Abstract
The release of cytochrome c from mitochondria, which leads to activation of the intrinsic apoptotic pathway, is regulated by interactions of Bax and Bak with antiapoptotic Bcl-2 family members. The factors that regulate these interactions are, at the present time, incompletely understood. Recent studies showing preferences in binding between synthetic Bcl-2 homology domain 3 and antiapoptotic Bcl-2 family members in vitro have suggested that the antiapoptotic proteins Mcl-1 and Bcl-xL, but not Bcl-2, restrain proapoptotic Bak from inducing mitochondrial membrane permeabilization and apoptosis. Here we show that Bak protein has a much higher affinity than the 26-amino acid Bak Bcl-2 homology domain 3 for Bcl-2, that some naturally occurring Bcl-2 allelic variants have an affinity for full-length Bak that is only 3-fold lower than that of Mcl-1, and that endogenous levels of these Bcl-2 variants (which are as much as 40-fold more abundant than Mcl-1) restrain part of the Bak in intact lymphoid cells. In addition, we demonstrate that Bcl-2 variants can, depending on their affinity for Bak, substitute for Mcl-1 in protecting cells. Thus, the ability of Bcl-2 to protect cells from activated Bak depends on two important contextual variables, the identity of the Bcl-2 present and the amount expressed.
The release of cytochrome c from mitochondria, which leads to activation of the intrinsic apoptotic pathway, is regulated by Bcl-2 family members (1–5). This group of proteins consists of three subgroups: Bax and Bak, which oligomerize upon death stimulation to form a putative pore in the outer mitochondrial membrane, thereby allowing efflux of cytochrome c and other mitochondrial intermembrane space components; Bcl-2, Bcl-xL, Mcl-1, and other antiapoptotic homologs, which antagonize the effects of Bax and Bak; and BH3-only proteins2 such as Bim, Bid, and Puma, which are proapoptotic Bcl-2 family members that share only limited homology with the other two groups in a single 15-amino acid domain (the BH3 domain, see Ref. 6). Although it is clear that BH3-only proteins serve as molecular sensors of various stresses and, when activated, trigger apoptosis (3, 6–11), the mechanism by which they do so remains incompletely understood. One current model suggests that BH3-only proteins trigger apoptosis solely by binding and neutralizing antiapoptotic Bcl-2 family members, thereby causing them to release the activated Bax and Bak that are bound (reviewed in Refs. 9 and 10; see also Refs. 12 and 13), whereas another current model suggests that certain BH3-only proteins also directly bind to and activate Bax (reviewed in Ref. 3; see also Refs. 14–17). Whichever model turns out to be correct, both models agree that certain antiapoptotic Bcl-2 family members can inhibit apoptosis, at least in part, by binding and neutralizing activated Bax and Bak before they permeabilize the outer mitochondrial membrane (13, 18, 19).
Much of the information about the interactions between pro- and antiapoptotic Bcl-2 family members has been derived from the study of synthetic peptides corresponding to BH3 domains. In particular, these synthetic peptides have been utilized as surrogates for the full-length proapoptotic proteins during structure determinations (20–22) as well as in functional studies exploring the effect of purified BH3 domains on isolated mitochondria (14, 23) and on Bax-mediated permeabilization of lipid vesicles (15).
Recent studies using these same peptides have suggested that interactions of the BH3 domains of Bax, Bak, and the BH3-only proteins with the “BH3 receptors” of the antiapoptotic Bcl-2 family members are not all equivalent. Surface plasmon resonance, a technique that is widely used to examine the interactions of biomolecules under cell-free conditions (24–26), has demonstrated that synthetic BH3 peptides of some BH3-only family members show striking preferences, with the Bad BH3 peptide binding to Bcl-2 and Bcl-xL but not Mcl-1, and the Noxa BH3 peptide binding to Mcl-1 but not Bcl-2 or Bcl-xL (27). Likewise, the Bak BH3 peptide exhibits selectivity, with high affinity for Bcl-xL and Mcl-1 but not Bcl-2 (12). The latter results have led to a model in which Bcl-xL and Mcl-1 restrain Bak and inhibit Bak-dependent apoptosis, whereas Bcl-2 does not (10).
Because the Bak protein contains multiple recognizable domains in addition to its BH3 motif (28, 29), we compared the binding of Bak BH3 peptide and Bak protein to Bcl-2. Surface plasmon resonance demonstrated that Bcl-2 binds Bak protein with much higher affinity than the Bak 26-mer BH3 peptide. Further experiments demonstrated that the KD for Bak differs among naturally occurring Bcl-2 sequence variants but is only 3-fold higher than that of Mcl-1 in some cases. In light of previous reports that Bcl-2 overexpression contributes to neoplastic transformation (30–33) and drug resistance (34–36) in lymphoid cells, we also examined Bcl-2 expression and Bak binding in a panel of neoplastic lymphoid cell lines. Results of these experiments demonstrated that Bcl-2 expression varies among different lymphoid cell lines but is up to 40-fold more abundant than Mcl-1. In lymphoid cell lines with abundant Bcl-2, Bak is detected in Bcl-2 as well as Mcl-1 immunoprecipitates; and Bak-dependent apoptosis induced by Mcl-1 down-regulation can be prevented by Bcl-2 overexpression. Collectively, these observations shed new light on the role of Bcl-2 in binding and neutralizing Bak.
EXPERIMENTAL PROCEDURES
Materials
Reagents were obtained from the following suppliers: the broad spectrum caspase inhibitor Q-VD-OPhe from Biomol (Plymouth Meeting, MA); polysorbate 20 from Biacore AB (Uppsala, Sweden); CM5 biosensor chips from GE Healthcare; glutathione and dimethyl pimelimidate from Sigma; and APC-conjugated annexin V from Pharmingen. The 26-mer Bak peptide (12) was produced by solid phase synthesis in the Mayo Clinic Protein Chemistry Shared Resource. All other reagents were obtained as described previously (37, 38).
Antibodies for immunoblotting or flow cytometry were obtained from the following suppliers: murine monoclonal antibodies that recognize Mcl-1 or Bcl-2 from BD Biosciences; murine Ab-1 monoclonal antibody to active Bak from Calbiochem; murine monoclonal anti-Bcl-2 from Dako (Carpinteria, CA); goat anti-actin from Santa Cruz Biotechnology (Santa Cruz, CA); rabbit anti-Bak and anti-Bax from Millipore (Danvers, MA); rabbit anti-Puma from ProSci (San Diego, CA); and rabbit antibodies to Mcl-1, Bcl-xL, and Bim from Cell Signaling Technology (Beverly, MA). Antibodies to poly(ADP-ribose) polymerase and Hsp90 were kind gifts from Guy Poirier (Laval University, Ste.-Foy, Quebec) and David Toft (Mayo Clinic, Rochester, MN), respectively.
Protein Expression and Purification
cDNAs encoding Bcl-2, Bcl-xL, or Bax lacking the transmembrane domain were cloned in-frame with GST in pGEX-4T-1. cDNA encoding Mcl-1 lacking the transmembrane domain was cloned into pET29b(+) to yield an S peptide-tagged construct. Plasmid encoding BakΔTM in pET29b(+) (22) was a kind gift from Qian Liu and Kalle Gehring (McGill University, Montreal, Canada). Plasmid encoding Bak N85A/I86AΔTM was generated from BakΔTM using site-directed mutagenesis. All plasmids were subjected to automated sequencing to verify the described alteration and confirm that no additional mutations were present.
Plasmids were transformed into Rosetta cells (BaxΔTM, Bcl-2ΔTM, Bcl-xLΔTM, and Mcl-1ΔTM) or BL21 (BakΔTM and Bak N85AI86AΔTM) by heat shock. After cells were grown to an optical density of 0.6, isopropyl 1-thio-β-d-galactopyranoside was added to 1 mm, and incubation was continued for 24 h at 16 °C (Bcl-2ΔTM) or 4 h at 37 °C (Bcl-xLΔTM, Mcl-1ΔTM, BaxΔTM, BakΔTM, and Bak N85AI86AΔTM). Bacteria were then washed and sonicated intermittently on ice in calcium- and magnesium-free Dulbecco's phosphate-buffered saline (PBS) containing 1 mm PMSF (GST-tagged proteins and S peptide-Mcl-1ΔTM) or TS buffer (150 mm NaCl containing 10 mm Tris-HCl (pH 7.4) and 1 mm PMSF, His6-tagged BakΔTM, and Bak N85AI86AΔTM). All further steps were performed at 4 °C. After His6-tagged proteins were applied to Ni2+-nitrilotriacetic acid-agarose (Novagen, La Jolla, CA), columns were washed with 20 volumes of TS buffer followed by 10 volumes of TS buffer containing 40 mm imidazole and then eluted with TS buffer containing 200 mm imidazole. His6-BakΔTM and Bak N85AI86AΔTM were further purified by fast protein liquid chromatography on a Superdex S200 size exclusion column and concentrated by ultrafiltration.
S peptide-tagged Mcl-1ΔTM was incubated with S protein-agarose for 4 h at 4 °C. After beads were washed twice with 10 volumes of PBS containing 1 mm PMSF, bound polypeptide was eluted with PBS containing 3 m MgCl2. After GST-tagged proteins were incubated with GSH-agarose for 4 h at 4 °C, GSH-agarose resin was washed twice with 20–25 volumes of PBS and eluted with PBS containing 20 mm reduced glutathione for 30 min at 4 °C.
All eluted polypeptides were concentrated using Centricon YM-10 centrifugal concentrators (Millipore) and dialyzed against Biacore running buffer consisting of 10 mm HEPES (pH 7.4), 150 mm NaCl, 0.05 mm EDTA and 0.005% (w/v) Polysorbate 20. Polypeptides were stored at 4 °C for <48 h before use.
Affinity Measurements by Surface Plasmon Resonance
Measurements were performed at 25 °C on a Biacore 3000 biosensor (Biacore, Uppsala, Sweden). BakΔTM, Bak BH3 peptide, or GST-Bcl-2 was immobilized onto a CM5 sensorchip as instructed by the supplier. After washing with Biacore running buffer, Mcl-1ΔTM, GST/Bcl-2ΔTM, GST/Bcl-xLΔTM, or GST was injected at 30 μl/min for 1 min. Bound polypeptide was allowed to dissociate by injection of protein-free Biacore running buffer at 30 μl/min for 15 min. Residual bound proteins were desorbed with 150 mm NaCl containing 10 mm glycine (pH 2.0). Binding kinetics were derived from sensorgrams using BIAevaluation software (Biacore, Uppsala, Sweden). The binding of BakΔTM or Bak N85AI86AΔTM to immobilized GST-Bcl-2 was analyzed in the same fashion.
Cell Culture
Cell lines were obtained from the following sources: Jurkat (T cell acute lymphocytic leukemia) from Paul Leibson (Mayo Clinic, Rochester, MN); RL (B cell lymphoma) from Thomas Witzig (Mayo Clinic, Rochester, MN); Molt3 and CCFR-CEM (both T cell acute lymphocytic leukemia) from American Type Culture Collection (Manassas, VA); and Daudi (Burkitt B cell lymphoma) from Adele Fielding (Mayo Clinic). All lines were maintained at densities below 106 cells/ml in RPMI 1640 medium containing 10% heat-inactivated fetal bovine serum, 100 units/ml penicillin G, 100 μg/ml streptomycin, and 2 mm glutamine.
Quantitation of Bak Binding to Endogenous Antiapoptotic Bcl-2 Family Members
Log phase cells were washed with PBS, lysed in lysis buffer consisting of 1% (w/v) CHAPS, 20 mm HEPES (pH 7.4), 150 mm NaCl, 1% (v/v) glycerol, 1 mm PMSF, 10 μg/ml leupeptin, 10 μg/ml pepstatin, 100 mm NaF, 10 mm sodium pyrophosphate, 1 mm sodium vanadate, and 20 nm microcystin. After centrifugation at 14,000 × g for 15 min to sediment insoluble material, lysates were precleared by incubation for 1 h at 4 °C with protein A/G-agarose beads followed by sedimentation. Aliquots containing 200 μg of precleared extract were incubated for 1 h with anti-Bcl-2, anti-Mcl-1, or anti-Bcl-xL that was precoupled to protein A/G-agarose using dimethyl pimelimidate (39, 40). After sedimentation at 8000 × g for 2 min, beads were rapidly washed four times with wash buffer consisting of 1% CHAPS, 20 mm HEPES (pH 7.4), 150 mm NaCl, 1% (v/v) glycerol, 1 mm PMSF, 100 units/ml aprotinin, 100 mm NaF, 10 mm sodium pyrophosphate, and 1 mm sodium vanadate. Polypeptides bound to the beads were released by heating for 20 min at 65 °C in SDS sample buffer consisting of 4 m urea, 2% (w/v) SDS, 62.5 mm Tris-HCl (pH 6.8), 1 mm EDTA and 5% (v/v) 2-mercaptoethanol. Immunoprecipitated Bcl-2 family members and serial dilutions of precleared cell lysate were subjected to SDS-PAGE, transferred to nitrocellulose, and probed with antibodies to Bak, Bcl-2, Mcl-1, and Bcl-xL as indicated in the figure legends. Films from three independent experiments were scanned on a Hewlett Packard Scanjet 4C scanner and quantified using ImageJ software (//rsb.info.nih.gov).
siRNA
Short oligonucleotides targeting Bax (nucleotides 271–289, GenBankTM accession number NM_138761), Bak (nucleotides 913–931, GenBankTM accession number NM_001188), Bim (nucleotides 325–343, GenBankTM accession number NM_138621), and Puma (nucleotides 1222–1240, GenBankTM accession number NM_001127242) were from Dharmacon (Boulder, CO). On day 1 log phase Jurkat cells suspended in 400 μl of cytomix (41) containing 1000 nm final oligonucleotide concentration and plasmid encoding EBFP were subjected to electroporation using a BTX 830 square wave electroporator (BTX, San Diego, CA) delivering a single pulse at 280 mV for 10 ms. On day 3 cells were transfected with pCMS5A plasmids as described below. 48 h after transfection with empty pCMS5A vector or constructs to knock down Bcl-2, cells were reacted with APC-conjugated annexin V as described (42) and analyzed on an LSRII flow cytometer (BD Biosciences) using the following lasers and filters: EBFP, 407 nm laser, 450/50 filter; EGFP, 488 nm laser, 530/30 filter; APC, 633 nm laser, 660/20 filter. After collection of 40,000 events, APC-annexin V binding was determined on cells that were positive for both EGFP and EBFP.
Mammalian Expression Plasmids and Transfection
Plasmid encoding S peptide-tagged Bcl-2 was constructed by inserting cDNA containing nucleotides 4–720 of the human Bcl-2 open reading frame (GenBankTM accession number X06487) into the KpnI and EcoRI sites of pSPN (43). shRNA experiments were performed using the plasmids pCMS5A and pCMS5C. pCMS5A, which was derived from pCMS4.eGFP (44), contains an H1P promoter for shRNA silencing, a cytomegalovirus promoter for driving the expression of shRNA-resistant cDNAs, and an additional SV40 promoter for driving the expression of histone H2B fused to EGFP. This plasmid was modified to contain shRNA targeting nucleotides 907–925 of the Mcl-1 open reading frame or nucleotides 526–546 (Bcl-2 shRNA 1) or nucleotides 1–21 (Bcl-2 shRNA 2) of the Bcl-2 open reading frame. Rather than an empty multiple cloning site, rescue constructs contained the previously described shRNA-resistant Mcl-1 cDNA (45), full-length Bcl-xL, full-length Bcl-2 (variant 1) (46, 47), or full-length Bcl-2 mutated to TGGATGACAGAATATTTAAC at nucleotides 526–546 (underlined nucleotides represent silent mutations rendering cDNA resistant to Bcl-2 shRNA 1). Plasmids encoding different Bcl-2 variants were made by putting full-length Bcl-2 (variant 1, 2, or 3) behind the cytomegalovirus promoter of pCMS5A. Plasmid pCMS5C was derived from pCMS5A by site-directed mutagenesis. Two amino acid substitutions (Y66H and Y145F) were inserted to convert EGFP to EBFP. Once again, all plasmids were sequenced to verify the integrity of inserted shRNA and rescue constructs.
Log phase cells growing in antibiotic-free medium were transiently transfected with the indicated plasmid using a BTX 830 square wave electroporator delivering a single pulse at 280 mV for 10 ms. Cells were incubated for 48 h and analyzed for apoptosis using APC-coupled annexin V as described previously (42, 45). After 20,000 events (EGFP gating) or 40,000 events (EBFP and EGFP gating) were collected on a FACSCalibur (BD Biosciences) flow cytometer, data were analyzed by gating on EGFP-histone H2B+ cells (typically 60–70% of transfected Jurkat cells) or EGFP-histone H2B+/EBFP-histone H2B+ (typically 40–50% of transfected cells) and assessing APC-annexin V binding. Alternatively, after transfection, cells were incubated for 48 h and assayed for active Bak as described below.
Assays for Bak Activation
48 h after transfection with siRNA constructs as indicated in the individual figures, cells were stained with anti-active Bak Ab-1 and examined by flow cytometry as described previously (45). Alternatively, as described above for other immunoprecipitations, cells were washed with PBS and lysed in isotonic lysis buffer containing 1% CHAPS. After lysates were precleared, immunoprecipitations were performed for 1 h at 4 °C using aliquots containing 200 μg of lysate protein and 5 μg of anti-active Bak Ab-1 that was precoupled to protein A/G-agarose beads using dimethyl pimelimidate. Following four washes with isotonic wash buffer containing 1% CHAPS, bound polypeptides were solubilized in SDS sample buffer, subjected to SDS-PAGE, and probed with antibodies that recognize total Bak.
Immunoprecipitation
Log phase cells growing in antibiotic-free medium were transiently transfected with the indicated plasmid. 48 h after transfection, cells were washed and lysed in CHAPS lysis buffer. After centrifugation, 200 μg of precleared extract was incubated for 1 h with anti-Bcl-2, or anti-Mcl-1 that was precoupled to protein A/G-agarose (39, 40). After sedimentation at 8000 × g for 2 min, beads were rapidly washed four times with wash buffer. Polypeptides bound to the beads were released by heating for 20 min at 65 °C in SDS sample buffer. Immunoprecipitated Bcl-2 family members and one-fifth of the precleared cell lysate were subjected to SDS-PAGE, transferred to nitrocellulose, and probed with antibodies to Bak, Bax, Bim, Puma, Bcl-2, and Mcl-1 as indicated in the figure legends.
RESULTS
Bak Protein Binds to Bcl-2 More Tightly than to Bcl-xL
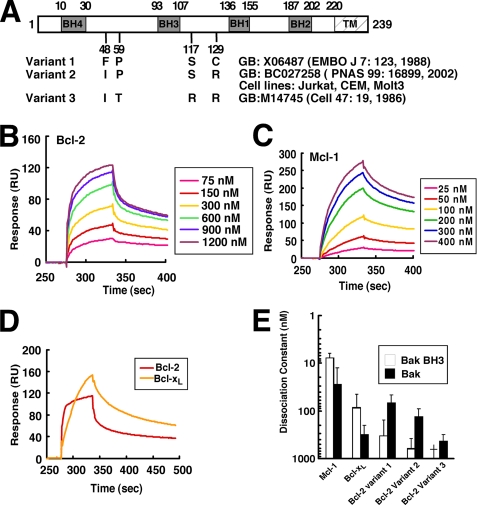
Models suggesting that Bcl-2 fails to bind Bak are based extensively on affinity measurements performed using a 26-mer peptide corresponding to the Bak BH3 domain. In this study, we immobilized this 26-mer peptide or Bak protein lacking the transmembrane domain (BakΔTM) and examined the binding of Bcl-2, as well as Mcl-1 and Bcl-xL, by surface plasmon resonance. Initial experiments utilized Bcl-2 variant 1 (Fig. 1 A, originally described in Ref. 46), a Bcl-2 sequence variant that has been widely used in the past to study Bcl-2 function.
FIGURE 1.
Affinities of Bak for different Bcl-2 variants, Mcl-1, and Bcl-xLin vitro. A, different Bcl-2 variants used in this study, with GenBankTM accession numbers and original citations. Amino acids that differ among the variants are shown. Our sequencing demonstrated that Jurkat, CEM, and Molt3 all express variant 2. B, surface plasmon resonance (relative units) observed when immobilized BakΔTM was exposed to different Bcl-2 (variant 1) concentrations. C, response of the same chip to different concentrations of Mcl-1. D, using the same chip as in B, responses of Bcl-2 (variant 1) and Bcl-xL at a saturating concentration of 2400 nm. E, based on the surface plasmon resonance assay, the affinities of Bak protein (B and C) and Bak BH3 peptide (Fig. S1, A–C) for different anti-apoptotic Bcl-2 family proteins were determined. Results are mean ± S.D. of three independent experiments using different chips and different protein batches. †, KD >1000 nm. GB, GenBankTM.
Consistent with previously reported results (12), binding of Mcl-1ΔTM (supplemental Fig. S1A) or Bcl-xLΔTM (supplemental Fig. 1B) to the Bak 26-mer peptide was readily detected. Further analysis demonstrated dissociation constants (KD values) of 8 and 80 nm, respectively (Fig. 1E). In contrast, Bcl-2ΔTM displayed a much lower affinity for this peptide (supplemental Fig. S1, C and D, and Fig. 1E), again confirming previous results.
A different picture emerged when binding to BakΔTM protein was analyzed using the same method. Binding of Bcl-2ΔTM was readily observed (Fig. 1B), with a KD of ∼70 nm (Fig. 1E, variant 1). This binding appeared qualitatively similar to that observed when Mcl-1ΔTM (Fig. 1C) or Bcl-xLΔTM (supplemental Fig. S2A) bound to BakΔTM. Indeed, the binding of BakΔTM to Bcl-2ΔTM was 2-fold tighter than the binding of BakΔTM to Bcl-xLΔTM (KD ∼200 nm; supplemental Fig. S2, A and B, and Fig. 1E). Direct comparison revealed that Bcl-2 bound Bak more rapidly (Fig. 1D) and more avidly (Fig. 1E) than Bcl-xL did.
Because of concern that these observations might differ from earlier studies as a result of sequence variation in Bcl-2 (48, 49), we examined not only Bcl-2 variant 1, but also the Bcl-2 variant endogenously expressed by many of our lymphoid cell lines (Fig. 1A, variant 2) and the Bcl-2 variant used in previous affinity measurements (variant 3, see Refs. 12, 13). Although the three Bcl-2 variants displayed differences in affinity for BakΔTM, in each case the Bcl-2 variant bound to Bak protein more tightly than to Bak 26-mer BH3 peptide (Fig. 1E). In addition, the Bcl-2 variant expressed in many of our cell lines (variant 2), like variant 1, bound Bak more tightly than Bcl-xL did (Fig. 1E). In contrast, variant 3 showed a 7-fold lower affinity for Bak.
Binding of Bak Mutant to Bcl-2
A previous study suggested that mutation of Ile-82 and Asn-83 in the BH3 domain of murine Bak to alanine abolishes binding to Bcl-xL and Mcl-1 (50). After the corresponding mutations were introduced into human Bak, purified wild type and mutant Bak constructs were assayed for their ability to bind Bcl-2 variant 2. As indicated in supplemental Fig. S3A, binding of the mutant BakΔTM polypeptide to Bcl-2 was readily detected, although the affinity was 2-fold lower than that of the wild type construct (supplemental Fig. S3, B and C).
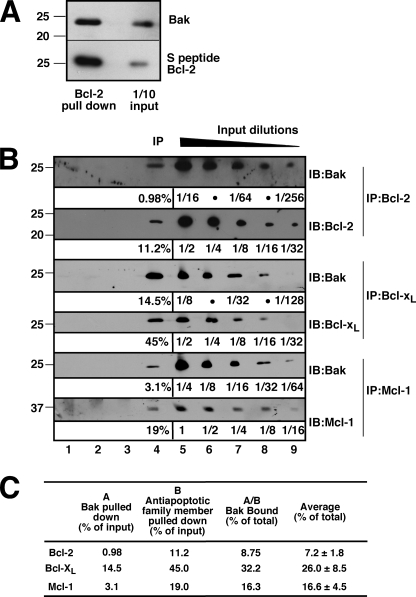
Recovery of Bak with Endogenous Bcl-2 from Jurkat Cells
Based on the preceding results, which suggest that Bak protein can bind to Bcl-2 under cell-free conditions, further experiments sought evidence that Bak and Bcl-2 interact in intact cells. When Jurkat cells transfected with S peptide-tagged Bcl-2 were lysed, ∼20% of the total cellular Bak was recovered in the Bcl-2 pulldowns (Fig. 2A). Because of concern that these results might result from tagging or overexpression of Bcl-2, further experiments examined the recovery of Bak when endogenous Bcl-2, Bcl-xL, and Mcl-1 were immunoprecipitated from lysates of untransfected cells. To provide an opportunity for quantitation, the amounts of antiapoptotic Bcl-2 family members as well as Bak in the immunoprecipitates were compared with serial dilutions of lysates on the same blots. This analysis (Fig. 2, B and C) demonstrated that, after correction for recovery of the antiapoptotic Bcl-2 family members, ∼7% of the total cellular Bak was recovered with endogenous Bcl-2, 26% with Bcl-xL and 17% with Mcl-1. As discussed below, we were able to account for ∼50% of the total cellular Bak in this manner, possibly reflecting the binding and neutralization of the remainder of the Bak by other polypeptides such as VDAC2 (51). Additional experiments indicated that recovery of Bak with endogenous Bcl-2 was similar after extensive versus limited washing of the immunoprecipitates and after immunoprecipitation for 12 h rather than 1 h.3
FIGURE 2.
Bak binds to Bcl-2 in pulldown and immunoprecipitation assays. A, 24 h after transient transfection with S peptide-tagged Bcl-2, cell lysates prepared in isotonic buffer containing 1% CHAPS (see “Experimental Procedures”) were reacted with S protein-agarose to recover Bcl-2 complexes. Pull downs and 1/10 of inputs were probed with Bak antibodies. B, Jurkat cell lysates prepared in 1% CHAPS were immunoprecipitated (IP) with Bcl-2, Bcl-xL, or Mcl-1 antibody (lane 4). Serial dilutions of lysates (lanes 5–9) were used for quantitation of Bak binding. Three different controls were examined at the same time: control antibody (isotype control, lane 1), beads + lysate without antibody (lane 2), and no lysate control (lane 3). IB, immunoblot. C, from the experiments shown in B, the percentages of Bak bound by Bcl-2, Bcl-xL, and Mcl-1 in Jurkat cells were calculated using ImageJ to quantify the amounts of Bak (column A) and anti-apoptotic Bcl-2 family member (column B) in the immunoprecipitates relative to a standard curve constructed from serial dilutions of lysates on the same blot. Results are mean ± S.D. of three independent experiments.
Bcl-2 Rescues Cells from Mcl-1 Knockdown
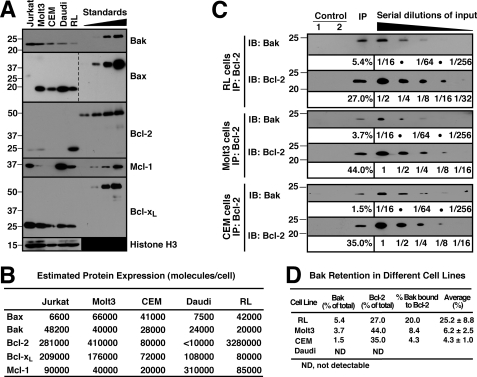
To assess the potential biological significance of the Bcl-2/Bak binding, further experiments examined the effect of Bcl-2 overexpression on Bak-dependent apoptosis. These studies utilized Jurkat cells, which express ∼7-fold more Bak than Bax (see Fig. 6, A and B). To avoid the use of pharmacological agents, which could activate multiple BH3-only family members, apoptosis was triggered by down-regulating Mcl-1, a change that has previously been reported to induce apoptosis in human leukemia cells (52) and in normal murine lymphohematopoietic cells (53). The externalization of phosphatidylserine, a hallmark feature of apoptotic cells (54, 55), was examined as an indicator of cellular response in these experiments.
FIGURE 6.
Bak binding to Bcl-2 varies among lymphoid cell lines. A, whole cell lysates from 3 × 105 of Jurkat, Molt3, CEM, Daudi, and RL cells were probed for Bak, Bax, Bcl-2, Mcl-1, and Bcl-xL. Purified GST-BakΔTM (0.1, 0.2, 0.4, and 0.8 ng), GST-BaxΔTM (0.3, 0.6, 1.2, and 2.4 ng), GST-Bcl-2ΔTM (15, 30, 60, and 120 ng), S peptide-tagged Mcl-1ΔTM (0.25, 0.5, 1.0, and 2.0 ng), and GST-Bcl-xLΔTM (0.6, 1.2, 2.4, and 4.8 ng) were used for immunoblot quantitation. Histone H3 was used as loading control. Additional blots of whole cell lysates are presented in supplemental Fig. S7. B, from the experiments shown in A and supplemental Fig. S7, the Bcl-2 family (Bak, Bax, Bcl-2, Mcl-1, and Bcl-xL) numbers per cell were calculated in Jurkat, Molt3, CEM, Daudi, and RL cells. C, CHAPS lysates from RL, Molt3, and CEM cells were immunoprecipitated (IP) with anti-Bcl-2 antibody and compared with serial dilutions of the input. Two different controls were used at the same time: beads + lysate without antibody (Control 1) and no lysate (Control 2). IB, immunoblot. D, from the experiments shown in C, the percentage of Bak bound by Bcl-2 was calculated in RL, Molt3, and CEM cells. Mean ± S.D. from three independent experiments is also shown. For Daudi, Bcl-2 is not detectable (N.D.) in either whole cell lysates (supplemental Fig. S7A) or the immunoprecipitates (data not shown).
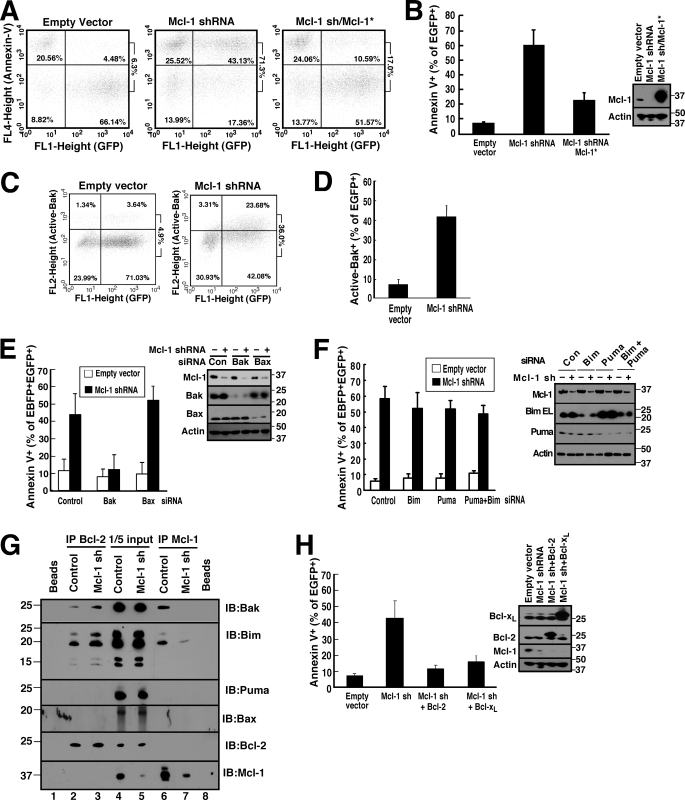
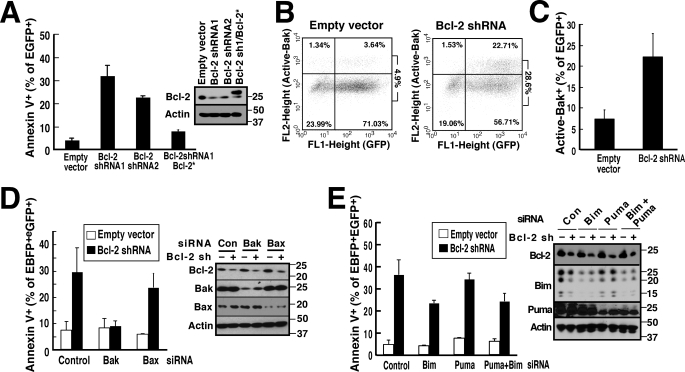
When Jurkat cells were transfected with Mcl-1 shRNA (45) and examined 48 h later, annexin V binding to phosphatidylserine was readily detected (Fig. 3, A and B). This apoptosis was accompanied by Bak activation as assessed by flow cytometry after staining with a previously described (56) conformation-sensitive Bak antibody (Fig. 3, C and D) or by immunoprecipitation with the same antibody (supplemental Fig. S4). Consistent with the predominant expression of Bak in this cell line, the Mcl-1 shRNA-induced apoptosis was prevented by Bak down-regulation but not by Bax down-regulation (Fig. 3E). This apoptosis was not diminished by down-regulation of the potent and promiscuous BH3-only proteins Bim and/or Puma (Fig. 3F) even though these same constructs inhibited apoptosis induced by various anticancer drugs,4 suggesting that the apoptosis is triggered by release of Bak rather than release of Bim and/or Puma. Consistent with this conclusion, we observed displacement of Bak upon Mcl-1 down-regulation (Fig. 3G). Bim was also displaced to Bcl-2 (Fig. 3G), although the shRNA result (Fig. 3F) did not suggest a critical role for this displacement. Further analysis demonstrated that expression of shRNA-resistant Mcl-1 (Mcl-1*, Fig. 3, A and B) or wild type Bcl-xL (Fig. 3H) abrogated the apoptosis resulting from Mcl-1 down-regulation. Importantly, expression of wild type Bcl-2 also rescued Jurkat cells from Mcl-1 shRNA-induced apoptosis (Fig. 3H), indicating that Bcl-2 can substitute for Mcl-1.
FIGURE 3.
Mcl-1 knockdown induces apoptosis in Jurkat cells that can be rescued by Bcl-2 or Bcl-xL overexpression. A and B, 48 h after transfection with empty vector (pCMS5A expressing EGFP-histone H2B), pCMS5A-Mcl-1-shRNA, or pCMS5A/Mcl-1-shRNA/Mcl-1* (shRNA-resistant Mcl-1), Jurkat cells were collected and stained with APC-annexin V. A representative experiment (A) and summarized results (B) indicating the percentage of EGFP+ cells stained with annexin V are indicated. Numbers at right of dot plots in this and subsequent figures are as follows: ratio of events in upper right panel to sum of lower right and upper right. Error bars in this and subsequent figures: mean ± S.D. of three independent experiments. Inset, whole cell lysates subjected to immunoblotting. C and D, 48 h after transfection with pCMS5A or Mcl-1 shRNA, Jurkat cells were collected, fixed, and stained with Ab-1, which recognizes active Bak. A representative experiment (C) and summarized results (D) indicating the percentage of EGFP+ cells stained with Ab-1 are shown. E, 48 h after co-transfection of control (Con) siRNA, Bak siRNA, or Bax siRNA along with EBFP-histone H2B, Jurkat cells were transfected with pCMS5A (encoding EGFP-histone H2B) or pCMS5A/Mcl-1-shRNA for another 48 h and then collected and stained with APC-annexin V. The percentage of EBFP+EGFP+ cells stained with annexin V is indicated. Cell lysates were also subjected to immunoblotting to confirm knockdown. F, 48 h after transfection of control siRNA, Bim siRNA, Puma siRNA, or Bim siRNA + Puma siRNA along with EBFP-histone H2B, Jurkat cells were transfected with pCMS5A or pCMS5A/Mcl-1 shRNA (encoding EGFP-histone H2B) for another 48 h, then collected and stained with APC-annexin V. The percentage of EBFP+EGFP+ cells that stained with annexin V is indicated. Inset, cell lysates probed with antibodies to the indicated Bcl-2 family members to confirm knockdown. G, 48 h after Jurkat cells transfected with pCMS5A (control) or pCMS5A/Mcl-1-shRNA (Mcl-1 sh), CHAPS lysates were immunoprecipitated (IP) with antibodies to Bcl-2 (IP Bcl-2) or Mcl-1 (IP Mcl-1). Proteins in the immunoprecipitates and 1/5 of the lysates (1/5 input) were probed for Bak, Bax, Bim, Puma, Bcl-2, and Mcl-1. Beads + lysate without antibody (Beads) served as a control. IB, immunoblot. H, 48 h after transfection with pCMS5A, pCMS5A/Mcl-1shRNA, pCMS5A/Mcl-1shRNA/Bcl-2, or pCMS5A/Mcl-1 shRNA/Bcl-xL, Jurkat cells were collected and stained with APC-annexin V. The percentage of EGFP+ cells stained with annexin V is indicated. Immunoblotting was performed to confirm knockdown.
Apoptosis Induced by Bcl-2 Down-regulation Is Bak-dependent but not Bim- or Puma-dependent
To complement and extend the observations in Fig. 3, subsequent experiments examined the effect of Bcl-2 down-regulation in the same cell line. Treatment with two different Bcl-2 shRNAs resulted in induction of apoptosis (Fig. 4A) that was likewise accompanied by activation of Bak (Fig. 4, B and C). This Bcl-2 down-regulation-induced apoptosis was abolished by down-regulation of Bak (Fig. 4D) but only slightly inhibited by down-regulation of the Bcl-2 binding partners Bim and/or Puma (Fig. 4E), again suggesting that Bak release is the major event that triggers this apoptosis.
FIGURE 4.
Bcl-2 knockdown-induced apoptosis in Jurkat cells can be completely inhibited by Bak knockdown but only partially inhibited by Bim and/or Puma knockdown. A, 48 h after transfection with pCMS5A (empty vector, which encodes EGFP-histone H2B), pCMS5A/Bcl-2 shRNA 1, pCMS5A/Bcl-2 shRNA 2, or pCMS5A/Bcl-2 shRNA #1/Bcl-2* (shRNA-resistant Bcl-2), Jurkat cells were collected and stained with APC-annexin V. The percentage of EGFP+ cells that stained with APC-annexin V is indicated. Cells were also subjected to immunoblotting. B and C, 48 h after transfection with pCMS5A or pCMS5A/Bcl-2 shRNA, Jurkat cells were collected, fixed, and stained with Ab-1, which recognizes active Bak. A representative experiment (B) and summary of percentage of EGFP+ cells stained with Ab-1 (C) is shown. D, 48 h after co-transfection of control siRNA, Bak siRNA, or Bax siRNA with EBFP-histone H2B, Jurkat cells were collected and transfected with pCMS5A vector or pCMS5A/Bcl-2 shRNA for another 48 h and then stained with APC-annexin V. The percentage of EBFP+EGFP+ cells that stained with annexin V is indicated. Cell lysates were probed to confirm knockdown. E, 48 h after co-transfection of control siRNA, Bim siRNA, Puma siRNA, or Bim siRNA + Puma siRNA with EBFP-histone H2B, Jurkat cells were collected and transfected with pCMS5A or pCMS5A/Bcl-2 shRNA for another 48 h and then collected and stained with APC-annexin V. The percentage of EBFP+EGFP+ cells that stained with annexin V is indicated. Cell lysates were probed to confirm knockdown.
Bcl-2 Variants Differ in Ability to Bind Bak and Prevent Apoptosis in Jurkat Cells
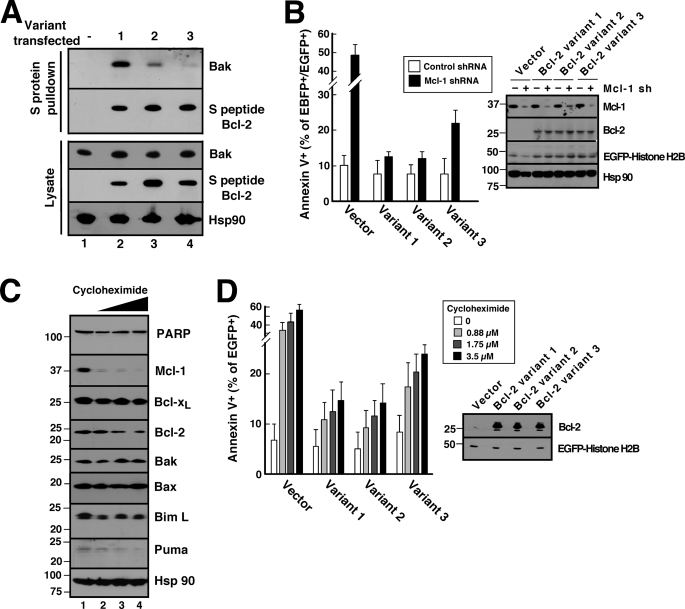
In further experiments, we utilized the experimental strategies shown in Figs. 2–4 to assess the potential biological impact of the Bcl-2 sequence variation demonstrated in Fig. 1. To evaluate potential differences in the ability of Bcl-2 variants to bind Bak in a cellular context, Jurkat cells were transfected plasmids encoding S peptide-tagged Bcl-2 variants 1–3. Despite almost equal expression of the variants (Fig. 5A, lower panel), variants 1 and 2 bound more Bak than variant 3 (Fig. 5A, upper panel).
FIGURE 5.
Different Bcl-2 variants exhibit different abilities to protect against Mcl-1 knockdown- and cycloheximide-induced apoptosis in Jurkat cells. A, 24 h after transient transfection with S peptide-tagged Bcl-2 variants, cell lysates prepared in CHAPS buffer were reacted with S protein-agarose to recover Bcl-2 complexes. Pulldowns were probed with anti-Bak antibody and, as a control, anti-S peptide antibody. B, 48 h after co-transfection with empty vector (pCMS5C expressing EBFP-histone H2B) or pCMS5C-Mcl-1-shRNA and pCMS5A vector (pCMS5A expressing EGFP-histone H2B), pCMS5A-Bcl-2 variant 1, pCMS5A-Bcl-2 variant 2, or pCMS5A-Bcl-2 variant 3, Jurkat cells were collected and stained with APC-annexin V. The percentage of EBFP+EGFP+ cells stained with annexin V is indicated. Error bars, mean ± S.D. of three independent experiments. A representative experiment is shown in supplemental Fig. S5. Inset, whole cell lysates from one of the experiments subjected to immunoblotting. C, after Jurkat cells were treated for 24 h with diluent (0.1% DMSO, lane 1) or cycloheximide at 0.88, 1.75, and 3.5 μm (lanes 2–4, respectively) in the presence of 5 μm Q-VD-OPhe to inhibit caspase activation, whole cells lysates were subjected to SDS-PAGE, transferred to nitrocellulose, and probed with antibodies to the indicated polypeptide. Hsp90 served as a loading control. PARP, poly(ADP-ribose) polymerase. D, 24 h after transfection with pCMS5A, pCMS5A-Bcl-2 variant 1, pCMS5A-Bcl-2 variant 2, or pCMS5A-Bcl-2 variant 3, Jurkat cells were treated with the indicated concentration of cycloheximide for another 24 h and then stained with APC-annexin V. The percentage of EGFP+ cells stained with annexin V is indicated. Error bars, mean ± S.D. of three independent experiments. A representative experiment is shown in supplemental Fig. S6. Inset, whole cell lysates from one of the experiments subjected to immunoblotting.
To determine whether this difference in binding resulted in a difference in protection, cells were co-transfected with a plasmid encoding both Mcl-1 shRNA and EBFP-histone H2B as well as a second plasmid encoding Bcl-2 variants 1–3 and EGFP-histone H2B. Despite equal expression of the three Bcl-2 variants (Fig. 5B, inset), the Bcl-2 variant that exhibited lower affinity for Bak (Fig. 1E, variant 3) also displayed diminished ability to protect cells from this apoptotic stimulus (Fig. 5B and supplemental Fig. S5).
To determine whether similar results would be observed when apoptosis is triggered by a chemical stimulus, Jurkat cells were treated with varying concentrations of cycloheximide for 24 h. Consistent with a previous report (57), this treatment was accompanied by dose-dependent Mcl-1 down-regulation (Fig. 5C). In this cell line, cycloheximide also induced down-regulation of Bcl-2 as well as Puma and Bim but not Bak (Fig. 5C), again suggesting that release of Bak is the critical event for the induction of apoptosis in these cells. When the Bcl-2 variants were overexpressed prior to cycloheximide addition, variants 1 and 2 again protected more completely than variant 3 did (Fig. 5D; for primary data, see supplemental Fig. S6).
Bcl-2/Bak Binding Varies among Different Cell Lines
Previous studies from our laboratory (58) and others (59) have demonstrated that the relative levels of Bcl-2 vary widely among different cells. To assess the impact of this quantitative variation, Bcl-2/Bak binding was examined in four additional human lymphoid cell lines with varying Bcl-2 content (Fig. 6, A and B, and supplemental Fig. S7A). Immunoprecipitation of endogenous Bcl-2 from these cell lines demonstrated that Bak was readily detectable in Bcl-2 immunoprecipitates from RL cells, which endogenously express very high levels of Bcl-2 and, to a lesser extent, in Bcl-2 pulldowns from Molt3 and CEM cells (Fig. 6, C and D). In contrast, Bak was not detected in pulldowns from Daudi cells (Fig. 6D), which lack detectable Bcl-2 (Fig. 6A and supplemental Fig. S7A).
DISCUSSION
Results of this study demonstrate that Bcl-2 exhibits a much higher affinity for Bak protein than for the synthetic Bak BH3 domains. Additional observations indicate that different naturally occurring Bcl-2 sequence variants have different affinities for Bak protein, with some of these variants binding to Bak more tightly than Bcl-xL does and almost as tightly as Mcl-1. Finally, estimates of the absolute abundance of Bax, Bak, and antiapoptotic Bcl-2 family members suggest that Bcl-2 levels in some lymphoid cell lines are as much as 40-fold higher than Bcl-xL or Mcl-1. Consistent with these results, we observed that Bcl-2 binds and appears to neutralize Bak in intact cells, although the percentage of Bak neutralized by Bcl-2 varies depending on the amount of Bcl-2 expressed. Each of these observations provides new insight into Bcl-2 family member interactions that regulate cellular life and death decisions.
To our knowledge, this study provides the first comparison of the affinity of an isolated BH3 peptide and the corresponding proapoptotic protein for antiapoptotic Bcl-2 family members using the same technique. Although affinity measurements using isolated BH3 peptides have provided substantial insight into the roles of BH3-only proteins (see Introduction), our results suggest that these methods can underestimate the strength of Bak/Bcl-2 interactions. When the interaction of BakΔTM and Bcl-2ΔTM was examined by surface plasmon resonance, the affinity was much stronger than previously reported using the Bak BH3 peptide (Fig. 1E). This difference in binding between the Bak peptide and Bak protein likely reflects the limited ability of certain BH3 peptides to assume the appropriate conformation for binding (60) in the absence of conformational constraint supplied by the remainder of the polypeptide. Whether other affinity measurements that utilized BH3 peptides as surrogates for full-length proapoptotic Bcl-2 family members might have similarly underestimated the strengths of Bcl-2 family member interactions remains to be determined.
Further experiments demonstrated that Bak could be pulled down with Bcl-2 from cell lysates. To minimize the possibility that complexes of Bak and Bcl-2 might be forming during the cell lysis and immunoprecipitation procedures, cellular contents were solubilized using CHAPS rather than a nonionic detergent (61, 62); and immunoprecipitations were performed at 4 °C using the shortest possible incubation that allowed us to bind and pull down part of the total cellular Bcl-2, Bcl-xL, and Mcl-1. This approach suggested that ∼50% of the total cellular Bak in Jurkat cells is bound to Bcl-2, Bcl-xL, or Mcl-1 (Fig. 2C) and ∼1/6 of this is bound to Bcl-2. Larger percentages of Bak were pulled down with Bcl-2 in RL cells (Fig. 6, C and D), which endogenously express more Bcl-2 (Fig. 6, A and B).
Several previous studies have also demonstrated that Bak can be pulled down with tagged, overexpressed Bcl-2 (28, 29, 63), providing a precedent for our observation that Bak can be immunoprecipitated with endogenous Bcl-2. Other investigations, however, have failed to detect Bak in Bcl-2 pulldowns (12, 51). To investigate these divergent results further, the binding of Bak to two additional naturally occurring Bcl-2 sequence variants was examined by surface plasmon resonance. These Bcl-2 allelic variants exhibited different affinities for BakΔTM (Fig. 1E), suggesting a potential explanation for some of the earlier divergent results. Further analysis demonstrated that Bak protein binds more tightly to some of these Bcl-2 variants than to Bcl-xL (Fig. 1, D and E).
To assess the potential biological significance of the Bak/Bcl-2 interaction, we examined the effect of Bcl-2 overexpression on Bak-mediated apoptosis. This analysis demonstrated that a tight-binding Bcl-2 variant can compensate for Mcl-1 during Bak-mediated apoptosis in intact Jurkat cells (Fig. 3H). Conversely, Bcl-2 down-regulation in these same cells induced Bak-dependent apoptosis (Fig. 4). Although one potential explanation for this Bcl-2 shRNA-induced apoptosis would be the release of the proapoptotic Bcl-2 family members Bim and Puma, which have been implicated in Bax activation in some studies (15–17), the observation that the Bcl-2 shRNA-induced apoptosis was scarcely affected by siRNA-mediated down-regulation of Bax (Fig. 4D) or Bim and Puma (Fig. 4E) argues against this possibility. Instead, the appearance of active Bak after Bcl-2 down-regulation (Fig. 4, B and C) and the ability of Bak down-regulation to abolish the effect of Bcl-2 shRNA (Fig. 4D) are most consistent with the suggestion that Bcl-2 can restrain Bak in a cellular context.
In further experiments, we examined the ability of the Bcl-2 variants to inhibit apoptosis. These studies examined Jurkat cells after treatment with Mcl-1 shRNA, a treatment that again induced Bak-dependent apoptosis that was unaffected by Bim and Puma down-regulation (Fig. 3F). Although all three Bcl-2 variants were able to inhibit this Bak-dependent apoptosis, the variant with lower affinity (Fig. 1E, variant 3) was less effective (Fig. 5B). Similar results were observed after treatment with cycloheximide, which simultaneously down-regulates Mcl-1 and Bcl-2 in Jurkat cells (Fig. 5, C and D). These observations suggest that the differences in affinity of Bcl-2 for Bak can result in differences in antiapoptotic potency in certain contexts.
On the other hand, it is also important to recognize that binding to Bak is only one aspect of Bcl-2 function. Variant 3, like variant 1, was originally isolated from a t(14;18)-associated lymphoma, suggesting that variant 3 is also able to contribute to cellular transformation. These observations raise the possibility that variant 3 might have evolved other properties that compensate for its diminished Bak binding, at least under certain conditions.
The preceding studies were performed using Mcl-1 or Bcl-2 shRNA to induce apoptosis. These experiments were not designed to distinguish between current models of BH3-only protein action. While performing these studies, however, we made several observations that might be pertinent to contemporary discussions of Bax/Bak activation. First, we observed that apoptosis in Jurkat cells is inhibited by Bak siRNA but not Bax siRNA (Figs. 3E and 4D), consistent with the much higher expression of Bak in this cell line (Fig. 6B). These observations recommend Jurkat cells as a somewhat simplified model for studying Bcl-2 family member interactions. Second, we observed that Mcl-1 knockdown was associated with increased binding of Bak and Bim but not Puma to Bcl-2 (Fig. 3G), suggesting that Bak and Bim but not Puma might be restrained, at least in part, by Mcl-1 in unstressed Jurkat cells. Third, we observed that the ability of Mcl-1 shRNA to induce apoptosis was inhibited by Bak siRNA but not Bim and Puma knockdown (Fig. 3, E and F). We cannot rule out the possibility that Puma or Bim levels remaining after siRNA were sufficient to trigger apoptosis upon Mcl-1 down-regulation even though the same Bim and Puma siRNA constructs markedly diminished drug-induced apoptosis in the same cells.4 Likewise, we cannot rule out the possibility that some other BH3-only family member is involved in Bak activation in this cell line. Barring these eventualities, however, the present results are consistent with previous suggestions that a fraction of Bak in certain cells might be activated unless restrained by antiapoptotic family members (12). Such a model would also be consistent with earlier indications that apoptosis is the default pathway in some cells unless antiapoptotic proteins intervene (64). The suggestion that Mcl-1 is preventing Bak from initiating apoptosis in Jurkat cells is also consistent with recent results showing that, in addition to binding BH3-only proteins, Bcl-xL is able to restrain Bax even after it is activated (17, 19). On the other hand, these results certainly do not rule out the possibility that BH3-only proteins might bind and directly activate Bax and/or Bak under other circumstances or in other cells as suggested by other recent studies (15–17).
During the course of the present studies, we also examined a Bak mutant analogous to one described by Kim et al. (50). Previous studies demonstrated that I82A/N83A mouse Bak was unable to kill Bax−/−Bak−/− mouse embryo fibroblasts unless a BH3-only protein was also transduced. These results have been widely interpreted as providing support for the direct activation model of BH3-only protein action. Although I82A/N83A mouse Bak was unable to pull down Mcl-1 or Bcl-xL (50), binding to Bcl-2 was not examined. Our demonstration that the human homolog of this construct binds Bcl-2 (supplemental Fig. S3) provides an alternative explanation for the reported inability of this mutant to induce apoptosis unless a promiscuous BH3-only protein is expressed. On the other hand, because we have examined the human Bak protein rather than mouse protein, further studies are required to distinguish between these two potential interpretations.
In addition to affinity, interactions depend on the relative abundances of polypeptides. Although multiple studies have previously demonstrated that the relative levels one or more Bcl-2 family members vary across panels of cell lines, to our knowledge this study presents the first attempt to estimate the absolute amounts of multiple Bcl-2 family members. This analysis suggests that Bcl-2 is more abundant than Mcl-1 or Bcl-xL in certain lymphoid lines. Jurkat cells, for example, contain ∼90,000 Mcl-1 and 280,000 Bcl-2 molecules per cell (Fig. 6, A and B, and supplemental S7A). Even though Bcl-2 binds Bak with lower affinity than Mcl-1 (Fig. 1E), the greater abundance of Bcl-2 in Jurkat cells increases its potential contribution to Bak neutralization, with a net result that endogenous Bcl-2 binds roughly half as much Bak as endogenous Mcl-1 (Fig. 2C). Even more Bak was recovered with Bcl-2 in RL cells, which express much higher levels of Bcl-2 and lower levels of Bcl-xL and Mcl-1 (Fig. 6). In contrast, there was lower Bak recovery in Bcl-2 immunoprecipitates from cell lines with lower Bcl-2 content (Fig. 6).
In summary, the results described above are not consistent with current models suggesting that Bak is bound and restrained exclusively by Bcl-xL and Mcl-1. Instead, the present experiments indicate that human cells express a variety of Bcl-2 alleles that display differing affinities for Bak protein. In addition, these studies indicate that levels of Bcl-2 and Mcl-1 vary over at least a 10-fold range across different cell lines from one lineage. Collectively, these results suggest a model in which Bak is restrained by multiple antiapoptotic Bcl-2 family members, including Bcl-xL, Mcl-1, and Bcl-2 itself, to varying extents depending upon the abundances and identity of the sequence variants expressed in various cells.
Supplementary Material
Acknowledgments
We gratefully thank Alyson Smith, Nga Dai, Andrew Badley, Qian Liu, and Kalle Gehring for plasmids; Guy Poirier and David Toft for kind gifts of antibodies; Georges Mer for use of the fast protein liquid chromatography; Muthu Ramakrishnan for advice regarding Biacore experiments; David Huang and Jerry Adams for comments on an early version of the manuscript; the anonymous reviewer for helpful suggestions; Kevin Peterson for assistance with the blots shown in Fig. 5C; and Deb Strauss for secretarial assistance. Flow cytometry was performed in the Mayo Clinic Flow Cytometry and Optical Morphology Shared Resource.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 CA69008. This work was also supported by a Mayo Foundation for Education and Research predoctoral fellowship (to S.-H. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7.
H. Dai and S. H. Kaufmann, unpublished observations.
S.-H. Lee and S. H. Kaufmann, unpublished observations.
- BH3-only proteins
- proteins whose homology to other Bcl-2 family members is confined to the BH3 domain
- BH3
- Bcl-2 homology domain 3
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate
- ΔTM
- lacking a transmembrane domain
- EBFP
- enhanced blue fluorescent protein
- EGFP
- enhanced green fluorescent protein
- PMSF
- α-phenylmethylsulfonyl fluoride
- shRNA
- short hairpin RNA
- siRNA
- small interfering RNA
- GST
- glutathione S-transferase
- APC
- allophycocyanin
- PBS
- calcium- and magnesium-free Dulbecco's phosphate-buffered saline
- PMSF
- phenylmethylsulfonyl fluoride.
REFERENCES
- 1.Jiang X., Wang X. ( 2004) Annu. Rev. Biochem. 73, 87– 106 [DOI] [PubMed] [Google Scholar]
- 2.Adams J. M., Cory S. ( 2007) Curr. Opin. Immunol. 19, 488– 496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chipuk J. E., Green D. R. ( 2008) Trends Cell Biol. 18, 157– 164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Youle R. J., Strasser A. ( 2008) Nat. Rev. Mol. Cell Biol. 9, 47– 59 [DOI] [PubMed] [Google Scholar]
- 5.Letai A. G. ( 2008) Nature Rev. Cancer 8, 121– 132 [DOI] [PubMed] [Google Scholar]
- 6.Puthalakath H., Strasser A. ( 2002) Cell Death Differ. 9, 505– 512 [DOI] [PubMed] [Google Scholar]
- 7.Cory S., Adams J. M. ( 2002) Nat. Rev. Cancer 2, 647– 656 [DOI] [PubMed] [Google Scholar]
- 8.Chittenden T. ( 2002) Cancer Cell 2, 165– 166 [DOI] [PubMed] [Google Scholar]
- 9.van Delft M. F., Huang D. C. ( 2006) Cell Res. 16, 203– 213 [DOI] [PubMed] [Google Scholar]
- 10.Willis S. N., Adams J. M. ( 2005) Curr. Opin. Cell Biol. 17, 617– 625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lessene G., Czabotar P. E., Colman P. M. ( 2008) Nat. Rev. Drug Discov. 7, 989– 1000 [DOI] [PubMed] [Google Scholar]
- 12.Willis S. N., Chen L., Dewson G., Wei A., Naik E., Fletcher J. I., Adams J. M., Huang D. C. ( 2005) Genes Dev. 19, 1294– 1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Willis S. N., Fletcher J. I., Kaufmann T., van Delft M. F., Chen L., Czabotar P. E., Ierino H., Lee E. F., Fairlie W. D., Bouillet P., Strasser A., Kluck R. M., Adams J. M., Huang D. C. ( 2007) Science 315, 856– 859 [DOI] [PubMed] [Google Scholar]
- 14.Letai A., Bassik M. C., Walensky L. D., Sorcinelli M. D., Weiler S., Korsmeyer S. J. ( 2002) Cancer cell 2, 183– 192 [DOI] [PubMed] [Google Scholar]
- 15.Kuwana T., Bouchier-Hayes L., Chipuk J. E., Bonzon C., Sullivan B. A., Green D. R., Newmeyer D. D. ( 2005) Mol. Cell 17, 525– 535 [DOI] [PubMed] [Google Scholar]
- 16.Gavathiotis E., Suzuki M., Davis M. L., Pitter K., Bird G. H., Katz S. G., Tu H. C., Kim H., Cheng E. H., Tjandra N., Walensky L. D. ( 2008) Nature 455, 1076– 1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lovell J. F., Billen L. P., Bindner S., Shamas-Din A., Fradin C., Leber B., Andrews D. W. ( 2008) Cell 135, 1074– 1084 [DOI] [PubMed] [Google Scholar]
- 18.Fletcher J. I., Meusburger S., Hawkins C. J., Riglar D. T., Lee E. F., Fairlie W. D., Huang D. C., Adams J. M. ( 2008) Proc. Natl. Acad. Sci. U. S. A. 105, 18081– 18087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Billen L. P., Kokoski C. L., Lovell J. F., Leber B., Andrews D. W. ( 2008) PLoS Biol. 6, e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Czabotar P. E., Lee E. F., van Delft M. F., Day C. L., Smith B. J., Huang D. C., Fairlie W. D., Hinds M. G., Colman P. M. ( 2007) Proc. Natl. Acad. Sci. U. S. A. 104, 6217– 6222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sattler M., Liang H., Nettesheim D., Meadows R. P., Harlan J. E., Eberstadt M., Yoon H. S., Shuker S. B., Chang B. S., Minn A. J., Thompson C. B., Fesik S. W. ( 1997) Science 275, 983– 986 [DOI] [PubMed] [Google Scholar]
- 22.Moldoveanu T., Liu Q., Tocilj A., Watson M., Shore G., Gehring K. ( 2006) Mol. Cell 24, 677– 688 [DOI] [PubMed] [Google Scholar]
- 23.Certo M., Del, Gaizo, Moore V., Nishino M., Wei G., Korsmeyer S., Armstrong S. A., Letai A. ( 2006) Cancer Cell 9, 351– 365 [DOI] [PubMed] [Google Scholar]
- 24.Stevenson R. ( 1991) Am. Biotechnol. Lab. 9, 36. [PubMed] [Google Scholar]
- 25.Jason-Moller L., Murphy M., Bruno J. ( 2006) Curr. Protoc. Protein Sci. Unit 19.13 [Google Scholar]
- 26.Berggård T., Linse S., James P. ( 2007) Proteomics 7, 2833– 2842 [DOI] [PubMed] [Google Scholar]
- 27.Chen L., Willis S. N., Wei A., Smith B. J., Fletcher J. I., Hinds M. G., Colman P. M., Day C. L., Adams J. M., Huang D. C. ( 2005) Mol. Cell 17, 393– 403 [DOI] [PubMed] [Google Scholar]
- 28.Chittenden T., Harrington E. A., O'Connor R., Flemington C., Lutz R. J., Evan G. I., Guild B. C. ( 1995) Nature 374, 733– 736 [DOI] [PubMed] [Google Scholar]
- 29.Chittenden T., Flemington C., Houghton A. B., Ebb R. G., Gallo G. J., Elangovan B., Chinnadurai G., Lutz R. J. ( 1995) EMBO J. 14, 5589– 5596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsujimoto Y., Cossman J., Jaffe E., Croce C. M. ( 1985) Science 228, 1440– 1443 [DOI] [PubMed] [Google Scholar]
- 31.Cleary M. L., Smith S. D., Sklar J. ( 1986) Cell 47, 19– 28 [DOI] [PubMed] [Google Scholar]
- 32.Vaux D. L., Cory S., Adams J. M. ( 1988) Nature 335, 440– 442 [DOI] [PubMed] [Google Scholar]
- 33.Strasser A., Harris A. W., Bath M. L., Cory S. ( 1990) Nature 348, 331– 333 [DOI] [PubMed] [Google Scholar]
- 34.Miyashita T., Reed J. C. ( 1992) Cancer Res. 52, 5407– 5411 [PubMed] [Google Scholar]
- 35.Maung Z. T., MacLean F. R., Reid M. M., Pearson A. D., Proctor S. J., Hamilton P. J., Hall A. G. ( 1994) Br. J. Haematol. 88, 105– 109 [DOI] [PubMed] [Google Scholar]
- 36.Shivakumar L., Armitage J. O. ( 2006) Clin. Lymphoma Myeloma 6, 455– 457 [DOI] [PubMed] [Google Scholar]
- 37.Meng X. W., Heldebrant M. P., Kaufmann S. H. ( 2002) J. Biol. Chem. 277, 3776– 3783 [DOI] [PubMed] [Google Scholar]
- 38.Meng X. W., Chandra J., Loegering D., Van, Becelaere K., Kottke T. J., Gore S. D., Karp J. E., Sebolt-Leopold J., Kaufmann S. H. ( 2003) J. Biol. Chem. 278, 47326– 47339 [DOI] [PubMed] [Google Scholar]
- 39.Harlow E., Lane D. ( 1988) Antibodies: A Laboratory Model, pp. 521– 523, Cold Spring Harbor Laboratory, New York [Google Scholar]
- 40.Cliby W. A., Lewis K. A., Lilly K. K., Kaufmann S. H. ( 2002) J. Biol. Chem. 277, 1599– 1606 [DOI] [PubMed] [Google Scholar]
- 41.van, den, Hoff M. J., Moorman A. F., Lamers W. H. ( 1992) Nucleic Acids Res. 20, 2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mesa R. A., Loegering D., Powell H. L., Flatten K., Arlander S. J., Dai N. T., Heldebrant M. P., Vroman B. T., Smith B. D., Karp J. E., Eyck C. J., Erlichman C., Kaufmann S. H., Karnitz L. M. ( 2005) Blood 106, 318– 327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hackbarth J. S., Lee S. H., Meng X. W., Vroman B. T., Kaufmann S. H., Karnitz L. M. ( 2004) BioTechniques 37, 835– 839 [PubMed] [Google Scholar]
- 44.Gomez T. S., McCarney S. D., Carrizosa E., Labno C. M., Comiskey E. O., Nolz J. C., Zhu P., Freedman B. D., Clark M. R., Rawlings D. J., Billadeau D. D., Burkhardt J. K. ( 2006) Immunity 24, 741– 752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meng X. W., Lee S. H., Dai H., Loegering D., Yu C., Flatten K., Schneider P., Dai N. T., Kumar S. K., Smith B. D., Karp J. E., Adjei A. A., Kaufmann S. H. ( 2007) J. Biol. Chem. 282, 29831– 29846 [DOI] [PubMed] [Google Scholar]
- 46.Seto M., Jaeger U., Hockett R. D., Graninger W., Bennett S., Goldman P., Korsmeyer S. J. ( 1988) EMBO J. 7, 123– 131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yin X. M., Oltvai Z. N., Korsmeyer S. J. ( 1994) Nature 369, 321– 323 [DOI] [PubMed] [Google Scholar]
- 48.Petros A. M., Medek A., Nettesheim D. G., Kim D. H., Yoon H. S., Swift K., Matayoshi E. D., Oltersdorf T., Fesik S. W. ( 2001) Proc. Natl. Acad. Sci. U. S. A. 98, 3012– 3017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Joseph M. K., Solomon L. R., Petros A. M., Cai J., Simmer R. L., Zhang H., Rosenberg S., Ng S. C. ( 2004) Oncogene 23, 835– 838 [DOI] [PubMed] [Google Scholar]
- 50.Kim H., Rafiuddin-Shah M., Tu H. C., Jeffers J. R., Zambetti G. P., Hsieh J. J., Cheng E. H. ( 2006) Nat. Cell Biol. 8, 1348– 1358 [DOI] [PubMed] [Google Scholar]
- 51.Cheng E. H., Sheiko T. V., Fisher J. K., Craigen W. J., Korsmeyer S. J. ( 2003) Science 301, 513– 517 [DOI] [PubMed] [Google Scholar]
- 52.Moulding D. A., Giles R. V., Spiller D. G., White M. R., Tidd D. M., Edwards S. W. ( 2000) Blood 96, 1756– 1763 [PubMed] [Google Scholar]
- 53.Opferman J. T., Iwasaki H., Ong C. C., Suh H., Mizuno S., Akashi K., Korsmeyer S. J. ( 2005) Science 307, 1101– 1104 [DOI] [PubMed] [Google Scholar]
- 54.Koopman G., Reutelingsperger C. P., Kuijten G. A., Keehnen R. M., Pals S. T., van, Oers M. H. ( 1994) Blood 84, 1415– 1420 [PubMed] [Google Scholar]
- 55.Brumatti G., Sheridan C., Martin S. J. ( 2008) Methods 44, 235– 240 [DOI] [PubMed] [Google Scholar]
- 56.Griffiths G. J., Corfe B. M., Savory P., Leech S., Esposti M. D., Hickman J. A., Dive C. ( 2001) Oncogene 20, 7668– 7676 [DOI] [PubMed] [Google Scholar]
- 57.Adams K. W., Cooper G. M. ( 2007) J. Biol. Chem. 282, 6192– 6200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kaufmann S. H., Karp J. E., Svingen P. A., Krajewski S., Burke P. J., Gore S. D., Reed J. C. ( 1998) Blood 91, 991– 1000 [PubMed] [Google Scholar]
- 59.Amundson S. A., Myers T. G., Scudiero D., Kitada S., Reed J. C., Fornace A. J., Jr. ( 2000) Cancer Res. 60, 6101– 6110 [PubMed] [Google Scholar]
- 60.Walensky L. D., Kung A. L., Escher I., Malia T. J., Barbuto S., Wright R. D., Wagner G., Verdine G. L., Korsmeyer S. J. ( 2004) Science 305, 1466– 1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hsu Y. T., Youle R. J. ( 1997) J. Biol. Chem. 272, 13829– 13834 [DOI] [PubMed] [Google Scholar]
- 62.Hsu Y. T., Youle R. J. ( 1998) J. Biol. Chem. 273, 10777– 10783 [DOI] [PubMed] [Google Scholar]
- 63.Zhai D., Jin C., Huang Z., Satterthwait A. C., Reed J. C. ( 2008) J. Biol. Chem. 283, 9580– 9586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Raff M. C. ( 1992) Nature 356, 397– 400 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.