Abstract
APC/Cdh1 is a major cell cycle regulator and its function has been implicated in DNA damage repair; however, its exact role remains unclear. Using affinity purification coupled with mass spectrometry, we identified Claspin as a novel Cdh1-interacting protein and further demonstrated that Claspin is a novel Cdh1 ubiquitin substrate. As a result, inactivation of Cdh1 leads to activation of the Claspin/Chk1 pathway. Previously, we demonstrated that Rb interacts with Cdh1 to influence its ability to degrade Skp2. Here, we report that Cdh1 reciprocally regulates the Rb pathway through competing with E2F1 to bind the hypophosphorylated form of Rb. Although inactivation of Cdh1 in HeLa cells, with defective p53/Rb pathways, led to premature S phase entry, acute depletion of Cdh1 in primary human fibroblasts resulted in premature senescence. Acute loss of many other major tumor suppressors, including PTEN and VHL, also induces premature senescence in a p53- or Rb-dependent manner. Similarly, we showed that inactivation of the p53/Rb pathways by overexpression of SV40 LT-antigen partially reversed Cdh1 depletion–induced growth arrest. Therefore, loss of Cdh1 is only beneficial to cells with abnormal p53 and Rb pathways, which helps explain why Cdh1 loss is not frequently found in many tumors.
INTRODUCTION
In normally dividing cells, the cell cycle is tightly controlled and defective cell cycle regulation can lead to genomic instability and ultimately, cancer development. Selective degradation of key cell cycle regulators by the ubiquitin-proteasome system has recently been shown to be a major regulatory mechanism for ensuring ordered and coordinated cell cycle progression (Cardozo and Pagano, 2004; Petroski and Deshaies, 2005). Two related, multiunit E3 ubiquitin ligase enzymes, anaphase-promoting complex (APC) and Skp1-Cullin1-F-box complex (SCF), are thought to be the major driving forces governing cell cycle progression (Nakayama et al., 2001). We previously demonstrated that Cdh1 could control the degradation of the SCF component Skp2, a known oncoprotein (Gstaiger et al., 2001), in the early G1 phase. Furthermore, many known Cdh1 substrates including Securin (Zou et al., 1999), Cyclin A, Polo-like kinase 1 (Takai et al., 2005), and Aurora Kinase (Giet et al., 2005) have been found to possess oncogenic activity/potential and are overexpressed in various types of cancer. The function of Cdh1 as a tumor suppressor was confirmed recently by mouse modeling (Garci-Higuera et al., 2008). Nevertheless, it remains unclear why in contrast to the frequently observed loss of other tumor suppressor genes including p53, PTEN, or VHL, loss of Cdh1 expression is not found frequently in various tumors.
In response to DNA damage signals, cells will arrest either in the G1 phase to prevent the replication of the damaged DNA or arrest in the G2 phase to prevent cells from segregating defective chromosomes (Weinert, 1998). It is widely assumed that activation of the ATM/ATR/Chk kinase pathway is the initial response to damaged DNA, resulting in the phosphorylation and stabilization of p53. The p53 tumor suppressor protein is considered the major effector of DNA damage signals, eliciting a cellular response by activating either a G1 or G2 arrest through its transcriptional targets p21 or Gadd45a (Lakin and Jackson, 1999; Meek, 1999). The p21 protein is an universal Cdk inhibitor, and many reports have demonstrated that the p53/p21 pathway is required for establishing G1 arrest after genotoxic stress (Waldman et al., 1995). On the other hand, it is known that cyclin, the regulatory subunit of various Cdk complexes, is extremely unstable and that destruction of cyclin by either the APC or SCF complexes is considered an efficient way to inhibit Cdk activity, thereby facilitating cell cycle arrest. It is noteworthy that APC/Cdh1 activity can be activated by DNA damage signals, resulting in enhanced degradation of its substrates including cyclin B, cyclin D (Agami and Bernards, 2000), Cdc25A (Donzelli et al., 2002), and Securin (Romero et al., 2001). Thus, Cdh1 activity may also play an important role in mediating cell cycle arrest after DNA damage signals. In support of this assertion, Cdh1−/− chick DT40 cells failed to arrest in either the G1 or G2 phase after DNA damage (Sudo et al., 2001). Furthermore, it was demonstrated recently that loss of Cdh1 in both human and mouse fibroblasts leads to accumulation of DNA damage signals (Engelbert et al., 2008; Garci-Higuera et al., 2008). However, the exact mechanism by which Cdh1 participates in the DNA damage repair process is still unknown. Using affinity purification coupled with mass spectrometry, we found that Claspin associates with Cdh1. In agreement with a recent report (Bassermann et al., 2008), we further demonstrated that Claspin is a novel substrate for Cdh1. Claspin is a positive regulator of Chk1 activity and is a key player in the DNA damage repair pathway (Sar et al., 2004). Thus our finding provides a bridge for further understanding the function of Cdh1 in the DNA-damage repair pathway.
We previously showed that Rb specifically interacts with Cdh1 and thereby influences its ability to degrade its substrate Skp2 (Binne et al., 2007). Here, we continue to demonstrate that by specifically interacting with the hypophosphorylated (active) form of Rb, Cdh1 can regulate the activity of the E2F1 transcription factor (Sorensen et al., 2000). As a result, in normal human fibroblasts, depletion of Cdh1 resulted in premature senescence, whereas inactivation of both the p53 and Rb pathways by overexpressing SV40 LT-antigen partially reversed this phenotype. Interestingly, inactivation of other major tumor suppressors including PTEN (Chen et al., 2005) and VHL (Young et al., 2008) is also reported to induce premature senescence. The ability of normal cells to establish premature senescence in response to various stresses, including DNA damage (Robles and Adami, 1998), oxidative stress (Chen et al., 1998), and expression of active oncogenic proteins (Bartkova et al., 2006; Koutsami et al., 2006; Mallette et al., 2007) has been proposed as a built-in fail-safe mechanism against cancer development (Campisi and d'Adda di Fagagna, 2007). In agreement with this model, our finding that normal human fibroblasts undergo premature senescence after acute loss of Cdh1 provides the possible underlying molecular mechanism for the less frequently observed Cdh1 loss in tumor cells and further implies that loss of Cdh1 occurs late in tumor development.
MATERIALS AND METHODS
Plasmids
A claspin construct was obtained from Michele Pagano (New York University School of Medicine, New York, NY), and subcloned into the pCDNA3.5Xmyc vector. Claspin mutants and Cdh1 mutants were generated using the QuikChange XL Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions. Full-length Cdh1 cDNA was subcloned using the Pfu polymerase (Stratagene) into the pGEX vector to create a GST-Cdh1 in-frame fusion protein. HA-E2F1 and Flag-Rb vectors were kind gifts from Dr. William Kaelin (Dana-Farber Cancer Institute, Boston, MA). The c-terminal of E2F1 (amino acids 382-437; Hsieh et al., 2002) was subcloned using the Pfu polymerase (Stratagene) into the pGEX vector to create GST-E2F1 in frame fusion protein. Various Cdh1 constructs were described previously. HA-Cdh1 and HA-Cdc20 were obtained from Dr. Peter Jackson (Genentech, South San Francisco, CA). The Fbw4, Fbw5, Fbw7, and Skp2 constructs were described previously (Wei et al., 2005). HA-Trcp1 vector was obtained from Dr. Wade Harper (Harvard Medical School, Boston, MA). Lentiviral short hairpin RNA (shRNA) vectors against GFP or Cdh1 were obtained from Dr. William Hahn (Dana-Farber Cancer Institute, Boston, MA).
Antibodies and Reagents
Anti-Chk1 antibody (SC-8408), anti-p21 antibody (sc-397), anti c-Myc antibody (sc-40), anti-p53 antibody (sc-126), anti-p27 antibody (SC-528), polyclonal anti- HA antibody (SC-805), anti-cyclin A antibody (SC-751), anti-cyclin B antibody (SC-245), anti-Cdc20 antibody (SC-8358), anti-Plk1 antibody (SC-17783), anti-cyclin E antibody (SC-247), and anti-Ets-2 antibody (SC-22803) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-tubulin antibody (T-5168), polyclonal anti-FLAG antibody (F2425), monoclonal anti-FLAG antibody (F-3165), anti-vinculin antibody (V4505), peroxidase-conjugated anti-mouse secondary antibody (A4416), and peroxidase-conjugated anti-rabbit secondary antibody (A4914) were purchased from Sigma (St. Louis, MO). Monoclonal anti-HA antibody (MMS-101P) was purchased from Covance (Madison, WI). Anti-GFP antibody (632380) and monoclonal anti-Skp2 antibody (32-3400) was purchased from Invitrogen (Carlsbad, CA). Monoclonal anti-Cdh1 (CC43) antibody was purchased from Calbiochem (San Diego, CA). Anti-Claspin antibody (A267A) was purchased from Bethyl Labs (Montgomery, TX). Anti-pSer317-Chk1 antibody (2344), anti-pSer15-p53 antibody (9284), anti-pSer20-p53 antibody (9287), and anti-pSer807pSer811-Rb antibody (9308) were purchased from Cell Signaling (Beverly, MA). Anti-E2F1 antibody (05-379) was purchased from Upstate Biotechnology (Lake Placid, NY). Anti-p16INK4a antibody was purchased from BD Pharmingen (San Diego, CA).
Small Interfering RNAs
Human small interfering RNA (siRNA) oligos which can deplete both β-Trcp1 and β-Trcp2 (sense, 5′-AAGUGGAAUUUGUG GAACAUC-3′) have been validated previously (Jin et al., 2003) and were purchased from Dharmacon (Boulder, CO). Human siRNA oligos against Fbw7, Skp2, and Cdh1 have been described previously (Wei et al., 2004, 2005). A luciferase GL2 siRNA oligo was purchased from Dharmacon. As described previously, siRNA oligos were transfected into subconfluent cells using Oligofectamine or Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions (Wei et al., 2004, 2005).
Cell culture and Cell Synchronization
Cell culture, including synchronization and transfection, has been described (Wei et al., 2004). Retrovirus packaging and subsequent infection of various cell lines were performed according to the protocol described previously (Wei et al., 2003). Lentiviral shRNA virus packaging and subsequent infection of various cell lines were performed according to the protocol described previously (Boehm et al., 2005).
Immunoblots and Immunoprecipitation
Cells were lysed in EBC (50 mM Tris, pH 7.5, 120 mM NaCl, and 0.5% NP-40) buffer supplemented with protease inhibitors (Complete Mini, Roche) and phosphatase inhibitors (phosphatase inhibitor cocktail set I and II, Calbiochem). The protein concentrations of the lysates were measured using the Bio-Rad protein assay reagent on a Beckman Coulter DU-800 spectrophotometer (Fullerton, CA). The lysates were then resolved by SDS-PAGE and immunoblotted with indicated antibodies. For immunoprecipitation, 800 μg lysates were incubated with the appropriate antibody (1–2 μg) for 3–4 h at 4°C followed by a 1-h incubation with protein A Sepharose beads (GE Healthcare, Waukesha, WI). Immunocomplexes were washed five times with NETN buffer (20 mM Tris, pH 8.0, 100 mM NaCl, 1 mM EDTA, and 0.5% NP-40) before being resolved by SDS-PAGE and immunoblotted with indicated antibodies.
Rb Binding Assays
Binding to immobilized GST proteins was performed as described before (Wei et al., 2004).
Protein Degradation and In Vitro Ubiquitination Analysis
Cells were transfected with a plasmid encoding a Myc-tagged version of Claspin along with HA-Cdh1 or HA-Cdc20, and a plasmid encoding GFP as a negative control. Forty hours after transfection, cells were lysed and protein abundances were measured by immunoblot analysis. In vitro ubiquitination assay was performed as described previously (Wei et al., 2004).
Bromodeoxyuridine Labeling and Senescence-associated β-Galactosidase Staining
Bromodeoxyuridine (BrdU) labeling and senescence-associated β-galactosidase (β-Gal) staining were performed using the protocol described previously (Wei et al., 2001). Experiments were repeated twice to generate error bars.
Fly Work
A null mutation for Fzr (fzrie28) was obtained from C. Lehner (University of Bayreuth, Bayreuth, Germany) (Jacobs et al., 2002). Mitotic clones were induced using the flippase recombination enzyme/flippase recognition target (FLP/FRT) technique (Xu and Rubin, 1993). Therefore, the mutant allele was recombined with an FRT-chromosome (FRT19A) to generate fzrie28, FRT19A, and ey-FLP was used to induce the clones in the eye. Immunostaining of eye imaginal disks was performed as previously described (Moon et al., 2006), using antibodies against green fluorescent protein (GFP; rabbit polyclonal, 1:2000, Invitrogen), Cyclin B (mouse monoclonal, 1:100, Developmental Studies Hybridoma Bank) and dE2F1 (guinea pig polyclonal, 1:200, a gift from T. Orr-Weaver, Whitehead Institute for Biomedical Research, Cambridge, MA).
RESULTS
Claspin Is Identified as a Specific Cdh1-interacting Protein
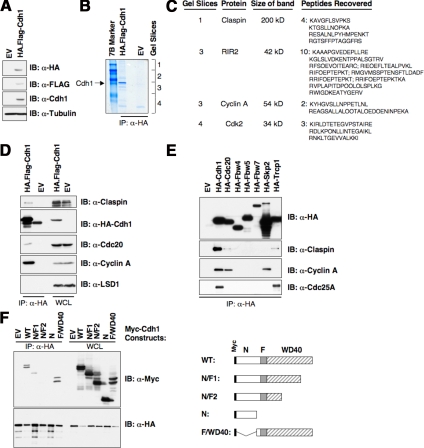
The interaction of Cdh1 with its substrates does not require any prior modification of the substrate (Harper et al., 2002; Peters, 2002). This feature suggests that an affinity-purification approach is a promising way to screen for putative Cdh1 substrates. Toward this end, we constructed a T98G cell line that overexpresses hemagglutinin (HA)- and Flag-tagged Cdh1 protein (Figure 1A). After affinity purification of the HA-Cdh1 protein, we utilized mass spectrometry to identify Cdh1-associated proteins (Figure 1B). From this analysis, 10 peptides were identified for a previously described Cdh1 substrate, RIR2 (Chabes et al., 2003), and two peptides were identified from the well-known Cdh1 substrate Cyclin A (Pfleger et al., 2001). These results clearly demonstrated that this method is a valid and practical approach to search for novel Cdh1 substrates. In addition to these two proteins, four peptides were identified for the Claspin protein (Figure 1C), which is an upstream regulator of the Chk1 kinase (Kumagai et al., 2004; Wang et al., 2006). The interaction between the Cdh1 and Claspin proteins was validated using immunoprecipitation analysis (Figure 1D). Furthermore, we showed that the interaction between Cdh1 and Claspin is specific: Claspin only weakly interacts with Cdc20 and does not interact with other F-box proteins we tested, except β-Trcp, which has been identified recently as an upstream E3 ligase that controls Claspin stability in the M phase (Figure 1E; Peschiaroli et al., 2006). Additionally, we also mapped the interaction domain on the Cdh1 protein required for interaction with Claspin (Figure 1F). We found that Cdh1 utilizes its WD-40 repeats motif to interact with Claspin. Previous studies suggested that most Cdh1 substrates interact with the WD-40 repeats-domain of Cdh1 (Harper et al., 2002); therefore this result supports our hypothesis that Claspin might be a novel Cdh1 ubiquitin substrate.
Figure 1.
Identification of Claspin as a novel Cdh1-interacting protein. (A) Immunoblot analysis of the T98G cell lines infected with control (EV) or pBabe-retroviral vector expressing HA.Flag-tagged Cdh1. (B) Cellular lysates were prepared from both cell lines, and subjected to HA immunoprecipitation. After extensive washes, the recovered proteins were separated by SDS-PAGE and stained with Gel-code blue reagent. (C) Illustration of the peptide sequences identified in the Cdh1-IP by mass spectrometry analysis. (D) Cell lysates were prepared from the two generated T98G cells, and after immunoprecipitation with HA antibody, immunoblot analyses with indicated antibody were performed to detect their ability to interact with ectopically expressed Cdh1 protein. (E) 293T cells were transiently transfected with various constructs as indicated. Forty-eight hours after transfection, cell lysates were recovered, and HA immunoprecipitation was performed. The immunoprecipitates were denatured in SDS-containing sample buffer and separated by SDS-PAGE before immunoblot analysis with indicated antibodies. (F) HA-Claspin and various Myc-tagged Cdh1 constructs were cotransfected into 293T cells. Forty-eight hours after transfection, cell lysates were recovered, and HA immunoprecipitation was performed. The immunoprecipitates were denatured in SDS-containing sample buffer and separated by SDS-PAGE before immunoblot analysis with HA and Myc antibodies.
Claspin Abundance Is Controlled by Cdh1
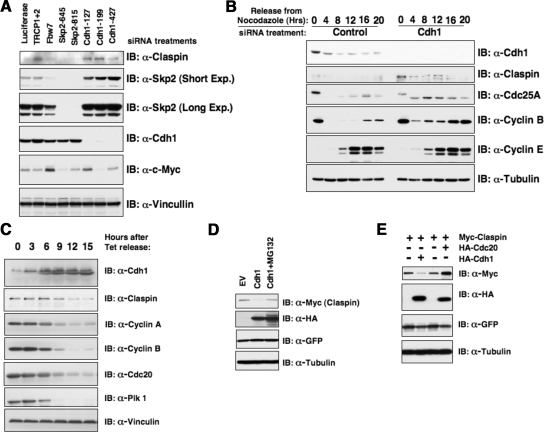
Consistent with the hypothesis that Claspin is a novel ubiquitin substrate for APC/Cdh1, depletion of Cdh1 by RNAi treatment led to an up-regulation of Claspin expression levels in both synchronized and asynchronized HeLa cells (see Figure 2, A and B), indicating that Cdh1 controls Claspin destruction. It is worth noting that depletion of β-Trcp1, another known upstream E3 ligase, but not other F-box proteins, also led to a significant up-regulation of Claspin abundance (Figure 2A). Conversely, we also found that overexpression of Cdh1 led to a significant reduction of Claspin protein abundance (Figure 2, C and D), which can be reversed by treatment with the proteasome inhibitor MG132 (Figure 2D). In keeping with our previous finding, Cdc20, which did not strongly interact with Claspin (as illustrated in Figure 1E), was also unable to promote Claspin degradation (Figure 2E). Sequence analysis revealed that similar to other known Cdh1 downstream substrates including Cyclin B1, Securin, and Skp2, Claspin contains a canonical destruction box as well as other types of degrons including A-box and O-boxes that can be recognized by the APC/Cdh1 E3 ubiquitin ligase (Supplemental Figure S1, A and B). However, to our surprise, after mutation of all the identified possible degron sequences, the mutant Claspin was still unstable in the early G1 phase, where APC/Cdh1 activity is high (Supplemental Figure S1D). This indicates that the Claspin protein might contain a novel degron sequence that has not yet been identified.
Figure 2.
Cdh1 controls Claspin stability. (A and B) Depletion of Cdh1 by siRNA treatments leads to significant induction of Claspin protein in both asynchronized (A) and synchronized (B) HeLa cells. (C) Immunoblot analysis of the U2OS cells, engineered to overexpress Myc-Cdh1 after removal of tetracycline to induce Cdh1, with indicated antibodies. (D–E) Myc-Claspin and HA-Cdh1 or HA-Cdc20 constructs were cotransfected into HeLa cells as indicated, and the differences in Claspin expression levels were detected by anti-Myc immunoblot analysis. MG132 was added in D as indicated to block the proteasomal degradation pathway.
The Claspin Protein Contains a Novel Degron at its N-Terminus That Governs Its Destruction by Cdh1
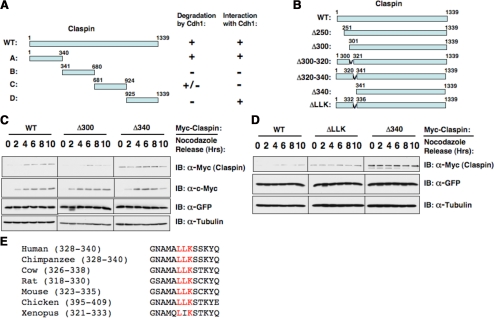
To identify the degron sequence critical for Cdh1-mediated Claspin destruction, we generated four Claspin constructs that contained four nonoverlapping fragments (Figure 3A). Further studies revealed that although both fragment A and fragment D could interact with Cdh1 (Supplemental Figure S2B), only fragment A could be efficiently degraded by overexpressing Cdh1 (Supplemental Figure S2A). This implied that the potential unidentified degron was localized within the first 340 amino acids (Figure 3A). Therefore, we made a series of N-terminal deletion Claspin constructs (Figure 3B). As illustrated previously (Bennett and Clarke, 2006), wild-type Claspin is unstable in the early G1 phase, where APC/Cdh1 is active (Figure 3C). Deletion of the first 300 amino acids did not affect its stability in early G1 phase. However, further deletion of the next 40 amino acids created a mutant Claspin that is stable in the early G1 phase (Figure 3C). To further pinpoint the exact location of the putative degron, we generated two internal deletion constructs, Δ300–320 and Δ320–340 (Supplemental Figure S2C). We found that the Δ320–340 mutant Claspin became stabilized in early G1 phase. This might be due to reduced ubiquitination by Cdh1 because after deletion of the degron sequence, the resulting mutant cannot be efficiently ubiquitinated by Cdh1 in vitro (Supplemental Figure S2D). After careful examination of the protein sequence in this region, we found that two motifs, LLK and KYQ, that were conserved across different species (Figure 3E). Further studies clearly showed that mutation or deletion of the KYQ motif did not affect the stability of Claspin (data not shown), whereas deletion of the LLK motif created a Claspin mutant that could not be degraded at the early G1 phase (Figure 3D). Furthermore, we found that deletion of the LLK motif also abolished its interaction with Cdh1 (Supplemental Figure S2E). Although it is distinct from all known Cdh1 degron motifs, it seems to be derived from a hybrid of the O-box and KEN box.
Figure 3.
Claspin contains a novel degron at its N-terminus that governs its destruction by Cdh1. (A) Summary of the nonoverlapping Claspin fragments and their interaction with and destruction by Cdh1. (B) Illustration of the various Claspin mutants to map the location of the C-box. (C and D) Immunoblot analysis of HeLa cells transfected with the indicated Myc-Claspin plasmid, synchronized by growth in nocodazole, and then released for the indicated periods of time. (E) Sequence alignment of the novel degron sequence among different species.
Loss of Cdh1 Activates the Claspin/Chk1 Pathway
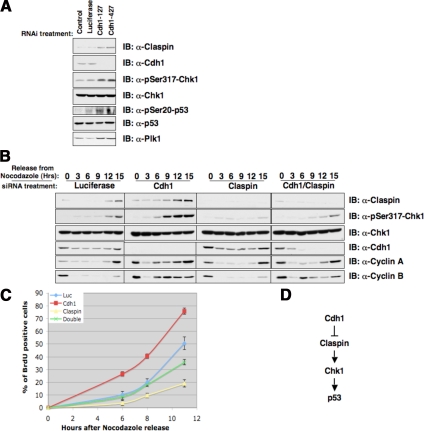
The Claspin protein plays a critical role in activating Chk1 kinase activity in response to various stresses including DNA-damage signals (Kumagai et al., 2004; Wang et al., 2006). We found that depletion of Cdh1 in HeLa, U2OS, and primary human fibroblasts led to an up-regulation of Claspin, and subsequent increase in activation of Chk1 activity, as determined by the p-Ser317-Chk1 antibody (Figure 4A, Supplemental Figures S3 and S5C). Claspin/Chk1 activation was typically seen after cells entered the S phase (Bennett and Clarke, 2006), and recent studies indicated that Claspin/Chk1 activity is important for proper DNA replication (Petermann et al., 2008). We found that overexpression of Claspin promotes BrdU incorporation (Supplemental Figure S3E), whereas depletion of Cdh1 in HeLa cells resulted in an unscheduled premature induction of Claspin/Chk1 activity, as evidenced by enhanced Ser317-Chk1 phosphorylation in the G1 phase (Figure 4B); and subsequent premature entry into S phase as determined by enhanced BrdU labeling, a phenotype which can be reversed by further inactivation of the Claspin protein (Figure 4C). Chk1 has been shown to phosphorylate p53 at the Ser-20 site to activate the p53 pathway (Shieh et al., 2000). Correspondingly, we found that inactivation of Cdh1 in U2OS cells led to a moderate increase in p53 Ser-20 phosphorylation (Figure 4A), indicating that activation of the Claspin/Chk1 pathway might contribute to the observed increase in p53 activity after Cdh1 loss (Engelbert et al., 2008; Garci-Higuera et al., 2008; Figure 4D). However, further studies are needed to dissect whether the increased p53 phosphorylation status is a cause or consequence of the accumulated DNA damage-like signals induced by Cdh1-loss (Engelbert et al., 2008; Garci-Higuera et al., 2008).
Figure 4.
Depletion of Cdh1 leads to unscheduled activation of Chk-1 activity in the G1 phase. (A) Depletion of Cdh1 by siRNA treatments leads to a significant induction of Claspin protein and activation of the Chk1/p53 pathway in asynchronized U2OS cells. (B) HeLa cells were transfected with indicated siRNA oligos, and 6 h after transfection, cells were synchronized with nocodazole. Eighteen hours later, cells were released back into the G1 phase, and at the various indicated time points, cells were lysed for immunoblot analysis using the indicated antibodies. (C) HeLa cells were transfected with the indicated siRNA oligos; 6 h after transfection, cells were synchronized with nocodazole. Eighteen hours later, cells were released back into the G1 phase. At various indicated time points, cells were pulsed with BrdU for 30 min, and afterward immunohistochemistry staining was performed with anti-BrdU antibody to detect the percentage of BrdU-positive cells. (D) Illustration of the proposed model by which Cdh1 could potentially activate Chk1 and p53 activity by controlling Claspin destruction.
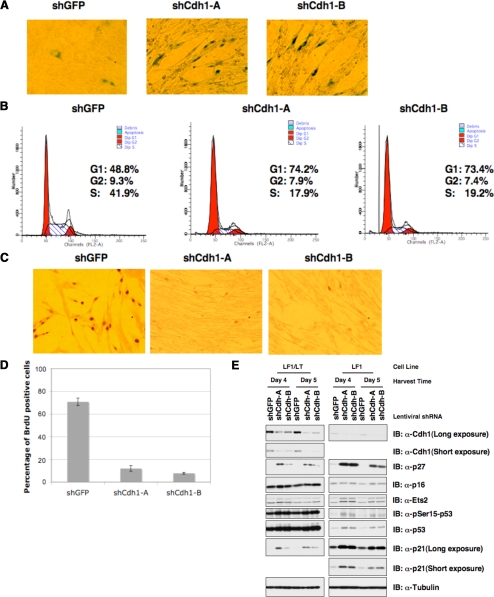
Depletion of Cdh1 Resulted in Premature Senescence in Primary Human Fibroblasts
Previous reports demonstrated that overexpression of strong oncogenes, such as Ha-Ras or Stat3, likewise led to activation of the ATM/ATR DNA damage pathway and premature senescence (Mallette et al., 2007), and that inactivation of both the p53 and Rb pathways is required for bypassing the growth arrest phenotype (Serrano et al., 1997; Wei et al., 2003). In keeping with previous reports (Bashir et al., 2004; Wei et al., 2004), we found that depletion of Cdh1 in HeLa cells, whose p53 and Rb pathways are defective due to the presence of HPV E6 and E7 proteins, did not cause growth arrest (Supplemental Figure S4A); instead, it resulted in early S phase entry as evidenced by enhanced BrdU labeling (Figure 4C).
In contrast to our observations in HeLa cells, we found that depletion of Cdh1 with lenti-viral shRNA vectors resulted in retarded growth in U2OS cells, which possess relatively normal p53 pathway, but defective Rb pathway due to loss of p16 expression (Stott et al., 1998; Supplemental Figure S4, B and E). A similar growth retardation phenotype is observed in mouse embryonic fibroblasts when Cdh1 is depleted (Garci-Higuera et al., 2008; Li et al., 2008). We further found that normal human fibroblasts (Wei et al., 2003) responded more severely to acute loss of Cdh1, undergoing the premature senescence program as evidenced by elevated β-Gal staining (Figure 5A) and ceased cell proliferation (Supplemental Figure S4F). This observation was supported by FACS analysis, which showed that the S-phase cell populations were significantly decreased after depletion of Cdh1 (Figure 5B). Additionally, we found that after Cdh1 depletion, there was almost no BrdU incorporation even after 6-h incubation, whereas more than 60% of the control cells were BrdU positive (Figure 5, C and D). In agreement with the growth arrest phenotype, we found that many senescence-associated molecular markers, including p21, p27, and to a lesser degree p16, were also induced after acute loss of Cdh1 in primary human fibroblasts (Figure 5E). p21 is a well-characterized p53 downstream target; thus, the significant induction of p21 provides further support for the notion that loss of Cdh1 activates the p53 pathway. On the other hand, consistent with a recent report (Li et al., 2008), we found that elevated Ets-2 expression might contribute to the moderate increase in p16 expression (Figure 5E).
Figure 5.
Acute loss of Cdh1 expression resulted in the onset of premature senescence in primary human fibroblasts. (A–C) Primary human lung fibroblast cells were infected with the indicated lentiviral shRNA constructs. Twenty-four hours after infection, cells were selected with 1 μg/ml puromycin to eliminate the noninfected cells. Seven days after puromycin selection, cells were fixed and stained for senescence-associated β-Gal activity (A). Six days after puromycin selection, cells were fixed by 70% ethanol and the cell cycle distribution was examined by FACS analysis (B). Six days after puromycin selection, cells were incubated with 1 μg/μl BrdU and 100 μg/μl uridine for 6 h before being fixed with cold methanol and subjected to immunohistochemical analysis using anti-BrdU antibody (C). (D) Quantification of the percentage of BrdU-positive cells in C. (E) Immunoblot analysis of primary human lung fibroblasts (LF1) and SV40 LT-antigen expressing LF1 (LF1/LT) cells infected with the indicated shRNA lentiviral vectors. Twenty-four hours after infection, cells were selected with 1 μg/ml puromycin to eliminate the noninfected cells. Whole-cell lysates were collected at the indicated times after infection.
Acute inactivation of other major tumor suppressor pathways such as PTEN or VHL has also been reported to induce the onset of premature senescence. Next, we used the matched HCT116 WT and HCT116 p53−/− cells to examine whether the p53 pathway plays a critical role in this process. To our surprise, depletion of Cdh1 in HCT116 p53−/− cells still resulted in retarded growth arrest (data not shown). Inactivation of both the p53 and Rb pathways has been shown to be required for bypassing oncogene-induced growth arrest (Serrano et al., 1997). Therefore, we next examined whether inactivation of both the Rb and p53 pathways is required to bypass Cdh1 depletion-induced growth arrest.
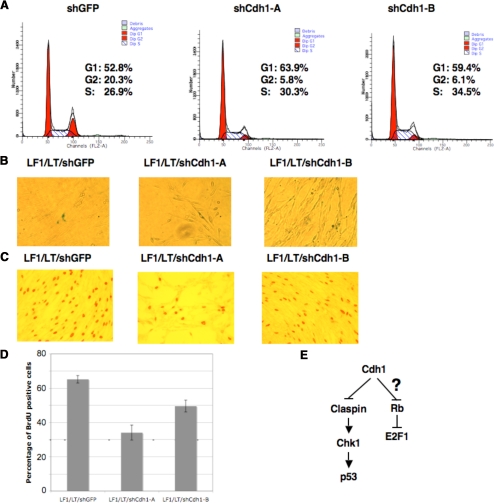
Inactivation of the p53 and Rb Pathways by Overexpressing SV40-LT Antigen Partially Bypassed Cdh1 Depletion–induced Premature Senescence in Primary Human Fibroblasts
To investigate the underlying molecular mechanisms for Cdh1 depletion–induced premature senescence, we generated LT-antigen expressing LF1 and foreskin human fibroblasts (FF1) human fibroblasts (Supplemental Figure S5, A and B). We found a dramatic induction of p53 (Borger and DeCaprio, 2006) and E2F1 (Nemethova et al., 2004) abundance after overexpressing LT-ag, and more importantly, the expression levels of most of the examined E2F1 targets including Cdh1, Chk1, Rb, Plk1, cyclin A, and Skp2 were induced as well (Supplemental Figure S5, A and B). In contrast to the dramatic decrease in S phase cells after Cdh1 depletion in normal LF1 cells (Figure 5B), the S phase population of LF1/LT cells did not decrease after depletion of Cdh1 (Figure 6A). In addition, compared with the total loss of BrdU incorporation in LF1 cells after depleting Cdh1 (Figure 5, C and D), there was only a 1.5- to 2-fold decrease in the percentage of BrdU-positive cells after depletion of Cdh1 in LF1/LT cells (Figure 6, C and D). These results indicated that the growth arrest phenotype was abolished after inactivation of the p53 and Rb pathways. Consistent with this finding, we found that loss of Cdh1 in LF1/LT did not significantly induce the staining of senescence-associated β-Gal (Figure 6B). In addition, we found that the induction of many senescence-associated markers including p21 and p27 in response to Cdh1 loss is largely reduced after overexpression of SV40 LT-antigen (Figure 5E), which might contribute to the bypass of the growth-arrest phenotype induced by acute Cdh1 loss. Moreover, we found that overexpression of SV40 LT-antigen in foreskin human fibroblast cells also partially rescued the onset of premature senescence induced by Cdh1 loss (Supplemental Figure S6).
Figure 6.
Overexpression of LT-ag partially blocked the premature senescence phenotype induced by depletion of Cdh1 in primary human fibroblasts. (A–C) LF1/LT cells were infected with the indicated lentiviral shRNA constructs. Twenty-four hours after infection, cells were selected with 1 μg/ml puromycin to eliminate the noninfected cells. Six days after puromycin selection, cells were fixed by 70% ethanol, and the cell cycle distribution was examined by FACS analysis (A). Seven days after puromycin selection, cells were fixed and stained for senescence-associated β-Gal activity (B). Six days after puromycin selection, cells were incubated with 1 μg/μl BrdU and 100 μg/μl uridine for 6 h before being fixed with cold methanol and subjected to immunohistochemical analysis using anti-BrdU antibody (C). (D) Quantification of the percentage of BrdU-positive cells in C. (E) Illustration of the proposed model by which Cdh1 could potentially regulate the activity of both the Claspin/Chk1/p53 and Rb/E2F1 pathways.
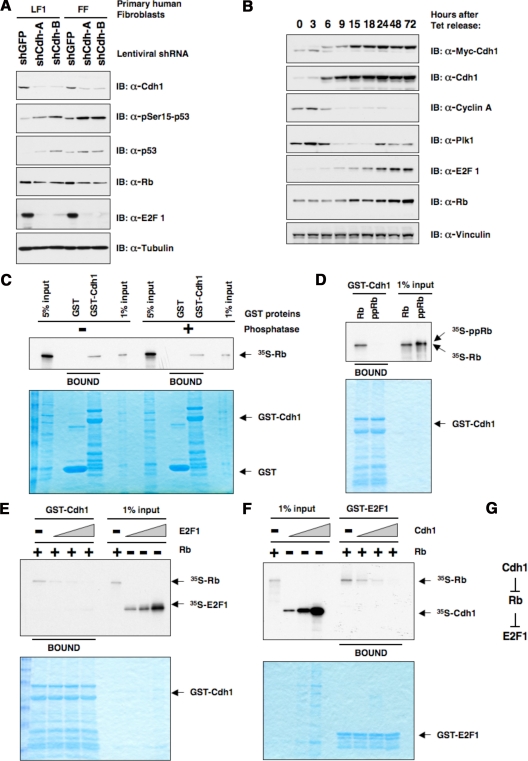
Cdh1 Regulates the Rb/E2F1 Pathway
The fact that inactivation of the p53 and Rb pathways allows the human primary fibroblasts to abrogate Cdh1 depletion–induced premature senescence implied that in addition to the Claspin/Chk1/p53 pathway, Cdh1 regulates the Rb pathway as well (Figure 6E). In support of this notion, we found that in addition to the activation of the p53 pathway as evidenced by an enhanced pSer15-p53 signal, inactivation of Cdh1 in both primary lung fibroblasts and primary foreskin fibroblasts resulted in a marked decrease in E2F1 abundance (Figure 7A). On the other hand, we also found that overexpression of Cdh1 in U2OS cells led to a significant increase of E2F1 levels concomitant with Cdh1 induction (Figure 7B). Next, we sought to investigate mechanistically how Cdh1 controls Rb/E2F1 activity. Previously we reported that Rb interacts with Cdh1 and influences Cdh1-induced Skp2 degradation (Binne et al., 2007). Here, we continued this study to further demonstrate that Cdh1 only interacts with the hypophosphorylated (active) form of Rb and that phosphorylation of Rb with the Cyclin A/Cdk2 complex abolished their interaction (Figure 7, C and D). Furthermore, we found that Cdh1 did not interact with E2F1 (Supplemental Figure S7, A and B). Because E2F1 only interacts with the hypophosphorylated form of Rb as well, we hypothesized that Cdh1 might compete with E2F1 for interaction with the active form of Rb, thus indirectly affecting E2F1 activity. Indeed, we found that an increased amount of E2F1 blocked the interaction between Cdh1 and Rb (Figure 7E) while an increased amount of Cdh1 was also able to block the interaction between E2F1 and Rb (Figure 7F and Supplemental Figure S7D). These results indicate that Cdh1 and E2F1 are mutually exclusive in interacting with Rb. As a result, overexpression of Cdh1 leads to increased levels of free E2F1, whereas depletion of Cdh1 leads to decreased levels of free E2F1 (Figure 7G).
Figure 7.
Cdh1 regulates the Rb/E2F1 pathway. (A) Immunoblot analysis of primary human lung fibroblasts and primary human foreskin fibroblasts infected with indicated shRNA lentiviral vectors. Twenty-four hours after infection, cells were selected with 1 μg/ml puromycin to eliminate the noninfected cells. Whole-cell lysates were harvested 6 d after puromycin selection. (B) Immunoblot analysis of the U2OS cells that were engineered to overexpress Myc-Cdh1 after removal of tetracycline with indicated antibodies. (C–E) Autoradiograms showing recovery of 35S-labeled Rb protein bound to GST-Cdh1 fusion proteins. Where indicated, in vitro–translated Rb protein was incubated with phosphatase (C) or cyclinA/Cdk2 kinase (D) before the pulldown assays. (E) Increased amount of in vitro–translated E2F1 protein was added to the binding reactions (3, 9, and 27 μl, as indicated by the triangles). (F) Autoradiograms showing recovery of 35S-labeled Rb protein bound to GST-E2F1 fusion proteins. Where indicated, increased amount of in vitro-translated Cdh1 protein was added to the binding reactions (3, 9, and 27 μl, as indicated by the triangles). (G) Illustration of the proposed model by which Cdh1 could potentially regulate the abundance of E2F1 through the Rb protein.
Interestingly, we found that deletion of the C-box motif, which has been reported to disrupt the interaction of Cdh1 with the APC core complex (Schwab et al., 2001; Burton et al., 2005), also abolished the interaction between Cdh1 and Rb (Supplemental Figure S8, A and B). Consistent with the model that Cdh1 competes with E2F for Rb interaction, the ΔC-box Cdh1 mutant, which has lost the ability to interact with Rb, also failed to induce E2F1 expression in U2OS cells (Supplemental Figure S8C). Additionally, in agreement with a previous report (Marti et al., 1999), we found that depletion of Skp2 resulted in elevated E2F1 levels (Supplemental Figure S9). Because Skp2 levels are up-regulated after Cdh1 depletion (Supplemental Figure S6D), it is possible that enhanced E2F1 destruction by Skp2 contributes to the decreased E2F1 levels as well.
APC/Cdh1 and Rb/E2F1 pathway functions are highly conserved between mammalian cells and the fruit fly Drosophila melanogaster. To investigate a possible role for Cdh1 loss on Rb/E2F1 pathway activity, we took advantage of Drosophila eye imaginal disks as a model system for the in vivo study of cell cycle regulation. During the third instar larval stage of Drosophila development, a wave of differentiation, controlled by the concerted action of a variety of signaling pathways, moves from the posterior toward the anterior end of the eye disk (Brennan and Moses, 2000). The edge of this wave is apparent as an indentation in the disk and is referred to as morphogenetic furrow (MF). Cells anterior to the MF divide asynchronously, whereas cells within the MF get synchronized in the G1 phase of the cell cycle. They go through one additional cell division immediately posterior to the MF, before they finally exit the cell cycle and differentiate. Homozygous mutant clones for Fizzy related (Fzr), the fly orthologue of Cdh1, were induced in eye imaginal disks using an FLP transgene under the control of an eye-specific promoter, ey-FLP, and identified by the absence of GFP signal. As expected, immunostaining using an antibody to Cyclin B, a known degradation target of APC/Cdh1, revealed elevated Cyclin B levels in fzr mutant clones compared with surrounding wild-type tissue (Supplemental Figure S10, A and B). However, Drosophila E2F1 (dE2F1) levels were markedly down-regulated in fzr mutant clones (Supplemental Figure S10, C and D). The most dramatic decrease in dE2F1 levels was observed in G1 phase cells within the MF, whereas cells posterior to the MF, which have exited the cell cycle, showed only a modest reduction in dE2F1 staining. However, dE2F1 protein decreased to similar overall levels in both regions of the eye disk upon inactivation of Fzr, suggesting that the observed differences in dE2F1 down-regulation are a result of the higher basal dE2F1 levels in the MF compared with the posterior part of the disk. Taken together, these data suggest that Cdh1/Fzr affects the activity of the Rb/E2F1 pathway in vivo in Drosophila.
DISCUSSION
Collectively, these results support the hypothesis that Cdh1 could potentially affect cell cycle progression through regulating the functions of both the Claspin/Chk1 and Rb/E2F1 pathways. In keeping with previous reports (Bassermann et al., 2008), we found that loss of Cdh1 also resulted in activation of the Claspin/Chk1 pathway (Figure 4). It has been proposed that activation of DNA damage checkpoints is a critical mediator for oncogene-induced premature senescence (Bartkova et al., 2006; Di Micco et al., 2006; Mallette et al., 2007). This indicates that in response to acute loss of Cdh1, a similar surveillance mechanism might be activated to trigger premature senescence that normally protects cells from overexpression of various oncogenic proteins including Ha-Ras, Raf, and E2F1 (Mallette et al., 2007).
We combined immunoprecipitation with mass spectrometry to identify Cdh1-interacting proteins. As shown in Figure 1C, we were able to pulldown known Cdh1 substrates, indicating that we developed a novel method for identifying potential Cdh1 substrates. Claspin was found to be a novel Cdh1 interactor, and further biochemical analysis confirmed that Claspin is indeed a novel Cdh1 substrate. Claspin is a well-known upstream activator of Chk1 (Kumagai et al., 2004; Wang et al., 2006). After DNA damage or replication stress, phosphorylation of Claspin by ATR promotes its interaction with Chk1, which subsequently promotes Chk1 activation by ATR (Yoo et al., 2006). Therefore, our results suggest that Cdh1 could modulate Chk1 activity by regulating Claspin stability. Further studies revealed that Cdh1 degrades Claspin mainly in the early G1 phase (Figure 3). Chk1 was reported to phosphorylate p53 at the Ser20 site (Shieh et al., 2000); we found that phosphorylation of the Ser20 site of p53 is moderately increased in Cdh1-depleted U2OS cells (Figure 4A). Loss of Cdh1 has been reported to create a DNA damage-like response (Engelbert et al., 2008; Garci-Higuera et al., 2008). Because loss of Cdh1 led to enhanced Claspin abundance, it is reasonable to assume that elevated Claspin levels will eventually activate the Chk1/p53 pathway to create the DNA damage-like response (Figure 4D), although further studies are required to pinpoint whether increased p53 activity is a cause or consequence of accumulated DNA damage-like signals. Alternatively, because loss of Cdh1 results in up-regulation of its downstream targets, many of which including Skp2, Plk1, and Aurora Kinase A have strong oncogenic activity, this could potentially result in activation of DNA damage checkpoints, which is currently considered as a protection mechanism for cells in response to abnormal oncogenic stress (Serrano et al., 1997; Mallette et al., 2007).
When Ha-Ras is overexpressed in normal human fibroblasts, instead of enhancing cell proliferation, it results in acute growth arrest, a phenotype called premature senescence (Serrano et al., 1997). Furthermore, it has been shown that inactivation of both the p53 and Rb pathways are required to bypass Ha-Ras–induced growth arrest. Therefore, it has been proposed that functionally intact p53 and Rb pathways are natural barriers to inhibit the cellular transformation process, which is typically linked with abnormal induction of one or multiple oncogenes (Lundberg et al., 2000). Interestingly, we found that depletion of Cdh1 in HeLa cells, which have defective p53 and Rb pathways, led to early S phase entry and enhanced BrdU labeling (Figure 4C). On the other hand, depletion of Cdh1 in both primary lung fibroblasts and primary foreskin fibroblasts led to premature senescence (Figure 5 and Supplemental Figure S6). In support of our findings, inactivation of Cdh1 in MEFs led to premature senescence as well (Li et al., 2008). Inactivation of other major tumor suppressors including PTEN and VHL was also reported to cause a growth arrest phenotype and the p53 or Rb pathways were found to be critical for the phenotype, respectively (Chen et al., 2005; Young et al., 2008). We found that inactivation of Cdh1 led to a marked decrease in E2F1 abundance in both lung fibroblasts and foreskin fibroblasts. This result indicated that in addition to affecting the Claspin/Chk1/p53 pathway, Cdh1 could modulate Rb/E2F1 activity as well.
We previously demonstrated that Rb could physically interact with many subunits of the APC core complex as well as Cdh1 (Binne et al., 2007). Here, we continued to show that similar to E2F1, Cdh1 only interacts with the hypophosphorylated (active) form of Rb, and phosphorylation of Rb by cyclin A/Cdk2 kinase abolished the ability of Rb to interact with both Cdh1 (Figure 7D) and E2F1 (Supplemental Figure S7C). Because both E2F1 and Cdh1 bind to the hypophosphorylated form of Rb in a similar manner, we found that they might compete with each other for Rb interaction (Figure 7, E and F). Therefore, it is plausible that in the late G1 phase, the free Cdh1 population is greatly enhanced after its dissociation from the APC core subunit, which might promote E2F1 release by competing for Rb interaction. As a result, we found that when Cdh1 is overproduced in U2OS cells, there is increased E2F1 abundance (Figure 7B). On the other hand, when Cdh1 is depleted, Rb will interact with E2F1 more strongly, which leads to repression of E2F1 activity (Figure 7A).
The finding that loss of Cdh1 in normal human fibroblasts leads to activation of both the p53 and Rb pathways and that the growth arrest phenotype can be overcome only after inactivation of both pathways (Figure 6 and Supplemental Figure S6) implies that loss of Cdh1 is not beneficial to cells with functional p53/Rb pathways. On one hand, it helps to explain why loss of Cdh1 is observed at a very low frequency in various tumors. On the other hand, it also indicates that in cells with defective Rb/p53 pathways, such as HeLa cells, depletion of Cdh1 leads to enhanced S phase entry and a proliferative advantage. Thus, the additional loss of Cdh1 in tumors with defective p53/Rb pathways is likely to promote tumorigenesis. In this case, loss of Cdh1 might be a relatively late event during the multistep tumorigenic process, which occurs after the disruption of the p53/Rb tumor suppressor pathways.
Supplementary Material
ACKNOWLEDGMENTS
We thank Alex Toker, Christoph Schorl, Shavali Shaik, and Susan Glueck for critical reading of the manuscript; James DeCaprio (Dana-Farber Cancer Institute, Boston, MA), Christoph Geisen (Dana-Farber Cancer Institute, Boston, MA), William Hahn, Bert Vogelstein (Johns Hopkins University, Baltimore, MD), Michele Pagano, and Peter Jackson for providing reagents; Michele Pagano for sharing unpublished data, and members of the Wei and Dyson labs for useful discussions. W.W. is V Scholar and Kimmel Scholar. M.K. is supported by the postdoctoral fellowship from the Deutsche Forschungsgemeinschaft (German Research Foundation). This work is supported in part by the Harvard Medical School Milton Fund, the start-up package provided by the Beth Israel Deaconess Medical Center, and National Institutes of Health Grant R01 GM89763 to W.W, and National Institutes of Health Grant R01 GM53203 to N.D.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-01-0092) on May 28, 2009.
REFERENCES
- Agami R., Bernards R. Distinct initiation and maintenance mechanisms cooperate to induce G1 cell cycle arrest in response to DNA damage. Cell. 2000;102:55–66. doi: 10.1016/s0092-8674(00)00010-6. [DOI] [PubMed] [Google Scholar]
- Bartkova J., et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- Bashir T., Dorrello N. V., Amador V., Guardavaccaro D., Pagano M. Control of the SCF(Skp2-Cks1) ubiquitin ligase by the APC/C(Cdh1) ubiquitin ligase. Nature. 2004;428:190–193. doi: 10.1038/nature02330. [DOI] [PubMed] [Google Scholar]
- Bassermann F., Frescas D., Guardavaccaro D., Busino L., Peschiaroli A., Pagano M. The Cdc14B-Cdh1-Plk1 axis controls the G2 DNA-damage-response checkpoint. Cell. 2008;134:256–267. doi: 10.1016/j.cell.2008.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett L. N., Clarke P. R. Regulation of Claspin degradation by the ubiquitin-proteosome pathway during the cell cycle and in response to ATR-dependent checkpoint activation. FEBS Lett. 2006;580:4176–4181. doi: 10.1016/j.febslet.2006.06.071. [DOI] [PubMed] [Google Scholar]
- Binne U. K., Classon M. K., Dick F. A., Wei W., Rape M., Kaelin W. G., Jr, Naar A. M., Dyson N. J. Retinoblastoma protein and anaphase-promoting complex physically interact and functionally cooperate during cell-cycle exit. Nat. Cell Biol. 2007;9:225–232. doi: 10.1038/ncb1532. [DOI] [PubMed] [Google Scholar]
- Boehm J. S., Hession M. T., Bulmer S. E., Hahn W. C. Transformation of human and murine fibroblasts without viral oncoproteins. Mol. Cell. Biol. 2005;25:6464–6474. doi: 10.1128/MCB.25.15.6464-6474.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borger D. R., DeCaprio J. A. Targeting of p300/CREB binding protein coactivators by simian virus 40 is mediated through p53. J. Virol. 2006;80:4292–4303. doi: 10.1128/JVI.80.9.4292-4303.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan C. A., Moses K. Determination of Drosophila photoreceptors: timing is everything. Cell Mol. Life Sci. 2000;57:195–214. doi: 10.1007/PL00000684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton J. L., Tsakraklides V., Solomon M. J. Assembly of an APC-Cdh1-substrate complex is stimulated by engagement of a destruction box. Mol. Cell. 2005;18:533–542. doi: 10.1016/j.molcel.2005.04.022. [DOI] [PubMed] [Google Scholar]
- Campisi J., d'Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 2007;8:729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- Cardozo T., Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat. Rev. Mol. Cell Biol. 2004;5:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- Chabes A. L., Pfleger C. M., Kirschner M. W., Thelander L. Mouse ribonucleotide reductase R2 protein: a new target for anaphase-promoting complex-Cdh1-mediated proteolysis. Proc. Natl. Acad. Sci. USA. 2003;100:3925–3929. doi: 10.1073/pnas.0330774100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q. M., Bartholomew J. C., Campisi J., Acosta M., Reagan J. D., Ames B. N. Molecular analysis of H2O2-induced senescent-like growth arrest in normal human fibroblasts: p53 and Rb control G1 arrest but not cell replication. Biochem. J. 1998;332(Pt 1):43–50. doi: 10.1042/bj3320043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–730. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Micco R., et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- Donzelli M., Squatrito M., Ganoth D., Hershko A., Pagano M., Draetta G. F. Dual mode of degradation of Cdc25 A phosphatase. EMBO J. 2002;21:4875–4884. doi: 10.1093/emboj/cdf491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelbert D., Schnerch D., Baumgarten A., Wasch R. The ubiquitin ligase APC(Cdh1) is required to maintain genome integrity in primary human cells. Oncogene. 2008;27:907–917. doi: 10.1038/sj.onc.1210703. [DOI] [PubMed] [Google Scholar]
- Garci-Higuera I., Manchado E., Dubus P., Canamero M., Mendez J., Moreno S., Malumbres M. Genomic stability and tumour suppression by the APC/C cofactor Cdh1. Nat. Cell Biol. 2008;10:802–811. doi: 10.1038/ncb1742. [DOI] [PubMed] [Google Scholar]
- Giet R., Petretti C., Prigent C. Aurora kinases, aneuploidy and cancer, a coincidence or a real link? Trends Cell Biol. 2005;15:241–250. doi: 10.1016/j.tcb.2005.03.004. [DOI] [PubMed] [Google Scholar]
- Gstaiger M., Jordan R., Lim M., Catzavelos C., Mestan J., Slingerland J., Krek W. Skp2 is oncogenic and overexpressed in human cancers. Proc. Natl. Acad. Sci. USA. 2001;98:5043–5048. doi: 10.1073/pnas.081474898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper J. W., Burton J. L., Solomon M. J. The anaphase-promoting complex: it's not just for mitosis any more. Genes Dev. 2002;16:2179–2206. doi: 10.1101/gad.1013102. [DOI] [PubMed] [Google Scholar]
- Hsieh J. K., Yap D., O'Connor D. J., Fogal V., Fallis L., Chan F., Zhong S., Lu X. Novel function of the cyclin A binding site of E2F in regulating p53-induced apoptosis in response to DNA damage. Mol. Cell. Biol. 2002;22:78–93. doi: 10.1128/MCB.22.1.78-93.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs H., Richter D., Venkatesh T., Lehner C. Completion of mitosis requires neither fzr/rap nor fzr2, a male germline-specific Drosophila Cdh1 homolog. Curr. Biol. 2002;12:1435–1441. doi: 10.1016/s0960-9822(02)01074-6. [DOI] [PubMed] [Google Scholar]
- Jin J., Shirogane T., Xu L., Nalepa G., Qin J., Elledge S. J., Harper J. W. SCFbeta-TRCP links Chk1 signaling to degradation of the Cdc25A protein phosphatase. Genes Dev. 2003;17:3062–3074. doi: 10.1101/gad.1157503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutsami M. K., et al. Centrosome abnormalities are frequently observed in non-small-cell lung cancer and are associated with aneuploidy and cyclin E overexpression. J. Pathol. 2006;209:512–521. doi: 10.1002/path.2005. [DOI] [PubMed] [Google Scholar]
- Kumagai A., Kim S. M., Dunphy W. G. Claspin and the activated form of ATR-ATRIP collaborate in the activation of Chk1. J. Biol. Chem. 2004;279:49599–49608. doi: 10.1074/jbc.M408353200. [DOI] [PubMed] [Google Scholar]
- Lakin N. D., Jackson S. P. Regulation of p53 in response to DNA damage. Oncogene. 1999;18:7644–7655. doi: 10.1038/sj.onc.1203015. [DOI] [PubMed] [Google Scholar]
- Li M., Shin Y. H., Hou L., Huang X., Wei Z., Klann E., Zhang P. The adaptor protein of the anaphase promoting complex Cdh1 is essential in maintaining replicative lifespan and in learning and memory. Nat. Cell Biol. 2008;10:1083–1089. doi: 10.1038/ncb1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg A. S., Hahn W. C., Gupta P., Weinberg R. A. Genes involved in senescence and immortalization. Curr. Opin. Cell Biol. 2000;12:705–709. doi: 10.1016/s0955-0674(00)00155-1. [DOI] [PubMed] [Google Scholar]
- Mallette F. A., Gaumont-Leclerc M. F., Ferbeyre G. The DNA damage signaling pathway is a critical mediator of oncogene-induced senescence. Genes Dev. 2007;21:43–48. doi: 10.1101/gad.1487307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti A., Wirbelauer C., Scheffner M., Krek W. Interaction between ubiquitin-protein ligase SCFSKP2 and E2F-1 underlies the regulation of E2F-1 degradation. Nat. Cell Biol. 1999;1:14–19. doi: 10.1038/8984. [DOI] [PubMed] [Google Scholar]
- Meek D. W. Mechanisms of switching on p 53, a role for covalent modification? Oncogene. 1999;18:7666–7675. doi: 10.1038/sj.onc.1202951. [DOI] [PubMed] [Google Scholar]
- Moon N. S., Di Stefano L., Dyson N. A gradient of epidermal growth factor receptor signaling determines the sensitivity of rbf1 mutant cells to E2F-dependent apoptosis. Mol. Cell. Biol. 2006;26:7601–7615. doi: 10.1128/MCB.00836-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama K. I., Hatakeyama S., Nakayama K. Regulation of the cell cycle at the G1-S transition by proteolysis of cyclin E and p27Kip1. Biochem. Biophys. Res. Commun. 2001;282:853–860. doi: 10.1006/bbrc.2001.4627. [DOI] [PubMed] [Google Scholar]
- Nemethova M., Smutny M., Wintersberger E. Transactivation of E2F-regulated genes by polyomavirus large T antigen: evidence for a two-step mechanism. Mol. Cell. Biol. 2004;24:10986–10994. doi: 10.1128/MCB.24.24.10986-10994.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschiaroli A., Dorrello N. V., Guardavaccaro D., Venere M., Halazonetis T., Sherman N. E., Pagano M. SCFbetaTrCP-mediated degradation of Claspin regulates recovery from the DNA replication checkpoint response. Mol. Cell. 2006;23:319–329. doi: 10.1016/j.molcel.2006.06.013. [DOI] [PubMed] [Google Scholar]
- Petermann E., Helleday T., Caldecott K. W. Claspin Promotes Normal Replication Fork Rates in Human Cells. Mol. Biol. Cell. 2008;19:2373–2378. doi: 10.1091/mbc.E07-10-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J. M. The anaphase-promoting complex: proteolysis in mitosis and beyond. Mol Cell. 2002;9:931–943. doi: 10.1016/s1097-2765(02)00540-3. [DOI] [PubMed] [Google Scholar]
- Petroski M. D., Deshaies R. J. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- Pfleger C. M., Lee E., Kirschner M. W. Substrate recognition by the Cdc20 and Cdh1 components of the anaphase-promoting complex. Genes Dev. 2001;15:2396–2407. doi: 10.1101/gad.918201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robles S. J., Adami G. R. Agents that cause DNA double strand breaks lead to p16INK4a enrichment and the premature senescence of normal fibroblasts. Oncogene. 1998;16:1113–1123. doi: 10.1038/sj.onc.1201862. [DOI] [PubMed] [Google Scholar]
- Romero F., Multon M. C., Ramos-Morales F., Dominguez A., Bernal J. A., Pintor-Toro J. A., Tortolero M. Human securin, hPTTG, is associated with Ku heterodimer, the regulatory subunit of the DNA-dependent protein kinase. Nucleic Acids Res. 2001;29:1300–1307. doi: 10.1093/nar/29.6.1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sar F., Lindsey-Boltz L. A., Subramanian D., Croteau D. L., Hutsell S. Q., Griffith J. D., Sancar A. Human claspin is a ring-shaped DNA-binding protein with high affinity to branched DNA structures. J. Biol. Chem. 2004;279:39289–39295. doi: 10.1074/jbc.M405793200. [DOI] [PubMed] [Google Scholar]
- Schwab M., Neutzner M., Mocker D., Seufert W. Yeast Hct1 recognizes the mitotic cyclin Clb2 and other substrates of the ubiquitin ligase APC. EMBO J. 2001;20:5165–5175. doi: 10.1093/emboj/20.18.5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M., Lin A. W., McCurrach M. E., Beach D., Lowe S. W. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- Shieh S. Y., Ahn J., Tamai K., Taya Y., Prives C. The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes Dev. 2000;14:289–300. [PMC free article] [PubMed] [Google Scholar]
- Sorensen C. S., Lukas C., Kramer E. R., Peters J. M., Bartek J., Lukas J. Nonperiodic activity of the human anaphase-promoting complex-Cdh1 ubiquitin ligase results in continuous DNA synthesis uncoupled from mitosis. Mol. Cell. Biol. 2000;20:7613–7623. doi: 10.1128/mcb.20.20.7613-7623.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stott F. J., et al. The alternative product from the human CDKN2A locus, p14(ARF), participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 1998;17:5001–5014. doi: 10.1093/emboj/17.17.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudo T., Ota Y., Kotani S., Nakao M., Takami Y., Takeda S., Saya H. Activation of Cdh1-dependent APC is required for G1 cell cycle arrest and DNA damage-induced G2 checkpoint in vertebrate cells. EMBO J. 2001;20:6499–6508. doi: 10.1093/emboj/20.22.6499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai N., Hamanaka R., Yoshimatsu J., Miyakawa I. Polo-like kinases (Plks) and cancer. Oncogene. 2005;24:287–291. doi: 10.1038/sj.onc.1208272. [DOI] [PubMed] [Google Scholar]
- Waldman T., Kinzler K. W., Vogelstein B. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res. 1995;55:5187–5190. [PubMed] [Google Scholar]
- Wang X., Zou L., Lu T., Bao S., Hurov K. E., Hittelman W. N., Elledge S. J., Li L. Rad17 phosphorylation is required for claspin recruitment and Chk1 activation in response to replication stress. Mol. Cell. 2006;23:331–341. doi: 10.1016/j.molcel.2006.06.022. [DOI] [PubMed] [Google Scholar]
- Wei W., Ayad N. G., Wan Y., Zhang G. J., Kirschner M. W., Kaelin W. G., Jr Degradation of the SCF component Skp2 in cell-cycle phase G1 by the anaphase-promoting complex. Nature. 2004;428:194–198. doi: 10.1038/nature02381. [DOI] [PubMed] [Google Scholar]
- Wei W., Hemmer R. M., Sedivy J. M. Role of p14(ARF) in replicative and induced senescence of human fibroblasts. Mol. Cell. Biol. 2001;21:6748–6757. doi: 10.1128/MCB.21.20.6748-6757.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W., Jin J., Schlisio S., Harper J. W., Kaelin W. G., Jr The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer Cell. 2005;8:25–33. doi: 10.1016/j.ccr.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Wei W., Jobling W. A., Chen W., Hahn W. C., Sedivy J. M. Abolition of cyclin-dependent kinase inhibitor p16Ink4a and p21Cip1/Waf1 functions permits Ras-induced anchorage-independent growth in telomerase-immortalized human fibroblasts. Mol. Cell. Biol. 2003;23:2859–2870. doi: 10.1128/MCB.23.8.2859-2870.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert T. DNA damage and checkpoint pathways: molecular anatomy and interactions with repair. Cell. 1998;94:555–558. doi: 10.1016/s0092-8674(00)81597-4. [DOI] [PubMed] [Google Scholar]
- Xu T., Rubin G. M. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117:1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- Yoo H. Y., Jeong S. Y., Dunphy W. G. Site-specific phosphorylation of a checkpoint mediator protein controls its responses to different DNA structures. Genes Dev. 2006;20:772–783. doi: 10.1101/gad.1398806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young A. P., Schlisio S., Minamishima Y. A., Zhang Q., Li L., Grisanzio C., Signoretti S., Kaelin W. G., Jr VHL loss actuates a HIF-independent senescence programme mediated by Rb and p400. Nat. Cell Biol. 2008;10:361–369. doi: 10.1038/ncb1699. [DOI] [PubMed] [Google Scholar]
- Zou H., McGarry T. J., Bernal T., Kirschner M. W. Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science. 1999;285:418–422. doi: 10.1126/science.285.5426.418. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.