Abstract
Background and Aims
The diploid goat grass Aegilops tauschii (2n = 2x = 14) is native to the Middle East and is the D-genome donor to hexaploid bread wheat. The aim of this study was to measure the diversity of different subspecies and varieties of wild Ae. tauschii collected across the major areas where it grows in Iran and to examine patterns of diversity related to the taxa and geography.
Methods
Inter-retroelement amplified polymorphism (IRAP) markers were used to analyse the biodiversity of DNA from 57 accessions of Ae. tauschii from northern and central Iran, and two hexaploid wheats.
Key Results
Eight IRAP primer combinations amplified a total of 171 distinct DNA fragments between 180 and 3200 bp long from the accessions, of which 169 were polymorphic. On average, about eight fragments were amplified with each primer combination, with more bands being amplified from accessions from the north-west of the country than from other accessions.
Conclusions
The IRAP markers showed high levels of genetic diversity. Analysis of all accessions together did not allow the allocation of individuals to taxa based on morphology, but showed a tendency to put accessions from the north-west apart from others regions. It is speculated that this could be due to different activity of retroelements in the different regions. Within the two taxa with most accessions, there was a range of IRAP genotypes that could be correlated closely with geographical origin. This supports suggestions that the centre of origin of the species is towards the south-east of the Caspian Sea. IRAP is an appropriate marker system to evaluate genetic diversity and evolutionary relationships within the taxa, but it is too variable to define the taxa themselves, where more slowly evolving morphological, DNA sequence or chromosomal makers may be more appropriate.
Key words: Aegilops squarrosa, Aegilops tauschii, biodiversity, fertile crescent, IRAPs, phylogeography, plant breeding, retrotransposons, Triticum
INTRODUCTION
As has been pointed out by Bennett (1982), understanding the characteristics of the plant cell nucleus – its nucleotype – is critical to learning about the genome, its behaviour, modulation and generation of biodiversity. This information is of value for both fundamental reasons and for applications in plant breeding, ecology, phylogeography and genetics (Bennett, 1984). Measurement of the diversity of a group of related taxa allows geneticists to understand evolution and plant breeders to exploit wider pools of diversity. The present genetic variation and genetic structure of a species reflects not only the patterns of genetic exchange, but also the history of gene flow, range fragmentation and isolation among population lineages: as Kihara (1947) noted, ‘the history of the earth is recorded in the layers of its crust. The history of all organisms is inscribed in the chromosomes’. Historical events such as population expansion over wide geographical areas and recurrent gene flow between lineages have been important evolutionary forces (Ellstrand et al., 1999). Studying the geographical distribution of plant lineages can help define evolutionary history and genetic exchange, providing insights into the factors that shape the genetic diversity of a taxon (Avise, 2000; Morrell et al., 2003) and crop origins (Eyre-Walker et al., 1998; Olsen and Schaal, 1999; Takahashi et al., 1999; Olsen and Purugganan, 2002).
The diploid goatgrass Aegilops tauschii Coss. [syn. Triticum tauschii (Coss.) Schmahlh., Aegilops squarrosa auct. non L.] (2n = 2x = 14, genome constitution DD) is the D-genome donor of bread wheat (Triticum aestivum L., 2n = 6x = 42, genome composition AABBDD). Aegilops tauschii, a well-demarcated species, grows wild in various regions of Iran and has a centre of distribution in the north of country. A number of infraspecific taxa including the subspecies tauschii Eig (subsp. eusquarroa sensu Kihara et al., 1965), with three varieties tauschii L. (the typica of Kihara et al., 1965), anathera (Eig) Hammer, and meyeri (Griseb.) Tzvelev, and subsp. strangulata Eig, have been noted since the 1920s based on morphological characters (Eig, 1929; Kihara et al., 1965), although the status of these taxa remains under discussion (Kihara et al., 1965; Hammer, 1980; Tanaka, 1983) and it is likely that some are not separated genetically (Lubbers et al., 1991; van Slageren, 1994; see also Saeidi et al., 2006). High polymorphism levels have been reported in RFLPs and microsatellites between Ae. tauschii accessions (Dvorak et al., 1998; Lelley et al., 2000), in particular between accessions from Iran (Lubbers et al., 1991; Pestsova et al., 2000; Saeidi et al., 2006). Over the last two decades, studies of genetic diversity in wild species have been revolutionized by the availability of DNA-based molecular methods, increasing the number of markers available for analysis of diversity from a small number of phenotypic or morphological variants that can be used as markers to potentially hundreds of thousands of polymorphisms.
The major part of plant genomes is made up of various classes of repetitive DNA (Flavell et al., 1974; see Kubis et al., 1998). In the last 5 years, knowledge about repetitive DNA dispersed in the genome has been exploited by the application of PCR methods to analyse the diversity of insertion sites of these elements between different accessions. Furthermore, crop work has been extended to their wild relatives where markers are transferable between related taxa (Cao et al., 2006), in contrast to SSR markers that are often specific to one taxon, particularly in cereals. Retrotransposons (Flavell et al., 1992; Voytas et al., 1992; Heslop-Harrison et al., 1997) and their degenerate relatives are often the most abundant class of repetitive DNA, and are rapidly evolving. As a consequence of their mode of amplification through an RNA intermediate, reverse transcription and insertion of the DNA copy into the nuclear genome, many copies of retroelements are present in genomes (although copies may be clustered in pericentromeric regions; Boyko et al., 2002). As multicopy, polymorphic sequences, they can be used as molecular markers, and hence for phylogenetic analyses (Shimamura et al., 1997). Retrotransposon insertional polymorphisms can be detected by a variety of PCR-based techniques such as inter-retrotransposon amplified polymorphisms (IRAPs), where outward-facing primers are designed for conserved domains such as long terminal repeats (LTRs) within the elements (Waugh et al., 1997; Flavell et al., 1998; Kalendar et al., 1999; Boyko et al., 2002). The polymorphisms may then be used to model the temporal sequence of insertion events in a lineage and to establish phylogenetic hypotheses. There are conserved regions within the LTRs of retrotransposons that contain sequences essential for expression (promoter and processing signals) and integration. From these conserved LTR regions, outward-facing primers for PCR amplification are designed that amplify the genomic DNA fragment lying between closely spaced retrotransposons (Teo et al., 2005). Variant primers can be used to amplify genomic DNA between members of different retrotransposon families (Kalendar et al., 1999; Boyko et al., 2002). Li et al. (2004) have analysed the sequence composition of Ae. tauschii and demonstrated that as much as 68·2 % of the genome is represented by transposable elements, more than the 50 % in maize or 14 % in rice. In the D-genome 55 % of the elements are retrotransposons, and Li et al. showed that most transposable elements were capable of transcription and amplified in the polyploid species of Triticeae.
Here, the IRAP method was tested and applied to characterize the diversity of Ae. tauschii collected across Iran. We also evaluated the method for analysing the relationships at infraspecific (subspecies and varieties) level to develop phylogeographic models for the distribution of these taxa.
MATERIALS AND METHODS
Fifty-seven accessions of Aegilops tauschii Coss. [syn. Triticum tauschii (Coss.) Schmahlh., Aegilops squarrosa auct. non L.] were collected from various regions of Iran by two of the authors (H.S. and M.R.R.) between May and July in 2002 and 2003 and maintained at the University of Isfahan (collection sites and altitudes are given Supplementary Information available online; Saeidi et al., 2006). Accessions were identified morphologically according to Eig (1929) and van Slageren (1994). Accessions were divided into four groups according to their geographical origin [north west (NW) 17 accessions; north (N) including south Caspian Sea shore area (30 accessions); north-east (NE) including east and south slopes of Alborz mountains (six accessions); and just west of the centre of the country (C; four accessions); see Fig. 2]. One accession of Iranian T. aestivum (a landrace collected from the central region) and the reference wheat cultivar ‘Chinese Spring’ were also included. From each accession, 20–30 seeds were grown in an experimental field (University of Isfahan, Iran) and DNA was isolated from 1·0–1·5 g fresh leaves from the plants of each accession following standard methods.
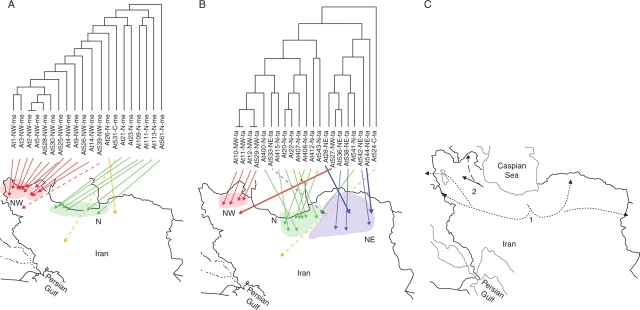
Fig. 2.
UPGMA dendrograms of the relationships based on IRAP analysis of (A) accessions of Ae. tauschii subsp. tauschii var. tauschii; and (B) accessions of Ae. tauschii subsp. tauschii var. meyeri superimposed on their geographic origins; thick black lines show political boundaries; major hydrographic features are indicated in light grey. (C) A possible phylogeography of Ae. tauschii supported by the IRAP data. The species originates from the north of Iran and is distributed in two directions. The populations possessing the subsp. tauschii genepool are suggested to have expanded from the central Alborz Mountains and then distributed eastward and westward (direction 1); populations possessing the subsp. strangulata genepool are suggested to have been distributed along the Caspian Sea shore (direction 2).
IRAP analysis used LTR primers derived from barley (Hordeum vulgare; (Kalendar et al., 1999, 2000; Manninen et al., 2000; Boyko et al., 2002) and banana (Reverse TY1; Teo et al., 2005). The primer sequences, retrotransposon source and orientation are shown in Table 1 Genomic DNA samples were diluted with sterile deionized water to 50 ng μL–1. The IRAP PCR was performed in a 20-μL reaction mixture containing 50 ng DNA, 1× PCR buffer (Promega, USA), 2 mm MgCl2, 5 pmol of each primer, 200 µm dNTP mix and 1 U Taq polymerase (Promega, USA). The annealing temperature was optimized using gradient PCR. The PCR reaction parameters consisted of: 95 °C, 2 min; 30 cycles of 95 °C, 60 s, annealing at the Ta specified in Table 2 for 60 s, ramp +0·5 °C s–1 to 72 °C, and 72 °C for 2 min adding 3 s per cycle, with a final extension at 72 °C for 10 min. PCR products were analysed by electrophoresis on 2 % (w/v) agarose gels (preferably using a 1 : 3 mixture of high resolution : normal agarose) and detected by ethidium bromide staining.
Table 1.
Sequences of primers used for IRAP, their retrotransposon source and direction (→ or ←)
| Primer | Retrotransposon source and direction | Sequence |
|---|---|---|
| LTR6150 | BARE-1 → | CTGGTTCGGCCCATGTCTAT |
| GTATCCACACATGGTA | ||
| LTR6149 | BARE-1 ← | CTCGCTCGCCCACTACA |
| TCAACCGCGTTTATT | ||
| 5′LTR2 | BARE-1 ← | ATCATTGCCTCTAGG |
| GCATAATTC | ||
| 3′LTR | BARE-1 → | TGTTTCCCATGCGA |
| CGTTCCCCAACA | ||
| Sukkula | Sukkula → | GATAGGGTCGCATC |
| TTGGGCGTGAC | ||
| Nikita | Nikita → | CGCATTTGTTCA |
| AGCCTAAACC | ||
| Reverse Ty1 | W1, W3, W7, W8 ← | CCYTGNAYYAANGCNGT* |
* Y = C + T, N = A + G + C + T.
Table 2.
Primer combinations, annealing temperature (Ta), percentage polymorphism, total and average band number used for IRAP
| Primer combination | Ta | Total bands | Polymorphic bands | Percentage polymorphism | Band size (bp) | Maximum band number |
Minimum band number |
Average bands in Ae. tauschii* | Average bands in T. aestivum* | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| LTR6150 + 3′LTR | 62·6 | 18 | 18 | 100 | 310–1450 | At536 | 11 | At531 | 2 | 7 ± 1·7 | 6·5 ± 0·7 |
| LTR6149 + Sukula | 47 | 9 | 9 | 100 | 200–1200 | At1 | 9 | At25 | 1 | 5 ± 0·5 | 2 |
| LTR6150 + Nikita | 59·5 | 16 | 15 | 93·7 | 180–3100 | At526 | 10 | Several accessions | 1 | 5·5 ± 2·8 | 11·5 ± 2·1 |
| LTR6150 + Sukula | 62·3 | 35 | 35 | 100 | 320–3100 | At524 | 16 | At407 | 5 | 10·9 ± 2·5 | 14 ± 1 |
| At544 | At408 | ||||||||||
| 5′LTR2 + Sukula | 65·6 | 34 | 34 | 100 | 320–3200 | At535 | 19 | At20 | 5 | 13·3 ± 3·4 | 6·5 ± 0·7 |
| At532 | |||||||||||
| 5′LTR2 + 3′ | 66 | 17 | 17 | 100 | 240–2500 | At25 | 8 | Several accessions | 1 | 3·5 ± 2·1 | 5 ± 2·8 |
| 3′LTR + RTY1 | 54·9 | 16 | 16 | 100 | 310–1050 | At28 | 11 | At537 | 1 | 6·4 ± 2 | 4 |
| At538 | 2 | ||||||||||
| 3′LTR + 3′LTR | 64 | 26 | 25 | 95·3 | 200–1600 | At9 | 14 | At25 | 6 | 10·1 ± 2·1 | 11·5 ± 0·7 |
| Total | 171 | 169 | 98·8 | 7·85 ± 3·31 | 8·12 ± 3·7 | ||||||
* ± s.d.
Analysis of diversity
The presence (1) or absence (0) of clear and distinguishable fragments of particular mobility was scored from the gels for each accession. Genetic similarities were calculated using NTSYSpc software (version 02e) and Powermarker (Liu and Muse, 2005) from the data tables. Dendrograms showing similarities between different accessions of Ae. tauschii were then constructed by a similarity-based method. The cophenetic (COPH) value matrix was computed for each tree matrix generated based on a particular similarity coefficient, and matrix correlation (r) and Mantel test statistics (z) (Mantel, 1967) were computed to measure the degree of relationship between the cophenetic matrix and similarity matrix. The dendrograms of simple matching (SM) coefficient showed the highest value of r (92·97 %). A dendrogram was constructed using the UPGMA method implemented in NTSYSpc software, version 02e. Data were also analysed with a principle component analysis method using a standardized data matrix (Darroch and Mosimann, 1985) and Projection (PROJ) implemented in NTSYSpc, and ordination plots were drawn. Two hexaploid wheat lines, ‘Chinese Spring’ and an Iranian landrace, were included. In the SSR analysis (Saeidi et al., 2006), mostly targets homologous to the Ae. tauschii sequences would be analysed from the D genome of the hexaploid, so these accessions acted as true outgroups; in IRAP analyses, sequences from all three genomes would be analysed so only a proportion of the bands would be homologous (a third if all three genomes had similar retroelement numbers and IRAP amplification in the hexaploid occurred with the same efficiency as in the diploid; in practice similar numbers of bands were amplified in both species) and hence the hexaploids cannot be used as outgroups in the same way.
RESULTS
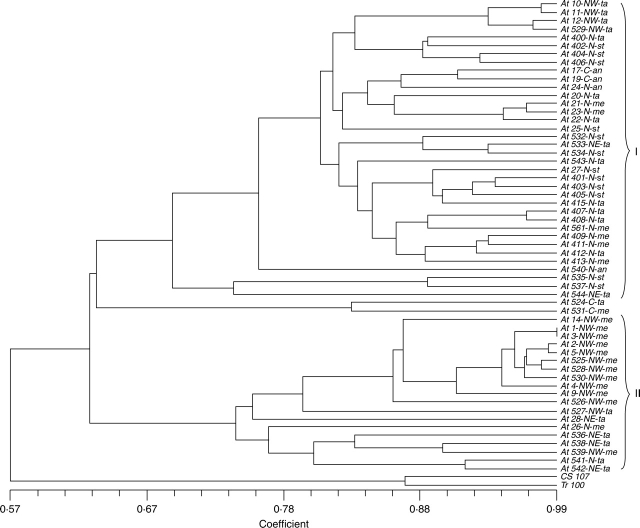
All eight IRAP primer combinations yielded multiple DNA fragments from genomic DNA of all 57 accessions of Ae. tauschii and two accessions of T. aestivum, with a high polymorphism level between accessions (Table 2). Of the 156 bands analysed from Ae. tauschii, 154 (98·8 %) were polymorphic. Karyotyping (data not shown) showed that all accessions were diploid. There were no notable differences between IRAP dendrograms generated using Jaccard (1908), simple matching and dice (Nei and Li, 1979) coefficients. Within the Ae. tauschii accessions, two pairs and a group of three accessions differed by less than 3 % of markers, whereas the lowest similarity (50 %) was between At543 (var. tauschii from the north region) and At530 (var. meyeri from the north-west). Fifteen bands were found only in T. aestivum (one or both accessions); the similarity between the two bread wheat accessions was 87·1%. Because different retrotransposons may show different patterns of insertional polymorphisms, dendrograms based on each primer combination were constructed; the branching of these dendrograms was similar to that of the pooled IRAP data.
In dendrograms of IRAP data, the two T. aestivum accessions were placed apart (100 % bootstrap support based on 1000 replicates). There were no significant differences in average band number within and between subspecies of Ae. tauschii. The average band number from the 40 C, N and NE accessions was 57·7 ± 7·6, whereas the 17 north-west accessions averaged 71·5 ± 8·4 bands (P < 0·001). With all individuals considered, groupings related to subspecies and varieties of Ae. tauschii were only weakly supported. The high polymorphism level within the subspecies and varieties meant that the IRAP markers were unable to allocate accessions to a taxon. However, it was noted that some deep branches related to the geographic origin of accessions; 32 of the 34 accessions collected from north and centre of the country lay on a single branch, and 13 of the 17 accessions from the north-west were placed on another branch (group I, Fig. 1). In the north-western–north-eastern group (group II, Fig. 1), the accessions collected from the north-east were clustered and of 17 accessions collected from the north-west, 13 accessions belonged to var. meyeri and were collected from near the border of Iran and Turkey and along the Aras. The four remaining north-western accessions (At-NW 10, 11, 12, 529) belonging to var. tauschii were grouped with northern accessions, a grouping evident in all dendrograms from different primer combinations.
Fig. 1.
UPGMA dendrogram generated using IRAP data and simple matching similarity coefficient. The relationships between 57 accessions of Ae. tauschii and two accessions of T. aestivum based on bands from this marker type are shown. The two bread wheat accessions are separated from the Ae. tauschii accessions, which are divided into two groups (Clusters I and II). Subspecies or varieties: st = subsp. strangulata; ta = subsp. tauschii var. tauschii; me = subsp. tauschii var meyeri; an = subsp. tauschii var. anathera. Geographical origin in Iran: C = centre; N = north; NW = north-west; NE = north-east.
To examine relationships within the varieties meyeri and tauschii (where most accessions were available), dendrograms were generated separately based on the known taxonomy. The IRAP similarity followed a geographical pattern within each variety (Fig. 2).
DISCUSSION
The retroelement insertional polymorphisms analysed here using IRAPs showed a high level of polymorphism, as expected from analysis of diversity in Ae. tauschii (Lubbers et al., 1991; Pestsova et al., 2000; Saeidi et al., 2006) and the use of IRAPs as one class of marker in a mapping population made by crossing two Ae. tauschii taxa (Boyko et al., 2002). The species is distributed in the north, north-east, north-west and just south of the centre of Iran (van Slageren, 1994) with subsp. strangulata distributed only in the north. Our collection included major taxonomic subgroups within the species, and covered nearly all regions of the species, in Iran. The populations studied from the northern region showed high levels of morphological and taxonomic diversity; the lowest morphological diversity was observed in populations studied from the north-west, mainly belonging to subsp. tauschii var. meyeri.
A few accessions within taxa were relatively similar in the IRAP analysis (short branch lengths in Figs 1 and 2), indicating that some samples originated from related populations. All the north-western accessions, regardless of taxon, had a higher average band number than the other accessions. Environment has a major effect on the amplification of transposable elements (Li et al., 2004; Altinkut et al., 2006) and particularly retroelements, so it is possible that the conditions in this region favour activity and dispersion of the elements. The increased band number, with more non-homologous bands having identical sizes, might lead to the similarity coefficients clustering by geographical origin rather than by taxa (Fig. 1), although when taxa are separated based on morphology the new bands correctly reflect genetic distances within the taxa (Fig. 2A, B). Lubbers et al. (1991) used RFLP approaches to look at diversity across 102 Ae. tauschii accessions from a geographically wide range across the Middle East. They found a large genetic distance between the two groups of var. tauschii (var. typica in their paper) with var. anathera, and subsp. strangulata with var. meyeri, but high similarity within these two groups. We suggest that there is high activity and rapid diversification of the IRAP markers so bands are not identical-by-descent at the level of the whole species; sequencing of the IRAP bands would be needed to show that products or exact lengths were different between subspecies. The recognition of the distinctness of the two groups of taxa at the RFLP level (Lubbers et al., 1991) supports our separate analysis of diversity in var. tauschii (Fig. 2A) and var. meyeri (Fig. 2B) taxa.
Given that increased numbers of bands may correlate with amplification of retroelements (shown to occur in the species by Li et al., 2004) and hence increased genome size, it will be important to measure genome sizes in Ae. tauschii accessions from different geographical regions, investigating whether there are similar trends across subspecies and maybe other species with equivalent geographical distributions. In the grass species Koeleria macrantha and Festuca pallens, Pecinka et al. (2006) and Smarda and Bures (2006), respectively, found intraspecific, interpopulation differences in genome size. However, Smarda et al. (2007) argued that the observed genome size variability in Festuca was not related to habitat, vegetation or microclimate, and hence is not a selected (or presumably inducible) character. Leitch et al. (2005) developed a model of evolution of DNA amounts across all land plants, and examined data showing the dynamic increases and decreases in genome size that have occurred across families and genera. The change in number of IRAP bands in the subspecies of Ae. tauschii in different geographical regions provides another example of a nucleotypic character, and further investigation of genome size will be valuable.
The case for unification or division of the subtaxa analysed here based on morphology has been discussed (Hammer, 1980; van Slageren, 1994). Badaeva et al. (2002), based on cytotaxonomy and the C-banding patterns of chromosomes, recognized strangulata and tauschii forms; Lubbers et al. (1991) used RFLP markers to recognize the meyeri/strangulata and tauschii/anathera forms. Taxon identification is not based on single-gene characters, although in the wild intercrossing can presumably occur. Gene flow between populations of the subspecies would give the pattern of relationships shown in Fig. 1. Even low rates of gene flow could homogenize molecular markers, although it is not clear how this would allow the taxa to remain morphologically distinct. Further analysis of large collections from across the range, using a combination of molecular and morphological approaches, is clearly needed to resolve the relationships within Ae. tauschii.
Aegilops tauschii subsp. strangulata is largely restricted to the Caspian Sea shore where the environmental conditions are milder than those beyond the Alborz Mountains. As has been suggested by Dudnikov (1998) and Boguslavsky (1981), the variation and distribution of subsp. tauschii (‘tauschii sub-genepool’) had an environmental adaptation base and could pass beyond the Alborz Mountains to the south slope, with subsequent westward and mainly eastward dispersal. Based on variation of RFLP markers, Lubbers et al. (1991) reported a high level of genetic similarity between accessions collected from Turkey, Afghanistan and Pakistan, indicating the distribution of the Ae. tauschii sub-genepool in these countries. Their results showed that accessions from Iran and west Caucasia had the highest levels of genetic diversity, and the highest probability of having a unique genotype, characteristic of an ancestral geographical region. Subspecies strangulata has been considered as the ancestral type within Ae. tauschii (Nakai, 1979; Jaaska, 1981) and the northern region of Iran as the centre of diversity and probably the centre of origin of the species. This study showed decreases in genetic diversity in parallel with taxonomic diversity of Ae. tauschii from the north towards the east and west. A trend to decreasing genetic diversity along with an increase in IRAP band number from the north spreading towards the east and particularly the west supports the presence of more ancestral populations in the northern region and derived populations distributed around this centre of diversity, and would support a distribution model as indicated in Fig 2C. The status of populations growing around the Aras River and near the border of Iran and Turkey is of interest; these had the maximum band number (75·4 ± 4·8) and the highest similarity average (78·4 ± 11·8 %). These populations were morphologically a relatively homogeneous group with slender and shorter spikes, smaller seeds, a lower seed number per spike and shorter stems compared with populations growing in other regions. Perhaps specific environmental conditions or founder effects and genetic drift have caused recent establishment of a particular genotype in this area.
In the longer term, analysis of genes related to adaptation, deeper analysis of nucleotypic and other measures of genome variation (Bennett and Leitch, 2005a, b), and additional quantitative analysis of chromosomal and morphological diversity is required to analyse and understand the events of speciation and evolution of genomes (Leitch et al., 2005). Anonymous marker types such as SSRs (Saeidi et al., 2006) or IRAPs (Figs 1 and 2) are appropriate for detecting polymorphisms at different levels. SSRs are particularly valuable at the individual level and within populations, as emphasized by their use in varietal identification within crops, whereas retroelement polymorphisms give results that can be interpreted within taxa. Higher levels of classification may require use of selected SSR or IRAP markers, or markers derived from chromosomes or DNA sequences.
SUPPLEMENTARY INFORMATION
Supplementary Information is available online at www.aob.oxfordjournals.org/ and lists the origin and taxonomic grouping of the 57 Aegilops tauschii accessions used in this study.
LITERATURE CITED
- Altinkut A, Kotseruba V, Kirzhner VM, Nevo E, Raskina O, Belyayev A. Ac-like transposons in populations of wild diploid Triticeae species: comparative analysis of chromosomal distribution. Chromosome Research. 2006;14:307–317. doi: 10.1007/s10577-006-1048-3. [DOI] [PubMed] [Google Scholar]
- Avise JC. Phylogeography. Cambridge, MA: Harvard University Press; 2000. [Google Scholar]
- Badaeva ED, Amosova AV, Muravenko OV, Samatadze TE, Chikida NN, Zelenin AV, Friebe B, Gill BS. Genome differentiation in Aegilops. Evolution of D-genome cluster. Plant Systematics and Evolution. 2002;231:163–190. [Google Scholar]
- Bennett MD. Nucleotypic basis of the spatial ordering of the chromosomes in eukaryotes and implications of the order for genome evolution and phenotypic variation. In: Dover GA, Flavell RB, editors. Genome evolution. London: Academic Press; 1982. pp. 239–261. [Google Scholar]
- Bennett MD. The genome, the natural karyotype and biosystematics. In: Grant WF, editor. Plant biosystematics. Toronto, Canada: Academic Press; 1984. pp. 41–66. [Google Scholar]
- Bennett MD, Leitch IJ. Plant genome size research: a field in focus. Annals of Botany. 2005;a 95:1–6. doi: 10.1093/aob/mci001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MD, Leitch IJ. Nuclear DNA amounts in angiosperms: progress, problems and prospects. Annals of Botany. 2005;b 95:45–90. doi: 10.1093/aob/mci003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boguslavsky RL. Evolution of the pollination type in the genus Aegilops L. in: Botanical and genetic resources of Dagestan flora. Mahachkala. 1981:50–53. (in Russian) 1981. [Google Scholar]
- Boyko E, Kalendar R, Korzun V, Gill B, Schulman AH. Combined mapping of Aegilops tauschii by retrotransposon, microsatellite, and gene markers. Plant Molecular Biology. 2002;48:767–790. doi: 10.1023/a:1014831511810. [DOI] [PubMed] [Google Scholar]
- Cao Q, Lu BR, Xia H, Rong J, Sala F, Spada A, Grassi F. Genetic diversity and origin of weedy rice (Oryza sativa f. spontanea) populations found in north-eastern China revealed by simple sequence repeat (SSR) markers. Annals of Botany. 2006;98:1241–1252. doi: 10.1093/aob/mcl210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darroch JN, Mosimann JE. Canonical and principal components of shape. Biometrika. 1985;72:241–252. (cited and implemented in NTSYSpc version 02e) [Google Scholar]
- Dudnikov AJ. Allozyme variation in transcaucasian populations of Aegilops squarrosa. Heredity. 1998;80:248–258. [Google Scholar]
- Dvorak J, Luo M-C, Yang Z-L, Zhang H-B. The structure of the Aegilops tauschii genepool and the evolution of hexaploid wheat. Theoretical and Applied Genetics. 1998;97:657–670. [Google Scholar]
- Eig A. Monographisch-kritische übersicht der Gattung Aegilops. Feddes Repertorium Specierum novarum regni vegetabilis. 1929;55:1–228. [Google Scholar]
- Ellstrand NC, Prentice HC, Hancock JF. Gene flow and introgression from domesticated plants into their wild relatives. Annual Review of Ecology and Systematics. 1999;30:539–563. [Google Scholar]
- Eyre-Walker A, Gaut RL, Hilton H, Feldman DL, Gaut BS. Investigation of the bottleneck leading to the domestication of maize. Proceedings of National Academy of Sciences of the USA; 1998. pp. 4441–4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell AJ, Dunbar E, Anderson R, Pearce SR, Hartley R, Kumar A. Tyl-copia group retrotransposons are ubiquitous and heterogeneous in higher plants. Nucleic Acids Research. 1992;20:3639–3644. doi: 10.1093/nar/20.14.3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell AJ, Knox MR, Pearce SR, Ellis THN. Retrotransposon-based insertion polymorphisms (RBIP) for high throughput marker analysis. Plant Journal. 1998;16:643–650. doi: 10.1046/j.1365-313x.1998.00334.x. [DOI] [PubMed] [Google Scholar]
- Flavell RB, Bennett MD, Smith JB, Smith DB. Genome size and the proportion of repeated nucleotide sequence DNA in plants. Biochemical Genetics. 1974;12:257–269. doi: 10.1007/BF00485947. [DOI] [PubMed] [Google Scholar]
- Hammer K. Vorarbeiten zur monographischen Darstellung von Wildpflanzensortimenten. Aegilops L. Kulturpflanze. 1980;28:33–180. [Google Scholar]
- Heslop-Harrison JS, Brandes A, Taketa S, Schmidt T, Vershinin AV, Alkhimova EG, et al. The chromosomal distributions of Ty1-copia group retrotransposable elements in higher plants and their implications for genome evolution. Genetica. 1997;100:197–204. [PubMed] [Google Scholar]
- Jaaska V. Aspartate aminotransferase and alcohol dehydrogenase isoenzyme: intraspecific differentiation in Aegilops tauschii and the origin of the D genome polyploids in the wheat group. Plant Systematics and Evolution. 1981;137:259–273. [Google Scholar]
- Jaccard P. Nouvelles recherches sur la distribution florale. Bulletin de la Société Vaudoise des Sciences Naturelles. 1908;44:223–270. (cited and implemented in NTSYSpc program, version 02e) [Google Scholar]
- Kalendar R, Grab T, Regina M, Suoniemi A, Schulman AH. IRAP and REMAP: two new retrotransposon-based DMA fingerprinting techniques. Theoretical and Applied Genetics. 1999;98:704–711. [Google Scholar]
- Kalendar R, Tanskanen J, Immonen S, Nevo E, Schulman AH. Genome evolution of wild barley (Hordeum spontaneum) by BARE-1 retrotransposon dynamics in response to sharp microclimatic divergence. Proceedings of National Academy of Sciences of the USA; 2000. pp. 6603–6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara H. In: Kimura M, Ohta T., editors. Ancestors of common wheat; Proceedings of the National Academy of Sciences of the USA; Tokyo: Sogensha Tokyo (in Japanese); 1947. pp. 2848–2852. Cited by On some principles governing molecular evolution. [Google Scholar]
- Kihara H, Yamashita K, Tanaka M. Results of the Kyoto University Scientific Expedition to the Karakoram and Hindukush. Kyoto, Japan: Kyoto University; 1965. Morphological, physiological, genetical, and cytological studies in Aegilops and Triticum collected in Pakistan, Afghanistan, and Iran; pp. 1–118. Cited by Tanaka (1983) [Google Scholar]
- Kubis S, Schmidt T, Heslop-Harrison JS. Repetitive DNA elements as a major component of plant genomes. Annals of Botany. 1998;82(Supp.A):45–55. [Google Scholar]
- Leitch IJ, Soltis DE, Soltis PS, Bennett MD. Evolution of DNA amounts across land plants (Embryophyta) Annals of Botany. 2005;95:207–217. doi: 10.1093/aob/mci014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelley T, Stachel M, Grausgruber H, Vollmann J. Analysis of relationships between Aegilops tauschii and the D genome of wheat utilizing microsatellites. Genome. 2000;43:661–668. [PubMed] [Google Scholar]
- Li W, Zhang P, Fellers JP, Friebe B, Gill BS. Sequence composition, organization, and evolution of the core Triticeae genome. Plant Journal. 2004;40:500–511. doi: 10.1111/j.1365-313X.2004.02228.x. [DOI] [PubMed] [Google Scholar]
- Liu K, Muse SV. PowerMarker: integrated analysis environment for genetic marker data. Bioinformatics. 2005;21:2128–2129. doi: 10.1093/bioinformatics/bti282. [DOI] [PubMed] [Google Scholar]
- Lubbers EL, Gill KS, Cox TS, Gill BS. Variation of molecular markers among geographically diverse accessions of Triticum tauschii. Genome. 1991;34:354–361. [Google Scholar]
- Manninen O, Kalendar R, Robinson J, Schulman AH. Application of BAR/M retrotransposon markers to the mapping of a major resistance gene for net blotch in barley. Molecular and General Genetics. 2000;264:325–334. doi: 10.1007/s004380000326. [DOI] [PubMed] [Google Scholar]
- Mantel NA. The detection of disease clustering and a generalized regression approach. Cancer Research. 1967;27:209–220. (cited and implemented in NTSYSpc version 02e) [PubMed] [Google Scholar]
- Morrell PL, Lundy KE, Clegg MT. Distinct geographic patterns of genetic diversity are maintained in wild barley (Hordeum vulgare ssp. spontaneum) despite migration. Proceedings of National Academy of Sciences of the USA; 2003. pp. 10812–10817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai Y. Isosyme variation in Aegilops and Triticum, IV. The origin of the common wheats revealed from the study on esterase isozymes in synthesized wheats. Japanese Journal of Genetics. 1979;54:175–189. [Google Scholar]
- Nei M, Li WH. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proceedings of National Academy of Sciences of the USA; 1979. pp. 5269–5273. (cited and implemented in NTSYSpc program, version 02e) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen KM, Purugganan MD. Molecular evolution on the origin and evolution of glutinous rice. Genetics. 2002;162:941–950. doi: 10.1093/genetics/162.2.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen KM, Schaal BA. Evidence on the origin of cassava: phylogeography of Manihot esculenta. Proceedings of National Academy of Sciences of the USA; 1999. pp. 5586–5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecinka A, Suchánková P, Lysak MA, Trávnícek B, Dolezel J. Nuclear DNA content variation among central European Koeleria taxa. Annals of Botany. 2006;98:117–122. doi: 10.1093/aob/mcl077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestsova E, Korzun V, Goncharov NP, Hammer K, Ganal MW, Röder MS. Microsatellite analysis of Aegilops tauschii germplasm. Theoretical and Applied Genetics. 2000;101:100–106. [Google Scholar]
- Saeidi H, Rahiminejad MR, Vallian S, Heslop-Harrison JS. Biodiversity of diploid D-genome Aegilops tauschii Coss. in Iran measured using microsatellites. Genetic Resources and Crop Evolution. 2006;53:1477–1484. [Google Scholar]
- Shimamura M, Yasue H, Ohshima K, Abe H, Kato H, Kishiro T, et al. Molecular evidence from retroposons that whales form a clade within even-toed ungulates. Nature. 1997;388:666–670. doi: 10.1038/41759. [DOI] [PubMed] [Google Scholar]
- van Slageren MW. Wild wheats: a monograph of Aegilops L. and Amblyopyrum (Jaub. & Spach) Eig. Wageningen, The Netherlands: Wageningen Agricultural University; 1994. [Google Scholar]
- Smarda P, Bures P. Intraspecific DNA content variability in Festuca pallens on different geographical scales and ploidy levels. Annals of Botany. 2006;98:665–678. doi: 10.1093/aob/mcl150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smarda P, Bures P, Horová L. Random distribution pattern and non-adaptivity of genome size in a highly variable population of Festuca pallens. Annals of Botany. 2007;100:141–150. doi: 10.1093/aob/mcm095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi C, Marshal JA, Bennett MD, Leitch IJ. Genomic relationships between maize and its wild relatives. Genome. 1999;42:1201–1207. doi: 10.1139/gen-42-6-1201. [DOI] [PubMed] [Google Scholar]
- Tanaka M. Geographical distribution of Aegilops species based on the collections at the Plant Germplasm Institute, Kyoto University. In: Sakamoto S, editor. Proceedings of the 6th International Wheat Genetics Symposium; 1983. pp. 1009–1024. [Google Scholar]
- Teo CH, Tan SH, Ho CL, Faridah QZ, Othman YR, Heslop-Harrison JS, Kalendar R, Schulman AH. Genome constitution and classification using retrotransposon-based markers in the orphan crop banana. Journal of Plant Biology. 2005;48:96–105. [Google Scholar]
- Voytas DF, Cummings MP, Konieczny AK, Ausubel FM, Rodermel SR. Copia-like retrotransposons are ubiquitous among plants. Proceedings of National Academy of Sciences of the USA; 1992. pp. 7124–7128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waugh R, McLean K, Flavell AJ, Pearce SR, Kumar A, Thomas BBT, Powell W. Genetic distribution of BARE-1-like retrotransposable elements in the barley genome revealed by sequence-specific amplification polymorphisms (S-SAP) Molecular and General Genetics. 1997;253:687–694. doi: 10.1007/s004380050372. [DOI] [PubMed] [Google Scholar]