Abstract
The vitamin D endocrine system is essential for calcium and phosphate homeostasis and skeletal mineralization. The 1,25-dihydroxyvitamin D3 (1,25(OH)2D3) hormone binds to the vitamin D receptor (VDR) to regulate gene expression. These gene products in turn mediate the actions of 1,25(OH)2D3 in mineral-regulating target cells such as the osteoblast. We showed previously that meningioma 1 (MN1) is a novel target of 1,25(OH)2D3 in MG-63 osteoblastic cells and that it is a coactivator for VDR-mediated transcription (Sutton, A. L., Zhang, X., Ellison, T. I., and MacDonald, P. N. (2005) Mol. Endocrinol. 19, 2234–2244). However, the functional significance of MN1 in osteoblastic cell biology is largely unknown. Here, we demonstrate that MN1 expression is increased dramatically during differentiation of primary osteoblastic cells. Using calvarial osteoblasts derived from wild-type and MN1 knock-out mice, we provide data supporting an essential role of MN1 in maintaining appropriate osteoblast proliferation, differentiation, and function. MN1 knock-out osteoblasts displayed altered morphology, decreased growth rate, impaired motility, and attenuated 1,25(OH)2D3/VDR-mediated transcription as well as reduced alkaline phosphatase activity and mineralized nodule formation. MN1 null osteoblasts were also impaired in supporting osteoclastogenesis in co-culture studies presumably because of marked reduction in the RANKL:OPG ratio in the MN1 null cells. Mechanistic studies supported a transcriptional role for MN1 in controlling RANKL gene expression through activation of the RANKL promoter. Cumulatively, these studies indicate an important role for MN1 in maintaining the appropriate maturation and function of calvarial osteoblasts.
The skeleton is a dynamic organ that undergoes a continuous remodeling process mediated by bone-forming osteoblasts and bone-resorbing osteoclasts. Osteoblast precursors originate from mesenchymal stem cells, which are thought to serve as common progenitors for osteoblasts, adipocytes, and chondrocytes in developing skeletal tissues (1, 2). Osteoblasts promote bone formation by secreting extracellular matrix proteins, such as collagen and non-collagenous proteoglycans, glycoproteins, and γ-carboxylated proteins (3). Osteoblasts are also required to regulate the formation of mature osteoclasts by producing essential cytokines and regulatory proteins, such as receptor activator of NF-κB ligand (RANKL),2 which stimulates osteoclastogenesis and osteoprotegerin (OPG), which inhibits this process (4). In hematopoietic osteoclast precursors, RANKL binds to the cell surface receptor, RANK, and activates an intracellular signaling pathway by recruiting adaptor molecules, such as tumor necrosis factor receptor-associated factors, c-src non-receptor-type tyrosine kinase, and the c-Fos family proteins (5). These adaptor proteins mediate the activation of NF-κB, c-Jun NH2-terminal kinase, and mitogen-activated protein kinase (6–8). In contrast, OPG is an osteoclastogenesis inhibiting factor that functions as a decoy receptor, binding to RANKL and blocking the activation of the RANK signaling cascade (9). The osteoblastic RANKL:OPG expression ratio is a major determinant of osteoclastogenesis. It is enhanced by numerous factors that promote osteoclastogenesis, such as glucocorticoids and parathyroid hormone (10, 11), or reduced by factors that inhibit osteoclastogenesis, such as estrogen and growth hormone (12, 13).
The classic role of the vitamin D endocrine system is to regulate proper calcium and phosphate absorption in the intestine, which is essential for maintaining appropriate bone mineralization and skeletal development (14, 15). The active form of vitamin D is 1,25-dihydroxyvitamin D3 (1,25(OH)2D3). Its physiological functions are mediated through the vitamin D receptor (VDR). Ligand binding induces VDR association with retinoid X receptor, binding to DNA, and the recruitment of coactivators to modulate the transcription of target genes. These gene products mediate the activities of 1,25(OH)2D3 in vivo. In the skeletal system both osteoblasts and osteoclasts are responsive to 1,25(OH)2D3. In osteoblasts, 1,25(OH)2D3 promotes the synthesis of bone matrix proteins and regulates the expression of osteoclastogenic factors. For example, 1,25(OH)2D3 enhances RANKL synthesis and inhibits OPG secretion, generating a RANKL:OPG ratio that favors osteoclastogenesis (16). In osteoclasts, 1,25(OH)2D3 enhances bone resorption activity and promotes calcium release. This is critical in maintaining mineral homeostasis when serum calcium and phosphate levels are low. The lack of active hormone or functional receptor results in undermineralized skeletal disorders, including rickets in children and osteomalacia in adults (14, 15).
We used gene expression array analysis to identify novel 1,25(OH)2D3-regulated genes to better understand the role of 1,25(OH)2D3 in bone (17). Meningioma 1 (MN1) was identified as one novel 1,25(OH)2D3 target gene in the MG-63 osteoblastic cell system. MN1 was originally discovered as a gene that is disrupted by a balanced translocation in meningioma (18). However, subsequent studies showed that overexpression of MN1 in bone marrow cells induces myeloid malignancy in mice and predicts poor treatment outcome in acute myeloid leukemia patients (19–21). In addition, a recurrent translocation resulting in mis-expression of an MN1-TEL fusion protein was identified in human myeloid diseases. Forced expression of MN1-TEL or MN1 in multipotent progenitor cells causes T-lymphoid tumors as well as acute myeloid leukemia (22–26). Although MN1 overexpression is highly correlated with myeloid disorders, studies with the MN1 knock-out mouse point to additional fundamental biologies. Targeted deletion of MN1 is post-natal lethal due to severe cleft palate (27). Moreover, multiple craniofacial bones that form by intramembranous ossification processes are either undermineralized or absent in MN1-null mice. The mechanisms underlying these defects in MN1 null mice are unknown.
The ossification defect in the cranial skeleton of MN1 knock-out mice (27) combined with the expression and regulation of MN1 in osteoblasts (17) suggest a potential role for MN1 in osteoblast function. Because little is known about the function and regulation of MN1 in bone cells, we initiated a series of studies to test the putative role of MN1 in osteoblastic cell function. Our data provide new insight, collectively showing a profound significance for MN1 in maintaining appropriate proliferation, differentiation, and function of the craniofacial osteoblast.
EXPERIMENTAL PROCEDURES
Cell Culture
MC3T3-E1 mouse fetal calvarial cells were grown in α-MEM (Invitrogen) with 10% fetal bovine serum. For differentiation studies, MC3T3-E1 cells were grown to confluence, and then the growth medium was supplemented for 2 weeks with 50 μg/ml l-ascorbic acid and 10 mm β-glycerophosphate to promote osteoblast differentiation. Medium was replenished twice per week during the differentiation period. All cell cultures were supplemented with 100 units/ml penicillin and 100 μg/ml streptomycin.
Northern Analysis
Total cellular RNA was extracted with RNA-Bee isolation reagent according to the manufacturer's recommendations (Tel-Test, Friendswood, TX). For Northern blot analysis, 15–20 μg of total RNA were separated on a formaldehyde/agarose gel and transferred to Magna nylon membrane (GE Osmanics, Minnetonka, MN) by capillary action. The blots were hybridized with α-32P-labeled probes synthesized using a Prime-A-Gene kit (Promega, Madison, WI). Mouse probes were generated by reverse transcription-PCR from mouse osteoblast RNA using the amplification primers listed in supplemental Table 1. Autoradiograms were quantitated using scanning densitometry and normalized using 18 S RNA.
Real-time PCR Analysis
Total RNA was reverse-transcribed using the High Capacity RNA-to-cDNA kit from Applied Biosystems as described by the manufacturer (ABI, Foster City, CA). First strand cDNA was amplified and quantitated using an ABI real time thermocycler coupled with Step-One Plus software and specific Taqman Gene Expression Assays for 18 S ribosomal RNA, murine Runx2, osterix, integrin binding bone sialoprotein (BSP2), and dentin matrix-binding protein (DMP1).
Reporter Gene Analysis
The 1,25(OH)2D3-responsive reporter gene constructs cyp24(-1200)-luc and VDRE4-TATA-luc were described previously (17, 28). To create the hRANKL(−1kb)-luc reporter vector, the human RANKL promoter (−922 to +23 relative to the transcriptional start site) was amplified by PCR and subcloned into the pGL3 basic vector (Promega). The appropriate firefly luciferase reporter construct (150 ng/well) together with the cytomegalovirus-Renilla plasmid (5 ng/well) were transfected in quadruplicate into osteoblastic cells plated in 24-well culture plates. Cells were treated as indicated, lysed with 80 μl per well of passive lysis buffer (Promega), and assayed for the firefly and renilla luciferase activities with the Dual Luciferase Reporter Assay System (Promega) using an LMax microplate luminometer (Molecular Devices, Sunnyvale, CA). All values are presented as the normalized firefly:renilla luciferase activity. In siRNA knockdown experiments, On-Target plus SMARTpool (Dharmacon Inc., Chicago, IL) siRNAs specific for mouse MN1 or nonspecific controls (siCONTROL) (Dharmacon Inc.) were transfected in triplicate into MC3T3-E1 cells with Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions. Twenty-four hours later the hRANKL(-1 kb)-luc or pGL3 control reporter constructs were transfected, and the reporter gene expression was analyzed after 48 h.
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium Bromide (MTT) Assay
Primary osteoblasts were seeded at a density of 1 × 103 cells/cm2 in 24-well culture plates and treated as indicated. At various times 0.1 ml of MTT (5 mg/ml) was added to 0.4 ml of culture media for each well, and cells were incubated for another 3 h. Then the media were removed, and the purple formazan product was dissolved in isopropanol containing 0.04 m HCl for 10 min at 37 °C. The absorbance at 570 nm of each sample was determined with a scanning multiwell spectrophotometer. Experiments were performed in quadruplicate.
BrdUrd Labeling Assay
BrdUrd labeling and analysis of primary osteoblast cultures was similar to previous studies (17). Briefly, primary osteoblasts were seeded in 4-well lab-Tak chamber slides and treated as indicated. Cells were pulse-labeled for 4 h with media containing 10 μm BrdUrd, fixed with 4% paraformaldehyde for 10 min, and permeabilized by 0.1% Triton X-100 in phosphate-buffered saline for 5 min. The BrdUrd incorporation sites were exposed by treating the monolayers with 100 units/ml DNase I (Amersham Biosciences) for 30 min. The monolayers were then blocked with 1.5% normal goat serum and incubated with anti-BrdUrd mouse monoclonal antibody (BD Biosciences) followed by Alexa Fluorophore 594-conjugated goat anti-mouse secondary antibody (Molecular Probes, Invitrogen). ProLong Antifade reagent with 4′,6-diamidino-2-phenylindole (Vector Laboratories, Burlingame, CA) was used to mount glass coverslips, and the samples were visualized using fluorescence microscopy. Cells from three different areas (80–120 cells each) were counted, and the ratio of BrdUrd-positive cells versus total cell number was determined.
To measure BrdUrd incorporation in embryonic calvaria, BrdUrd (0.1 mg/g body weight) was injected into pregnant (18.5 day post-conception) MN1 heterozygous mice, and then the E18.5 calvaria were isolated 3 h later. These calvaria were fixed overnight in 10% neutral phosphate-buffered formalin and decalcified overnight in Immunocal formic acid solution (American Mastertech Scientific, Lodi, CA). After an overnight equilibration in phosphate-buffered saline-buffered 30% sucrose, the calvaria were embedded in Tissue-Tek O.C.T. frozen tissue matrix (Sakura Finetek, USA, Torrance, CA). Replicate 5-μm cryostat sections were cut from three different areas of each calvaria separated by 200–250 μm. The sections were immersed in 2 n HCl for 15 min at room temperature to expose the BrdUrd incorporation sites and then blocked and stained as described above except that Alexa Fluorophore 488 conjugated goat anti-mouse secondary antibody (Molecular Probes) was used. The sections were then counterstained with 0.5 μg/ml 4′,6-diamidino-2-phenylindole and mounted with ProLong Antifade reagent (Vector Laboratories). Overlapping images were taken to cover the entire calvarial slice, cells from the three different areas of each calvarium were counted, and the ratio of BrdUrd-positive cells versus total cell number was determined. Five animals from each genotype were analyzed.
In Vitro Wound Repair Assay
In vitro wound repair assays were performed as described (29). Briefly, confluent primary osteoblast monolayers were wounded by creating a linear scratch using a sterile pipette tip. The cells were washed with phosphate-buffered saline to remove debris, and then fresh media were replaced. Images were taken 0 and 16 h after injury, and the percentage of wound closure was determined.
Primary Osteoblast Culture
MN1 heterozygous breeding pairs were kindly provided by Gerard C. Grosveld (St. Jude Children's Research Hospital). All animals were maintained in a 12-h light, 12-h dark cycle following the rules of Institutional Animal Care and Use Committee at Case Western Reserve University. Primary osteoblasts were isolated from E18.5 calvaria as described (30). Briefly, connective tissue was removed from the isolated calvaria, and the osteoblasts were released through sequential 15-min digestions in 60% α-MEM supplemented with 0.024 mg/ml type 1 collagenase (Worthington Biochemical Corp., Lakewood, NJ), 0.072 mg/ml type 2 collagenase (Worthington), 0.1 mm CaCl2, 0.06% bovine serum albumin, and 15 mm Hepes (pH 7.4) at 37 °C. The first two digestions were discarded, and the next four digestions were collected and combined. Collagenase was removed with sequential media washes, and the osteoblasts were maintained in α-MEM supplemented with 15% fetal bovine serum. Osteoblasts from different animals with the same genotype were combined for experiments. Individual embryos were genotyped by PCR of tail digests as described (27). For differentiation experiments, the osteoblasts were seeded at a density of 6 × 103 cells/cm2, grown to confluence within 48 h, and replaced with growth media supplemented with 10 mm β-glycerophosphate and 50 μg/ml ascorbic acid. Media was changed every 3–4 days, and differentiating osteoblasts were collected at the indicated time for individual analysis.
Osteoclast Differentiation and Tartrate-resistant Phosphatase (TRAP) Activity Staining
Primary osteoblasts were isolated and differentiated for 15 days as described above. Spleen cells were isolated from 7–10-week-old female wild-type mice as described previously (31) and plated overnight in phenol red-free α-MEM (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA) and 10 ng/ml macrophage colony-stimulating factor (R & D Systems, Inc., Minneapolis, MN). Non-adherent cells containing osteoclast precursors were added to differentiated osteoblasts at a density of 500,000 cells per cm2 in phenol red-free α-MEM containing 10% heat-inactivated fetal bovine serum, 100 nm dexamethasone, and the indicated concentrations of 1,25(OH)2D3. The co-cultures were incubated for 8 days, and the medium was changed twice during this time. Mature osteoclasts were analyzed for enzymatic activity of TRAP using the leukocyte acid phosphatase kit (Sigma-Aldrich). TRAP-positive multinucleated cells with three or more nuclei were counted in triplicate wells.
Alkaline Phosphatase Activity Analysis
Primary osteoblast cultures were isolated and differentiated as described above. To stain for alkaline phosphatase activity, the cultures were fixed with 4% paraformaldehyde and then incubated for 20 min at room temperature with freshly prepared 0.4 mg/ml Fast Red TR in 0.2 mg/ml naphthol AS-MX phosphate alkaline solution (Sigma-Aldrich). Quantitative analysis of soluble alkaline phosphatase activity in cell extracts was performed using a colorimetric kit (Sigma-Aldrich) that measures the conversion of p-nitrophenyl phosphate to p-nitrophenol according to the manufacturer's instructions. Alkaline phosphatase activity was normalized to protein concentration of the lysate as measured by the bicinchoninic acid (BCA) assay (Pierce).
Mineralization Assay
Primary osteoblasts were isolated and differentiated for 31 days as described above. To stain for mineralized nodules, cells were fixed with 70% ethanol for 10 min at room temperature and then stained with freshly prepared 1% Alizarin Red S solution for 10 min. The numbers of mineralized nodules were counted under microscope. The Alizarin Red S stain was extracted with 100 mm cetylpyridinium chloride as described (32), and the absorbance at 500 nm was determined with a scanning multi-well spectrophotometer. All experiments were performed in triplicate.
RESULTS
MN1 mRNA Transcripts Are Induced by 1,25(OH)2D3 and by Osteoblastic Cell Differentiation
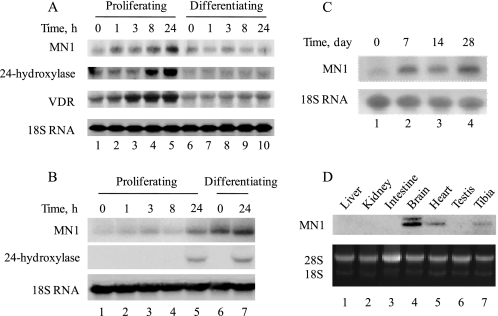
We previously characterized MN1 as a 1,25(OH)2D3-induced gene in the human MG-63 osteosarcoma cell line (17). However, this is an immortalized, transformed cell line representing a limited window of the osteoblast life cycle, namely that of the immature osteoblast. To more firmly establish the relevance of MN1 in osteoblastic cell biology, we examined more native osteoblastic cell systems, including the MC3T3-E1 mouse fetal calvarial cell line and primary osteoblasts obtained from embryonic mouse calvaria. Both models undergo a well defined differentiation program that progresses from immature proliferating cells to post-confluence mature, mineralized-matrix secreting cells in vitro. Thus, we could monitor 1,25(OH)2D3-activated expression of MN1 in immature proliferating osteoblasts as well as more fully differentiated, mature osteoblasts. More significantly, we could address whether MN1 expression levels were impacted by the process of osteoblastic cell differentiation in these systems. Thus, MC3T3-E1 cells or primary murine calvarial osteoblasts were treated with 1,25(OH)2D3 at various times throughout their in vitro differentiation program, and the steady-state levels of MN1 were examined. As shown in Fig. 1A, 1,25(OH)2D3 enhanced MN1 transcript levels in proliferating MC3T3-E1 cells in a time-dependent manner with increases evident as early as 1 h with a maximal increase of 4.2-fold at 24 h. This increase preceded the up-regulation of 24-hydroxylase, a well established primary VDR target gene, and paralleled increases of the VDR transcript. The up-regulation of MN1 by 1,25(OH)2D3 was not as profound in differentiating MC3T3-E1 cells, possibly due to the dramatic down-regulation of VDR in this system (compare VDR transcripts in lanes 6–10 with lanes 1–5 in Fig. 1A). Indeed, 1,25(OH)2D3-induced expression of 24-hydroxylase was also dramatically reduced in differentiating MC3T3-E1 cells.
FIGURE 1.
MN1 mRNA transcripts are induced by 1,25(OH)2D3 and by osteoblastic cell differentiation. MC3T3-E1 cells (A) or primary calvarial osteoblasts (B) were maintained at the proliferating stage or differentiated for 2 weeks as described under “Experimental Procedures.” Proliferating or differentiating cells were treated with 10 nm 1,25(OH)2D3 for the indicated times, and the expression of MN1, 24-hydroxylase, VDR, or 18 S RNA was examined by Northern blot analysis. C, primary osteoblasts were differentiated for the indicated times, and the expression of MN1 or 18 S RNA was examined by Northern blot analysis. D, total RNA was extracted from indicated tissues of 6-week-old wild-type mice, and the expression of MN1 was determined by Northern blot analysis. Ethidium bromide staining of 18 S/28 S ribosomal RNA was used to normalize loading differences.
Similar to MC3T3-E1 cells, steady-state levels of MN1 mRNA in proliferating primary calvarial osteoblasts were increased by 1,25(OH)2D3 in a time-dependent manner with a maximal effect of 7-fold at 24 h (Fig. 1B, lanes 1–5). Again, we observed only modest regulation (1.4-fold) of MN1 transcripts by 1,25(OH)2D3 in more differentiated osteoblasts (Fig. 1B, lanes 6 and 7). Importantly, differentiating primary osteoblasts expressed a higher basal level of MN1 than did their proliferating counterparts (Fig. 1B, compare lanes 1 and 6). Elevated MN1 mRNA levels were apparent after 7 days of differentiation and continued to increase modestly throughout the 28-day differentiation regimen (Fig. 1C, lanes 1–4). Finally, endogenous MN1 transcripts are readily detected in extracts of bone obtained from mouse tibia (Fig. 1D, lane 7), which represents one of the richest tissue sources of MN1 transcripts in addition to brain and cardiac muscle. Overall, these expression studies provide support for a potential significance of MN1 in skeletal biology and in the process of osteoblast cell differentiation and/or function.
MN1 Knock-out Osteoblasts Have Diminished VDR-mediated Transcriptional Responses
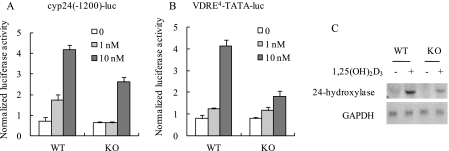
In a previous study we characterized MN1 as a coactivator for VDR-mediated transcription using ectopic expression approaches in heterologous cell lines (17). To evaluate the significance of endogenous MN1 in VDR/1,25(OH)2D3-mediated transcription in the osteoblast, we examined the consequence of MN1 ablation on VDR/1,25(OH)2D3-activated reporter gene activity in primary osteoblastic cell cultures derived from MN1 knock-out mice. As shown in Fig. 2A, 1,25(OH)2D3-activated expression of the cyp24(-1200)-luc reporter containing the native promoter of the human 24-hydroxylase gene was dramatically reduced in MN1 knock-out osteoblasts compared with the wild-type cells. Similar results were obtained with a more defined promoter consisting of a minimal TATA element and four tandem copies of a VDRE (Fig. 2B). We also consistently observed impaired activation of the native 24-hydroxylase gene by 1,25(OH)2D3 in the MN1 knock-out cells compared with wild-type controls (Fig. 2C). These data strongly support a significance for MN1 in 1,25(OH)2D3/VDR-mediated transactivation in osteoblasts.
FIGURE 2.
MN1 knock-out osteoblasts have diminished VDR-mediated transcriptional responses. Subconfluent WT and MN1 KO primary osteoblasts were transiently transfected with a 1.2-kb human 24-hydroxylase promoter reporter (cyp24(-1200)-luc in A) or a multicopy VDRE-driven reporter (VDRE4-TATA-luc in B). Cells were treated with the indicated concentration of 1,25(OH)2D3 for 24 h, and the normalized luciferase activity was measured as described under “Experimental Procedures.” Data represent the mean ± S.D. (n = 4 in each group). C, RNA was isolated from MN1 WT or KO cells that were treated ethanol vehicle (−) or 10 nm 1,25(OH)2D3 (+) for 24 h. The expression of 24-hydroxylase and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was examined by Northern blot analysis.
MN1 Knock-out Osteoblasts Have a Reduced Growth Rate
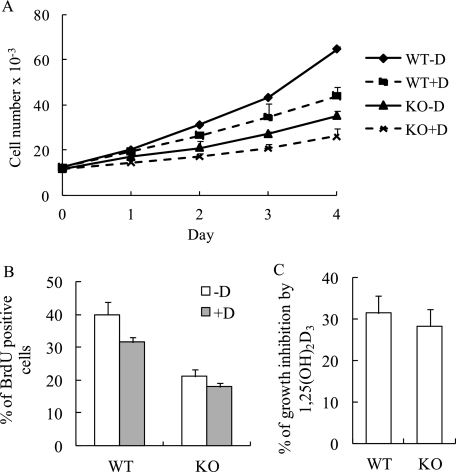
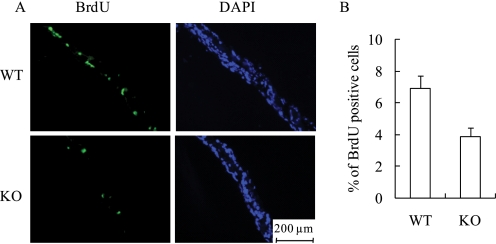
The cellular basis for the undermineralized skeletal phenotype in the MN1 knock-out mice is unresolved. Thus, to explore the potential significance of MN1 in bone cell function, we examined a variety of osteoblastic cell parameters using calvarial osteoblasts derived from E18.5 MN1 knock-out and wild-type embryos. Cell proliferation, as determined by counting trypan blue-excluding cells, indicated that MN1-null primary osteoblasts grew at a much reduced rate compared with wild-type cells (Fig. 3A). Moreover, BrdUrd-labeling analysis consistently showed that MN1 knock-out osteoblasts exhibited lower BrdUrd incorporation, indicating reduced S-phase entry in MN1 knock-out cells (Fig. 3B). Cell cycle analysis revealed that MN1 null osteoblasts have a higher percentage of G0/G1 cells (66%) compared with the wild-type cells (56%) (data not shown). The antiproliferative effects of 1,25(OH)2D3 were modest in these primary osteoblasts, but there was no statistically significant difference in the response of wild-type cells or MN1 null cells to 1,25(OH)2D3 as measured by multiple MTT assays (Fig. 3C), indicating that MN1 may not be required for the antiproliferative effects of 1,25(OH)2D3 in this cell system. To explore the in vivo significance of MN1 in controlling osteoblast proliferation, BrdUrd incorporation was determined in embryonic calvaria. Consistent with in vitro observations, MN1 knock-out calvaria had 40% less BrdUrd-labeled cells compared with the wild-type controls (Fig. 4). These studies indicate that MN1 is required for appropriate osteoblast proliferation in vitro and in vivo.
FIGURE 3.
MN1 knock-out osteoblasts have reduced growth rate. A, WT and MN1 KO primary osteoblasts were plated at a density of 1 × 103 cells/cm2 and treated with ethanol vehicle (-D) or 10 nm 1,25(OH)2D3 (+D) at day 0. Trypan blue-excluding cells were counted at the indicated time points. Data represent the mean ± S.D. (n = 3 in each group). B, MN1 WT and KO primary osteoblasts were plated at equal densities and treated with ethanol vehicle (-D) or 10 nm 1,25(OH)2D3 (+D) for 72 h. Cells were labeled with BrdUrd, and the BrdUrd-positive cells were quantitated. Data represent the mean ± S.D. (n = 3 in each group). C, the percentage of growth inhibition by 4-day treatment of 10 nm 1,25(OH)2D3 from three independent MTT assays. Data represent the mean ± S.D. (n = 3 in each group).
FIGURE 4.
MN1 knock-out calvarial cells have reduced proliferation in vivo. The BrdUrd-labeled calvaria from WT or MN1 KO E18.5 embryos were prepared as described under “Experimental Procedures.” Three distinct sections from different areas of each calvarium were analyzed for BrdUrd (BrdU) incorporation and counterstained with 4′,6-diamidino-2-phenylindole (DAPI). A, representative images of BrdUrd and 4′,6-diamidino-2-phenylindole stained sections. B, the percentage of BrdUrd-positive cells was quantitated. Five embryos from each genotype were analyzed. Data represent the mean ± S.D. (n = 15 individual sections in each group).
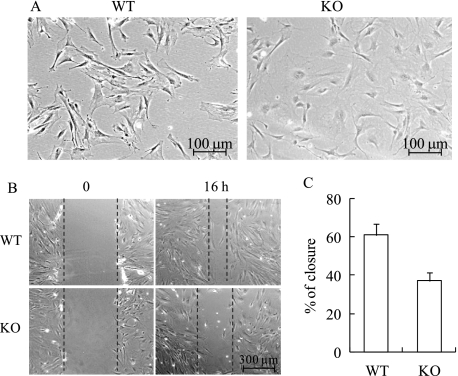
MN1 Knock-out Osteoblasts Show Altered Morphology and Reduced Motility
In proliferating osteoblast cultures, we observed a dramatic difference in cell morphology between MN1 knock-out and wild-type cells (Fig. 5A). In general, subconfluent MN1 knock-out calvarial osteoblasts appeared abnormal, displaying a large, flattened morphology. When observed using phase contrast microscopy, the MN1 knock-out cells were less refractile along their edges compared with the wild-type cells. This morphological difference indicates potential changes in cell adhesion state and cytoskeletal structure, suggesting a possible role for MN1 in cell migration. Thus, the motility of wild-type and MN1 knock-out osteoblasts was analyzed. Confluent osteoblast monolayers were “wounded” with a pipette tip to create a linear scratch, and the movement of cells into the wound area was determined. Compared with the wild-type controls, the wound closure in MN1 knock-out cells was markedly impaired (Fig. 5, B and C). Delayed wound closure in this model may result from impaired cell migration or proliferation. However, the 16-h time frame in which these migration studies were conducted was significantly less than the 40- and 60-h doubling times of the wild-type and knock-out cells, respectively (Fig. 3A). Thus, contributions of cell proliferation were minimized under these conditions.
FIGURE 5.
MN1 knock-out osteoblasts have altered morphology and decreased motility. A, representative phase contrast images of sub-confluent WT and MN1 KO primary osteoblasts. B, MN1 wild-type or knock-out cells were grown to confluence, and linear scratches were created. Pictures were taken 0 and 16 h after injury. The black dashed lines mark the wound edges. C, the percentage of wound closure was quantified and presented as the mean ± S.D. (n = 9 in each group).
MN1 Knock-out Calvarial Cells Show Reduced Differentiation and Mineralization
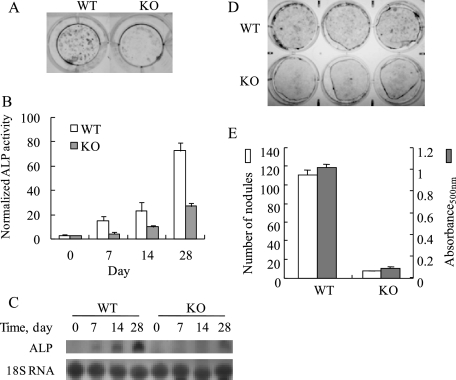
Mature primary calvarial osteoblasts express alkaline phosphatase and produce a mineralized matrix in vitro. To investigate the functional roles of MN1 in osteoblast maturation, MN1 wild-type or knock-out osteoblasts were differentiated in vitro and analyzed for alkaline phosphatase expression as well as calcium deposition. To minimize the effects of proliferation differences on this analysis, MN1 wild-type or knock-out osteoblasts were seeded at a high initial density (6 × 103 cells/cm2). Under these conditions proliferative differences were minimal, and both cultures reached a state of confluence within 48 h. As shown in Fig. 6, A–C, both mRNA expression and enzymatic activity of alkaline phosphatase were reduced in MN1 knock-out osteoblasts compared with wild-type cells. In addition, MN1 knock-out osteoblasts formed fewer and smaller Alizarin Red S-stained mineralized nodules (Fig. 6, D and E). These data indicate a potential defect in differentiation of MN1−/− osteoblasts.
FIGURE 6.
MN1 knock-out osteoblasts show decreased differentiation and mineralization. A, WT and MN1 KO osteoblasts were cultured for 28 days in differentiation media and stained in situ for alkaline phosphatase (ALP) enzyme activity. Representative images of alkaline phosphatase stained cultures are shown. Color images are shown in supplemental Fig. 1. B, MN1 WT or KO osteoblasts were cultured under osteogenic conditions for the indicated times. Alkaline phosphatase activity was determined with a colorimetric assay as described under “Experimental Procedures.” C, Northern analysis of alkaline phosphatase transcript levels in differentiating cultures of WT and MN1 KO calvarial osteoblasts. D and E, WT and MN1 KO osteoblasts were cultured for 31 days in differentiation media and stained for mineralized nodules with Alizarin Red S. D, representative images of Alizarin Red S stained cultures. Darkened areas are mineralized nodules. Color images are shown in supplemental Fig. 1. E, mineralized nodules were quantitated and compared (unfilled bars). The Alizarin Red S stain was extracted with 100 mm cetylpyridinium chloride, and the absorbance at 500 nm (absorbance500 nm) was measured (filled bars). Data represent the mean ± S.D. (n = 3 in each group).
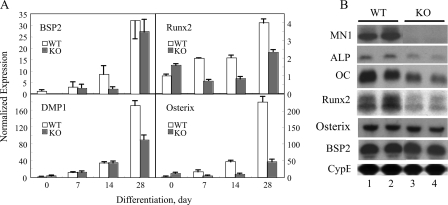
In this regard the expression of several key osteoblastic marker genes was either attenuated or delayed as MN1−/− calvarial cells progressed through the in vitro osteoblast differentiation system compared with the MN1+/+ cells (Fig. 7A). The osteoblast matrix proteins BSP2 and DMP1 showed significant decreases in expression in the MN1−/− cultures at 14- and 28-days of differentiation, respectively. Expression of osterix and Runx2, two obligatory factors that are required for appropriate osteoblast differentiation, are dramatically attenuated in differentiating MN1 null osteoblasts compared with WT controls (Fig. 7A). These data indicate that MN1 is required for appropriate differentiation and mineralized matrix production in this in vitro osteoblast culture system. To evaluate the significance of MN1 in osteoblastic maturation in vivo, the expression of osteoblast marker genes was examined in extracts obtained from MN1 wild-type and knock-out calvaria. As shown in Fig. 7B, MN1 transcripts were readily detected in WT calvaria and, as expected, were undetectable in MN1−/− mice. We observed only modest, if any, differences in the expression of BSP2 and osterix in the WT and KO calvarial extracts. This may be due to the mixed population of pre-osteoblastic and mature osteoblasts present in these calvarial extracts compared with the more synchronous differentiation program of the in vitro system. Importantly, the expression levels of alkaline phosphatase, osteocalcin, and Runx2 were markedly reduced in MN1 knock-out calvaria compared with the wild-type controls. Cumulatively, these data strongly suggest that intact E18.5 calvaria from the MN1 knock-out mice have fewer or less differentiated osteoblasts in vivo and that MN1 is an important gene that controls osteoblast differentiation and mineralization in vitro and in vivo.
FIGURE 7.
MN1 knock-out calvarial cells have reduced expression of osteoblastic genes in vitro and in vivo. A, WT and MN1 KO osteoblasts were cultured under osteogenic conditions for the indicated times. The expression of BSP2, DMP1, Runx2, osterix, and 18 S RNA were determined by TaqMan real-time reverse transcription-PCR approaches using 18 S RNA as an internal control. B, total RNA was extracted from WT and MN1 KO E18.5 calvaria. The expression of MN1, Runx2, osterix, BSP2, alkaline phosphatase (ALP), osteocalcin (OC), and cyclophilin B (CypB) was examined by Northern blot analysis. Cyclophilin B expression was used as a loading control.
MN1 Knock-out Osteoblasts Have Reduced Ability to Support 1,25(OH)2D3-induced Osteoclastogenesis
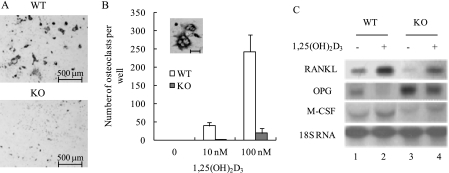
Mature osteoblasts secrete essential factors that regulate osteoclastogenesis, and a key function of 1,25(OH)2D3 in osteoblasts is to stimulate the expression of genes that support osteoclast differentiation. To determine the significance of osteoblastic MN1 in VDR/1,25(OH)2D3-stimulated osteoclastogenesis, wild-type or MN1 knock-out osteoblasts were used in spleenocyte co-culture studies, and TRAP-positive multinucleated osteoclast formation was assessed as described under “Experimental Procedures” (Fig. 8, A and B). As expected, no TRAP-positive multinucleated cells were detected in the absence of 1,25(OH)2D3. In the presence of 10 or 100 nm 1,25(OH)2D3, the numbers of TRAP-positive, multinucleated osteoclasts increased dramatically in the wild-type cells. In contrast, osteoclastogenesis was severely impaired in the MN1 knock-out cultures. These data suggest that osteoblast-derived MN1 plays an important role in supporting 1,25(OH)2D3-stimulated osteoclast differentiation. However, these studies do not exclude the possibility that intrinsic defects may also exist in osteoclast progenitors in the MN1 knock-out mouse.
FIGURE 8.
MN1 knock-out osteoblasts have reduced ability to support 1,25(OH)2D3-stimulated osteoclastogenesis. A, osteoclastogenic co-culture analysis was performed as described under “Experimental Procedures” using WT and MN1 KO osteoblasts. Co-cultures were stained for TRAP, an osteoclast-selective marker. Representative images of TRAP-stained co-cultures are shown. Color images are shown in supplemental Fig. 1. B, TRAP-positive multinucleated cells (≥3 nuclei) were counted and compared. Data represent the mean ± S.D. (n = 3 in each group). The inset shows a high magnification image of TRAP positive, multinucleated osteoclasts. The scale bar represents 60 μm. C, MN1 WT and KO osteoblasts were differentiated for 15 days and treated with ethanol (−) or 10 nm 1,25(OH)2D3 (+) for 24 h. Northern blot analysis was performed to examine mRNA expression levels of OPG, RANKL, macrophage colony-stimulating factor (M-CSF), or 18 S RNA.
To investigate the mechanism underlying the reduced ability of MN1 knock-out osteoblasts to support 1,25(OH)2D3-induced osteoclastogenesis, osteoblast expression of key osteoclastogenic factors were compared in wild-type and MN1 knock-out cells. RANKL and macrophage colony-stimulating factor are two factors that promote osteoclast formation. MN1 knock-out osteoblasts expressed lower levels of RANKL and macrophage colony-stimulating factor in both the basal and 1,25(OH)2D3-activated states when compared with the wild-type osteoblasts (Fig. 8C). In contrast, MN1 knock-out osteoblasts produced higher levels of OPG, a factor that inhibits osteoclast differentiation, in both the absence and presence of 1,25(OH)2D3 (Fig. 8C). These data indicate that MN1 is important for maintaining an appropriate RANKL:OPG ratio to support optimal osteoblast-mediated osteoclastogenesis.
MN1 Stimulates RANKL Promoter Activity
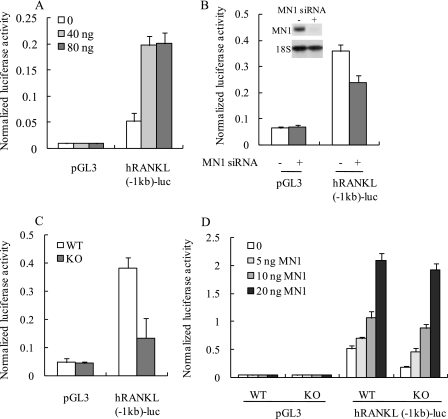
The previous data (Fig. 8C) suggest that MN1 may stimulate RANKL promoter activity and enhance the synthesis of RANKL mRNA. To test this hypothesis, the activity of a −1 kb human RANKL promoter was examined under conditions of MN1 overexpression or MN1 depletion. As shown in Fig. 9A, the RANKL promoter was stimulated by ectopic expression of MN1 in primary osteoblasts. Furthermore, decreased RANKL promoter activity was observed when MN1 was knocked down by siRNA (Fig. 9B) in MC3T3-E1 osteoblasts. RANKL promoter activity was also significantly lower in MN1 knock-out osteoblasts compared with the wild-type cells (Fig. 9C). Re-expression of MN1 rescued the RANKL promoter activity in MN1 knock-out cells in a dose-dependent manner (Fig. 9D). Importantly, expression of the promoter-less pGL3 control reporter gene was unaffected by MN1 overexpression or MN1 ablation (Fig. 9). The stimulation of RANKL promoter activity by MN1 supports a potential mechanism in which MN1 promotes osteoclastogenesis via increased transcription of the RANKL gene in osteoblasts.
FIGURE 9.
MN1 stimulates RANKL promoter activity. A, MC3T3-E1 cells were transiently transfected with a 1-kb human RANKL promoter reporter gene (hRANKL(−1kb)-luc) or pGL3 control reporter gene. The indicated amounts of the MN1 expression plasmid were cotransfected with the total molar amount of expression plasmid DNA kept constant by the addition of empty vector. The normalized luciferase activity was measured after 48 h. Data represent the mean ± S.D. (n = 3 in each group). B, control siRNA (−) or MN1 siRNA (+) was transfected into MC3T3-E1 cells 24 h before pGL3 or hRANKL(−1kb)-luc was transfected. The normalized luciferase activity was measured 48 h later. Data represent the mean ± S.D. (n = 3 in each group). The inset shows a Northern blot analysis of MN1 mRNA expression 24 h after siRNA transfection. C, WT or MN1 KO primary osteoblasts were transfected with pGL3 or hRANKL(−1kb)-luc. The normalized luciferase activity was measured after 48 h. Data represent the mean ± S.D. (n = 3 in each group). D, WT or KO primary osteoblasts were transiently transfected with pGL3 or hRANKL(−1kb)-luc. The indicated amounts of the MN1 expression plasmid were cotransfected with the total molar amount of expression plasmid DNA kept constant by the addition of empty vector. The normalized luciferase activity was measured after 48 h. Data represent the mean ± S.D. (n = 3 in each group).
DISCUSSION
We identified MN1 as an osteoblast-expressed gene that is up-regulated by 1,25(OH)2D3 and that positively influences VDR-activated transcription in the human MG-63 osteosarcoma cell line (17). Those studies implied that MN1 has a previously unrecognized significance in the osteoblast. The studies presented here provide strong support for that significance. The major findings in the present study are that primary calvarial osteoblasts derived from the MN1 knock-out mouse are defective in VDR-mediated gene expression and in osteoblast proliferation, migration, differentiation, and mineralization as well as in osteoblast-mediated osteoclastogenesis. We further provide a mechanistic basis for the involvement of MN1 in osteoblast-directed osteoclastogenesis that likely involves transcriptional regulation of the RANKL promoter, resulting in enhanced RANKL:OPG ratios that favor osteoclast formation. These new findings provide strong support for a role of MN1 in maintaining appropriate calvarial osteoblast function.
MN1 is a transcription factor that functions in part as a nuclear receptor coactivator protein. Heterologous expression studies in Hep3B and COS7 cells show that MN1 selectively augments VDR- and retinoic acid receptor-activated transcription, perhaps through direct interactions with the receptor and/or with p160 and p300 coactivator proteins (17, 33). Limitations of these past studies on nuclear receptor pathways include a reliance on overexpression of MN1 and a lack of approaches addressing a functional role for MN1 in appropriate target cells. Thus, the observations that MN1 ablation reduces both VDR-activated reporter gene expression as well as 1,25(OH)2D3-activated expression of the native cyp24 gene (a direct VDR target gene) in primary osteoblastic cells (Fig. 2) provides important support for the significance of MN1 in the mechanism of VDR-mediated gene expression in osteoblasts. MN1 ablation also dramatically reduces 1,25(OH)2D3-stimulated, osteoblast-mediated osteoclastogenesis (Fig. 8), thus showing a requirement for MN1 in critical VDR/1,25(OH)2D3-mediated biological activities in the osteoblast. Although these in vitro studies provide a solid foundation for the significance of MN1 in VDR-mediated transcription, it will be important to explore the in vivo requirement of MN1 in the vitamin D endocrine system and the maintenance of global mineral homeostasis or hair follicle cycling. Currently, this cannot be addressed because the MN1 knock-out mice do not survive postnatally where disruptions in the vitamin D endocrine system become evident. Tissue-directed MN1 null models that survive postnatally, perhaps targeting intestine or bone, need to be developed to tackle this important issue.
In addition to interacting with nuclear receptors and coactivator proteins to influence their transcriptional activities, MN1 also binds DNA directly via a CACCCAC promoter sequence to increase transcription of the IGFBP5 gene as well as a variety of other genes (34, 35). Our studies indicate that ectopic expression of MN1 stimulates RANKL promoter activity in osteoblastic cells, and knockdown or ablation of MN1 attenuates it. Thus, these observations support a transcriptional role for MN1 in the regulation of RANKL gene expression in the osteoblast. Indeed, several CACCCAC-like elements exist in the ∼1 kb of the 5′-flanking region of the human RANKL gene that was used here. Clearly, additional studies are needed to test whether this transcriptional role for MN1 is direct and mediated through binding of MN1 to the RANKL promoter or whether alternative mechanisms exist. It is unlikely that these effects are mediated through the VDR as the vitamin D-responsive elements of the RANKL gene reside at extreme upstream distal regions (at 16–76 kb upstream from the transcriptional start site) (36), far outside the immediate promoter regions tested here. However, based on the established role of MN1 in VDR-activated transcription, it would be intriguing to test the interplays between MN1 and these distal regulatory regions as 1,25(OH)2D3-mediated expression of RANKL is also impacted in MN1 knock-out osteoblasts. Finally, the combination of defective osteoblast differentiation, mineralization, and impaired osteoblast-directed, RANKL-stimulated, osteoclastogenesis has the potential to produce a global bone cell imbalance in the cranial vault that may explain in part the undermineralized phenotype in the MN1 knock-out mice.
The craniofacial phenotype of the MN1 knock-out embryo highlights the additional concept that MN1 also functions in critical developmental pathways that are independent of the VDR/1,25(OH)2D3 endocrine system. This is perhaps best illustrated by the lack of overt defects in cranial skeletal development or palatal defects in the 1α-hydroxylase or VDR knock-out mice which lack 1,25(OH)2D3 or its receptor, respectively, or in humans with inactivating mutations in these gene (37, 38). Thus, VDR is unlikely to play a major role in the function that MN1 exerts during mouse development. Interestingly, the MN1 knock-out skeletal defects are restricted to cranial skeletal elements, many of which form by intramembranous ossification processes (27). Long bones of the appendicular skeleton that form by endochondral ossification appear to develop normally in the MN1 knock-out embryos when analyzed for gross mineralization using Alcian Blue and Alizarin Red whole skeletal stains (27). Indeed, we observed no differences in bone mineral densities and morphological parameters of tibia from E18.5 MN1+/+ and MN1−/− embryos using more refined quantitative micro-computed tomography imaging technology (supplemental Table S2). Meester-Smoor et al. (27) suggested that MN1 was selectively involved in intramembranous bone formation. However, there are several important exceptions to this selectivity issue. Not all bones that form by intramembranous ossification are affected in the MN1 null embryos. The mandible and clavicle develop mainly through intramembranous ossification, and their development is intact in the MN1 knock-out embryo. Moreover, MN1 knock-out embryos display anomalies in numerous skeletal elements that form by endochondral ossification processes including the agenic, delayed, deformed, and/or undermineralized development of the pterygoid, basisphenoid, alisphenoid, supraoccipital, exoccipital, and sternum (Ref. 26 and data not shown). This indicates that the selectivity of the skeletal elements that require MN1 for normal development may not be related to the processes through which they form (e.g. intramembranous versus endochondral) but, instead, may arise through other, as yet unknown mechanisms.
The vertebrate skeleton is derived from three distinct sources. In general, the craniofacial skeleton originates from cranial neural crest cells, the axial skeleton is formed by paraxial mesoderm (somites), and the appendicular skeleton is derived from lateral plate mesodermal cells (39). The restricted craniofacial defects of the MN1 knock-out mice indicate the potential involvement of MN1 in neural crest cell migration and/or differentiation. Indeed, a recent study revealed that MN1 acts upstream of Tbx22, a gene that is mutated in X-linked cleft palate in humans (40). This supports an important role for MN1 in regulating the expression of genes involved in neural crest-initiated, palatal shelf development. This combined with the defects in osteoblast function in MN1 knock-out calvarial cells point to more global roles for MN1 in early development of neural crest-derived skeletal structures, in the subsequent differentiation and mineralization of osteoblasts during embryonic development of the skull and perhaps in postnatal skeletal homeostasis. Similar multifaceted roles in embryonic development and in embryonic and/or postnatal osteoblast dynamics exist for a variety of other genes including several of the Distal-less (Dlx) homeobox transcription factors (41), several of the hox family of transcription factors including hoxA10 (42), SATB2 (43), Connexin43 (44), Odd-skipped related 2 (45), transforming growth factor-β, bone morphogenic protein family members (46), and numerous others. The significance of osteoblastic MN1 in postnatal skeletal dynamics can only be revealed with the development of an osteoblast-targeted MN1 gene knock-out that survives postnatally. The profound osteoblastic defects uncovered here provide strong justification for the development of such a model.
In summary, our studies provide key evidence for a significance of MN1 in vitamin D-mediated transcription and in numerous other critical aspects of osteoblast biology. Our findings help to establish MN1 as a critical protein that is needed for appropriate osteoblast growth, differentiation, activity, and function. Understanding detailed molecular and cellular mechanisms underlying these functional aspects of osteoblastic MN1 is an important area of future research efforts.
Supplementary Material
Acknowledgment
We thank Dr. Gerard Grosveld (St. Jude Children's Research Hospital, Memphis, TN) for providing MN1 knock-out mice breeding pairs.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1DK53980 and RO1DK071769 (to P. N. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Fig. 1.
- RANKL
- receptor activator of NF-κB ligand
- α-MEM
- α-minimum essential medium
- 1,25(OH)2D3
- 1,25-dihydroxyvitamin D3
- BSP2
- integrin binding bone sialoprotein
- DMP1
- dentin matrix protein 1
- MN1
- meningioma 1
- OPG
- osteoprotegerin
- TRAP
- tartrate-resistant phosphatase
- VDR
- vitamin D receptor
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- WT
- wild type
- KO
- knock-out
- siRNA
- small interfering RNA
- BrdUrd
- bromodeoxyuridine
- kb
- kilobase pair(s)
- VDRE
- vitamin D response element.
REFERENCES
- 1.Bianco P., Riminucci M., Gronthos S., Robey P. G. ( 2001) Stem Cells 19, 180– 192 [DOI] [PubMed] [Google Scholar]
- 2.Kassem M., Abdallah B. M., Saeed H. ( 2008) Arch. Biochem. Biophys. 473, 183– 187 [DOI] [PubMed] [Google Scholar]
- 3.Aubin J. E. ( 1998) J. Cell. Biochem. Suppl. 30–31, 73– 82 [PubMed] [Google Scholar]
- 4.Miyamoto T., Suda T. ( 2003) Keio J. Med. 52, 1– 7 [DOI] [PubMed] [Google Scholar]
- 5.Teitelbaum S. L., Ross F. P. ( 2003) Nat. Rev. Genet. 4, 638– 649 [DOI] [PubMed] [Google Scholar]
- 6.Darnay B. G., Haridas V., Ni J., Moore P. A., Aggarwal B. B. ( 1998) J. Biol. Chem. 273, 20551– 20555 [DOI] [PubMed] [Google Scholar]
- 7.Galibert L., Tometsko M. E., Anderson D. M., Cosman D., Dougall W. C. ( 1998) J. Biol. Chem. 273, 34120– 34127 [DOI] [PubMed] [Google Scholar]
- 8.Kim H. H., Lee D. E., Shin J. N., Lee Y. S., Jeon Y. M., Chung C. H., Ni J., Kwon B. S., Lee Z. H. ( 1999) FEBS Lett. 443, 297– 302 [DOI] [PubMed] [Google Scholar]
- 9.Lacey D. L., Timms E., Tan H. L., Kelley M. J., Dunstan C. R., Burgess T., Elliott R., Colombero A., Elliott G., Scully S., Hsu H., Sullivan J., Hawkins N., Davy E., Capparelli C., Eli A., Qian Y. X., Kaufman S., Sarosi I., Shalhoub V., Senaldi G., Guo J., Delaney J., Boyle W. J. ( 1998) Cell 93, 165– 176 [DOI] [PubMed] [Google Scholar]
- 10.Xing L., Boyce B. F. ( 2005) Biochem. Biophys. Res. Commun. 328, 709– 720 [DOI] [PubMed] [Google Scholar]
- 11.Yasuda H., Shima N., Nakagawa N., Yamaguchi K., Kinosaki M., Mochizuki S., Tomoyasu A., Yano K., Goto M., Murakami A., Tsuda E., Morinaga T., Higashio K., Udagawa N., Takahashi N., Suda T. ( 1998) Proc. Natl. Acad. Sci. U. S. A. 95, 3597– 3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bord S., Ireland D. C., Beavan S. R., Compston J. E. ( 2003) Bone 32, 136– 141 [DOI] [PubMed] [Google Scholar]
- 13.Mrak E., Villa I., Lanzi R., Losa M., Guidobono F., Rubinacci A. ( 2007) J. Endocrinol. 192, 639– 645 [DOI] [PubMed] [Google Scholar]
- 14.Jurutka P. W., Bartik L., Whitfield G. K., Mathern D. R., Barthel T. K., Gurevich M., Hsieh J. C., Kaczmarska M., Haussler C. A., Haussler M. R. ( 2007) J. Bone Miner. Res. 22, Suppl. 2, V2– 10 [DOI] [PubMed] [Google Scholar]
- 15.Sutton A. L., MacDonald P. N. ( 2003) Mol. Endocrinol. 17, 777– 791 [DOI] [PubMed] [Google Scholar]
- 16.Horwood N. J., Elliott J., Martin T. J., Gillespie M. T. ( 1998) Endocrinology 139, 4743– 4746 [DOI] [PubMed] [Google Scholar]
- 17.Sutton A. L., Zhang X., Ellison T. I., Macdonald P. N. ( 2005) Mol. Endocrinol. 19, 2234– 2244 [DOI] [PubMed] [Google Scholar]
- 18.Lekanne Deprez R. H., Riegman P. H., Groen N. A., Warringa U. L., van Biezen N. A., Molijn A. C., Bootsma D., de Jong P. J., Menon A. G., Kley N. A. ( 1995) Oncogene 10, 1521– 1528 [PubMed] [Google Scholar]
- 19.Carella C., Bonten J., Sirma S., Kranenburg T. A., Terranova S., Klein-Geltink R., Shurtleff S., Downing J. R., Zwarthoff E. C., Liu P. P., Grosveld G. C. ( 2007) Leukemia 21, 1679– 1690 [DOI] [PubMed] [Google Scholar]
- 20.Heuser M., Argiropoulos B., Kuchenbauer F., Yung E., Piper J., Fung S., Schlenk R. F., Dohner K., Hinrichsen T., Rudolph C., Schambach A., Baum C., Schlegelberger B., Dohner H., Ganser A., Humphries R. K. ( 2007) Blood 110, 1639– 1647 [DOI] [PubMed] [Google Scholar]
- 21.Heuser M., Beutel G., Krauter J., Döhner K., von Neuhoff N., Schlegelberger B., Ganser A. ( 2006) Blood 108, 3898– 3905 [DOI] [PubMed] [Google Scholar]
- 22.Buijs A., Sherr S., van Baal S., van Bezouw S., van der Plas D., Geurts van Kessel A., Riegman P., Lekanne Deprez R., Zwarthoff E., Hagemeijer A. ( 1995) Oncogene 10, 1511– 1519 [PubMed] [Google Scholar]
- 23.Carella C., Bonten J., Rehg J., Grosveld G. C. ( 2006) Leukemia 20, 1582– 1592 [DOI] [PubMed] [Google Scholar]
- 24.Kawagoe H., Grosveld G. C. ( 2005) Blood 106, 4269– 4277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawagoe H., Grosveld G. C. ( 2005) Blood 106, 4278– 4286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mrózek K., Döhner H., Bloomfield C. D. ( 2007) Curr. Opin. Hematol. 14, 106– 114 [DOI] [PubMed] [Google Scholar]
- 27.Meester-Smoor M. A., Vermeij M., van Helmond M. J., Molijn A. C., van Wely K. H., Hekman A. C., Vermey-Keers C., Riegman P. H., Zwarthoff E. C. ( 2005) Mol. Cell. Biol. 25, 4229– 4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ellison T. I., Eckert R. L., MacDonald P. N. ( 2007) J. Biol. Chem. 282, 10953– 10962 [DOI] [PubMed] [Google Scholar]
- 29.Bove P. F., Hristova M., Wesley U. V., Olson N., Lounsbury K. M., van der Vliet A. ( 2008) J. Biol. Chem. 283, 17919– 17928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Divieti P., Lanske B., Kronenberg H. M., Bringhurst F. R. ( 1998) J. Bone Miner. Res. 13, 1835– 1845 [DOI] [PubMed] [Google Scholar]
- 31.Sutton A. L., Zhang X., Dowd D. R., Kharode Y. P., Komm B. S., Macdonald P. N. ( 2008) Mol. Endocrinol. 22, 1370– 1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gregory C. A., Gunn W. G., Peister A., Prockop D. J. ( 2004) Anal. Biochem. 329, 77– 84 [DOI] [PubMed] [Google Scholar]
- 33.van Wely K. H., Molijn A. C., Buijs A., Meester-Smoor M. A., Aarnoudse A. J., Hellemons A., den Besten P., Grosveld G. C., Zwarthoff E. C. ( 2003) Oncogene 22, 699– 709 [DOI] [PubMed] [Google Scholar]
- 34.Meester-Smoor M. A., Janssen M. J., Grosveld G. C., de Klein A., van IJcken W. F., Douben H., Zwarthoff E. C. ( 2008) Carcinogenesis 29, 2025– 2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meester-Smoor M. A., Molijn A. C., Zhao Y., Groen N. A., Groffen C. A., Boogaard M., van Dalsum-Verbiest D., Grosveld G. C., Zwarthoff E. C. ( 2007) J. Mol. Endocrinol. 38, 113– 125 [DOI] [PubMed] [Google Scholar]
- 36.Kim S., Yamazaki M., Zella L. A., Shevde N. K., Pike J. W. ( 2006) Mol. Cell. Biol. 26, 6469– 6486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dardenne O., Prud'homme J., Arabian A., Glorieux F. H., St-Arnaud R. ( 2001) Endocrinology 142, 3135– 3141 [DOI] [PubMed] [Google Scholar]
- 38.Li Y. C., Pirro A. E., Amling M., Delling G., Baron R., Bronson R., Demay M. B. ( 1997) Proc. Natl. Acad. Sci. U. S. A. 94, 9831– 9835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olsen B. R., Reginato A. M., Wang W. ( 2000) Annu. Rev. Cell Dev. Biol. 16, 191– 220 [DOI] [PubMed] [Google Scholar]
- 40.Liu W., Lan Y., Pauws E., Meester-Smoor M. A., Stanier P., Zwarthoff E. C., Jiang R. ( 2008) Development 135, 3959– 3968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Samee N., de Vernejoul M. C., Levi G. ( 2007) Crit. Rev. Eukaryotic Gene Expression 17, 173– 186 [DOI] [PubMed] [Google Scholar]
- 42.Hassan M. Q., Tare R., Lee S. H., Mandeville M., Weiner B., Montecino M., van Wijnen A. J., Stein J. L., Stein G. S., Lian J. B. ( 2007) Mol. Cell. Biol. 27, 3337– 3352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dobreva G., Chahrour M., Dautzenberg M., Chirivella L., Kanzler B., Fariñas I., Karsenty G., Grosschedl R. ( 2006) Cell 125, 971– 986 [DOI] [PubMed] [Google Scholar]
- 44.Lecanda F., Warlow P. M., Sheikh S., Furlan F., Steinberg T. H., Civitelli R. ( 2000) J. Cell Biol. 151, 931– 944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kawai S., Yamauchi M., Wakisaka S., Ooshima T., Amano A. ( 2007) J. Bone Miner. Res. 22, 1362– 1372 [DOI] [PubMed] [Google Scholar]
- 46.Sanford L. P., Ormsby I., Gittenberger-de Groot A. C., Sariola H., Friedman R., Boivin G. P., Cardell E. L., Doetschman T. ( 1997) Development 124, 2659– 2670 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.