Abstract
Simian immunodeficiency virus (SIV) infection of macaques may result in encephalitis, a feature more commonly observed in macaques with rapid progressive disease than in those with conventional disease. This is the first report of two conventional progressors (H631 and H636) with encephalitis in rhesus macaques inoculated with a derivative of SIVsmE543-3. Phylogenetic analyses of viruses isolated from the cerebral spinal fluid (CSF) and plasma from both animals demonstrated tissue compartmentalization. Furthermore, these viruses appear to have undergone adaptive evolution to preferentially replicate in their respective cell targets in infectivity assays with monocyte derived macrophages and peripheral blood mononuclear cells. Analysis of the number of potential N-linked glycosylation sites (PNGS) in gp160 showed that there was a statistically significant loss of PNGS in viruses isolated from CNS in both macaques compared to SIVsmE543-3. These viruses provide a relevant model to study the adaptations required for SIV to induce encephalitis.
Keywords: SIV, neuroAIDS, encephalitis, adaptive evolution, preferential cell tropism, compartmentalization, potential N-linked glycosylation sites
INTRODUCTION
Human immunodeficiency virus type 1 (HIV-1) has been shown to invade the central nervous system (CNS) early after systemic infection and neurological complications caused or contributed by HIV-1 infection have been observed in patients with acquired immunodeficiency syndrome (AIDS). The introduction of highly active antiretroviral therapy (HAART) in 1996 has decreased the occurrence of many neurologic diseases including HIV-associated dementia (HAD) [1]. However, despite the initial success of HAART, the incidence rate of HAD has risen again due to the increase in the number of people living with HIV/AIDS and the prolonged life span of these individuals [2]. Clinically, HAD or AIDS dementia complex (ADC), is defined by the impairment of cognitive, behavioral, and motor function [3, 4]. Major neuropathological features characterizing HIV-1 infection in the brain are microglial nodules, reactive astrocytes, perivascular infiltrates, white matter pallor [5], and the presence of syncytia or multinucleated giant cells (MNGC) consisting of macrophages [5, 6]. This latter feature is considered the hallmark of HIV encephalitis (HIVE) [7]. In addition, perivascular cuffing, the accumulation of perivascular macrophages around blood vessels, is also observed [5]. The presence of HIVE is consistent with the diagnosis of ADC [3, 5].
Similar to HIV infection in humans, simian immunodeficiency viruses (SIV) can cause AIDS in macaques. The survival time for the majority of SIV-infected macaques that develop AIDS range from one to three years [8, 9]; however, a small percentage of infected animals will develop a rapid disease progression with death at approximately three to six months [9]. This shortened survival time of rapid progressor (RP) animals has been correlated with the presence of SIV-induced encephalitis (SIVE) [10, 11]. Histopathological changes in the brain such as MNGC, microglial nodules, and perivascular infiltrates identical to those seen in humans with HIVE, have also been observed in macaques with encephalitis [12, 13]. Indeed, SIV neuropathogenesis can recapitulate key elements of HIV-1 neuropathogenesis [14]. Therefore, the SIV-infected macaque is an excellent animal model to study the pathogenesis of HIV-associated encephalitis [12, 13].
Recently, we have observed SIVE in two rhesus macaques with conventional progressive disease, surviving greater than a year after inoculation. This situation more likely parallels that of HIV-infected patients who do not develop HIVE/dementia until late stage AIDS [15, 16] than do RP animals with SIVE. In the present study, we evaluated the sequential genetic and biologic evolution of SIV in the plasma, cerebral spinal fluid (CSF), and brain from these conventional progressors with encephalitis. Analyses of isolated viruses suggested diverging viral evolution and different selective pressures in the plasma and CSF resulting in compartmentalization of these viruses. Additionally, we observed a preferential infection of primary rhesus monocyte-derived macrophages (MDM) versus peripheral blood mononuclear cells (PBMC) by viruses isolated from CSF and brain. Conversely, viruses isolated from plasma more efficiently infected PBMC than did viruses isolated from CSF or brain.
MATERIALS AND METHODS
Macaques
All animals were housed in accordance with the NRC Guide for the Care and Use of Laboratory Animals [17].
In situ hybridization, immunohistochemistry, and confocal microscopy
Analysis of formalin fixed, paraffin embedded tissues for SIV RNA and the different cell surface markers was performed utilizing a method previously described [18].
Viral RNA quantification
Viral RNA, isolated from either CSF or plasma samples, were quantified as previously described [19].
Virus isolation and replication assay
To isolate virus, samples of cryopreserved CSF and plasma collected at week 76 (H636) and at euthanasia (week 116, H631) were incubated with primary pig-tail macaque PBMC. Additionally, for H631, virus was isolated from brain samples obtained during euthanasia. PBMC culture supernatant was filtered to generate cell-free virus stocks and RT assay was performed to normalize all virus stock. For virus replication assay, Ficoll-Hypaque density gradient centrifugation of EDTA-treated whole blood from four different rhesus macaques was performed to isolate PBMC.
Reverse Transcriptase Assay
An aliquot of cell-free virus-containing supernatant was removed every three days from infected cells and frozen at −80°C. Measurement of virion-associated reverse transcriptase (RT) activity was performed using P32 dTTP and all assays were done at the same time in order to ensure that the results could be compared with each other.
Sequence and Phylogenetic Analyses
Viral RNA was isolated from sequential CSF and plasma samples, as well as from isolated viruses. RT-PCR was performed as previously described [19]. To avoid amplification error bias of the template, the PCR reaction was performed three to four times and several clones were chosen for sequencing from each PCR reaction.
Nucleotide alignment of the gp120 or gp160 region sequences from sequential tissue samples or isolated virus, respectively, was done using MUSCLE and manually edited with BioEdit v5.0.9. Gaps in the alignment were stripped prior to phylogenetic analyses. A maximum likelihood tree incorporating site-specific nucleotide substitution rates was constructed using PHYLIP and DNArates 1.1.0. In addition, a transition/transversion ratio of 1.5 was used in the analyses. The tree was rooted on the parental SIVsmE543-3 sequence (accession no. U72748). Maximum likelihood bootstrapping support values were calculated with PHYLIP. Synonymous/non-synonymous analysis program (SNAP), http://www.hiv.lanl.gov/content/hiv-db/SNAP/WEBSNAP/SNAP.html was used to calculate the synonymous and non-synonymous substitution rates. Potential N-linked glycosylation sites (PNGS) were analyzed by N-Glycosite program (http://www.hiv.lanl.gov/content/hivdb/GLYCOSITE/glycosite.html). Comparison of the number of PNGS in Env of H631 virus isolates and H636 CSF virus isolates to SIVsmE543-3 Env was performed using one sample T test, two tailed. Comparison of the number of PNGS in Env of H631 brain virus isolates to that of H631 CSF or plasma virus isolates was performed with unpaired two sample T test, two tailed. P values < 0.05 were reported as significant.
RESULTS
Clinical and pathological features
As part of prior ongoing studies, rhesus macaque H445 was inoculated intravenously with the molecular clone, SIVsmE543-3 [20]. H445 developed a transient antibody and cytotoxic T lymphocyte (CTL) response, had high plasma and tissue viral load, and rapidly progressed to AIDS with euthanasia at week 16 post inoculation (p.i.) [21]. Previous studies have suggested that virus passaged in macaques have evolved to become more virulent [22–26] Therefore, we sought to evaluate the pathogenicity of SIV isolates from this rapid progressor. Uncloned virus was isolated from H445 mesenteric lymph node and inoculated intravenously into a cohort of six rhesus macaques [19].
All six animals became infected and had clinical and pathological symptoms characteristic of SIV-related disease [19]. Although six animals were inoculated with virus isolated from a RP, only H635 progressed rapidly [27] and was subsequently euthanized at week 9 p.i. [19]. The survival time of the other five conventional progressor animals ranged from 52 to 116 weeks p.i. [19].
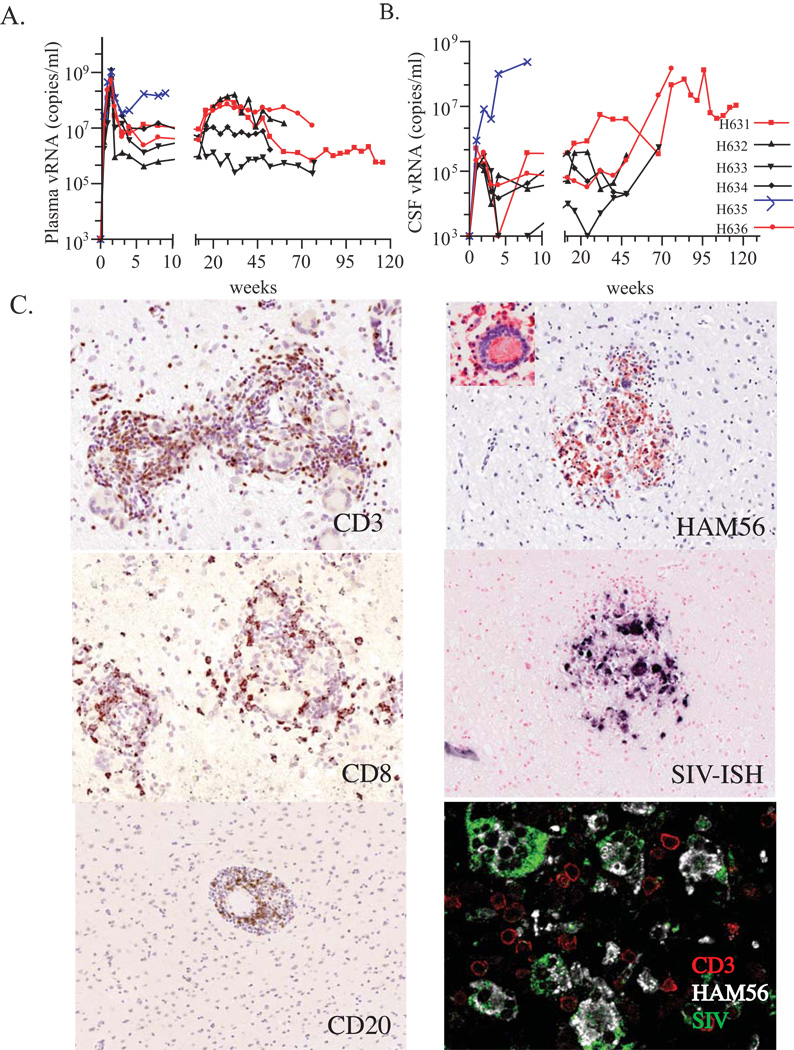
High levels of plasma viral RNA were detected in all animals during primary viremia. These levels generally decreased by one to two logs by week 4 p.i. and were maintained at these levels until death (figure 1A). The rapid progressor macaque, H635 was the one exception, showing increasing viral load until death. Detection of viral RNA in the CSF was observed by week 1 p.i. Only CSF viral RNA levels for H631, H635, and H636, increased terminally (figure 1B).
Fig. 1.
Clinical and pathological features. (A) Plasma and (B) CSF viral RNA levels. Samples were obtained at sequential time points post inoculation. Y axis represents viral RNA load (copies/ml) and the X axis represents weeks post inoculation. Macaques H631 (square symbol) and H636 (circle symbol) are shown in red and the rapid progressor macaque H635 is shown in blue. (C) SIV-specific in situ hybridization and immunohistochemistry of infiltrating cell populations in the brain of conventional progressor macaques with SIVE, H631 and H636. Prominent perivascular mononuclear infiltrates stained specifically with antibodies to CD3 (H631), CD8 (H631), and CD20 (H636) on the left identified by DAB substrate (brown). The right panels show the infiltration of macrophages as evident by staining with HAM56 in red (H636), with an inset of a characteristic HAM56+ multinucleated giant cell. A serial section shows the expression of SIV RNA in these infiltrating cells in blue. The bottom right panel shows confocal microscopy of the brain of H636 showing that SIV-expressing cells (green), co-expressed HAM56 (white) consistent with their identification as macrophages although uninfected CD3+ T cells (red) were also present.
CD4+ T cell levels gradually declined in all animals with a mean of 198 cells/µl at the time of euthanasia. The only exception was H635 where the number of CD4+ T cells recovered back to normal levels at death. Moderate to robust responses to SIV-Gag by week eight and to SIV-Env (gp160) by week 32 were observed in all animals except for H635 (data not shown).
Encephalitis Observed in Two Conventional Progressors
Histological examination of brain obtained during either necropsy or autopsy of all six animals revealed that half of the cohort developed SIV meningoencephalitis [19]. SIVE in these three animals was characterized by the presence of MNGC. Perivascular lymphocytic cuffing was observed in the brains of the two conventional progressors, H631 and H636, a pathologic feature not observed in the RP brain sections. Immunohistochemistry using anti-CD20 and anti-CD3 antibodies, demonstrated that the infiltrating cells consisted of B and T cells, respectively, and that these latter cells were predominantly CD8+ T cells (figure 1C). In contrast, only occasional T cells and no B cells were observed in the brain of H635. Additionally, infiltrating macrophages and MNGC were identified by HAM56, a macrophage marker, and these cells co-localized in lesions with SIV expression, as shown by in situ hybridization (ISH) (figure 1C). Triple label confocal microscopy using ISH for SIV RNA and immunofluorescence for HAM56 and anti-CD3 demonstrated that the SIV-expressing cells in the brain were macrophages (figure 1C).
Molecular evolution of SIV
To determine whether specific SIV variants were associated with the development of SIVE in these three animals, we examined the Env sequence of viral RNA amplified directly from CSF and plasma of H631, H636, and H635 and compared it to the inoculum, SIVsmH445. The SIVsmH445 virus inoculum stock showed few changes in the gp120 region of Env as compared to the parental SIVsmE543-3 [19]. Of the few mutations observed, the majority were characteristic RP-specific mutations [19, 28] that we have described previously in RP macaques. However, clones with these mutations were a minority population. Analyses of sequences obtained directly from CSF (data not shown) and plasma [19] of H635 at the time of death showed that those RP-specific mutations predominated in both compartments, consistent with specific selection of this minority viral variant from the inoculum. In contrast, sequencing of CSF and plasma from H631 and H636 did not reveal RP-specific mutations in any of the clones analyzed at any time point. Consequently, we have used the parental SIVsmE543-3 sequence for comparisons.
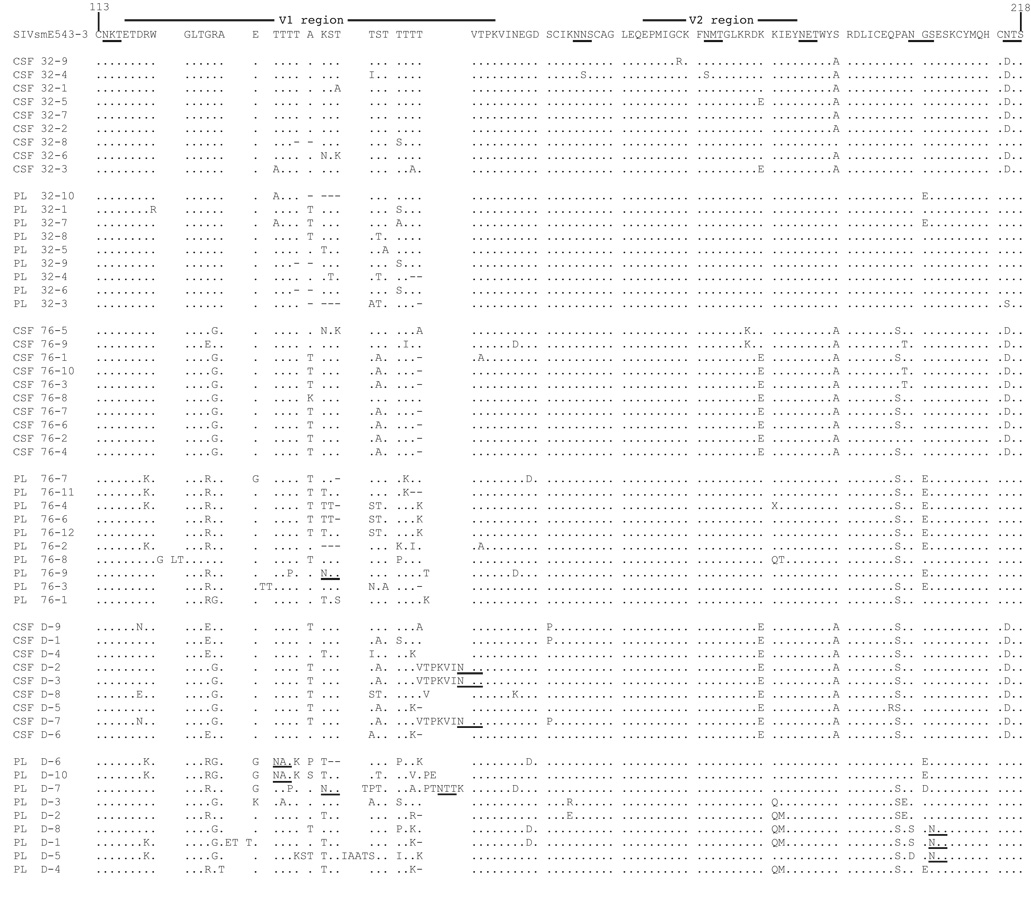
Sequential samples obtained during primary viremia and up to and including death at week 116 (H631) were analyzed. Animal H636 died unexpectedly; consequently, only samples from the last time point at week 76 p.i. were available. Sequencing analysis of the gp120 region revealed that virus during primary viremia in both compartments for both animals had few and random mutations compared to that of SIVsmE543-3. However, by week 32, mutations in the V1/V2 regions of gp120 could be observed (figure 2) with extensive changes seen by week 76. By week 32, in H631 CSF and plasma clones (figure 2A), except for two substitutions (S193A and N215D) in CSF, there were no consistent mutations observed. Interestingly, these two substitutions were maintained through death. The latter mutation resulted in a loss of a potential N-linked glycosylation site (PNGS). This suggests that these mutations may have conferred a selective advantage for viral fitness to the virus in H631 CSF.
Fig. 2.
Sequence analysis of the V1/V2 region of gp120 clones directly from CSF and plasma viral RNA in H631 (A) and H636 (B) obtained from weeks 32 and 76 post inoculation. In addition, samples obtained at week 116 (death) for H631 are shown in (A). Amino acid numbering is based on SIVsmE543-3 sequence. Amino acid substitutions are indicated, dashes indicate deletions, a dot indicates identity, and potential N-linked glycosylation sites are underlined.
In contrast, there appeared to be fewer total mutations in H636 clones (figure 2B) than in H631, and there were no consistent changes that were maintained from week 32 to week 76 as seen in H631. Overall, these data suggest that virus in H636 was possibly under different selective pressures at these two time points.
Phylogenetic analyses were then performed to study the relationship of these sequences (figure 3). Sequences obtained from H631 CSF and plasma viral RNA (figure 3A) were found to be clustered based on their time points at week 1 and 32. At week one, CSF and plasma sequences were still closely related to one another and to SIVsmE543-3, indicating that very little evolution occurred. At week 32, CSF and plasma sequences for the most part, formed their own clade suggesting that selective pressures in these compartments may have been different from that of week 1. Viral sequences isolated at week 76 and at death continued to diverge from week 32, but virus from CSF and virus from plasma remained clustered based on their respective compartment.
Similar to what was observed at week 1 in H631, CSF and plasma sequences in H636 were also very similar to one another and to SIVsmE543-3 (figure 3B). By week 32, while virus could be seen to have diverged from SIVsmE543-3, CSF and plasma sequences did not form their own clades. Half of the sequences from plasma at week 76 appear to be direct descendents of virus from plasma at week 32. In contrast, the other half, possibly under different selective pressures, had formed a different clade with virus from CSF.
To determine if the changes observed in the gp120 sequences were due to random mutations or due to positive selection from immune pressure, the rate of nonsynonymous (dN) to synonymous (dS) change was calculated for each clone at sequential timepoints and the average ratio is shown in Table 1. Positive selection is evidenced by a dN/dS ratio > 1. In H631, increasing positive selection could be seen in both CSF and plasma gp120 sequences throughout the course of infection, with positive selection occurring by week 76. Strong positive selection was evident by death indicating that immune pressures in these compartments drove the evolution of these viruses in H631. In the case of H636, increasing positive selection was also observed for plasma sequences; however, with sequences obtained from CSF, only slight positive selection was seen by week 32, but had weakened slightly by week 76.
Table 1.
Analysis of Nonsynonymous to Synonymous Changes in gp120 at Sequential Timepoints
| Average dN/dSa | ||||
|---|---|---|---|---|
| H631 | H636 | |||
| Week post inoculation | CSF | Plasma | CSF | Plasma |
| 1 | 0.043 (9)b | 0.231 (10) | 0.208 (8) | 0.156 (9) |
| 32 | 0.796 (9) | 0.696 (9) | 1.035 (4) | 0.990 (7) |
| 76 | 1.607 (10) | 1.488 (10) | 0.821 (10) | 1.355 (10) |
| 116 (death) | 2.427 (9) | 2.746 (9) | NAc | NA |
Average ratio of nonsynonymous (dN) to synonymous (dS) change as calculated by SNAP.
Number of clones used to determine the average dN/dS ratio.
Not applicable.
Replicative abilities of viruses isolated from H631 and H636
To study the biological consequences of the observed mutations, we isolated virus from CSF and plasma from both animals at the last available time point. Additionally, for H631, virus was isolated from the brain. Virus isolates from H631 brain, CSF, and plasma (Fig. 4A), as well as from H636 CSF and plasma (Fig. 4B) were all able to infect donor PBMC. However, the replication efficiency varied among the viruses. Virus from both H631 plasma and H636 plasma were found to replicate much more efficiently than the corresponding virus from the central nervous system (CNS) of the same animal.
Based on our earlier observation of SIV-infected macrophages in the brain, we then wanted to evaluate viral replication in MDM (figure 4C–D). Again, all viruses from both animals were able to replicate in MDM. However, in contrast to what was observed in the PBMC infections, infectivity of MDM by virus isolated from CNS of both macaques was much higher than that of virus isolated from PBMC.
To ensure that there were no donor biases, as different macaques may have variable susceptibilities to infection [21], this infectivity assay was repeated using an additional three different donor macaques. The same donor macaques were used in both the PBMC and MDM experiments. Data from the four different infection assays were then combined to obtain an average, and the experiment was then independently repeated a second time. These experiments (data not shown) confirmed the initial results and demonstrated a marked trend for virus isolated from plasma of either H631 or H636 to replicate in PBMC better than their counterpart virus isolated from the CNS. Similarly, replication of viruses isolated from CNS was observed to infect MDM better than virus isolated from plasma. Thus, our data suggests that there is a preference for cell tropism that is dependant on the origin of the virus compartment.
Phylogenetic analyses of isolated viruses
Based on the observed differences in biological phenotypes, we wanted to examine the gp160 sequences to evaluate the phylogenetic relationships of the isolated viruses. Additionally, bootstrapping support tests were done to examine the reliability of the topology or branching order of the tree. Bootstrap support values were based on 1000 replicates and only values greater than 70% at the major nodes are shown. With the exception of one envelope clone obtained from a plasma virus, all sequences from H631 brain, CSF, and plasma clustered within their respective groups (figure 5A). Although the viruses were quite distinct and formed their own clades, bootstrap values did not support compartmentalization of the viruses. However, phylogenetic analyses and bootstrap support tests indicated that H636 viruses from the CSF and plasma compartments were indeed compartmentalized (bootstrap values 916 and 736, respectively) (figure 5B). In contrast to H631 and H636, gp160 sequences amplified directly from CSF and plasma viral RNA of H635 showed that there was intermingling of virus between plasma and CSF and no tissue-specific localization of virus (figure 5C).
The rates of nonsynonymous to synonymous changes were also calculated for H631 and H636 isolated viruses. For all cases, the dN/dS ratio was greater than one, indicating that positive selection was responsible for the evolution of these viruses (data not shown).
Loss of PNGS
Sequencing analyses revealed that there were changes in the PNGS pattern (figure 2 and data not shown) resulting in a statistically significant loss of PNGS in gp160 of all H631 virus isolates and H636 CSF virus isolates compared to E543-3 (P< 0.0001 to P<0.0054) (figure 6). Furthermore, the number of PNGS in Env of H631 brain virus isolates was found to be lower than that of H631 CSF and plasma virus (P< 0.0001 and P<0.0005, respectively).
DISCUSSION
In terms of SIV neuropathogenesis studies, there are primarily three macaque models that have been shown to reliably induce neuroAIDS [29–31]. These studies have appeared to focus mainly on macaques with rapid progressive disease and encephalitis. In contrast, in the current study, we present data on two animals with conventional progressive disease having SIVE.
We studied the evolution of actively replicating viruses and determined that during the acute phase of infection, viruses from CSF and plasma in their respective hosts were phylogenetically very similar to one another. This indicated a systemic spread by the inoculum, with seeding from the periphery to the CNS since the animals were inoculated by intravenous route. With the exception of H636 CSF, increasingly positive selection was observed for the changes seen in the genotypes, suggesting that immune selection pressures or lack thereof, continued to drive the evolution of SIV in these macaques. Indeed, divergent evolution among the different compartments was observed with, bootstrap support values indicating definite compartmentalization of H636 viruses. Despite H631 viruses clustering together (with the exception of one clone from plasma) in their own clade, bootstrap values did not indicate compartmentalization. This is not altogether surprising as we expected brain and CSF viral sequences to be more closely related to one another and therefore were not supported by bootstrap analyses. However, when the phylogenetic relationship was re-analyzed but excluded sequences from brain, similar to what was observed with H636 viruses, bootstrap values did support compartmentalization between viruses isolated from CSF and plasma (722 and 943, respectively, data not shown).
The lack of compartmentalization in H635 is consistent with rapid disease progression and with major dysfunction of the blood-brain barrier in RP macaques with SIVE where the distribution of SIV can be found throughout the brain parenchyma (unpublished data) [29]. In contrast, SIV was primarily localized to the perivascular regions in H631 and H636. Furthermore, the pathology of the brain of these two conventional progressors was significantly different from that of H635 (which did not have lymphocytic infiltrates), with the presence of prominent perivascular mononuclear infiltrates containing both T and B cells. This is the first report to observe the presence of B cell infiltrates in SIV-infected monkeys. The presence of lymphocytes is more similar to the pathology of HIVE [32–36]. These differences support a SIV/macaque model for neuroAIDS using conventional progressors with SIVE rather than RP with SIVE.
From our phylogenetic analyses, our data strongly suggest that the inoculum evolved over time in different compartments under different local selective pressures resulting in divergent evolution of virus. Consequently, virus from brain/CSF adapted to preferentially infect MDM and virus from plasma was better suited to infect PBMC. Perivascular macrophages have been shown to be the primary cell type productively infected by HIV-1 and SIV in the brains of humans and macaques with lentivirus-induced encephalitis [37, 38]. Thus our observations extend and support the notion that viruses isolated from CNS would evolve to selectively infect macrophages.
Given that expression of CD4 levels on human and macaque macrophages are low to undetectable [38–40], it is possible that this adaptation may be due to the selection of viral strains that have less dependence on using CD4 for entry. Indeed, both SIV and HIV viral variants have been reported previously to be CD4-independent by infectivity assays [41, 42]. Moreover, CD4 independence has been associated with a loss of PNGS [43, 44]. The finding that there was a statistically significant loss of PNGS in gp160 in H631 and H636 CSF virus isolates, and in particular, in H631 brain virus isolates, may be explained by the fact that the brain is a selectively immune-privileged site. Therefore, it is reasonable to assume that H631 virus in this compartment may adapt to decrease the number of PNGS given that they are less likely to encounter humoral immune pressure.
In summary, we have isolated viruses from the brain and CSF of two conventional progressor macaques with neuroAIDS. Our data demonstrated the importance of the environment on the selection of particular genotypes from quasispecies. Indeed, these viruses appear to have undergone adaptive evolution to efficiently replicate in the CNS. Additionally, our data suggests that neurovirulence may be in part due to selective adaptation and is not inherited since the inoculum was derived from the mesentery lymph node and three other animals from the same cohort had conventional disease progression but did not develop SIVE.
Supplementary Material
Acknowledgements
We thank Sonya Whitted and Christopher Erb (LMM, NIAID) for technical assistance.
This research is supported by the Division of Intramural Research, NIAID, NIH.
Footnotes
The authors do not have any commercial or other association that may pose a conflict of interest.
This work has been previously presented at the Palm Springs Symposium on HIV/AIDS in March 2007 in Palm Springs, Ca.
References
- 1.Sacktor N, Lyles RH, Skolasky R, et al. HIV-associated neurologic disease incidence changes: Multicenter AIDS Cohort Study, 1990–1998. Neurology. 2001;56:257–260. doi: 10.1212/wnl.56.2.257. [DOI] [PubMed] [Google Scholar]
- 2.McArthur JC. HIV dementia: an evolving disease. J Neuroimmunol. 2004;157:3–10. doi: 10.1016/j.jneuroim.2004.08.042. [DOI] [PubMed] [Google Scholar]
- 3.Nomenclature and research case definitions for neurologic manifestations of human immunodeficiency virus-type 1 (HIV-1) infection. Report of a Working Group of the American Academy of Neurology AIDS Task Force. Neurology. 1991;41:778–785. doi: 10.1212/wnl.41.6.778. [DOI] [PubMed] [Google Scholar]
- 4.Navia BA, Jordan BD, Price RW. The AIDS dementia complex: I. Clinical features. Ann Neurol. 1986;19:517–524. doi: 10.1002/ana.410190602. [DOI] [PubMed] [Google Scholar]
- 5.Navia BA, Cho ES, Petito CK, Price RW. The AIDS dementia complex: II. Neuropathology. Ann Neurol. 1986;19:525–535. doi: 10.1002/ana.410190603. [DOI] [PubMed] [Google Scholar]
- 6.Pumarola-Sune T, Navia BA, Cordon-Cardo C, Cho ES, Price RW. HIV antigen in the brains of patients with the AIDS dementia complex. Ann Neurol. 1987;21:490–496. doi: 10.1002/ana.410210513. [DOI] [PubMed] [Google Scholar]
- 7.Sharer LR, Cho ES, Epstein LG. Multinucleated giant cells and HTLV-III in AIDS encephalopathy. Hum Pathol. 1985;16:760. doi: 10.1016/s0046-8177(85)80245-8. [DOI] [PubMed] [Google Scholar]
- 8.Daniel MD, Letvin NL, Sehgal PK, et al. Long-term persistent infection of macaque monkeys with the simian immunodeficiency virus. J Gen Virol. 1987;68(Pt 12):3183–3189. doi: 10.1099/0022-1317-68-12-3183. [DOI] [PubMed] [Google Scholar]
- 9.Hirsch VM, Johnson PR. Pathogenic diversity of simian immunodeficiency viruses. Virus Res. 1994;32:183–203. doi: 10.1016/0168-1702(94)90041-8. [DOI] [PubMed] [Google Scholar]
- 10.Baskin GB, Murphey-Corb M, Roberts ED, Didier PJ, Martin LN. Correlates of SIV encephalitis in rhesus monkeys. J Med Primatol. 1992;21:59–63. [PubMed] [Google Scholar]
- 11.Westmoreland SV, Halpern E, Lackner AA. Simian immunodeficiency virus encephalitis in rhesus macaques is associated with rapid disease progression. J Neurovirol. 1998;4:260–268. doi: 10.3109/13550289809114527. [DOI] [PubMed] [Google Scholar]
- 12.Ringler DJ, Hunt RD, Desrosiers RC, Daniel MD, Chalifoux LV, King NW. Simian immunodeficiency virus-induced meningoencephalitis: natural history and retrospective study. Ann Neurol. 1988;23 Suppl:S101–S107. doi: 10.1002/ana.410230726. [DOI] [PubMed] [Google Scholar]
- 13.Sharer LR, Baskin GB, Cho ES, Murphey-Corb M, Blumberg BM, Epstein LG. Comparison of simian immunodeficiency virus and human immunodeficiency virus encephalitides in the immature host. Ann Neurol. 1988;23 Suppl:S108–S112. doi: 10.1002/ana.410230727. [DOI] [PubMed] [Google Scholar]
- 14.Lackner AA, Dandekar S, Gardner MB. Neurobiology of simian and feline immunodeficiency virus infections. Brain Pathol. 1991;1:201–212. doi: 10.1111/j.1750-3639.1991.tb00660.x. [DOI] [PubMed] [Google Scholar]
- 15.Price RW, Brew B, Sidtis J, Rosenblum M, Scheck AC, Cleary P. The brain in AIDS: central nervous system HIV-1 infection and AIDS dementia complex. Science. 1988;239:586–592. doi: 10.1126/science.3277272. [DOI] [PubMed] [Google Scholar]
- 16.Welch K, Morse A. The clinical profile of end-stage AIDS in the era of highly active antiretroviral therapy. AIDS Patient Care STDS. 2002;16:75–81. doi: 10.1089/10872910252806126. [DOI] [PubMed] [Google Scholar]
- 17.NRC. Guidelines for the care and use of laboratory animals. Washington, D.C.: National Academy Press; 1996. [Google Scholar]
- 18.Dehghani H, Brown CR, Plishka R, Buckler-White A, Hirsch VM. The ITAM in Nef influences acute pathogenesis of AIDS-inducing simian immunodeficiency viruses SIVsm and SIVagm without altering kinetics or extent of viremia. J Virol. 2002;76:4379–4389. doi: 10.1128/JVI.76.9.4379-4389.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuwata T, Dehghani H, Brown CR, et al. Infectious molecular clones from a simian immunodeficiency virus-infected rapid-progressor (RP) macaque: evidence of differential selection of RP-specific envelope mutations in vitro and in vivo. J Virol. 2006;80:1463–1475. doi: 10.1128/JVI.80.3.1463-1475.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirsch V, Adger-Johnson D, Campbell B, et al. A molecularly cloned, pathogenic, neutralization-resistant simian immunodeficiency virus, SIVsmE543-3. J Virol. 1997;71:1608–1620. doi: 10.1128/jvi.71.2.1608-1620.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldstein S, Brown CR, Dehghani H, Lifson JD, Hirsch VM. Intrinsic susceptibility of rhesus macaque peripheral CD4(+) T cells to simian immunodeficiency virus in vitro is predictive of in vivo viral replication. J Virol. 2000;74:9388–9395. doi: 10.1128/jvi.74.20.9388-9395.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edmonson P, Murphey-Corb M, Martin LN, et al. Evolution of a Simian Immunodeficiency Virus Pathogen. J. Virol. 1998;72:405–414. doi: 10.1128/jvi.72.1.405-414.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirsch VM. Evolution of the fittest ends in tragedy. Nat Med. 1999;5:488–489. doi: 10.1038/8369. [DOI] [PubMed] [Google Scholar]
- 24.Holterman L, Niphuis H, ten Haaft PJ, Goudsmit J, Baskin G, Heeney JL. Specific passage of simian immunodeficiency virus from end-stage disease results in accelerated progression to AIDS in rhesus macaques. J Gen Virol. 1999;80(Pt 12):3089–3097. doi: 10.1099/0022-1317-80-12-3089. [DOI] [PubMed] [Google Scholar]
- 25.Kimata JT, Kuller L, Anderson DB, Dailey P, Overbaugh J. Emerging cytopathic and antigenic simian immunodeficiency virus variants influence AIDS progression. Nat Med. 1999;5:535–541. doi: 10.1038/8414. [DOI] [PubMed] [Google Scholar]
- 26.Tao B, Fultz PN. Molecular and biological analyses of quasispecies during evolution of a virulent simian immunodeficiency virus, SIVsmmPBj14. J Virol. 1995;69:2031–2037. doi: 10.1128/jvi.69.4.2031-2037.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirsch VM, Santra S, Goldstein S, et al. Immune failure in the absence of profound CD4+ T-lymphocyte depletion in simian immunodeficiency virus-infected rapid progressor macaques. J Virol. 2004;78:275–284. doi: 10.1128/JVI.78.1.275-284.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dehghani H, Puffer BA, Doms RW, Hirsch VM. Unique pattern of convergent envelope evolution in simian immunodeficiency virus-infected rapid progressor macaques: association with CD4-independent usage of CCR5. J Virol. 2003;77:6405–6418. doi: 10.1128/JVI.77.11.6405-6418.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Neil SP, Suwyn C, Anderson DC, et al. Correlation of acute humoral response with brain virus burden and survival time in pig-tailed macaques infected with the neurovirulent simian immunodeficiency virus SIVsmmFGb. Am J Pathol. 2004;164:1157–1172. doi: 10.1016/S0002-9440(10)63204-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roberts ES, Zandonatti MA, Watry DD, et al. Induction of pathogenic sets of genes in macrophages and neurons in NeuroAIDS. Am J Pathol. 2003;162:2041–2057. doi: 10.1016/S0002-9440(10)64336-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zink MC, Amedee AM, Mankowski JL, et al. Pathogenesis of SIV encephalitis. Selection and replication of neurovirulent SIV. Am J Pathol. 1997;151:793–803. [PMC free article] [PubMed] [Google Scholar]
- 32.Anthony IC, Crawford DH, Bell JE. Effects of human immunodeficiency virus encephalitis and drug abuse on the B lymphocyte population of the brain. J Neurovirol. 2004;10:181–188. doi: 10.1080/13550280490444100. [DOI] [PubMed] [Google Scholar]
- 33.Katsetos CD, Fincke JE, Legido A, et al. Angiocentric CD3(+) T-cell infiltrates in human immunodeficiency virus type 1-associated central nervous system disease in children. Clin Diagn Lab Immunol. 1999;6:105–114. doi: 10.1128/cdli.6.1.105-114.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller RF, Isaacson PG, Hall-Craggs M, et al. Cerebral CD8+ lymphocytosis in HIV-1 infected patients with immune restoration induced by HAART. Acta Neuropathol (Berl) 2004;108:17–23. doi: 10.1007/s00401-004-0852-0. [DOI] [PubMed] [Google Scholar]
- 35.Petito CK, Adkins B, McCarthy M, Roberts B, Khamis I. CD4+ and CD8+ cells accumulate in the brains of acquired immunodeficiency syndrome patients with human immunodeficiency virus encephalitis. J Neurovirol. 2003;9:36–44. doi: 10.1080/13550280390173391. [DOI] [PubMed] [Google Scholar]
- 36.Petito CK, Torres-Munoz JE, Zielger F, McCarthy M. Brain CD8+ and cytotoxic T lymphocytes are associated with, and may be specific for, human immunodeficiency virus type 1 encephalitis in patients with acquired immunodeficiency syndrome. J Neurovirol. 2006;12:272–283. doi: 10.1080/13550280600879204. [DOI] [PubMed] [Google Scholar]
- 37.Fischer-Smith T, Croul S, Sverstiuk AE, et al. CNS invasion by CD14+/CD16+ peripheral blood-derived monocytes in HIV dementia: perivascular accumulation and reservoir of HIV infection. J Neurovirol. 2001;7:528–541. doi: 10.1080/135502801753248114. [DOI] [PubMed] [Google Scholar]
- 38.Williams KC, Corey S, Westmoreland SV, et al. Perivascular macrophages are the primary cell type productively infected by simian immunodeficiency virus in the brains of macaques: implications for the neuropathogenesis of AIDS. J Exp Med. 2001;193:905–915. doi: 10.1084/jem.193.8.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mori K, Rosenzweig M, Desrosiers RC. Mechanisms for adaptation of simian immunodeficiency virus to replication in alveolar macrophages. J Virol. 2000;74:10852–10859. doi: 10.1128/jvi.74.22.10852-10859.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williams K, Bar-Or A, Ulvestad E, Olivier A, Antel JP, Yong VW. Biology of adult human microglia in culture: comparisons with peripheral blood monocytes and astrocytes. J Neuropathol Exp Neurol. 1992;51:538–549. doi: 10.1097/00005072-199209000-00009. [DOI] [PubMed] [Google Scholar]
- 41.Edinger AL, Mankowski JL, Doranz BJ, et al. CD4-independent, CCR5-dependent infection of brain capillary endothelial cells by a neurovirulent simian immunodeficiency virus strain. Proc Natl Acad Sci U S A. 1997;94:14742–14747. doi: 10.1073/pnas.94.26.14742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kolchinsky P, Mirzabekov T, Farzan M, et al. Adaptation of a CCR5-using, primary human immunodeficiency virus type 1 isolate for CD4-independent replication. J Virol. 1999;73:8120–8126. doi: 10.1128/jvi.73.10.8120-8126.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kolchinsky P, Kiprilov E, Bartley P, Rubinstein R, Sodroski J. Loss of a single N-linked glycan allows CD4-independent human immunodeficiency virus type 1 infection by altering the position of the gp120 V1/V2 variable loops. J Virol. 2001;75:3435–3443. doi: 10.1128/JVI.75.7.3435-3443.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morikawa Y, Moore JP, Wilkinson AJ, Jones IM. Reduction in CD4 binding affinity associated with removal of a single glycosylation site in the external glycoprotein of HIV-2. Virology. 1991;180:853–856. doi: 10.1016/0042-6822(91)90106-l. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.