Abstract
Myo4p, a single-headed and nonprocessive class V myosin in budding yeast, transports >20 different mRNAs asymmetrically to the bud. Here, we determine the features of the Myo4p motor that are necessary for correct localization of ASH1 mRNA to the daughter cell, a process that also requires the adapter protein She3p and the dimeric mRNA-binding protein She2p. The rod region of Myo4p, but not the globular tail, is essential for correct localization of ASH1 mRNA, confirming that the rod contains the primary binding site for She3p. The requirement for both the rod region and She3p can be bypassed by directly coupling the mRNA-binding protein She2p to Myo4p. ASH1 mRNA was also correctly localized when one motor was bound per dimeric She2p, or when two motors were joined together by a leucine zipper. Because multiple mRNAs are cotransported to the bud, it is likely that this process involves multiple motor transport regardless of the number of motors per zip code. Our results show that the most important feature for correct localization is the retention of coupling between all the members of the complex (Myo4p–She3p–She2p–ASH1 mRNA), which is aided by She3p being a tightly bound subunit of Myo4p.
INTRODUCTION
Class V myosins are involved in cellular processes including organelle and vacuole transport, mRNA localization, and spindle positioning (reviewed in Reck-Peterson et al., 2000; Trybus, 2008). Of the five myosins in the budding yeast Saccharomyces cerevisiae, two are members of class V. Myo2p transports membranous cargo, including vacuoles, mitochondria, and secretory vesicles (reviewed in Weisman, 2006). The major role of the other class V myosin, Myo4p, is to asymmetrically transport >20 different mRNAs (Shepard et al., 2003; Jambhekar et al., 2005), a widely used and efficient strategy to spatially restrict a protein to a region of interest within the cell (reviewed in Tekotte and Davis, 2002; Condeelis and Singer, 2005). Another cargo of Myo4p is cortical endoplasmic reticulum (Estrada et al., 2003; Schmid et al., 2006). Unlike Myo2p, Myo4p is not essential for viability.
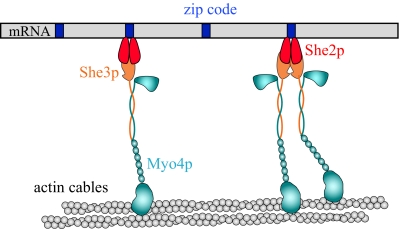
The most well-studied transported mRNA in yeast is ASH1, which is moved by Myo4p to the bud tip to repress mating type switching in the daughter cell (Haarer et al., 1994; Sil and Herskowitz, 1996; Long et al., 1997; Takizawa et al., 1997). The connection of myosin to mRNA occurs via the adapter proteins She2p and She3p (reviewed in Beach and Bloom, 2001; Cosma, 2004; Gonsalvez et al., 2005; Bullock, 2007; Muller et al., 2007). She3p binds to Myo4p, whereas She2p binds ASH1 mRNA at specific localization elements referred to as zip codes (Figure 1) (Bohl et al., 2000; Long et al., 2000). The transport complex is completed when Myo4p/She3p binds to She2p/mRNA. In vertebrates, myosin Va has also been implicated in mRNA transport in the dendritic spines of neuronal cells (Yoshimura et al., 2006).
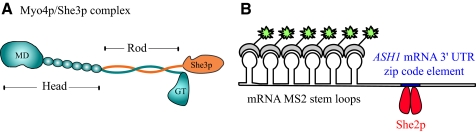
Figure 1.
(A) Schematic of the proposed complex between Myo4p (cyan) and She3p (orange). The Myo4p head consists of the motor domain (MD) and six IQ motifs to which calmodulin is bound. The rod region is proposed to be a heterocoiled-coil between Myo4p and the adapter protein She3p (Hodges et al., 2008). The globular tail of Myo4p follows the rod. (B) Diagram of the reporter system used to observe fluorescently labeled ASH1 mRNA in living yeast (Bertrand et al., 1998). The 3′ UTR region of ASH1 is joined to six MS2 stem loop binding motifs for the MS2 coat protein. Each stem loop binds one MS2-GFP molecule. Unbound MS2-GFP is confined to the nucleus. The region of ASH1 mRNA used in this reporter contains one zip code element, which binds one homodimeric molecule of She2p (red), based on the work of Niessing et al. (2004).
A feature long thought to be a hallmark of all class V myosins is their ability to move processively, which means that the motor takes multiple steps on actin before dissociating. Recent evidence has shown, however, that not all class V myosins are processive. The kinetic cycle of Drosophila myosin V (Toth et al., 2005) and human myosin Vc (Takagi et al., 2008; Watanabe et al., 2008) differs from that established for processive myosin Va, such that ADP release is no longer the rate-limiting step of the ATPase cycle. These myosins thus have a low-duty cycle, meaning that they are attached to actin in a strong binding state for only a small fraction of their overall ATPase cycle time, which precludes their ability to move on actin processively as a single molecule. These motors can still be cargo transporters, because processivity is only necessary to achieve continuous movement if a single motor is attached to the cargo, a situation that may rarely occur in vivo (Gross et al., 2002).
Both class V myosins in budding yeast are nonprocessive, but each for a different reason (Reck-Peterson et al., 2001; Dunn et al., 2007; Hodges et al., 2008). It was inferred from motility assays that the Myo2p motor domain has a low-duty cycle and thus cannot support continuous cargo movement as a single two-headed molecule (Reck-Peterson et al., 2001; Dunn et al., 2007). Although Myo4p has a high-duty cycle motor domain (Krementsova et al., 2006), it is not processive because the Myo4p rod shows little tendency to dimerize with itself, and a single-headed motor cannot move processively (Dunn et al., 2007; Heuck et al., 2007; Hodges et al., 2008). We proposed that the adapter protein She3p binds tightly to the rod region of Myo4p and forms a heterocoiled-coil, making it an intrinsic part of the Myo4p motor complex (Hodges et al., 2008) (Figure 1A). This interpretation accounts both for the high affinity of She3p for Myo4p and provides a mechanism by which She3p inhibits dimerization of Myo4p with itself. All cargoes of Myo4p require She3p; thus, it makes biological sense to have the adapter protein become a subunit of the single-headed motor complex. Myo2p, in contrast, uses many different adapter proteins to select a distinct cargo as required. The binding site for the adapter proteins for vacuole and secretory vesicle transport map to distinct binding sites in the globular tail of Myo2p (reviewed in Weisman, 2006). Our observation that She3p binds tightly to the rod region of Myo4p (Hodges et al., 2008) differs from the more typical observation that the globular tail of class V myosins provides the primary binding site for cargo adapter proteins.
Here, we expressed chimeric and deletion constructs in budding yeast to show that the rod region of Myo4p is necessary to bind She3p and localize ASH1 mRNA to the bud tip. We also designed a minimal motor construct that bypasses the requirement for the rod region and She3p, and correctly localizes ASH1 mRNA. The extent to which correct mRNA localization is affected by motor number, speed, and duty cycle was also investigated. We conclude that the most important feature for correct localization of ASH1 mRNA is the retention of coupling between all the members of the complex.
MATERIALS AND METHODS
Chimeras of Myo4p and Myo2p
Three domains from either Myo2p or Myo4p were interchanged to create full-length chimeras. The three domains were as follows: head (motor domain and 6 IQ motifs), rod, and globular tail. The chimeras are named by the source from which each of these three domains is obtained. For example, m4m2m4 contains the head and globular tail from Myo4p and the rod from Myo2p. The motor domain ended after the sixth IQ motif at Leu926 for Myo2p and Leu922 for Myo4p. The rod domain consisted of residues Lys927-Thr1090 for Myo2p and Gln923-Thr1067 for Myo4p. The globular tail domain consisted of residues Thr1091-His1574 for Myo2p and Thr1068-Lys1471 for Myo4p. In a truncated Myo4p construct that lacked the rod region (m4Δrod), residues Gln923-Thr1067 were deleted. A stop codon was inserted after Thr1067 to create the construct that lacked the globular tail (m4ΔGT). A C-terminal FLAG-tagged (DYKDDDDK) version of the wild-type Myo4p, and a chimera containing the rod from Myo2p (m4m2m4) were also cloned for immunoblotting purposes to assess relative expression levels.
Chimeras of Myo4p and She2p
The Myo4p motor domain and complete neck containing six IQ motifs were directly fused to She2p (m4 6IQ-She2p). The last amino acid of Myo4p was Arg929, followed by She2p which started at Ser2. A similar construct with a shorter neck that contained one IQ motif was also engineered (m4 1IQ-She2p). The last amino acid of Myo4p was Leu808, followed by She2p starting at Lys3.
Chimeras Containing Myo4p and Two Copies of She2p
To obtain one single-headed motor per She2p dimer, two She2p molecules separated by a linker were cloned after the motor. The crystal structure of She2p shows that the N and C termini of She2p are at opposite ends of the molecule (Niessing et al., 2004). To bring the C terminus of one molecule into proximity of the N terminus of a second molecule, the fifth helix of the first She2p was deleted. This was possible because the crystal structure of She2p indicates that only the first four of its five helices are involved in mRNA binding and homodimerization (Niessing et al., 2004). This deletion brings the C terminus of the first She2p closer to the N terminus of the second She2p molecule.
The resulting chimera consists of Myo4p joined to the first four helices of She2p (ending at Glu205), followed by a poly-Gly10 linker, followed by a full copy of She2p. The poly-Gly10 linker between the tandem She2p genes should provide the necessary flexibility to permit homodimerization of the two molecules of She2p. This chimera was functional based on its ability to rescue myo4Δ and she2Δ mutants (see Results). Two chimeras containing different length myosin constructs were created. The She2p dimer described above was joined to the C terminus of a full-length Myo4p construct by a poly-Gly10 linker (m4-She2p-She2p). Alternatively, it was joined directly after Arg929 of the Myo4p head (m4 head-She2p-She2p).
Constructs with the GCN4 Leucine Zipper
Two Myo4p constructs were engineered to contain a 32-amino acid leucine zipper after the predicted coiled-coil region in the rod. For the full-length zippered construct (m4-Zip), the leucine zipper was inserted between Leu1024 and Ile1025. For a shorter construct that lacks the GT, called m4ΔGT-Zip, the leucine zipper was added after Leu1024, followed by a stop codon.
Plasmid Construction
Myosin constructs were cloned into the vector pRS314 with TRP1 selection behind the native Myo4p promoter (800 nucleotides upstream of the MYO4 gene). Plasmid templates containing MYO2 and MYO4 were gifts from Brian Haarer (SUNY Medical Center, Syracuse, NY) and Karen Beningo (Wayne State University, Detroit, MI). The SHE2 and SHE3 genes were amplified by polymerase chain reaction (PCR) from S. cerevisiae genomic DNA and verified by DNA sequencing and coding sequences found at the Saccharomyces genome database. The full-length genes were cloned into a pRS314 derivative with ADE2 selection behind the inducible GAL1 promoter.
ASH1 mRNA Reporter System
A two-plasmid system was used to follow ASH1 mRNA localization. The two plasmids pG14-MS2-GFP with LEU2 selection, and YEplac195 lacZ-MS2-ASH1 with URA3 selection, were generous gifts from Roy Long (Medical College of Wisconsin, Milwaukee, WI) (Bertrand et al., 1998). pG14-MS2-GFP expresses an MS2 coat protein-green fluorescent protein (GFP) under a constitutive promoter. The MS2-GFP chimera contains a nuclear-location signal to confine unbound MS2-GFP to the nucleus. YEplac195 lacZ-MS2-ASH1 expresses the ASH1 mRNA along with six stem loop binding motifs for the MS2 coat protein under control of a galactose-inducible promoter. The URA3 marker in YEplac195 lacZ-MS2-ASH1 was replaced with HIS3.
Cortical Endoplasmic Reticulum (ER)
Cortical ER movement was tracked in cells by using an Hmg1p-GFP plasmid (NH2-terminal transmembrane domain of HMG-CoA reductase isozyme 1 fused to GFP) (Du et al., 2001; Estrada et al., 2003). The myo4Δ yeast strain was cotransformed with the Hmg1p-GFP plasmid and a plasmid encoding either for wild-type (WT) Myo4p, m4ΔGT, or a plasmid with no insert as a negative control. The live cells were mounted on a slide in the appropriate liquid medium and viewed with an epifluorescence microscope (Nikon, Tokyo, Japan) by using a 100× objective. Actively budding yeast were scored according to whether fluorescent cortical ER was detected in the bud tip. At least 500 cells were observed per sample. The plasmid containing Hmg1p-GFP was a generous gift from Susan Ferro-Novick (University of California, San Diego, La Jolla, CA).
Yeast Strains
Three mutant strains (W303a background) were used: K5209, MATa ade2 his3 leu2 trp1 ura3 can1-100 myo4::URA3; K5477, MATα ade2 his3 leu2 trp1 ura3 can1-100 she2::URA); and K5234, MATa ade2 his3 leu2 trp ura3 can1-100 she3::URA3).
Visualization of Transformed Yeast
Yeast strains (myo4Δ, she2Δ, or she3Δ) were each transformed with three plasmids. Two were required for the ASH1 reporter system. The third plasmid contained either Myo4p, She2p, or She3p as positive controls for the corresponding deletion strain, no insert for the negative control, or one of the various Myo4 constructs as indicated. Transformants were grown on the appropriate complete synthetic medium lacking the nutrients needed to maintain the plasmids. To induce expression of the reporter mRNA, a single colony was grown overnight in the same synthetic medium containing 2% galactose as the sole carbon source. Four hours before visualization, the yeast cells were streaked in a fresh patch to ensure exponential growth when viewed and scored. The live cells were mounted on a slide in the appropriate liquid medium containing galactose and viewed with a TE2000-E2 inverted microscope (Nikon) using a Plan Apo60× oil objective lens, and differential interference contrast (DIC) and fluorescein isothiocyanate filters. An EXFO X-Cite 120 fluorescence illuminator and a CoolSNAP HQ2 14-bit camera (Photometrics, Tucson, AZ) were used. Images were processed using NIS Elements software (Nikon) and ImageJ (National Institutes of Health, Bethesda, MD). Actively budding yeast were scored according to the location and number of fluorescent particles seen. The data used to determine whether the particle was correctly localized from the mother to the bud included only those cells that contained a single fluorescent particle, unless otherwise noted.
Observation of Particle Movement
Cells were mounted on agarose containing complete synthetic medium with 2% glucose and observed at room temperature by epifluorescence microscopy with a 60× objective plus 1.5× auxiliary magnification. Exposures (200 ms) were collected every 2 s. Images were processed using NIS Elements software and a tracking plug-in for ImageJ. Velocities were measured as displacement of the particle image over time. Velocities, although measured in a consistent manner, are probably underestimated because the particles took brief pauses and because we did not take into account movement perpendicular to the focal plane. At least 25 particles were analyzed per sample.
Immunoblots
C-Terminal FLAG-tagged versions of WT Myo4p and the chimeric m4m2m4 construct, under the control of the native Myo4p promoter, were transformed into myo4Δ cells. A 500-ml culture was grown in YPDA to a final OD595 of 0.23 (9 × 106 cells/ml). The cells were pelleted, washed once with FLAG lysis buffer (10 mM imidazole, pH∼7.0, 0.3 M NaCl, 1 mM EGTA, 5 mM MgCl2, 7% sucrose, 1 mM dithiothreitol, and 10 μl/ml protease inhibitor cocktail [catalog no. P8215; Sigma-Aldrich, St. Louis, MO]), and resuspended in 1 ml of FLAG lysis buffer. Cells were lysed using an MP FastPrep bead beater for 4 min total, cooling on ice between every minute of beating. The supernatant was diluted to 5 mg/ml, 0.5% octyl-glucoside was added, and the lysate incubated for 30 min at 4°C. After addition of 2 mM MgATP, the lysate was clarified at 12,000 rpm for 10 min at 4°C. Then, 500 μl of the supernatant was added to 30 μl of FLAG resin in an Eppendorf tube, and the slurry was incubated at 4°C for 1.5 h. The resin was pelleted at 6000 rpm for 1 min, the lysate was removed, and the resin washed six times with FLAG buffer. Then, 30 μl of 2× sample buffer was added, the sample was boiled, and 12 μl was run on a 3–8% gradient gel (Invitrogen, Carlsbad, CA). The protein was transferred to nitrocellulose, blocked with 3% powered milk, and then incubated overnight with 4.4 μg/ml mouse monoclonal anti-FLAG M2 antibody (catalog no. A8592; Sigma-Aldrich). After washing, the blot was incubated with a goat anti-mouse immunoglobulin G-HRP conjugate (Bio-Rad Laboratories, Hercules, CA), washed, and then incubated with a chemiluminescent substrate (SuperSignal West Pico; Pierce Chemical, Rockford, IL) for detection.
RESULTS
Reporter System for ASH1 mRNA Localization in Living Yeast
Mutations in five genes, SHE1–SHE5, cause defects in localization of the ASH1 mRNA transcript in budding yeast. Here, we focus on three of the proteins encoded by these genes: the class V myosin Myo4p (She1p) and two adapter proteins (She2p and She3p). The Myo4p–She3p complex is linked to ASH1 mRNA via the RNA-binding protein She2p (Figure 1). The goal is to further understand the features of the Myo4p motor and the adapter protein She3p that are essential for mRNA transport. If the connection between Myo4p, She3p, and She2p is abolished, ASH1 mRNA will not be localized to the bud tip.
An in vivo assay was used to track ASH1 mRNA localization to the bud tip of living yeast. It involves a two-plasmid reporter system designed and validated in previous studies (Bertrand et al., 1998). One plasmid codes for the 3′ untranslated region (UTR) of ASH1 mRNA fused to six MS2 binding sites, whereas a second plasmid codes for the MS2-binding protein joined to GFP (Figure 1B). The 3′ untranslated region of ASH1 contains one of the four zip codes found in ASH1, which is sufficient for its translocation to the bud tip by the She protein complex (Chartrand et al., 1999).
The Rod, but Not the Globular Tail, Is Sufficient to Bind She3p to Myo4p
Our in vitro studies with baculovirus-expressed constructs suggested that She3p binds tightly to the rod region of Myo4p (Hodges et al., 2008). Here, we show that the rod region of Myo4p is sufficient to ensure She3p binding in vivo, by using a series of deletion mutants and chimeric constructs. Chimeras were made by swapping domains between Myo4p and Myo2p, the two class V myosins in budding yeast. Three domains of the molecule were interchanged: the head (i.e., motor domain with 6IQ motifs), the rod, and the globular tail. The chimeras are named based on the origin of the three domains, e.g., a construct called m2m4m4 contains the head from Myo2p and the rod and globular tail from Myo4p (Figure 2).
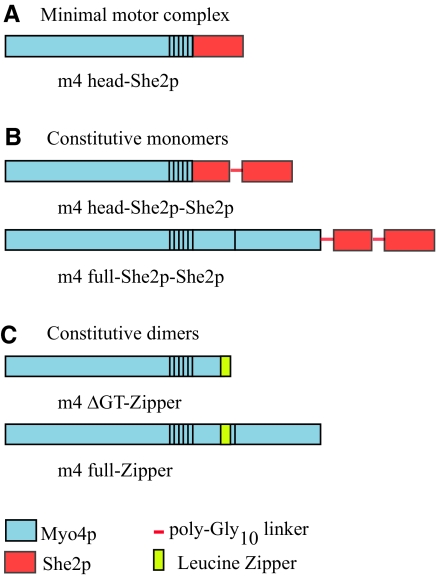
Figure 2.
Schematic showing chimeric and truncated constructs used to transform a myo4Δ strain. The Myo4p molecule consists of three domains: head (motor domain with 6 IQ motifs), rod, and GT. Domains derived from Myo4p are indicated in cyan. Domains derived from Myo2p, the other class V myosin from budding yeast, are shown in yellow. Chimeric constructs are named by the origin of each of the three domains. For example, m2m4m4 contains the head of Myo2p and the rod and globular tail from Myo4p. Two deletion constructs, one construct lacking the globular tail (m4ΔGT) and one construct lacking the rod (m4Δrod), were also constructed.
Yeast myo4Δ cells were cotransformed with a plasmid containing the Myo4p construct of interest under the control of the native Myo4p promoter and the two reporter plasmids described above. Actively budding yeast cells that contained one fluorescent mRNA particle were scored according to whether the particle was located in the bud tip (correct localization) or the mother (incorrect localization) (Figure 3). Transformation of the myo4Δ cells with wild-type Myo4p resulted in 98% of the particles being correctly delivered to the bud tip (Figure 4A). A negative control consisting of plasmid with no insert showed only 5% localization at the bud tip. Thus, the assay system is robust, as expected from a system that either does, or does not, retain the correct linkage between motor and cargo.
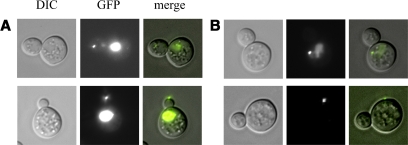
Figure 3.
Representative light microscopy and fluorescent images of correctly and incorrectly localized ASH1 mRNA. (A) Images of a single particle (small bright spot) correctly localized to the bud tip by Myo4p. The MS2-GFP chimera contains a nuclear location signal to confine unbound MS2-GFP to the nucleus (large bright spot). (B) A nontranslocated particle that remains in the mother cell. Left, DIC. Middle, epifluorescence to visualize GFP. Right, merged images.
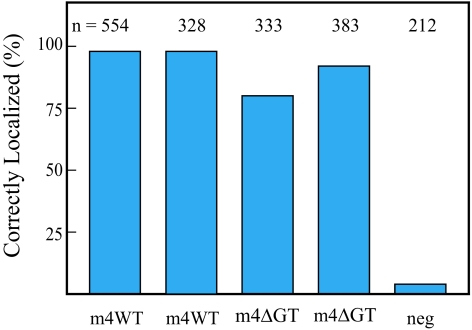
Figure 4.
Percentage of correctly localized ASH1 mRNA particles for the chimeric and truncation constructs illustrated in Figure 2. (A) The indicated constructs were transformed into a myo4Δ strain. Constructs lacking the Myo4p rod do not localize ASH1 mRNA. The negative control (neg) is a plasmid lacking an insert. The number of cells counted is indicated above each bar. Data were derived from at least two independent transformations per construct. (B) Relative expression levels of WT Myo4p, and the m4m2m4 chimera (which did not correctly localize ASH1 mRNA), are qualitatively similar. Both constructs were FLAG tagged.
Strikingly, only chimeric constructs that contained the rod region from Myo4p retained the ability to correctly deliver the particle to the bud tip (Figure 4A). These included two full-length chimeras, one chimera that contained both the rod and globular tail from Myo4p (m2m4m4), and another chimera that had only the rod from Myo4p (m2m4m2). These two chimeras were almost equally effective (95 vs. 90%) with regard to their ability to transport ASH1 mRNA to the bud.
Complementary full-length chimeras that contained the rod from Myo2p (m4m2m4), or the head and rod from Myo2p (m2m2m4), were unable to transport the particle from the mother cell to the developing bud tip (Figure 4A). Expression levels of FLAG-tagged versions of the WT Myo4p and the chimera m4m2m4 were qualitatively similar, showing that the lack of correct localization was not due to a lack of expression of the chimeric construct (Figure 4B). These constructs show that the Myo4p globular tail by itself is not sufficient to bind She3p in vivo, even when present as a pair at the C terminus of the dimeric α-helical coiled-coil rod of Myo2p (Dunn et al., 2007).
Results with two truncated mutants were also consistent with the observation that the rod region is the primary binding site for She3p (Figure 4A). A construct containing the head and globular tail, but lacking the rod region (m4Δrod), did not translocate ASH1 mRNA. In contrast, a Myo4p construct containing the head and rod, but lacking the globular tail (m4ΔGT), delivered the particle to the bud tip. The efficiency of transport by m4ΔGT (80%) was lower than the full-length chimera containing the Myo4p rod (m2m4m2, 90%) or wild-type Myo4p (98%). This observation suggests that the presence of a globular tail, even a heterologous one from Myo2p, may contribute to stabilizing the connection between Myo4p/She3p and She2p/ASH1 mRNA.
Myo4p also transports cortical ER in a She3p-dependent manner (Estrada et al., 2003). To test whether a construct lacking the globular tail can deliver cortical ER into the emerging bud, localization of a GFP-tagged version of the endoplasmic reticulum marker HMG-CoA reductase was followed in myo4Δ cells. The WT construct gave 97% (n = 777) correct localization versus 71% (n = 924) for m4ΔGT and 50% (n = 905) for the negative control, a plasmid lacking an insert. Thus, the globular tail is also not required for cortical ER transport, but as with the ASH1 mRNA cargo, it may help stabilize the transport complex.
A Myo4p Head Fused to She2p Localizes ASH1 mRNA
If the primary role of She3p is to link the motor complex to the She2p–mRNA complex, then the ability to localize mRNA to the bud in she3Δ cells should be retained by a construct in which the Myo4p head is directly attached to She2p (m4 head-She2p) (Figure 5A). Successful ASH1 mRNA transport was evident in both myo4Δ and she3Δ cells containing the m4 head-She2p construct (Table 1). Thus, the chimera overcomes the requirement for either endogenous Myo4p or She3p in vivo.
Figure 5.
Schematic of Myo4p–She2p chimeras and Myo4p dimers. (A) The Myo4p head is directly joined to the mRNA binding protein She2p, thus bypassing the requirement for the rod and She3p. (B) The Myo4p head, or full-length Myo4p, is joined to two copies of She2p. The tandem copies of She2p are separated by a poly-Gly10 linker to allow intramolecular dimerization. These constructs ensure one motor head per She2p dimer. (C) Constitutive dimers were engineered by adding a leucine zipper immediately after the predicted α-helical coiled coil region in either a construct lacking the GT, or full-length Myo4p.
Table 1.
Percentage of cells with a correctly localized particle of ASH1 mRNA
| Yeast strain | % correct localization of ASH1 mRNA by the indicated construct |
||||
|---|---|---|---|---|---|
| Negative controla | Positive controlb | m4 head-She2p | m4 head-She2p-She2p | m4 full-She2p-She2p | |
| myo4Δ | 5 (n = 212) | 98 (n = 554) | 93 (n = 325) | 95 (n = 464) | 92 (n = 273) |
| she2Δ | 6 (n = 212) | 97 (n = 387) | 58c (n = 100) | 87 (n = 344) | 85 (n = 204) |
| she3Δ | 0 (n = 92) | 94 (n = 267) | 93 (n = 274) | 97 (n = 336) | 94 (n = 274) |
The numbers in parentheses indicates the number of cells counted. Data were derived from at least two independent transformations per construct.
a Negative control is transformation with a plasmid containing no insert.
b Positive control is a plasmid encoding for the deleted protein, i.e., Myo4p for myo4Δ, She2p for she2Δ,and She3p for she3Δ.
c Less than 10% of the she2Δ cells contained a fluorescent particle compared with the myo4Δ cells in which 85% of the cells displayed one or more particles.
In contrast, in a she2Δ background, the chimera only partially supported ASH1 mRNA transport, with 58% of the cells showing a single particle that was correctly localized (Table 1). Even more striking, only 10% of the she2Δ cells, compared with 85% of the myo4Δ cells, contained a single fluorescent particle. She2p dimerization is required for it to bind to a zip code in the mRNA (Niessing et al., 2004). Thus, a likely explanation for our observation is that when She2p is coupled directly to Myo4p, it dimerizes very poorly with itself. The she2Δ strain contains no endogenous She2p to complete the dimer. This explanation also implies that the m4 head-She2p chimera successfully dimerizes with an endogenous She2p monomer in the myo4Δ and she3Δ cells.
mRNA Transport by One Motor per She2p Dimer
That She2p forms a homodimer raises the question of whether one or two Myo4p–She3p complexes are bound per She2p dimer during mRNA translocation. Filter binding experiments suggest that approximately one She2p dimer binds per zip code (Niessing et al., 2004). We designed a construct to address the question of whether mRNA transport can be accomplished by one single-headed motor complex per She2p dimer. Our strategy was to clone two copies of She2p after the Myo4p heavy chain, so that the tandem She2p molecules would dimerize (see Materials and Methods).
Two different-length Myo4p constructs were joined to the tandem copies of She2p: a Myo4p head (m4 head-She2p-She2p) and a full length Myo4p (m4 full-She2p-She2p) (Figure 5B). Both constructs supported correct ASH1 mRNA localization in all strains tested, including the she2Δ strain, suggesting that a single motor head per She2p dimer is sufficient for transport (Table 1). How do we know that the tandemly cloned She2p molecules formed a functional dimer in vivo? The evidence supporting this comes from a comparison with the construct that contained only one She2p, described in the previous section (Table 1). Although the Myo4p head joined to only one molecule of She2p is defective in mRNA transport in a she2Δ background, the tandem She2p construct is fully functional.
It should be noted that although some data support the idea that one She2p dimer binds per zip code (Niessing et al., 2004), this conclusion is tempered by the observation by the same authors that She2p has a tendency to form higher oligomers, which was minimized by mutation of 4 Cys residues to Ser. We thus compared the ability of the mutant She2p (C14S/C68S/C106S/C180S) and WT She2p to support correct localization of ASH1 mRNA in a she2Δ background. The WT She2p showed 97% (n = 387) correct localization, indistinguishable from the 96% (n = 215) correct localization supported by the mutant She2p. A negative control, showed 6% (n = 212) correct localization. Thus, either the oligomers/aggregates observed in vitro do not occur in vivo, or they are present but have no effect on ASH1 mRNA localization.
Effect of Joining Two Single-headed Motor Complexes Together
A complementary experiment to that described above is to test whether two Myo4p–She3p complexes, tethered together by a leucine zipper, can correctly localize mRNA (Figure 5C). The leucine zipper is a short ∼30-amino acid segment that forms a stable α-helical coiled coil and has been widely used as a dimerization domain. A leucine zipper was added to the full length Myo4p construct after the predicted coiled-coil region in the rod. The inclusion of the zipper had no detrimental effect on ASH1 mRNA localization compared with wild-type Myo4p (Figure 6). The truncated construct lacking the globular tail (m4ΔGT) also showed no negative effect from being tethered together by a leucine zipper (Figure 6), and in fact increased the number of cells with correctly localized mRNA from 80 to 92% (Figure 6). The lack of any negative effect from putting two motors in proximity leaves open the possibility that multiple motors may be bound per She2p dimer in vivo.
Figure 6.
Percentage of correctly localized ASH1 mRNA particles for constructs dimerized with a leucine zipper (see Figure 5C). Two zippered dimeric constructs, one full-length and the other lacking the globular tail, were transformed into a myo4Δ strain and compared with the corresponding construct lacking the leucine zipper. The negative control (neg) is a plasmid lacking an insert. The number of cells counted is indicated above each bar. Data are derived from at least two independent transformations per construct.
Does the Speed or Duty Cycle of the Motor Matter for mRNA Localization?
To establish whether a certain motor speed is optimal for translocation of the particle to the bud tip, we compared two constructs with different numbers of IQ motifs. Based on the lever arm model for movement, truncation of the neck from six IQ motifs to one IQ motif should decrease the motor speed (Schott et al., 2002; Moore et al., 2004). Although the m4 head-She2p chimera containing 6IQ motifs correctly localized particles 93% of the time (n = 325), an m4(1-IQ)-She2p chimera localized them correctly 82% of the time (n = 556). The average speed of particle movement by the m4 head-She2p chimera which contained six IQ motifs was 0.138 ± 0.045 μm/s (n = 29) versus 0.099 ± 0.035 μm/s (n = 25) for the m4(1-IQ)-She2p chimera, values that were statistically different (p < 0.05). This result suggests that a slower moving motor is less effective at correctly localizing ASH1 mRNA. A full-length WT construct, which has 6IQ motifs, showed a rate of particle movement (0.160 ± 0.055 μm/s) that was statistically the same (p < 0.05) as the m4 head-She2p chimera, which also has 6 IQ motifs.
A chimeric construct made up of the faster Myo2p motor, and the rod and globular tail of Myo4p, was also tested (m2m4m4; see Figure 4). Myo2p has a lower duty cycle than Myo4p and supports rates of actin filament gliding in a motility assay that are two- to fourfold faster motor than Myo4p (Reck-Peterson et al., 2001; Krementsova et al., 2006; Dunn et al., 2007). The m2m4m4 chimera correctly localized the particle 95% of the time, indistinguishable from the wild-type Myo4p construct (98%). Thus, movement by a motor with a low duty cycle and faster speed had no detrimental effect on correctly localizing mRNA to the bud tip.
DISCUSSION
Here, we establish several features of the class V Myo4p motor that are essential to correctly localize ASH1 mRNA to the bud tip. First, the rod region of Myo4p is critical. Based on the present in vivo study with native Myo4p, along with our previous studies with expressed chimeric constructs containing the rod and globular tail from Myo4p (Hodges et al., 2008), we conclude that the rod region is the primary binding site for the adapter protein She3p. Constructs that lack the rod region from Myo4p cannot transport ASH1 mRNA, even if they contain the globular tail from Myo4p. These studies do not rule out the possibility that the globular tail contains a weak secondary binding site for She3p in wild-type Myo4p.
Most adapter proteins bind to the globular tail of class V myosins and not the rod. Myo2p, the other class V myosin in budding yeast, binds adapter proteins for secretory vesicles and vacuoles at two distinct, nonoverlapping regions of the globular tail (reviewed in Weisman, 2006). With mammalian myosin Va, the adapter protein melanophilin has binding sites that require both the globular tail and an alternatively spliced exon in the rod region (Wu et al., 2002). Inclusion of an alternatively spliced exon in the binding site for an adapter protein increases the diversity of potential cargoes that a particular myosin isoform can bind.
Why did Myo4p make its adapter protein She3p a subunit of the motor complex? We proposed that because all cargoes of Myo4p (i.e., mRNA and cortical endoplasmic reticulum) require She3p, there is no advantage for this adapter protein to cycle between bound and unbound states (Hodges et al., 2008). Neither the rod region nor She3p is needed for ASH1 mRNA transport if the head of Myo4p is directly joined to the mRNA-binding protein She2p. This observation provides further support for the idea that the primary role of the rod and She3p is to tightly interact with each other and thus provide a robust coupling between motor and downstream cargo adapter proteins.
Slowing the speed at which ASH1 mRNA is transported decreased the percentage of ASH1 mRNA that was correctly localized to the bud, whereas substituting a two- to fourfold faster motor had no detrimental effect on localization. Thus, slower motors may not be as efficient as faster motors, but a range of speeds is tolerated. Likewise, substituting the low-duty cycle head from Myo2p in place of the high-duty cycle Myo4p did not decrease the percentage of correctly localized particles, a result also observed using a different reporter system (Dunn et al., 2007). This result is consistent with the idea that many motors are involved in moving the particle, which overrides the necessity for a high-duty cycle motor.
The protein BLOS2 has been identified as a putative She3p homologue by sequence alignment (Felten et al., 2007). A direct interaction of BLOS2 with the globular tail of mammalian myosin V was not detected by immunoprecipitation, despite observing colocalization with myosin V in vesicle-like structures in the cell (Felten et al., 2007). A potential reconciliation of these observations is that BLOS2, like She3p, binds to the rod and not the globular tail of mammalian myosin V.
Optimal Number of Motors per Zip Code
An unanswered question is how many motors are bound per zip code. Filter binding experiments suggest that one homodimeric She2p binds per zip code element (Niessing et al., 2004). Assuming that this stoichiometry holds in vivo, a situation with either one or two motors per zip code could arise depending on whether She2p is capable of binding one or two Myo4p–She3p complexes (Figure 7). If She2p forms higher oligomers under cellular conditions, the potential for even more motors per zip code exists.
Figure 7.
Model for how the Myo4p–She3p complex transports mRNA on actin cables in budding yeast. For illustrative purposes, a single mRNA (gray bar) with four zip code elements (blue rectangles) is shown. In vivo, many mRNA molecules are transported together to the bud tip. The number of zip code localization elements per mRNA also varies for different mRNAs. One homodimeric She2p binds per zip code element, based on in vitro binding data from Niessing et al. (2004). It is not known whether one or two Myo4p–She3p motor complexes can bind per She2p dimer. Assuming that one She2p dimer binds per zip code in vivo, our data imply that mRNA is transported equally well when one or more motor heads are bound per zip code; thus, both possibilities are shown. The two coiled coil regions of adjacent motor complexes are shown separated, but it is possible that they can form a helical bundle that may serve to coordinate the two heads. Actin cables are depicted because all the motor heads do not need to travel along a single actin filament in vivo.
We tested whether one single-headed motor complex per She2p dimer could transport ASH1 mRNA. This was achieved by cloning two copies of She2p after either the Myo4p head or the full-length molecule. This experiment was performed in a she2Δ strain, so that endogenous She2p cannot complete the dimer. Both constructs with tandem She2p molecules showed ≥85% correct localization in a she2Δ background, suggesting that one single-headed motor complex per She2p dimer is capable of correctly localizing ASH1 mRNA. Assuming that only one She2p dimer binds per zip code in vivo, this would correspond to one motor head per zip code.
Next, we tethered two single-headed motor complexes together by a leucine zipper. This does not cause the rod region of Myo4p to dimerize with itself and displace She3p but joins together two Myo4p–She3p heterocoiled-coil complexes (Hodges et al., 2008). Our results show no decrease in correct localization when two single-headed motor complexes are tethered in this manner. Closely abutting motors do not, therefore, interfere with each others' function. Assuming that one She2p dimer binds per zip code, this scenario reflects movement driven by two to four motors per zip code (depending on whether a She2p dimer binds 1 or 2 of the zippered motor complexes).
These experiments imply that motor number per zip code does not need to be tightly regulated for optimal transport function in vivo. This may reflect the fact that most mRNAs have multiple zip code elements, and even more importantly, that the single particle that gets translocated contains multiple copies of mRNA (Lange et al., 2008). Both of these situations would contribute to multiple motor transport. Implicit in this explanation is the assumption that multiple high-duty cycle monomeric motors are as efficient at achieving continuous transport as a single processive dimer. Experimental evidence in favor of this idea was obtained in vitro. A Quantum dot “cargo,” to which 3–4 Myo4p/She3p high-duty cycle single-headed motors were bound, moved continuously on actin filaments with run lengths that were similar (∼1 μm) to those supported by a single molecule of processive mouse myosin Va (Hodges et al., 2008).
Comparison with Other Studies
The observation that She3p copurifies with Myo4p when isolated from budding yeast (Dunn et al., 2007) agrees with similar experiments that we performed in Sf9 cells (Hodges et al., 2008) and suggests that the two proteins bind to each other with high affinity. There also seems to be general agreement, based on different approaches, that Myo4p can exist as a single-headed motor (Dunn et al., 2007; Heuck et al., 2007; Hodges et al., 2008).
Our interpretation of the role of She3p differs, however, from that of another recent study (Heuck et al., 2007). Our data suggest that Myo4p is monomeric because it forms a complex with She3p instead of with another molecule of Myo4p. In contrast, Heuck et al. (2007) suggest that Myo4p is monomeric without She3p and that She3p mediates dimerization of Myo4p. Their basic premise is that to obtain continuous motion, Myo4p must be dimerized when incorporated into the cargo–translocation complex. Given this, they propose that She3p-mediated dimerization will increase the processivity of Myo4p. However, the notion that a processive dimeric motor is needed for continuous mRNA transport only applies if one motor is transporting a cargo. Many Myo4p–She3p–She2p–ASH1 mRNA complexes, along with additional proteins, coalesce into the particle that is transported to the developing bud tip, and thus there are likely to be many motors involved in transport, negating the need for processive dimers.
Some of the difference in interpretation is based on their observation that She3p binds to a dimeric but not a monomeric globular tail and that She3p was localized at the bud tip 49% of the time with a construct that contains only the globular tail of Myo4p, and the rest of the molecule from Myo2p (Heuck et al., 2007). Our comparable construct (m2m2m4) with the Myo4p globular tail shows no localization of ASH1 mRNA, consistent with the idea that She3p does not bind to this construct because it lacks the Myo4p rod. One difference between the two studies is that Heuck et al. (2007) observed localization of the She3p protein at the bud tip, whereas our assay measured localization of the cargo ASH1 mRNA at the bud tip. Our in vivo findings and previous studies with purified recombinant proteins (Hodges et al., 2008) suggest that the rod of Myo4p is the most important determinant for She3p binding and that the globular tail plays a secondary role.
CONCLUSIONS
ASH1 mRNA localization seems to be relatively independent of motor speed, the duty cycle of the motor, and motor number (Figure 7). This is likely due to many motors being involved in particle translocation and given that Myo4p is a high-duty cycle motor, only a few are needed to support continuous motion. The only modification that completely prevented ASH1 mRNA localization was abolishing the coupling of Myo4p to the mRNA by virtue of removing the rod, the primary binding site for the adapter protein She3p. Given this result, we propose that yeast has optimized its mRNA transport system by making She3p a subunit of Myo4p, thus ensuring that these two essential proteins are always in place when it is time to move mRNA or cortical endoplasmic reticulum.
ACKNOWLEDGMENTS
We thank Usha Nair for designing the linked She2p dimer and Alex Hodges, Elena Krementsova, and Susan Lowey for critical reading of the manuscript. We also thank Lois Weisman and Yui Jin for help in optimizing conditions to obtain the immunoblot. We thank Roy Long for providing the yeast strains and the reporter plasmids for ASH1 mRNA, Susan Ferro-Novick for the Hmg1p-GFP plasmid, and Brian Haarer and Karen Beningo for providing plasmids containing MYO2 and MYO4. This work was funded by National Institutes of Health grant GM-078097 (to K.M.T.) and American Heart Association Scientist Development grant 0835236N (to M. L.).
Abbreviations used:
- ER
endoplasmic reticulum
- GT
globular tail
- MD
motor domain.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-08-0801) on May 28, 2009.
REFERENCES
- Beach D. L., Bloom K. ASH1 mRNA localization in three acts. Mol. Biol. Cell. 2001;12:2567–2577. doi: 10.1091/mbc.12.9.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand E., Chartrand P., Schaefer M., Shenoy S. M., Singer R. H., Long R. M. Localization of ASH1 mRNA particles in living yeast. Mol. Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- Bohl F., Kruse C., Frank A., Ferring D., Jansen R. P. She2p, a novel RNA-binding protein tethers ASH1 mRNA to the Myo4p myosin motor via She3p. EMBO J. 2000;19:5514–5524. doi: 10.1093/emboj/19.20.5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock S. L. Translocation of mRNAs by molecular motors: think complex? Semin. Cell Dev. Biol. 2007;18:194–201. doi: 10.1016/j.semcdb.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Chartrand P., Meng X. H., Singer R. H., Long R. M. Structural elements required for the localization of ASH1 mRNA and of a green fluorescent protein reporter particle in vivo. Curr. Biol. 1999;9:333–336. doi: 10.1016/s0960-9822(99)80144-4. [DOI] [PubMed] [Google Scholar]
- Condeelis J., Singer R. H. How and why does beta-actin mRNA target? Biol. Cell. 2005;97:97–110. doi: 10.1042/BC20040063. [DOI] [PubMed] [Google Scholar]
- Cosma M. P. Daughter-specific repression of Saccharomyces cerevisiae HO: Ash1 is the commander. EMBO Rep. 2004;5:953–957. doi: 10.1038/sj.embor.7400251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y., Pypaert M., Novick P., Ferro-Novick S. Aux1p/Swa2p is required for cortical endoplasmic reticulum inheritance in Saccharomyces cerevisiae. Mol. Biol. Cell. 2001;12:2614–2628. doi: 10.1091/mbc.12.9.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn B. D., Sakamoto T., Hong M. S., Sellers J. R., Takizawa P. A. Myo4p is a monomeric myosin with motility uniquely adapted to transport mRNA. J. Cell Biol. 2007;178:1193–1206. doi: 10.1083/jcb.200707080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada P., Kim J., Coleman J., Walker L., Dunn B., Takizawa P., Novick P., Ferro-Novick S. Myo4p and She3p are required for cortical ER inheritance in Saccharomyces cerevisiae. J. Cell Biol. 2003;163:1255–1266. doi: 10.1083/jcb.200304030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felten A., Leister P., Burgdorf S., Uhlmann L., Scheidtmann K. H. Characterization of rat BLOS2/Ceap, a putative yeast She3 homolog, as interaction partner of apoptosis antagonizing transcription factor/Che-1. Biol. Chem. 2007;388:569–582. doi: 10.1515/BC.2007.073. [DOI] [PubMed] [Google Scholar]
- Gonsalvez G. B., Urbinati C. R., Long R. M. RNA localization in yeast: moving towards a mechanism. Biol. Cell. 2005;97:75–86. doi: 10.1042/BC20040066. [DOI] [PubMed] [Google Scholar]
- Gross S. P., Tuma M. C., Deacon S. W., Serpinskaya A. S., Reilein A. R., Gelfand V. I. Interactions and regulation of molecular motors in Xenopus melanophores. J. Cell Biol. 2002;156:855–865. doi: 10.1083/jcb.200105055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarer B. K., Petzold A., Lillie S. H., Brown S. S. Identification of MYO4, a second class V myosin gene in yeast. J. Cell Sci. 1994;107:1055–1064. doi: 10.1242/jcs.107.4.1055. [DOI] [PubMed] [Google Scholar]
- Heuck A., Du T. G., Jellbauer S., Richter K., Kruse C., Jaklin S., Muller M., Buchner J., Jansen R. P., Niessing D. Monomeric myosin V uses two binding regions for the assembly of stable translocation complexes. Proc. Natl. Acad. Sci. USA. 2007;104:19778–19783. doi: 10.1073/pnas.0706780104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges A. R., Krementsova E. B., Trybus K. M. She3p binds to the rod of yeast myosin V and prevents it from dimerizing, forming a single-headed motor complex. J. Biol. Chem. 2008;283:6906–6914. doi: 10.1074/jbc.M708865200. [DOI] [PubMed] [Google Scholar]
- Jambhekar A., McDermott K., Sorber K., Shepard K. A., Vale R. D., Takizawa P. A., DeRisi J. L. Unbiased selection of localization elements reveals cis-acting determinants of mRNA bud localization in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 2005;102:18005–18010. doi: 10.1073/pnas.0509229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krementsova E. B., Hodges A. R., Lu H., Trybus K. M. Processivity of chimeric class V myosins. J. Biol. Chem. 2006;281:6079–6086. doi: 10.1074/jbc.M510041200. [DOI] [PubMed] [Google Scholar]
- Lange S., Katayama Y., Schmid M., Burkacky O., Brauchle C., Lamb D. C., Jansen R. P. Simultaneous transport of different localized mRNA species revealed by live-cell imaging. Traffic. 2008;9:1256–1267. doi: 10.1111/j.1600-0854.2008.00763.x. [DOI] [PubMed] [Google Scholar]
- Long R. M., Gu W., Lorimer E., Singer R. H., Chartrand P. She2p is a novel RNA-binding protein that recruits the Myo4p-She3p complex to ASH1 mRNA. EMBO J. 2000;19:6592–6601. doi: 10.1093/emboj/19.23.6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long R. M., Singer R. H., Meng X., Gonzalez I., Nasmyth K., Jansen R. P. Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science. 1997;277:383–387. doi: 10.1126/science.277.5324.383. [DOI] [PubMed] [Google Scholar]
- Moore J. R., Krementsova E. B., Trybus K. M., Warshaw D. M. Does the myosin V neck region act as a lever? J. Muscle Res. Cell Motil. 2004;25:29–35. doi: 10.1023/b:jure.0000021394.48560.71. [DOI] [PubMed] [Google Scholar]
- Muller M., Heuck A., Niessing D. Directional mRNA transport in eukaryotes: lessons from yeast. Cell Mol. Life Sci. 2007;64:171–180. doi: 10.1007/s00018-006-6286-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niessing D., Huttelmaier S., Zenklusen D., Singer R. H., Burley S. K. She2p is a novel RNA binding protein with a basic helical hairpin motif. Cell. 2004;119:491–502. doi: 10.1016/j.cell.2004.10.018. [DOI] [PubMed] [Google Scholar]
- Reck-Peterson S. L., Provance D. W., Jr, Mooseker M. S., Mercer J. A. Class V myosins. Biochim. Biophys. Acta. 2000;1496:36–51. doi: 10.1016/s0167-4889(00)00007-0. [DOI] [PubMed] [Google Scholar]
- Reck-Peterson S. L., Tyska M. J., Novick P. J., Mooseker M. S. The yeast class V myosins, Myo2p and Myo4p, are nonprocessive actin-based motors. J. Cell Biol. 2001;153:1121–1126. doi: 10.1083/jcb.153.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M., Jaedicke A., Du T. G., Jansen R. P. Coordination of endoplasmic reticulum and mRNA localization to the yeast bud. Curr. Biol. 2006;16:1538–1543. doi: 10.1016/j.cub.2006.06.025. [DOI] [PubMed] [Google Scholar]
- Schott D. H., Collins R. N., Bretscher A. Secretory vesicle transport velocity in living cells depends on the myosin-V lever arm length. J. Cell Biol. 2002;156:35–39. doi: 10.1083/jcb.200110086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard K. A., Gerber A. P., Jambhekar A., Takizawa P. A., Brown P. O., Herschlag D., DeRisi J. L., Vale R. D. Widespread cytoplasmic mRNA transport in yeast: identification of 22 bud-localized transcripts using DNA microarray analysis. Proc. Natl. Acad. Sci. USA. 2003;100:11429–11434. doi: 10.1073/pnas.2033246100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sil A., Herskowitz I. Identification of asymmetrically localized determinant, Ash1p, required for lineage-specific transcription of the yeast HO gene. Cell. 1996;84:711–722. doi: 10.1016/s0092-8674(00)81049-1. [DOI] [PubMed] [Google Scholar]
- Takagi Y., Yang Y., Fujiwara I., Jacobs D., Cheney R. E., Sellers J. R., Kovacs M. Human myosin Vc is a low duty ratio, nonprocessive molecular motor. J. Biol. Chem. 2008;283:8527–8537. doi: 10.1074/jbc.M709150200. [DOI] [PubMed] [Google Scholar]
- Takizawa P. A., Sil A., Swedlow J. R., Herskowitz I., Vale R. D. Actin-dependent localization of an RNA encoding a cell-fate determinant in yeast. Nature. 1997;389:90–93. doi: 10.1038/38015. [DOI] [PubMed] [Google Scholar]
- Tekotte H., Davis I. Intracellular mRNA localization: motors move messages. Trends Genet. 2002;18:636–642. doi: 10.1016/s0168-9525(02)02819-6. [DOI] [PubMed] [Google Scholar]
- Toth J., Kovacs M., Wang F., Nyitray L., Sellers J. R. Myosin V from Drosophila reveals diversity of motor mechanisms within the myosin V family. J. Biol. Chem. 2005;280:30594–30603. doi: 10.1074/jbc.M505209200. [DOI] [PubMed] [Google Scholar]
- Trybus K. M. Myosin V from head to tail. Cell Mol. Life Sci. 2008;65:1378–1389. doi: 10.1007/s00018-008-7507-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S., Watanabe T. M., Sato O., Awata J., Homma K., Umeki N., Higuchi H., Ikebe R., Ikebe M. Human myosin Vc is a low duty ratio nonprocessive motor. J. Biol. Chem. 2008;283:10581–10592. doi: 10.1074/jbc.M707657200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisman L. S. Organelles on the move: insights from yeast vacuole inheritance. Nat. Rev. Mol. Cell Biol. 2006;7:243–252. doi: 10.1038/nrm1892. [DOI] [PubMed] [Google Scholar]
- Wu X. S., Rao K., Zhang H., Wang F., Sellers J. R., Matesic L. E., Copeland N. G., Jenkins N. A., Hammer J. A., 3rd. Identification of an organelle receptor for myosin-Va. Nat. Cell Biol. 2002;4:271–278. doi: 10.1038/ncb760. [DOI] [PubMed] [Google Scholar]
- Yoshimura A., Fujii R., Watanabe Y., Okabe S., Fukui K., Takumi T. Myosin-Va facilitates the accumulation of mRNA/protein complex in dendritic spines. Curr. Biol. 2006;16:2345–2351. doi: 10.1016/j.cub.2006.10.024. [DOI] [PubMed] [Google Scholar]