] [Google Scholar]
- 19.Dietzek B., Tschierlei S., Hermann G., Yartsev A., Pascher T., Sundström V., Schmitt M., Popp J. ( 2009) Chemphyschem. 10, 144– 150 [DOI] [PubMed] [Google Scholar]
- 20.Oppermann U., Filling C., Hult M., Shafqat N., Wu X., Lindh M., Shafqat J., Nordling E., Kallberg Y., Persson B., Jörnvall H. ( 2003) Chem. Biol. Interact. 143–144, 247– 253 [DOI] [PubMed] [Google Scholar]
- 21.Hammes-Schiffer S. ( 2002) Biochemistry 41, 13335– 13343 [DOI] [PubMed] [Google Scholar]
- 22.Benkovic S. J., Hammes-Schiffer S. ( 2003) Science 301, 1196– 1202 [DOI] [PubMed] [Google Scholar]
- 23.Boehr D. D., McElheny D., Dyson H. J., Wright P. E. ( 2006) Science 313, 1638– 1642 [DOI] [PubMed] [Google Scholar]
- 24.Kohen A., Cannio R., Bartolucci S., Klinman J. P. ( 1999) Nature 399, 496– 499 [DOI] [PubMed] [Google Scholar]
- 25.Kohen A., Klinman J. P. ( 2000) J. Am. Chem. Soc. 122, 10738– 10739 [Google Scholar]
- 26.Sutcliffe M. J., Scrutton N. S. ( 2000) Trends Biochem. Sci. 25, 405– 408 [DOI] [PubMed] [Google Scholar]
- 27.Townley H. E., Sessions R. B., Clarke A. R., Dafforn T. R., Griffiths W. T. ( 2001) Proteins 44, 329– 335 [DOI] [PubMed] [Google Scholar]
- 28.Ensor C. M., Tai H. H. ( 1991) Biochem. Biophys. Res. Commun. 176, 840– 845 [DOI] [PubMed] [Google Scholar]
- 29.Obeid J., White P. C. ( 1992) Biochem. Biophys. Res. Commun. 188, 222– 227 [DOI] [PubMed] [Google Scholar]
- 30.Chen Z., Jiang J. C., Lin Z. G., Lee W. R., Baker M. E., Chang S. H. ( 1993) Biochemistry 32, 3342– 3346 [DOI] [PubMed] [Google Scholar]
- 31.Varughese K. I., Xuong N. H., Kiefer P. M., Matthews D. A., Whiteley J. M. ( 1994) Proc. Natl. Acad. Sci. U.S.A. 91, 5582– 5586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lebedev N., Karginova O., McIvor W., Timko M. P. ( 2001) Biochemistry 40, 12562– 12574 [DOI] [PubMed] [Google Scholar]
- 33.Heyes D. J., Hunter C. N. ( 2002) Biochem. Soc. Trans. 30, 601– 604 [DOI] [PubMed] [Google Scholar]
- 34.Heyes D. J., Menon B. R., Sakuma M., Scrutton N. S. ( 2008) Biochemistry 47, 10991– 10998 [DOI] [PubMed] [Google Scholar]
- 35.McFarlane M. J., Hunter C. N., Heyes D. J. ( 2005) Photochem. Photobiol. Sci. 4, 1055– 1059 [DOI] [PubMed] [Google Scholar]
- 36.Henneke C. M., Wedding R. T. ( 1975) Arch. Biochem. Biophys. 168, 436– 442 [DOI] [PubMed] [Google Scholar]
- 37.Jiménez M. C., Miranda M. A., Tormos R. ( 2005) Chem. Soc. Rev. 34, 783– 796 [DOI] [PubMed] [Google Scholar]
- 38.Deonarain M. P., Berry A., Scrutton N. S., Perham R. N. ( 1989) Biochemistry 28, 9602– 9607 [DOI] [PubMed] [Google Scholar]
- 39.Zheng Y. J., Ornstein R. L. ( 1997) J. Am. Chem. Soc. 119, 1523– 1528 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.