Abstract
Mycobacterium tuberculosis uses multiple mechanisms to avoid elimination by the immune system. We have previously shown that M. tuberculosis can inhibit selected macrophage responses to IFN-γ through TLR2-dependent and -independent mechanisms. To specifically address the role of TLR2 signaling in mediating this inhibition, we stimulated macrophages with the specific TLR2/1 ligand Pam3CSK4 and assayed responses to IFN-γ. Pam3CSK4 stimulation prior to IFN-γ inhibited transcription of the unrelated IFN-γ-inducible genes, CIITA and CXCL11. Surface expression of MHC class II and secretion of CXCL11 were greatly reduced as well, indicating that the reduction in transcripts had downstream effects. Inhibition of both genes required new protein synthesis. Using chromatin immunoprecipitation, we found that TLR2 stimulation inhibited IFN-γ-induced RNA polymerase II binding to the CIITA and CXCL11 promoters. Furthermore, TATA binding protein was unable to bind the TATA box of the CXCL11 promoter, suggesting that assembly of transcriptional machinery was disrupted. However, TLR2 stimulation affected chromatin modifications differently at each of the inhibited promoters. Histone H3 and H4 acetylation was reduced at the CIITA promoter but unaffected at the CXCL11 promoter. In addition, NF-κB signaling was required for inhibition of CXCL11 transcription, but not for inhibition of CIITA. Taken together, these results indicate that TLR2-dependent inhibition of IFN-γ-induced gene expression is mediated by distinct, gene-specific mechanisms that disrupt binding of the transcriptional machinery to the promoters.
Introduction
Macrophages are important mediator cells during the immune response to invading pathogens. They are able to recognize a variety of pathogens through cell surface receptors, including members of the Toll-like receptor (TLR) family [1]. Among these receptors, TLR2 and TLR4 specifically recognize bacteria-derived lipopeptides and LPS, respectively. Engagement of TLRs results in activation of MAPK and NF-κB signaling pathways, culminating in the expression of proinflammatory cytokines and antimicrobial effector molecules [2], [3], as well as in the induction of apoptosis [4].
Macrophages also function as effector cells in the adaptive immune response. While macrophages play an important part in controlling infections as part of the innate immune response, full activation of their antimicrobial capacity and antigen presentation function only occurs after stimulation with the Th1 cytokine IFN-γ [5]. IFN-γ is essential for the control of Mycobacterium tuberculosis [6]–[9] and the clearance of other intracellular pathogens [10]–[13].
IFN-γ acts by binding to the heterodimeric IFN-γ receptor. Receptor binding and dimerization leads to the recruitment of JAKs 1 and 2 and ultimately to tyrosine and serine phosphorylation of the transcription factor STAT1 [14]. Phospho-STAT1 dimers then drive gene expression by binding gamma-activated sites (GAS) in the promoters of a large number of genes.
While exposure to a TLR agonist and IFN-γ can have synergistic effects and enhance activation of some IFN-γ-induced genes [15], a number of studies have shown that LPS [16], [17], whole mycobacteria [18]–[22], the mycobacterial lipoglycan phosphatidylinositol mannan [23], and mycobacterial lipoproteins [24], [25] can have inhibitory effects on a subset of IFN-γ-induced genes. This appears to be of special relevance in the context of infections with M. tuberculosis where, even in the presence of a strong adaptive immune response, clearance of bacteria from the infected tissue is not achieved [6], [26], [27].
Although inhibition of IFN-γ-induced gene expression by M. tuberculosis occurs by both TLR2-dependent and -independent mechanisms in vitro [24] and in vivo [28], we focused on the contribution of TLR2 signaling to inhibition in the experiments reported here. Pam3CSK4, a synthetic triacylated hexapeptide and specific TLR2/1 ligand [29], has been found to mimic the inhibitory effects of mycobacterial lipoproteins in macrophages [23]. Inhibition of class II transactivator (CIITA), a gene required for antigen presentation via MHC class II to CD4+ T cells [30], has been well characterized [18], [20], [24], [25], [31]. We wanted to extend these findings and compare inhibition of CIITA with that of CXCL11, another IFN-γ-inducible gene that we found to be strongly inhibited by Pam3CSK4 through microarray analysis. CXCL11 is a member of the CXC chemokine family and ligand for CXCR3, which is expressed on activated CD4+ T cells [32]. CXCL11 acts as a chemoattractant to recruit these cells to the site of inflammation [33]. Although studied during chronic M. tuberculosis infection [34], the role of CXCL11 during early infection is unknown.
We found that TLR2 inhibition of IFN-γ-induced transcription of CXCL11 and CIITA required new protein synthesis, but that inhibition of each of these genes involved distinct downstream mechanisms.
Materials and Methods
Reagents and antibodies
(S)-[2,3,-Bis(palmitoyloxy)-(2-RS)-propyl]-N-palmitoyl-(R)-Cys-(S)-Ser-(S)-Lys4-OH, 3HCl (Pam3CSK4; Calbiochem) was stored at 1 mg/ml in endotoxin-tested water (Invitrogen Life Technologies). Cycloheximide (Calbiochem) was stored at 100 mg/ml (355 mM) in DMSO. Recombinant murine IFN-γ was purchased from BD Biosciences. Polyclonal anti-acetyl-histone H3 and H4 antibodies were from Millipore. Antibodies for RNA polymerase II (N-20) and TFIID (TBP) (N-12) were purchased from Santa Cruz Biotechnology.
Mice
C57BL/6 mice were purchased from The Jackson Laboratory, BALB/c mice were purchased from Taconic, and TNFκB (TNF−/−/RelA+/−) mice were purchased from Riken. All were maintained under specific pathogen-free conditions. All work with animals was approved by the New York University School of Medicine Institutional Animal Care and Use Committee.
Isolation and culture of bone marrow-derived macrophages
Bone marrow-derived macrophages (BMDM) were isolated and cultured as previously described [23]. All experiments were done with C57BL/6 BMDM unless otherwise specified.
RNA harvest and quantitative real-time PCR (qPCR)
BMDM from BALB/c mice (2×106) were incubated with Pam3CSK4 (10 ng/ml) for 8 h followed by IFN-γ (20 ng/ml) for 4, 8, and 12 h. For cycloheximide experiments, BMDM from C57BL/6 mice were pretreated with DMSO or cycloheximide (500 nM) for 1 h, followed by Pam3CSK4 (10 ng/ml) for 8 h followed by IFN-γ (20 ng/ml) for 4 h in the continued absence or presence of the inhibitor. For examining NF-κB, BMDM from TNF−/−/RelA+/+ and TNF−/−/RelA−/− mice were treated with Pam3CSK4 (10 ng/ml) for 8 h followed by IFN-γ (20 ng/ml) for 4 h. Total RNA was harvested using Qiagen RNeasy columns according to the manufacturer's directions. Genomic DNA contamination was removed by DNase treatment (Ambion). Total RNA yield was determined by nanodrop quantitation and 1 µg was reverse transcribed using the Reverse Transcription System (Promega). The cDNA equivalent of 10 ng (for GAPDH) or 50 ng (for CXCL11, CIITA, and NOS2) of total RNA was analyzed by quantitative PCR using FastStart Universal SYBR Green Master Mix (Roche) on an MJ Research Opticon 2. For quantitation, the relative values were determined by comparing the threshold cycle of each sample to a standard curve consisting of serial dilutions of a positive control cDNA sample and normalized to GAPDH. The following primers were used: CXCL11 sense, 5′-GCA CCT CTT TCA GTC TGT TTC CTG-3′; CXCL11 antisense, 5′-AGC CAT CCC TAC CAT TCA TTC AC-3′; CIITA pIV sense, 5′-GAA GTT CAC CAT TGA GCC ATT TAA-3′; CIITA pIV antisense, 5′-CTG GGT CTG CAC GAG ACG AT-3′; NOS2 sense, 5′-GTT CTC AGC CCA ACA ATA CAA GA-3′; NOS2 antisense, 5′-GTG GAC GGG TCG ATG TCA C-3′; GAPDH sense, 5′-TGT GTC CGT CGT GGA TCT GA-3′; GAPDH antisense, 5′-CCT GCT TCA CCA CCT TCT TGA-3′.
Flow cytometry
Macrophages (2×106) were plated on non-tissue culture treated plates and treated with Pam3CSK4 (10 ng/ml) for 12–15 h, followed by IFN-γ (20 ng/ml) for 24 h. Cells were harvested from plates by incubation in PBS containing 5 mM EDTA for 20 min at 4°C, then vigorous pipetting. Cells were stained with Alexa 647-conjugated anti-mouse I-A/I-E (Biolegend), washed, and resuspended in ice-cold FACS buffer (PBS, 0.1% sodium azide, 1% FCS, 500 µM EDTA). Cells were analyzed for MHC class II surface expression using a FACSCalibur (30,000 total events gated by forward and side scatter; BD Biosciences).
ELISA
Since the CXCL11 gene in C57BL/6 mice contains a point mutation that results in the lack of CXCL11 secretion [35], BMDM from BALB/c mice were treated with Pam3CSK4 (10 ng/ml) for 8 h, followed by IFN-γ (20 ng/ml) for 4, 8, and 12 h. Culture supernatants were harvested and assayed for murine CXCL11 by ELISA according to the manufacturer's directions (R & D Systems). Samples were used neat or diluted 1∶10 or 1∶50 to allow detection within the range of the assay. Results were quantitated using an ELx800UV spectrophotometer (Bio-Tek Instruments).
Chromatin immunoprecipitation (ChIP)
8−10×106 macrophages were treated with Pam3CSK4 (10 ng/ml) for 8–9 h, followed by IFN-γ (20 ng/ml) for 4, 8, and 12 h (for PolII and TBP binding) or 1, 2, 4, and 8 h (for histone acetylation). Cells were crosslinked by adding 1% formaldehyde for 10 min at 37°C, followed by addition of glycine (125 mM) for 5 min at room temperature. Cells were washed twice with ice-cold PBS and scraped in PBS containing protease inhibitors (Complete Mini, Roche). Fixed cells were pelleted and snap-frozen in liquid nitrogen. Samples were processed using the ChIP assay kit from Millipore (17–295). Cell pellets were thawed, resuspended in SDS lysis buffer, and lysed on ice for 10 min. Chromatin was fragmented using a Branson Digital Sonifier 250 (6 rounds at 20% amplitude for 40 s each round (0.5 s pulse, 1 s break)). One-third of the fragmented chromatin was diluted five-fold in ChIP dilution buffer. 1% of each sample was set aside as input DNA. Chromatin was immunoprecipitated overnight with anti-RNA polymerase II (10 µg), anti-TFIID (10 µg), or anti-acetylated histone H3 (5 µg) or H4 (4 µg) antibodies; the specificity of binding was determined using controls in which the primary antibody was omitted. Chromatin-antibody complexes were captured by incubation with protein A agarose beads for 1 h at 4°C, then chromatin-antibody-bead complexes were washed for 5 min at 4°C with 1 ml of each buffer in the following order: low salt immune complex wash buffer, high salt immune complex wash buffer, LiCl immune complex wash buffer, and TE (as described in the manufacturer's protocol). Chromatin was released from the beads with elution buffer (1% SDS, 0.1 M NaHCO3), and crosslinking was reversed by incubating input and sample chromatin in 0.2 M NaCl for 4 h at 65°C. Sample chromatin was incubated with proteinase K for 1 h at 45°C and ethanol-precipitated, then sample and input chromatin were diluted five-fold in PB buffer and purified with QiaQuick columns according to the manufacturer's protocol (Qiagen). Purified sample and input DNA was eluted with 50 µl EB buffer. 2.5 µl of eluted DNA were assayed by qPCR using primers specific for the promoter region of the assayed gene and genomic DNA as standard. ChIP output was normalized to the amount of input DNA. The resulting values for each gene were normalized using their corresponding GAPDH values. Fold enrichment is expressed in relation to the value determined for the untreated sample value. The primers used are listed in Table 1.
Table 1. Primers used for ChIPs.
| Primer name | Sequence (5′ to 3′) |
| CXCL11_HISF | TCT GCC CAG AAT CCC TAC AC |
| CXCL11_HISR | AGA AGC CAC TGG AAG GTG AA |
| GAPDH_HISF | GGT CCA AAG AGA GGG AGG AG |
| GAPDH_HISR | AGC TAC GTG CAC CCG TAA AG |
| CIITA_HISF | AGC AAA CTT GGG TTG CAT GT |
| CIITA_HISR | TCC TGG CAG CTA TCT CAC AA |
| NOS2_HISF | CAC TAT TCT GCC CAA GCT GAC TTA C |
| NOS2_HISR | CAA TAT TCC AAC ACG CCC AGG |
| CXCL11_POLF | ACT GCC TGA AGA TTG CTG GT |
| CXCL11_POLR | ATA TTG CAG CCA GGG CTA TG |
| GAPDH_POLF | CCG CAT CTT CTT GTG CAG T |
| GAPDH_POLR | TCC CTA GAC CCG TAC AGT GC |
| CIITA_POLF | GAT AGC TGC CAG GAG ACT GC |
| CIITA_POLR | CAA ACG GGA TCT TGG AGA CA |
| NOS2_POLF | CCC TTT GGG AAC AGT TAT GC |
| NOS2_POLR | CCA AGG TGG CTG AGA AGT TT |
| CXCL11_TBPF | GCT GAG TGC TTT CAC CTT CC |
| CXCL11_TBPR | GGC TGA ACC TGA GGA GTC TG |
Results
TLR2 stimulation inhibits IFN-γ-induced transcription of CXCL11 and CIITA, but not NOS2
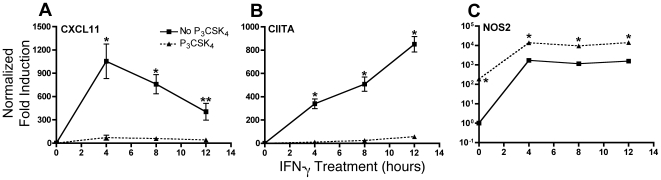
In the context of initial infection with M. tuberculosis, lung macrophages are likely to encounter the bacteria before being stimulated by IFN-γ. Therefore, we studied the effects of TLR2 stimulation by Pam3CSK4 prior to stimulation of macrophages with IFN-γ. We first analyzed the kinetics of TLR2-initiated inhibition of IFN-γ induction of CXCL11 and CIITA. Bone marrow-derived macrophages (BMDM) were treated with Pam3CSK4 for 8 h then stimulated with IFN-γ for 4, 8, and 12 h. Although CIITA and CXCL11 were both induced by IFN-γ, the kinetics of their induction differed. CXCL11 mRNA levels peaked after 4 h of IFN-γ stimulation whereas CIITA mRNA peaked after 12 h (Figs. 1A and B). Transcription of both CXCL11 and CIITA was fully inhibited in cells previously exposed to Pam3CSK4, regardless of the length of IFN-γ stimulation. As a control, we examined transcription of NOS2, an IFN-γ-inducible gene that has been found not to be inhibited by M. tuberculosis or mycobacterial lipoproteins [21], [24]. We found that Pam3CSK4 treatment prior to IFN-γ stimulation enhanced transcription of NOS2 in macrophages (Fig. 1C). These data show that TLR2 stimulation affects IFN-γ-induced transcription in a gene-specific manner.
Figure 1. TLR2 stimulation inhibits IFN-γ-induced transcription of a subset of genes.
BALB/c BMDM were treated with 10 ng/ml Pam3CSK4 for 8 h, and then 20 ng/ml IFN-γ for 4, 8, and 12 h. Total RNA was harvested, reverse transcribed, and CXCL11 (A), CIITA (B), and NOS2 (C) expression analyzed by quantitative real-time PCR (qPCR). All values were normalized to GAPDH. Results are shown as fold induction compared to untreated sample without Pam3CSK4 or IFN-γ. *, p<0.01, **, p<0.05 comparing IFN-γ alone samples with those treated with Pam3CSK4 prior to IFN-γ (as determined by two-tailed t test). Results in (A) are expressed as means±SEM of three independent experiments. Results in (B) and (C) are representative of three independent experiments. Similar results were obtained with C57BL/6 BMDM.
TLR2 stimulation inhibits CXCL11 protein production and MHC class II surface expression
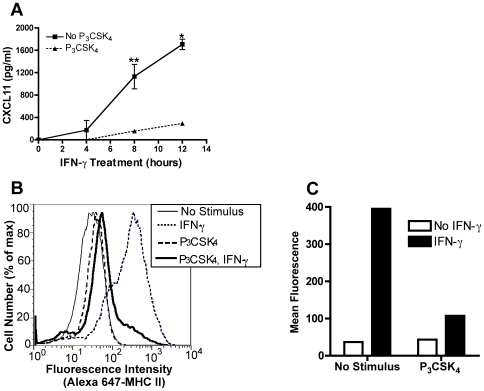
Since transcription of CXCL11 and CIITA was significantly reduced in TLR2 stimulated macrophages, we determined whether inhibition was reflected by reduction of the protein products of these genes. To examine the effect on CXCL11, we treated BALB/c BMDM with Pam3CSK4 for 8 h followed by IFN-γ for 4, 8, and 12 h. Culture supernatants were harvested and assayed for CXCL11 by ELISA. Stimulation with IFN-γ resulted in secretion of CXCL11 after 8 and 12 h of treatment (Fig. 2A). However, prior TLR2 stimulation inhibited IFN-γ-induced CXCL11 protein levels by over 80% at both of these time points.
Figure 2. TLR2-mediated inhibition of IFN-γ induction of CXCL11 and CIITA decreases expression of protein products.
A. BALB/c BMDM were treated with 10 ng/ml Pam3CSK4 for 8 h followed by 20 ng/ml IFN-γ for 4, 8, and 12 h. Culture supernatants were collected and assayed for CXCL11 protein by ELISA. *, p<0.01, **, p<0.05 comparing IFN-γ alone with Pam3CSK4 and IFN-γ treated samples (as determined by two-tailed t-test). B and C. BMDM were treated with 10 ng/ml Pam3CSK4 for 12–15 h prior to stimulation with 20 ng/ml IFN-γ for 24 h. Cells were stained with Alexa 647-conjugated anti-mouse I-A/I-E and analyzed by flow cytometry. Data shown are fluorescence intensity vs. cell number (B) and mean I-A/I-E fluorescence (C). Results are expressed as means±SEM from two independent experiments (A) and are representative of at least five independent experiments (B and C).
To examine the downstream effect of TLR2 stimulation on CIITA, we measured surface expression of MHC class II, whose expression is regulated by, and depends on, CIITA. BMDM were treated with Pam3CSK4 for 12–15 h then stimulated with IFN-γ for 24 h (the minimal time needed for increased MHC class II surface expression). IFN-γ stimulation alone caused an eleven-fold increase of MHC class II on the cell surface (Fig. 2B). However, Pam3CSK4 treatment prior to IFN-γ inhibited this upregulation by 73%, as assessed by mean fluorescent intensity (Fig. 2C). Pam3CSK4 alone (without IFN-γ) had no effect on surface MHC class II levels. These results indicate that TLR2-mediated inhibition of CXCL11 and CIITA transcription has downstream consequences at the protein level.
TLR2-mediated inhibition of CXCL11 and CIITA requires new protein synthesis
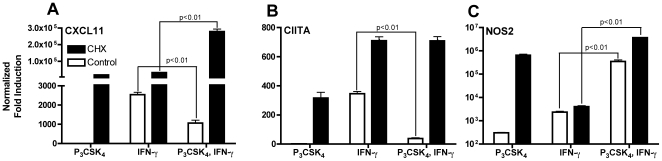
We have previously shown that M. tuberculosis-mediated inhibition of CIITA induction by IFN-γ requires new protein synthesis [23]. We extended these findings and examined the effect of cycloheximide, a pharmacological inhibitor of protein synthesis, on induction of CXCL11, CIITA, and NOS2 mRNA by IFN-γ in TLR2 stimulated macrophages. In control samples, TLR2 stimulation prior to IFN-γ inhibited CXCL11 and CIITA induction, but enhanced NOS2 induction, as previously seen. However, treatment with cycloheximide fully reversed inhibition of CXCL11 and CIITA and further enhanced NOS2 expression (Fig. 3). These results indicate that TLR2 stimulation induces production of one or more proteins that are required for inhibition of macrophage gene expression in response to IFN-γ. Cycloheximide treatment also greatly enhanced expression of all three genes, with the strongest effect on CXCL11, presumably due to the lack of a negative regulatory protein.
Figure 3. New protein synthesis is required for inhibition of CIITA and CXCL11.
BMDM were pretreated with 500 nM cycloheximide or DMSO for 1 h prior to treatment with 10 ng/ml Pam3CSK4 for 8 h followed by 20 ng/ml IFN-γ for 4 h in the continued presence or absence of inhibitor. Total RNA was harvested after IFN-γ stimulation. CXCL11 (A), CIITA (B), and NOS2 (C) expression was assayed by qPCR and normalized to GAPDH and untreated samples. The concentration of cycloheximide used inhibited TNF production (as a measure of protein synthesis) by over 90% with minimal cell death. Statistical significance between IFN-γ alone samples and those treated with Pam3CSK4 prior to IFNγ was determined by two-tailed t-test.
TLR2 stimulation prevents RNA polymerase II from binding the CXCL11 and CIITA promoters
To further characterize the mechanism of inhibition of transcriptional responses to IFN-γ by prior TLR2 stimulation, we determined whether RNA polymerase II (PolII) binds the promoters of the affected genes. We treated BMDM with Pam3CSK4 for 8 h then stimulated with IFN-γ for 4 h, followed by chromatin immunoprecipitation (ChIP) to assay binding of PolII to the CXCL11, CIITA, and NOS2 promoters. Primers that specifically amplified regions flanking the transcriptional start site of each gene were designed to detect initial binding of PolII to the promoters. IFN-γ stimulation induced binding of PolII to the CXCL11 and CIITA promoters, as indicated by a three-fold increase in pulldown of promoter fragments of these genes (Fig. 4A). However, stimulation of TLR2 prior to IFN-γ inhibited binding of PolII by 66% (CXCL11) and 76% (CIITA) compared with IFN-γ alone. The effect of TLR2 stimulation was not merely to delay IFN-γ-stimulated PolII binding at these promoters, as similar inhibition of binding was seen after 8 and 12 h of IFN-γ stimulation (data not shown). As a control, we assayed PolII binding at the NOS2 promoter, since TLR2 stimulation did not decrease NOS2 mRNA induction by IFN-γ (Fig. 1C). IFN-γ stimulation alone caused a two-fold increase in binding and prior Pam3CSK4 treatment resulted in a further increase in binding (Fig. 4A). This was similar to the effect observed at the level of transcription (Fig. 1C). These results indicate that TLR2 stimulation prevents PolII from binding to the CXCL11 and CIITA promoters, but does not affect binding to the NOS2 promoter. The effect of TLR2 stimulation on transcription of IFN-γ-responsive genes correlates with PolII binding at the respective promoters.
Figure 4. TLR2 stimulation prevents binding of general transcriptional machinery to the CIITA and CXCL11 promoters.
BMDM were treated with 10 ng/ml Pam3CSK4 for 8–9 h, then 20 ng/ml IFN-γ for 4 h. Cross-linked DNA was sheared and immunoprecipitated with anti-PolII (A) or anti-TBP (B) antibodies. Precipitated and input DNA for each sample were assayed by qPCR with primers specific for the transcriptional start site in the promoters of CXCL11, CIITA, and NOS2 (A) or the TATA box of the CXCL11 promoter (B). All values were normalized to GAPDH. Results are expressed as fold increase over untreated controls and are the mean of triplicate samples±SD. Statistical significance between IFN-γ alone samples and Pam3CSK4 prior to IFN-γ treated samples was determined by two-tailed t-test. Results are representative of at least two independent experiments.
TLR2 stimulation prevents binding of TBP to the CXCL11 promoter
Since we found that TLR2 stimulation prevents PolII from binding the CXCL11 and CIITA promoters, we determined whether another member of the general transcriptional machinery was similarly affected. We examined the ability of TATA binding protein (TBP) to bind the TATA box in the CXCL11 promoter following Pam3CSK4 and IFN-γ stimulation. The CIITA promoter lacks a TATA box (our unpublished observation), so we did not include it in our experiments. IFN-γ stimulation resulted in a five to seven-fold increase in TBP binding to the CXCL11 promoter after 4, 8, and 12 h (Fig. 4B and data not shown). However, TLR2 stimulation prior to IFN-γ decreased TBP binding to the CXCL11 promoter by 65%. This suggests that the lack of transcription is not due to a deficiency in PolII binding alone, but that prolonged TLR2 signaling also inhibits binding of other members of the general transcriptional machinery.
TLR2 signaling inhibits histone acetylation at the CIITA promoter, but not at the CXCL11 promoter
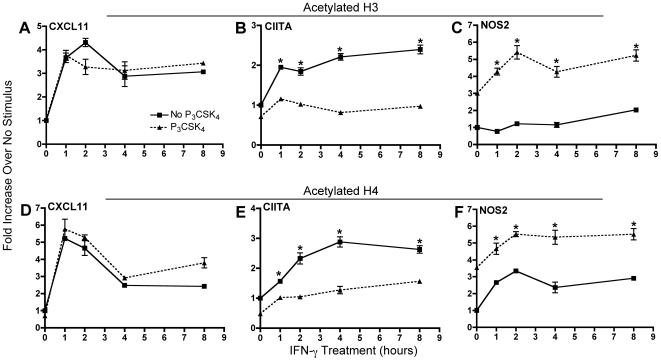
Transcriptional activity is usually associated with increased acetylation of histones H3 and H4 in the promoter region of transcribed genes [36], and TLR2 stimulation by the mycobacterial lipoprotein LpqH inhibits acetylation of histones H3 and H4 at the CIITA promoter after 4 h of IFN-γ stimulation in macrophages [25]. We extended these findings by examining earlier IFN-γ time points and by determining whether TLR2 stimulation also inhibited histone acetylation at the CXCL11 promoter. BMDM were treated with Pam3CSK4 for 8 h followed by IFN-γ for 1, 2, 4, and 8 h. ChIPs were then performed using antibodies against either acetylated histone H3 or acetylated histone H4. At the CXCL11 promoter, we observed a rapid increase in acetylation of both histones after 1 h of IFN-γ stimulation, which persisted for as long as IFN-γ was present (Figs. 5A and D). TLR2 stimulation prior to IFN-γ did not inhibit histone acetylation at the CXCL11 promoter at any time point after IFN-γ stimulation. In contrast, TLR2 stimulation significantly inhibited histone H3 and H4 acetylation at the CIITA promoter by 35–63% at all IFN-γ time points (Figs. 5B and E). For comparison, we also examined histone acetylation at the NOS2 promoter. TLR2 stimulation prior to IFN-γ resulted in an increase of histone acetylation over IFN-γ stimulation alone (Figs. 5C and F). These data indicate that TLR2-mediated inhibition of IFN-γ induction of CIITA and CXCL11 occurs by at least two distinct mechanisms, one that affects histone acetylation and one that does not.
Figure 5. TLR2 stimulation inhibits histone acetylation at the CIITA promoter, but not the CXCL11 promoter.
BMDM were treated with 10 ng/ml Pam3CSK4 for 8 h prior to stimulation with 20 ng/ml IFN-γ for 1, 2, 4, and 8 h. Cross-linked, sheared DNA was immunoprecipitated with antibodies against either acetylated histone H3 (A, B, C) or H4 (D, E, F). Precipitates were analyzed by qPCR using primers specific for the CXCL11 (A, D), CIITA (B, E), and NOS2 (C, F) promoters. Values were normalized to GAPDH and untreated controls and are the mean of triplicate samples±SD. *, p<0.01 comparing IFN-γ alone with Pam3CSK4 prior to IFN-γ treated samples (as determined by two-tailed t-test). Results are representative of two independent experiments.
NF-κB is required for TLR2-mediated inhibition of CXCL11, but not CIITA, transcription
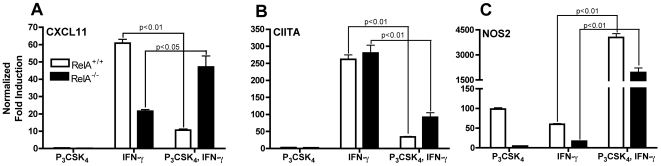
Transcriptional responses to TLR2 activation are largely mediated by the transcription factor NF-κB, a heterodimer commonly consisting of the RelA and p50 subunits. RelA deficiency is embryonic lethal but can be rescued by deletion of TNF [37], [38]. We examined the role of NF-κB in TLR2-mediated inhibition of responses to IFN-γ using macrophages from TNF−/−/RelA−/− and TNF−/−/RelA+/+ mice. BMDM were treated with Pam3CSK4 for 8 h followed by IFN-γ for 4 h. TLR2 stimulation prior to IFN-γ inhibited induction of CXCL11 and CIITA in RelA+/+ macrophages (Figs. 6A and B). In RelA−/− macrophages, CXCL11 induction by IFN-γ was fully restored despite prior TLR2 stimulation (Fig. 6A). However, CIITA induction was still significantly reduced in these cells (Fig. 6B). TNF deficiency did not affect transcriptional responses of CXCL11 or CIITA, as results with TNF−/−/RelA+/+ macrophages were similar to C57BL/6 macrophages (data not shown). However, lack of TNF did result in lower NOS2 expression that was further decreased in RelA−/− macrophages (Fig. 6C). These data provide further support that CXCL11 and CIITA are differentially regulated upon TLR2 stimulation. Inhibition of CXCL11 induction requires NF-κB whereas inhibition of CIITA does not.
Figure 6. IFN-γ-induced transcription of CXCL11, but not CIITA, is restored in TLR2 stimulated RelA−/− macrophages.
BMDM from TNF−/−/RelA+/+ and TNF−/−/RelA−/− mice were treated with 10 ng/ml Pam3CSK4 for 8 h followed by 20 ng/ml IFN-γ for 4 h. Total RNA was harvested, reverse transcribed, and CXCL11 (A), CIITA (B), and NOS2 (C) expression analyzed by qPCR. All values were normalized to GAPDH and shown as fold induction compared to untreated samples without Pam3CSK4 or IFN-γ. Statistical significance was determined by two-tailed t-test between IFN-γ alone with Pam3CSK4 prior to IFN-γ samples. C57BL/6 BMDM showed similar results to those obtained with TNF−/−/RelA+/+ BMDM.
Discussion
M. tuberculosis survives in macrophages, even when they are stimulated with IFN-γ [39], [40]. We and others have found that M. tuberculosis blocks selected macrophage responses to IFN-γ by inhibiting transcription of a subset of IFN-γ-inducible genes [18], [20]–[22]. At least two proximal mechanisms are involved. One is initiated by mycobacterial peptidoglycan in a TLR2- and MyD88-independent manner [24], while the other requires TLR2 and MyD88 and is initiated by lipoproteins and phosphatidylinositol mannan [23], [24]. In the experiments presented here, we examined the mechanisms downstream of TLR2 in mediating this inhibition, by using the specific TLR2/1 agonist Pam3CSK4.
This work expands on previous studies that examined inhibition of CIITA transcription [18], [20], [24], [25], [31], by comparing the kinetics of inhibition of CIITA with that of CXCL11, an unrelated IFN-γ-inducible gene, as well as with NOS2, an IFN-γ-inducible gene that is not inhibited by M. tuberculosis [21], [24]. We found that TLR2 stimulation inhibited IFN-γ-induced transcription of CIITA and CXCL11 to similar levels over a time course of IFN-γ stimulation (Figs. 1A and B), indicating that macrophages are unable to recover the ability to respond to IFN-γ, regardless of the length of stimulation. This reduction in transcription resulted in decreased CXCL11 secretion and decreased MHC class II on the macrophage surface (Fig. 2). However, TLR2 stimulation resulted in an increase in NOS2 mRNA (Fig. 1C), indicating that TLR2-mediated transcriptional inhibition is gene specific, and therefore not mediated by inhibition of a proximal signaling step such as STAT1 activation.
One of the initial steps in transcription initiation is acetylation of lysine residues within the N-terminal tails of core histones. This decreases their affinity for DNA, allowing a more permissive chromatin structure for transcription factors and other proteins to bind to DNA [41]. This process is tightly controlled by two counteracting enzymatic activities: the histone deacetylases (HDACs) and the histone acetyltransferases (HATs). HDACs repress transcription by removing acetyl groups whereas HATs acetylate critical lysines.
Mammalian HDACs fall into three classes (I, II, and III) based on sequence homology to yeast HDACs [42]. Class II and III HDACs are expressed in a limited number of tissues. However, HDACs 1 and 2 (of class I) appear to be constitutively expressed [43]–[47]. Therefore, their activity must be tightly regulated. They are the enzymatically active components of multi-protein complexes, which include DNA binding proteins and corepressors. These complexes target the HDACs to the promoters of genes by interactions with sequence-specific transcription factors, leading to transcriptional repression of select genes. HDACs require complex formation for enzymatic activity, as most purified recombinant HDACs are inactive [48], [49]. In addition to regulation by protein-protein interactions, HDACs can be post-translationally modified. Casein kinase 2 (CK2), a ubiquitously expressed protein kinase, has been identified as a key regulator of class I HDACs [50], [51]. CK2 activity is induced by a number of stimuli including IFN-γ and the TLR4 ligand LPS [52]. CK2 phosphorylation of S421/S422 and S423/S424 at the C-terminal region of HDACs 1 and 2, respectively, is important for complex formation and enzymatic activity. Although more light is being shed on how HDAC activity is regulated, many of the signaling pathways involved remain to be elucidated.
HATs are a diverse group of enzymes that regulate transcription by rendering the chromatin more accessible via acetylation of histone tails. Gene transcription in response to IFN-γ involves CREB binding protein (CBP) and/or p300, coactivators that have HAT activity [53]. Following IFN-γ stimulation, phosphorylated STAT1 associates with CBP/p300, which is thought to facilitate contact with transcriptional machinery at the promoter regions of IFN-γ-inducible genes [54]. Similar to HDACs, the HAT activity of CBP and p300 is tightly regulated via interactions with other proteins as well as by phosphorylation by a number of kinases. Phosphorylation by p42/p44 MAPK, CDK2, protein kinase A, and IKKα upregulate HAT activity [53], [55] whereas phosphorylation by protein kinase Cδ reduces HAT activity [56].
Transcriptional repressors also regulate gene expression. General transcriptional repression occurs when a repressor sequesters or modifies a member of the general transcriptional machinery or PolII itself [57]. Expression of all genes transcribed by PolII will then be inhibited. Gene-specific repression occurs when a repressor targets a specific coactivator or interacts in a promoter-specific manner with members of the general transcriptional machinery or PolII. Therefore, only a subset of genes will be inhibited.
Previously published data suggest that gene-specific repressors may contribute to TLR2-initiated inhibition of transcriptional responses to IFN-γ [31]. The mycobacterial lipoprotein and TLR2 agonist LpqH induces expression of the transcription factor C/EBPβ, which can act as a transcriptional activator or repressor, depending on the promoter and stimulus. This increased expression correlated with inhibition of IFN-γ-induced CIITA transcription, and macrophages stimulated with LpqH and IFN-γ exhibited increased C/EBPβ binding to the CIITA promoter. The NOS2 promoter also has a C/EBPβ binding site that is involved in gene induction in response to TLR and IFN-γ stimulation [58]. The CXCL11 promoter has two potential C/EBPβ binding sites from −52 to −44 (TGCCTGAAG) and from −24 to −16 (TCCTCAGAC), although the functionality of these sites remains to be determined. It is therefore possible that C/EBPβ or a related protein may contribute to negative regulation of IFN-γ-induction of both CIITA and CXCL11. However, C/EBPβ−/− macrophages were found to remain sensitive to LpqH-mediated transcriptional inhibition of CIITA [31], suggesting that additional factors are involved. One such factor may be C/EBPδ, whose expression is induced by TLR4 stimulation and has been shown to regulate genes involved in the innate immune response as part of a circuit with other transcription factors [59]. TLR2 stimulation also induces C/EBPδ expression and binding to the CIITA promoter [31]. However, the potential inhibitory function of C/EBPδ on IFN-γ-induced transcription needs to be explored.
We attempted to identify other potential transcription factor binding sites that might be responsible for TLR2-mediated inhibition by comparing the promoter sequences of several IFN-γ-inducible, Pam3CSK4-inhibited genes. Computer-based comparison of these promoter sequences with those from a control group of unaffected, IFN-γ-inducible genes did not yield an over-represented transcription factor binding motif, indicating that more than one signaling pathway is involved, or that inhibition is mediated by one or proteins that do not bind promoter elements directly (data not shown).
However, when specifically examining the CXCL11 and CIITA promoters, we found that CXCL11 has an NF-κB binding site at −68 to −59 (GGGGAATTCC) that is missing in CIITA. Further investigation of the role of NF-κB in TLR2-mediated inhibition of these genes using RelA−/− macrophages showed that inhibition of CXCL11, but not CIITA, is NF-κB dependent (Fig. 6). We could not detect binding of p65 or p50 to this site in the CXCL11 promoter (data not shown), suggesting that NF-κB most likely has an indirect inhibitory effect, possibly by inducing expression of a protein that blocks CXCL11 transcription upon TLR2 stimulation. The reversal of transcriptional inhibition seen when new protein synthesis is blocked is concordant with this mechanism (Fig. 3A).
We also examined potential epigenetic mechanisms as TLR2 stimulation has been shown to inhibit IFN-γ-induced histone acetylation at the promoters of genes involved in MHC class II antigen presentation [25], [43]. ChIP experiments done in murine macrophages stimulated with LpqH followed by IFN-γ showed that acetylation of histones H3 and H4 was reduced at the CIITA promoter compared to IFN-γ stimulation alone [25]. This inhibition was abrogated with pharmacological blockade of MAPKs. In addition, this inhibition was partially reversed in the presence of the HDAC inhibitor sodium butyrate, suggesting that inhibition of histone acetylation is one mechanism by which TLR2 stimulation prevents CIITA expression.
We elaborated on those experiments to determine if inhibition of histone acetylation was a common mechanism for TLR2-mediated inhibition of other IFN-γ-inducible genes. In contrast to the inhibition of IFN-γ-induced histone acetylation at the CIITA promoter (Fig. 5B and E), histone acetylation at the CXCL11 promoter was unaffected by TLR2 stimulation (Fig. 5A and D). Concordant with the transcriptional data, histone acetylation at the NOS2 promoter increased with TLR2 stimulation (Fig. 5C and F). This indicates that a decrease in modifications that make the chromatin more accessible to transcription factors and coactivators may be involved, but this is not the sole mechanism responsible for TLR2-mediated inhibition of transcriptional responses to IFN-γ.
In contrast to the gene-selective requirement for NF-κB and different effects of TLR2 stimulation on histone acetylation, we found that TLR2 stimulation decreased IFN-γ-induced binding of RNA polymerase II at both the CIITA and CXCL11 promoters, but not at the NOS2 promoter (Fig. 4A). This indicates that while the intermediate signaling steps may vary for distinct genes, TLR2 stimulation interrupts a crucial step in transcription initiation at specific IFN-γ-responsive genes.
In addition to M. tuberculosis, other pathogens have developed mechanisms to evade immune responses through disruption of host gene transcription. The intracellular bacteria Listeria monocytogenes induces a reduction in total cellular histone acetylation early after infection, mediated partially by listeriolysin O [60]. The opportunistic pathogen Mycobacterium avium inhibits histone acetylation at the HLA-DRα promoter, possibly through recruitment of HDAC corepressor mSin3a, which was found to bind the promoter following infection [43]. In those studies, infection also led to a reduction in CBP recruitment to the HLA-DRα promoter.
Viruses also disrupt host gene transcription by preventing transcriptional machinery assembly at gene promoters. Poliovirus cleaves general transcription factors [61], Rift Valley Fever virus blocks transcription factor assembly [62], and vesicular stomatitis virus targets TBP using an unknown mechanism [63]. These all prevent general PolII transcription. However, murine cytomegalovirus has been shown to inhibit specific gene transcription by targeting IFN-γ-inducible genes without affecting JAK/STAT activation [64]. PolII binding to the promoters of the affected genes was significantly reduced, suggesting that disruption of transcriptional machinery assembly was responsible for this inhibition.
The findings reported here extend the understanding of the mechanisms that may be used by at least one pathogen, Mycobacterium tuberculosis, to evade elimination by adaptive immune responses. Developing the means to increase the efficacy of adaptive immune responses in order to better control infection with M. tuberculosis will require additional investigation; increasing the efficacy of IFN-γ by restoring macrophage transcriptional responses to this cytokine may be one effective approach, but will require further study.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: These studies were supported by a grant from the National Institute of Health (RAI046097). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Aderem A, Ulevitch RJ. Toll-like receptors in the induction of the innate immune response. Nature. 2000;406:782–787. doi: 10.1038/35021228. [DOI] [PubMed] [Google Scholar]
- 2.Dunne A, O'Neill LA. The interleukin-1 receptor/Toll-like receptor superfamily: signal transduction during inflammation and host defense. Sci STKE. 2003;2003:re3. doi: 10.1126/stke.2003.171.re3. [DOI] [PubMed] [Google Scholar]
- 3.Foster SL, Hargreaves DC, Medzhitov R. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature. 2007;447:972–978. doi: 10.1038/nature05836. [DOI] [PubMed] [Google Scholar]
- 4.Lopez M, Sly LM, Luu Y, Young D, Cooper H, et al. The 19-kDa Mycobacterium tuberculosis protein induces macrophage apoptosis through Toll-like receptor-2. J Immunol. 2003;170:2409–2416. doi: 10.4049/jimmunol.170.5.2409. [DOI] [PubMed] [Google Scholar]
- 5.Mosser DM. The many faces of macrophage activation. J Leukoc Biol. 2003;73:209–212. doi: 10.1189/jlb.0602325. [DOI] [PubMed] [Google Scholar]
- 6.Mogues T, Goodrich ME, Ryan L, LaCourse R, North RJ. The relative importance of T cell subsets in immunity and immunopathology of airborne Mycobacterium tuberculosis infection in mice. J Exp Med. 2001;193:271–280. doi: 10.1084/jem.193.3.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cooper AM, Dalton DK, Stewart TA, Griffin JP, Russell DG, et al. Disseminated tuberculosis in interferon gamma gene-disrupted mice. J Exp Med. 1993;178:2243–2247. doi: 10.1084/jem.178.6.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flynn JL, Chan J, Triebold KJ, Dalton DK, Stewart TA, et al. An essential role for interferon gamma in resistance to Mycobacterium tuberculosis infection. J Exp Med. 1993;178:2249–2254. doi: 10.1084/jem.178.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flynn JL, Chan J. Immunology of tuberculosis. Annu Rev Immunol. 2001;19:93–129. doi: 10.1146/annurev.immunol.19.1.93. [DOI] [PubMed] [Google Scholar]
- 10.Murray HW, Granger AM, Teitelbaum RF. Gamma interferon-activated human macrophages and Toxoplasma gondii, Chlamydia psittaci, and Leishmania donovani: antimicrobial role of limiting intracellular iron. Infect Immun. 1991;59:4684–4686. doi: 10.1128/iai.59.12.4684-4686.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murray HW, Rubin BY, Rothermel CD. Killing of intracellular Leishmania donovani by lymphokine-stimulated human mononuclear phagocytes. Evidence that interferon-gamma is the activating lymphokine. J Clin Invest. 1983;72:1506–1510. doi: 10.1172/JCI111107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhardwaj N, Nash TW, Horwitz MA. Interferon-gamma-activated human monocytes inhibit the intracellular multiplication of Legionella pneumophila. J Immunol. 1986;137:2662–2669. [PubMed] [Google Scholar]
- 13.Skerrett SJ, Martin TR. Intratracheal interferon-gamma augments pulmonary defenses in experimental legionellosis. Am J Respir Crit Care Med. 1994;149:50–58. doi: 10.1164/ajrccm.149.1.8111597. [DOI] [PubMed] [Google Scholar]
- 14.Levy DE, Darnell JE., Jr Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 15.Kovarik P, Stoiber D, Novy M, Decker T. Stat1 combines signals derived from IFN-gamma and LPS receptors during macrophage activation. Embo J. 1998;17:3660–3668. doi: 10.1093/emboj/17.13.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stoiber D, Kovarik P, Cohney S, Johnston JA, Steinlein P, et al. Lipopolysaccharide induces in macrophages the synthesis of the suppressor of cytokine signaling 3 and suppresses signal transduction in response to the activating factor IFN-gamma. J Immunol. 1999;163:2640–2647. [PubMed] [Google Scholar]
- 17.Yao Y, Xu Q, Kwon MJ, Matta R, Liu Y, et al. ERK and p38 MAPK signaling pathways negatively regulate CIITA gene expression in dendritic cells and macrophages. J Immunol. 2006;177:70–76. doi: 10.4049/jimmunol.177.1.70. [DOI] [PubMed] [Google Scholar]
- 18.Kincaid EZ, Ernst JD. Mycobacterium tuberculosis exerts gene-selective inhibition of transcriptional responses to IFN-gamma without inhibiting STAT1 function. J Immunol. 2003;171:2042–2049. doi: 10.4049/jimmunol.171.4.2042. [DOI] [PubMed] [Google Scholar]
- 19.Lafuse WP, Alvarez GR, Curry HM, Zwelling BS. Mycobacterium tuberculosis and Mycobacterium avium inhibit IFN- gamma -induced gene expression by TLR2-dependent and independent pathways. J Interferon Cytokine Res. 2006;26:548–561. doi: 10.1089/jir.2006.26.548. [DOI] [PubMed] [Google Scholar]
- 20.Pai RK, Convery M, Hamilton TA, Boom WH, Harding CV. Inhibition of IFN-gamma-induced class II transactivator expression by a 19-kDa lipoprotein from Mycobacterium tuberculosis: a potential mechanism for immune evasion. J Immunol. 2003;171:175–184. doi: 10.4049/jimmunol.171.1.175. [DOI] [PubMed] [Google Scholar]
- 21.Pai RK, Pennini ME, Tobian AA, Canaday DH, Boom WH, et al. Prolonged toll-like receptor signaling by Mycobacterium tuberculosis and its 19-kilodalton lipoprotein inhibits gamma interferon-induced regulation of selected genes in macrophages. Infect Immun. 2004;72:6603–6614. doi: 10.1128/IAI.72.11.6603-6614.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ting LM, Kim AC, Cattamanchi A, Ernst JD. Mycobacterium tuberculosis inhibits IFN-gamma transcriptional responses without inhibiting activation of STAT1. J Immunol. 1999;163:3898–3906. [PubMed] [Google Scholar]
- 23.Banaiee N, Kincaid EZ, Buchwald U, Jacobs WR, Jr, Ernst JD. Potent inhibition of macrophage responses to IFN-gamma by live virulent Mycobacterium tuberculosis is independent of mature mycobacterial lipoproteins but dependent on TLR2. J Immunol. 2006;176:3019–3027. doi: 10.4049/jimmunol.176.5.3019. [DOI] [PubMed] [Google Scholar]
- 24.Fortune SM, Solache A, Jaeger A, Hill PJ, Belisle JT, et al. Mycobacterium tuberculosis inhibits macrophage responses to IFN-gamma through myeloid differentiation factor 88-dependent and -independent mechanisms. J Immunol. 2004;172:6272–6280. doi: 10.4049/jimmunol.172.10.6272. [DOI] [PubMed] [Google Scholar]
- 25.Pennini ME, Pai RK, Schultz DC, Boom WH, Harding CV. Mycobacterium tuberculosis 19-kDa lipoprotein inhibits IFN-gamma-induced chromatin remodeling of MHC2TA by TLR2 and MAPK signaling. J Immunol. 2006;176:4323–4330. doi: 10.4049/jimmunol.176.7.4323. [DOI] [PubMed] [Google Scholar]
- 26.Barnes PF, Mistry SD, Cooper CL, Pirmez C, Rea TH, et al. Compartmentalization of a CD4+ T lymphocyte subpopulation in tuberculous pleuritis. J Immunol. 1989;142:1114–1119. [PubMed] [Google Scholar]
- 27.Gomez JE, McKinney JD. M. tuberculosis persistence, latency, and drug tolerance. Tuberculosis (Edinb) 2004;84:29–44. doi: 10.1016/j.tube.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 28.Kincaid EZ, Wolf AJ, Desvignes L, Mahapatra S, Crick DC, et al. Codominance of TLR2-Dependent and TLR2-Independent Modulation of MHC Class II in Mycobacterium tuberculosis Infection In Vivo. J Immunol. 2007;179:3187–3195. doi: 10.4049/jimmunol.179.5.3187. [DOI] [PubMed] [Google Scholar]
- 29.Jin MS, Kim SE, Heo JY, Lee ME, Kim HM, et al. Crystal Structure of the TLR1-TLR2 Heterodimer Induced by Binding of a Tri-Acylated Lipopeptide. Cell. 2007;130:1071–1082. doi: 10.1016/j.cell.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 30.Steimle V, Siegrist CA, Mottet A, Lisowska-Grospierre B, Mach B. Regulation of MHC class II expression by interferon-gamma mediated by the transactivator gene CIITA. Science. 1994;265:106–109. doi: 10.1126/science.8016643. [DOI] [PubMed] [Google Scholar]
- 31.Pennini ME, Liu Y, Yang J, Croniger CM, Boom WH, et al. CCAAT/enhancer-binding protein beta and delta binding to CIITA promoters is associated with the inhibition of CIITA expression in response to Mycobacterium tuberculosis 19-kDa lipoprotein. J Immunol. 2007;179:6910–6918. doi: 10.4049/jimmunol.179.10.6910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loetscher M, Gerber B, Loetscher P, Jones SA, Piali L, et al. Chemokine receptor specific for IP10 and mig: structure, function, and expression in activated T-lymphocytes. J Exp Med. 1996;184:963–969. doi: 10.1084/jem.184.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cole KE, Strick CA, Paradis TJ, Ogborne KT, Loetscher M, et al. Interferon-inducible T cell alpha chemoattractant (I-TAC): a novel non-ELR CXC chemokine with potent activity on activated T cells through selective high affinity binding to CXCR3. J Exp Med. 1998;187:2009–2021. doi: 10.1084/jem.187.12.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fuller C, Flynn JL, Reinhart TA. In situ study of abundant expression of proinflammatory chemokines and cytokines in pulmonary granulomas that develop in cynomolgus macaques experimentally infected with Mycobacterium tuberculosis. Infect Immun. 2003;71:7023–7034. doi: 10.1128/IAI.71.12.7023-7034.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sierro F, Biben C, Martínez-Muñoz L, Mellado M, Ransohoff RM, et al. Disrupted cardiac development but normal hematopoiesis in mice deficient in the second CXCL12/SDF-1 receptor, CXCR7. Proc Natl Acad Sci U S A. 2007;104:14759–14764. doi: 10.1073/pnas.0702229104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 37.Beg AA, Baltimore D. An essential role for NF-kappaB in preventing TNF-alpha-induced cell death. Science. 1996;274:782–784. doi: 10.1126/science.274.5288.782. [DOI] [PubMed] [Google Scholar]
- 38.Doi TS, Marino MW, Takahashi T, Yoshida T, Sakakura T, et al. Absence of tumor necrosis factor rescues RelA-deficient mice from embryonic lethality. Proc Natl Acad Sci U S A. 1999;96:2994–2999. doi: 10.1073/pnas.96.6.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Douvas GS, Looker DL, Vatter AE, Crowle AJ. Gamma interferon activates human macrophages to become tumoricidal and leishmanicidal but enhances replication of macrophage-associated mycobacteria. Infect Immun. 1985;50:1–8. doi: 10.1128/iai.50.1.1-8.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rook GA, Steele J, Ainsworth M, Champion BR. Activation of macrophages to inhibit proliferation of Mycobacterium tuberculosis: comparison of the effects of recombinant gamma-interferon on human monocytes and murine peritoneal macrophages. Immunology. 1986;59:333–338. [PMC free article] [PubMed] [Google Scholar]
- 41.Lee KK, Workman JL. Histone acetyltransferase complexes: one size doesn't fit all. Nat Rev Mol Cell Biol. 2007;8:284–295. doi: 10.1038/nrm2145. [DOI] [PubMed] [Google Scholar]
- 42.Sengupta N, Seto E. Regulation of histone deacetylase activities. J Cell Biochem. 2004;93:57–67. doi: 10.1002/jcb.20179. [DOI] [PubMed] [Google Scholar]
- 43.Wang Y, Curry HM, Zwilling BS, Lafuse WP. Mycobacteria inhibition of IFN-gamma induced HLA-DR gene expression by up-regulating histone deacetylation at the promoter region in human THP-1 monocytic cells. J Immunol. 2005;174:5687–5694. doi: 10.4049/jimmunol.174.9.5687. [DOI] [PubMed] [Google Scholar]
- 44.Zhu P, Zhou W, Wang J, Puc J, Ohgi KA, et al. A histone H2A deubiquitinase complex coordinating histone acetylation and H1 dissociation in transcriptional regulation. Mol Cell. 2007;27:609–621. doi: 10.1016/j.molcel.2007.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pluemsampant S, Safronova OS, Nakahama K, Morita I. Protein kinase CK2 is a key activator of histone deacetylase in hypoxia-associated tumors. Int J Cancer. 2008;122:333–341. doi: 10.1002/ijc.23094. [DOI] [PubMed] [Google Scholar]
- 46.Yang SR, Chida AS, Bauter MR, Shafiq N, Seweryniak K, et al. Cigarette smoke induces proinflammatory cytokine release by activation of NF-kappaB and posttranslational modifications of histone deacetylase in macrophages. Am J Physiol Lung Cell Mol Physiol. 2006;291:L46–57. doi: 10.1152/ajplung.00241.2005. [DOI] [PubMed] [Google Scholar]
- 47.Ito K, Ito M, Elliot WM, Cosio B, Caramori G, et al. Decreased histone deacetylase activity in chronic obstructive pulmonary disease. N Engl J Med. 2005;352:1967–1976. doi: 10.1056/NEJMoa041892. [DOI] [PubMed] [Google Scholar]
- 48.Hu E, Chen Z, Fredrickson T, Zhu Y, Kirkpatrick R, et al. Cloning and characterization of a novel human class I histone deacetylase that functions as a transcription repressor. J Biol Chem. 2000;275:15254–15264. doi: 10.1074/jbc.M908988199. [DOI] [PubMed] [Google Scholar]
- 49.Lee H, Rezai-Zadeh N, Seto E. Negative regulation of histone deacetylase 8 activity by cyclic AMP-dependent protein kinase A. Mol Cell Biol. 2004;24:765–773. doi: 10.1128/MCB.24.2.765-773.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pflum MK, Tong JK, Lane WS, Schreiber SL. Histone deacetylase 1 phosphorylation promotes enzymatic activity and complex formation. J Biol Chem. 2001;276:47733–47741. doi: 10.1074/jbc.M105590200. [DOI] [PubMed] [Google Scholar]
- 51.Tsai SC, Seto E. Regulation of histone deacetylase 2 by protein kinase CK2. J Biol Chem. 2002;277:31826–31833. doi: 10.1074/jbc.M204149200. [DOI] [PubMed] [Google Scholar]
- 52.Singh NN, Ramji DP. Protein kinase CK2, an important regulator of the inflammatory response? J Mol Med. 2008;86:887–897. doi: 10.1007/s00109-008-0352-0. [DOI] [PubMed] [Google Scholar]
- 53.Kalkhove E. CBP and p300: HATs for different occasions. Biochem Pharmacol. 2004;68:1145–1155. doi: 10.1016/j.bcp.2004.03.045. [DOI] [PubMed] [Google Scholar]
- 54.Zhang JJ, Vinkemeier U, Gu W, Chakravarti D, Horvath CM, et al. Two contact regions between Stat1 and CBP/p300 in interferon gamma signaling. Proc Natl Acad Sci U S A. 1996;93:15092–15096. doi: 10.1073/pnas.93.26.15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang WC, Ju TK, Hung MC, Chen CC. Phosphorylation of CBP by IKKalpha promotes cell growth by switching the binding preference of CBP from p53 to NF-kappaB. Mol Cell. 2007;26:75–87. doi: 10.1016/j.molcel.2007.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yuan LW, Soh JW, Weinstein IB. Inhibition of histone acetyltransferase function of p300 by PKCdelta. Biochim Biophys Acta. 2002;1592:205–211. doi: 10.1016/s0167-4889(02)00327-0. [DOI] [PubMed] [Google Scholar]
- 57.Gaston K, Jayaraman PS. Transcriptional repression in eukaryotes: repressors and repression mechanisms. Cell Mol Life Sci. 2003;60:721–741. doi: 10.1007/s00018-003-2260-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gupta AK, Kone BC. CCAAT/enhancer binding protein-beta trans-activates murine nitric oxide synthase 2 gene in an MTAL cell line. Am J Physiol. 1999;276:F599–605. doi: 10.1152/ajprenal.1999.276.4.F599. [DOI] [PubMed] [Google Scholar]
- 59.Litvak V, Ramsey SA, Rust AG, Zak DE, Kennedy KA, et al. Function of C/EBPdelta in a regulatory circuit that discriminates between transient and persistent TLR4-induced signals. Nat Immunol. 2009;10:437–443. doi: 10.1038/ni.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hamon MA, Batsche E, Regnault B, Tham TN, Seveau S, et al. Histone modifications induced by a family of bacterial toxins. Proc Natl Acad Sci U S A. 2007;104:13467–13472. doi: 10.1073/pnas.0702729104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weidman MK, Sharma R, Raychaudhuri S, Kundu P, Tsai W, et al. The interaction of cytoplasmic RNA viruses with the nucleus. Virus Res. 2003;95:75–85. doi: 10.1016/s0168-1702(03)00164-3. [DOI] [PubMed] [Google Scholar]
- 62.Le May N, Dubaele S, Proietti De Santis L, Billecocq A, Bouloy M, et al. TFIIH transcription factor, a target for the Rift Valley hemorrhagic fever virus. Cell. 2004;116:541–550. doi: 10.1016/s0092-8674(04)00132-1. [DOI] [PubMed] [Google Scholar]
- 63.Yuan H, Yoza BK, Lyles DS. Inhibition of host RNA polymerase II-dependent transcription by vesicular stomatitis virus results from inactivation of TFIID. Virology. 1998;251:383–392. doi: 10.1006/viro.1998.9413. [DOI] [PubMed] [Google Scholar]
- 64.Popkin DL, Watson MA, Karaskov E, Dunn GP, Bremner R, et al. Murine cytomegalovirus paralyzes macrophages by blocking IFN gamma-induced promoter assembly. Proc Natl Acad Sci U S A. 2003;100:14309–14314. doi: 10.1073/pnas.1835673100. [DOI] [PMC free article] [PubMed] [Google Scholar]