Abstract
Glycosylation affects the physical properties of proteins in a number of ways including solubility and aggregation behavior. To elucidate the mechanism underlying these effects, we have measured second virial coefficients (A2) of the heavily glycosylated pheniophora lycii phytase (Phy) and its enzymatically deglycosylated counterpart (dgPhy) in native and in denatured form by means of small angle x-ray scattering. The measured A2-values show that the native forms of Phy and dgPhy are equally repulsive at the studied pH 8 where A2 equals 10.9 ± 0.1 × 104 mL mol g−2. However, when thermally denatured, the A2 of dgPhy decreases to 9.0 ± 0.2 × 104 mL mol g−2 whereas it remained unchanged for Phy. In accord with earlier investigations, the p(r)-function measured here suggested that the glycans did not affect the peptide structure of the native protein. Conversely, glycosylation markedly changed the structure of thermally denatured protein. This was evident from the radius of gyration, which increased by 32% for Phy and only 11% for dgPhy on denaturation. We suggest that this expanding effect of the glycans on the denatured protein conformation relies on steric hindrance that limits the range of torsion angles available to the polypeptide.
Introduction
Glycosylation, the most abundant post-translational modification of eukaryotic proteins, has a wide variety of roles including immune regulation (1–3) and providing additional recognition epitopes for receptors (4). These functions depend on the detailed three-dimensional structure of the glycan. On the other hand, glycosylation also brings about a number of less specific biophysical changes that seem to depend on the bulk of the glycan, rather, than its specific three-dimensional structure. These effects include improved solubility (5) and reduced aggregation propensity (6). In fact, one of the major roles of glycosylation seems to be suppression of aggregation of newly synthesized protein in the endoplasmatic reticulum (ER) (7–9). Although much is known about the structure and biosynthesis of glycans in the ER, it is not understood how glycosylation suppresses aggregation of unfolded and partial unfolded aggregation prone species. Various explanations are proposed in literature. Although earlier reports have emphasized the hydrophilic nature of the glycan (5,10–14), more recent studies suggest amphiphilic properties of the glycan to be important for refolding. This latter interpretation is in accord with biophysical investigations that show that the glycans interacts less favorably with water (i.e., are less hydrophilic) than the peptide moiety (15,16). These lines of evidence show that suppression of protein aggregation by glycosylation can not simply be rationalized in terms of solubilizing the protein by attaching a more hydrophilic component. Instead, the studies suggest that the mechanism underlying the increased solubility and reduced aggregation of glycoproteins is likely to be more complex than what have been recognized previously.
To elucidate this mechanism, we have probed the intermolecular interactions of the heavily glycosylated enzyme pheniophora lycii phytase (Phy) and the enzymatically deglycosylated counterpart (dgPhy) in native as well as denatured form by small angle x-ray scattering (SAXS). Furthermore, the use of SAXS allowed us to obtain information on the effect of glycosylation on the structure transitions when going from native to the thermally denatured state.
Materials and Methods
Purification and sample preparation
The heavily glycosylated P. lycii phytase (65 kD) was expressed in Aspergillus oryzae and purified according to procedures published previously (17). The purified product (>95% pure as assessed by SDS-PAGE) was stored in 50 mM sodium acetate, pH 5.5 at −25°C. A portion of this was enzymatically deglycosylated by Endo-β-N-acetylglycosaminidase F1 (Endo F1, EC 2.2.1.96) following procedures described previously (6). This endoglycosidase removes the glycans by hydrolyzing the glycoside bond between the two N-acetylglycosamine groups (17) linking the glycan to the polypeptide, thus, the deglycosylated protein carries one N-acetylglycosamine group at each of the 10 glycosylation sites. Subsequent to deglycosylation reaction only one band was detected on SDS-PAGE at 47–48 kDa and no reduction in enzymatic activity was detected using an enzymatic assay procedure published previously (17). The deglycosylated phytase (dgPhy) as well as glycosylated phytase (Phy) were extensively dialyzed in Spectra/Pore dialyzing membranes (12–14 kDa cutoff) against 50 mM HEPES, pH 8. The protein concentration was determined by UV spectroscopy at 280 nm (ɛ = 1.0 M−1 cm−1), diluted to the appropriate concentration with HEPES, pH 8 and stored at 5°C. Immediately before use, each sample was individually centrifuged for 2 min at 10,000 rpm and the molar concentration of the diluted samples was determined again on a NanoDrop spectrophotometer (Wilmington, DE).
SAXS measurements
The scattering experiments were carried out at The EMBL X33 beamline at the DORIS storage ring (DESY, Hamburg) following standard procedures. The data were collected on a two-dimensional image plate with a momentum transfer range 0.08 < q < 4.96 nm−1 (q = 4 π sin θ/λ, where 2θ is the scattering angle and λ is the x-ray wavelength). Background buffers were measured prior and subsequent to each sample and the averaged background scattering were subtracted from the scattering of the sample. Absolute calibration of the scattering intensity into units of scattering cross section per unit volume (1/cm) was carried out using water as a secondary standard (18). An additional measurement was carried out at MAXlab (Lund, Sweden) on the beamline I711. In this experiment the scattered beam was registered on a MAR165 CCD detector in the q-range 0.1–3.3 nm−1.
SAXS data analysis
The forward scattering (I(0)) was estimated with the Guinier approximation (19):
Where I(q) is the scattering intensity at q, I(0) the forward scattering (scattering intensity extrapolated to zero q) and Rg the apparent radius of gyration. The Guinier fits were made with the data reduction and processing program Primus (20) and with the aim of obtaining accurate I(0) estimates rather than reliable Rg-values. For the lowest concentrations a standard fitting region of q × Rg < 1.3 could be used, whereas, for the concentrated samples the fits were restricted to the lowermost q-range in which a well-defined straight line could be identified. This gave precise estimates of the I(0) for all concentrations, however, the obtained Rg values of the concentrated samples were obviously only apparent values as they were systematically suppressed by particle-particle interaction effects. As a independent control of this approach the I(0) was determined by indirect Fourier transformation using a purposely too large Dmax as transformation parameter. The two methods returned the same I(0)-values within the precision of the data.
The scattering intensity of a solution with finite concentration may be different from that of an ideal dilute solution. The Zimm approximation (21), which is based on the concentration dependence of the osmotic pressure, takes into account this concentration dependence:
Here I(q,c) is the apparent scattering function at concentration c, P(q) is the form factor i.e., scattering contribution from the molecule under ideal dilute conditions (no interparticle interference), M is the molecular mass of the protein and Ai is the ith virial coefficient. For relatively dilute protein solutions (<20 mg/mL) the concentration dependence is normally described adequately by the use of A2 only (22). Therefore, by carrying out the experiments at several concentrations, A2 can be deduced from a linear fit in a Zimm plot.
The molecular weights were calculated using values for the excess scattering length density per unit mass (Δρm) of 1.97 × 1010 cm/g and 2.93 × 1010 cm/g for the peptide and the carbohydrate part, respectively. These values were calculated relative to water and by assuming the mass densities of the peptide, carbohydrate and water to be, respectively, 1.35 g/cm3, 1.60 g/cm3, and 1.00 g/cm3 and their electron densities to be, respectively, 433 e × nm−3, 501 e × nm−3, and 333 e × nm−3, and by using the electron scattering length of 2.81 × 10−13 cm (19). The electron density of the carbohydrate component was calculated based on a glycan containing six mannose and two N-acetylglycosamine groups. Therefore, Δρm = 1.97 × 1010 cm/g was used to calculate the molecular weight of dgPhy whereas Δρm = 2.19 × 1010 cm/g was used to calculate the molecular weight of Phy. The later value was calculated by combining the excess scattering length of the glycan and the peptide Δρ according to their relative mass composition of Phy.
As an independent control, the molecular weight of dgPhy was calculated relative to that of a reference solution of bovine serum albumin (A-0281, Sigma-Aldrich, Steinheim, Germany) of known concentration (∼5 mg/mL). The sample was freshly prepared in cold HEPES buffer and spun at 10,000 rpm for 10 min immediately before use. The deviation of calculated molecular masses found by these two methods was within 2%. Due to the different Δρm-value for glycosylated proteins this method can only be used for nonglycosylated proteins.
The recorded scattering profiles were transformed into real space representation in terms of the so-called pair distance distribution function p(r), by means of indirect Fourier transformation (23) using a Bayesian analysis method (24):
The p(r) may be regarded as a one-dimensional representation of the distances within the particles weighted by the product of their excess scattering length relative to the average of the sample. The height of the p(r) is therefore proportional to the number of possible connection lines with the length r and the product of the excess scattering lengths of the endpoints.
For the purpose of presenting a scattering curve with low noise at high-q and without contribution of particle-particle interference effects at low-q scattering curves of different concentrations were merged. This was done by merging three curves of ∼4 mg/mL, ∼8 mg/mL, and ∼12 mg/mL with the q-ranges 0.081–0.993 nm−1, 0.878–2.363 nm−1, and 2.249–4.954 nm−1, respectively.
Results
Structure of Phy and dgPhy
Representative examples of merged scattering profiles of Phy and dgPhy are shown in Fig. 1. It seems that the glycan mantel of Phy affected the scattering profile of the native protein in several ways. First, this is seen from lowest q-values, where the scattering intensity I(q) of Phy is clearly higher than that of dgPhy. Second, the scattering curve of Phy contains a small concentration independent bump at intermediate q-values (2–3 nm−1), corresponding to real space distances of ∼2.5 nm and we attribute the bump as a form factor contribution from the glycans.
Figure 1.
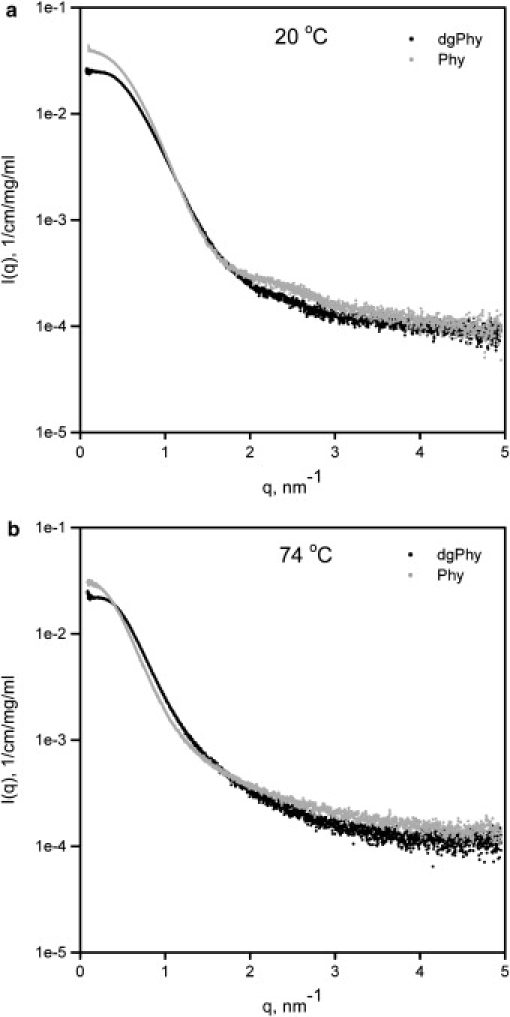
Merged scattering curves (I(q)[1/cm/(mg/mL)] vs. q [nm−1]) of Phy and dgPhy at (a) 20°C and (b) 74°C. Note the small bump in the scattering curve (2–3 nm−1) of Phy at 20°C that originates from the polysaccharides. Also note the curvature Phy that is steeper than that of dgPhy around q = 1 nm−1, i.e., the dimensions of Phy is greater than those of dgPhy.
However, the presence of the glycans is more easily observed in Fig. 2, which illustrates the corresponding pair distance distribution functions (p(r), and clearly shows that Phy contains additional interatomic distances at all length scales. This difference is also reflected in the output parameters from the indirect Fourier transformation listed in Table 1, where the Rg of native Phy and dgPhy were 2.55 and 2.50 nm, respectively. The additional interatomic distances of Phy, although small, are also manifested in axial ratios as output by the indirect Fourier transformation T that were 1.66 and 1.76, respectively (Table 1). This rather elongated shape of dgPhy was supported by additional SAXS-analysis with the program BODIES (20) that showed that Phy was most precisely described by a geometrical object as a prolate ellipsoid with a axial ratio ∼1.7. The observation that deglycosylation did not decrease the maximum dimension (Dmax) of the protein suggests that the glycans are primarily situated in a belt around the longest axis where they do not increase Dmax.
Figure 2.
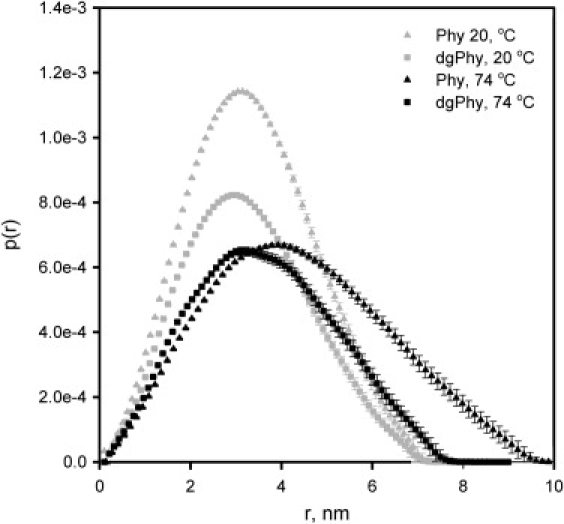
Pair distances distribution functions obtained by indirect Fourier formation of the scattering data displayed in Fig. 1. Phy: triangles; dgPhy: squares. At 20°C (gray color) Phy and dgPhy are clearly more alike than at 74°C (black color). Also note the increase in Dmax when the temperature is raised.
Table 1.
Key parameters of the SAXS data analysis
| Temperature°C | Sample | Rg (nm) | Axial ratio | Dmax (nm) | I(0)/C 1/cm/(mg/mL) | Mw (kD) | A2 × 10 × mL mol g−2 |
|---|---|---|---|---|---|---|---|
| 20 | Phy | 2.55 | 1.66 | 7.03 | 4.68 × 10−2 | 62 | 10.9 ± 0.1 |
| dgPhy | 2.50 | 1.76 | 6.99 | 2.31 × 10−2 | 48 | 10.9 ± 0.1 | |
| 74 | Phy | 3.38 | 1.99 | 9.46 | 3.55 × 10−2 | 47 | 11.0 ± 0.3 |
| dgPhy | 2.77 | 1.81 | 7.46 | 2.21 × 10−2 | 46 | 9.0 ± 0.2 |
The A2-values are based on the linear fits displayed in Fig. 6. A total of 7 and 10 concentrations were used at 20°C and 74°C, respectively. The Rg-values are zero concentrations extrapolated values obtained by Guinier analysis. The molecular weights are calculated based on the I(0) from the Guinier analysis and the Dmax-values and the axial ratios are output parameters from the indirect Fourier transformation.
The experimental temperature, 74°C was chosen such that both glycoforms would be completely denatured. To confirm this, the thermal stability was measured by differential scanning calorimetry using the same buffer conditions, 50 mM HEPES, pH 8 (data not shown). The transition temperature (where 50% of the population of protein molecules is denatured) of Phy and dgPhy were determined to 30.9°C and 27.0°C, respectively. Furthermore, in the case of both glycoforms the denaturation transitions were completed below 50°C, thus, confirming that both glycoforms are completely denatured at 74°C. Due to the rather low transition temperature of dgPhy an extra SAXS measurement was carried out at 3.0°C to verify that this glycoform is in its native form at 20°C. This control measurement showed that the two scattering curves as well as their corresponding p(r)-functions are practically identical at 3.0°C and 20°C, which strongly suggests that dgPhy is, in fact, native at 20°C (see Supplementary Material).
When thermally denatured at 74°C the scattering curve of Phy in Fig. 1 was changed in several ways, the most noticeable being disappearance of the feature originating from the glycans at intermediate distances (∼2.5 nm−1), which probably reflects increased structural flexibility. However, when comparing with dgPhy it is also clear that the scattering curve of Phy had a much steeper curvature showing that Phy had considerably larger dimensions at high temperatures. This is also seen in the corresponding p(r)-function in Fig. 2 were the Dmax, which is 7.0 nm in the native state for both glyco-variants, changes to 9.5 nm for Phy but only to 7.5 nm for dgPhy. Similar differences were observed in the Rg-values that increased 33% on denaturation (from 2.55 nm to 3.38 nm) for Phy and only 11% (from 2.50 nm to 2.77 nm) for dgPhy. The impact of glycosylation on the structure of the denatured protein was also seen in the axial ratios as determined by indirect Fourier transformation where that of Phy increased relatively more than that of dgPhy (Table 1). We note that the changes in dimensions associated with thermal denaturation of dgPhy are considerably smaller than that typically seen for chemically denatured proteins without prosthetic-groups or disulfide bonds. When chemically denatured, this group of proteins basically behave like random coiled polymers with excluded volume effects, such that there is a power-law relationship between polymer length and the ensemble averaged Rg (25):
Where N is the number of residues in the polypeptide chain. R0 is a constant that reflects the persistence length of the polymer chain. Nu is the exponential scaling factor that for chemically denatured protein is 0.588 in accordance with renormalization group theory predictions (26). If R0 is taken as an average of the chemically denatured proteins reviewed by Kohn et al. (25) (R0 = 2.02 ± 0.21), one can calculate that dgPhy (n = 439) should have a R0 of 7.2 ± 0.7 nm if it was to be a random coil. The thermally denatured form of dgPhy has a Rg of only 2.8 nm that show clearly that the unfolded conformers of dgPhy are considerably more compact than that of a random coil, despite the fact, that the protein is completely denatured as measured by differential scanning calorimetry. The observation is in accord with a significant content of residual secondary structure in the denatured state, as seen by synchrotron radiation circular dichroism (27).
To further highlight these structural differences, Fig. 3 shows the SAXS data in a Kratky plot, i.e., I(q) × q−2 vs. q, which is a convenient way to gauge the degree of unfolding. A globular protein scatters as q−4 yielding a Kratky plot that is proportional to q−2 whereas an unfolded and fully flexible polypeptide chain will scatter like a Gaussian random coil, i.e., q−2, thus giving a straight line in a Kratky plot. Therefore, the relative degree of unfolding may be assessed by comparing the heights of the parabola shaped curves in a Kratky plot (28).
Figure 3.
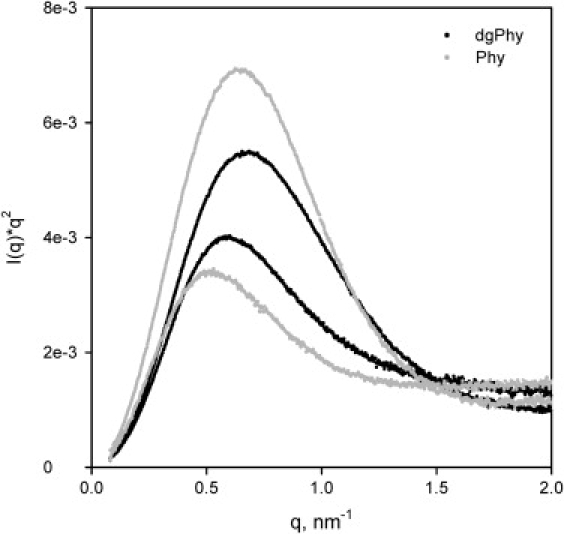
Kratky plots, I(q)q−2 vs. q, of Phy (gray) and dgPhy (black). The upper (gray or black) curves are the scattering profiles recorded at 20°C whereas the two lower curves are obtained at 74°C. The relatively larger shift in the heights of the parabola shaped curves Phy suggest the Phy unfolds to a more gaussian random coil-like structure at higher temperatures.
Raising the temperature from 20°C to 74°C cause considerable unfolding of both Phy and dgPhy, although, a significant degree of globular structure still remains (Fig. 3). However, when comparing the relative shift in heights of the parabolas it is also apparent that Phy was unfolded to a higher degree than dgPhy. Furthermore, at 74°C the peak of the parabola of Phy is clearly shifted toward lower q-values that reflect an increase in the radius of gyration (Rg).
Interparticle interactions
To assess the effect of the glycans on the intermolecular interactions second virial coefficients (A2) of Phy and dgPhy were measured. The SAXS-measurements were carried out at 7 and 10 different concentrations at 20°C and 74°C, respectively, in the concentration range of 1.5–15 mg/mL. No indications of aggregation were detected during data-collections, even at high concentrations. Nonetheless, consecutive data-collections were made on selected samples to ensure that no aggregation or radiation damage occurred. Guinier plots (ln I(q) vs. q2) in Fig. 4 show linear relationships within the Guinier region and the extrapolations of the fits were used to determine the forward scattering (I(0)[1/cm]) as explained previously.
Figure 4.
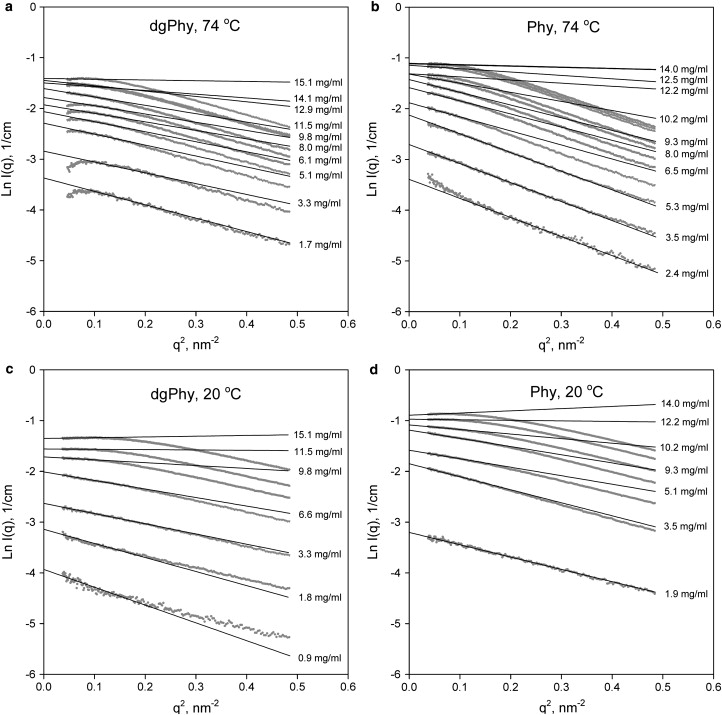
Guinier plots, ln I(q) vs. q2, of the scattering profiles of Phy (a and c) and dgPhy (b and d) from a series of concentration in range 1.5–15 mg/mL. (a and b) 74°C; (c and d) 20°C. The fits are restricted to the linear part of the Guinier range within q × Rg < 1.3.
The values of A2 can be determined from the slope of a plot of c/I(0,c) versus the protein concentration as illustrated in Fig. 5. As can be seen from the quality of fits, the non-ideality of the solutions in this concentration range is described satisfactorily by A2 without the use of higher order terms.
Figure 5.
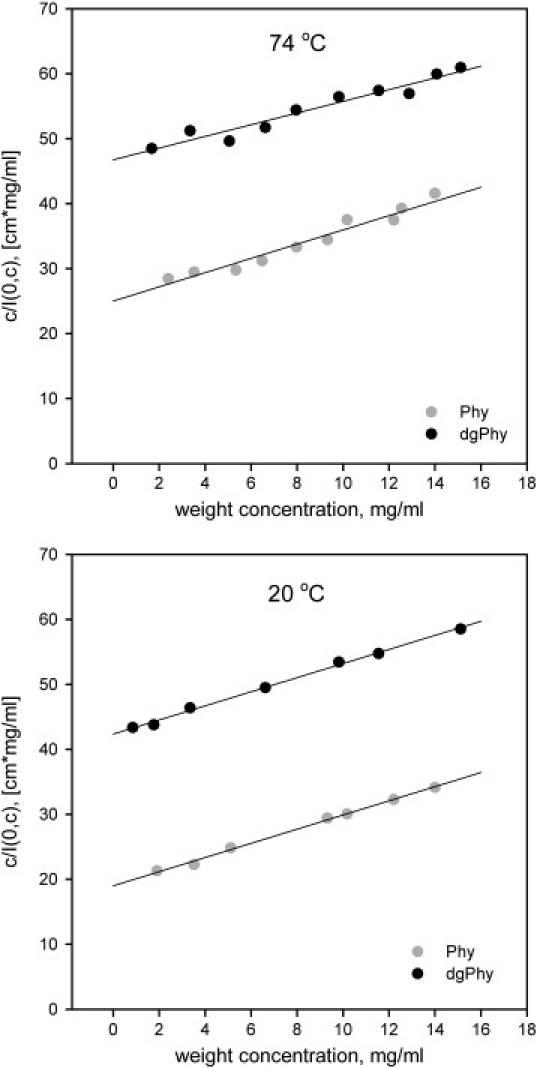
Dependence on protein concentration of c/I(0,c). The units of the ordinate, cm × mg/mL, originates from the protein concentration (mg/mL) divided by the absolute scaled scattering-data (1/cm). The A2-values can be deduced from the slope of the fitted straight lines in this plot. Note that the slope of dgPhy is less steep than that of Phy at 74°C whereas the slopes are nearly identical at 20°C.
Both glycoforms had an A2 of 10.9 ± 0.1 × 104 mL mol g−2 at 20°C showing that both forms are highly repulsive, however, at 74°C the A2 of dgPhy decreased to 9.0 ± 0.2 × 104 mL mol g−2 whereas A2 of Phy remained unchanged (11.0 ± 0.3 × 104 mL mol g−2) (Table 1).
The calculated molecular masses at low temperature listed in Table 1 are close to the ones determined previously by SDS-PAGE (Phy: ∼65 kD and dgPHY: ∼48 kD) (6), however, at high temperature the calculated molecular masses deviates considerably and especially so in the case of Phy. Presumably this discrepancy is consequence of the protein scattering length density used in these calculations because this is an average value found for native proteins at room temperatures. However, even though the isothermal volume change accompanying unfolding of proteins are generally small (29), unfolded protein states are known to have considerably higher partial specific expansivities (∼5.5 × 10−4 cm3 g−1 K−1) than native (∼3.0 × 10−4 cm3 g−1 K−1) (30). For this reason, the partial specific volume at 74°C is likely to be higher than the value actually used. Therefore, if this volume expansion is taken into account excess scattering length density per unit mass Δρm of dgPhy is decreased from 1.97 × 10−10 cm/g to 1.87 × 10−10 cm/g, which, theoretically, would give rise to a 10% decrease in the forward scattering in line with our observations of an apparent decrease in the molar mass with increasing temperatures.
The schematic representation in Fig. 6 sums up the results presented in this study. As seen in the figure, the shape of dgPhy can be satisfactorily described as a prolate ellipsoid with an axial ratio of ∼1.7. The glycans appear to be located as a belt around the longest axis of Phy where their effect on Dmax is limited. When denatured, the glycans induce a considerably more expanded structure of Phy than that of dgPhy and the presence of the glycans also make mutual interactions of the protein more repulsive.
Figure 6.
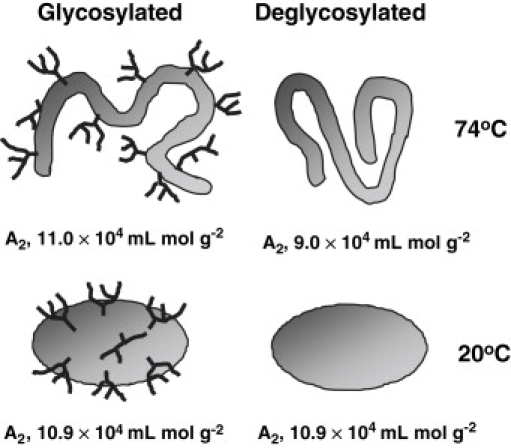
Schematic representation of the structure of Phy (left) and dgPhy (right) at 20°C (bottom) and 74°C (top). The measured A2-values are listed underneath each figure. Note that the glycans are located along the longest axis of Phy and that Phy is more expanded than dgPhy at high temperatures.
Discussion
Glycosylation and structure
It is generally accepted that glycosylation reduces the backbone dynamics in denatured proteins (31–34). Although this effect may be rather pronounced, the covalent attachment of glycans to peptides seems to affect the conformation of only the first few neighboring residues of the glycan (35–38) and in most cases glycosylation does not affect the backbone conformation of natively folded proteins (32,39).
The recorded scattering profiles of Phy and dgPhy clearly show that the presence of the glycans significantly alters the structure of the denatured protein, making it more expanded than its unglycosylated counterpart (Figs. 1 and 2). In contrast, the peptide structure of the native protein seems to be unaffected by glycosylation. This is in accordance with earlier spectroscopic investigations, which showed that the secondary structure of phytase was unaffected by the glycans (27). These conclusions are based on the p(r) functions (Fig. 2) that are similar for the native glyco-variants, but markedly different for the thermally denatured proteins. Also, the glycans give rise to significantly larger maximal dimension of the denatured molecule, and this structure expanding effect is also clearly reflected in Rg, which increases three times more for Phy on denaturation (Table 1).
To our knowledge, this is the first time that N-glycosylation has been reported to perturb the denatured structure of a natural protein, but similar effect of glycosylation have been reported previously for heavily O-glycosylated mucins (40,41). Interestingly, single site glycosylation (O- as well as N-glycosylation) often have little or no effect on the global conformation of peptides (38,42,43), although, in some cases single site N-glycosylation have been reported to promote locally more compact β-turn conformations (44–46) presumably via steric interactions between the glycan and peptide moiety. This suggests that the structure expanding effect of the glycans is dependent on the presence of multiple glycosylation sites and that steric hindrance of the glycans play an important role for this effect. In support of this idea, a large majority of studies have shown that attaching glycans to either peptides or proteins has little effect on the glycan conformation showing that glycans are generally solvent-exposed and not tightly bound to the peptide surface (39,47–50). However, the two innermost GlcNAc residues of the glycan may be an important exception to this general rule as they are found frequently in contact with aromatic residues (51–53). Based on computer simulations, Hoffman and Florke have argued that the presence of a glycan can amplify the preference for certain peptide conformation by selectively increasing the entropy of these conformations (54). Effects of this type along with steric hindrance from the bulky glycans are likely to reduce the range of available torsion angles in the denatured polypeptide and hence promote elongated conformations.
Glycosylation and interparticle interactions
To get insight into the interprotein interactions of the two glycoforms both in the native and denatured states, second virial coefficients (A2) were measured at 20°C and 74°C. The thermodynamic quantity, A2, is a measure of the non-ideality of the solution arising e.g., from the finite size of the dissolved particles, the interparticle interactions, and the particle-solvent interactions. The values 10.9 ± 0.1 × 104 mL mol g−2 for both Phy and dgPhy at 20°C show that both glycoforms are highly repulsive, i.e., particle-solvent are far more favorable than particle-particle interactions. We note that these values are in line with the values reported previously for native Phy and dgPhy at lower pH (6) and they seem to support the trend (6) that the difference in A2 between Phy and dgPhy diminishes as pH is shifted away from pI, which is ∼3.6 (6). Measurements of A2 for denatured phytase at lower pH seems to have a potential to highlight the role of glycans on protein repulsion, but they are experimentally precluded by rapid protein aggregation. As it turned out, the differences in A2 between Phy and dgPhy was overshadowed by electrostatic repulsion at 20°C, however, clear differences were observed at 74°C, where A2 of dgPhy decreased to 9.0 ± 0.2 × 104 mL mol g−2 whereas the value of Phy did not change. This difference in A2, even small, reflects the crucial impact of the glycan on the colloidal stability of unfolded and partial unfolded forms of the protein that helps disfavor the process of irreversible aggregation and increase the likelihood of refolding. In fact, previous studies have shown that at lower pH this effect of the glycans becomes so dominant that it reduces the thermally-induced aggregation of Phy by a factor of >200 (6).
Implications for aggregation and folding
It is starting to become recognized that that N-glycans, and in particularly the GlcNAc unit, have amphiphilic properties that contribute to hydrophobic N-glycan-protein interactions and greatly promote protein folding (55-57). This observation combined with the work presented in this study points toward a dual function of N-glycans. Hence, they may promote intramolecular peptide association and disfavors intermolecular peptide interactions. Thus, whereas the two innermost GlcNAc residues may promote intramolecular contacts and act as nucleation site for folding, the inherent flexibility and steric crowding of the remaining part of the glycan probably play an equally important role in directing folding and ensuring stable interparticle interactions. This ingenious two-way effect of the N-glycosylation, therefore, helps to tip the balance form irreversible aggregation toward reversible folding.
Conclusion
The SAXS-data presented in this study shows that whereas the glycans has virtually no effect on the intermolecular interactions of Phy in native form, they promote the colloidal stability of aggregation prone unfolded forms of the protein. This observation strongly suggests that the glycans does not prevent aggregation by increasing the overall hydrophilicity of protein because this would in general increase A2 and impact the native form of the protein as well. The glycan mantle of Phy does not seem to affect the native peptide structure; however, the presence of the glycans significantly perturbs the unfolded structure of Phy that make the peptide structure become considerably more expanded. This perturbation may be rationalized as steric hindrance exerted by the glycans, which forces the denatured protein into more elongated conformers.
Supplementary Material
Two figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(08)00009-X.
Supplementary Material
Acknowledgments
We gratefully acknowledge use of the EMBL X31 beamline at the DORIS storage ring, DESY, Hamburg as well as the help of D. Svergun and M. Roessle. We are also grateful for the fast access to beamtime at beamline I711 at MAX-lab, Lund, Sweden. Novozymes is gratefully acknowledged for providing us with Endo-β-N-acetylglycosaminidase F1 and plenty of Peniophora lycii phytase.
References
- 1.Varki A. Biological roles of oligosaccharides—all of the theories are correct. Glycobiology. 1993;3:97–130. doi: 10.1093/glycob/3.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lowe J.B., Marth J.D. A genetic approach to mammalian glycan function. Annu. Rev. Biochem. 2003;72:643–691. doi: 10.1146/annurev.biochem.72.121801.161809. [DOI] [PubMed] [Google Scholar]
- 3.Hounsell E.F., Davies M.J. Role of protein glycosylation in immune regulation. Ann. Rheum. Dis. 1993;52:S22–S29. doi: 10.1136/ard.52.suppl_1.s22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lis H., Sharon N. Lectins: carbohydrate-specific proteins that mediate cellular recognition. Chem. Rev. 1998;98:637–674. doi: 10.1021/cr940413g. [DOI] [PubMed] [Google Scholar]
- 5.Tams J.W., Vind J., Welinder K.G. Adapting protein solubility by glycosylation. N-glycosylation mutants of Coprinus cinereus peroxidase in salt and organic solutions. Biochim. Biophys. Acta. 1999;1432:214–221. doi: 10.1016/s0167-4838(99)00103-x. [DOI] [PubMed] [Google Scholar]
- 6.Hoiberg-Nielsen R., Fuglsang C.C., Arleth L., Westh P. Interrelation ships of glycosylation and aggregation kinetics for Peniophora lycii phytase. Biochemistry. 2006;45:5057–5066. doi: 10.1021/bi0522955. [DOI] [PubMed] [Google Scholar]
- 7.Paulson J.C. Glycoproteins—what are the sugar chains for. Trends Biochem. Sci. 1989;14:272–276. doi: 10.1016/0968-0004(89)90062-5. [DOI] [PubMed] [Google Scholar]
- 8.Helenius A. How N-linked oligosaccharides affect glycoprotein folding in the endoplasmic-reticulum. Mol. Biol. Cell. 1994;5:253–265. doi: 10.1091/mbc.5.3.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trombetta E.S. The contribution of N-glycans and their processing in the endoplasmic reticulum to glycoprotein biosynthesis. Glycobiology. 2003;13:77R–91R. doi: 10.1093/glycob/cwg075. [DOI] [PubMed] [Google Scholar]
- 10.Jafari-Aghdam J., Khajeh K., Ranjbar B., Nemat-Gorgani M. Deglycosylation of glucoamylase from Aspergillus niger: effects on structure, activity and stability. Biochim. Biophys. Acta. 2005;1750:61–68. doi: 10.1016/j.bbapap.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 11.Creighton, TE . W.H. Freeman; New York: 1996. Proteins. [Google Scholar]
- 12.Cavaille D., Combes D. Effect of temperature and pressure on yeast invertase stability: a kinetic and conformational study. J. Biotechnol. 1995;43:221–228. doi: 10.1016/0168-1656(95)00145-x. [DOI] [PubMed] [Google Scholar]
- 13.Schulke N., Schmid F.X. Effect of glycosylation on the mechanism of renaturation of invertase from yeast. J. Biol. Chem. 1988;263:8832–8837. [PubMed] [Google Scholar]
- 14.Tani F., Shirai N., Nakanishi Y., Yasumoto K., Kitabatake N. Role of the carbohydrate chain and two phosphate moieties in the heat-induced aggregation of hen ovalbumin. Biosci. Biotechnol. Biochem. 2004;68:2466–2476. doi: 10.1271/bbb.68.2466. [DOI] [PubMed] [Google Scholar]
- 15.Bagger H.L., Fuglsang C.C., Westh P. Preferential binding of two compatible solutes to the glycan moieties of Peniophora lycii phytase. Biochemistry. 2003;42:10295–10300. doi: 10.1021/bi034693i. [DOI] [PubMed] [Google Scholar]
- 16.Bagger H.L., Fuglsang C.C., Westh P. Hydration of a glycoprotein: relative water affinity of peptide and glycan moieties. Eur. Biophys. J. 2006;35:367–371. doi: 10.1007/s00249-005-0035-5. [DOI] [PubMed] [Google Scholar]
- 17.Lassen S.F., Breinholt J., Ostergaard P.R., Brugger R., Bischoff A. Expression, gene cloning, and characterization of five novel phytases from four basidiomycete fungi: Peniophora lycii, Agrocybe pediades, a Ceriporia sp., and Trametes pubescens. Appl. Environ. Microbiol. 2001;67:4701–4707. doi: 10.1128/AEM.67.10.4701-4707.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orthaber D., Bergmann A., Glatter O. SAXS experiments on absolute scale with Kratky systems using water as a secondary standard. J. Appl. Cryst. 2000;33:218–225. [Google Scholar]
- 19.Lindner P., Zemb Th. Elsevier Science B.V; Amsterdam, The Netherlands: 2002. Neutrons, X-Rays and Light: Scattering Methods Applied to Soft Condensed Matter. [Google Scholar]
- 20.Konarev P.V., Volkov V.V., Sokolova A.V., Koch M.H.J., Svergun D.I. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Cryst. 2003;36:1277–1282. [Google Scholar]
- 21.Zimm B.H. The scattering of light and the radial distribution function of high polymer solutions. J. Chem. Phys. 1948;16:1093–1099. [Google Scholar]
- 22.Receveur V., Durand D., Desmadril M., Calmettes P. Repulsive interparticle interactions in a denatured protein solution revealed by small angle neutron scattering. FEBS Lett. 1998;426:57–61. doi: 10.1016/s0014-5793(98)00309-3. [DOI] [PubMed] [Google Scholar]
- 23.Glatter O. A new method for the evaluation of small-angle scattering data. J. Appl. Crystallogr. 1977;10:415–421. [Google Scholar]
- 24.Hansen S. Bayesian estimation of hyperparameters for indirect Fourier transformation in small-angle scattering. J. Appl. Crystallogr. 2000;33:1415–1421. [Google Scholar]
- 25.Kohn J.E., Millett I.S., Jacob J., Zagrovic B., Dillon T.M., N Random-coil behavior and the dimensions of chemically unfolded proteins. Proc. Natl. Acad. Sci. USA. 2004;101:12491–12496. doi: 10.1073/pnas.0403643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leguillou J.C., Zinnjustin J. Critical exponents for N-vector model in 3 dimensions from field-theory. Phys. Rev. Lett. 1977;39:95–98. [Google Scholar]
- 27.Bagger H.L., Hoffmann S.V., Fuglsang C.C., Westh P. Glycoprotein-surfactant interactions: a calorimetric and spectroscopic investigation of the phytase-SDS system. Biophys. Chem. 2007;129:251–258. doi: 10.1016/j.bpc.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 28.Glatter O., Kratky O. Academic Press; New York: 1982. Small Angle X-Ray Scattering. [Google Scholar]
- 29.Murphy L.R., Matubayasi N., Payne V.A., Levy R.M. Protein hydration and unfolding—insights from experimental partial specific volumes and unfolded protein models. Fold. Des. 1998;3:105–118. doi: 10.1016/S1359-0278(98)00016-9. [DOI] [PubMed] [Google Scholar]
- 30.Chalikian T.V. Volumetric properties of proteins. Annu. Rev. Biophys. Biomol. Struct. 2003;32:207–235. doi: 10.1146/annurev.biophys.32.110601.141709. [DOI] [PubMed] [Google Scholar]
- 31.Joao H.C., Dwek R.A. Effects of glycosylation on protein-structure and dynamics in ribonuclease-B and some of its individual glycoforms. Eur. J. Biochem. 1993;218:239–244. doi: 10.1111/j.1432-1033.1993.tb18370.x. [DOI] [PubMed] [Google Scholar]
- 32.Rudd P.M., Joao H.C., Coghill E., Fiten P., Saunders M.R. Glycoforms modify the dynamic stability and functional-activity of an enzyme. Biochemistry. 1994;33:17–22. doi: 10.1021/bi00167a003. [DOI] [PubMed] [Google Scholar]
- 33.Joao H.C., Scragg I.G., Dwek R.A. Effects of glycosylation on protein conformation and amide proton-exchange rates in Rnase-B. FEBS Lett. 1992;307:343–346. doi: 10.1016/0014-5793(92)80709-p. [DOI] [PubMed] [Google Scholar]
- 34.Imperiali B., Rickert K.W. Conformational implications of asparagine-linked glycosylation. Proc. Natl. Acad. Sci. USA. 1995;92:97–101. doi: 10.1073/pnas.92.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lommerse J.P., Kroonbatenburg L.M., Kroon J., Kamerling J.P., Vliegenthart J.F. Conformations and internal mobility of a glycopeptide derived from bromelain using molecular dynamics simulations and NOESY analysis. J. Biomol. NMR. 1995;6:79–94. doi: 10.1007/BF00417494. [DOI] [PubMed] [Google Scholar]
- 36.Live D.H., Kumar R.A., Beebe X., Danishefsky S.J. Conformational influences of glycosylation of a peptide: a possible model for the effect of glycosylation on the rate of protein folding. Proc. Natl. Acad. Sci. USA. 1996;93:12759–12761. doi: 10.1073/pnas.93.23.12759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carotenuto A., D'Ursi A.M., Nardi E., Papini A.M., Rovero P. Conformational analysis of a glycosylated human myelin oligodendrocyte glycoprotein peptide epitope able to detect antibody response in multiple sclerosis. J. Med. Chem. 2001;44:2378–2381. doi: 10.1021/jm010811t. [DOI] [PubMed] [Google Scholar]
- 38.Hashimoto Y., Toma K., Nishikido J., Yamamoto K., Haneda K. Effects of glycosylation on the structure and dynamics of eel calcitonin in micelles and lipid bilayers determined by nuclear magnetic resonance spectroscopy. Biochemistry. 1999;38:8377–8384. doi: 10.1021/bi983018j. [DOI] [PubMed] [Google Scholar]
- 39.Wormald M.R., Petrescu A.J., Pao Y.L., Glithero A., Elliott T. Conformational studies of oligosaccharides and glycopeptides: complementarity of NMR, x-ray crystallography, and molecular modelling. Chem. Rev. 2002;102:371–386. doi: 10.1021/cr990368i. [DOI] [PubMed] [Google Scholar]
- 40.Gerken T.A., Butenhof K.J., Shogren R. Effects of glycosylation on the conformation and dynamics of O-linked glycoproteins—C-13 Nmr-studies of ovine submaxillary mucin. Biochemistry. 1989;28:5536–5543. doi: 10.1021/bi00439a030. [DOI] [PubMed] [Google Scholar]
- 41.Shogren R., Gerken T.A., Jentoft N. Role of glycosylation on the conformation and chain dimensions of O-linked glycoproteins—light-scattering-studies of ovine submaxillary mucin. Biochemistry. 1989;28:5525–5536. doi: 10.1021/bi00439a029. [DOI] [PubMed] [Google Scholar]
- 42.Wilce J.A., Craik D.J., Ede N., Jackson D.C., Schreiber G. H-1 NMR studies of peptide fragments from the N-terminus of chicken and human transthyretin. Biochem. Mol. Biol. Int. 1995;36:1153–1159. [PubMed] [Google Scholar]
- 43.Kirnarsky L., Prakash O., Vogen S.M., Nomoto M., Hollingsworth M.A. Structural effects of O-glycosylation on a 15-residue peptide from the mucin (MUC1) core protein. Biochemistry. 2000;39:12076–12082. doi: 10.1021/bi0010120. [DOI] [PubMed] [Google Scholar]
- 44.O'Connor S.E., Imperiali B. A molecular basis for glycosylation-induced conformational switching. Chem. Biol. 1998;5:427–437. doi: 10.1016/s1074-5521(98)90159-4. [DOI] [PubMed] [Google Scholar]
- 45.O'Connor S.E., Imperiali B. Conformational switching by asparagine-linked glycosylation. J. Am. Chem. Soc. 1997;119:2295–2296. [Google Scholar]
- 46.Andreotti A.H., Kahne D. Effects of glycosylation on peptide backbone conformation. J. Am. Chem. Soc. 1993;115:3352–3353. [Google Scholar]
- 47.Imberty A., Perez S. Stereochemistry of the N-glycosylation sites in glycoproteins. Protein Eng. 1995;8:699–709. doi: 10.1093/protein/8.7.699. [DOI] [PubMed] [Google Scholar]
- 48.Wyss D.F., Choi J.S., Li J., Knoppers M.H., Willis K.J. Conformation and function of the N-linked glycan in the adhesion domain of human Cd2. Science. 1995;269:1273–1278. doi: 10.1126/science.7544493. [DOI] [PubMed] [Google Scholar]
- 49.Wormald M.R., Wooten E.W., Bazzo R., Edge C.J., Feinstein A. The conformational effects of N-glycosylation on the tailpiece from serum Igm. Eur. J. Biochem. 1991;198:131–139. doi: 10.1111/j.1432-1033.1991.tb15995.x. [DOI] [PubMed] [Google Scholar]
- 50.Weller C.T., Lustbader J., Seshadri K., Brown J.M., Chadwick C.A. Structural and conformational analysis of glycan moieties in situ on isotopically C-13,N-15-enriched recombinant human chorionic gonadotropin. Biochemistry. 1996;35:8815–8823. doi: 10.1021/bi960432f. [DOI] [PubMed] [Google Scholar]
- 51.Thijssen-van Zuylen C.W.E.M., de Beer T., Leeflang B.R., Boelens R., Kaptein R. Mobilities of the inner three core residues and the Man(alpha 1 ↠ 6) branch of the glycan at Asn78 of the alpha-subunit of human chorionic gonadotropin are restricted by the protein. Biochemistry. 1998;37:1933–1940. doi: 10.1021/bi9718548. [DOI] [PubMed] [Google Scholar]
- 52.Petrescu A.J., Milac A.L., Petrescu S.M., Dwek R.A., Wormald M.R. Statistical analysis of the protein environment of N-glycosylation sites: implications for occupancy, structure, and folding. Glycobiology. 2004;14:103–114. doi: 10.1093/glycob/cwh008. [DOI] [PubMed] [Google Scholar]
- 53.De Beer T., VanZuylen C.W.E.M., Leeflang B.R., Hard K., Boelens R., R NMR studies of the free alpha subunit of human chorionic gonadotropin—structural influences of N-glycosylation and the beta subunit on the conformation of the alpha subunit. Eur. J. Biochem. 1996;241:229–242. doi: 10.1111/j.1432-1033.1996.0229t.x. [DOI] [PubMed] [Google Scholar]
- 54.Hoffmann D., Florke H. A structural role for glycosylation: lessons from the hp model. Fold. Des. 1998;3:337–343. doi: 10.1016/S1359-0278(98)00046-7. [DOI] [PubMed] [Google Scholar]
- 55.Nishimura I., Uchida M., Inohana Y., Setoh K., Daba K. Oxidative refolding of bovine pancreatic RNases A and B promoted by Asn-glycans. J. Biochem. (Tokyo) 1998;123:516–520. doi: 10.1093/oxfordjournals.jbchem.a021966. [DOI] [PubMed] [Google Scholar]
- 56.Jitsuhara Y., Toyoda T., Itai T., Yamaguchi H. Chaperone-like functions of high-mannose type and complex-type N-glycans and their molecular basis. J. Biochem. (Tokyo) 2002;132:803–811. doi: 10.1093/oxfordjournals.jbchem.a003290. [DOI] [PubMed] [Google Scholar]
- 57.Yamaguchi H., Uchida M. A chaperone-like function of intramolecular high-mannose chains in the oxidative refolding of bovine pancreatic RNase B. J. Biochem. (Tokyo) 1996;120:474–477. doi: 10.1093/oxfordjournals.jbchem.a021435. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


