Abstract
Nuclear receptors constitute a large family of ligand-modulated transcription factors that mediate cellular responses to small lipophilic molecules, including steroids, retinoids, fatty acids, and exogenous ligands. Orphan nuclear receptors with no known endogenous ligands have been discovered to regulate drug-mediated induction of cytochromes P450 (CYP), the major drug-metabolizing enzymes. Here, we report the cloning of an orphan nuclear receptor from chicken, termed chicken xenobiotic receptor (CXR), that is closely related to two mammalian xenobiotic-activated receptors, the pregnane X receptor (PXR) and the constitutive androstane receptor (CAR). Expression of CXR is restricted to tissues where drug induction of CYPs predominantly occurs, namely liver, kidney, small intestine, and colon. Furthermore, CXR binds to a previously identified phenobarbital-responsive enhancer unit (PBRU) in the 5′-flanking region of the chicken CYP2H1 gene. A variety of drugs, steroids, and chemicals activate CXR in CV-1 monkey cell transactivation assays. The same agents induce PBRU-dependent reporter gene expression and CYP2H1 transcription in a chicken hepatoma cell line. These results provide convincing evidence for a major role of CXR in the regulation of CYP2H1 and add a member to the family of xenobiotic-activated orphan nuclear receptors.
Members of the cytochrome P450 (CYP) gene superfamily encode for heme proteins involved in the oxidative metabolism of xenobiotic and endogenous substrates in all species (1). In vertebrates, this biotransformation of drugs, steroids, and other compounds occurs predominantly in the liver, whereas extrahepatic tissues such as intestine, kidney, skin, lung, or brain play a less prominent role (2, 3). Expression of some of these CYPs can be transcriptionally regulated by their substrates or other compounds (2–5). Phenobarbital (PB) and other drugs of this class of prototypical inducers display a distinct activation pattern of CYPs. PB-type inducers are a family of compounds that are structurally diverse but share the common general characteristics of being small and lipophilic molecules (3).
Steroid hormones possess similar properties in regard to size and hydrophobicity, which enable them to diffuse into cells and interact with specific intracellular receptors (6, 7). Steroid hormone receptors belong to the superfamily of nuclear receptors with conserved structural features. Also included in this superfamily are receptors with nonsteroid ligands such as thyroid hormone, retinoids, or fatty acids (8–10). Nuclear receptors with known endogenous ligands are distinguished from so-called orphan nuclear receptors, for which true endogenous ligands have not been identified (6, 8, 11). Steroid hormone receptors bind to cognate DNA recognition elements as homodimers, in contrast to most of the nonsteroid nuclear receptors, which heterodimerize with the 9-cis-retinoic acid receptor (RXR, NR2B group) to bind to DNA (7, 11). The recognition elements of steroid hormone receptors consist usually of two hexamer half-sites arranged as a palindrome. Nonsteroid nuclear receptors preferentially bind to two hexamer half-sites organized as direct repeats (DRs) spaced by a variable number of nucleotides (8, 11).
A link between CYP regulation and nuclear receptors has been established with the discovery of the role of several orphan nuclear receptors in the induction of CYPs by xenobiotics. In particular, pregnane X receptor (PXR; NR1I2) and constitutive androstane receptor (CAR; NR1I3) have been shown to play crucial roles in induction of members of the CYP3A and CYP2B subfamilies in human and mouse liver, respectively (reviewed in refs. 12–15). PXR and CAR are activated by the same compounds that induce the respective CYPs and bind as heterodimers with RXR to response elements in 5′-enhancer regions of these genes. Xenobiotic-responsive elements have previously been defined in rat CYP2B2 and CYP3A1, mouse Cyp2b10, and human CYP2B6 and CYP3A4 (reviewed in refs. 13, 15, and 16). These elements are characterized by putative nuclear receptor recognition sites.
Chicken hepatocytes and, more recently, chicken hepatoma cells have been used extensively as a model for elucidating the molecular mechanism of xenobiotic induction (for recent examples, see refs. 17 and 18). In chicken liver, CYP2H1 has been identified as one of the major PB-inducible genes, and a PB-responsive enhancer unit (PBRU) in its 5′-flanking region has been characterized (17, 19). We have shown that a DR-4 element in combination with a nuclear factor-1 (NF-1) site within the CYP2H1 264-bp PBRU is required for the response to drugs (19). Similar arrangements of response elements are conserved in mouse, rat, and human PBRUs. Moreover, drug activation of human and mouse PBRUs in a chicken hepatoma cell line (LMH) strongly suggests conservation of the molecular induction mechanisms between chicken and mammals (19).
In this report, we describe the cloning and functional analysis of a chicken orphan nuclear receptor that is closely related to the NR1I subfamily of mammalian xenobiotic receptors. We have designated the receptor “chicken xenobiotic receptor” (CXR) because it responds to numerous drugs and chemicals and activates a xenobiotic-metabolizing enzyme, chicken CYP2H1. We demonstrate that this receptor mediates the transcriptional control of CYP2H1 by various drugs and chemicals by means of the recently described enhancer unit. Sequence comparison and activation profiles suggest a close relationship of this transcription factor to the mammalian drug-sensing orphan nuclear receptors PXR and CAR.
Materials and Methods
Reagents.
Dexamethasone, metyrapone (2-methyl-1,2-di-3-pyridyl-1-propanone), 5-pregnen-3β-ol-20-one-16α-carbonitrile (PCN), rifampicin, and clotrimazole (1-[o-chlorotrityl]-imidazole) were obtained from Sigma. Propylisopropylacetamide (PIA) was generously provided by P. Sinclair (Veterans Affairs Hospital, White River Junction, VT). Glutethimide and β-naphthoflavone were purchased from Aldrich. RU-486 (mifepristone) was obtained from Roussel-UCLAF, Paris. 1,4-Bis[2-(3,5-dichloropyridyloxy)]benzene (TCPOBOP) was a gift from U. Schmidt (Institute of Toxicology, Bayer, Wuppertal, Germany). PB sodium salt (5-ethyl-5-phenyl-barbituric acid sodium salt) was purchased from Fluka. Poly(dI-dC)⋅poly(dI-dC) was from Amersham Pharmacia Biotech. All other reagents and supplies were obtained from standard sources. Cell culture media, sera, and tissue culture reagents were purchased from Life Technologies, Basel, unless noted otherwise.
Plasmids.
Generation of the reporter plasmid pBLCAT5 containing either the CYP2H1 264-bp PBRU or the CYP2H1 264-bp double mutant has previously been described (19). CXR and chicken RXRγ coding regions (20) were generated by PCR, sequenced, and subsequently cloned into the expression vector pSG5 (Stratagene Europe, Amsterdam).
Culture and Transfection of LMH Cells.
LMH cells were obtained from the American Type Culture Collection and thawed immediately after arrival. Cultivation in William's E medium and transfection with FuGENE 6 Transfection Reagent (Roche Molecular Biochemicals) were performed as described (19).
Isolation of Total and Poly(A)+ RNA.
Tissues from an adult female chicken were frozen in liquid nitrogen and homogenized with a glass–Teflon homogenizer at 4°C in Trizol reagent (Life Technologies), and total RNA from both chicken tissues and LMH cells was obtained according to the supplier's manual. Poly(A)+ RNA was subsequently prepared from total RNA with the Oligotex mRNA Midi Kit (Qiagen, Basel), following the protocol provided by the supplier.
Northern Analysis.
The 820-bp fragment that was used in screening the chicken liver cDNA library described below was 32P-labeled with the Random Primed DNA Labeling Kit (Roche Molecular Biochemicals) in a Klenow reaction using random hexamer primers, followed by purification over a Biospin 6 chromatography column (Bio-Rad). RNA was spectrophotometrically quantified with the GeneQuant II DNA/RNA calculator (Amersham Pharmacia Biotech). Twenty micrograms of total RNA or 15 μg of poly(A)+ RNA was loaded onto a formaldehyde/agarose gel. Northern blotting was carried out according to standard techniques (21). Transfer efficiency and equal RNA loading were controlled by methylene blue staining of the nitrocellulose membrane.
Gel-Mobility-Shift Assays.
Chicken CXR and chicken RXRγ were synthesized in vitro by using the TNT T7 Quick Coupled Transcription/Translation System (Promega) according to the manufacturer's instructions. Probes were labeled with the Klenow fragment of DNA polymerase in the presence of radiolabeled [α-32P]ATP, and the probe was purified over a Biospin 6 chromatography column. A volume of labeled oligonucleotide corresponding to 100,000 cpm was used for each reaction in 10 mM Tris⋅HCl, pH 8.0/40 mM KCl/0.05% Nonidet P-40/6% (vol/vol) glycerol/1 mM DTT containing 0.2 μg of poly(dI-dC)⋅poly(dI-dC) and 2.5 μl of the in vitro synthesized proteins as described previously (22). To test for supershifts, 0.5 μl of monoclonal anti-mouse-RXR rabbit antibody (kindly provided by P. Chambon, Institut de Génétique et de Biologie Moléculaire et Cellulaire, Université Louis Pasteur, Illkirch, France) were added to the reaction mix. This antibody has been positively tested for cross-reaction with the chicken RXRγ in Western blots (data not shown). Unlabeled oligonucleotides for competition experiments were added at a 20-fold molar excess. The mix was incubated for 20 min at room temperature and subsequently electrophoresed on a 6% polyacrylamide gel in 0.5× Tris/borate/EDTA buffer followed by autoradiography at −70°C.
Transcriptional Activation and Chloramphenicol Acetyltransferase (CAT) Reporter Gene Assays.
To perform transactivation assays, CV-1 monkey kidney cells were kept in DMEM/F12 medium without phenol red, supplemented with 10% charcoal-stripped FBS, and were plated in six-well dishes at a density of 625,000 cells per well. A total of 2.5 μg of DNA per well, including 150 ng of receptor expression vector, 400 ng of CAT reporter gene plasmid, 800 ng of pSV-β-galactosidase expression vector (Promega), and carrier plasmid were transfected, and cells were exposed to drugs as described in ref. 22. Cell extracts were prepared and assayed for CAT by using a CAT-ELISA kit (Roche Molecular Biochemicals). β-Galactosidase activities were determined according to ref. 23. CAT concentrations were then normalized against β-galactosidase values to compensate for varying transfection efficiencies.
Semiquantitative PCR with the Taqman System.
One microgram of total RNA was reverse-transcribed with the Maloney murine leukemia virus (MMLV) reverse transcriptase (Roche Molecular Biochemicals). PCR was performed with the Taqman PCR Core Reagent Kit (PE Applied Biosystems, Rotkreuz, Switzerland) and the transcript level was quantitated with an ABI Prism 7700 Sequence Detection System (PE Applied Biosystems, Rotkreuz, Switzerland). CYP2H1 and glyceraldehyde-3-phosphate dehydrogenase transcript levels were measured in separate tubes, and glyceraldehyde-3-phosphate dehydrogenase was used for normalization of the CYP2H1 values as described previously (19).
Results
Isolation and Classification of the Chicken Orphan Nuclear Receptor CXR.
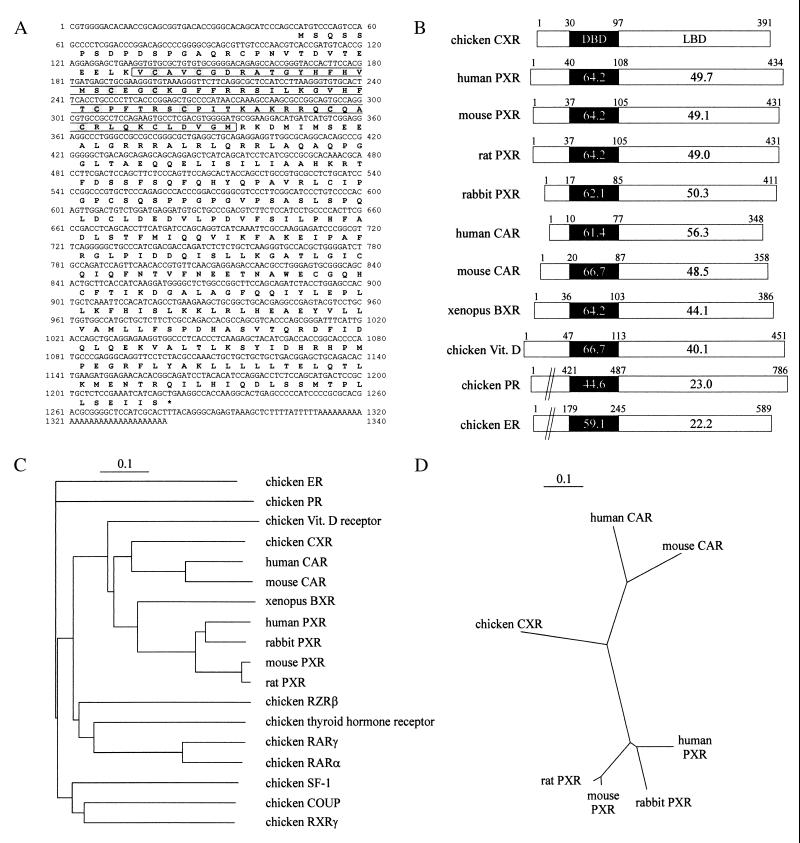
The cDNA sequences of human and mouse PXR and CAR, respectively, were aligned by using the multiple alignment algorithm blocks (24) to identify conserved sequence stretches. With the codehop program, we subsequently screened the aligned sequence stretches for degenerate, 5′-clamped primers accounting for chicken codon usage (25). The primers 5′-GCC TGC CGG CTG CAR AAR TGY YT-3′ and 5′-GGC CCA TCA GCT TGG CRT ANA RRA A-3′ amplified an 820-bp fragment in a hot-start, touchdown PCR with LMH cDNA as template. This fragment was subcloned and used as a probe in the screening of a chicken liver Lambda ZAP phage cDNA library (Stratagene). Two different clones were obtained, subcloned, and sequenced. One clone contained a partial sequence with high homology to mammalian orphan nuclear receptors, whereas the other clone consisted of a different, full-length orphan nuclear receptor cDNA. This full-length clone was denominated “CXR” and its DNA and deduced amino acid sequences are depicted in Fig. 1A where the DNA-binding domain (DBD) is boxed and the conserved cysteines responsible for forming zinc-finger domains (26) are shaded. The DBD and ligand-binding domain (LBD) of CXR were compared with those of other receptors by using the gap algorithm included in the Wisconsin Package Version 10.0 software [Genetics Computer Group (GCG)]. Similarity between DBDs of CXR and other nuclear receptors, including mammalian PXRs (27) and CARs (28, 29), Xenopus benzoate X receptor (BXR, NR1I2, ref. 30) and chicken 1,25-dihydroxyvitamin D3 receptor (NR1I1, ref. 31) was between 61% and 67% amino acid identity (Fig. 1B). By comparison, the DBDs of chicken progesterone receptor (PR; NR3C3) and estrogen receptor (ER; NR3A1) revealed 44.6% and 59.1% amino acid identity with CXR, respectively. The highest similarity to the LBD of CXR was observed in human CAR, with 56.3% amino acid identity. All of the PXRs and mouse CAR exhibit a similarity of about 50% to CXR. When the CXR LBD was compared with the Xenopus benzoate X receptor and the chicken 1,25-dihydroxyvitamin D3 receptor, similarities dropped to 44.1% and 40.1%, respectively. Moreover, the chicken progesterone and estrogen receptor LBDs displayed weak similarity of about 23%. These comparisons position the chicken CXR into the NR1 subfamily of orphan nuclear receptors (8, 10). The same conclusion was reached when generating phylogenetic trees from multiple alignments of full-length amino acid sequences with the ClustalW program (Fig. 1 C and D). Apart from the receptors used in Fig. 1B, the chicken RZRβ (NR1F2), thyroid hormone receptor (NR1A group), RARγ (NR1B3), RARα (NR1B1), SF-1 (NR5A1), COUP (NR2F group), and RXRγ (NR2B3) were included in the trees. The branch lengths of the phylogram (Fig. 1C) and the unrooted tree (Fig. 1D) were calculated from the number of amino acid substitutions per site. The phylogram depicting the chicken CXR within a selection of nuclear receptors also positions CXR into the nuclear receptor subfamily NR1 (8, 10). Together, these results suggest that CXR is closely related to mammalian drug-responsive orphan nuclear receptors of the NR1I group (10).
Figure 1.
CXR is closely related to mammalian drug-responsive orphan nuclear receptors. (A) Nucleotide and predicted amino acid sequence of CXR. The putative DBD is boxed and the conserved cysteines responsible for forming zinc-finger domains are shaded. (B) Amino acid comparison between CXR and a selection of nuclear receptors. The similarity of the DBDs and LBDs between CXR and the selected nuclear receptors is indicated as percentage amino acid identity. (C) Phylogram of full-length amino acid sequences of CXR and related nuclear receptors. (D) Unrooted phylogenetic tree of full-length amino acid sequences of CXR and mammalian CARs and PXRs, respectively. The scale bars for distance measurements represent 0.1 amino acid substitution per site.
Chicken CXR Is Expressed in the Main Drug-Metabolizing Tissues.
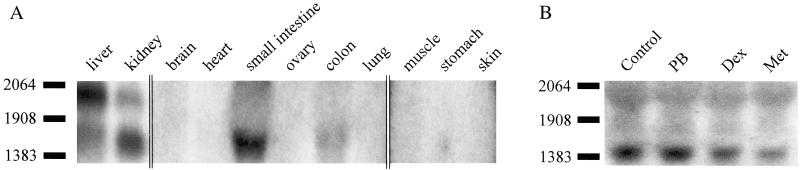
CXR tissue expression was examined by Northern blot analysis of various adult chicken tissues (Fig. 2A). Expression of CXR was detected in liver, kidney, small intestine, and colon but not in any of the other tissues (Fig. 2A). Expression levels in liver and kidney were high, whereas in small intestine and colon, only a faint signal could be detected. Two bands with sizes of about 2 kb and 1.4 kb, respectively, could be observed in both liver and kidney, whereas only the 1.4-kb band was observed in small intestine and colon. In liver, the 2-kb transcript was more abundant than the 1.4-kb mRNA, while in kidney, the 1.4-kb band was more prominent than the 2-kb mRNA. The chicken LMH hepatoma cell line exhibited similar expression levels compared with adult chicken kidney (Fig. 2B). The 1.4-kb transcript was clearly detectable in total RNA of LMH cells, whereas only a very weak signal could be detected for the 2-kb band. No change in the level of CXR mRNA was observed in LMH cells after 24 h of drug induction with vehicle, 400 μM PB, 50 μM dexamethasone, or 400 μM metyrapone (Fig. 2B).
Figure 2.
Expression pattern of CXR. (A) Northern blot analysis of adult chicken tissue. Fifteen micrograms of poly(A)+ RNA was used for all tissues. The three panels separated by bars represent different exposure times of the blot to optimize contrast and background for lanes containing bands with different intensities. (B) Northern blot analysis of LMH cells. LMH cells were induced with vehicle (Contr), PB, dexamethasone (Dex), or metyrapone (Met), and total RNA was isolated, of which 20 μg was used for Northern blots. RNA size markers in number of bases are shown to the left of the blots.
CXR Binds to the Wild-Type Chicken CYP2H1 264-bp PBRU.
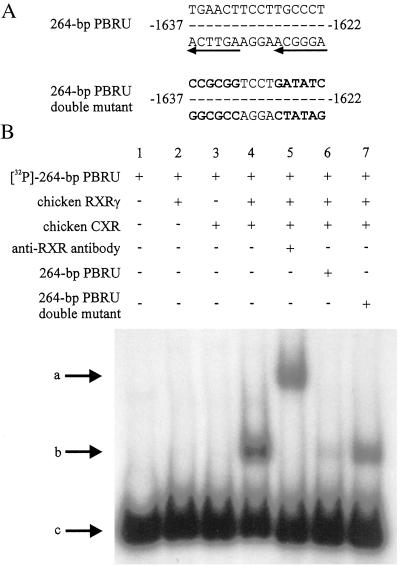
A PB-responsive unit has been identified in the chicken CYP2H1 5′-flanking region, and a conserved nuclear receptor-recognition element has been shown to confer PB inducibility in this element (19). This nuclear receptor-recognition element consists of two hexameric core motifs separated by four nucleotides, a so-called direct repeat (DR-4). Mutations in either hexamer half-site or in both half-sites of this DR-4 abolish PB induction by this element in reporter gene assays (19). The sequences of both the wild-type DR-4 and the double mutation within the 264-bp PBRU are shown in Fig. 3A. As nuclear receptors of the NR1 subfamily have been shown to interact predominantly with DR elements (8) and CXR was found to be a member of this class, a gel-mobility-shift assay was used to determine whether CXR binds to the PB-responsive element from chicken CYP2H1. Neither in vitro transcribed/translated chicken CXR nor chicken RXRγ alone bound to the radiolabeled 264-bp PBRU (Fig. 3B, lanes 2 and 3). Only the CXR-RXRγ heterodimer binds to the PBRU, and this complex could be supershifted with antibodies against RXR (Fig. 3B, lanes 4 and 5). A 20-fold molar excess of unlabeled 264-bp PBRU competed with the radiolabeled probe for CXR/RXRγ binding and therefore reduced the shift. Unlabeled double mutant 264-bp PBRU competed only weakly with the wild-type 264-bp PBRU for CXR/RXRγ binding (Fig. 3B, lanes 6 and 7), further substantiating the importance of the DR-4 element in the 264-bp PBRU.
Figure 3.
CXR binds to the CYP2H1 264-bp PBRU. (A) Sequences of the wild-type DR-4 element within the 264-bp PBRU and the double mutant with knocked-out hexamer half-sites as adapted from ref. 19. (B) Gel-mobility-shift assay. Radiolabeled 264-bp PBRU was incubated with in vitro transcribed/translated CXR (lane 3–7), chicken RXRγ (lanes 2 and 4–7), anti-RXR antibody (lane 5), unlabeled 264-bp PBRU in 20-fold molar excess (lane 6), and unlabeled 264-bp PBRU double mutant in 20-fold molar excess as competitors (lane 7), as indicated. Arrows depict the unbound probe (c), the complex of CXR, RXRγ, and the probe leading to a shift (b), and the complex of CXR, RXRγ, anti-RXR antibody, and the probe resulting in a supershift (a).
Chicken CXR Activates the CYP2H1 264-bp PBRU in CV-1 Cells.
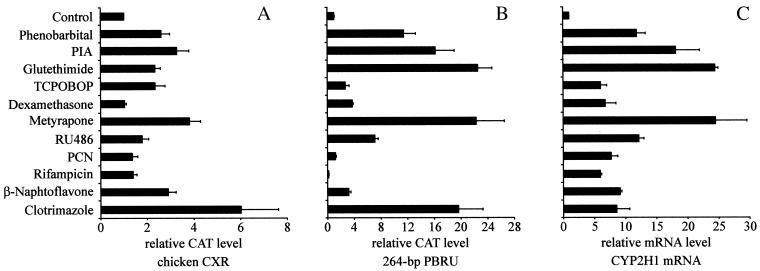
Transactivation assays using a selection of prototypical inducers drugs in CV-1 cells were performed to test whether CXR activates the 264-bp PBRU in correlation with the results from gel-mobility-shift assays. Previous studies have used a wide variety of drugs to investigate nuclear receptor and CYP activation in chicken and mouse (19, 32). Accordingly, we chose PB (400 μM) and the PB-like inducers PIA (250 μM), glutethimide (500 μM), and TCPOBOP (10 μM), a potent inducer in mouse. For comparison, dexamethasone (50 μM), metyrapone (400 μM), and RU-486 (10 μM), three common CYP3A inducers, and the prototypical CYP1A1 inducer β-naphthoflavone (10 μM) were also tested. Furthermore, we were interested in the effects of PCN (50 μM) and rifampicin (100 μM) because of their species-specific effects on PXR activation and CYP3A expression. Clotrimazole (10 μM) was chosen because it has profound effects on mouse and human PXR and CAR (32). CV-1 cells were transfected with chicken CXR and the 264-bp PBRU in a CAT reporter gene vector and treated with drugs for 24 h. None of the drugs had an effect on reporter gene expression in the absence of CXR (data not shown). The classical PB-type inducers all activated CXR and thereby the reporter gene constructs between 2.3- and 3.3-fold (Fig. 4A). Metyrapone was the most potent inducer from the class of typical CYP3A inducers, showing 3.8-fold induction. RU-486, dexamethasone, PCN, and rifampicin, however, were limited to less than 1.8-fold activation. The known CYP1A1 inducer β-naphthoflavone also strongly activated CXR 2.9-fold, which exceeds the effect found after PB treatment. Finally, clotrimazole was the most potent inducer of CXR in these assays, resulting in 6-fold induction (Fig. 4A).
Figure 4.
Activation of CXR by different drugs. (A) CV-1 cells were cotransfected with an expression plasmid for CXR together with a CAT reporter gene plasmid containing the CYP2H1 264-bp PBRU. Cells were then treated with either vehicle or drugs for 24 h. Cell extracts were analyzed for CAT expression normalized against β-galactosidase levels. (B) Activation pattern of the CYP2H1 264-bp PBRU. LMH cells were transfected with the 264-bp PBRU in a CAT reporter gene plasmid and induced for 16 h with the same compounds as in A. CAT levels were determined and normalized against nontreated control cells. Except for the previously unpublished results of clotrimazole, RU-486, and TCPOBOP, data are adapted from ref. 19. (C) Activation pattern of CYP2H1 mRNA. LMH cells were treated with the compounds used in A and B for 16 h. Total RNA was isolated and reverse-transcribed. Relative mRNA levels of CYP2H1 standardized against glyceraldehyde-3-phosphate dehydrogenase levels were obtained by semiquantitative PCR using the Taqman system. Values are the average of three independent experiments and error bars represent standard deviations.
Next, we wanted to investigate the role of CXR on the response element by comparing its activation pattern in CV-1 cells to activation of PBRU reporter gene and transcriptional activation of CYP2H1 mRNA in a physiological environment. LMH cells were therefore transiently transfected with a 264-bp PBRU reporter gene plasmid and were treated with the same drugs as mentioned above for 16 h. CAT levels were measured and normalized against total protein content of the differently treated LMH cells. Relative induction levels of CAT revealed high activation of the 264-bp PBRU with PB, metyrapone, PIA, and glutethimide. Dexamethasone, β-naphthoflavone, PCN, and rifampicin had only minor or no effects. These results have in part been published previously (19) and are listed here for comparison. In addition, clotrimazole was one of the most potent inducers of the 264-bp PBRU in LMH cells, mediating a 20-fold induction. These findings are similar to those for metyrapone, PIA, and glutethimide. Moreover, RU-486 was a weaker inducer (7-fold) compared with clotrimazole, whereas TCPOBOP elevated reporter gene levels only 2.7-fold (Fig. 4B).
With the 264-bp PBRU being only a small part of the CYP2H1 5′-flanking region, overall response of these inducers on CYP2H1 mRNA levels was of interest. All of the compounds induced CYP2H1 mRNA after 16 h of drug exposure. Metyrapone, PIA, and glutethimide were the strongest activators, mediating an 18- to 25-fold increase in CYP2H1 mRNA (Fig. 4C). PB and RU-486 moderately elevated CYP2H1 transcript levels 12-fold, and the remainder of the drugs displayed resulted in only about 6-fold activation (Fig. 4C). Taken together, the transactivation assays provide unequivocal evidence that CXR is a xenobiotic-responsive receptor and that CXR is the major mediator of xenobiotic response through the CYP2H1 PBRU.
Discussion
In this report, we describe the cloning and functional analysis of CXR, a chicken xenobiotic-sensing receptor not previously described. CXR shows all of the structural features typical for a member of the nuclear receptor superfamily, including a conserved DBD with two zinc-finger domains (26) and a conserved LBD (8). CXR heterodimerizes with chicken RXRγ to bind to DNA, as evidenced in gel-mobility-shift assays. This binding occurs on a repeat of hexamer-half-sites derived from the AG(G/T)TCA consensus sequence characteristic for all nuclear receptors (8), and binding is abolished by site-directed mutagenesis of these half-sites. Moreover, these hexamer half-sites are organized as a DR with a 4-bp spacer (DR-4), preferentially bound by nonsteroid liganded nuclear receptors (11). The apparent lack of an endogenous ligand specifies the CXR at this time to be an orphan nuclear receptor (8, 11).
In adult chicken, CXR is expressed in liver, kidney, small intestine, and colon. The same tissues are known for their role in drug metabolism and xenobiotic induction of multiple CYP genes which constitute the major drug-metabolizing enzyme system (2, 3). In the Northern blot, two mRNA bands were detected that showed different distribution in liver compared with kidney and intestine. In liver, a 2-kb transcript was more abundant compared with a 1.4-kb mRNA, whereas the ratio was reversed in kidney. Whether these two bands are splice variants of CXRs or represent different, but closely related, receptors remains to be investigated. Interestingly, a distribution pattern similar to that observed in kidney and intestine was observed in the chicken hepatoma cell line LMH. This might be because of the transformed properties of the cell line or because of its clonal selection. In contrast, chicken liver obviously represents a mixture of different cell types of which only a fraction are drug responsive.
The binding of CXR as heterodimer with chicken RXRγ to a recently identified PBRU from chicken CYP2H1 (19) suggests an essential role of CXR in CYP regulation. Therefore, in search of activators of CXR, we tested a variety of compounds known to induce different CYPs. The activation pattern of CXR in CV-1 cell transactivation assays closely correlated with the activation of the 264-bp PBRU in reporter gene assays in LMH cells and paralleled drug effects on CYP2H1 mRNA. Thus, CXR clearly represents the major receptor involved in activating the CYP2H1 264-bp PBRU. CYP2H1 mRNA was also induced by some compounds that were inactive in reporter gene assays, suggesting additional transcriptional control of CYP2H1 by other orphan nuclear receptors and other recognition sites in the CYP2H1 5′-flanking region.
Sequence comparison of the DBD and LBD of CXR with other nuclear receptors surprisingly revealed almost equal amino acid identity to several receptors of the group NR1I. This group presently consists of the 1,25-dihydroxyvitamin D3 receptor, the Xenopus benzoate X receptor, and the mammalian xenobiotic-sensing receptors PXR and CAR. Among these, CXR displays higher similarity to PXRs and CARs when the full-length amino acid sequences are compared. Calculation of the average evolutionary distance between CXR and human and mouse CARs revealed 0.474 amino acid substitution per site and was slightly smaller than the distance between CXR and mammalian PXRs (0.52 amino acid substitution per site). By comparison, the average distance between PXRs and CARs is 0.55 amino acid substitution per site. Therefore, CXR is almost equally related to PXRs and CARs at the amino acid sequence level. This raises the question of whether CXR represents an orthologue of mammalian PXRs or CARs or, alternatively, a new class of xenobiotic-sensing receptors. Usually, nuclear receptor orthologues share more than 90% amino acid identity in their DBDs and LBDs (8). This is different for the xenobiotic-sensing receptors PXRs and CARs, where the LBDs share only about 70% identity among mammals alone (32). The CXR sequence revealed 61% to 67% amino acid identity in the DBD and 40% to 56% identity in the LBD compared with other members of the NR1I group of nuclear receptors.
However, the evolutionary distance between CXR, PXR, and CAR deviates considerably from that of other chicken orthologues of nuclear receptors. Thus, the similarities between the chicken and mammalian farnesoid X receptor (FXR) and liver X receptor (LXR), the receptors for bile acids and oxysterols, respectively, are around 94% and 86% amino acid identity in the DBD and about 86% and 83% identity in the LBD, respectively (R. Amherd and U.A.M., unpublished results).
We conclude that CXR may well be a new type of xenobiotic-sensing receptor with potential orthologues in mammalian genomes. Alternatively, CXR might represent the ancestral gene that diverged into CAR and PXR in the course of evolution as an adaptive response to different environmental and nutritional pressures. In any case, the basic mechanism of drug induction of enzymes involved in drug metabolism by orphan nuclear receptors apparently is evolutionary conserved over the 300 million years that separate birds from humans.
Acknowledgments
We thank Linda B. Moore and Steven A. Kliewer for helpful suggestions concerning the CV-1 transactivation assays, Francine Hoffmann and Remo Amherd for cloning and providing the chicken RXRγ, and Roland Geiser for help in preparation of the adult chicken tissues. This work was supported by the Swiss National Science Foundation.
Abbreviations
- CYP
cytochrome P450
- CXR
chicken xenobiotic receptor
- PXR
pregnane X receptor
- CAR
constitutive androstane receptor
- PB
phenobarbital
- PBRU
PB-responsive unit
- RXR
9-cis-retinoic acid receptor
- DR
direct repeat
- PIA
propylisopropylacetamide
- PCN
5-pregnen-3β-ol-20-one-16α-carbonitrile
- TCPOBOP
1,4-bis[2-(3,5-dichloropyridyloxy)]benzene
- CAT
chloramphenicol acetyltransferase
- DBD
DNA-binding domain
- LBD
ligand-binding domain
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF276753).
References
- 1.Nelson D R, Koymans L, Kamataki T, Stegeman J J, Feyereisen R, Waxman D J, Waterman M R, Gotoh O, Coon M J, Estabrook R W, Gunsalus I C, Nebert D W. Pharmacogenetics. 1996;6:1–42. doi: 10.1097/00008571-199602000-00002. [DOI] [PubMed] [Google Scholar]
- 2.Okey A B. Pharmacol Ther. 1990;45:241–298. doi: 10.1016/0163-7258(90)90030-6. [DOI] [PubMed] [Google Scholar]
- 3.Waxman D J, Azaroff L. Biochem J. 1992;281:577–592. doi: 10.1042/bj2810577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Denison M S, Whitlock J P., Jr J Biol Chem. 1995;270:18175–18178. doi: 10.1074/jbc.270.31.18175. [DOI] [PubMed] [Google Scholar]
- 5.Dogra S C, Whitelaw M L, May B K. Clin Exp Pharmacol Physiol. 1998;25:1–9. doi: 10.1111/j.1440-1681.1998.tb02135.x. [DOI] [PubMed] [Google Scholar]
- 6.Evans R M. Science. 1988;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beato M, Herrlich P, Schütz G. Cell. 1995;83:851–857. doi: 10.1016/0092-8674(95)90201-5. [DOI] [PubMed] [Google Scholar]
- 8.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schütz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans R M. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kastner P, Mark M, Chambon P. Cell. 1995;83:859–869. doi: 10.1016/0092-8674(95)90202-3. [DOI] [PubMed] [Google Scholar]
- 10.Nuclear Receptors Nomenclature Committee. Cell. 1999;97:161–163. doi: 10.1016/s0092-8674(00)80726-6. [DOI] [PubMed] [Google Scholar]
- 11.Mangelsdorf D J, Evans R M. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 12.Waxman D J. Arch Biochem Biophys. 1999;369:11–23. doi: 10.1006/abbi.1999.1351. [DOI] [PubMed] [Google Scholar]
- 13.Savas U, Griffin K J, Johnson E F. Mol Pharmacol. 1999;56:851–857. doi: 10.1124/mol.56.5.851. [DOI] [PubMed] [Google Scholar]
- 14.Kliewer S A, Lehmann J M, Willson T M. Science. 1999;284:757–760. doi: 10.1126/science.284.5415.757. [DOI] [PubMed] [Google Scholar]
- 15.Honkakoski P, Negishi M. Biochem J. 2000;347:321–337. doi: 10.1042/0264-6021:3470321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodwin B, Hodgson E, Liddle C. Mol Pharmacol. 1999;56:1329–1339. doi: 10.1124/mol.56.6.1329. [DOI] [PubMed] [Google Scholar]
- 17.Dogra S C, Davidson B P, May B K. Mol Pharmacol. 1999;55:14–22. doi: 10.1124/mol.55.1.14. [DOI] [PubMed] [Google Scholar]
- 18.Ourlin J C, Baader M, Fraser D, Halpert J R, Meyer U A. Arch Biochem Biophys. 2000;373:375–384. doi: 10.1006/abbi.1999.1566. [DOI] [PubMed] [Google Scholar]
- 19.Handschin C, Meyer U A. J Biol Chem. 2000;275:13362–13369. doi: 10.1074/jbc.275.18.13362. [DOI] [PubMed] [Google Scholar]
- 20.Rowe A, Eager N S C, Brickell P M. Development (Cambridge, UK) 1991;111:771–778. doi: 10.1242/dev.111.3.771. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 22.Kliewer S A, Moore J T, Wade L, Staudinger J L, Watson M A, Jones S A, McKee D D, Oliver B B, Willson T M, Zetterstrom R H, et al. Cell. 1998;92:73–82. doi: 10.1016/s0092-8674(00)80900-9. [DOI] [PubMed] [Google Scholar]
- 23.Iniguez-Lluhi J A, Lou D Y, Yamamoto K R. J Biol Chem. 1997;272:4149–4156. doi: 10.1074/jbc.272.7.4149. [DOI] [PubMed] [Google Scholar]
- 24.Pietrokovski S, Henikoff J G, Henikoff S. Trends Genet. 1998;14:162–163. doi: 10.1016/s0168-9525(98)01421-8. [DOI] [PubMed] [Google Scholar]
- 25.Rose T M, Schultz E R, Henikoff J G, Pietrokovski S, McCallum C M, Henikoff S. Nucleic Acids Res. 1998;26:1628–1635. doi: 10.1093/nar/26.7.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rastinejad F, Perlmann T, Evans R M, Sigler P B. Nature (London) 1995;375:203–211. doi: 10.1038/375203a0. [DOI] [PubMed] [Google Scholar]
- 27.Jones S A, Moore L B, Shenk J L, Wisely G B, Hamilton G A, McKee D D, Tomkinson N C, LeCluyse E L, Lambert M H, Willson T M, et al. Mol Endocrinol. 2000;14:27–39. doi: 10.1210/mend.14.1.0409. [DOI] [PubMed] [Google Scholar]
- 28.Baes M, Gulick T, Choi H S, Martinoli M G, Simha D, Moore D D. Mol Cell Biol. 1994;14:1544–1552. doi: 10.1128/mcb.14.3.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choi H S, Chung M, Tzameli I, Simha D, Lee Y K, Seol W, Moore D D. J Biol Chem. 1997;272:23565–23571. doi: 10.1074/jbc.272.38.23565. [DOI] [PubMed] [Google Scholar]
- 30.Blumberg B, Kang H, Bolado J, Chen H, Craig A G, Moreno T A, Umesono K, Perlmann T, De Robertis E M, Evans R M. Genes Dev. 1998;12:1269–1277. doi: 10.1101/gad.12.9.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McDonnell D P, Mangelsdorf D J, Pike J W, Haussler M R, O'Malley B W. Science. 1987;235:1214–1217. doi: 10.1126/science.3029866. [DOI] [PubMed] [Google Scholar]
- 32.Moore L B, Parks D J, Jones S A, Bledsoe R K, Consler T G, Stimmel J B, Goodwin B, Liddle C, Blanchard S G, Willson T M, et al. J Biol Chem. 2000;275:15122–15127. doi: 10.1074/jbc.M001215200. [DOI] [PubMed] [Google Scholar]