Abstract
Background and Aims
In published studies, positive relationships between nucleotype and the duration of the mitotic cell cycle in angiosperms have been reported but the highest number of species analyzed was approx. 60. Here an analysis is presented of DNA C-values and cell cycle times in root apical meristems of angiosperms comprising 110 measurements, including monocots and eudicots within a set temperature range, and encompassing an approx. 290-fold variation in DNA C-values.
Methods
Data for 110 published cell cycle times of seedlings grown at temperatures between 20–25 °C were compared with DNA C-values (58 values for monocots and 52 for eudicots). Regression analyses were undertaken for all species, and separately for monocots and eudicots, diploids and polyploids, and annuals and perennials. Cell cycle times were plotted against the nuclear DNA C-values.
Key Results
A positive relationship was observed between DNA C-value and cell cycle time for all species and for eudicots and monocots separately, regardless of the presence or absence of polyploid values. In this sample, among 52 eudicots the maximum cell cycle length was 18 h, whereas the 58 monocot values ranged from 8–120 h. There was a striking additional increase in cell cycle duration in perennial monocots with C-values greater than 25 pg. Indeed, the most powerful relationship between DNA C-value and cell cycle time and the widest range of cell cycle times was in perennials regardless of ploidy level.
Conclusions
DNA replication is identified as a rate limiting step in the cell cycle, the flexibility of DNA replication is explored, and we speculate on how the licensing of initiation points of DNA replication may be a responsive component of the positive nucleotypic effect of C-value on the duration of the mitotic cell cycle.
Key words: Cell cycle times, DNA C-value, nucleotype, ploidy level
INTRODUCTION
When meiocytes produce gametes, nuclear DNA amount drops from 4C to 1C. The latter is defined as the amount of nuclear DNA in the unreplicated genome of a gamete. Syngamy results in a 2C-zygote, which then replicates its nuclear DNA to 4C, which then falls to 2C in each daughter cell as a result of mitosis. Hence, the DNA C-value alternates between 2C and 4C in proliferative cells, mainly located in the root and shoot apical meristems, together with cells of the pericycle and cambia. DNA C-value continues to increase in endoreduplicating cells in which nuclear DNA replication is uncoupled from mitosis. In eukaryotes, DNA C-values are not correlated with genic complexity. Indeed, evolution has permitted very large amounts of non-genic DNA to become interspersed between comparatively small amounts of genic DNA along eukaryote chromosomes. This evolutionary enigma (see Gregory, 2001) was defined by Thomas (1971) as the DNA C-value paradox.
The first published analysis of the influence of DNA C-value on cell cycle time was by Van't Hof and Sparrow (1963). Six diploid species grown at 23 °C revealed a positive relationship between DNA amount and cell cycle time. In a separate study, seven eudicots and eight monocots also showed a strong correlation between cycle time and nuclear DNA amount (Evans et al., 1972). Bennett defined the physical effect of the mass of nuclear DNA (comprising both genic and non-genic DNA) on the phenotype as the nucleotype and authored/co-authored a series of seminal studies showing that nuclear DNA amount was positively related to a number of whole-organism and cellular traits. In one such study, Bennett (1972) reported on cell cycle duration and DNA C-value for 31 angiosperms grown at 23–25 °C (Bennett, 1972). Nineteen annuals had a significantly lower mean cell cycle time than eight obligate perennials, but not when compared with the cycle times of four facultative perennials.
A more recent analysis encompassed 25 monocot and 36 eudicot species (Ivanov, 1978) from a compilation of known cell cyle times and C-values (Grif and Ivanov, 1975). Once again, a clear positive relationship was established between DNA C-value and cell cycle time in separate regression analyses of data from monocots and eudicots. In this paper, Ivanov emphasized the need to examine narrower taxonomic groups than encompassed by the 61 species in his analysis. Indeed, a recurrent observation in these analyses was that combining data from diploid and polyploid species tended to undermine the positive effect of DNA C-value on cell cycle time. Since then, few studies have focused on this relationship between cell cycle duration and DNA C-value. However, Grif and Ivanov (1980) compiled further cell cycle data but appear not to have related these to DNA C-value. Additionally, a data set from 44 species was reviewed by Delbos (1983), but no new conclusions were drawn.
The number of C-values reported for angiosperms now stands at approximately 4400 (www.rbgkew.org.uk/cval/database1.html, Release 4·0; October 2005) out of approximately 250 000 taxonomically identified flowering plant species world-wide, but there are far fewer published cell cycle times for angiosperms.
The aim of the work reported here was to collate as many angiosperm cell cycle times as possible and analyze the relationship between DNA C-value and cell cycle time. We (a) present 110 separate cell cycle times spanning a 290-fold variation in DNA C-values, which to our knowledge becomes the largest cell cycle time survey to date, (b) revisit nucleotypic effects on these cycle times, and (c) indicate the extent to which different regulatory components of the plant cell cycle might be important factors in mediating nucleotypic effects. All genome size data are presented as the holoploid genome size or C-value (see Greilhuber et al., 2005).
The most frequently used technique for measuring cell cycle duration is the fraction (or percentage) of labelled mitoses (FLM/PLM) method (Quastler and Sherman, 1959). Radioactive thymidine is used to track labelled cells from one mitosis to the next. The root apical meristem (RAM) has been a popular choice for measuring cell cycle length because it can easily be pulsed with [methyl-3H] thymidine (3H-TdR) for a short time (mostly 15–60 min), and then transferred to non-radioactive medium. The plant material is then sampled at regular (hourly) intervals following the radioactive pulse.
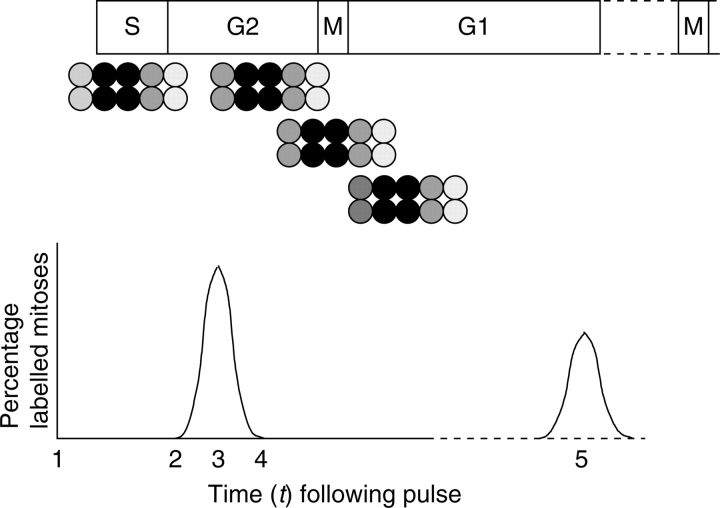
Roots are fixed and, most commonly, are stained by the Feulgen reaction (Feulgen and Rossenbeck, 1924). The RAM is then either squashed to become a cellular monolayer or wax-embedded and sectioned. The resulting squash preparations or de-waxed sections are dipped into photographic emulsion; Ilford K2 is particularly sensitive to the β-emissions from tritium (Rogers, 1979). Alternatively sections are overlaid with AR10 stripping film. After 10–14 days at 5 °C in darkness, the slides are developed and fixed as permanent autoradiographs. By pulse-labelling, β-emissions can be tracked from labelled nuclei throughout the cell cycle (Fig. 1). Proliferative cells labelled in S-phase that progress to mitosis will show chromosomes covered by silver grains. If, following the pulse with 3H-TdR, RAM cells are monitored over a period of time, the PLM data will conform to a curve that begins to rise when cells labelled at the end of S-phase traverse G2 and enter prophase of mitosis (Fig. 1). The curve peaks when the bulk of labelled cells enter mitosis, plateaus when the rate of entry and exit of labelled mitotic cells is similar, and plummets as the final cells labelled in early S complete mitosis (Fig. 1). If sampled for long enough, the labelled cohort progresses through interphase and exhibits a second peak of labelled mitoses (Fig. 1) so that the interval between the first and second peaks of the PLM curve represents one cell cycle time (CCT; Quastler and Sherman, 1959).
Fig. 1.
Idealised population of cells. At time t1, the cells are exposed to and incorporate [methyl-3H] thymidine (for 1 h) and are then pulse-chased with cold (non-radioactive) thymidine. Note that the cells at either end of the cluster will be lightly labelled as they move out of, or into, S-phase during the course of the pulse label. The most heavily labelled cells (e.g. in a 5-h S-phase) will be those that were in early-to-mid- S-phase during the pulse. The PLM (percentage of labelled mitoses) curve rises as the leading cells of the cluster arrive at mitosis (t2), peaks when the bulk of labelled cells are in mitosis (t3) and begins to fall as the final labelled cells exit mitosis (t4). A second peak (t5) would occur when the labelled cohort arrive at the next mitosis.
Table 1 presents the temperature range over which CCT would be expected to double (Q10) for temperatures between 20–25 °C for a few species, and the range of these coefficients implies that they might affect CCT(s). However, the mean Q10 of 2·498 ± 0·175 s.e. (±s.d. of 0·525) indicates that variation within the data set as a whole is relatively small.
Table 1.
The relationship between temperature (20–25 °C) and cell cycle time (CCT) expressed as a respiratory quotient (Q10) in root apical meristems
| Species | Q10 of CCT | Estimated CCT at 0 °C | Author* |
|---|---|---|---|
| Allium cepa | 2·54 | 129·2 | 1 |
| A. cepa | 2·22 | 106·1 | 2 |
| Helianthus annuus | 3·30 | 145·3 | 3 |
| Pisum sativum | 1·98 | 82·8 | 4 |
| Triticum aestivum | 1·69 | 37·1 | 5 |
| Vicia faba | 3·02 | 186·3 | 6 |
| V. faba | 3·00a | 269·5 | 7 |
| Zea mays | 2·36 | 79·7 | 8 |
| Z. mays | 2·37 | 90·2 | 9 |
* Key to authors: 1, López-Sáez et al. (1966); 2, González-Fernández et al. (1971); 3, Burholt and Van't Hof (1971); 4, Gudkov et al. (1974); 5, Bayliss (unpubl. res., but quoted by Bennett and Finch, 1972); 6, Murín (1966); 7, Evans and Savage (1959); 8, Verma and Walden (1969); 9, Verma, 1980.
a This value is for the cycle duration calculated from the rate of accumulation of colchicine-induced metaphases and therefore represents the potential doubling time; all other estimates for Q10 apply to the cell cycle duration measured directly. The above authors reported results from which these estimates of Q10 were made.
The PLM technique has faced some criticisms over the years (e.g. Green, and Bauer, 1977; Baskin, 2000; Tardieu and Granier, 2000). Space does not permit a detailed critique of the PLM method compared with other chemical or non-invasive techniques. However, autoradiography is excellent in tracking cells in S-phase and mitosis. Here we discuss briefly some of the criticisms.
Supplying a radioactively labelled nucleoside such as 3H-TdR to a biological system has its perils (see De la Torre and Clowes, 1974; Stetka and Webster, 1975). Indeed, some of the early measures of cell cycles involved high levels of radioactivity. Over the years, the specific activity of 3H-TdR used has lessened as autoradiographic techniques became more sensitive. Also, in many cases, the RAMs were pulsed and then transferred to (chased with) cold (non-radioactive) thymidine. Certainly the pulse–chase is a good way of flushing out uncombined radioactivity. Other markers of DNA synthesis, such as 5'-bromodeoxyuridine coupled with Giemsa staining, track labelled mitoses in the same way as the PLM technique and arrive at cycle time estimates for RAMs (Evans and Filion, 1980). However, to our knowledge, this technique has seldom been used for measuring plant cell cycle times.
Concern has been expressed that PLM data could ignore the passage of cells through the meristem during the post-labelling interval (Green and Bauer, 1977). Indeed, some PLM data have been collected at a fixed distance from the root tip. This means that different cohorts of cells will have been scored over the course of the experiment. However, PLMs used to determine ‘average’ cycle times, such as those discussed here, are constructed by analysing cells from the entire meristem and do not include those that fall outside of the meristematic domain.
Because PLM curves never rise to 100 %, and because the second peak is always smaller than the first, it has been suggested by some workers that labelled cells drop out of the cell cycle over the period of the experiment but are retained within the RAM. However, the RAM is largely made up of files of cells developed according to geometric doubling, i.e. these files consist of contiguous packets of (for example) 2, 4, 8, 16, 32, 64 cells (Lück et al., 1994). At each cycle, about half of all the meristematic cells are lost to the elongation zone, and the same number of new cells is created. If half are lost, then the second peak would be expected to have half the area (or maybe height) of the first peak. We contend, therefore, that the PLM technique, while imperfect, provides reliable estimates of cell cycle times, and that these are suitable for comparative assessments.
ANALYTICAL METHODS USED HERE
We compared 110 published cell cycle times with DNA C-values (58 values for monocots and 52 for eudicots). We focused on data for CCTs in RAMs, believing that the cells in question constitute a relatively uniform population from which legitimate comparative analyses can be made. In order to investigate the relationship between DNA C-value and CCT, only cell cycle times obtained from the same (PLM) method for RAMs of seedlings grown at temperatures between 20 and 25 °C were collected. This temperature range also enables more species to be included compared with data from, say, those grown at 20 °C. For certain species (e.g. Vicia faba), a number of CCT values have been published, but in others only a single value is known. This led to some repetition of values for a given species due to the various reports from independent researchers. In other instances (e.g. Dactylis glomerata), measurements made on different natural populations with differing C-values are included as well as data from different cultivars. However, our analyses do not extend to component phase durations and no attempt is made to re-interpret any of the data reported on here. Regression analyses were undertaken for all species, and separately for monocots and eudicots, diploids and polyploids, and annuals and perennials. Cell cycle times were plotted against the nuclear DNA C-values (see Table 3).
Table 3.
Species, DNA C-value (pg), cell cycle time in hours (CCT), temperature used (°C), eudicot (D) or monocot (M), annual (A) or perennial (P), and ploidy level
| Species | C-value | CCT | °C | D/M | A/P | Ploidy | Author |
|---|---|---|---|---|---|---|---|
| 1. Aegilops squarrosa | 5·10 | 11·4 | 20 | M | A | 2 | Davies and Rees (1975) |
| 2. Ae. umbellulata | 5·05 | 10·7 | 20 | M | A | 2 | Kidd (1987) |
| 3. Agoserris heterophylla6 | 1·15 | 8·8 | 24 | D | A | 2 | Price and Bachmann (1976) |
| 4. A. retrorsa6 | 3·20 | 9·0 | 24 | A | 2 | Price and Bachmann (1976) | |
| 5. Agrostis capillaris ‘Parys’ | 4·09 | 12·0 | 20 | M | P | 2 | Walker (1993) |
| 6. Allium carinatum | 11·20 | 9·2 | 22 | M | P | 2 | Bösen and Nagl (1978) |
| 7. A. cepa | 16·40 | 18·8 | 20 | M | P | 2 | Van't Hof and Bjerknes (1981) |
| 8. A. cepa | 16·75 | 24·0 | 25 | M | P | 2 | Grant (1964) |
| 9. A. fistulosum | 12·38 | 18·8 | 23 | M | P | 2 | Van't Hof (1965) |
| 10. A. tuberosum8 | 17·40 | 20·6 | 23 | M | P | 2 | Van't Hof (1965), Matagne (1968), Bryant (1969) |
| 11. Anacyclus clavatus | 5·25 | 11·0 | 22 | D | A | 2 | Nagl (1974, 1978) |
| 12. A. depressus | 6·20 | 16·0 | 22 | D | P | 2 | Nagl (1974, 1978) |
| 13. A. radiatus | 8·45 | 14·0 | 22 | D | A | 2 | Nagl (1974, 1978) |
| 14. Anthemis austriaca | 4·80 | 7·0 | 22 | D | A | 2 | Nagl (1974, 1978) |
| 15. A. cota | 7·90 | 6·0 | 22 | D | A | 2 | Nagl (1974, 1978) |
| 16. A. tinctoria | 3·75 | 12·0 | 22 | D | P | 2 | Nagl (1974, 1978) |
| 17. Arabidopsis thaliana | 0·16 | 8·5 | 23 | D | A | 2 | Van't Hof et al. (1978) |
| 18. Artemisia absinthum6 | 3·65 | 9·5 | 22 | D | P | 2 | Nagl (1974, 1978) |
| 19. A. annua | 1·88 | 8·0 | 22 | D | A | 2 | Nagl (1974, 1978) |
| 20. Avena pilosa | 4·73 | 8·9 | 25 | M | A | 2 | Yang and Dodson (1970) |
| 21. A. strigosa | 4·00 | 9·5 | 20 | M | A | 2 | Yang and Dodson (1970) |
| 22. A. strigosa | 4·006 | 10·0 | 20 | M | A | 2 | Yang and Dodson (1970) |
| 23. Bellavalia romana6 | 12·00 | 21·0 | 25 | M | P | 2 | Jona (1966) |
| 24. Beta vulgaris | 1·25 | 16·0 | 21 | D | P | 2 | Tiţu and Popovici (1970) |
| 25. Chamerion angustifolium | 0·40 | 10·0 | 20 | D | P | 2 | Thomas (1992) |
| 26. Crepis capillaris8 | 2·10 | 10·0 | 23 | D | A | 2 | Langridge et al. (1970), Van't Hof (1965), Abraham et al. (1968) |
| 27. C. neglecta | 2·90 | 10·0 | 23 | D | A | 2 | Langridge et al. (1970) |
| 28. C. tectorum | 3·38 | 12·0 | 23 | D | A | 2 | Langridge et al. (1970) |
| 29. Cucurbita pepo | 0·55 | 18·0 | 23 | D | A | 4 | Marciniak et al. (1978) |
| 20. Dactylis glomerata ‘Sylvan’ | 3·30 | 9·0 | 20 | M | P | 2 | Creber (1994) |
| 31. D. glomerata pop. A1 | 3·30 | 7·0 | 20 | M | P | 2 | Creber et al. (1993) |
| 32. D. glomerata pop. B1 | 5·39 | 10·0 | 20 | M | P | 4 | Creber et al. (1993) |
| 33. D. glomerata pop. C1 | 4·40 | 14·0 | 20 | M | P | 4 | Creber et al. (1993) |
| 34. D. glomerata pop. D1 | 4·71 | 21·0 | 20 | M | P | 4 | Creber et al. (1993) |
| 35. D. glomerata pop. E1 | 4·56 | 14·0 | 20 | M | P | 4 | Creber et al. (1993) |
| 36. D. glomerata pop. F1 | 4·74 | 10·0 | 20 | M | P | 4 | Creber et al. (1993) |
| 37. Daucus carota | 1·00 | 8·0 | 25 | D | A-P2 | 2 | Bayliss (1975) |
| 38. Epilobium hirsutum | 0·30 | 7·0 | 20 | D | P | 2 | Thomas (1992) |
| 39. Eragrostis tef | 0·70 | 9·7 | 20 | M | A | 4 | Kidd (1987) |
| 40. Festuca rubra ‘Merlin’ | 4·73 | 16·0 | 20 | M | A | 2 | Powell et al. (1986) |
| 41. F. rubra ‘S59’ | 4·73 | 16·4 | 20 | M | A | 2 | Powell et al. (1986) |
| 42. Glycine max (‘AGS 292 Taisho Shiroge’) | 1·13 | 7·0 | 23 | M | A | 2 | Reckless (1995) |
| 43. Glycine max (‘AGS 129 Shih’ × ‘SRF 400’) | 1·13 | 8·0 | 23 | M | A | 2 | Reckless (1995) |
| 44. Haplopappus gracilis | 2·05 | 12·0 | 20 | D | A | 2 | Sparvoli et al. (1966) |
| 45. Helianthus annuus | 2·43 | 12·0 | 23 | D | A | 2 | Burholt and Van't Hof (1971) |
| 46. Impatiens balsamina | 1·33 | 9·0 | 23 | D | A | 2 | Van't Hof (1965) |
| 47. Hordeum bulbosum7 | 8·28 | 14·0 | 20 | M | A | 2 | Kidd (1987) |
| 48. H. vulgare | 5·50 | 13·0 | 20 | M | P | 2 | Kidd (1987) |
| 49. Humulus lupulus | 2·90 | 10·0 | 20 | D | A | 2 | Yamamoto and Yamaguchi (1969) |
| 50. Lathyrus tingitanus | 9·05 | 16·8 | Ng9 | D | A | 2 | Evans et al. (1972) |
| 51. L. hirsutus | 9·98 | 18·0 | Ng9 | D | A | 2 | Evans et al. (1972) |
| 52. Lactuca sativa | 2·65 | 10·0 | 20 | D | A | 2 | Mazzuca et al. (2000) |
| 53. Lilium longiflorum | 35·2 | 24·0 | 20 | M | P | 2 | Kidd (1987) |
| 54. Linum usitatissimum | 0·70 | 14·0 | 22 | D | A | 2 | Evans et al. (1972) |
| 55. Lolium perenne | 2·08 | 8·1 | 20 | M | P | 2 | Evans et al. (1972) |
| 56. Lycopersicum esculentum4 | 2·55 | 11·0 | 23 | D | A | 2 | Van't Hof (1965) |
| 57. L. esculentum4 | 2·55 | 15·0 | 20 | D | A | 2 | Tiţu (1967) |
| 58. L. esculentum4 | 2·55 | 13·0 | 20 | D | A | 2 | Tiţu (1967) |
| 59. Melandrium album5 | 2·88 | 15·5 | 25 | D | P | 2 | Choudhury (1969) |
| 60. Microseris bigelovii | 1·50 | 8·0 | 24 | D | A | 2 | Price and Bachmann (1976) |
| 61. M. douglassii | 1·20 | 8·0 | 24 | D | A | 2 | Price and Bachmann (1976) |
| 62. M. lacianata lacianata | 3·35 | 9·0 | 24 | D | P | 2 | Price and Bachmann (1976) |
| 63. M. laciniata leptosepala6 | 3·40 | 10 | 24 | D | P | 2 | Price and Bachmann (1976) |
| 64. Nicotiana tabacum | 5·85 | 9·0 | 22 | D | A | 2 | Gupta (1969) |
| 65. N. plumbaginifolia | 4·15 | 11·0 | 22 | D | A | 4 | Gupta (1969) |
| 66. Nigella damascena | 10·55 | 16·5 | Ng9 | D | A | 2 | Evans et al. (1972) |
| 67. Ornithogalum umbellatum6 | 24·55 | 14 | 22 | M | P | 2 | Tagliaschi et al. (1983) |
| 68. Oryza sativa | 0·50 | 10·8 | 20 | M | A | 2 | Kidd (1987) |
| 69. Pennisetum americanum | 2·40 | 12·4 | 20 | M | A | 2 | Kidd (1987) |
| 70. Pisum sativum | 4·88 | 11·0 | 20–24 | D | A | 2 | Gudkov et al. (1974) |
| 71. P. sativum ‘Pioneer Gribovskoi’6 | 5·25 | 19·5 | 22·5 | D | A | 2 | Bogdanov et al. (1967) |
| 72. Pyrrhopappus caroliniana | 6·28 | 12·0 | 24 | D | B | 2 | Price and Bachmann (1976) |
| 73. P. multicaulis | 6·65 | 12·0 | 24 | D | A | 2 | Price and Bachmann (1976) |
| 74. Scilla sibirica | 31·65 | 67 | 23 | M | P | 3 | Baumann (1972) |
| 75. S. sibirica6 | 47·55 | 70 | 23 | M | P | 3 | Baumann (1972) |
| 76. Secale cereale | 8·30 | 12·4 | 20 | M | A | 2 | Wimber (1966) |
| 77. S. cereale ‘UC 90’ | 8·30 | 12 | 20 | M | A | 2 | Kidd (1987) |
| 78. S. cereale | 8·30 | 13·4 | 20–25 | M | A | 2 | Kidd (1987) |
| 79. S. cereale8 | 16·60 | 13·0 | 22 | M | A | 4 | Karpovskaya and Balyaeva (1973), Fadeyeva and Shakhla (1974) |
| 80. Sorghum bicolor | 1·68 | 13·9 | 20 | M | A | 4 | Kidd (1987) |
| 81. Tradescantia paludosa | 20·63 | 20·1 | 20–22 | M | P | 2 | Wimber (1966) |
| 82. Trillium erectum | 42·25 | 84·0 | 20 | M | P | 2 | Boothroyd and Mark (1980) |
| 83. T. erectum | 42·25 | 29·0 | 20 | M | P | 2 | Van't Hof and Sparrow (1963) |
| 84. T. grandiflorum | 46·00 | 120 | 25 | M | P | 2 | Van't Hof and Sparrow (1963) |
| 85. ×Triticosecale T7'6 | 16·80 | 11·0 | 20 | M | A | 6 | Kidd (1987) |
| 86. ×Triticosecale ‘Cocorit’ × ‘UC90’6 | 16·80 | 12·0 | 20 | M | A | 6 | Kidd (1987) |
| 87. ×Triticosecale | 19·80 | 12·1 | 20 | M | A | 6 | Kaltsikes (1971) |
| 88. Triticum aestivum | 17·30 | 10·6 | 20 | M | A | 6 | Francis and Barlow (1988) |
| 89. T. aestivum ‘Lyallpur’ | 17·30 | 14·0 | 20 | M | A | 6 | Hanif (1993) |
| 90. T. aestivum ‘Lu-26-sg’ | 17·30 | 12·0 | 20 | M | A | 6 | Hanif (1993) |
| 91. T. dicoccoides | 12·28 | 12·7 | 20 | M | A | 4 | Davies and Rees (1975) |
| 92. T. monococcum | 6·23 | 11·5 | 20 | M | A | 2 | Davies and Rees (1975) |
| 93. T. monococcum6 | 7·30 | 12·5 | 20 | M | A | 2 | Davies and Rees (1975) |
| 94. T. spelta6 | 18·90 | 19·7 | 20 | M | A | 6 | Davies and Rees (1975) |
| 95. T. timopheevi | 11·30 | 15·0 | 20 | M | A | 4 | Davies and Rees (1975) |
| 96. T. turgidum ‘Cocorit’ | 12·28 | 12·0 | 20 | M | A | 4 | Kidd (1987) |
| 97. T. turgidum var. durum | 10·95 | 11·0 | 20 | M | A | 4 | Kaltsikes (1971) |
| 98. T. turgidum var. durum8 | 10·95 | 13·9 | 20 | M | A | 4 | Valovich (1974), Boothroyd and Mark (1980) |
| 99. Tulipa kaufmaniana | 22·58 | 30·0 | 20 | M | P | 2 | Kidd (1987) |
| 100. T. kaufmanniana | 22·55 | 16·1 | 23 | M | P | 2 | Tagliasacchi and Vocaturo (1977) |
| 101. Vicia faba 0–13 | 13·33 | 14·0 | 20 | D | A | 2 | Taha (1989) |
| 102. V. faba 1–23 | 13·33 | 12·0 | 20 | D | A | 2 | Taha (1989) |
| 103. V. faba 2–33 | 13·33 | 12·0 | 20 | D | A | 2 | Taha (1989) |
| 104. V. faba 3–43 | 13·33 | 14·0 | 20 | D | A | 2 | Taha (1989) |
| 105. V. faba 4–53 | 13·33 | 14·0 | 20 | D | A | 2 | Taha (1989) |
| 106. V. faba | 13·33 | 18·0 | 20–25 | D | A | 2 | Essad (1973) |
| 107. V. faba | 13·33 | 13·0 | 22–24 | D | A | 2 | Schubert and Meister (1977) |
| 108. V. sativa | 2·25 | 12·0 | 20 | D | A | 2 | Essad (1973) |
| 109. Zea mays | 2·73 | 11·9 | 20 | M | A | 2 | Kidd (1987) |
| 110. Z. mays | 2·73 | 13·0 | 20 | M | A | 2 | Marciniak et al. (1978) |
Prime DNA C-values were obtained from the Plant DNA C-values database, Royal Botanic Gardens, Kew (release 4·0, Oct. 2005, http://www.kew.org/cvalues/homepage.html).
1Natural populations of Dactylis glomerata from different regions of Europe.
2Facultative biennial classed here as a perennial.
3A basipetal sequence of cell cycle times measured in different 1-mm segments of primary roots of 7-d-old seedlings at 20 °C, as measured from the junction between root cap and meristem.
4Now treated as Solanum lycopersicum.
5Now treated as Silene latifolia.
6C-values, listed neither as prime nor median on the C-values database, taken from the original paper/review.
7A median C-value as presented on the DNA C-values database.
8CCTs sourced to several different authors are means from each lab.
9Ng, not given.
RESULTS AND DISCUSSION
DNA C-value is positively related to cell cycle time
In the past there has been the troubling question of whether the inclusion of cell cycle times from both diploid and polyploid species disrupted or confounded correlations between C-value and cycle time (see Evans et al., 1972). The present, largest-ever collection of cell cycle times in angiosperms is inconsistent with nucleotypic effects that are confounded by ploidy level because there was a highly significant regression (P < 0·001) for all species (Fig. 2A; Table 2). However, the slope of the regression was unduly influenced by estimates of CCT in monocots with DNA C-values greater than 25 pg (Fig. 2A). Excluding these outliers from the regression reduced the slope, b, to a value of 0·873 ( P < 0·001). Hence, a strong nucleotypic effect on cell cycle time occurs regardless of phylogeny, although it was not linear across the entire range of C-values (Fig. 2A). When the 58 monocot measurements were considered, the slope was b = 1·29 (Table 2), but it reduced to 0·873 if CCTs of the six monocots with C-values > 25 pg were removed from the analysis (P < 0·001). Hence, another interesting feature of these data is that the distinctive kinetic change in the gradient of cycle time on C-value (b = 3·12, n = 5) is accounted for solely by monocots with C-values > 25 pg (Fig. 2A). However, the limitation of a sample size of n = 6 for the monocot outliers renders this regression non-significant. More cell cycle time data would be required to test the idea of a kinetic change to cell cycle times at a threshold C-value of 25 pg. A similar analysis based on eight monocots failed to show a positive relationship between cycle time and DNA C-value (Kidd et al., 1987). However, the current data confirm earlier analyses of the positive relationship between cell cycle time and DNA C-value in diploid populations of angiosperms (Van't Hof and Sparrow, 1963; Bennett, 1971; Ivanov, 1978).
Fig. 2.
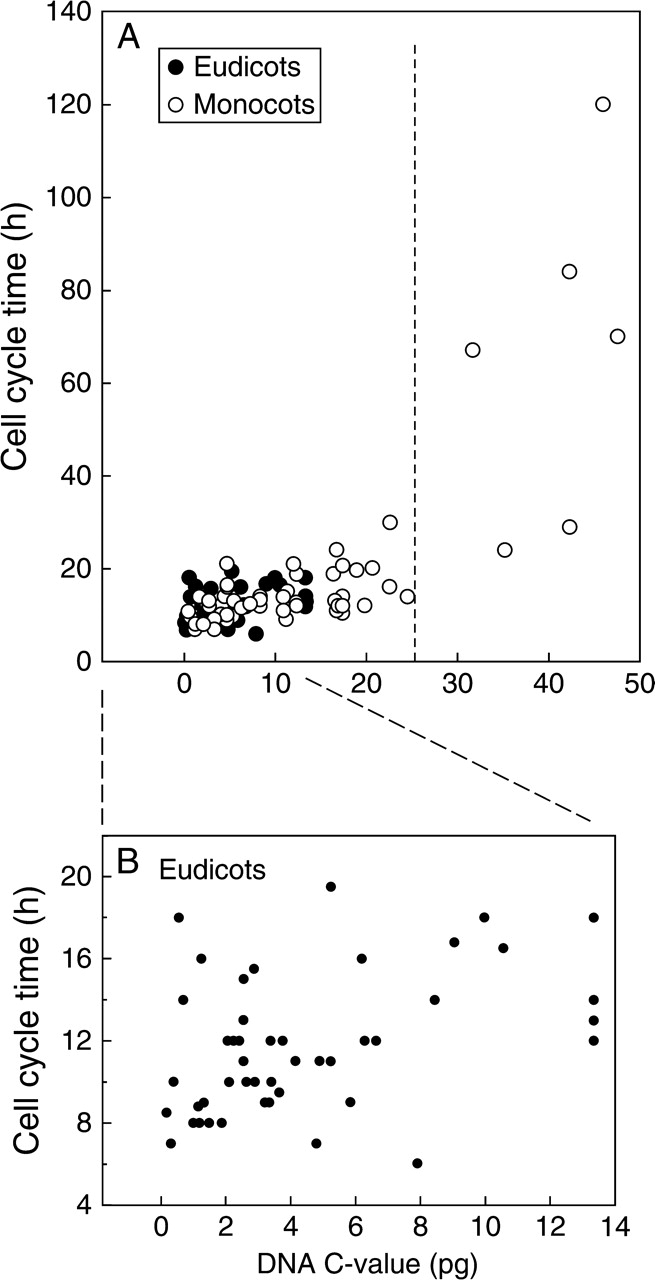
DNA C-value (pg) and cell cycle time (h) in the root apical meristem of a range of (A) eudicots and monocots (n = 110), and (B) eudicots (n = 60). See Table 2 for regression analyses.
Table 2.
Regression analyses of all data presented in Figs. 2–4 together with the percentage variance accounted for by the regression (R2), the level of probability (P) for each regression
| Regression (y = bx + a) | R2 | P | n | |
|---|---|---|---|---|
| All measurements | y = 1·09x + 5·39 | 54·2 | *** | 110 |
| Monocots | y = 1·29x + 2·44 | 58·7 | *** | 48 |
| Eudicots | y = 0·32x + 10·2 | 15·4 | *** | 62 |
| Diploids | y = 1·04x + 4·95 | 49·86 | *** | 86 |
| Polyploids | y = 1·14x + 3·12 | 56·3 | *** | 24 |
| Annuals | y = 0·20x + 10·7 | 19·9 | *** | 75 |
| Perennials | y = 1·37x + 4·13 | 63·6 | *** | 35 |
*** P < 0·001; n, number of replicates.
The spread of the plot for C-values and cell cycle times is much narrower for eudicots than for monocots (Fig. 2B); CCTs did not exceed 20 h for eudicots occurring, as they do, within a narrow window of C-values (∼1–14 pg; Fig. 2B). However, analyses of C-value in monocots closest to this limit began to include species in which cell cycle time exceeded 20 h (e.g. Bellavalia romana, 1C = 12 pg, cycle time = 21 h; Table 3). The data reveal a hitherto unsuspected tight clustering of eudicot cell cycle times ranging from 8 to 20 h (Fig. 2B), and a minimum cycle time that did not exceed 20 h compared with a much greater spread of cycle times for the monocots. If DNA mass per se is the limiting factor for cell cycle time, we hypothesize that cycle times would be the same for dicots and monocots of comparable C-value. This is so even if the data for Scilla sibirica and Trillium grandiflorum are excluded. Indeed, if we ignore the marked discontinuity of the y-axis caused by their inclusion, then the nucleotypic effect is strong for all species regardless of phylogeny. To test the rigour of these hypotheses would require data to plug the gap between Trillium grandiflorum and the majority of C-value/cell cycle times analysed here.
Separate plots for diploids and polyploids show a strong nucleotypic effect on CCT in diploids (Fig. 3; Table 2). Removing the five diploid outliers (>25 pg) reduced the slope (b = 0·27) by approximately four-fold but the regression continued to be significant (P < 0·001). For the polyploids, a nucleotypic effect on CCT was also detected (Fig. 3; Table 2); however, removing the two polyploid outliers rendered the regression non-significant (y = 0·03x − 13·5). This confirms previous work in which the slope/rate of increase in CCT with increasing DNA was higher in diploids than in autopolyploids (Evans et al., 1972). With the exception of Scilla sibirica, CCT in polyploids is generally more buffered than in diploids (Fig. 3).
Fig. 3.
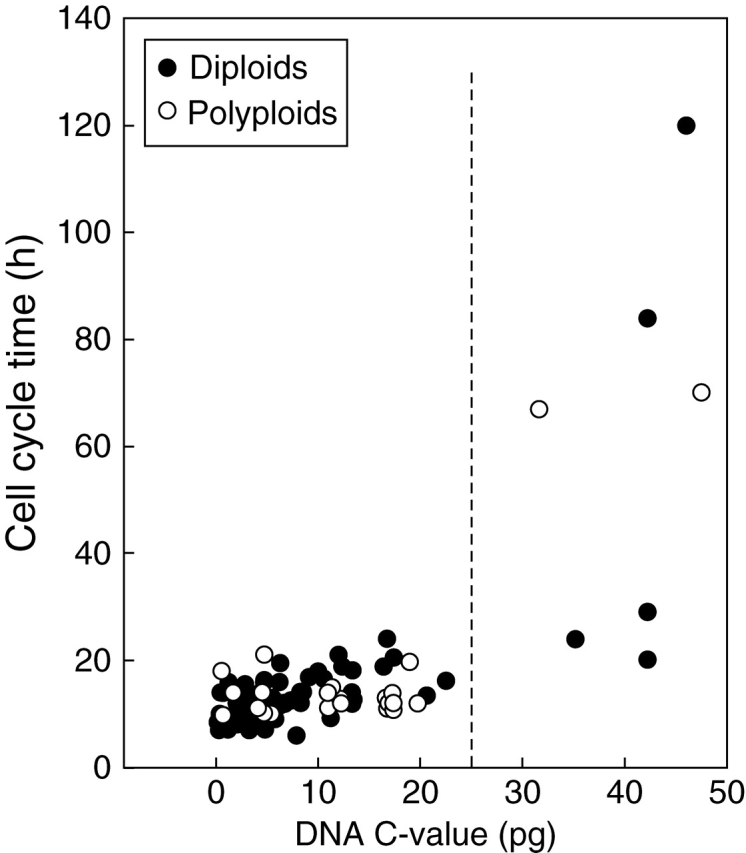
DNA C-value (pg) and cell cycle time (h) in the root apical meristem of a range of diploid and polyploid angiosperms. See Table 2 for regression analyses.
We acknowledge that some traditionally classified diploids are not necessarily so (see Soltis and Soltis, 1999). For example, there are strong arguments that Zea mays is actually an allotetraploid (2n = 4x = 20; Gaut and Doebly, 1997). However, in the data reported here we have assigned ploidy level as listed by the authors of the papers and reviews we have consulted.
The longest CCTs (>20 h) are exhibited by the perennials (Fig. 4). Indeed, the data for perennials overall had a nearly seven-fold steeper slope (b = 1·37) than a comparable regression for annuals (b = 0·20; Table 2). These data are consistent with findings of Bennett (1972) where the mean CCT in 19 annuals was significantly shorter than in eight obligate perennials. Where our analyses differ from Bennett (1972) is in relation to the broad range of CCTs shown by perennials compared with annuals (Fig. 4). However, in Fig. 4 the longer CCTs (>60 h) were recorded in perennials with C-values >25 pg. If they are removed from the regression analysis, the slope decreases (b = 0·70) but is still three-fold greater than that for the annuals (b = 0·20); some of the fastest CCTs are features of diploid perennials.
Fig. 4.
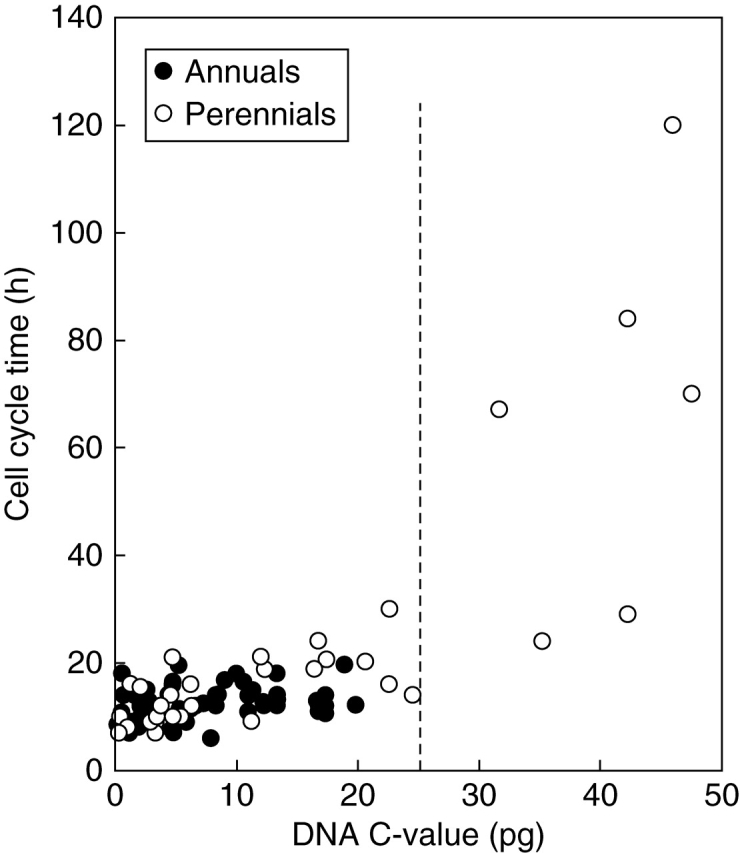
DNA C-value (pg) and cell cycle time (h) in the root apical meristem of a range of annuals and perennials. See Table 2 for regression analyses.
A change in kinetics of CCTs beyond a C-value threshold of 25 pg is once again observed in the perennial subset (>25 pg: b = 3·02, n = 6, P > 0·05; <25 pg: b = 0·70, n = 29, P < 0·001; Fig. 4). However, more values for perennials with C-values greater than 25 pg would be necessary to test this trend.
Nucleotypic effects and cyclin-dependent kinases: a theoretical assessment
Could there be a molecular basis for the nucleotypic correlations reported here? Many CCT data were collected prior to the application of molecular techniques to the cell cycle. Here we present a theoretical discussion of C-values in relation to the regulation of the cell cycle.
The cell cycle is carefully regulated at G2/M and G1/S, in which cyclin-dependent kinases (CDKs) are themselves activated by binding with their partner cyclins; this dual complex drives cells into mitosis and into S-phase (reviewed by Francis, 2007). At G2/M, two types of CDKs, A and B, are important to drive cells into mitosis, whereas only the A group is required at G1/S (Joubes et al., 2000). Hence, rates of cell division could be regulated by the expression of the A and B-type CDK genes. However, the plant cell cycle is remarkably plastic. For example, premature cell division at a small cell size can occur because CDKB activity is premature and G2 shortens. However, this is often followed by a compensatory increase in the length of G1 phase so that cell cycle duration remains constant (e.g. Orchard et al., 2005). Moreover, in RAMs a bootstrap L-system used to model the division patterns along cell files predicts that any premature induction of cell division has a compensatory mechanism that resets cell size for the next division (Lück et al., 1994), although in this model the cells divide at a constant size: it is the degree of symmetry of division that is the variable in shortening or lengthening cell cycles. If such compensation for asymmetric divisions does not occur, some cells would eventually disappear in a lethal down-spiralling of cell size (see Sveiczer et al., 1996).
One is reminded of the lack of correlation between nuclear DNA amount and organismal complexity, which is certainly the case for the genes required for the G2/M transition. For example, Arabidopsis thaliana, Arath;CDKA;1 exhibits an amino acid motif in its ά1 helix, PSTAIRE, which is very well conserved in cdc2 genes in a range of unrelated species, from fission yeast to A. thaliana.
A phylogenetic tree of plant CDKs was produced by Renaudin et al. (1996) that shows clustering of CDKAs according to family: Amaranthaceae, Apiaceae, Brassicaceae, Fabaceae, Plantaginaceae, Poaceae and Solanaceae. However, the number of higher plant species in which CDKA and CDKB types have been cloned is approximately 30. Nine CDKB types have been cloned, but some of these clones are for the same species, A. thaliana (e.g. Arath;CDKB;1 : 1, 1 : 2, 2 : 1 and 2 : 2). Moreover in Brassicaceae, there is a clustering together of CDKs for the ephemeral A. thaliana, and the facultative biennial Brassica napus. Hence, it seems unlikely that the molecular network of genes required for the G2/M transition shows sufficient interspecific variation to act as a sensitive component of a nucleotypic effect of DNA C-value on cell cycle time. Indeed, it also unlikely that interspecific differences between genes that regulate the G1/S transition show any relationship with nucleotypic effects. Note, genes that induce DNA replication, CDKAs, D-type cyclins and the S-phase transcription factor, E2F, have been cloned in A. thaliana, Medicago sativa, Oryza sativa and Zea mays (DeWitte and Murray, 2003).
In species within a ploidy level, the time taken to replicate nuclear DNA is invariably longer than the interval required to separate chromatids during mitosis. Hence, we presume that DNA replication is one rate-limiting factor of CCT. Indeed, the duration of S-phase was positively related to DNA C-value in a survey of eight diploid eudicot species at 20°C (Van't Hof, 1965) and in seven eudicots and eight monocot species (Evans and Rees, 1970). This was also the case in a separate study of 15 monocots (ten diploids + five polyploids) all grown at 20 °C (Fig. 5; Kidd, 1987).
Fig. 5.
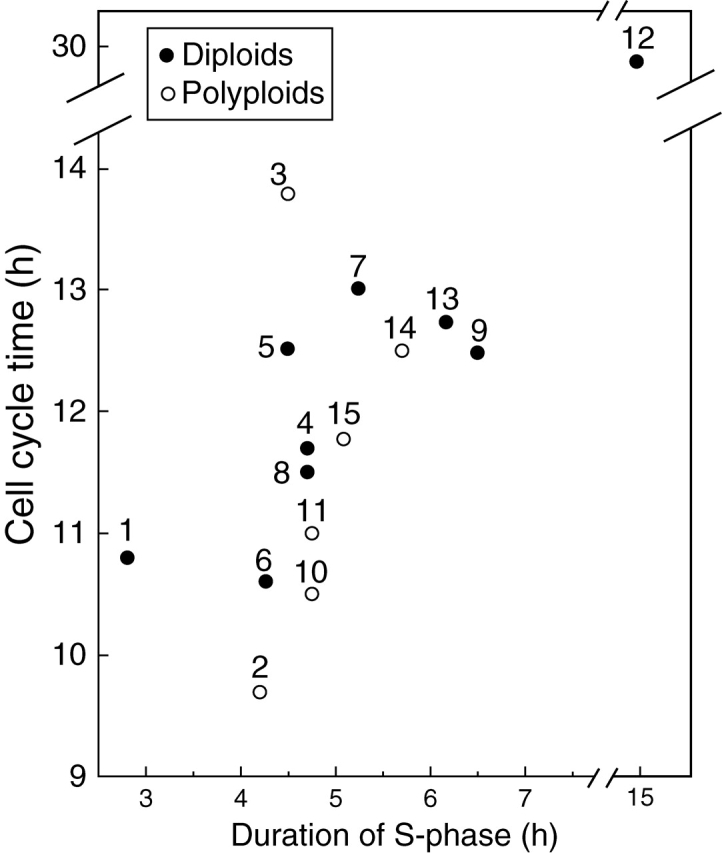
Cell cycle time (h) and the duration of S-phase (h) in root apical meristems of ten diploid and five polyploid monocots. Key: 1, Oryza sativa; 2, Eragrostis tef; 3, Sorghum bicolor; 4, Zea mays; 5, Pennisetum americanum; 6, Aegilops umbellulata; 7, Hordeum vulgare; 8, Triticum monococcum; 9, Secale cereale ‘Dominant’; 10, Triticum aestivum; 11, Triticosecale ‘T7’; 12, Tulipa kaufmanniana; 13, Secale cereale ‘UC 90’; 14, Triticum turgidum ‘Cocorit’; 15, “Triticosecale ‘Cocorit’” UC 90. y = 1·69x + 3·31 (n = 15, P < 0·001); minus outlier (12), y = 0·92x + 7·07 (n = 14, P < 0·001). Adapted from Kidd (1987).
DNA replication occurs when chromosomal DNA unwinds (albeit transiently) and replication is initiated from a replication origin. Initiation points are spaced at regular intervals throughout each chromosome but they fire asynchronously (Blow et al., 2001). In Xenopus embryos, DNA replication can begin anywhere, regardless of base sequence (Blow et al., 2001). This is different from the situation in Saccharomyces cerevisiae where initiation points are characterized as well-defined DNA sequences (Diffley and Labib, 2002).
In A. thaliana there are 30 000 replicons that function in two overlapping replication families (Van't Hof et al., 1978). However, little is known about how plant replicons are regulated, although we know something of their plasticity. For example, secondary initiation points could be induced by exposing cells of garden pea to UVB and a DNA cross-linker, psoralen (Francis et al., 1985b). Conversely, in lettuce, trigonelline treatment rendered some initiation sites dormant; this treatment lengthened S-phase and cell cycle time (Mazzuca et al., 2000).
Replication origins in diploid rye (2n = 2x = 14) are spaced at ∼60-kb intervals (Francis and Bennett, 1982) whereas in bread wheat (2n = 6x = 42) the distance reduces to 15-kb intervals (Francis et al., 1985a). In allohexaploid triticale obtained by crossing diploid rye with tetraploid wheat (2n = 6x = 42) initiation sites were detected every 15 kb (Kidd et al., 1992). Thus replicons spaced at 60-kb intervals in rye chromosomes in a rye background occur at the same frequency (15-kb intervals) as those of wheat chromosomes in the triticale background (15-kb intervals). It is the control of replicon activation that is important for regulating the cell cycle time. When we analysed different aspects of DNA replication in a range of monocot species, a striking positive correlation was observed between the synchrony of replicon activation and the rate of DNA replication per replication fork. In other words, as DNA amount increases, the synchrony of replicon activation becomes less and less (Kidd et al., 1989).
We regarded 60- and 15-kb intervals as strong and weak sites, respectively, and noted that S-phase need not lengthen because of an increased nuclear DNA amount per se. Note that whereas there is a two-to-three-fold increase in nuclear DNA amount in allohexaploid triticale compared with its parental diploid and tetraploids at 20°C, S-phase takes 5 h in each species mainly because, in the primary allohexaploid hybrid (triticale), weaker origins of DNA replication are brought into play in the replication process and hence moderate the effect of a large C-value (Kidd et al., 1992).
In Saccharomyces cerevisiae, replication initiation sites are known as autonomous replicating sequences (ARSs), which can initiate DNA replication in origin-less plasmids. ARSs have a conserved sequence (A/TTTTATG/ATTTA/T) and this provides a binding site for the origin recognition complex (ORC; Bell and Stillman, 1992; Cocker et al., 1996). Somewhat surprisingly, ARS assays have not proved useful in identifying replication origins in higher eukaryotes.
ORCs can comprise up to six polypeptides. They bind strongly to the replication origin (Bell and Stillman, 1992) and remain in place from cell cycle to cell cycle. The ORCs, CDC6/Cdc18 and Cdt1 promote the loading of MCM2-7 complexes onto replication origins during late mitosis and G1, thereby ‘licensing’ the origin for use in the subsequent S phase (Diffley, 2004; Blow and Dutta, 2005). As initiation of DNA replication occurs at each origin, MCM2-7 in conjunction with DNA topoisomerase II facilitate the unwinding of DNA in a gyrase-like activity either side of the replication origin; CDC45 stabilizes the gyrase complex (reviewed by Francis, 2007). The MCM complex moves ahead of the replication fork thereby ensuring that an initiation site fires only once in each cell cycle (Blow and Dutta, 2005). In A. thaliana, ORC genes (Gavin et al., 1995) CDT1 (Lin et al., 1999 ), CDC6 (Ramos et al., 2001), MCMs (Springer et al., 1995; Sabelli et al., 1996) and CDC45 (Stevens et al., 2004) have been cloned. In Pisum sativum, Pissa;MCM3 shows about 50 % homology with human MCM3 (Bryant et al., 2001). In Nicotiana tabacum, Nicta;CDC6 is expressed maximally in early S-phase, and its promoter contains an E2F consensus site (Drambruskas et al., 2003). Hence the plant genome is equipped with a genetic tool kit for successful licensing of DNA replication origins.
In diploids, as cell cycle times lengthen with increasing DNA C-value, S-phase also lengthens. The positive nucleotypic effect of DNA C-value on S-phase and on the cell cycle is particularly strong among diploid perennials and suggests a decrease in the frequency of licensing DNA replication origins. This may well occur while the chromosomes are in mitosis.
One question raised by these data is whether disproportionate increases in heterochromatin that occur as C-value increases (Flavell, 1980) alter the conformation of DNA and, in effect, shield these regions from licensing factors? In Xenopus, chromatin remodelling is a prerequisite for sperm nuclei to become licensed for nuclear DNA replication (Gillespie and Blow, 2000). Certainly the species with the highest DNA amount and the longest cell cycle included in our analysis, Scilla sibirica, is well endowed with heterochromatin, located mainly in intercalary and terminal regions of the chromosomes (Deumling and Greilhuber, 1982). Moreover, although the restriction enzyme, HaeIII, is able to cut heterochromatic regions of S. sibirica chromosomes, the DNA remains firmly enmeshed with chromosomal proteins making chromosomal DNA extraction extremely difficult (Lozano et al., 1992). Hence, the proportion of nuclear DNA classified as hetero- as opposed to euchromatic may well have an important bearing on the question of chromatin topology in relation to regulation of nuclear DNA replication.
B chromosomes cause a lengthening of cycle time in both S. cereale and Z. mays (Ayonoadou and Rees, 1968; Jones and Rees, 1968). Whether late replication of the B chromosomes in S-phase is responsible for increasing the duration of S-phase is unclear, because in S. cereale and Z. mays B chromosomes are late-labelling whereas in Lolium they are not so (Evans et al., 1972).
In conclusion, we report a strong and positive correlation between DNA C-value and cell cycle time, regardless of ploidy level and regardless of life cycle. However, the relationship is strongest among diploid perennials, suggesting that species with high DNA C-values within this group of plants exhibit proportionately more heterochromatin, which increases DNA mass and which may have an additive effect on the length of S-phase. We also hypothesize that licensing of DNA replication origins occurs at a lesser frequency in species carrying more heterochromatin. However, part of the mechanism is likely to depend on the pattern of replicon activation in relation to the rate of DNA replication fork movement. The tool kit necessary to further investigate nucleotypic effects on nuclear DNA replication and how this might influence cell cycle time is accumulating. However, more data, particularly from species with C-values >25 pg would be required to understand further nucleotypic effects on cell cycle times in angiosperms.
ACKNOWLEDGEMENTS
We thank those diligent scientists who strove through sleepless nights to obtain the cell cycle times that we discuss here. Thanks also to anonymous referees whose suggestions helped to improve the present paper.
LITERATURE CITED
- Abraham S, Ames IH, Smith HH. Autoradiographic studies of DNA synthesis in the B chromosomes of Crepis capillaris. Journal of Heredity. 1968;59:297–299. doi: 10.1093/oxfordjournals.jhered.a107724. [DOI] [PubMed] [Google Scholar]
- Ayonoadu U, Rees H. The influence of B chromosomes on chiasma frequency in Black Mexican sweet corn. Genetica. 1968;39:75–81. [Google Scholar]
- Baskin TI. On the constancy of cell division rate in the root meristem. Plant Molecular Biology. 2000;43:545–554. doi: 10.1023/a:1006383921517. [DOI] [PubMed] [Google Scholar]
- Baumann TW. Der Mitosezyklus im diploiden und tetraploiden Wurzelmeristem von Scilla sibirica. Experientia. 1972;28:860–862. [Google Scholar]
- Bayliss MW. The duration of the cell cycle of Daucus carota L. in vivo and in vitro. Experimental Cell Research. 1975;92:31–38. doi: 10.1016/0014-4827(75)90633-3. [DOI] [PubMed] [Google Scholar]
- Bell SP, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multi-protein complex. Nature. 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- Bennett MD. The duration of meiosis. Proceedings of the Royal Society of London, B. 1971;178:277–299. [Google Scholar]
- Bennett MD. Nuclear DNA content and minimum generation time in herbaceous plants. Proceedings of the Royal Society of London B. 1972;181:109–135. doi: 10.1098/rspb.1972.0042. [DOI] [PubMed] [Google Scholar]
- Bennett MD, Finch RA. The mitotic cycle time of root meristem cells of Hordeum vulgare. Caryologia. 1972;25:439–444. [Google Scholar]
- Blow JJ, Dutta A. Preventing re-replication of chromosomal DNA. Nature Reviews in Molecular and Cellular Biology. 2005;6:476–486. doi: 10.1038/nrm1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow JJ, Gillespie PJ, Francis D, Jackson DA. Replication origins in Xenopus egg extract are 5–15 kb apart and are activated in clusters that fire at different times. Journal of Cell Biology. 2001;152:15–25. doi: 10.1083/jcb.152.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanov Yu.F, Liapunova NA, Sherudilo AI. Cell population in pea embryos and root tip meristem. Microphometric and autoradiographic studies. Tsitologiya. 1967;9:569–576. (in Russian) [Google Scholar]
- Boothroyd ER, Mark NM. Mitotic cycles in root tip cells of two species of Trillium (Liliaceae) Canadian Journal of Genetics and Cytology. 1980;12:750–758. [Google Scholar]
- Bösen H, Nagl W. Short duration of the mitotic and endomitotic cell cycle in the heterochromatin-rich monocot Allium carinatum. Cell Biology International Reports. 1978;2:565–571. doi: 10.1016/0309-1651(78)90065-6. [DOI] [PubMed] [Google Scholar]
- Bryant JA, Moore K, Aves SJ. Origins and complexes: the initiation of DNA replication. Journal of Experimental Botany. 2001;52:193–202. [PubMed] [Google Scholar]
- Bryant TR. DNA synthesis and cell division in germinating onion II. Mitotic cycle and DNA content. Caryologia. 1969;22:139–148. [Google Scholar]
- Burholt DR, Van't Hof J. Quantitative thermal-induced changes in growth and cell population kinetics of Helianthus roots. American Journal of Botany. 1971;58:386–393. [Google Scholar]
- Choudhury HC. Late DNA replication pattern in sex chromosomes of Melandrium. Canadian Journal of Genetics and Cytology. 1969;11:192–198. doi: 10.1139/g69-023. [DOI] [PubMed] [Google Scholar]
- Cocker JH, Piatti S, Santocanale C, Nasmyth K, Diffley JF. An essential role for the Cdc6 protein in forming the pre-replicative complexes of budding yeast. Nature. 1996;379:180–182. doi: 10.1038/379180a0. [DOI] [PubMed] [Google Scholar]
- Creber HMC. The effects of temperature on cell dividsion and root and leaf morphology in natural populations of Dactylis glomerata L. of diverse geographical origin. Cardiff: University of Wales; 1994. PhD Thesis. [Google Scholar]
- Creber HMC, Davies MS, Francis D. Effects of temperature on cell division in root meristems of natural populations of Dactylis glomerata of contrasting latitudinal origins. Environmental and Experimental Botany. 1993;33:433–442. [Google Scholar]
- Davies POL, Rees H. Mitotic cycles in Triticum species. Heredity. 1975;35:337–345. [Google Scholar]
- De la Torre C, Clowes FAL. Thymidine and the measurement of rates of mitosis in meristems. New Phytologist. 1974;73:919–925. [Google Scholar]
- Delbos M. Le cycle mitotique chez les plantes: révue bibliographique. Agronomie. 1983;3:595–605. [Google Scholar]
- Deumling B, Greilhuber J. Characterisation of heterochromatin in different species of the Scilla sibirica group (Liliaceae) by in-situ hybridisation of satellite DNAs and fluorochrome banding. Chromosoma. 1982;84:535–555. [Google Scholar]
- DeWitte W, Murray JA. The plant cell cycle. Annual Review of Plant Biology. 2003;54:235–264. doi: 10.1146/annurev.arplant.54.031902.134836. [DOI] [PubMed] [Google Scholar]
- Diffley JF. Regulation of early events in chromosome replication. Current Biology. 2004;14:R778–786. doi: 10.1016/j.cub.2004.09.019. [DOI] [PubMed] [Google Scholar]
- Diffley JF, Labib K. The chromosome replication cycle. Journal of Cell Science. 2002;115:869–872. doi: 10.1242/jcs.115.5.869. [DOI] [PubMed] [Google Scholar]
- Drambruskas G, Bryant JA, Aves SJ, Rogers HJ, Francis D. MCM and CDC6 homologues in the tobacco TBY-2 cell line. Journal of Experimental Botany. 2003;54:699–706. doi: 10.1093/jxb/erg079. [DOI] [PubMed] [Google Scholar]
- Essad S. Variations du cycle mitotique et des teneurs en ADN chez Vicia sativa L. Mise en évidence de liaisons entre les durées des phases du cycle mitotique dans différent groupes botaniques. Caryologia. 1973;26:357–374. [Google Scholar]
- Evans GM, Rees H. Mitotic cycles in dicotyledons and monocotyledons. Nature. 1971;233:350–351. doi: 10.1038/233350a0. [DOI] [PubMed] [Google Scholar]
- Evans GM, Rees H, Snell CL, Sun S. The relation between nuclear DNA amount and the duration of the mitotic cycle. Chromosomes Today. 1972;3:24–31. [Google Scholar]
- Evans HJ, Savage JRK. The effect of temperature on mitosis and on the action of colchicine in root meristems of Vicia faba. Experimental Cell Research. 1959;18:51–61. doi: 10.1016/0014-4827(59)90290-3. [DOI] [PubMed] [Google Scholar]
- Evans KJ, Filion WG. Determination of cell cycle in plants using BrdU-FPG. Canadian Journal of Genetics and Cytology. 1980;22:305–308. [Google Scholar]
- Fadeyeva TS, Shakhla Z. Study of mitotic-cycle in roots of one tetraploid and two diploid forms of rye. Tsitologiya. 1974;16:698–703. (in Russian) [Google Scholar]
- Feulgen R, Rossenbeck H. Mikroskopisch-chemischer Nachweis einer Nucleinsäure vom Typus der Thymonucleinsäure und die darauf beruhende selektive Färbung von Zellkernen in mikroskopischen Präparaten. Hope-Seylers Zeitschrift für Physiologische Chemie. 1924;135:203–248. [Google Scholar]
- Flavell R. The molecular characterisation and organisation of plant chromosomal DNA sequences. Annual Review of Plant Physiology. 1980;31:569–596. [Google Scholar]
- Francis D. The plant cell cycle–15 years on. New Phytologist. 2007;174:261–278. doi: 10.1111/j.1469-8137.2007.02038.x. [DOI] [PubMed] [Google Scholar]
- Francis D, Barlow PW. Temperature and the cell cycle. Symposia for the Society for Experimental Biology. 1988;42:181–202. [PubMed] [Google Scholar]
- Francis D, Bennett MD. Replicon size and mean rate of DNA synthesis in rye (Secale cereale L. cv. Petkus Spring) Chromosoma. 1982;86:115–122. [Google Scholar]
- Francis D, Kidd AD, Bennett MD. DNA replication in relation to DNA C values. In: Bryant JA, Francis D., editors. The cell division cycle in plants. a. Cambridge: Cambridge University Press; 1985. pp. 61–81. [Google Scholar]
- Francis D, Davies ND, Bryant JA, Hughes SG, Sibson DR, Fitchett PN. Effects of psoralen on replicon size and mean rate of DNA synthesis in partially synchronized cells of Pisum sativum L. Experimental Cell Research. 1985;b 158:500–508. doi: 10.1016/0014-4827(85)90473-2. [DOI] [PubMed] [Google Scholar]
- Gaut BS, Doebly JF. DNA sequence evidence for the segmental allopolyploid origin of maize. Proceedings of the National Academy of Sciences of the USA. 1997;94:6809–6814. doi: 10.1073/pnas.94.13.6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin KA, Hidaka M, Stillman B. Conserved initiator proteins in eukaryotes. Science. 1995;270:1667–1771. doi: 10.1126/science.270.5242.1667. [DOI] [PubMed] [Google Scholar]
- Gillespie PJ, Blow JJ. Nucleoplasmin-mediated chromatin remodelling is required for Xenopus sperm nuclei to become licensed for DNA replication. Nucleic Acids Research. 2000;28:472–480. doi: 10.1093/nar/28.2.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Fernández A, Giménez-Martín G Díez JL, de la Torre C, López-Sáez JF. Interphase development and beginning of mitosis in the different nuclei of polynucleate homokaryotic cells. Chromosoma. 1971;36:100–111. [Google Scholar]
- Grant CJ. Induced chromosome breakage and the chromosome cycle. Oxford University; 1964. D. Phil. Thesis. [Google Scholar]
- Green PB, Bauer K. Analyzing changing cell cycle. Journal of Theoretical Biology. 1977;68:299–315. doi: 10.1016/0022-5193(77)90167-9. [DOI] [PubMed] [Google Scholar]
- Gregory TR. Coincidence, coevolution, or causation? DNA content, cell size and the C-value enigma. Biological Reviews. 2001;76:65–101. doi: 10.1017/s1464793100005595. [DOI] [PubMed] [Google Scholar]
- Greilhuber J, Doležel J, Lysák M, Bennett MD. The origin, evolution and proposed stabilization of the terms ‘genome size’ and ‘C-value’ to describe nuclear DNA amounts. Annals of Botany. 2005;95:255–260. doi: 10.1093/aob/mci019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grif VG, Ivanov VB. Temporary pattern of the mitotic cycle in flowering plants. Tsitologiya. 1975;17:694–717. (in Russian) [Google Scholar]
- Grif VG, Ivanov VB. The data on the temporary patterns of the mitotic cycle in flower plants. Tsitologiya. 1980;22:107–120. (in Russian) [Google Scholar]
- Gudkov IN, Petrova SA, Zezina NV. The effect of temperature on the duration of the mitotic stages in cells of the pea root meristem and its radioresistance. Fiziologiya i biokhimiya kul'turnykh rastenii. 1974;6:257–262. (in Russian) [Google Scholar]
- Gupta SB. Duration of mitotic cycle and regulation of DNA replication in Nicotiana plumbaginifolia and a hybrid derivative of N. tabacum showing chromosome instability. Canadian Journal of Genetics and Cytology. 1969;11:133–142. [Google Scholar]
- Hanif M. The effect of salt on cellular aspects of root growth in contrasting cereal cultivars. Cardiff: University of Wales; 1993. PhD Thesis. [Google Scholar]
- Ivanov VB. DNA content in the nucleus and rate of development in plants. Soviet Journal of Developmental Biology. 1978;9:39–53. [Google Scholar]
- Jona A. La durata del ciclo mitotico nelle Bellavalia romana determinate per ira autoradiografica mediante l'impiego della timidina 3H. Caryologia. 1966;19:429–442. [Google Scholar]
- Jones RN, Rees H. The influence of B chromosomes upon the nuclear phenotype. Chromosoma. 1968;24:158–176. [Google Scholar]
- Joubes J, Chevalier C, Dudits D, Heberle-Bors E, Inzé D, Umeda M, Renaudin JP. CDK-related protein kinases in plants. Plant Molecular Biology. 2000;43:607–620. doi: 10.1023/a:1006470301554. [DOI] [PubMed] [Google Scholar]
- Kaltsikes PJ. The mitotic cycle in an amphidiploid (triticale) and its parental species. Canadian Journal of Genetics and Cytology. 1971;13:656–662. [Google Scholar]
- Karpovskaya EV, Belyaeva ES. The duration of the period of DNA synthesis in the cells of diploid and tetraploid rye plants. Tsitologiya. 1973;15:104–107. (in Russian) [Google Scholar]
- Kidd AD. Studies on DNA replication and the cell cycle in the root meristem of fifteen monocotyledonous angiosperms of heterogeneous DNA C values. Cardiff: University of Wales; 1987. PhD Thesis. [Google Scholar]
- Kidd AD, Francis D, Bennett MD. Replicon size, mean rate of DNA replication and the duration of the cell cycle and its component phases in eight monocotyledonous species of contrasting C values. Annals of Botany. 1987;59:603–609. [Google Scholar]
- Kidd AD, Francis D, Bennett MD. Replicon size, and rate of DNA replication fork movement are correlated in grasses. Experimental Cell Research. 1989;184:262–267. doi: 10.1016/0014-4827(89)90385-6. [DOI] [PubMed] [Google Scholar]
- Kidd AD, Francis D, Bennett MD. Replicon size, rate of DNA replication and the cell cycle in primary hexaploid triticale and its parents. Genome. 1992;135:126–132. [Google Scholar]
- Langridge WHR, O'Malley TA, Wallace H. Neutral amphiplasticity and regulation of the cell cycle in Crepis herbs. Proceedings of the National Academy of Sciences of the USA. 1970;67:1894–1900. doi: 10.1073/pnas.67.4.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Kaul S, Rounsley S, Shea TP, Benito MI, Town CD, et al. Sequence and analysis of chromosome 2 of the plant Arabidopsis thaliana. Nature. 1999;402:761–768. doi: 10.1038/45471. [DOI] [PubMed] [Google Scholar]
- López-Sáez JF, Giménez-Martín G, González-Fernández A. Duration of the cell division cycle and its dependence on temperature. Zeitschrift für Zellforschung. 1966;75:591–600. doi: 10.1007/BF00341516. [DOI] [PubMed] [Google Scholar]
- Lozano R, Sentís C, Fernández-Piqueras J, Ruiz Rejón M. In situ digestion of satellite DNA of Scilla sibirica. Chromosoma. 1992;100:439–442. [Google Scholar]
- Lück J, Barlow PW, Lück HB. Cell genealogies in a plant meristem deduced with the aid of a. Cell Proliferation. 1994;27:1–21. doi: 10.1111/j.1365-2184.1994.tb01402.x. ‘bootstrap’ L-system. [DOI] [PubMed] [Google Scholar]
- Marciniak K, Olszewska MJ, Osiecka R, Bialas J. Relation between nuclear-DNA content and rate of cell-growth during interphase in 4 species of Angiospermae. Acta Societatis Botanicorum Poloniae. 1978;47:297–305. [Google Scholar]
- Matagne R. Duration of mitotic cycle and patterns of DNA replication in chromosomes of Allium cepa. Caryologia. 1968;21:209–224. [Google Scholar]
- Mazzuca S, Bitonti MB, Innocenti AM, Francis D. Inactivation of DNA replication origins by the cell cycle regulator, trigonelline, in root meristems of Lactuca sativa. Planta. 2000;211:127–132. doi: 10.1007/s004250000272. [DOI] [PubMed] [Google Scholar]
- Murín A. The effect of temperature on the mitotic cycle and its time parameters in root tips of Vicia faba. Naturwissenschaften. 1966;53:312–313. doi: 10.1007/BF00712233. [DOI] [PubMed] [Google Scholar]
- Nagl W. Mitotic cycle time in perennial and annual plants with various amounts of DNA and heterochromatin. Developmental Biology. 1974;39:342–346. doi: 10.1016/0012-1606(74)90248-6. [DOI] [PubMed] [Google Scholar]
- Nagl W. Endopolyploidy and polyteny in differentiation and evolution. The Netherlands: North-Holland Publishing Company; 1978. [Google Scholar]
- Orchard CB, Siciliano I, Sorrell Marchbank A, Rogers HJ, Francis D, et al. Tobacco BY-2 cells expressing fission yeast cdc25 bypass a G2/M block on the cell cycle. Plant Journal. 2005;44:290–299. doi: 10.1111/j.1365-313X.2005.02524.x. [DOI] [PubMed] [Google Scholar]
- Powell MJ, Davies MS, Francis D. The influence of zinc on the cell cycle in the root meristem of a zinc-tolerant and a non-tolerant cultivar of Festuca rubra. New Phytologist. 1986;102:419–428. doi: 10.1111/j.1469-8137.1986.tb00668.x. [DOI] [PubMed] [Google Scholar]
- Price HJ, Bachmann K. Mitotic cycle time and DNA content in annual and perennial Microseridinae (Compositae, Cichoriaceae) Plant Systematics and Evolution. 1976;126:323–330. [Google Scholar]
- Quastler H, Sherman FG. Cell population kinetics in the intestinal epithelium of the mouse. Experimental Cell Research. 1959;17:420–438. doi: 10.1016/0014-4827(59)90063-1. [DOI] [PubMed] [Google Scholar]
- Ramos GB, de Almeida Engler J, Ferreira PC, Hemerly AS. DNA replication in plants: characterization of a cdc6 homologue from Arabidopsis thaliana. Journal of Experimental Botany. 2001;52:2239–2240. doi: 10.1093/jexbot/52.364.2239. [DOI] [PubMed] [Google Scholar]
- Reckless DM. Cardiff: University of Wales; 1995. The effect of temperature on growth and development and cell division of Vicia faba and Glycine max roots. PhD Thesis. [Google Scholar]
- Renaudin JP, Doonan JH, Freeman D, Hashimoto J, Hirt H, Inzé D, et al. Plant cyclins: a unified nomenclature for plant A-, B- and D-type cyclins based on sequence organization. Plant Molecular Biology. 1996;32:1003–1018. doi: 10.1007/BF00041384. [DOI] [PubMed] [Google Scholar]
- Rogers AW. Techniques of autoradiography. 3rd edn. Amsterdam: Elsevier; 1979. [Google Scholar]
- Sabelli PA, Burgess SR, Kush AK, Young MR, Shewry PR. cDNA cloning and characterisation of a maize homologue of the MCM proteins required for the initiation of DNA replication. Molecular and General Genetics. 1996;252:125–136. doi: 10.1007/BF02173212. [DOI] [PubMed] [Google Scholar]
- Schubert I, Meister A. Zellzyklusuntersuchungen an Wurzelspitzenmeristemen rekonstruierter Karyotypen von Vicia faba. Biologisches Zentralblatt. 1977;96:183–201. [Google Scholar]
- Soltis DE, Soltis PS. Polyploidy: recurrent formation and genome evolution. Trends in Ecology and Evolution. 1999;14:348–352. doi: 10.1016/s0169-5347(99)01638-9. [DOI] [PubMed] [Google Scholar]
- Sparvoli E, Gay H, Kaufman BP. Duration of the mitotic cell cycle in Haplopappus gracilis. Caryologia. 1966;19:65–71. [Google Scholar]
- Springer PS, McCombie WR, Sundaresan V, Martienssen RA. Gene trap tagging of PROLIFERA, an essential MCM2-3–5-like gene in Arabidopsis. Science. 1995;268:877–880. doi: 10.1126/science.7754372. [DOI] [PubMed] [Google Scholar]
- Stetka DG, Webster PL. Tritiated-thymidine induced changes in cell population kinetics in root meristems of Pisum sativum. Radiation Research. 1975;64:475–484. [PubMed] [Google Scholar]
- Stevens R, Grelon M, Vezon D, Oh J, Meyer P, Perennes C, Domenichini S, Bergounioux C. A CDC45 homolog in Arabidopsis is essential for meiosis, as shown by RNA interference-induced gene silencing. Plant Cell. 2004;16:99–113. doi: 10.1105/tpc.016865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sveiczer A, Novak B, Mitchison JM. The size control of fission yeast revisited. Journal of Cell Science. 1996;109:2947–2957. doi: 10.1242/jcs.109.12.2947. [DOI] [PubMed] [Google Scholar]
- Tagliasacchi A, Vocaturo R. Effect of the seed ageing of Triticum durum cv. Cappelli on the length of mitotic cycle as measured by 3H-thymidine incorporation in the root meristem. Caryologia. 1977;30:225–230. [Google Scholar]
- Tagliasacchi A, Berta G, Fusconi A. Duration of the mitotic cycle of Ornithogalum umbellatum as measured by 3H-thymidine in the root meristem. Caryologia. 1983;36:189–193. [Google Scholar]
- Taha RM. Studies on the cellular behaviour of roots of Vicia faba L. in vivo and in vitro. Cardiff: University of Wales; 1989. PhD Thesis. [Google Scholar]
- Tardieu F, Granier G. Quantitative analysis of cell division in leaves: methods, developmental patterns and effects of environmental conditions. Plant Molecular Biology. 2000;43:555–567. doi: 10.1023/a:1006438321386. [DOI] [PubMed] [Google Scholar]
- Thomas CA. The genetic organisation of chromosomes. Annual Review of Genetics. 1971;5:1–27. doi: 10.1146/annurev.ge.05.120171.001321. [DOI] [PubMed] [Google Scholar]
- Thomas HC. Effects of manganese and phosphorus on cellular aspects of growth in contrasting species. Cardiff: University of Wales; 1992. PhD Thesis. [Google Scholar]
- Tiţu H. Durata cicluli mitotic şi a perioadei de sinteză a AND la tomatele haploide şi diploide. (Lycopersicon esculentum Mill.) Studii şi cercatări de Biologie, seria botanică. 1967;19:165–172. [Google Scholar]
- Tiţu H, Popovici I. Duration of mitotic cycle phases in the radicular meristem of diploid and tetraploid sugar beet (Beta vulgaris) Révue Roumaine de Biologie série de Botanique. 1970;15:51–56. [Google Scholar]
- Valovich EM. Action of low-temperature upon chromosomes of plants. Tsitologiya. 1974;16:33–37. (in Russian) [PubMed] [Google Scholar]
- Van't Hof J. Relationship between mitotic cell cycle duration and S period duration and the average rate of DNA synthesis in the root meristem cells of several plants. Experimental Cell Research. 1965;39:48–54. doi: 10.1016/0014-4827(65)90006-6. [DOI] [PubMed] [Google Scholar]
- Van't Hof J, Bjerknes CA. Similar replicon properties of higher plant cells with different S period and genome properties. Experimental Cell Research. 1981;136:461–465. doi: 10.1016/0014-4827(81)90027-6. [DOI] [PubMed] [Google Scholar]
- Van't Hof J, Sparrow AH. A relationship between DNA content, nuclear volume and minimal cycle time. Proceedings of the National Academy of Sciences of the USA. 1963;49:897–902. doi: 10.1073/pnas.49.6.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van't Hof J, Kuniyuki A, Bjerknes CA. The size and number of replicon families of chromosomal DNA of Arabidopsis thaliana. Chromosoma. 1978;68:269–285. [Google Scholar]
- Verma RS. The duration of G1, S, G2, and mitosis at four different temperatures in Zea mays L. as measured by 3H-thymidine. Cytologia. 1980;45:327–333. [Google Scholar]
- Verma RS, Walden DB. Temperature and nuclear cycle duration in Zea mays L. Agronomy Abstracts 1969 (61st Annual Meeting of the American Society of Agronomy, Detroit, MI) 1969:20. section C. [Google Scholar]
- Walker HD. Effects of N-form and concentration on root growth and meristematic activity in two contrasting grass species. Cardiff: University of Wales; 1993. PhD Thesis. [Google Scholar]
- Wimber DE. Duration of the nuclear cycle in Tradescantia root tips at three temperatures as measured with H3-thymidine. American Journal of Botany. 1966;53:21–24. [PubMed] [Google Scholar]
- Yamamoto K, Yamaguchi H. Chromosome duplication in root tip cells of the female plant of Humulus lupulus. Radioisotopes. 1969;18:91–96. [Google Scholar]
- Yang D-P, Dodson EO. The amounts of nuclear DNA and the duration of DNA synthetic period (S) in related diploid and autotetraploid species of oats. Chromosoma. 1970;31:309–320. [Google Scholar]