Abstract
Purpose
Elevated levels of dietary histidine have previously been shown to prevent or mitigate cataract formation in farmed Atlantic salmon (Salmo salar L). The aim of this study was to shed light on the mechanisms by which histidine acts. Applying microarray analysis to the lens transcriptome, we screened for differentially expressed genes in search for a model explaining cataract development in Atlantic salmon and possible markers for early cataract diagnosis.
Methods
Adult Atlantic salmon (1.7 kg) were fed three standard commercial salmon diets only differing in the histidine content (9, 13, and 17 g histidine/kg diet) for four months. Individual cataract scores for both eyes were assessed by slit-lamp biomicroscopy. Lens N-acetyl histidine contents were measured by high performance liquid chromatography (HPLC). Total RNA extracted from whole lenses was analyzed using the GRASP 16K salmonid microarray. The microarray data were analyzed using J-Express Pro 2.7 and validated by quantitative real-time polymerase chain reaction (qRT–PCR).
Results
Fish developed cataracts with different severity in response to dietary histidine levels. Lens N-acetyl histidine contents reflected the dietary histidine levels and were negatively correlated to cataract scores. Significance analysis of microarrays (SAM) revealed 248 significantly up-regulated transcripts and 266 significantly down-regulated transcripts in fish that were fed a low level of histidine compared to fish fed a higher histidine level. Among the differentially expressed transcripts were metallothionein A and B as well as transcripts involved in lipid metabolism, carbohydrate metabolism, regulation of ion homeostasis, and protein degradation. Hierarchical clustering and correspondence analysis plot confirmed differences in gene expression between the feeding groups. The differentially expressed genes could be categorized as “early” and “late” responsive according to their expression pattern relative to progression in cataract formation.
Conclusions
Dietary histidine regimes affected cataract formation and lens gene expression in adult Atlantic salmon. Regulated transcripts selected from the results of this genome-wide transcription analysis might be used as possible biological markers for cataract development in Atlantic salmon.
Introduction
A cataract is defined as the loss of transparency of the eye lens. The eye lens is composed of two types of cells, an outer monolayer of epithelial cells and underlying fiber cells, which are nourished by the outer monolayer. As the lens grows, epithelial cells differentiate into fiber cells covering the older layers of fiber cells like the skins of an onion. The fiber cells eventually lose their nuclei and other organelles. The further the fiber cells are from the epithelial cells, the lower the metabolic activity. The fiber cells contain the major lens proteins, the crystallins. These proteins are highly ordered and tightly packed, which enables light to pass through the clear lens and to be absorbed by the retina where vision occurs [1]. Cataracts can be caused by a variety of factors including physical damage, oxidative stress, age, and genetic predisposition. Several nutrient deficiencies have been found to provoke cataracts. Since cataracts are a major problem for humans, especially elderly people, several mammalian models, mostly rodents, have been developed to study the disease. However, cataracts are not unique to mammals. They have also been observed in populations of wild and farmed fish, mainly Atlantic salmon (Salmo salar L) [2]. For the fish farming industry, this constitutes a serious problem with the potential for economic losses. Affected fish have reduced growth rates and increased susceptibility to secondary diseases compared to healthy fish [3]. Numerous nutritional factors have been related to cataract formation in farmed fish [4], and during the last few years, advances in feed composition have reduced both the incidence and severity of cataract outbreaks.
Dietary levels of the essential amino acid histidine (His) above the suggested minimum requirement for salmonids of 7 g His/kg diet [5] have been found to prevent or slow the progression of cataract development in Atlantic salmon smolts [6-9]. The His derivative N- acetyl histidine (NAH) is a major component of the salmon lens free amino acid pool. Lens NAH concentrations directly reflect dietary His levels, and NAH has therefore been established as a lens-specific marker for the His status of salmon [6,9]. It has been proposed that NAH may act as an osmolyte in the goldfish lens, transporting water out of the cell along the NAH gradient followed by immediate hydrolysis and active uptake of acetate and His back into the cell [10]. Studies with Atlantic salmon have supported a role of NAH in lens water homeostasis, although the exact mechanism remains unknown [6]. Additional possible cataract preventative functions of His and His-related compounds include anti-oxidation [11,12], anti-inflammation [13], anti-glycation [14], and buffering capacity [15].
However, at present, it is still unclear how His prevents or mitigates cataract development in salmon, and the molecular basis of cataractogenesis in the salmon lens is unclear. Increased knowledge of these underlying mechanisms would enable us to better advise the fish farming industry on how to eliminate risk factors leading to cataract development, especially in connection with the increased inclusion of alternative feed resources in aquaculture. This would not only improve fish welfare but may also increase fish production with low additional cost. Research performed in teleost fish may also contribute to our understanding of cataract development in higher vertebrates including humans.
The aim of this study was to shed light on the mechanisms by which dietary His prevents or delays cataract development in Atlantic salmon. Using microarray analysis of the transcriptome in lenses of salmon that were fed diets with different His content, we screened for differentially expressed genes in search for a model explaining cataract development in salmon and possible markers for early cataract diagnosis.
Methods
Fish feeding experiment
The feeding experiment was performed at Lerang Research Station (Lerang, Norway). The experimental procedures were approved by and animals handled according to the guidelines of the Norwegian State Commission for Laboratory Animals. Atlantic salmon in their second year in sea with a mean start weight of 1,662 g (n=1,834) were fed three diets containing low (L), medium (M), or high (H) levels of His (L: 9 g/kg diet; M: 13 g/kg diet; H: 17 g/kg diet) in duplicate sea net pens. The diets were based on a commercial feed and had a similar overall composition (protein: 375 g/kg; fat: 342 g/kg; ash: 73 g/kg; moisture 83 g/kg). The trial, which was run from June to October, 2006, was divided into three experimental periods defined by two intermediate sampling points in July and September in addition to start and end point sampling. At all sampling points, tissue was sampled and cataract status diagnosed by slit-lamp biomicroscopic inspection of both eyes. The cataract score per lens was assessed on a scale from zero (clear lens) to four (completely clouded lens), summing up to a possible maximum score of eight per fish [16]. We screened for differences in the lens transcriptome in two selected dietary groups, the low-His group LLL (diet L during all three experimental periods; sampled after the third period) and the medium-His group MMM (diet M during all three experimental periods; sampled after the third period). Each dietary group contained 11 biological replicates.
Tissue sampling
The fish were anesthetized with metacaine and killed by a blow to the head. The lens was dissected quickly after opening the cornea by an incision along the limbus. Muscle tissue attached to the lens was removed, and the lens was cleaned of aqueous humor by rolling it gently on bench paper. The lens was then immediately frozen in a 2 ml RNase-free microcentrifuge tube by placing the tube on dry ice. Of each sampled fish, the right eye lens was used for RNA extraction while the left eye lens was used for NAH analysis. The lenses were stored at -80 °C until RNA isolation.
NAH analysis
Lens NAH concentrations were analyzed by isocratic reverse phase high performance liquid chromatography (HPLC) with ultraviolet (UV) absorbance at 210 nm using external standard calibration as previously described by Breck and coworkers [9].
RNA purification
The samples were homogenized on day one using a Retsch MM 301 homogenizer (Retsch Gmbh, Haan, Germany) and were then further processed on the four successive days in randomized order. The number of samples belonging to each group was balanced for each of the four days. Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA). Genomic DNA was eliminated from the samples by DNase treatment (DNA-free; Ambion, Austin, TX). RNA for microarray analysis was further purified using the RNeasy MinElute Cleanup Kit (Qiagen, Hilden, Germany). The amount and purity of the isolated RNA was measured with a NanoDrop ND-1000 UV-Vis Spectrophotometer (NanoDrop Technologies, Wilmington, DE). The A260/A280 ratios lay between 2.08 and 2.12 for all RNA samples. RNA quality was determined with the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). One of the samples had a RNA integrity number (RIN) of 7.9, and the others lay between 8.1 and 9.2. The isolated RNA was stored at -80 °C.
Microarray experiment
A common reference design with a pool of all RNA samples as the reference was used for the two-channel microarray experiment. All samples were labeled with Cy5, and the reference was labeled with Cy3. The RNA was hybridized to 16K GRASP v. 2.0 arrays [17] on a Tecan HS 4800™ hybridization station (Tecan Group Ltd., Männedorf, Switzerland). The arrays were scanned with a Tecan LS Reloaded scanner (Tecan Group Ltd.) and analyzed using the Axon GenePix 5.1 software (MDS Inc., Toronto, Canada).
The raw data were filtered and normalized using J-Express Pro v.2.7 [18]. The foreground signal intensity values for each channel were extracted per spot from the data files, and all empty, flagged, and control spots were filtered out before the data were normalized using a nonlinear normalization method, global lowess [19]. After normalization, weak spots with a foreground signal intensity below the sum of the background signal intensity and 1.5 times the standard deviation of the background signal intensity in at least one channel were filtered out. All arrays were compiled into a single expression profile data matrix (gene by sample) containing the log ratio of the two foreground signal intensities. Eexpressed sequence tag (EST) clones with more than 30% missing values were removed from the analysis. Missing values were estimated and replaced using the method introduced by Bø et al. [20], LSimpute_adaptive.
Correspondence analysis (CA) [21], significance analysis of microarrays (SAM) [22], and hierarchical clustering of samples and transcripts were performed on the sub-data sets in J-Express. Functional annotation of the transcripts in the data sets was done using the Blast2GO platform [23]. The Gossip tool [24] integrated in Blast2GO was used for functional enrichment analysis applying Fisher's exact test. The microarray experiment was designed to comply with the Minimum Information about a Microarray Experiment (MIAME) guidelines [25]. The applied protocols and final results were uploaded to BASE. MIAME-compliant microarray data were finally uploaded to the ArrayExpress database (accession number: E-TABM-678).
Quantitative real-time PCR
The results of the microarray experiment were validated by two-step quantitative real-time polymerase chain reaction (qRT–PCR) of selected transcripts that were up-regulated or down-regulated in the low-His group. Primers were designed within the coding sequences using Primer3Plus [26]. Isoform-specific primers were used to amplify sodium/potassium-transporting ATPase subunit alpha-1C (ATPA1C) [27]. We tested four potential reference genes that had shown constant expression rates among the experimental groups in the microarray experiment. Three of them have been previously used as reference genes in qRT–PCR analysis in Atlantic salmon [28]. An overview over the target genes and the respective PCR primers is given in Table 1.
Table 1. Transcript names, short names, accession numbers, primer sequences, amplicon sizes, and fold change expression values obtained by microarray and qRT–PCR for selected transcripts.
| Transcript name | Short name | Accession Number | Forward primer | Reverse primer | Amplicon size | FC by microarray | FC by qRT–PCR |
|---|---|---|---|---|---|---|---|
| 40S ribosomal protein S20 |
RPS20 |
BG936672 |
GCAGACCTTATCCGTGGAGCTA |
TGGTGATGCGCAGAGTCTTG |
85 |
||
| HSP90-beta |
HSP90B |
AF135117 |
CTCTGGGATGAGCTCCTCACA |
CCTTTGACCTCTTTGAGAACAAGAA |
98 |
||
| Elongation factor 1AA |
EF1AA |
AF321836 |
CCCCTCCAGGACGTTTACAAA |
CACACGGCCCACAGGTACA |
57 |
||
| Elongation factor 1AB |
EF1AB |
BG933853 |
TGCCCCTCCAGGATGTCTAC |
CACGGCCCACAGGTACTG |
59 |
||
| Metallothionein B |
MT-B |
CK990996 |
TGAATAAAGAAGCGCGATCAAA |
CTGGTGCATGCGCAGTTG |
111 |
1.8 |
1.5* |
| Fatty acid binding protein |
FABP2 |
CB511332 |
TTACGGAAGGTGCTGGATTC |
GGGCATCAATGTGATGAAGA |
108 |
−2.2 |
−2.5* |
| Gamma crystallin M2 |
CryG M2 |
CB510188 |
GGAGAAGATGTCACGGTGGT |
CCCCCTACAGAGGAGCCTAC |
125 |
−1.8 |
−1.8* |
| Ependymin precursor |
EPN |
CA042089 |
TCCAGTTTAGCGTCCTGACA |
GACAAGACCGGCTGGATACT |
104 |
−1.7 |
−2.3* |
| Fructose-bisphosphate aldolase B |
Aldo b |
CA043730 |
CCTGTGTGGTCATCTCTCCAT |
TGACAAGGGTGTCCTCTTCC |
116 |
−1.7 |
−1.1 |
| Sodium/potassium-transporting ATPase subunit alpha-1 precursor |
ATPA1C |
CA054630 |
AGGGAGACGTACTACTAGAAAGCAT |
CAGAACTTAAAATTCCGAGCAGCAA |
85 |
1.4 |
1.5* |
| Calpain small subunit 1 |
CAPNS1 |
CB505442 |
AGGCACCCAATGAAGTTGTC |
CTCAGGGCTCATCTCCTCAC |
144 |
1.4 |
1.5* |
| Glyceraldehyde-3-phosphate dehydrogenase |
GAPDH |
CA051897 |
GGTATCCCTTCATGGGTCCT |
CGTGTCAGTGGTGGACCTAA |
104 |
1.2 |
1.8* |
| Ubiquitin-conjugating enzyme E2 D4 |
UbE2D4 |
CB508919 |
CATTGCATACTTTTGAGTCCAATC |
ACCCAGATGACCCCCTAGTC |
101 |
−1.3 |
−1.4* |
| Cold-inducible RNA-binding protein |
Cirbp |
CA062835 |
TGAGCTTCGACACCACAGAG |
ACGAGACCTTCCCGTTTCTT |
104 |
1.3 |
1.1 |
| Peroxiredoxin-6 |
PRDX6 |
CA062947 |
TCAGGGATGTTTGGAGGAAC |
GGGGCGTAACTTTGATGAGA |
126 |
−1.2 |
−1.1 |
| SPARC precursor |
SPARC |
CA052160 |
CCAAGGCGATGTACTTGTCA |
GGTTCCTGTCCCACACAGAG |
117 |
1.3 |
1.3* |
| Lens fiber membrane intrinsic protein |
LIM2 |
CA039129 |
CCAGGAAGTTCACCGTCACT |
TAATCGCAGGCATTCTCTCC |
147 |
−1.3 |
−1.3* |
| Heat shock cognate 70 kDa protein |
HSC71 |
CA767816 |
ACCTCGTTGCACTTCTCCAG |
GCAGCGTGACAAGGTCTCTT |
138 |
1.2 |
1.2* |
| Ferritin. heavy subunit |
Ferritin H |
CA052852 |
GTGTGGGTCGTTGTGTTCAG |
AGTGGGGTAGTGGTGTGGAG |
110 |
1.2 |
−1 |
| Phospholipid hydroperoxide glutathione peroxidase. mitochondrial precursor | GPX4 | CB505439 | GGCTGTTCCTTCATCCACTT | GCCAGGTACAGAGGTGGAAA | 126 | 1.2 | 1.1 |
The first four rows in the table contain the selected reference genes that are not differentially expressed in lenses from the different dietary groups. Expression of the 16 transcripts in the subsequent rows was significantly different between the dietary groups when analyzed by microarray and SAM, but only 11 of the transcripts were significantly different when analyzed by qRT–PCR analysis and Mann–Whitney test (FCs are marked with an asterisk in the table).
Total lens RNA (500 ng) was reverse transcribed to cDNA using TaqMan Reverse Transcription Reagents (Applied Biosystems, Foster City, CA). Each RNA sample was reverse transcribed in triplicates. A standard curve composed of a six-point twofold serial dilution (1,000–31.25 ng) of a pool of all RNA samples was run in triplicates to calculate real-time PCR efficiencies for each gene. All cDNA samples were diluted 1:4 in Milli-Q water (Millipore, Billerica, MA). Real-time PCR was performed on 384 well plates in a reaction volume of 10 μl containing 1X Light Cycler 480 SYBR Green I Master (Roche Applied Science, Basel, Switzerland), gene specific primers (0.5 μM each), and 2 µl cDNA template. A melting curve analysis was applied to confirm the amplification of the expected gene-specific product.
The second derivative maximum method was applied to calculate crossing point (CP) values using the Lightcycler 480 Software (Roche Applied Science). CP values were further converted into quantities using gene-specific efficiency values calculated from the standard curves according to the geNorm manual [29]. Dividing the mean of the triplicate quantities for each sample by a normalization factor led to mean normalized expression (MNE) values for the particular genes. The normalization factor was determined using the geNorm VBA applet for Microsoft Excel version 3.4 [29]. All four potential reference genes tested were highly stable with gene expression stability (M) values below 0.3, and hence all four were used to calculate the normalization factor.
Statistical analysis
Differences in the lens NAH contents between LLL and MMM fish were tested by t-test, and differences in the cataract scores between LLL and MMM fish were tested by the Mann–Whitney test. Individual lens NAH concentrations were correlated to cataract scores by Spearman rank order test. The qRT–PCR data were analyzed by Mann–Whitney test, and correlation between fold change (FC) values obtained by microarray and qRT–PCR was tested by Spearman rank order test using GraphPad Prism version 5.01 for Windows (GraphPad Software, San Diego, CA). Correlation between individual cataract scores and gene expression values was tested by Spearman rank order test using the Statistica data analysis software system version 7.1. (StatSoft Inc., Tulsa, OK).
Results
Cataract scores and lens NAH concentrations
During the second and third experimental period, the fish developed cataracts with different severity depending on the dietary His regimes. Fish that were fed the low-His diet during the first and second period had a higher cataract frequency and severity than fish that were fed the medium- and high-His diet (Figure 1) [30]. The medium-His group was selected for microarray analysis to avoid possible negative effects of too high His concentrations in the high-His group. At the end of the trial, when samples for the microarray experiment were taken, there were significant differences in both cataract severity (Mann–Whitney test, p=0.001) and lens NAH concentration (t-test, p<0.001) between the low-His group and the medium-His group. The mean NAH concentration was 4.4±0.8 μmol/g (mean±SEM) in the low-His group and 10.4±0.3 μmol/g in the medium-His group. The individual single lens cataract scores and lens NAH concentrations are shown in Figure 2A. Lens NAH concentrations were significantly negatively correlated to the respective cataract scores (Spearman rank test; r=-0.63, p<0.002, n=22), which is shown in Figure 2B.
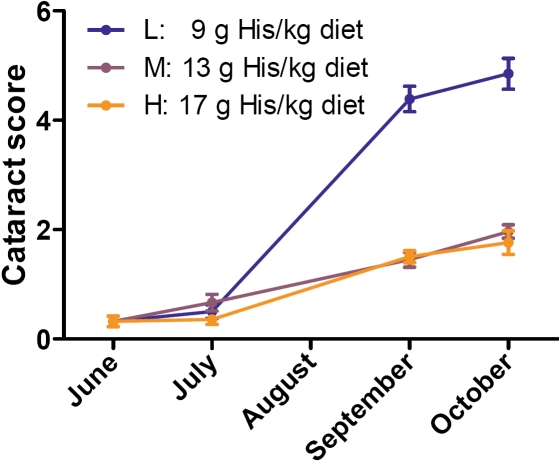
Figure 1.
Cataract scores in selected dietary groups throughout the experimental period. The cataract score for each dietary group is given as the mean of the sums of the scores for both eyes, resulting in a possible maximum score of 8 (4 for each lens). Error bars show the standard error of the mean (SEM). The number of fish per group (n) varied from 31 to 113.
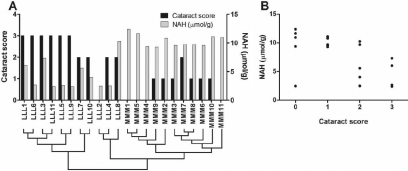
Figure 2.
Individual cataract scores and N-acetyl histidine (NAH) concentrations in lenses of the fish used for microarray analysis. The right lens of the fish was used for microarray analysis, and thus the cataract scores (on a scale from 0 to 4) of the right lens are presented in the graphs. The NAH concentrations were determined in the left lens of the same fish. A: Cataract scores and NAH concentrations for the individual samples are shown in this graph. Under the sample names, the sample clustering (obtained by hierarchical clustering of genes and samples, see Figure 4) is shown to relate individual cataract scores and NAH concentrations to gene expression patterns (the closer the samples are in the cluster tree, the more similar is the lens transcriptome). B: Lens NAH concentrations were significantly negatively correlated to the cataract scores of the right lens (Spearman rank test; r=−0.63, p<0.002, n=22).
Correspondence analysis plot
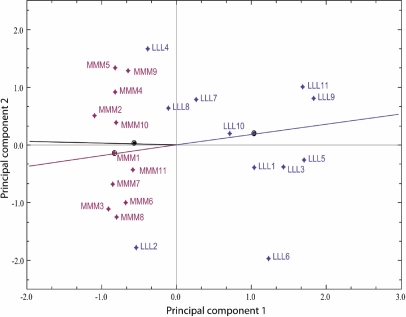
Global differences in lens gene expression between the dietary groups were analyzed by microarray. After the pre-processing and filtering steps, the data set contained 4,242 transcripts. Correspondence analysis (CA) [21] was applied to look for associations between the samples and expression levels of the transcripts in the data set. Deviations from the null hypothesis (no association between samples and expression levels) add to the total χ2. This total χ2 is decomposed in the CA plot shown in Figure 3 where the two largest dimensions, analogous to the principal components in factor analysis, are plotted on the x- and y-axis. The LLL and MMM samples were clearly separated along the first principal component (PC 1), which is the dimension explaining the largest amount of variance in the data set. The lines plotted from the point of origin through the group medians formed an angle of nearly 180°, indicating a clear separation of the dietary groups.
Figure 3.
Correspondence analysis plot. The principal components 1 and 2, which explain the highest amounts of variance in the data set, are shown on the x- and y-axis of the plot, respectively. The samples are colored according to the dietary groups. The low-His samples (LLL) are blue, and the medium-His samples (MMM) are dark red. The dark red and blue lines are plotted from the point of origin through the respective group medians, which are marked by an equally colored dot. The total variance retained in the plot is 16.349%, the x-axis component variance is 10.623%, and the y-axis component variance is 5.726%.
Significance analysis of microarrays
Significance analysis of microarrays (SAM) [22] ranks the transcripts in a data set according to the regularized t-score that it calculates. It also provides a q value, which is a measure of the statistical significance of the differences in expression levels between the compared groups. The q value is a false discovery rate, which states the expected number of false positives on the list. In other words, SAM ranks the transcripts according to the significance of the difference in expression levels between the two dietary groups. On top of the SAM ranking list (Appendix 1) were 514 transcripts with a significant q value below 5%. Of these transcripts, 248 were up-regulated and 266 were down-regulated in the low-His group (LLL) compared to the medium-His group (MMM). Furthermore, 145 of these 514 transcripts had a highly significant q value of 0% (Table 2). Of these 145 transcripts, 59 transcripts were up-regulated and 86 were down-regulated in the low-His group compared to the medium-His group. The highest FC was 2.1 for the strongest up-regulated transcript and −2.5 for the strongest down-regulated transcript.
Table 2. SAM ranked gene list.
| Rank | Accession Number | Transcript name | d[i] | de[i] | Fold Change | q-value | Regulation category* |
|---|---|---|---|---|---|---|---|
| 1 |
CK990996 |
Metallothionein B |
−7.557 |
−2.868 |
1.757 |
0 |
L |
| 2 |
CA062118 |
PREDICTED: similar to POMT2 [Danio rerio] |
5.977 |
2.89 |
−1.907 |
0 |
L |
| 3 |
CA051877 |
UNKNOWN |
5.144 |
2.66 |
−1.855 |
0 |
E |
| 4 |
CB507722 |
Metallothionein B |
−5.119 |
−2.649 |
1.58 |
0 |
E |
| 5 |
CB493454 |
Lipocalin precursor |
4.976 |
2.521 |
−1.666 |
0 |
E |
| 6 |
CB498358 |
Fatty acid-binding protein. intestinal |
4.875 |
2.439 |
−2.403 |
0 |
L |
| 7 |
CA054659 |
Fatty acid-binding protein. intestinal |
4.766 |
2.382 |
−2.539 |
0 |
L |
| 8 |
CB492836 |
Lipocalin precursor |
4.564 |
2.33 |
−1.639 |
0 |
L |
| 9 |
CA064204 |
Protein S100-B |
4.539 |
2.293 |
−1.579 |
0 |
L |
| 10 |
CN442545 |
Cytochrome c oxidase subunit 3 |
−4.529 |
−2.528 |
1.504 |
0 |
L |
| 11 |
CK990592 |
Metallothionein B |
−4.493 |
−2.447 |
1.549 |
0 |
L |
| 12 |
CB509992 |
Lipocalin precursor |
4.439 |
2.256 |
−1.527 |
0 |
E |
| 13 |
CB496407 |
Betaine aldehyde dehydrogenase |
4.406 |
2.225 |
−1.824 |
0 |
L |
| 14 |
CB515799 |
UNKNOWN |
−4.403 |
−2.385 |
1.638 |
0 |
L |
| 15 |
CA063208 |
UNKNOWN |
4.387 |
2.194 |
−1.951 |
0 |
E |
| 16 |
CA769320 |
Fatty acid-binding protein. intestinal |
4.361 |
2.167 |
−2.247 |
0 |
L |
| 17 |
CB498606 |
Fatty acid-binding protein. intestinal |
4.328 |
2.142 |
−2.372 |
0 |
L |
| 18 |
CB511332 |
Fatty acid-binding protein. intestinal |
4.297 |
2.119 |
−2.156 |
0 |
L |
| 19 |
CB492197 |
Metallothionein A |
−4.276 |
−2.336 |
1.487 |
0 |
L |
| 20 |
CB515213 |
B-cell linker protein |
4.275 |
2.098 |
−1.604 |
0 |
L |
| 21 |
CA046225 |
Metallothionein B |
−4.218 |
−2.294 |
1.512 |
0 |
L |
| 22 |
CA051958 |
Trafficking protein particle complex subunit 3 |
4.213 |
2.079 |
−1.6 |
0 |
L |
| 23 |
CA051480 |
UNKNOWN |
4.2 |
2.06 |
−2 |
0 |
L |
| 24 |
CA044316 |
Excluded (Chimera) |
4.196 |
2.044 |
−1.433 |
0 |
L |
| 25 |
CB494699 |
Gamma crystallin M2 |
4.192 |
2.028 |
−1.544 |
0 |
L |
| 26 |
CK990422 |
UNKNOWN |
4.172 |
2.013 |
−1.669 |
0 |
L |
| 27 |
CA059685 |
UNKNOWN |
−4.163 |
−2.258 |
1.713 |
0 |
E |
| 28 |
CB498630 |
Apolipoprotein Eb precursor |
−4.161 |
−2.225 |
1.818 |
0 |
L |
| 29 |
CA058895 |
Apolipoprotein Eb precursor |
−4.07 |
−2.195 |
2.113 |
0 |
E |
| 30 |
CA064247 |
CD9 antigen |
4.068 |
1.998 |
−1.866 |
0 |
L |
| 31 |
CA053993 |
UNKNOWN |
4.062 |
1.985 |
−1.736 |
0 |
L |
| 32 |
CB499689 |
Chromodomain-helicase-DNA-binding protein 2 |
4.059 |
1.971 |
−1.74 |
0 |
L |
| 33 |
CA055129 |
UNKNOWN |
4.045 |
1.958 |
−1.573 |
0 |
L |
| 34 |
CA042615 |
SH3 domain-binding glutamic acid-rich-like protein |
4.039 |
1.946 |
−1.528 |
0 |
L |
| 35 |
CB510889 |
Putative polypeptide N-acetylgalactosaminyltransferase-like protein 4 |
4.022 |
1.935 |
−1.461 |
0 |
L |
| 36 |
CA061651 |
PREDICTED: similar to calmodulin-dependent phosphodiesterase [Danio rerio] |
3.961 |
1.924 |
−1.81 |
0 |
L |
| 37 |
CA041385 |
Proactivator polypeptide precursor |
−3.888 |
−2.169 |
1.396 |
0 |
E |
| 38 |
CA042089 |
Ependymin precursor |
3.868 |
1.913 |
−1.684 |
0 |
E |
| 39 |
CB510842 |
Calcium-regulated heat stable protein 1 |
3.867 |
1.902 |
−1.952 |
0 |
L |
| 40 |
CB492748 |
pigment epithelium-derived factor [Paralichthys olivaceus] >gi|71063313|gb|AAZ22324.1| pigment epithelium-derived factor [Paralichthys olivaceus] |
3.866 |
1.892 |
−1.625 |
0 |
L |
| 41 |
CA058611 |
Clusterin precursor |
−3.863 |
−2.145 |
1.698 |
0 |
L |
| 42 |
CB510615 |
UNKNOWN |
3.852 |
1.883 |
−1.47 |
0 |
L |
| 43 |
CA063671 |
UNKNOWN |
−3.841 |
−2.123 |
1.751 |
0 |
L |
| 44 |
CA042930 |
UNKNOWN |
3.832 |
1.875 |
−1.612 |
0 |
L |
| 45 |
CA062117 |
Protein FAM44B |
3.795 |
1.865 |
−1.72 |
0 |
L |
| 46 |
CN442558 |
Cytochrome c oxidase subunit 3 |
−3.745 |
−2.103 |
1.433 |
0 |
L |
| 47 |
CA043114 |
Meprin A subunit alpha precursor |
3.744 |
1.855 |
−1.738 |
0 |
L |
| 48 |
CA043730 |
Fructose-bisphosphate aldolase B |
3.74 |
1.847 |
−1.674 |
0 |
L |
| 49 |
CB494396 |
Glycylpeptide N-tetradecanoyltransferase 1 |
3.717 |
1.839 |
−1.719 |
0 |
E |
| 50 |
CB494172 |
Keratin. type II cytoskeletal 8 |
−3.716 |
−2.086 |
1.533 |
0 |
L |
| 51 |
CA052024 |
UNKNOWN |
3.709 |
1.83 |
−1.796 |
0 |
L |
| 52 |
CB510653 |
Salvelinus alpinus metallothionein B gene. introns 1 and 2 and partial cds |
−3.708 |
−2.068 |
1.376 |
0 |
L |
| 53 |
CB506047 |
Thymosin beta-12 |
−3.674 |
−2.052 |
1.504 |
0 |
L |
| 54 |
CA052938 |
UNKNOWN |
3.663 |
1.822 |
−1.828 |
0 |
L |
| 55 |
CA054630 |
Sodium/potassium-transporting ATPase subunit alpha-1 precursor |
−3.66 |
−2.036 |
1.409 |
0 |
L |
| 56 |
CB511371 |
Gamma crystallin M3 |
3.654 |
1.814 |
−1.584 |
0 |
L |
| 57 |
CA063207 |
UNKNOWN |
3.653 |
1.806 |
−1.776 |
0 |
L |
| 58 |
CA055638 |
UNKNOWN |
3.651 |
1.799 |
−1.497 |
0 |
L |
| 59 |
CA044410 |
UNKNOWN |
3.65 |
1.792 |
−1.561 |
0 |
L |
| 60 |
CB498510 |
UNKNOWN |
3.644 |
1.785 |
−1.652 |
0 |
L |
| 61 |
CB510188 |
Gamma crystallin M2 |
3.643 |
1.778 |
−1.825 |
0 |
L |
| 62 |
CB511962 |
Salmo salar TNF-alpha 2 gene. complete cds |
3.613 |
1.77 |
−1.392 |
0 |
L |
| 63 |
CN442525 |
ATP synthase a chain |
−3.603 |
−2.02 |
1.298 |
0 |
L |
| 64 |
CB512365 |
UNKNOWN |
3.6 |
1.764 |
−1.451 |
0 |
E |
| 65 |
CA047979 |
UNKNOWN |
−3.599 |
−2.006 |
1.482 |
0 |
L |
| 66 |
CB508872 |
GDP-L-fucose synthetase |
−3.584 |
−1.992 |
1.388 |
0 |
L |
| 67 |
CA056904 |
UNKNOWN |
3.573 |
1.757 |
−1.75 |
0 |
L |
| 68 |
CK990888 |
UNKNOWN |
−3.567 |
−1.98 |
1.234 |
0 |
L |
| 69 |
CA052962 |
Pentraxin fusion protein precursor |
3.567 |
1.75 |
−1.639 |
0 |
L |
| 70 |
CA062371 |
UNKNOWN |
3.566 |
1.743 |
−1.622 |
0 |
L |
| 71 |
CB517893 |
UNKNOWN |
−3.559 |
−1.967 |
1.657 |
0 |
L |
| 72 |
CB514528 |
Annexin A2-A |
−3.543 |
−1.956 |
1.43 |
0 |
L |
| 73 |
CB493750 |
Thymosin beta-12 |
−3.541 |
−1.944 |
1.53 |
0 |
L |
| 74 |
CB505442 |
Calpain small subunit 1 |
−3.541 |
−1.932 |
1.395 |
0 |
L |
| 75 |
CB502127 |
UNKNOWN |
3.539 |
1.738 |
−1.836 |
0 |
L |
| 76 |
CA039176 |
Gamma crystallin M2 |
3.538 |
1.732 |
−1.321 |
0 |
L |
| 77 |
CB511903 |
Uncharacterized protein C1orf74 homolog |
3.535 |
1.725 |
−1.682 |
0 |
L |
| 78 |
CN442536 |
Cytochrome c oxidase subunit 3 |
−3.534 |
−1.921 |
1.3 |
0 |
L |
| 79 |
CA063802 |
UNKNOWN |
3.514 |
1.719 |
−1.455 |
0 |
L |
| 80 |
CA047126 |
Glyceraldehyde-3-phosphate dehydrogenase |
−3.499 |
−1.911 |
1.378 |
0 |
L |
| 81 |
CA051479 |
UNKNOWN |
3.47 |
1.714 |
−1.928 |
0 |
L |
| 82 |
CA050751 |
Asparaginyl-tRNA synthetase. cytoplasmic |
−3.47 |
−1.899 |
1.39 |
0 |
L |
| 83 |
CA061252 |
Pleckstrin homology-like domain family A member 1 |
3.457 |
1.708 |
−1.62 |
0 |
L |
| 84 |
CB502503 |
Cathepsin L precursor |
3.453 |
1.702 |
−1.306 |
0 |
L |
| 85 |
CB509531 |
Clusterin precursor |
−3.447 |
−1.89 |
1.626 |
0 |
L |
| 86 |
CA057824 |
Apolipoprotein Eb precursor |
−3.436 |
−1.88 |
1.626 |
0 |
E |
| 87 |
CA059732 |
Oncorhynchus mykiss SYPG1 (SYPG1). PHF1 (PHF1). and RGL2 (RGL2) genes. complete cds; DNaseII pseudogene. complete sequence; LGN-like. PBX2 (PBX2). NOTCH-like. TAP1 (TAP1). and BRD2 (BRD2) genes. complete cds; and MHCII-alpha and Raftlin-like pseudogenes. complete sequence |
3.423 |
1.697 |
−1.716 |
0 |
L |
| 88 |
CB513063 |
Perforin-1 precursor |
3.417 |
1.692 |
−1.463 |
0 |
L |
| 89 |
CA768741 |
UNKNOWN |
−3.413 |
−1.871 |
1.357 |
0 |
L |
| 90 |
CK990741 |
60S acidic ribosomal protein P1 |
3.398 |
1.687 |
−1.421 |
0 |
L |
| 91 |
CB512134 |
Myosin light polypeptide 6 |
−3.388 |
−1.863 |
1.423 |
0 |
E |
| 92 |
CB498494 |
Gamma crystallin M2 |
3.363 |
1.681 |
−1.345 |
0 |
L |
| 93 |
CB502483 |
Fructose-bisphosphate aldolase B |
3.358 |
1.676 |
−1.485 |
0 |
L |
| 94 |
CA047466 |
UNKNOWN |
3.344 |
1.671 |
−1.338 |
0 |
L |
| 95 |
CB496999 |
UNKNOWN |
−3.34 |
−1.854 |
1.273 |
0 |
L |
| 96 |
CA060333 |
Hypoxanthine-guanine phosphoribosyltransferase |
3.339 |
1.666 |
−1.267 |
0 |
L |
| 97 |
CA062021 |
UNKNOWN |
−3.323 |
−1.845 |
1.348 |
0 |
L |
| 98 |
CA768027 |
Ubiquitin carboxyl-terminal hydrolase 32 |
3.321 |
1.661 |
−1.436 |
0 |
L |
| 99 |
CA063261 |
Hexokinase-2 |
3.316 |
1.656 |
−1.633 |
0 |
L |
| 100 |
CN442552 |
Cytochrome c oxidase subunit 3 |
−3.312 |
−1.837 |
1.334 |
0 |
L |
| 101 |
CA042961 |
Eukaryotic peptide chain release factor subunit 1 |
−3.309 |
−1.829 |
1.345 |
0 |
L |
| 102 |
CA057781 |
Nascent polypeptide-associated complex subunit alpha |
3.308 |
1.651 |
−1.241 |
0 |
L |
| 103 |
CA045638 |
Ubiquitin-conjugating enzyme E2 D4 |
3.304 |
1.646 |
−1.39 |
0 |
L |
| 104 |
CA052800 |
THO complex subunit 1 |
3.285 |
1.641 |
−1.274 |
0 |
L |
| 105 |
CB511609 |
Cathepsin L precursor |
−3.279 |
−1.821 |
1.367 |
0 |
L |
| 106 |
CB511219 |
Gamma crystallin M2 |
3.279 |
1.636 |
−1.504 |
0 |
E |
| 107 |
CK990761 |
Cytochrome b |
−3.275 |
−1.813 |
1.387 |
0 |
L |
| 108 |
CA056981 |
UNKNOWN |
3.274 |
1.631 |
−1.37 |
0 |
E |
| 109 |
CA063526 |
Oncorhynchus mykiss G-protein (P-ras) mRNA. complete cds |
3.263 |
1.627 |
−1.681 |
0 |
L |
| 110 |
CN442497 |
Cytochrome c oxidase subunit 3 |
−3.259 |
−1.805 |
1.403 |
0 |
L |
| 111 |
CA051104 |
Salmo salar TNF-alpha 2 gene. complete cds |
3.247 |
1.622 |
−1.472 |
0 |
L |
| 112 |
CA055845 |
UNKNOWN |
3.244 |
1.618 |
−1.653 |
0 |
L |
| 113 |
CA056167 |
Clusterin precursor |
−3.209 |
−1.798 |
1.697 |
0 |
L |
| 114 |
CA768207 |
Barrier-to-autointegration factor |
3.206 |
1.613 |
−1.557 |
0 |
L |
| 115 |
CA043681 |
UNKNOWN |
−3.186 |
−1.791 |
1.25 |
0 |
L |
| 116 |
CA047249 |
Excluded (Chimera) |
−3.185 |
−1.784 |
1.63 |
0 |
L |
| 117 |
CA052125 |
UNKNOWN |
−3.185 |
−1.777 |
1.441 |
0 |
L |
| 118 |
CB488101 |
Cold-inducible RNA-binding protein |
−3.184 |
−1.77 |
1.35 |
0 |
L |
| 119 |
CB515267 |
Integral membrane protein 2C |
3.18 |
1.609 |
−1.436 |
0 |
L |
| 120 |
CB499782 |
ADP-ribosylation factor 4 |
3.177 |
1.605 |
−1.585 |
0 |
L |
| 121 |
CB492597 |
Ependymin precursor |
3.17 |
1.6 |
−1.405 |
0 |
E |
| 122 |
CK990775 |
UNKNOWN |
−3.16 |
−1.763 |
1.291 |
0 |
L |
| 123 |
CA051121 |
Homo sapiens calcium homeostasis endoplasmic reticulum protein (CHERP). mRNA |
−3.157 |
−1.756 |
1.308 |
0 |
L |
| 124 |
CB496460 |
hyperosmotic glycine rich protein [Salmo salar] |
−3.147 |
−1.749 |
1.353 |
0 |
L |
| 125 |
CB497026 |
Cathepsin L2 precursor |
−3.146 |
−1.742 |
1.256 |
0 |
L |
| 126 |
CA064587 |
UNKNOWN |
3.146 |
1.596 |
−1.282 |
0 |
L |
| 127 |
CA051534 |
UNKNOWN |
−3.141 |
−1.736 |
1.391 |
0 |
L |
| 128 |
CA052327 |
Mus musculus 10 days neonate skin cDNA. RIKEN full-length enriched library. clone:4732428C20 product:unclassifiable. full insert sequence |
−3.138 |
−1.73 |
1.225 |
0 |
L |
| 129 |
CB512146 |
B-cell linker protein |
3.135 |
1.592 |
−1.468 |
0 |
L |
| 130 |
CA041767 |
UNKNOWN |
3.13 |
1.588 |
−1.399 |
0 |
L |
| 131 |
CB497995 |
Eukaryotic translation initiation factor 4 gamma 1 |
3.127 |
1.584 |
−1.399 |
0 |
L |
| 132 |
CA062835 |
Cold-inducible RNA-binding protein |
−3.122 |
−1.725 |
1.297 |
0 |
L |
| 133 |
CB514425 |
UNKNOWN |
3.114 |
1.58 |
−1.383 |
0 |
L |
| 134 |
CA037913 |
UNKNOWN |
−3.096 |
−1.718 |
1.489 |
0 |
E |
| 135 |
CB501344 |
Uncharacterized protein C8orf4 homolog |
−3.095 |
−1.713 |
1.53 |
0 |
L |
| 136 |
CA039269 |
UNKNOWN |
3.094 |
1.576 |
−1.282 |
0 |
L |
| 137 |
CK990381 |
Beta crystallin B3 |
3.092 |
1.572 |
−1.312 |
0 |
L |
| 138 |
CK990533 |
Keratin. type II cytoskeletal 8 |
−3.077 |
−1.706 |
1.619 |
0 |
L |
| 139 |
CA052374 |
UNKNOWN |
3.071 |
1.568 |
−1.41 |
0 |
L |
| 140 |
CA063499 |
UNKNOWN |
3.055 |
1.564 |
−1.327 |
0 |
L |
| 141 |
CA043660 |
Nuclear receptor 0B2 |
−3.044 |
−1.701 |
1.378 |
0 |
E |
| 142 |
CA064148 |
UNKNOWN |
−3.038 |
−1.696 |
1.271 |
0 |
L |
| 143 |
CB494413 |
UNKNOWN |
−3.009 |
−1.69 |
1.345 |
0 |
L |
| 144 |
CB498981 |
RAB3A-interacting protein |
3.003 |
1.561 |
−1.661 |
0 |
L |
| 145 | CA046217 | UNKNOWN | −3.002 | −1.685 | 1.357 | 0 | L |
The transcripts shown are those with a q-value of 0% with the most significantly differentially expressed transcripts on top of the list. The complete list is given in Appendix 1. Abbreviations used in the table header are: d[i], SAM score for a transcript i; de[i], expected SAM score for a transcript i. * The term "Regulation category" in the header of the last column refers to a categorization of the transcripts determined by appearance of the graphs resulting from correlation of expression levels to individual cataract scores (see Results section). The two categories are named E (“early” regulated transcripts), and L (“late” regulated transcripts).
Hierarchical clustering
Hierarchical clustering of samples and transcripts was performed with the most significantly differentially expressed transcripts in the SAM top list including transcripts with q=0% (Figure 4). The transcripts (on the left side of the heat map) are clustered into two main groups, transcripts up-regulated in the low-His group and transcripts down-regulated in the low-His group. The samples (on top of the heat map) are arranged into three main clusters, representing the two dietary groups. The samples LLL2, LLL4, and LLL8 formed a main cluster together with the MMM samples with many transcripts displaying similar expression levels in this main cluster, which is shown by similar colors. In Figure 2A, the sample clustering is shown in relation to individual cataract scores and lens NAH concentrations to visualize the interactions between lens His status, cataract scores, and gene expression patterns in individual fish of the two dietary groups. There are three main clusters, cluster 1, cluster 2, and cluster 3. While cluster 1 including samples LLL1, LLL6, LLL3, LLL11, LLL5, LLL9, LLL7, and LLL10 is relatively uniform with high cataract scores and low NAH concentrations, cluster 2 with samples LLL2, LLL4, and LLL8 shows both high and low cataract scores and high and low NAH concentrations. In cluster 3 containing all MMM samples, NAH concentrations are equally high, and cataract scores are relatively low.
Figure 4.
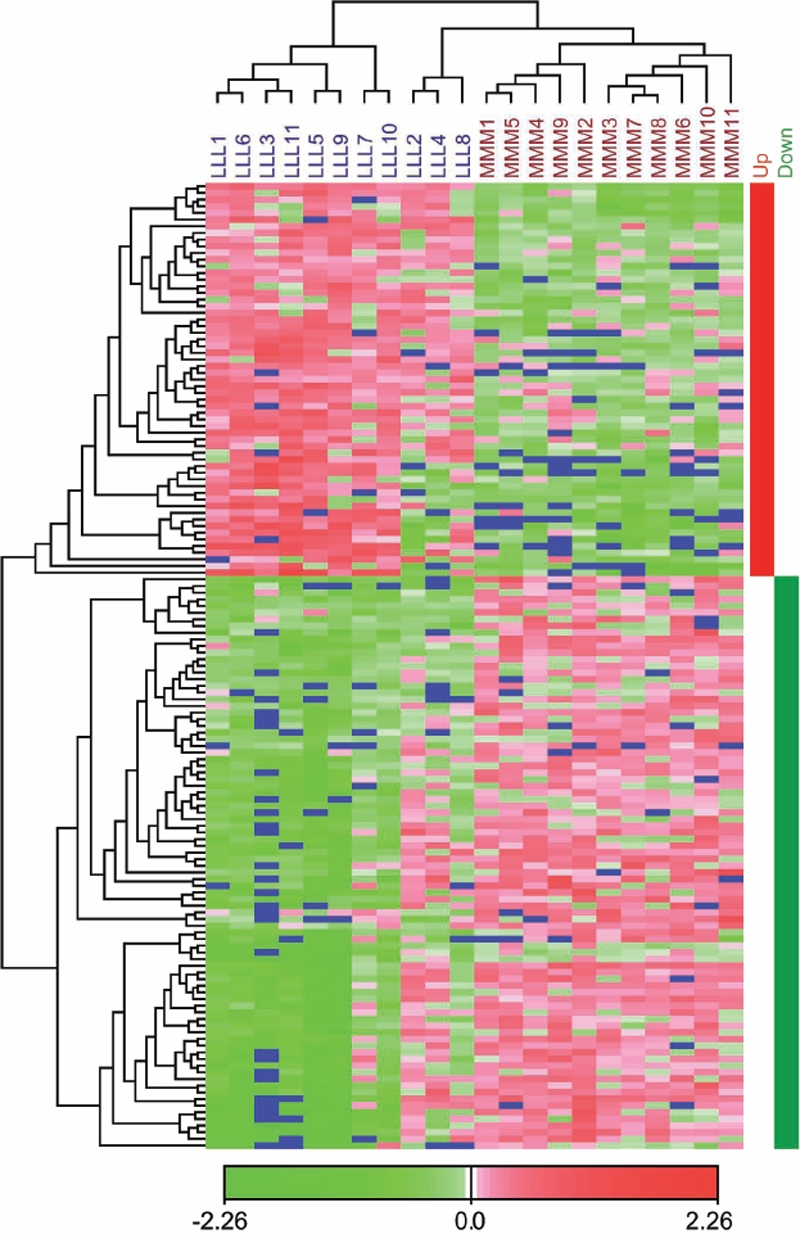
Hierarchical clustering of samples and transcripts. The samples are arranged in columns, and the transcripts are arranged in rows. Only the transcripts with a q-value of 0% in the SAM list were clustered. Negative log intensity ratios are shown in green and positive log intensity ratios are shown in red in the heat map as indicated by the color bar. The blue color represents missing values. The transcripts divide into two distinct clusters. The first cluster contains the transcripts that are up-regulated in the low-His group compared to the medium-His group and is marked by a red bar at the right side of the heat map. The second cluster contains the down-regulated transcripts and is marked by a green bar at the right side of the heat map. The samples divide into three main clusters, reflecting the His feeding regimes. Low-His samples are clearly separated from medium-His samples.
Functional enrichment analysis using Blast2GO
Functional enrichment analysis was performed using the Blast2GO platform with the aim to see if groups of transcripts belonging to the same functional classes were enriched among the most significantly differentially expressed transcripts. The top of the SAM list (including transcripts with q<5%) was compared to the complete SAM list. The complete analysis results can be found in (Appendix 2). Among others, the functional categories described by the following Gene Ontology (GO) terms were enriched with a false discovery rate (FDR) of less than or equal to 5% (with the respective transcript names): “Cysteine-type endopeptidase activity” (Calpain small subunit 1, Cathepsin L precursor, Ubiquitin carboxyl-terminal hydrolase 32, Cathepsin L2 precursor, Calpain-2 catalytic subunit precursor, Cathepsin B precursor, Calpain-2 catalytic subunit), “Glycolysis” (Fructose-bisphosphate aldolase B, Glyceraldehyde-3-phosphate dehydrogenase, Hexokinase-2, Triose phosphate isomerase), and “Lipid metabolic process” (Lipocalin precursor, Clusterin precursor, Sodium/potassium-transporting ATPase subunit alpha-1 precursor, Peroxiredoxin-6, Proactivator polypeptide precursor, Fatty acid binding protein 3 (FABP3), Phospholipid hydroperoxide glutathione peroxidase, mitochondrial precursor, Triose phosphate isomerase, Acyl-CoA-binding protein, Prostaglandin E synthase 3, Diacylglycerol O-acyltransferase 2).
Correlation of gene expression to cataract score and lens NAH concentrations
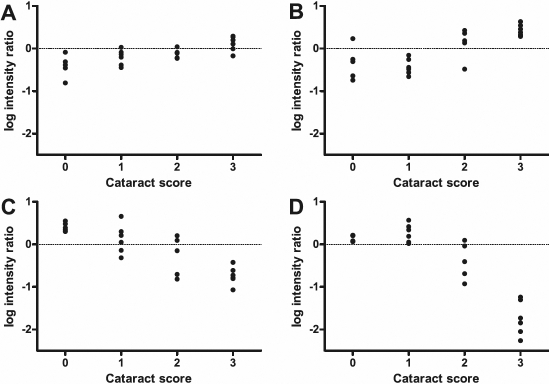
In the microarray experiment, we statistically compared samples from different dietary groups. To further elaborate the results of the microarray experiment, we correlated individual gene expression data of the 145 highly significantly differentially expressed transcripts (q=0% in the SAM top list) to the cataract score and the NAH concentration of the respective lens. The expression of most of the transcripts (99%) was significantly correlated to the cataract score (Spearman rank test; p<0.05, n=22). Similarly, expression of 94% of the transcripts was significantly correlated to the lens NAH concentration (Spearman rank test; p<0.05, n=22). According to their expression pattern relative to the cataract score, the transcripts could roughly be divided into two regulation categories, “early” regulated and “late” regulated transcripts. To illustrate the observed patterns, Figure 5 shows graphs for SPARC precursor (SPARC), metallothionein B (MT-B), ependymin (EPN), and fatty acid binding protein 2 (FABP2).
Figure 5.
Examples of transcripts with different expression patterns related to cataract score. For four selected significantly differentially expressed transcripts, the log intensity ratios are plotted against the cataract score of the respective sample, not taking into account which dietary group the samples belong to. For a certain transcript, if the difference between the mean log intensity ratios of the lenses with a score of 0 and the lenses with a score of 1 was 0.2 or greater, this transcript was classified as “early” regulated. If this difference was less than 0.2, the transcript was classified as “late” regulated. A: SPARC precursor (SPARC; CA052160) was chosen as an example for “early” up-regulated transcripts. B: Metallothionein B (MT-B; CK990996) was chosen as an example of “late” up-regulated transcripts. C: Ependymin (EPN; CA042089) was chosen as an example of “early” down-regulated transcripts. D: Fatty acid binding protein 2 (FABP2; CA054659) was chosen as an example of “late” down-regulated transcripts.
For the “early” regulated transcripts, the expression levels changed continuously from lenses with cataract score 0 to the highest observed cataract score, which was 3. To distinguish between “early” and “late” regulation for a transcript, we used the difference between the mean log intensity ratios of the lenses that scored 0 and the lenses that scored 1. For “early” regulated transcripts, we defined this difference to be 0.2 or greater. “Early” regulated transcripts had either consistently increasing (Figure 5A) or decreasing (Figure 5C) expression levels, or had a maximum in lenses with cataract score 1 and decreasing expression levels at the higher cataract scores (data not shown). In contrast, for the “late” regulated transcripts, there were no apparent differences in expression levels between lenses with a score of 0 and lenses with a score of 1 (the differences in log intensity ratios between the mean of the lenses with score 0 and the mean of the lenses with score 1 were less than 0.2). With more severe cataracts, i.e., higher cataract scores, the expression levels increased (Figure 5B) or decreased (Figure 5D). Appendix 1 and Table 2 summarize which type of regulation category the transcripts with q=0% in the SAM top list could be assigned to. The majority of the transcripts (88%) were found to be “late” regulated.
Validation
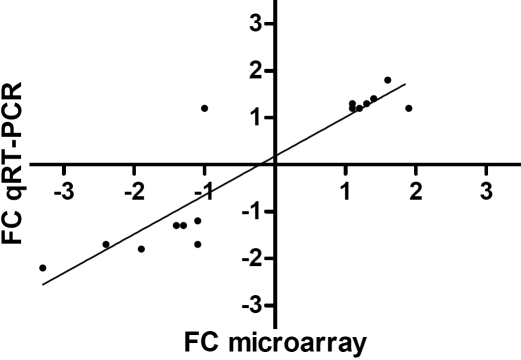
From the transcripts that were significantly up-regulated or down-regulated (q<5%) in the low-His group when compared to the medium-His group in the microarray experiment, we selected sixteen EST clones for qRT–PCR validation. Eleven of these sixteen transcripts were significantly differentially expressed between the two dietary groups when tested by qRT–PCR and Mann–Whitney test, thereby confirming the microarray results. The FC values obtained by microarray and qRT–PCR analysis are listed in Table 1. The FC values of the qRT–PCR results were calculated based on the median of the MNE values of the samples in both dietary groups. There was a significant correlation between the FC values obtained by microarray and qRT–PCR analysis (Spearman rank test; r=0.89, p<0.0001, n=16; Figure 6).
Figure 6.
Correlation between fold change values obtained by microarray analysis and qRT–PCR for 16 selected transcripts. Fold change (FC) values obtained by microarray analysis were significantly correlated to those obtained by qRT–PCR (Spearman rank test; r=0.89, p<0.0001, n=16).
Discussion
The occurrence of cataracts in Atlantic salmon is related to dietary histidine
The present feeding experiment showed that adult Atlantic salmon in sea water that were fed a low-His diet during the first experimental period from June to July and/or the second period from July to September developed severe cataracts, appearing mainly after the second period (Figure 1). However, the levels of dietary His did not affect the growth of the fish in the different feeding groups during the trial [30]. Since there were no other known variables in this feeding experiment, the differences in cataract development and gene expression observed between the dietary groups are assumed to be solely due to dietary His feeding regimes. This assumption was supported by the concentration differences of the His derivative NAH in the lenses (Figure 2A), the concentration of imidazoles in muscle tissue [30], and the strong negative correlation between individual lens NAH concentrations and cataract scores (Figure 2B). A similar His-related cataract development was reported in younger Atlantic salmon smolt after sea transfer [9]. The current His minimum requirement for Atlantic salmon is estimated to be 7 g/kg diet [5]. All experimental diets contained more than 7 g/kg diet. Even so, the incidences of cataract observed during this trial strongly indicate that the current theoretical His minimum requirement level is not sufficient to prevent development of cataracts in adult Atlantic salmon in their second year in sea water.
Methodological considerations on the microarray experiment
The present microarray study was undertaken to explore molecular events connected to the observed dietary His-related cataract development in Atlantic salmon. Interpretation of the microarray data by correspondence analysis, SAM, and hierarchical clustering revealed clear differences in the lens transcriptome between the compared dietary groups. Both by CA of the whole data set (Figure 3) and hierarchical clustering of the significantly differentially expressed transcripts (Figure 4), the three samples LLL2, LLL4, and LLL8 were situated close to the MMM samples, showing that this relation is not only restricted to the highly significantly differentially expressed transcripts. In these three samples, the expression patterns of the significantly differentially expressed transcripts were similar (Figure 4), but the lens NAH concentrations and cataract scores were different (Figure 2A). This might indicate that gene expression is also influenced by individual predisposition.
Our findings clearly confirm that changed gene expression is involved in the process of His-related cataract development. Although the differences in the expression levels, given as fold change (FC) values, were generally low, a considerable number of transcripts were found to be significantly differentially expressed. The main cataract outbreak was registered in the period from July to September, while lenses for the microarray experiment were sampled in October. Some of the observed differences in gene expression levels might thus reflect secondary and compensatory reactions to pathophysiologic changes in the lenses over a longer period of time rather than processes directly involved in cataract development.
In contrast to the human genome and to model species like the mouse and rat, the annotation of the salmon genome is rather poor. The salmonid microarray used in the study is based on EST clones, and the annotation is mainly based on sequence similarities to other species [17]. Only few transcripts on the array are characterized in salmonids. The SAM list contained about 25% uncharacterized EST clones (named UNKNOWN in Appendix 1 and Table 2) while 75% had been identified and were annotated with a transcript name. Using Blast2GO, approximately 67% of these identified transcripts (about 50% of all transcripts in the SAM list) were functionally annotated with at least one GO term. This left about 25% of the transcripts in the SAM list identified but without functional annotation. These 25% were not included in the functional enrichment analysis. An example for this is the intestinal type fatty acid binding protein (FABP2), which was not included in the functional category “Lipid metabolic process”, although it is known to be involved in lipid metabolic processes. Thus, the functional enrichment analysis results do not give a complete view of the functional categories enriched in the data set.
Selected differentially expressed transcripts and their possible role in His-related cataract
The first transcript in the SAM list (Table 2) is an EST clone coding for metallothionein B (MT-B). Metallothionein A (MT-A) is number 19 in the list. Both isoforms are up-regulated in the low-His group. Metallothioneins are multifunctional stress proteins induced by a variety of stresses. They can take part in the detoxification of heavy metals, the regulation of zinc levels in the cell, and the protection against oxidative stress [31]. While heavy metals such as cadmium [32] and lead [33] have been linked to cataract development, oxidative stress is generally one of the major factors associated with cataracts [34,35]. Direct evidence of the involvement of metallothioneins in cataract-related processes has been given by the study of Kantorow et al. [36], who detected increased levels of metallothionein IIA transcripts in human age-related cataractous lenses relative to clear lenses by RT–PCR differential display. Hawse et al. [37] showed later that metallothionein IIA defended lens epithelial cells against oxidative stress induced by cadmium and tertiary butyl hydroperoxide.
We hypothesized that oxidative stress related to cataract triggered the increased expression of metallothioneins in the low-His group and assumed that other antioxidant genes present in the data set might be regulated similarly to MT-B and MT-A. Other transcripts with an antioxidant function that are present in the SAM list with q<5% were peroxiredoxin-6 (PRDX6), phospholipid hydroperoxide glutathione peroxidase (GPX4), selenoprotein Pb precursor, and thioredoxin (TRX; Appendix 1 and Table 2). Only GPX4 was up-regulated. Although significant with fold changes around 1.2, none of the above mentioned antioxidant genes were as clearly regulated as the metallothionein transcripts. In contrast to our first hypothesis, down-regulation of antioxidant genes could lead to decreased antioxidant capacity and increased oxidative stress, which might in turn lead to cataract development. This effect might have been observed in a study on the impact of elevated water oxygen levels on cataract formation in smolting salmon [38]. The authors found trends of down-regulation of the antioxidant genes Cu/Zn superoxide dismutase (Cu/Zn SOD) and glutathione S-transferase (GST) in lenses of the treatment groups that developed more severe cataracts. Despite the suggested antioxidative properties of imidazoles, we cannot conclude which role oxidative stress might play in the present His-related cataracts observed in Atlantic salmon. More confirmative work has to be done to address this question. It also has to be considered that the expression of the stress-responsive antioxidant genes is often rapidly regulated upon the inducing stress. The fact that the development of cataracts in our study probably was more like a chronic stress to the lens further complicates the interpretation of the results.
One of the functional categories of transcripts revealed by functional enrichment analysis was related to lipid metabolism. Among the strongest down-regulated transcripts in the low-His group were lipocalin precursor (presumably coding for a lipocalin-type prostaglandin-D synthase, Ptgds) and the intestinal type fatty acid binding protein (FABP2). Ptgds is one of the most abundantly expressed transcripts in human [39] and zebrafish (Danio rerio) [40] lenses. Ptgds has two functions, the synthesis of the prostaglandin PGD2 in several tissues and binding to a variety of lipophilic ligands like biliverdin, bilirubin, retinaldehyde, and retinoic acid [41]. PGD2 is the major prostaglandin in the central nervous system and is involved in numerous physiologic functions. In the eye, PGD2 lowers the intraocular pressure and triggers inflammatory effects on the conjunctiva [42]. Cataract formation in lens epithelial cells is preceded by programmed cell death (apoptosis) [43]. Ptgds has been shown to protect neurons and oligodendrocytes against apoptosis in a mouse model of a demyelinating disease [44], but also a pro-apoptotic function has been reported [45]. The exact role of Ptgds in the cataractous salmon lens remains to be identified, but it might be involved in lens compensation mechanisms and repair.
FABP2 belongs to the fatty acid binding proteins. The members of this protein family are generally thought to facilitate lipid transport in the cell but may also be involved in lipid signaling pathways [46]. Fatty acid binding protein subtypes are expressed in numerous tissues. The expression of more than one subtype in a cell type indicates specific functions of the subtypes. Our data indicates the expression of FABP2, FABP3, FABP4, and FABP7 in the salmon lens (Appendix 1). In bovine, human, and rat lenses, the expression of the epidermal type fatty acid binding protein (FABP5) has been demonstrated [47,48]. It has recently been shown that FABP2 stimulates mitochondrial β-oxidation and affects the cellular cholesterol transport in human intestine epithelial cells [49]. Given a similar function in the salmon lens, the decreased expression of FABP2 would enhance cholesterol absorption and decrease fatty acid oxidation, leading to decreased energy production in the lenses of fish in the low-His group.
In contrast to Ptgds and FABP2, apolipoprotein Eb (Apo Eb) and clusterin precursor were up-regulated in the low-His group. Apo Eb serves as an extracellular transport protein for cholesterol and other lipids via binding to low density lipoprotein (LDL) receptors on the target cell surface, but also functions in repair response to tissue injury, immunoregulation, and modulation of cell growth and differentiation have been reported [50]. Expression of Apo Eb is activated by peroxisome proliferator-activated receptor γ (PPARγ) [51]. Clusterin is associated with high density lipoprotein (HDL) in the plasma and is also called apolipoprotein J [52,53]. Clusterin is up-regulated in developmental remodeling, apoptotic states in neurodegeneration, response to injuries, and other stresses and interacts with a variety of molecules [54]. Its expression is regulated by the heat shock transcription factor, HSF-1, and clusterin was proposed as an extracellular chaperone [55]. A truncated form acts as a death signal in the nucleus [56] while the normal secreted form promotes cell survival [57,58]. In the cataractous salmon lens, Apo Eb and clusterin might possibly play a role in tissue repair, similar to what is observed in nerve tissue.
However, a pure lipid transporting role of this group of transcripts, which maintain the cellular lipid homeostasis, is supported by the fact that the water temperature at the research station rose from 10 °C to 20 °C from June to July. As an adaptation to the environmental temperature changes, the membrane lipid composition in poikilotherms is changed to maintain fluidity and thus proper function [59]. A rapid and strict regulation of the membrane lipid composition in the salmon lens is assumed to be essential to keep the crystalline lens clear. A temperature-induced change in expression levels, however, would be expected to have declined after three months. Nevertheless, there is some evidence in the literature for the importance of lipids in relation to cataracts. Atlantic salmon that were fed diets based on plant lipid sources in a full life cycle feeding experiment seemed to be more prone to cataract development than fish that were fed diets based on conventional lipids of marine origin [60]. Similarly, age-related cataracts in humans have been related to dietary fat intake. Elevated intakes of 18:2n-6 (linoleic acid) and 18:3n-3 (α-linolenic acid) may increase the risk for cataract [61], while higher intakes of n-3 polyunsaturated fatty acids (PUFA) from fatty fish consumption may contribute to cataract prevention [62]. The study of lipid-related processes in the Atlantic salmon lens will be important to resolve processes leading to cataract development, especially seen in the light of the increasing importance of alternative sustainable lipid sources like plant oils in fish feed.
Glucose is the main energy source for the lens [1]. Several transcripts involved in carbohydrate metabolism were differentially regulated in the low-His group: fructose-bisphosphate aldolase B (down), glyceraldehyde-3-phosphate dehydrogenase (up), hexokinase-2 (down), and triose phosphate isomerase (up). The encoded proteins are all part of the glycolytic pathway, and three of them can also catalyze the reverse reaction in gluconeogenesis. The fourth, hexokinase-2, catalyzes the first step in the glycolytic pathway, and its down-regulation thus indicates a decrease in the energy-producing glycolytic activity in the low-His group. A central enzyme in the non-oxidative pentose phosphate shunt, transaldolase, was also down-regulated in the low-His group. The pentose phosphate shunt is the main source for reduced coenzyme, nicotinamide adenine dinucleotide phosphate (NADPH), which is needed for lipid biosynthesis and to regenerate oxidized glutathione, the major antioxidant in the lens [63,64]. The lack of reduced glutathione might critically impair the redox state of the lens cells and thus promote oxidative damage leading to cataract.
One transcript up-regulated in the low-His group was an EST clone encoding the α-1 subunit of Na+/K+ ATPase. Na+/K+ ATPase plays an important role in the regulation of the Na+ and K+ ion balance and thus the osmotic balance in cells, which is especially important to maintain transparency in the lens. Different Na+/K+ ATPase subunit isoforms are expressed in lenses of different species, and the isoform expression pattern is also specific for cell type and localization in the lens [65]. There are at least five isoforms of the α subunit in Atlantic salmon [66]. In several studies, Na+/K+ ATPase has been found to be involved in cataract-related processes. Its activity was impaired by H2O2-induced oxidative stress in cultured bovine lenses [67] and by the lipid peroxidation product, 4-hydroxynonenal [68]. In cataractous human lenses, the Na+/K+ ATPase activity was found to be decreased [69], and inhibition of the Na+/K+ ATPase activity has been shown to increase opacity in cultured rat lenses [70]. Disturbance of the ion balance by the ionophore, amphotericin B, led to increased Na+/K+ ATPase α-2 expression in porcine lens epithelium [71]. The upregulation of Na+/K+ ATPase α-1 seen in our study might be a sign of disturbed lens ion balance in the low-His group. NAH has been suggested as a major osmolyte in the fish lens [7,10], and the low concentrations of NAH in the low-His group (Figure 2A) suggest a lower preparedness to osmotic challenges and impacts on other actors in osmoregulation. The activity of Na+/K+ ATPase is also very energy-demanding, and the increased expression might be an attempt to compensate for decreased enzymatic activity caused by the lack of energy, resulting from the indicated decrease in glycolytic activity in the low-His group.
Several proteases were up-regulated in the low-His group, the regulatory and catalytic subunits of calpain and cathepsin L and B. Calpain is a calcium-dependent neutral protease that plays a role in the process of apoptosis [72]. Apoptosis has been related to cataract [43], and calpain has been found to be activated in various types of cataracts in rodents [73-75]. Possible calpain substrates in the lens are β-crystallins [76] and aquaporin 0, the main water channel in the lens [77]. Cathepsins are lysosomal cysteine proteases that participate in the degradation of structural proteins in the post-mortem muscle of salmon [78]. Cathepsins also seem to be involved in cataract-related processes since cathepsin A activity in the aqueous humor of cataract patients was found to be increased when compared to the aqueous humor of patients with other ocular diseases [79]. These proteases most probably play roles in secondary repair processes in the cataractous salmon lenses.
Potential early cataractogenesis markers
In addition to conventional microarray data analysis, we explored our data further by correlating individual gene expression values directly to the respective cataract scores and lens NAH concentrations without considering the dietary background. The results of this approach strengthen our findings and confirm the role of lens NAH as a marker for dietary His levels and the impact of dietary His regimes on lens gene expression. According to their expression pattern relative to cataract score, the transcripts could roughly be divided into two regulation categories, “early” regulated and “late” regulated transcripts (Figure 5). “Early” regulated transcripts are probably more directly involved in or affected by cataract development and might be used as biological markers for early cataract detection in future experiments. “Late” regulated transcripts might be induced or repressed by secondary changes and compensatory mechanisms in the cataractous lens. One of the “early” up-regulated transcripts is SPARC, which is also among the most abundant transcripts in the zebrafish lens [40]. SPARC is an extracellular matrix-associated glycoprotein with multiple functions in tissue development and remodeling, cell turnover, and tissue repair [80,81]. Kantorow and coworkers [82] detected increased levels of SPARC transcripts in cataractous lenses when compared to normal lenses, and the same was shown on the protein level [83]. SPARC was also increased in cataractous lenses when compared to normal lenses as revealed by a microarray study [84]. Deletion of SPARC in mice leads to cataract development [85,86]. Emerson and coworkers [87] proposed a chaperone-like activity for SPARC.
One of the “early” down-regulated transcripts is ependymin. Ependymin is a glycoprotein and a major component of the brain extracellular fluid of goldfish (Carassius aureatus) and is involved in neuroplasticity, memory and learning, and tissue regeneration [88]. Its expression is induced by cold in zebrafish and carp (Cyprinus carpio) brain [89]. A trypsin-derived peptide fragment of ependymin activates the transcription factor, AP-1, in mouse neuroblastoma cells [90] and increases the expression of the antioxidant enzymes superoxide dismutase (SOD), catalase (CAT), and glutathione peroxidase (GPX) in rat primary cortical cultures [91]. As mentioned earlier, trends of down-regulation of the antioxidant genes, Cu/Zn SOD and GST, were observed in smolting salmon that were developing cataracts after exposure to elevated water oxygen levels [38]. Further dedicated experiments must be undertaken to strengthen and verify the indications we obtained by relating gene expression levels directly to cataract scores, and to establish the proposed transcripts (or corresponding functional analyses) as markers for early cataractogenesis.
Dietary histidine regimes affected cataract formation in adult Atlantic salmon and lens gene expression. Among the differentially expressed transcripts found in this study were metallothionein A and B as well as transcripts involved in lipid metabolism, carbohydrate metabolism, regulation of ion homeostasis, and protein degradation. In addition to providing new directions for cataract research in Atlantic salmon, the results of this genome-wide transcription analysis allowed us to suggest selected transcripts as possible biological markers for early cataract diagnosis in Atlantic salmon and with a potential for use in mammalian experiments.
Acknowledgments
Skretting ARC is thanked for providing the research facilities at Lerang, and the authors like to thank the staff for rearing the fish and their help with the samplings. Ben F. Koop and Willie Davidson of the Consortium of Genome Research on All Salmon Project at the University of Victoria, Canada (cGRASP) are acknowledged for providing the microarrays. Atle van Beelen Granlund of the Norwegian Microarray Consortium (NMC) is thanked for doing the microarray hybridizations at the national technology platform in Trondheim, supported by the Functional Genomics Program (FUGE) in the Norwegian Research Council (NRC). This research was supported by the NRC Grant 168435/S40.
Appendix 1. SAM complete ranked gene list.
To access the data, click or select the words “Appendix 1.” This will initiate the download of a compressed (pdf) archive that contains the file. The most significantly differentially expressed transcripts are on top of the list. Transcripts with a q-value under 5% are regarded significantly differentially expressed between the two dietary groups. Abbreviations used in the table header are: d[i], SAM score for a transcript i; de[i], expected SAM score for a transcript i. * The term "Regulation category" in the header of the last column refers to a categorization of the transcripts determined by appearance of the graphs resulting from correlation of expression levels to individual cataract scores (see Results section). The two categories are named E (“early” regulated transcripts), and L (“late” regulated transcripts).
Appendix 2. Functional enrichment analysis.
To access the data, click or select the words “Appendix 2.” This will initiate the download of a Microsoft Excel worksheet (xls) file. Functional annotations for the transcripts in the data set were assigned using the Blast2GO software. We compared transcripts with q-values less than 5% in the SAM top list to the complete SAM list to find functional categories overrepresented among the significantly differentially expressed transcripts. Listed are functional categories enriched with a false discovery rate (FDR) of 0.5 or below. In the table headings, “Test” stands for the tested SAM top list with q-values of less than 5%, and “Ref” stands for the remaining transcripts in the SAM list. FWER: family wise error rate.
References
- 1.Brown NAP, Bron AJ, editors. Lens disorders: a clinical manual of cataract diagnosis. Oxford: Butterworth-Heinemann; 1996. [Google Scholar]
- 2.Hughes SG. Nutritional Eye Diseases in Salmonids - a Review. Prog Fish-Cult. 1985;47:81–5. [Google Scholar]
- 3.Menzies FD, Crockford T, Breck O, Midtlyng PJ. Estimation of direct costs associated with cataracts in farmed Atlantic salmon (Salmo salar). Bull Eur Assoc Fish Pathol. 2002;22:27–32. [Google Scholar]
- 4.Bjerkås E, Breck O, Waagbo R. The role of nutrition in cataract formation in farmed fish. CAB Reviews: Perspectives in Agriculture, Veterinary Science, Nutrition and Natural Resources. 2006;1:1–16. [Google Scholar]
- 5.NRC. Nutrient requirements of fish. Washington DC: National Academy Press; 1993. [Google Scholar]
- 6.Breck O, Bjerkas E, Sanderson J, Waagbo R, Campbell P. Dietary histidine affects lens protein turnover and synthesis of N-acetylhistidine in Atlantic salmon (Salmo salar L.) undergoing parr-smolt transformation. Aquacult Nutr. 2005;11:321–32. [Google Scholar]
- 7.Breck O. Histidine nutrition and cataract development in Atlantic salmon, Salmo salar L. Bergen: National Institute of Nutrition and Seafood Research: Department of Fisheries and Marine Biology, University of Bergen; Ph.D. thesis. 2004. [Google Scholar]
- 8.Breck O, Bjerkas E, Campbell P, Arnesen P, Haldorsen P, Waagbo R. Cataract preventative role of mammalian blood meal, histidine, iron and zinc in diets for Atlantic salmon (Salmo salar L.) of different strains. Aquacult Nutr. 2003;9:341–50. [Google Scholar]
- 9.Breck O, Bjerkas E, Campbell P, Rhodes JD, Sanderson J, Waagbo R. Histidine nutrition and genotype affect cataract development in Atlantic salmon, Salmo salar L. J Fish Dis. 2005;28:357–71. doi: 10.1111/j.1365-2761.2005.00640.x. [DOI] [PubMed] [Google Scholar]
- 10.Baslow MH. Function of the N-acetyl-L-histidine system in the vertebrate eye - Evidence in support of a role as a molecular water pump. J Mol Neurosci. 1998;10:193–208. doi: 10.1007/BF02761774. [DOI] [PubMed] [Google Scholar]
- 11.Babizhayev MA, Deyev AI, Yermakova VN, Brikman IV, Bours J. Lipid Peroxidation and Cataracts: N-Acetylcarnosine as a Therapeutic Tool to Manage Age-Related Cataracts in Human and in Canine Eyes. Drugs R D. 2004;5:125–39. doi: 10.2165/00126839-200405030-00001. [DOI] [PubMed] [Google Scholar]
- 12.Babizhayev MA. Antioxidant Activity of L–Carnosine, a Natural Histidine-Containing Dipeptide in Crystalline Lens. Biochim Biophys Acta. 1989;1004:363–71. doi: 10.1016/0005-2760(89)90085-4. [DOI] [PubMed] [Google Scholar]
- 13.Wade AM, Tucker HN. Antioxidant characteristics of L-histidine. J Nutr Biochem. 1998;9:308–15. [Google Scholar]
- 14.Hobart LJ, Seibel I, Yeargans GS, Seidler NW. Anti-crosslinking properties of carnosine: Significance of histidine. Life Sci. 2004;75:1379–89. doi: 10.1016/j.lfs.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 15.Abe H, Dobson GP, Hoeger U, Parkhouse WS. Role of histidine-related compounds to intracellular buffering in fish skeletal muscle. Am J Physiol. 1985;249:R449–54. doi: 10.1152/ajpregu.1985.249.4.R449. [DOI] [PubMed] [Google Scholar]
- 16.Wall T, Bjerkas E. A simplified method of scoring cataracts in fish. Bull Eur Assoc Fish Pathol. 1999;19:162–5. [Google Scholar]
- 17.von Schalburg KR, Rise ML, Cooper GA, Brown GD, Gibbs AR, Nelson CC, Davidson WS, Koop BF. Fish and chips: various methodologies demonstrate utility of a 16,006-gene salmonid microarray. BMC Genomics. 2005;6:126. doi: 10.1186/1471-2164-6-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dysvik B, Jonassen I. J-Express: exploring gene expression data using Java. Bioinformatics. 2001;17:369–70. doi: 10.1093/bioinformatics/17.4.369. [DOI] [PubMed] [Google Scholar]
- 19.Cleveland WS, Devlin SJ. Locally weighted regression - an approach to regression-analysis by local fitting. J Am Stat Assoc. 1988;83:596–610. [Google Scholar]
- 20.Bo TH, Dysvik J, Jonassen I. LSimpute: accurate estimation of missing values in microarray data with least squares methods. Nucleic Acids Res. 2004;32:e34. doi: 10.1093/nar/gnh026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fellenberg K, Hauser NC, Brors B, Neutzner A, Hoheisel JD, Vingron M. Correspondence analysis applied to microarray data. Proc Natl Acad Sci USA. 2001;98:10781–6. doi: 10.1073/pnas.181597298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–6. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 24.Blüthgen N, Brand K, Cajavec B, Swat M, Herzel H, Beule D. biological profiling of gene groups utilizing gene ontology. Genome Inform. 2005;16:106–15. [PubMed] [Google Scholar]
- 25.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, Gaasterland T, Glenisson P, Holstege FC, Kim IF, Markowitz V, Matese JC, Parkinson H, Robinson A, Sarkans U, Schulze-Kremer S, Stewart J, Taylor R, Vilo J, Vingron M. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001;29:365–71. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 26.Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JAM. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007;35:W71–4. doi: 10.1093/nar/gkm306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nilsen TO, Ebbesson LOE, Madsen SS, McCormick SD, Andersson E, Bjornsson BT, Prunet P, Stefansson SO. Differential expression of gill Na+,K+-ATPase {alpha}- and {beta}-subunits, Na+,K+,2Cl- cotransporter and CFTR anion channel in juvenile anadromous and landlocked Atlantic salmon Salmo salar. J Exp Biol. 2007;210:2885–96. doi: 10.1242/jeb.002873. [DOI] [PubMed] [Google Scholar]
- 28.Olsvik PA, Lie KK, Jordal AE, Nilsen TO, Hordvik I. Evaluation of potential reference genes in real-time RT-PCR studies of Atlantic salmon. BMC Mol Biol. 2005;6:21. doi: 10.1186/1471-2199-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tröße C, Breck O, Koppe W, Fontanillas R, Waagbø R. Dietary histidine supplementation prevents cataract development in Atlantic salmon growers in their 2nd year in sea. XIII International Symposium on Fish Nutrition and Feeding; 2008 June 1–5; Florianopolis (Brazil). [Google Scholar]
- 31.Sato M, Bremner I. Oxygen free radicals and metallothionein. Free Radic Biol Med. 1993;14:325–37. doi: 10.1016/0891-5849(93)90029-t. [DOI] [PubMed] [Google Scholar]
- 32.Eriyamremu GE, Ojimogho SE, Asagba SO, Osagie VE. Palm oil induced changes in ocular tissue lipid peroxidation, antioxidant enzymes and ATPases of rabbits in cadmium toxicity. Food Chem Toxicol. 2008;46:3155–8. doi: 10.1016/j.fct.2008.06.088. [DOI] [PubMed] [Google Scholar]
- 33.Neal R, Aykin-Burns N, Ercal N, Zigler JJS. Pb2+ exposure alters the lens [alpha]A-crystallin protein profile in vivo and induces cataract formation in lens organ culture. Toxicology. 2005;212:1–9. doi: 10.1016/j.tox.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 34.Truscott RJW. Age-related nuclear cataract–oxidation is the key. Exp Eye Res. 2005;80:709–25. doi: 10.1016/j.exer.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 35.Williams DL. Oxidation, antioxidants and cataract formation: a literature review. Vet Ophthalmol. 2006;9:292–8. doi: 10.1111/j.1463-5224.2006.00498.x. [DOI] [PubMed] [Google Scholar]
- 36.Kantorow M, Kays T, Horwitz J, Huang Q, Sun J, Piatigorsky J, Carper D. Differential display detects altered gene expression between cataractous and normal human lenses. Invest Ophthalmol Vis Sci. 1998;39:2344–54. [PubMed] [Google Scholar]
- 37.Hawse JR, Padgaonkar VA, Leverenz VR, Pelliccia SE, Kantorow M, Giblin FJ. The role of metallothionein IIa in defending lens epithelial cells against cadmium and TBHP induced oxidative stress. Mol Vis. 2006;12:342–9. [PMC free article] [PubMed] [Google Scholar]
- 38.Waagbø R, Hosfeld CD, Fivelstad S, Olsvik PA, Breck O. The impact of different water gas levels on cataract formation, muscle and lens free amino acids, and lens antioxidant enzymes and heat shock protein mRNA abundance in smolting Atlantic salmon, Salmo salar L. Comp Biochem Physiol A Mol Integr Physiol. 2008;149:396–404. doi: 10.1016/j.cbpa.2008.01.034. [DOI] [PubMed] [Google Scholar]
- 39.Wistow G, Bernstein SL, Wyatt MK, Behal A, Touchman JW, Bouffard G, Smith D, Peterson K. Expressed sequence tag analysis of adult human lens for the NEIBank Project: over 2000 non-redundant transcripts, novel genes and splice variants. Mol Vis. 2002;8:171–84. [PubMed] [Google Scholar]
- 40.Vihtelic TS, Fadool JM, Gao J, Thornton KA, Hyde DR, Wistow G. Expressed sequence tag analysis of zebrafish eye tissues for NEIBank. Mol Vis. 2005;11:1083–100. [PubMed] [Google Scholar]
- 41.Urade Y, Hayaishi O. Biochemical, structural, genetic, physiological, and pathophysiological features of lipocalin-type prostaglandin D synthase. Biochim Biophys Acta. 2000;1482:259–71. doi: 10.1016/s0167-4838(00)00161-8. [DOI] [PubMed] [Google Scholar]
- 42.Woodward DF, Hawley SB, Williams LS, Ralston TR, Protzman CE, Spada CS, Nieves AL. Studies on the ocular pharmacology of prostaglandin D2. Invest Ophthalmol Vis Sci. 1990;31:138–46. [PubMed] [Google Scholar]
- 43.Li WC, Kuszak JR, Dunn K, Wang RR, Ma W, Wang GM, Spector A, Leib M, Cotliar AM, Weiss M, Espy J, Howard G, Farris RL, Auran J, Donn A, Hofeldt A, Mackay C, Merriam J, Mittl R, Smith TR. Lens epithelial cell apoptosis appears to be a common cellular basis for non-congenital cataract development in humans and animals. J Cell Biol. 1995;130:169–81. doi: 10.1083/jcb.130.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taniike M, Mohri I, Eguchi N, Beuckmann CT, Suzuki K, Urade Y. Perineuronal Oligodendrocytes Protect against Neuronal Apoptosis through the Production of Lipocalin-Type Prostaglandin D Synthase in a Genetic Demyelinating Model. J Neurosci. 2002;22:4885–96. doi: 10.1523/JNEUROSCI.22-12-04885.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ragolia L, Hall CE, Palaia T. Post-translational modification regulates prostaglandin D2 synthase apoptotic activity: Characterization by site-directed mutagenesis. Prostaglandins Other Lipid Mediat. 2007;83:25–32. doi: 10.1016/j.prostaglandins.2006.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Makowski L, Hotamisligil GS. Fatty acid binding proteins–the evolutionary crossroads of inflammatory and metabolic responses. J Nutr. 2004;134:2464S–8S. doi: 10.1093/jn/134.9.2464S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jaworski C, Wistow G. LP2, a differentiation-associated lipid-binding protein expressed in bovine lens. Biochem J. 1996;320:49–54. doi: 10.1042/bj3200049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wen Y, Li G-W, Chen P, Wong E, Bekhor I. Lens epithelial cell mRNA, II. Expression of a mRNA encoding a lipid-binding protein in rat lens epithelial cells. Gene. 1995;158:269–74. doi: 10.1016/0378-1119(95)00168-6. [DOI] [PubMed] [Google Scholar]
- 49.Montoudis A, Seidman E, Boudreau F, Beaulieu JF, Menard D, Elchebly M, Mailhot G, Sane AT, Lambert M, Delvin E, Levy E. Intestinal fatty acid binding protein regulates mitochondrion {beta}-oxidation and cholesterol uptake. J Lipid Res. 2008;49:961–72. doi: 10.1194/jlr.M700363-JLR200. [DOI] [PubMed] [Google Scholar]
- 50.Mahley RW. Apolipoprotein E: cholesterol transport protein with expanding role in cell biology. Science. 1988;240:622–30. doi: 10.1126/science.3283935. [DOI] [PubMed] [Google Scholar]
- 51.Galetto R, Albajar M, Polanco JI, Zakin MM, Rodriguez-Rey JC. Identification of a peroxisome-proliferator-activated-receptor response element in the apolipoprotein E gene control region. Biochem J. 2001;357:521–7. doi: 10.1042/0264-6021:3570521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Silva HV, Stuart WD, Park YB, Mao SJ, Gil CM, Wetterau JR, Busch SJ, Harmony JA. Purification and characterization of apolipoprotein J. J Biol Chem. 1990;265:14292–7. [PubMed] [Google Scholar]
- 53.de Silva HV, Stuart WD, Duvic CR, Wetterau JR, Ray MJ, Ferguson DG, Albers HW, Smith WR, Harmony JA. A 70-kDa apolipoprotein designated ApoJ is a marker for subclasses of human plasma high density lipoproteins. J Biol Chem. 1990;265:13240–7. [PubMed] [Google Scholar]
- 54.Jones SE, Jomary C. Clusterin. Int J Biochem Cell Biol. 2002;34:427–31. doi: 10.1016/s1357-2725(01)00155-8. [DOI] [PubMed] [Google Scholar]
- 55.Michel D, Chatelain G, North S, Brun G. Stress-induced transcription of the clusterin/apoJ gene. Biochem J. 1997;328:45–50. doi: 10.1042/bj3280045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang CR, Leskov K, Hosley-Eberlein K, Criswell T, Pink JJ, Kinsella TJ, Boothman DA. Nuclear clusterin/XIP8, an x-ray-induced Ku70-binding protein that signals cell death. Proc Natl Acad Sci USA. 2000;97:5907–12. doi: 10.1073/pnas.97.11.5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang H, Kim JK, Edwards CA, Xu Z, Taichman R, Wang CY. Clusterin inhibits apoptosis by interacting with activated Bax. Nat Cell Biol. 2005;7:909–15. doi: 10.1038/ncb1291. [DOI] [PubMed] [Google Scholar]
- 58.French LE, Wohlwend A, Sappino AP, Tschopp J, Schifferli JA. Human Clusterin Gene-Expression Is Confined to Surviving Cells during in-Vitro Programmed Cell-Death. J Clin Invest. 1994;93:877–84. doi: 10.1172/JCI117043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hazel JR, Williams EE. The role of alterations in membrane lipid composition in enabling physiological adaptation of organisms to their physical environment. Prog Lipid Res. 1990;29:167–227. doi: 10.1016/0163-7827(90)90002-3. [DOI] [PubMed] [Google Scholar]
- 60.Waagbø R, Breck O, Torstensen BE. Dietary lipid regimes can affect cataract development in Atlantic salmon (Salmo salar L.) growers. 11th International Symposium on Nutrition and Feeding in Fish; 2004 May 2-7; Phuket (Thailand). [Google Scholar]
- 61.Lu M, Taylor A, Chylack LT, Jr, Rogers G, Hankinson SE, Willett WC, Jacques PF. Dietary fat intake and early age-related lens opacities. Am J Clin Nutr. 2005;81:773–9. doi: 10.1093/ajcn/81.4.773. [DOI] [PubMed] [Google Scholar]
- 62.Lu M, Cho E, Taylor A, Hankinson SE, Willett WC, Jacques PF. Prospective Study of Dietary Fat and Risk of Cataract Extraction among US Women. Am J Epidemiol. 2005;161:948. doi: 10.1093/aje/kwi118. [DOI] [PubMed] [Google Scholar]
- 63.Lou MF. Redox regulation in the lens. Prog Retin Eye Res. 2003;22:657–82. doi: 10.1016/s1350-9462(03)00050-8. [DOI] [PubMed] [Google Scholar]
- 64.Giblin FJ. Glutathione: a vital lens antioxidant. J Ocul Pharmacol Ther. 2000;16:121–35. doi: 10.1089/jop.2000.16.121. [DOI] [PubMed] [Google Scholar]
- 65.Paterson CA, Delamere NA. ATPases and lens ion balance. Exp Eye Res. 2004;78:699–703. doi: 10.1016/j.exer.2003.09.018. [DOI] [PubMed] [Google Scholar]
- 66.Gharbi K, Ferguson M, Danzmann R. Characterization of Na, K-ATPase genes in Atlantic salmon (Salmo salar) and comparative genomic organization with rainbow trout (Oncorhynchus mykiss). Mol Genet Genomics. 2005;273:474–83. doi: 10.1007/s00438-005-1135-8. [DOI] [PubMed] [Google Scholar]
- 67.Garner WH, Garner MH, Spector A. H2O2-Induced Uncoupling of Bovine Lens Na+,K+-Atpase. Proc Natl Acad Sci USA. 1983;80:2044–8. doi: 10.1073/pnas.80.7.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Siems WG, Hapner SJ, Van Kuijk FJGM. 4-Hydroxynonenal inhibits Na+-K+-ATPase. Free Radic Biol Med. 1996;20:215–23. doi: 10.1016/0891-5849(95)02041-1. [DOI] [PubMed] [Google Scholar]
- 69.Kobatashi S, Roy D, Spector A. Sodium/potassium ATPase in normal and cataractous human lenses. Curr Eye Res. 1982;2:327–34. doi: 10.3109/02713688209000778. [DOI] [PubMed] [Google Scholar]
- 70.Pellegrino de Iraldi A, Peña C, Rodriguez de Lores Arnaiz G. The Effect of an Endogenous Na+, K+-ATPase Inhibitor on Rat Lens Transparency and Ultrastructure. Cell Mol Neurobiol. 2003;23:131–41. doi: 10.1023/A:1022941720546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Delamere NA, Dean WL, Stidam JM, Moseley AE. Influence of amphotericin B on the sodium pump of porcine lens epithelium. Am J Physiol. 1996;270:C465–73. doi: 10.1152/ajpcell.1996.270.2.C465. [DOI] [PubMed] [Google Scholar]
- 72.Squier MK, Miller AC, Malkinson AM, Cohen JJ. Calpain activation in apoptosis. J Cell Physiol. 1994;159:229–37. doi: 10.1002/jcp.1041590206. [DOI] [PubMed] [Google Scholar]
- 73.David LL, Shearer TR, Shih M. Sequence analysis of lens beta-crystallins suggests involvement of calpain in cataract formation. J Biol Chem. 1993;268:1937–40. [PubMed] [Google Scholar]
- 74.David LL, Calvin HI, Fu SCJ. Buthionine Sulfoximine Induced Cataracts in Mice Contain Insolubilized Crystallins with Calpain-Ii Cleavage Sites. Exp Eye Res. 1994;59:501–4. doi: 10.1006/exer.1994.1136. [DOI] [PubMed] [Google Scholar]
- 75.Kadoya K, Azuma M, David LL, Shearer TR. Role of calpain in hydrogen peroxide induced cataract. Curr Eye Res. 1993;12:341–6. doi: 10.3109/02713689308999458. [DOI] [PubMed] [Google Scholar]
- 76.David LL, Azuma M, Shearer TR. Cataract and the acceleration of calpain-induced beta-crystallin insolubilization occurring during normal maturation of rat lens. Invest Ophthalmol Vis Sci. 1994;35:785–93. [PubMed] [Google Scholar]
- 77.Ma H, Azuma M, Shearer TR. Degradation of human aquaporin 0 by m-calpain. FEBS Lett. 2005;579:6745–8. doi: 10.1016/j.febslet.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 78.Yamashita M, Konagaya S. Hydrolytic action of salmon cathepsins B and L to muscle structural proteins in respect of muscle softening. Nippon Suisan Gakkaishi. 1991;57:1917–22. [Google Scholar]
- 79.Obuchowska I, Bankowski E, Stankiewicz A. Cathepsin A activity in the aqueous humor in patients with cataract, absolute glaucoma and intraocular tumors. Klin Oczna. 1999;101:161–3. [PubMed] [Google Scholar]
- 80.Lane TF, Sage EH. The biology of SPARC, a protein that modulates cell-matrix interactions. FASEB J. 1994;8:163–73. [PubMed] [Google Scholar]
- 81.Yan Q, Sage EH. SPARC, a matricellular glycoprotein with important biological functions. J Histochem Cytochem. 1999;47:1495–506. doi: 10.1177/002215549904701201. [DOI] [PubMed] [Google Scholar]
- 82.Kantorow M, Horwitz J, Carper D. Up-regulation of osteonectin/SPARC in age-related cataractous human lens epithelia. Mol Vis. 1998;4:17. [PubMed] [Google Scholar]
- 83.Kantorow M, Huang Q, Yang XJ, Sage EH, Magabo KS, Miller KM, Horwitz J. Increased expression of osteonectin/SPARC mRNA and protein in age-related human cataracts and spatial expression in the normal human lens. Mol Vis. 2000;6:24–9. [PMC free article] [PubMed] [Google Scholar]
- 84.Hawse JR, Hejtmancik JF, Huang Q, Sheets NL, Hosack DA, Lempicki RA, Horwitz J, Kantorow M. Identification and functional clustering of global gene expression differences between human age-related cataract and clear lenses. Mol Vis. 2003;9:515–37. [PMC free article] [PubMed] [Google Scholar]
- 85.Norose K, Clark JI, Syed NA, Basu A, Heber-Katz E, Sage EH, Howe CC. SPARC deficiency leads to early-onset cataractogenesis. Invest Ophthalmol Vis Sci. 1998;39:2674–80. [PubMed] [Google Scholar]
- 86.Gilmour DT, Lyon GJ, Carlton MB, Sanes JR, Cunningham JM, Anderson JR, Hogan BL, Evans MJ, Colledge WH. Mice deficient for the secreted glycoprotein SPARC/osteonectin/BM40 develop normally but show severe age-onset cataract formation and disruption of the lens. EMBO J. 1998;17:1860–70. doi: 10.1093/emboj/17.7.1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Emerson RO, Sage EH, Ghosh JG, Clark JI. Chaperone-like activity revealed in the matricellular protein SPARC. J Cell Biochem. 2006;98:701–5. doi: 10.1002/jcb.20867. [DOI] [PubMed] [Google Scholar]
- 88.Shashoua VE. Ependymin, a Brain Extracellular Glycoprotein, and CNS Plasticity. Ann N Y Acad Sci. 1991;627:94–114. doi: 10.1111/j.1749-6632.1991.tb25916.x. [DOI] [PubMed] [Google Scholar]
- 89.Tang S-J, Sun K-H, Sun G-H, Lin G, Lin W-W, Chuang M-J. Cold-induced ependymin expression in zebrafish and carp brain: implications for cold acclimation. FEBS Lett. 1999;459:95–9. doi: 10.1016/s0014-5793(99)01229-6. [DOI] [PubMed] [Google Scholar]
- 90.Shashoua VE, Adams D, Boyer-Boiteau A. CMX-8933, a peptide fragment of the glycoprotein ependymin, promotes activation of AP-1 transcription factor in mouse neuroblastoma and rat cortical cell cultures. Neurosci Lett. 2001;312:103–7. doi: 10.1016/s0304-3940(01)02119-x. [DOI] [PubMed] [Google Scholar]
- 91.Shashoua VE, Adams DS, Volodina NV, Li H. New synthetic peptides can enhance gene expression of key antioxidant defense enzymes in vitro and in vivo. Brain Res. 2004;1024:34–43. doi: 10.1016/j.brainres.2004.06.086. [DOI] [PubMed] [Google Scholar]