Abstract
The tumor suppressor p53 plays a crucial role in cellular response to various stresses. Recent experiments have shown that p53 level exhibits a series of pulses after DNA damage caused by ionizing radiation (IR). However, how the p53 pulses govern cell survival and death remains unclear. Here, we develop an integrated model with four modules for the p53 network and explore the mechanism for cell fate decision based on the dynamics of the network. By numerical simulations, the following processes are characterized. First, DNA repair proteins bind to IR-induced double-strand breaks, forming complexes, which are then detected by ataxia telangiectasia mutated (ATM). Activated ATM initiates the p53 oscillator to produce pulses. Consequently, the target genes of p53 are selectively induced to control cell fate. We propose that p53 promotes the repair of minor DNA damage but suppresses the repair of severe damage. We demonstrate that cell fate is determined by the number of p53 pulses relying on the extent of DNA damage. At low damage levels, few p53 pulses evoke cell cycle arrest by inducing p21 and promote cell survival, whereas at high damage levels, sustained p53 pulses trigger apoptosis by inducing p53AIP1. We find that p53 can effectively maintain genomic integrity by regulating the efficiency and fidelity of DNA repair. We also show that stochasticity in the generation and repair of DNA damage leads to variability in cell fate. These findings are consistent with experimental observations and advance our understanding of the dynamics and functions of the p53 network.
Keywords: p53 network, DNA repair, apoptosis, variability
The tumor suppressor protein p53 is a key mediator of cellular response to various stresses (1), and it performs its function primarily as a transcription factor, controlling the expression of a number of target genes (2). In response to DNA damage, the p53 signaling network is activated to induce cell cycle arrest, DNA repair, and apoptosis (1). Previously, it was proposed that p53 responds to DNA damage in an analog mode, i.e., a low level of p53 promotes cell survival by inducing cell cycle arrest and facilitating DNA repair, whereas a high level of p53 induces apoptosis to destroy irreparably damaged cells (1). Recently, damped oscillations of p53 level have been observed upon IR at the population level in several human cell lines and transgenic mice (3–6), with the amplitude of oscillations depending on the IR dose (5). More interestingly, pulses of p53 level were revealed in individual MCF7 cells, and the mean number of pulses, rather than the amplitude, was found to be related to the IR dose (7). This was suggested as a digital mode of p53 response (7). Later, it was further found that some MCF7 cells exhibit permanent p53 pulses whereas a few cells still show a finite number of p53 pulses during long-term observations (8). Such permanent oscillations may be related to the deficiency in DNA repair and apoptosis induction in those transformed cells (9–11). Moreover, the damped oscillations observed in cell populations can be interpreted as a summation of p53 pulses of individual cells (8, 12). Thus, the above experimental observations clearly justify the presence of p53 pulses at the single-cell level. An issue then arises concerning how the p53 pulses govern cell fate decision between survival and death after DNA damage. It is also intriguing to explore the relationship between p53 pulses and IR dose in untransformed cells.
Because IR is one of the most frequently used techniques in cancer therapy, it is of great significance to investigate the response of p53 signaling network to IR-induced DNA damage. Previously, several models have been proposed to elucidate the generation of p53 oscillations (3, 8, 12–15). Especially, Ma et al. (12) constructed a three-module model and found that p53 pulses can be generated based on a negative-feedback loop between p53 and Mdm2 with a long time delay. Zhang et al. (15) further proposed four dual-feedback mechanisms for p53 digital oscillations and classified the p53 pulses into three forms to explore the functions of p53. Most recently, Batchelor et al. (16) reported a recurrent mechanism for the generation of p53 pulses by ATM pulses. However, to obtain a coherent picture of the p53-mediated cellular response to DNA damage, several important aspects need be considered. First, both the generation of p53 pulses and cell fate decision should be included in modeling. Second, the functions of p53 pulses should be further elucidated. Third, the variability between individual cells in p53 dynamics should be reflected in the variability in cell fate. Fourth, because p53 also plays a transcription-independent role in DNA repair (17), the link between p53-mediated regulation of the repair process and cell fate decision should be built. These are important for the clarification of how p53 dynamically controls cell survival and death after DNA damage.
Here, we build an integrated model for the p53 signaling network and study the whole process from generation of DNA damage to cell fate decision. The model is composed of four subsystems: a DNA repair module, an ATM switch, the p53–Mdm2 oscillator, and a cell fate decision module. We propose that p53 plays a dual, transcription-independent role in DNA repair, i.e., p53 promotes the repair of minor DNA damage but suppresses the repair of severe damage. The combined negative and positive feedback loops between p53 and Mdm2 are responsible for robust oscillatory dynamics. We find that cell fate is governed by the number of p53 pulses, i.e., few pulses promote cell survival, whereas sustained pulses induce apoptosis. We show that p53 subserves the maintenance of genomic integrity by regulating the fidelity and efficiency of DSB repair. These findings advance our understanding of the dynamics and functions of the p53 network and provide clues for therapeutic manipulation of the p53 pathway in the treatment of cancer. Our work presents a theoretical framework for further exploring cellular response to stress.
Model
p53 Signaling Network with Four Modules.
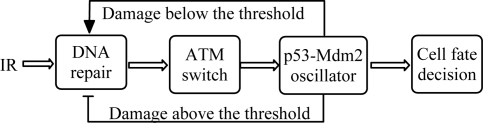
In this work, the p53 network is composed of four modules (Fig. 1). When cells are exposed to IR, a certain number of DSBs are first generated in every cell. With DNA repair proteins binding to the DSBs, DSB repair-protein complexes (DSBCs) form and the repair process ensues in the DNA repair module. Subsequently, ATM is activated by the DSBCs in the ATM switch module. Then p53 is activated by the active ATM, and the p53 level exhibits a series of pulses in the p53–Mdm2 oscillator module. Finally, in the cell fate decision module, p53 controls the expression of the target genes; cell cycle arrest and apoptosis are separately evoked by the induction of p21 and p53AIP1. Thus, the damage signal is transmitted forward until DNA repair is completed or apoptosis is initiated. In our model, it is assumed that p53 promotes repair when DNA damage is minor and suppresses it otherwise (Fig. 1). The details of each module are described in the following.
Fig. 1.
An integrated model of p53 signaling network. There are four modules: a DNA repair module, an ATM switch, the p53–Mdm2 oscillator, and a cell fate decision module. The p53 regulation of DNA repair is threshold-dependent and is denoted as feedback from the p53–Mdm2 oscillator to the DNA repair module.
DNA Repair.
The DNA repair module characterizes the generation and repair of IR-induced DSBs. For each cell, DSBs are first generated relying on the IR dose, DIR, and the numbers of DSBs for a population obey a Poisson distribution with a mean of 35 Gy−1 × DIR (18). The repair process then involves binding of repair proteins to the DSBs and releasing of the proteins from the fixed DSBs. Such a process proceeds in a stochastic way (see SI Appendix for details). Each locus in which a DSB is initially generated can be in one of three states corresponding to intact DSB, DSBC, and fixed DSB. Thus, the number of DSBs in each state can be used to describe the repair process. In the simulation, we adopt the two-lesion-kinetic model (19) (Fig. S1 and Eqs. 1–3 in SI Appendix) and characterize the dynamics of DSB repair by using a Monte Carlo method proposed by Ma et al. (12).
Generally, DSBs are repaired mainly by nonhomologous end-joining (NHEJ) in mammalian cells, especially in G1 phase of the cell cycle (20). Note that p53 plays an important role in DNA repair (17). However, the effect of p53 on NHEJ is complicated: a stimulatory effect was observed (21, 22), whereas an inhibitory effect was also reported (23, 24). To reconcile these findings, it was speculated that p53 may inhibit error-prone repair but promote error-free repair (17). Thus, p53 regulates the fidelity of DSB repair. It was also shown that the fidelity is related to the number of DSBs, i.e., the fidelity is high or low for few and excessive DSBs, respectively (25, 26). Therefore, we assume that p53 promotes the DSB repair when the initial number of DSBs is below a threshold and suppresses it otherwise (Fig. 1). In the model, the threshold for DSB repair is set to 175, i.e., the mean number of DSBs generated at DIR = 5 Gy.
ATM Switch.
The ATM switch acts as a detector for DSBs, involving three components: ATM dimer (ATM2), inactive ATM monomer (ATM), and active ATM monomer (ATM*) (Fig. S2 in SI Appendix). ATM exists predominantly as dimers in unstressed cells. After IR, DSBCs promote the conversion of inactive monomers to active ones, and a positive feedback further accelerates the activation of ATM through intermolecular autophosphorylation (27). Consequently, active monomers become dominant after IR.
In our model, the total concentration of ATM is assumed to be constant, and the rate of ATM activation is a function of the amounts of DSBC and ATM*. After IR, the concentration of ATM* rises from a low level to a high one; it returns to the low level when the number of DSBs becomes sufficiently small because of DNA repair. This behavior is characterized by a bistable switch with discrete “on” and “off” states. The switch transmits a switching signal to the downstream p53–Mdm2 oscillator, ensuring that it is initiated in an “all-or-none” manner. The “off” state of the switch leads to a low p53 level, whereas the “on” state drives a series of p53 pulses. The dynamics of the ATM switch are described by Eqs. 4–6 in SI Appendix.
p53–Mdm2 Oscillator.
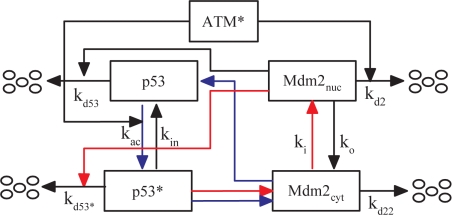
The p53–Mdm2 oscillator generates pulses after the activation of ATM. This module is composed of two core proteins: p53 and its negative regulator, Mdm2 (Fig. 2). Here, only nuclear p53 is considered, whereas Mdm2 is classified into nuclear and cytoplasmic forms. In the nucleus, p53 is activated by ATM* in two ways: ATM* promotes the phosphorylation of p53 on Ser-15 (28) and accelerates the degradation of Mdm2 through phosphorylation (29). Thus, ATM* induces a conversion of p53 from the inactive state to the active state (p53*) (29). Meanwhile, p53* is deactivated at a basal rate. Here, only p53* can induce production of Mdm2cyt, which promotes translation of p53 mRNA in the cytoplasm (30). For simplicity, we assume that both the degradation rate of Mdm2 and the activation rate of p53 increase linearly with ATM* level (Eqs. 7–8 in SI Appendix).
Fig. 2.
Model of the p53–Mdm2 oscillator. Two forms of p53 in the nucleus are considered, inactive p53 and active p53*. Mdm2 in the nucleus and cytoplasm is denoted as Mdm2nuc and Mdm2cyt, respectively. They can shuttle between the two compartments at rates ki and ko. Inactive p53 is degraded rapidly by Mdm2 at a rate kd53, whereas active p53* is degraded slowly at a rate kd53* because of its weak binding affinity to Mdm2nuc. Combined negative (red lines) and positive (blue lines) feedback loops are responsible for p53 oscillations. In the negative-feedback loop, p53* promotes production of Mdm2cyt, which then enters the nucleus to induce degradation of p53*. In the positive-feedback loop, p53* induces Mdm2 to synthesize Mdm2cyt, which promotes the translation of p53 mRNA to produce inactive p53.
Two coupled feedback loops are involved in the p53–Mdm2 oscillator. One is the negative-feedback loop between p53* and Mdm2nuc (the red lines in Fig. 2); the other is the positive one between p53* and Mdm2cyt (30) (the blue lines). In undamaged cells, p53 levels are kept low by Mdm2. After DNA damage, the p53–Mdm2 complex is dissociated because of activation of p53 by ATM*, with the negative feedback weakened. The levels of p53, p53*, and Mdm2cyt increase abruptly through the positive feedback. Subsequently, high-level Mdm2nuc enables the negative feedback to predominate again, and then p53 is degraded quickly. The alternate domination of the positive and the negative feedback can make p53 and Mdm2 levels oscillate periodically. Note that several theoretical studies have demonstrated that interlinked feedback loops are superior to a single feedback loop in response to various signals (14, 31, 32). It was argued that robust p53 oscillations can be accomplished through a combination of negative and positive feedback loops rather than a single negative feedback loop with a time delay (14, 32). The dynamics of this module are characterized by Eqs. 7–14 in SI Appendix.
Cell Fate Decision.
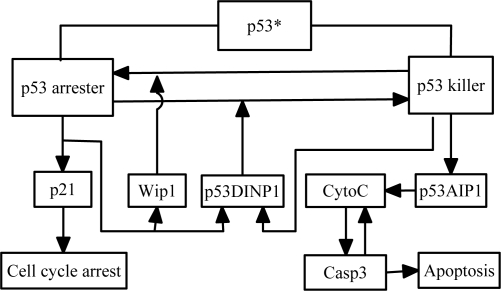
In this module, p53 coordinates cell cycle arrest and apoptosis to govern cell fate, in which the phosphorylation of p53 at distinct sites is a key factor. For example, the primary phosphorylation of p53 on Ser-15 leads to cell cycle arrest, whereas its further phosphorylation on Ser-46 promotes expression of pro-apoptotic genes such as p53AIP1 (33). Here, the above two forms of phosphorylated p53 are called p53 arrester and p53 killer, respectively (Fig. 3). Moreover, the product of p53 arrester-inducible gene p53DINP1 contributes to the formation of p53 killer (34), whereas that of another p53 arrester-inducible gene Wip1 promotes the reversion of p53 killer to p53 arrester (35). Clearly, p53 arrester and p53 killer play distinct roles in cell fate decision. p53 arrester induces cell cycle arrest through the transcriptional activation of p21. Differently, p53 killer promotes the expression of pro-apoptotic genes such as p53AIP1. Over-expression of p53AIP1 then induces the release of cytochrome c (CytoC) from mitochondrial caused by dissipation of mitochondrial membrane potential Δψm (33), and apoptosis rapidly ensues after the activation of Caspase 3 (Casp3). A positive-feedback loop between CytoC and Casp3 (36) underlies the apoptotic switch in our model (37). The dynamics of this module are described by Eqs. 15–22 in SI Appendix.
Fig. 3.
Model of cell fate decision. There are two forms of active p53: p53 arrester and p53 killer. p53 arrester regulates the expression of Wip1, p53DINP1, and p21, whereas p53 killer controls the expression of p53DINP1 and p53AIP1. CytoC and Casp3 are the pro-apoptotic components. There is a positive-feedback loop between CytoC and Casp3, which creates an apoptotic switch. The release of CytoC leads to activation of Casp3, which in turn cleaves the inhibitors of p53AIP1 (such as Bcl-2 and Bcl-xL) to enhance the release of CytoC.
Results
An Overview of Signal Transduction in the p53 Network.
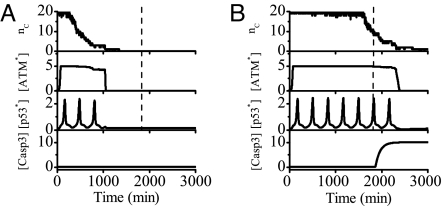
Fig. 4 shows two examples of the output of each module after IR. The number of DSBCs, nC, first rises rapidly to its maximum (here 20 DNA repair proteins per cell is assumed). Then DSBCs turn on the ATM switch, and the activated ATM initiates the p53–Mdm2 oscillator to produce p53 pulses. The number of p53 pulses is determined by the duration of ATM in the activated state. At the low IR dose of 3 Gy, transient p53 pulses cannot activate Casp3, and the cell recovers to normal growth after the damage is effectively repaired (Fig. 4A). At the high dose of 5 Gy, sustained p53 pulses lead to apoptosis through activation of Casp3 (Fig. 4B). This means that minor DNA damage only evokes protective cell cycle arrest and severe damage induces apoptosis. It is found that cell fate is governed by the number of p53 pulses, nP. In our model, when nP < 6, slightly damaged cells recover to normal growth; when nP ≥ 6, the concentration of p53AIP1 gradually accumulates to a high level and oscillates there until Casp3 level exceeds a threshold for apoptosis induction (see Fig. S11 in SI Appendix). That is, the critical pulse number for cell fate decision is 6. Note that such a value depends on the model and the parameter set. The dashed lines in Fig. 4 denote the threshold time taken for the number of p53 pulses to reach the critical number, i.e., cells recover to normal proliferation if DSB repair is finished before that time and otherwise are eliminated through apoptosis.
Fig. 4.
Overview of signal transduction in the p53 network. Shown are time courses of the output of each module at the IR dose of 3 Gy (A) or 5 Gy (B). Upon IR, a number of DSBCs, nC, are produced and then ATM is activated, initiating the p53 oscillations. When DNA damage is effectively repaired, the ATM switch is turned off and the concentration of p53* returns to a low level. At low IR doses, few p53 pulses cannot activate Casp3 and the cell recovers to normal growth. At high doses, sustained p53 pulses lead to activation of Casp3 and apoptosis ensues. The dashed lines denote a threshold time for cell fate decision.
DNA Damage Repair.
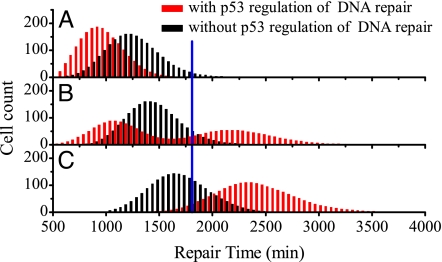
Because of stochasticity in the initial numbers of DSBs and repair dynamics, individual cells under identical conditions show considerable variability in the repair time, which is defined as the time taken for the number of DSBs to reduce below 2 (here the apoptotic effect is not involved). A histogram of the repair time for 2,000 cells is used to characterize the variability in repair time. In the presence of p53 regulation of DNA repair, the histograms are unimodal at low and high IR doses (Fig. 5 A and C), whereas the histogram becomes bimodal around DIR = 5 Gy (Fig. 5B). The mean repair time increases with IR dose, and the histogram moves rightward for higher dose (see Figs. S3 and S4 in SI Appendix for details). In the absence of p53 regulation, however, the histograms are all unimodal at various IR doses.
Fig. 5.
Histograms of repair time. The cell count vs. the repair time is plotted for a population of 2,000 cells with and without p53 regulation (red and black lines, respectively) at the IR dose of 4 Gy (A), 5 Gy (B), or 6 Gy (C). The blue line denotes the threshold time for cell fate decision. With p53 regulation, the repair process depends on the levels of p53 and DNA damage, i.e., p53 promotes the repair of minor damage but suppresses the repair of severe damage. A threshold of 175 DSBs is set to distinguish between low and high damage levels.
Note that p53 regulates the efficiency of DNA repair in a threshold-dependent manner: p53 promotes the repair when the initial number of DSBs is less than 175 and suppresses the repair otherwise. At low IR doses, say DIR = 4 Gy, p53 promotes the repair with high fidelity, and the histogram for the case with p53 regulation shifts leftward (Fig. 5A). At high doses, say DIR = 6 Gy, p53 represses the repair, and the histogram for the case with p53 regulation shifts rightward (Fig. 5C). Interestingly, at DIR = 5 Gy, the repair time for some cells decreases whereas that for the others increases, compared with the case without p53 regulation. As a result, the histogram becomes bimodal.
The histogram for the case without p53 regulation at DIR = 5 Gy is consistent with that in figure 5C in ref. 12. Differently, here the dual role of p53 in regulation of DNA repair has significant implications for the maintenance of genomic integrity. Because the repair of slight damage is strengthened and that of severe damage is repressed, p53 promotes the repair with high fidelity and suppresses the repair with low fidelity. As we shall see later, this is in concert with the role of p53 in cell fate decision, i.e., p53 promotes survival of reparably damaged cells or apoptosis of irreparably damaged cells. Therefore, DNA repair and cell fate decision are well coordinated by p53.
ATM Switch.
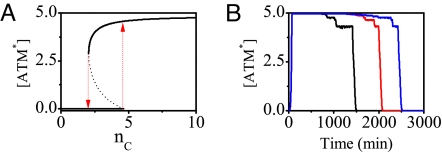
As a detector for DSBs, the activation of ATM exhibits a switch-like behavior (Fig. 6 and Fig. S5 in SI Appendix). Because nC varies on a time scale of hours but ATM equilibrates in minutes because of fast phosphorylation (Fig. 4), ATM* level follows the time course of nC adiabatically. The steady-state ATM* level versus nC is plotted in Fig. 6A. ATM activation starts with nC = 5, whereas ATM deactivation occurs when nC = 2 (i.e., the DNA damage is effectively repaired). Thus, ATM* level indeed behaves as a bistable switch, with the on (off) state corresponding to the upper (lower) branch in Fig. 6A.
Fig. 6.
Switching behavior of ATM. (A) Bifurcation diagram of ATM* level vs. the number of DSBCs, nC. The ATM switch turns on if nC > 4 (the upward arrow) or turns off if nC < 3 (downward arrow). (B) Time courses of ATM* level for 3 individual cells at DIR = 5 Gy. The ATM switch is on during the DSB repair process and is off only after DSBs are effectively repaired.
The time courses of ATM* level for 3 individual cells at DIR = 5 Gy are plotted in Fig. 6B (see also Fig. S6 in SI Appendix). After IR, ATM is activated rapidly and its level reaches a high plateau with [ATM*] ≈ 5. The width of the plateau is distinct for different cells, showing considerable variability. Besides that plateau, there exist two short lower plateaus separately corresponding to nC = 4 and nC = 3. Clearly, the duration of active ATM state is determined by the repair time and prolongs with increased IR dose (Fig. 4 and Fig. S6 in SI Appendix). Moreover, once nC < 3, ATM* level decays to zero quickly. This step-like response results from the positive feedback involved in the autophosphorylation of ATM (27). These results indicate that few DSBs are able to activate ATM rapidly and ATM remains active until DNA damage is effectively repaired. Therefore, ATM is a sensitive and reliable detector for DSBs. Furthermore, the ATM* activity relays an on–off switching signal to the p53–Mdm2 oscillator.
p53 Pulses.
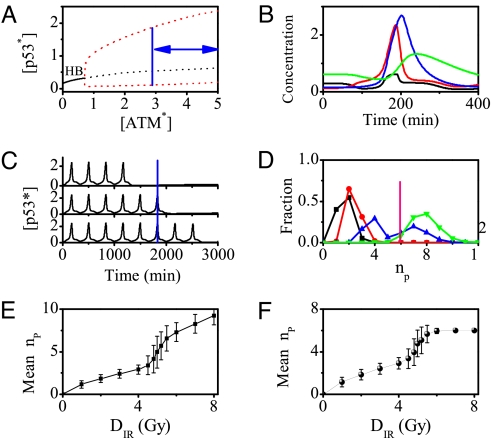
The generation of p53 pulses is controlled by active ATM. To show this, a bifurcation diagram of p53* level versus ATM* level is plotted in Fig. 7A. When [ATM*] < 0.7, p53 remains at a low level. There is a Hopf bifurcation (labeled as “HB”) at [ATM*] = 0.7, and beyond that p53* level undergoes periodical oscillations. As seen in Figs. 7A and 6A, the on state of ATM* evokes the p53 oscillations, whereas the off state terminates them. Thus, ATM* level is limited in a region between 2.9 and 5.0 for driving p53 oscillations.
Fig. 7.
p53 pulses. (A) Bifurcation diagram of p53* level vs. ATM* level. In unstressed cells, ATM is inactive and p53 remains in a low-level steady state (solid line). Upon IR, the steady state becomes unstable (black dashed line), and p53* level undergoes oscillations between the maxima and minima of the limit cycle (red dashed lines) when ATM level is in the region marked by the bidirectional arrows. (B) Time course of the levels of p53 (black), p53* (red), Mdm2cyt (blue), and Mdm2nuc (green) over one period. (C) A series of p53 pulses for three individual cells at DIR = 5 Gy. The blue line denotes the threshold time for cell fate decision (compare Fig. 4). (D) Fraction of cells with different numbers of pulses at IR doses of 1 Gy (black), 3 Gy (red), 5 Gy (blue), and 6 Gy (green). The pink line denotes the critical pulse number for cell fate decision. (E) The mean nP vs. DIR without the effect of apoptosis. (F) The mean nP vs. DIR with the effect of apoptosis. Here the mean number is obtained by averaging over 2,000 cells, and the error bars indicate the standard deviation.
Fig. 7B shows the time evolution of four components of the p53–Mdm2 oscillator, describing the generation of the pulses. After IR, p53* level rises abruptly owing to activation by ATM*. In contrast, Mdm2nuc level first decreases because of its accelerated degradation induced by ATM* and then increases after the rise of Mdm2cyt because of p53 induction. As a result, the translation rate of p53 mRNA rises, leading to a further increase in p53* level. Meanwhile, more Mdm2cyt enters the nucleus, and the p53* level drops when Mdm2nuc level is high enough. Subsequently, Mdm2 levels fall because of a reduction in synthesis. Thus, p53* level exhibits the pulse-like behavior. The oscillation period (≈5.5 h) and phase differences between components are in agreement with the experimental observations (8). Moreover, the parameter dependence of the period and amplitude of p53 oscillations is presented in Figs. S7 and S8 in SI Appendix.
Fig. 7C shows the time courses of p53* level for 3 cells at DIR = 5 Gy. The amplitudes and periods of p53 pulses are almost constant. However, the numbers of p53 pulses are variable because of the stochasticity in the generation and repair of DSBs as shown in Fig. 5. Sparse p53 pulses cease when DNA damage is quickly repaired; when the DSBs cannot be fixed within the threshold time, p53 pulses persist until apoptosis is induced. Moreover, the averaged p53* level over 2,000 individual cells exhibits damped oscillations (Fig. S9 in SI Appendix), consistent with the experimental observation (3). Note that if the cells are deficient in apoptosis induction and DNA repair, the p53 pulses may last permanently as observed in MCF7 cells (8).
To show variability between cells in p53 dynamics, the histograms of the number of p53 pulses are plotted for four IR doses in Fig. 7D. It is seen that the number of p53 pulses shows considerable variability for certain DIR, and most distributions are unimodal except a bimodal one for DIR = 5 Gy. The distributions here are correlated with those of the repair time shown in Fig. 5. Furthermore, the mean nP averaged over 2,000 cells versus DIR is presented in Fig. 7 E and F. When the apoptotic effect is not involved, i.e., the pulses persist until all DNA damage is fixed, the mean nP first increases linearly, then rapidly and finally linearly again with increasing DIR (Fig. 7E). The two linear regions are related to the unimodal distributions in Fig. 7D. There is a large fluctuation in nP around DIR = 5 Gy because of the bimodal distribution in Fig. 7D. Differently, when the effect of apoptosis is included, the mean nP becomes saturated at high DIR (Fig. 7F). Clearly, this is because most cells commit suicide when nP = 6. Taken together, a sigmoidal relationship between the mean pulse number and IR dose is found, and a digital mode of p53 response is presented.
Cell Fate Decision.
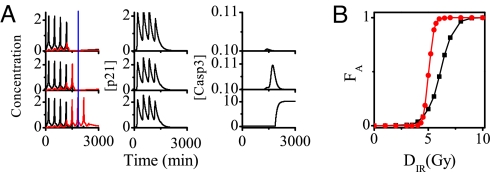
p53 pulses govern cell fate through selective induction of the target genes. Fig. 8A shows the time courses of the levels of p53 arrester, p53 killer, p21, and Casp3 after IR. The first four p53 pulses consist of p53 arrester, whereas the fifth and subsequent pulses are composed of p53 killer. A conversion from p53 arrester to p53 killer occurs because of the accumulation of p53DINP1 induced by p53 arrester (Fig. 3). If DNA damage is quickly repaired, only pulses of p53 arrester appear and the cell cycle inhibitor p21 is produced (the first row in Fig. 8A). Consequently, the cell recovers to normal proliferation after a transient cell cycle arrest. In contrast, if DNA damage is severe, the pulses of p53 killer become dominant after those of p53 arrester, inducing p53AIP1. Subsequently, Casp3 level quickly rises to a high value and remains there thereafter, behaving as a bistable switch (see also Figs. S10 and S11 in SI Appendix), and finally apoptosis ensues (the third row in Fig. 8A). Interestingly, a reversion of apoptosis occurs at the early stage of its onset because of the DNA repair (the middle row in Fig. 8A), which is consistent with the experimental observation (38). This happens when DNA damage is effectively repaired before the pro-apoptotic components are fully activated. Therefore, the conversion between p53 arrester and killer links the extent of DNA damage with cell fate.
Fig. 8.
p53-induced cell cycle arrest and apoptosis. (A) Time courses of the levels of p53 arrester and p53 killer (Left), p21 (Center), and Casp3 (Right) for 3 cells at the IR dose of 5 Gy. In Left, the blue line denotes the threshold time for cell fate decision and the black and red pulses represent p53 arrester and p53 killer, respectively. There exists remarkable variability in p53 dynamics and cell fate. (B) Fraction of apoptotic cells within a 2,000 cell population, FA, vs. DIR for the cases with (circles) and without (squares) p53 regulation of DNA repair.
Given stochasticity in the generation and repair of DSBs, there exists variability in p53 dynamics and cell fate. To quantify such variability, the fraction of apoptotic cells in a population, FA, versus DIR is shown in Fig. 8B. Compared with the case without p53 regulation of DNA repair, FA increasing with IR dose shows a switch-like behavior for the case with p53 regulation. This is because p53 promotes repair at low damage levels but suppresses it at high damage levels. Such a regulation can reduce cellular heterogeneity in cell fate, so that most cells either survive minor DNA damage or are destroyed because of severe damage. This may be important for cancer therapy (39). Thus, p53 can fine tune its regulation of DNA repair to maintain genomic integrity. These predictions can be tested by further experiments.
Discussion
Implications of p53 Pulses in Cell Fate Decision.
As mentioned in the beginning of the paper, p53 pulses were observed in individual MCF7 cells (7, 8), whereas damped p53 oscillations were also observed in other cell lines (4), implying the presence of pulses at the single-cell level (12, 13). But these damped oscillations can be disrupted by high-level Mdm2 attributable to a single-nucleotide polymorphism in the Mdm2 gene, together with the acceleration of tumor formation (4). These indicate that the pulsatile response of p53 is important in cellular response to DNA damage, as proposed by Lahav and coworkers (7).
Based on the presence of such p53 pulses, here we demonstrated how p53 pulses govern cell fate in putatively normal cells with intact DNA repair and apoptosis systems. We showed that the number of p53 pulses can be used to distinguish cell survival from apoptosis relying on the extent of DNA damage. We observed that there exists variability in the number of p53 pulses and the mean number has a sigmoidal relationship with the radiation dose. We found that the pulsatile response of p53 level enables the p53 network to repeatedly evaluate the damage signal and thus exerts a reliable and flexible control on cell fate decision. That is, p53 pulses can be terminated rapidly to avoid inappropriate death once DNA damage is effectively fixed, or apoptosis is triggered to maintain genomic integrity once the number of p53 pulses exceeds the critical number. Thus, our proposal that cell fate can be determined by counting the number of p53 pulses represents a reasonable mechanism.
Actually, there can exist considerable variability in the amplitudes of p53 pulses as shown experimentally (8). Providing that apoptosis is triggered when the levels of the pro-apoptotic factors are integrated over the cycles of p53 pulses to reach certain thresholds, the number of p53 pulses required for inducing apoptosis may depend on the amplitudes of p53 pulses. In this sense, cell fate decision is related to the amplitude of p53 level. In addition, as the mode of p53 response to DNA damage may be cell- and stress-type specific, different modes could be related to distinct mechanisms for cell fate decision. Clearly, our integrated model will be helpful to explore those mechanisms because the present framework describes a generic signaling pathway for the p53 response to stress, and each module can be separately and easily modified to accommodate new experimental observations.
p53 Pulses Act as a Molecular Timer.
In our model, p53 pulses are classified into p53 arrester and p53 killer according to their distinct functions. The p53 pulses can be considered as a molecular timer for downstream events such as cell cycle arrest and apoptosis. Cell cycle arrest is induced by the transcriptional activation of p21 during the first pulse of p53 arrester. That is, the first alarm of the timer is set for initiation of cell cycle arrest once DNA damage is produced. The cell cycle arrest persists during the pulses of p53 arrester. When DNA damage is severe, pulses of p53 killer gradually develop accompanying the pulses of p53 arrester. Thus, as the second alarm of the timer, the activation of p53 killer sends a warning signal for initiation of apoptosis, leading to expression of pro-apoptotic genes such as p53AIP1. Subsequently, the release of CytoC and activation of Casp3 lead to apoptosis. Note that, if the DNA repair is completed during the first pulse of p53 killer, the apoptotic alarm ceases and cells recover to normal proliferation.
Dual Role of p53 in DNA Repair.
We propose that p53 promotes the repair of minor DNA damage but represses the repair of severe damage. This ensures that reparably damaged cells are protected from apoptosis and irreparably damaged cells are more prone to commit suicide. Therefore, the dual role of p53 in DNA repair is consistent with its function as a tumor suppressor, and this can also reconcile the controversy about the effect of p53 on NHEJ (17, 22, 24). It would be interesting to test these predictions in future experiments. In addition, given the stochasticity in DSB generation and repair dynamics, whether damaged cells survive or die becomes probabilistic, and the probability for apoptosis is determined by radiation dose. The p53 regulation of DNA repair makes cell fates more homogeneous at the population level. Specially, the pro-apoptotic function of p53 is enhanced at high damage levels, decreasing cellular heterogeneity. This may be important to radiation therapy for cancer (39).
ATM Dynamics.
The p53 pulses can be evoked by ATM which shows a switch-like behavior as shown in this work and also in the previous work (12). This switch-like activation of ATM has been observed experimentally (27). Here, we further showed that the ATM switch, as a detector for DNA damage, is capable of initiating and terminating p53 oscillations. Once ATM is activated, the amplitude and period of p53 pulses are almost constant, guaranteeing an all-or-none response of p53 to DNA damage. There is only a feedforward connection between the ATM switch and downstream p53 oscillator. However, the p53 pulses can also be evoked by the ATM pulses in the recurrent initiation mechanism reported recently (16). Meanwhile, the ATM activity is suppressed by p53 pulses but reactivated by DNA damage, depending on a negative feedback loop between ATM and p53 via Wip1. These results mean that robust p53 pulses can be triggered by both switch- and pulse-like ATM dynamics, and which mode is in action may be cell-type dependent. No matter how p53 pulses are generated, we believe, cell fate decision can still be governed by the pulses through selectively inducing downstream target genes, provided that ATM acts as a reliable detector for DNA damage.
Remarks on the Model.
For simplicity, some other factors related to apoptosis, such as Bcl-2, Apaf-1, Bax, and Puma, are not involved in our model (40). Although these factors are important to the regulation of apoptosis under some conditions, there is no direct evidence that they are target genes of p53 killer. Moreover, constrained by available experimental data, we did not consider the intrinsic noise in gene expression. However, such noise may be important under some conditions. For example, nonadiabatic noise from binding and unbinding of proteins to DNA plays a key role in determining the region of the parameter space accommodating excitable behavior (41). Moreover, when the number of proteins involved in the gene network is very small, noise from extinction and resurrection of protein species can evoke bistability in the deterministically monostable system (42). It has also been demonstrated that potential energy landscape can be exploited to study the global features of cellular networks with intrinsic noise, such as stability and robustness (43–45). Presumably, adding intrinsic noise to our model will make the amplitude of p53 pulses more variable and result in more variability between individual cells in cell fate. Therefore, it would be intriguing to assess whether the proposed mechanism for cell fate decision is robust to noise.
Conclusion
The present work characterizes the whole process from generation of DNA damage to cell fate decision upon IR at the single-cell level. An integrated model with four modules has been proposed for the p53 network in response to DNA damage. The network exhibits rich dynamics: the switch-like activation of ATM, a series of discrete p53 pulses, and the apoptotic switch controlled by the pulses. It is shown that at low damage levels, few p53 pulses induce transient cell cycle arrest and augment DNA repair, whereas at high damage levels, sustained p53 pulses suppress DNA repair and induce apoptosis. This indicates a digital mode of p53 response. In addition, p53 can effectively maintain genomic integrity by regulating the efficiency and fidelity of DNA repair. Moreover, the stochasticity in DSB generation and repair dynamics is well responsible for the variability in cell fate. Our results are consistent with experimental observations, and our predictions are experimentally testable. The proposed theoretical framework can be used to further explore how the p53 network responds to stresses. Our work provides clues for designing more effective and less toxic treatments for cancer.
Materials and Methods
Details of the equations, parameter values, and methods for our model are presented in the SI Appendix. The stochastic DNA repair process was characterized by a Monte Carlo method. The units of time and radiation dose are minute and gray, respectively, and the other variables are dimensionless.
Supplementary Material
Acknowledgments.
We thank John J. Tyson, Galit Lahav, and Jun Wang for helpful comments on the manuscript. This work was supported by National Basic Research Program (China) Grants 2007CB814806 and 2006CB910302, NNSF (China) Grant 10834002, and NCET-08-0269.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0813088106/DCSupplemental.
References
- 1.Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8:275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 2.Harris SL, Levine AJ. The p53 pathway: positive and negative feedback loops. Oncogene. 2005;24:2899–2908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- 3.Lev Bar-Or R, et al. Generation of oscillations by the p53-Mdm2 feedback loop: A theoretical and experimental study. Proc Natl Acad Sci USA. 2000;97:11250–11255. doi: 10.1073/pnas.210171597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu W, et al. A single nucleotide polymorphism in the Mdm2 gene disrupts the oscillation of p53 and Mdm2 levels in cells. Cancer Res. 2007;67:2757–2765. doi: 10.1158/0008-5472.CAN-06-2656. [DOI] [PubMed] [Google Scholar]
- 5.Ramalingam S, et al. Quantitative assessment of the p53-Mdm2 feedback loop using protein lysate microarrays. Cancer Res. 2007;67:6247–6252. doi: 10.1158/0008-5472.CAN-07-0342. [DOI] [PubMed] [Google Scholar]
- 6.Hamstra DA, et al. Real-time evaluation of p53 oscillatory behavior in vivo using bioluminescent imaging. Cancer Res. 2006;66:7482–7489. doi: 10.1158/0008-5472.CAN-06-1405. [DOI] [PubMed] [Google Scholar]
- 7.Lahav G, et al. Dynamics of the p53-Mdm2 feedback loop in individual cells. Nat Genet. 2004;36:147–150. doi: 10.1038/ng1293. [DOI] [PubMed] [Google Scholar]
- 8.Geva-Zatorsky N, et al. Oscillations and variability in the p53 system. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100068. 2006.0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blanc C, et al. Caspase-3 is essential for procaspase-9 processing and cisplatin-induced apoptosis of MCF-7 breast cancer cells. Cancer Res. 2000;60:4386–4390. [PubMed] [Google Scholar]
- 10.Tyson JJ. Another turn for p53. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100060. 2006.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Francisco DC, et al. Induction and processing of complex DNA damage in human breast cancer cells MCF-7 and nonmalignant MCF-10A cells. Free Radic Biol Med. 2008;44:558–569. doi: 10.1016/j.freeradbiomed.2007.10.045. [DOI] [PubMed] [Google Scholar]
- 12.Ma L, et al. A plausible model for the digital response of p53 to DNA damage. Proc Natl Acad Sci USA. 2005;102:14266–14271. doi: 10.1073/pnas.0501352102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tiana G, Jensen MH, Sneppen K. Time delay as a key to apoptosis induction in the p53 network. Eur Phys J B. 2002;29:135–140. [Google Scholar]
- 14.Ciliberto A, Novak B, Tyson JJ. Steady states and oscillations in the p53/Mdm2 network. Cell Cycle. 2005;4:488–493. doi: 10.4161/cc.4.3.1548. [DOI] [PubMed] [Google Scholar]
- 15.Zhang T, Brazhnik P, Tyson JJ. Exploring mechanisms of the DNA-damage response: p53 pulses and their possible relevance to apoptosis. Cell Cycle. 2007;6:85–94. doi: 10.4161/cc.6.1.3705. [DOI] [PubMed] [Google Scholar]
- 16.Batchelor E, Mock CS, Bhan I, Loewer A, Lahav G. Recurrent initiation: a mechanism for triggering p53 pulses in response to DNA damage. Mol Cell. 2008;30:277–289. doi: 10.1016/j.molcel.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sengupta S, Harris CC. p53: traffic cop at the crossroads of DNA repair and recombination. Nat Rev Mol Cell Biol. 2005;6:44–55. doi: 10.1038/nrm1546. [DOI] [PubMed] [Google Scholar]
- 18.Rothkamm K, Lobrich M. Evidence for a lack of DNA double-strand break repair in human cells exposed to very low x-ray doses. Proc Natl Acad Sci USA. 2003;100:5057–5062. doi: 10.1073/pnas.0830918100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stewart RD. Two-lesion kinetic model of double-strand break rejoining and cell killing. Radiat Res. 2001;156:365–378. doi: 10.1667/0033-7587(2001)156[0365:tlkmod]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 20.Burma S, Chen BPC, Chen DJ. Role of non-homologous end joining (NHEJ) in maintaining genomic integrity. DNA Repair. 2006;5:1042–1048. doi: 10.1016/j.dnarep.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 21.Yang T, et al. p53 induced by ionizing radiation mediates DNA end-joining activity, but not apoptosis of thyroid cells. Oncogene. 1997;14:1511–1519. doi: 10.1038/sj.onc.1200979. [DOI] [PubMed] [Google Scholar]
- 22.Tang W, Willers H, Powell SN. p53 directly enhances rejoining of DNA double-strand breaks with cohesive ends in gamma-irradiated mouse fibroblasts. Cancer Res. 1999;59:2562–2565. [PubMed] [Google Scholar]
- 23.Bill CA, Yu Y, Miselis NR, Little JB, Nickoloff JA. A role for p53 in DNA end rejoining by human cell extracts. Mutat Res. 1997;385:21–29. doi: 10.1016/s0921-8777(97)00040-2. [DOI] [PubMed] [Google Scholar]
- 24.Akyuz N, et al. DNA substrate dependence of p53-mediated regulation of double-strand break repair. Mol Cell Biol. 2002;22:6306–6317. doi: 10.1128/MCB.22.17.6306-6317.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeggo PA. The fidelity of repair of radiation damage. Radiat Prot Dosimetry. 2002;99:117–122. doi: 10.1093/oxfordjournals.rpd.a006740. [DOI] [PubMed] [Google Scholar]
- 26.Brady N, Gaymes TJ, Cheung M, Mufti GJ, Rassool FV. Increased error-prone NHEJ activity in myeloid leukemias is associated with DNA damage at sites that recruit key nonhomologous end-joining proteins. Cancer Res. 2003;63:1798–1805. [PubMed] [Google Scholar]
- 27.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 28.Prives C. Signaling to p53: breaking the Mdm2–p53 circuit. Cell. 1998;95:5–8. doi: 10.1016/s0092-8674(00)81774-2. [DOI] [PubMed] [Google Scholar]
- 29.Stommel JM, Wahl GM. Accelerated Mdm2 auto-degradation induced by DNA-damage kinases is required for p53 activation. EMBO J. 2004;23:1547–1556. doi: 10.1038/sj.emboj.7600145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yin Y, Stephen CW, Luciani MG, Fahraeus R. p53 stability and activity is regulated by Mdm2-mediated induction of alternative p53 translation products. Nat Cell Biol. 2002;4:462–467. doi: 10.1038/ncb801. [DOI] [PubMed] [Google Scholar]
- 31.Zhang XP, Cheng Z, Liu F, Wang W. Linking fast and slow positive feedback loops creates an optimal bistable switch in cell signaling. Phys Rev E. 2007;76 doi: 10.1103/PhysRevE.76.031924. 031924. [DOI] [PubMed] [Google Scholar]
- 32.Pomerening JR, Kim SY, Ferrell JE., Jr Systems-level dissection of the cell-cycle oscillator: bypassing positive feedback produces damped oscillations. Cell. 2005;122:565–578. doi: 10.1016/j.cell.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 33.Oda K, et al. p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell. 2000;102:849–862. doi: 10.1016/s0092-8674(00)00073-8. [DOI] [PubMed] [Google Scholar]
- 34.Okamura S, et al. p53DINP1, a p53-inducible gene, regulates p53-dependent apoptosis. Mol Cell. 2001;8:85–94. doi: 10.1016/s1097-2765(01)00284-2. [DOI] [PubMed] [Google Scholar]
- 35.Fiscella M, et al. Wip1, a novel human protein phosphatase that is induced in response to ionizing radiation in a p53-dependent manner. Proc Natl Acad Sci USA. 1997;94:6048–6053. doi: 10.1073/pnas.94.12.6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kirsch DG, et al. Caspase-3-dependent cleavage of Bcl-2 promotes release of cytochrome c. J Biol Chem. 1999;274:21155–21161. doi: 10.1074/jbc.274.30.21155. [DOI] [PubMed] [Google Scholar]
- 37.Bagci EZ, Vodovotz Y, Billiar TR, Ermentrout GB, Bahar I. Bistability in apoptosis: roles of Bax, Bcl-2, and mitochondrial permeability transition pores. Biophys J. 2006;90:1546–1559. doi: 10.1529/biophysj.105.068122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Geske FJ, Lieberman R, Srtange R, Gerschenson LE. Early stages of p53-induced apoptosis are reversible. Cell Death Differ. 2001;8:182–191. doi: 10.1038/sj.cdd.4400786. [DOI] [PubMed] [Google Scholar]
- 39.Kitano H. Cancer as a robust system: implications for anticancer therapy. Nat Rev Cancer. 2004;4:227–235. doi: 10.1038/nrc1300. [DOI] [PubMed] [Google Scholar]
- 40.Cory S, Adams JM. The Bcl2 family: regulators of the cellular life-or-death switch. Nat Rev Cancer. 2002;2:647–656. doi: 10.1038/nrc883. [DOI] [PubMed] [Google Scholar]
- 41.Schultz D, Ben Jacob E, Onuchic JN, Wolynes PG. Molecular level stochastic model for competence cycles in Bacillus subtilis. Proc Natl Acad Sci USA. 2007;104:17582–17587. doi: 10.1073/pnas.0707965104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schultz D, Walczak AM, Onuchic JN, Wolynes PG. Extinction and resurrection in gene networks. Proc Natl Acad Sci USA. 2008;105:19165–19170. doi: 10.1073/pnas.0810366105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lapidus S, Han B, Wang J. Intrinsic noise, dissipation cost, and robustness of cellular networks: The underlying energy landscape of MAPK signal transduction. Proc Natl Acad Sci USA. 2008;105:6039–6044. doi: 10.1073/pnas.0708708105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lepzelter D, Kim KY, Wang J. Dynamics and intrinsic statistical fluctuations of a gene switch. J Phys Chem B. 2007;111:10239–10247. doi: 10.1021/jp071735u. [DOI] [PubMed] [Google Scholar]
- 45.Kim KY, Wang J. Potential energy landscape and robustness of a gene regulatory network: toggle switch. PLoS Comput Biol. 2007;3:e60. doi: 10.1371/journal.pcbi.0030060. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.